Summary
We consider analysis of clustered data with mixed bivariate responses, i.e., where each member of the cluster has a binary and a continuous outcome. We propose a new bivariate random effects model that induces associations among the binary outcomes within a cluster, among the continuous outcomes within a cluster, between a binary outcome and a continuous outcome from different subjects within a cluster, as well as the direct association between the binary and continuous outcomes within the same subject. For the ease of interpretations of the regression effects, the marginal model of the binary response probability integrated over the random effects preserves the logistic form and the marginal expectation of the continuous response preserves the linear form. We implement maximum likelihood estimation of our model parameters using standard software such as PROC NLMIXED of SAS. Our simulation study demonstrates the robustness of our method with respect to the misspecification of the regression model as well as the random effects model. We illustrate our methodology by analyzing a developmental toxicity study of ethylene glycol in mice.
Keywords: Bivariate binary and continuous responses, Bridge distribution, Logit link, Probability integral transformation
1. Introduction
In many biomedical studies, the basic sampling unit is a cluster of subjects, such as siblings in a family or rats in a litter. Further, each member of the cluster may have multiple outcomes, including both discrete and continuous endpoints. When the bivariate responses are both continuous and approximately Gaussian, the traditional random effects linear model framework can be used for inference. However, the presence of both categorical/ordinal as well as continuous outcomes, called mixed responses, complicates the situation due to the lack of a discrete multivariate analog of the multivariate Gaussian distribution (Fitzmaurice and Laird, 1995). For example, the developmental toxicity study of ethylene glycol (EG), a high-volume industrial chemical (Price et al., 1985), involves measurements of both binary and continuous responses, namely, (a) fetal malformation and (b) fetal weight of 1028 fetuses from 94 litters. In this study, one of the four levels of EG was administered to 94 pregnant mice over the period of major organogenesis, beginning just after implantation. In particular, 25 pregnant mice received 0 g/kg/day, 24 received 0.75 g/kg/day, 22 received 1.5 g/kg/day, and 23 received 3 g/kg/day. We expect a within cluster (“litter”) association, along with the direct association between the fetal malformation (binary) and fetal weight (continuous) of same fetus. We are primarily interested in modeling the effects of dose on the binary and continuous outcomes, as interpretable regression functions, as well as modeling the within subject and within litter associations between the binary and continuous outcomes.
A number of joint models have been proposed for developmental toxicity studies involving both discrete and continuous responses from each member of a cluster (Geys, Molenberghs, and Ryan, 1999; Regan and Catalano, 1999; Dunson, Chen, and Harry, 2003; Gueorguieva and Agresti, 2001). Catalano and Ryan (1992) suggested a model assuming that the discrete outcome is a discretization of an unobserved continuous latent variable and this latent variable and the continuous response within same subject have a bivariate normal distribution. Joint distribution of the two responses is then formed by a product of marginal distribution of the continuous response and conditional distribution of the binary response conditioned on the continuous response. The other type of models consider conditioning on the binary response. Fitzmaurice and Laird (1995) assumed a logit link function for marginal probability of binary response and a normal distribution for the continuous response given the binary response. Cox and Wermuth (1992) compared different joint distribution models for analyzing data with quantitative and qualitative responses. For more discussions related to conditional models, see Little and Schluchter (1985), Cox and Wermuth (1994), Krzanowski (1988), and Schafer (1997).
The model proposed by Fitzmaurice and Laird (1995), henceforth FL, focuses on estimating and interpreting the marginal regression functions of two responses, while treating the association between the discrete and continuous responses as a nuisance. FL models have the attractive feature that the marginal regression parameters of both responses have good physical interpretations. However, this model only considers the cluster effect on the continuous response given the binary responses from all cluster members. In this model, the specification of the regression model conditional on the cluster-specific random effects is completely unknown. The magnitude of the marginal regression parameters, particularly for the binary response, gets attenuated from that of the conditional regression parameters due to the heterogeneity of clusters. This degree of attenuation is important for future study design as well as understanding the expected effect of covariates on other populations and on individual subjects.
In this article, we avoid the above shortcomings of FL modeling. We propose a model with practical physical interpretations of the marginal as well as conditional (given the cluster-specific random effects) regression functions associated with both discrete and continuous responses. We specify a bivariate distribution of two cluster-specific random effects—presented as separate random intercepts for two types of responses. We use a separate parameter to account for the direct within-subject association between the two responses. Hence, association between the discrete and continuous responses from different subjects within the same cluster is induced by the two correlated random effects. Association between the binary and continuous outcomes on the same subject is induced by correlations between the two within-cluster random effects, as well as the direct dependence of the continuous response on the binary response from the same subject. We use a logit link for the conditional regression function of the binary response given cluster-specific random effect, and an identity link for the regression function of the continuous response conditioned on the binary response and the cluster random effects. The joint distribution of the discrete and continuous responses given the random effects is expressed by the product of the marginal distribution of the binary outcome and the conditional distribution of the continuous response given the binary response. In the FL model, they considered maximum likelihood (henceforth ML) estimation as quite complicated and intractable, and hence they proposed the generalized estimating equations (GEE) method for computational convenience. Contrary to that, our likelihood is much simpler to evaluate and amenable to maximization through routine nonadaptive Gaussian quadrature techniques, because conditional on the random effects, all outcomes are independent.
One drawback of the random effects models for clustered mixed responses is that the marginal regression model of the binary response (after integrating out the random effects) usually is not of logistic form when the subject-specific model conditional on the random effect is of logistic form. In our model, the bridge distribution (Wang and Louis, 2003) of the random effect for the binary response allows the marginal probability of the binary response, integrated over the random intercept, to have a logistic structure with an odds ratio interpretation of the marginal regression effect. The regression parameters in the marginal logistic regression model are proportional to the corresponding regression parameters in the subject-specific conditional logistic model. This is a major advantage over other existing models including the FL model.
The rest of the article is organized as follows. In Section 2, we begin with our random effects model of clustered data with bivariate mixed responses within each subject. We illustrate how different levels of the association between two random effects induce different Kendall's τ values for association between binary and continuous responses from same cluster. In Section 3, we extend this conditional model of the clustered mixed responses to accommodate a direct within subject association between two responses (even after adjusting for the within cluster association). We also give the likelihood representation of our model. In Section 4, we demonstrate the ML estimation of the regression parameters of our models via analyzing the data from a developmental toxicity experiment of EG on mice. Finally, Section 5 discusses the need and advantages of our model. We also study the robustness of our method to model and random effect distribution misspecification through simulation studies.
2. Random Effects Model for Mixed Responses
For subject j = 1,…, Mi from cluster i = 1,…, N, we have a binary response Y1ij, where Y1ij = 1 when subject j has response 1 and 0 otherwise, and a continuous response Y2ij. Each subject has a K × 1 covariate vector Xij. In this section, we use the usual linear model with normal random (cluster) effect Wi for the continuous outcomes Y2ij from cluster i. Furthermore, for the binary outcomes Y1ij from cluster i, we let the cluster random effects Bi have the bridge density of Wang and Louis (2003). The key to the new approach proposed here is a bivariate density for the joint distribution of the normal random effect Wi and the bridge random effect Bi. This bivariate distribution will induce an association between the vector of continuous and the vector of binary outcomes within the same cluster.
For the time being, we assume that given the cluster-specific random effects (Bi, Wi), the responses Ykij for k = 1, 2; j = 1,…, Mi within cluster i are mutually independent. We assume the conditional distribution of binary Y1ij given the random effects (Bi, Wi) to be Bernoulli with success probability pij, with
| (1) |
where the marginal distribution of the random intercept Bi of binary response is a bridge distribution (Wang and Louis, 2003) with unknown parameter ϕ (0 < ϕ < 1) and density
The bridge density is symmetric (mean of Bi is 0), and the variance of Bi is . It has a slightly heavier tail and is more peaked than the normal density. The marginal probability P[Y1ij = 1 |Xij] (when integrated over the random intercepts (Bi, Wi)) still has logistic form. The parameter ϕ of the bridge distribution being the same across all clusters assures the exchangeability of cluster random effects on the binary responses. The marginal regression parameter of the binary response is proportional to the conditional regression parameter β multiplied by the attenuation parameter ϕ, i.e.,
| (2) |
Small ϕ indicates the important facts: high correlation of the binary responses within a cluster; high heterogeneity among clusters related to the binary response; and a corresponding high degree of attenuation of β in the population due to cluster heterogeneity.
The continuous response Y2ij given the cluster random effect (Bi, Wi) is modeled by a normally distributed linear mixed effects model with mean
| (3) |
and variance . The marginal distribution of the random intercept Wi of the continuous response is normal with mean 0 and variance . The marginal distribution of Y2ij integrated over (Bi, Wi) is again normal with mean
| (4) |
In this model for the clustered mixed outcomes, both the conditional (on the cluster-specific random effects) and marginal regression functions in equations (1)–(4) have useful physical interpretations. As we describe below, the association between a binary and a continuous outcome within same cluster is induced by formulating a joint bivariate distribution for the cluster random effects (Bi, Wi) while preserving the desired forms of the marginal densities of Bi and Wi. This bivariate distribution is constructed from transformations of a pair of latent random variables (Z1i, Z2i) with a bivariate normal distribution with mean zero, unit standard deviations, and correlation ρ.
To formulate the bivariate distribution for (Bi, Wi), we use the probability integral transformation
| (5) |
where Φ(·) is the cumulative distribution function (c.d.f.) of standard normal, and (·) is the inverse cumulative distribution
| (6) |
of the bridge density for 0 < u < 1. This assures that the marginal density of Bi will be the bridge distribution with c.d.f.
Because both the bridge cumulative and inverse cumulative distributions have closed-form expressions, they can easily be implemented in standard statistical software and simulations are easy to perform. In order to obtain , we let Wi = σ1Z2i.
In an extensive simulation study, we find the correlation between the two random intercepts Bi and Wi are approximately the same as the correlation between Z1i and Z2i. To understand the associations within (Bi, Wi) and the association within (Y1ij, Y2ij′) induced via the correlation ρ of (Z1i, Z2i), we compute Kendall's τ (Hougaard, 2000) for (Bi, Wi) and Kendall's τ for (Y1ij, Y2ij′) for different values of ρ. The transformations (Φ(Z1i)) and Wi = σ1Z2i are both monotone increasing. Kendall's τ for bivariate normal random variables (Z1i, Z2i) with correlation ρ is τ = 2 arcsin(ρ)/π with −1 ≤ τ ≤ 1. This is also Kendall's τ for (Bi, Wi) because Kendall's τ is invaraint to increasing transformations (Hougaard, 2000). Kendall's τ for (Y1ij, Y2ij′), defined as E[sign{(Y1ij − Y1i* j*)(Y2ij′ − Y2i* j**)}], was simulated via Monte Carlo methods using equations (1), (3), (5), (6), and Wi = σ1Z2i. Figure 1 shows values of Kendall's τ of (Bi, Wi) for different values of ρ. Figure 2 shows Kendall's τ of the binary and continuous responses (Y1ij, Y2ij′) from the same cluster for different values of ρ and a fixed value of ϕ.
Figure 1.
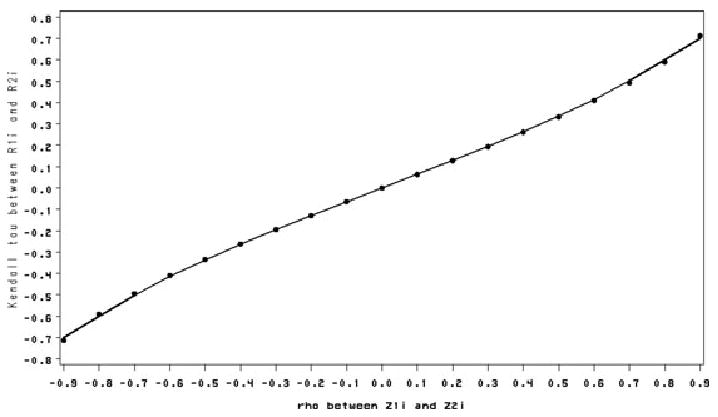
Kendall's τ of Bi and Wi for different ρ.
Figure 2.
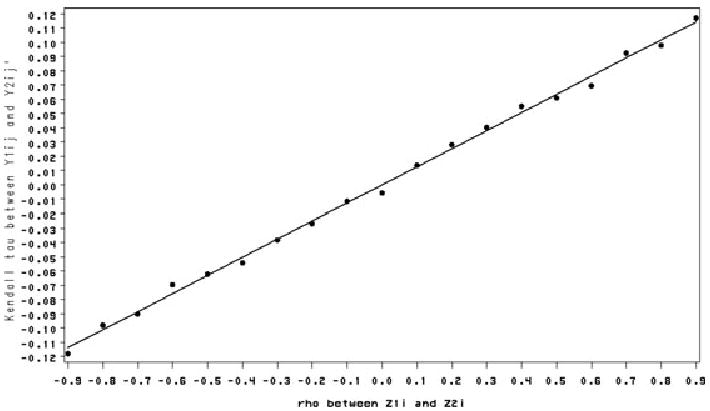
Kendall's τ of Y1ij and Y2ij′ with ϕ = 0.735 for different ρ.
3. Models for Bivariate Mixed Responses with Clustering
In Section 2, models (1) and (3) induce an exchangeable association structure among binary and continuous responses in the same cluster because given cluster-specific (Bi, Wi), responses within a subject are considered independent. However, we might expect the association between a binary Y1ij and a continuous Y2ij from the same subject to be stronger than the association between Y1ij and Y2ij′ from different subjects j and j′ within cluster i. In this section, we extend our model to accommodate the possibility of the direct association between binary and continuous outcomes from the same subject, even after adjusting for the within cluster association. To this end, we modify the model (3) by incorporating a direct effect of subject-specific binary Y1ij on the continuous Y2ij, such that
| (7) |
where , while keeping the regression model for Y1ij identical to that of equation (1). Hence, the association between Y1ij and Y2ij from the same subject is induced by the direct dependence of Y2ij on Y1ij measured by γ as well as the association parameter ρ of the random effects (Bi, Wi).
The conditional expectation of Y2ij integrated over Wi is
because E[Wi | Y1ij, Xij] = EE[Wi | Bi, Y1ij, Xij] = EE [Wi | Bi] = 0. Hence, marginal expectation is the linear function of Xij, same as in equation (4).
From equation (7), the covariance between responses Y1ij and Y2ij from the same subject is
| (8) |
The covariance of two responses from different subjects j and j′ within the same cluster i is
| (9) |
where Var(Y1ij) = π1ij(1 − π1ij). Our model ensures the anticipated result | Cov[Y1ij, Y2ij] | >| Cov[Y1ij, Y2ij′] because Var(Y1ij − Y1ij′) = 2Var(Y1ij) − 2Cov[Y1ij, Y1ij′] > 0. For covariate vectors Xij and Xij′, we have
| (10) |
| (11) |
where Eb and Ebw, respectively, are the expectation with respect to the marginal density of Bi and that with respect to the joint density of (Bi, Wi).
Now, the likelihood contribution from cluster i is proportional to
where fY1ij | Bi, Wi, Xij (y1ij | bi,wi, xij) is equal to , and fY2ij | Bi, Wi, Y1ij (y2ij | bi, wi, y 1ij, xij) is the normal density with variance and mean .
Unlike the FL model, our likelihood is tractable and maximization is straightforward. In order to use the ML method for estimation, the likelihood is obtained via taking the expectation with respect to the joint density of (Bi, Wi). These integrals are implemented using nonadaptive Gaussian quadrature techniques in PROC NLMIXED of SAS (v9.1) along with optimization tools such as Newton–Raphson with ridging.
4. Example: EG Data
We apply the method described above to analyze data from the development toxicity study using 94 pregnant mice randomized to receive one of the four different doses of EG, 0, 0.75, 1.5, or 3 g/kg/day. Fetal malformation and fetal weight of 1028 fetuses from the 94 litters/clusters, with cluster size ranging from 1 to 16, were recorded. A summary of the fetal malformations and fetal weights is given in Table 1. We clearly observe that the proportion of malformation increases from 0.34% to 57.08% and sample mean of weight decreases from 0.972 to 0.704 as dose level increases from 0 to 3. For more details on the specifics of this data set, see Catalano and Ryan (1992), Fitzmaurice and Laird (1995), and also the monograph by Aerts et al. (2002).
Table 1. Descriptive statistics of data from developmental toxicity study.
| Dose (g/kg) | Litters | Live fetuses | Fetal malformation No.(%) | Fetal weight(g) Mean (S.D.) |
|---|---|---|---|---|
| 0 | 25 | 297 | 1 (0.3) | 0.972 (0.098) |
| 0.75 | 24 | 276 | 26 (9.4) | 0.877 (0.104) |
| 1.5 | 22 | 229 | 89 (38.9) | 0.764 (0.107) |
| 3 | 23 | 226 | 129 (57.1) | 0.704 (0.124) |
Treating dose Xi(= Xij) assigned to the ith mice/litter as a covariate, the binary Y1ij (fetal malformation) and the continuous Y2ij (fetal weight) in model 1 are modeled as logit (E [Y1ij | Bi, Wi, Xi]) = β0 + β1Xi + Bi and E[Y2ij | Bi, Wi, Y1ij, Xi] = α0 + α1Xi + γ (Y1ij − π1i) + Wi, where π1i = exp{ϕ(β0 + β1Xi)}/[1 + exp{ϕ(β0 + β1Xi)}], and Y2ij given (Bi, Wi, Y1ij) is normally distributed with variance . The random effects (Bi, Wi) are assumed to have a bivariate density with the marginal density of Bi being the bridge with parameter ϕ, and the marginal density of Wi being normal with mean 0 and variance .
Table 2 shows the regression parameter estimates for the two responses from model 1. The positive estimate for the dose effect on fetal malformation and the negative estimate on fetal weight are consistent with what we observe from Table 1: the malformation rate increases and weight decreases as dose level increases. All of the marginal parameter estimates have slightly different point estimates than the FL model (Table 2 in Fitzmaurice and Laird, 1995), but the estimated standard errors of the parameter estimates are smaller than that of the FL model, especially for the binary response (approximately 20%), leading to tighter 95% confidence intervals.
Table 2. ML estimates of parameters from model 1 and model 2.
| Model 1 | Model 2 | |||
|---|---|---|---|---|
| Parameter | Estimate (S.E.) | P-Value | Estimate (S.E.) | P-Value |
| Fetal malformation | ||||
| ϕβ0 | −2.9737 (0.2307) | <0.0001 | −4.1643 (0.5450) | <0.0001 |
| ϕβ1 | 1.1634 (0.1182) | <0.0001 | 3.7180 (0.7258) | <0.0001 |
| ϕβ2 | −0.7645 (0.1928) | 0.0001 | ||
| Fetal weight | ||||
| α0 | 0.9531 (0.0122) | <0.0001 | 0.9687 (0.0120) | <0.0001 |
| α1 | −0.0843 (0.0060) | <0.0001 | −0.1750 (0.0271) | <0.0001 |
| α2 | 0.0298 (0.0087) | 0.0009 | ||
| γ | −0.0337 (0.0076) | <0.0001 | −0.0324 (0.0078) | <0.0001 |
| Other parameters | ||||
| σ0 | 0.0737 (0.0017) | <0.0001 | 0.0740 (0.0017) | <0.0001 |
| ϕ | 0.6618 (0.0415) | <0.0001 | 0.7262 (0.0519) | <0.0001 |
| σ1 | 0.0954 (0.0054) | <0.0001 | 0.0865 (0.0066) | <0.0001 |
| ρY1 | 0.3382 (0.0415) | <0.0001 | 0.2738 (0.0519) | <0.0001 |
| ρY2 | 0.6262 (0.0291) | <0.0001 | 0.5771 (0.0394) | <0.0001 |
| ρ | −0.6433 (0.0644) | <0.0001 | −0.6110 (0.0920) | <0.0001 |
The estimate of the attenuation parameter ϕ = 0.6618 confirms that heterogeneity of the clusters causes moderate attenuation of the estimated marginal dose effects. The intracluster correlation ρY1 between the binary responses is estimated by 1 − ϕ = 0.3382 (Wang and Louis, 2003). The intracluster correlation ρY2 between the continuous responses are estimated by the between-cluster variability divided by the sum of the within- and between-cluster variabilities . These estimates are somewhat greater than the estimates of the intracluster correlations obtained from the FL model. The negative value −0.6433 for estimate of association parameter ρ between the two cluster-specific random effects Bi and Wi, as well as −0.03369 for estimate of the within subject dependence coefficient γ of Y2ij on Y1ij, suggest reverse association among the two responses fetal malformation and fetal weight. For comparison with the results from model 1, we consider a more complex correlation structure. We change the parameter γ of model 1 to (γ1 + γ2Xi) to verify whether the dose effect on fetal weight is the same for both the normal and malformed fetus. We obtain the p-value for γ2 as 0.7154, which indicates that the direct effect of malformation on weight does not depend on dose.
In smoothed residual plots of model 1 obtained via the LOESS procedure in SAS (Cleveland, 1979), we observe some quadratic trends of dose for both the malformation and weight responses. We reanalyze the data by adding quadratic terms of dose effect for both responses (model 2). Estimates of the parameters of model 2 are in Table 2, where ϕβ2 and α2 are the marginal parameters of squared dose effect for the two responses. No further trends are detected from corresponding residual plots. The estimates of model 2 also suggest that the quadratic dose effect terms are necessary.
Our method allows fractional polynomial regression equations (Royston and Altman, 1994; Geys et al., 1999) for E(Y1ij| Xi) and E(Y2ij| Xi). We have fitted several such models including models using to our data; however, the corresponding Akaike's information criterion (AIC) and Bayesian information criterion (BIC) values are bigger than those of model 2. For experiments with more dose levels than we have, a fractional polynomial model may become more useful for the dose-response effect.
To see the impact of association between fetal malformation Y1ij and fetal weight Y2ij on the estimates of the regression parameters, we fit separate and independent regression models for Y1ij and Y2ij, i.e., ρ = 0 and γ = 0 (model 0a). We modify model 1 with γ = 0 (model 0b) to accommodate the special case no direct within-subject association between Y1ij and Y2ij. We also consider separate and independent regression models for Y1ij and Y2ij (ρ = 0 and γ = 0) with covariate vector (model 0c). No noticeable changes in point estimates are observed comparing results of model 0a and 0b with model 1, and model 0c with model 2. However, for model 0a, we observe approximately 35% and 25% larger estimated standard errors for parameters of the binary response Y1ij and continuous response Y2ij, respectively, compared to corresponding estimates from model 1. This indicates that substantially larger sample sizes are required while using model 0a to obtain similar standard errors as model 1. Similarly, 14% and 3% larger standard errors are observed when comparing results for the regression parameters of Y1ij and Y2ij from model 0b to model 1. This is also the case for model 0c compared to model 2. Particularly for this data set, the p-values for the association ρ between the two random effects and direct association γ of Y1ij on Y2ij are both less than 0.0001 implying statistically significant association between the two responses. AIC and BIC of the five fitted models are shown in Table 3. Model 2 has by far the smallest AIC and BIC values (with differences larger than 10 compared to model 1), which indicates it is the best fitted model for our data set.
Table 3. Fit statistics for five models.
| Fit statistics | Model 0a | Model 0b | Model 0c | Model 1 | Model 2 |
|---|---|---|---|---|---|
| −2 Log-likelihood | −1254 | −1278 | −1292 | −1311 | −1331 |
| AIC | −1240 | −1262 | −1274 | −1293 | −1309 |
| BIC | −1223 | −1242 | −1251 | −1271 | −1281 |
Using estimates from model 2 and expressions (8)–(11), we obtain from Monte Carlo simulation that, for dose level 0, the estimated correlation between two responses from the same fetus is , and the estimated correlation between two responses from different fetuses within the same cluster is . This shows that the former correlation is approximately 19% higher than the latter correlation.
5. Discussion
In this article, we have developed a regression model for bivariate discrete and continuous outcomes motivated by an application in developmental toxicology where the experiment measures the fetal malformation and fetal weight for different dose levels administered to different clusters. Our model is attractive in that it provides both conditional and marginal interpretation of the dose-response modeling, which happens to be the cornerstone of quantitative risk assessment. Similar to a generalized linear mixed model setup, the binary responses are related to the covariates through a logit link whereas the conditional distribution of the continuous responses given the binary responses are related to the covariates by an identity link. Our model can efficiently handle the effects of clustering through introduction of cluster-specific random effects in a regression setup, as well as the association between mixed outcomes on the same (different) fetus.
Our method of estimation is ML as opposed to the GEE methods adapted in the FL approach. With a correctly specified model, provided estimation techniques are computationally feasible, the ML method reigns supreme over any other estimation methods for its fidelity to the likelihood principle as well as for its asymptotic properties. There are clearly several advantages of our ML method over the GEE method used in the FL approach. Firstly, although ML estimation appears to be complicated in a clustered mixed responses setting, our model is easily amenable to ML estimation through routine optimization techniques readily available in standard software like SAS (v9.1). Secondly, ML estimates are asymptotically efficient (Lehmann and Casella, 1998). Thus, using our method we achieve smaller standard errors of the parameter estimates and consequently tighter confidence intervals over the estimates obtained by the GEE method. Thirdly, GEE methods provide only marginal models, thus we are unable to measure the attenuation of the dose effect to the population due to the heterogeneity induced by clustering (litter effect). Our model has both conditional and marginal interpretation in this context.
In theory, the only advantage GEE has over our full-likelihood approach is when the marginal regression model is correctly specified, but the full likelihood is misspecified; in this case, the GEE estimate of the marginal regression parameters will be asymptotically unbiased, but those from our full likelihood could be biased. To investigate the robustness of our ML approach, we performed small-scale simulation studies under misspecification of regression function as well as misspecification of the random effect distribution for binary response. Our current model (model 1) assumes that the parameter (γ) measuring the direct association between the two responses within the same subject does not depend on dose (or other covariates). We generate 50 data sets from a modification of this model with γ replaced by γ1 + γ2Xi assuming γ1 = γ2 for simplicity, and using estimated values from results of model 1. Each data set contains 100 clusters with fixed cluster size 10. Both our likelihood approach and FL's approach using a GEE method (one-step) are applied to the simulated data. The results for estimates of the marginal dose effect are shown in Table 4. We also fit our model to data simulated from an extension of model 1, in which the distribution of Bi of the binary response is standard normal instead of bridge. The average standard error (with corresponding relative bias) of the estimated dose effects on fetal malformation (conditional) and fetal weight (marginal) from model 1 are 0.1543 (0.0748) and 0.0079 (0.0997), respectively. In this small-scale simulation, we observe that our full likelihood approach is robust to regression model misspecifications as well as random effects distributional misspecification compared to the FL approach as far as the bias of regression estimates are concerned. This makes our approach an extremely attractive alternative to the GEE based FL approach.
Table 4. Simulation results of misspecification of regression function.
| Dose effect (malformation) |
Dose effect (weight) |
|||
|---|---|---|---|---|
| Mean S.E. | Rel. bias | Mean S.E. | Rel. bias | |
| FL model | 0.1565 | 0.1017 | 0.0078 | 0.0923 |
| Bridge model | 0.1361 | 0.1056 | 0.0087 | 0.1021 |
Supplementary Material
data EG_data; | |||
input Litter_ID Dose Fetal_Wt Fetal_Mal; | |||
dose=dose/1000; | |||
cards; | |||
60 |
0 |
0.903 |
0 |
60 |
0 |
0.828 |
0 |
60 |
0 |
0.953 |
0 |
60 |
0 |
0.954 |
0 |
60 |
0 |
1.070 |
0 |
60 |
0 |
1.065 |
0 |
60 |
0 |
0.959 |
0 |
61 |
0 |
1.025 |
0 |
61 |
0 |
1.006 |
0 |
61 |
0 |
1.069 |
0 |
61 |
0 |
0.825 |
0 |
61 |
0 |
1.013 |
0 |
61 |
0 |
0.994 |
0 |
61 |
0 |
1.059 |
0 |
61 |
0 |
1.041 |
0 |
61 |
0 |
1.020 |
0 |
61 |
0 |
1.041 |
0 |
61 |
0 |
0.911 |
0 |
61 |
0 |
0.988 |
0 |
61 |
0 |
0.947 |
0 |
61 |
0 |
0.976 |
0 |
68 |
0 |
1.058 |
0 |
68 |
0 |
0.950 |
0 |
68 |
0 |
0.860 |
0 |
68 |
0 |
1.015 |
0 |
68 |
0 |
0.907 |
0 |
68 |
0 |
0.856 |
0 |
68 |
0 |
1.075 |
0 |
68 |
0 |
0.958 |
0 |
68 |
0 |
1.082 |
0 |
68 |
0 |
0.916 |
0 |
68 |
0 |
1.054 |
0 |
68 |
0 |
0.921 |
0 |
68 |
0 |
0.874 |
0 |
69 |
0 |
0.999 |
0 |
69 |
0 |
0.950 |
0 |
69 |
0 |
1.065 |
0 |
69 |
0 |
0.944 |
0 |
69 |
0 |
0.949 |
0 |
69 |
0 |
1.044 |
0 |
69 |
0 |
1.025 |
0 |
69 |
0 |
0.987 |
0 |
69 |
0 |
0.879 |
0 |
69 |
0 |
0.908 |
0 |
76 |
0 |
1.071 |
1 |
76 |
0 |
1.056 |
0 |
76 |
0 |
1.084 |
0 |
76 |
0 |
0.851 |
0 |
76 |
0 |
0.925 |
0 |
76 |
0 |
0.832 |
0 |
76 |
0 |
0.979 |
0 |
76 |
0 |
1.096 |
0 |
76 |
0 |
0.954 |
0 |
76 |
0 |
0.875 |
0 |
76 |
0 |
1.057 |
0 |
76 |
0 |
1.245 |
0 |
76 |
0 |
1.018 |
0 |
76 |
0 |
1.123 |
0 |
76 |
0 |
1.030 |
0 |
76 |
0 |
0.968 |
0 |
77 |
0 |
0.852 |
0 |
77 |
0 |
0.940 |
0 |
77 |
0 |
0.905 |
0 |
77 |
0 |
0.787 |
0 |
77 |
0 |
0.918 |
0 |
77 |
0 |
0.745 |
0 |
77 |
0 |
0.907 |
0 |
77 |
0 |
0.818 |
0 |
77 |
0 |
0.896 |
0 |
77 |
0 |
0.858 |
0 |
77 |
0 |
0.935 |
0 |
77 |
0 |
0.669 |
0 |
77 |
0 |
0.930 |
0 |
77 |
0 |
0.890 |
0 |
84 |
0 |
0.983 |
0 |
84 |
0 |
0.974 |
0 |
84 |
0 |
1.005 |
0 |
84 |
0 |
0.897 |
0 |
84 |
0 |
1.110 |
0 |
84 |
0 |
1.035 |
0 |
84 |
0 |
0.997 |
0 |
84 |
0 |
1.059 |
0 |
84 |
0 |
1.067 |
0 |
84 |
0 |
0.911 |
0 |
85 |
0 |
1.122 |
0 |
85 |
0 |
1.134 |
0 |
85 |
0 |
1.117 |
0 |
85 |
0 |
0.999 |
0 |
85 |
0 |
1.200 |
0 |
85 |
0 |
1.154 |
0 |
85 |
0 |
1.077 |
0 |
85 |
0 |
0.980 |
0 |
85 |
0 |
0.989 |
0 |
85 |
0 |
1.161 |
0 |
85 |
0 |
1.101 |
0 |
85 |
0 |
1.153 |
0 |
93 |
0 |
1.051 |
0 |
93 |
0 |
1.082 |
0 |
93 |
0 |
1.062 |
0 |
93 |
0 |
0.955 |
0 |
93 |
0 |
1.095 |
0 |
93 |
0 |
0.994 |
0 |
93 |
0 |
1.093 |
0 |
93 |
0 |
1.002 |
0 |
93 |
0 |
1.038 |
0 |
93 |
0 |
1.084 |
0 |
93 |
0 |
1.026 |
0 |
100 |
0 |
0.975 |
0 |
100 |
0 |
1.110 |
0 |
100 |
0 |
1.120 |
0 |
100 |
0 |
1.170 |
0 |
100 |
0 |
1.130 |
0 |
100 |
0 |
1.070 |
0 |
100 |
0 |
1.100 |
0 |
100 |
0 |
1.020 |
0 |
101 |
0 |
1.190 |
0 |
101 |
0 |
1.040 |
0 |
101 |
0 |
0.821 |
0 |
101 |
0 |
1.070 |
0 |
101 |
0 |
0.665 |
0 |
101 |
0 |
1.120 |
0 |
106 |
0 |
1.044 |
0 |
106 |
0 |
0.933 |
0 |
106 |
0 |
0.978 |
0 |
106 |
0 |
0.981 |
0 |
106 |
0 |
0.958 |
0 |
106 |
0 |
1.005 |
0 |
106 |
0 |
1.053 |
0 |
106 |
0 |
1.059 |
0 |
106 |
0 |
0.917 |
0 |
106 |
0 |
1.079 |
0 |
106 |
0 |
1.077 |
0 |
106 |
0 |
1.060 |
0 |
106 |
0 |
1.031 |
0 |
106 |
0 |
1.006 |
0 |
106 |
0 |
1.007 |
0 |
107 |
0 |
0.960 |
0 |
107 |
0 |
0.844 |
0 |
107 |
0 |
0.833 |
0 |
107 |
0 |
0.870 |
0 |
107 |
0 |
0.887 |
0 |
107 |
0 |
0.835 |
0 |
107 |
0 |
0.716 |
0 |
107 |
0 |
0.964 |
0 |
107 |
0 |
0.933 |
0 |
107 |
0 |
0.753 |
0 |
107 |
0 |
0.803 |
0 |
107 |
0 |
0.742 |
0 |
114 |
0 |
0.885 |
0 |
114 |
0 |
0.903 |
0 |
114 |
0 |
0.815 |
0 |
114 |
0 |
0.937 |
0 |
114 |
0 |
0.936 |
0 |
114 |
0 |
0.984 |
0 |
114 |
0 |
0.835 |
0 |
114 |
0 |
0.954 |
0 |
114 |
0 |
0.890 |
0 |
114 |
0 |
0.930 |
0 |
114 |
0 |
0.924 |
0 |
114 |
0 |
0.873 |
0 |
115 |
0 |
1.045 |
0 |
115 |
0 |
1.087 |
0 |
115 |
0 |
1.021 |
0 |
115 |
0 |
0.999 |
0 |
115 |
0 |
0.908 |
0 |
115 |
0 |
1.027 |
0 |
115 |
0 |
1.041 |
0 |
115 |
0 |
0.968 |
0 |
115 |
0 |
0.971 |
0 |
115 |
0 |
1.029 |
0 |
115 |
0 |
0.923 |
0 |
115 |
0 |
0.944 |
0 |
115 |
0 |
0.953 |
0 |
122 |
0 |
1.032 |
0 |
122 |
0 |
1.045 |
0 |
122 |
0 |
0.967 |
0 |
122 |
0 |
1.065 |
0 |
122 |
0 |
0.981 |
0 |
122 |
0 |
0.964 |
0 |
122 |
0 |
1.080 |
0 |
122 |
0 |
1.100 |
0 |
122 |
0 |
0.994 |
0 |
122 |
0 |
0.949 |
0 |
123 |
0 |
1.080 |
0 |
123 |
0 |
1.081 |
0 |
123 |
0 |
1.012 |
0 |
123 |
0 |
0.856 |
0 |
123 |
0 |
0.915 |
0 |
123 |
0 |
0.952 |
0 |
123 |
0 |
0.983 |
0 |
123 |
0 |
0.872 |
0 |
123 |
0 |
0.950 |
0 |
123 |
0 |
0.939 |
0 |
130 |
0 |
1.014 |
0 |
130 |
0 |
0.907 |
0 |
130 |
0 |
0.841 |
0 |
130 |
0 |
0.892 |
0 |
130 |
0 |
0.868 |
0 |
130 |
0 |
0.837 |
0 |
130 |
0 |
0.931 |
0 |
130 |
0 |
0.871 |
0 |
130 |
0 |
0.901 |
0 |
130 |
0 |
0.987 |
0 |
130 |
0 |
0.986 |
0 |
131 |
0 |
0.933 |
0 |
131 |
0 |
0.935 |
0 |
131 |
0 |
1.033 |
0 |
131 |
0 |
1.081 |
0 |
131 |
0 |
0.773 |
0 |
131 |
0 |
1.054 |
0 |
131 |
0 |
0.946 |
0 |
131 |
0 |
0.994 |
0 |
131 |
0 |
0.724 |
0 |
131 |
0 |
0.883 |
0 |
131 |
0 |
1.047 |
0 |
131 |
0 |
0.951 |
0 |
138 |
0 |
0.806 |
0 |
138 |
0 |
0.858 |
0 |
138 |
0 |
0.815 |
0 |
138 |
0 |
0.914 |
0 |
138 |
0 |
0.841 |
0 |
138 |
0 |
0.866 |
0 |
138 |
0 |
0.792 |
0 |
138 |
0 |
0.773 |
0 |
138 |
0 |
0.882 |
0 |
138 |
0 |
0.887 |
0 |
138 |
0 |
0.847 |
0 |
138 |
0 |
1.035 |
0 |
138 |
0 |
0.918 |
0 |
139 |
0 |
0.886 |
0 |
139 |
0 |
0.923 |
0 |
139 |
0 |
1.027 |
0 |
139 |
0 |
0.991 |
0 |
139 |
0 |
0.909 |
0 |
139 |
0 |
1.061 |
0 |
139 |
0 |
1.054 |
0 |
139 |
0 |
1.018 |
0 |
139 |
0 |
1.029 |
0 |
139 |
0 |
1.100 |
0 |
139 |
0 |
0.932 |
0 |
139 |
0 |
0.886 |
0 |
139 |
0 |
1.024 |
0 |
139 |
0 |
0.930 |
0 |
146 |
0 |
1.025 |
0 |
146 |
0 |
1.014 |
0 |
146 |
0 |
0.954 |
0 |
146 |
0 |
1.081 |
0 |
146 |
0 |
0.967 |
0 |
146 |
0 |
0.912 |
0 |
146 |
0 |
1.030 |
0 |
146 |
0 |
0.871 |
0 |
146 |
0 |
0.940 |
0 |
146 |
0 |
0.996 |
0 |
146 |
0 |
1.028 |
0 |
146 |
0 |
0.794 |
0 |
146 |
0 |
0.879 |
0 |
147 |
0 |
1.058 |
0 |
147 |
0 |
1.100 |
0 |
147 |
0 |
1.121 |
0 |
147 |
0 |
1.093 |
0 |
147 |
0 |
1.017 |
0 |
147 |
0 |
1.058 |
0 |
147 |
0 |
1.090 |
0 |
147 |
0 |
1.052 |
0 |
147 |
0 |
1.046 |
0 |
147 |
0 |
1.103 |
0 |
147 |
0 |
1.026 |
0 |
147 |
0 |
1.076 |
0 |
147 |
0 |
1.095 |
0 |
153 |
0 |
1.037 |
0 |
153 |
0 |
0.987 |
0 |
153 |
0 |
1.025 |
0 |
153 |
0 |
0.967 |
0 |
153 |
0 |
0.785 |
0 |
153 |
0 |
0.861 |
0 |
153 |
0 |
0.893 |
0 |
153 |
0 |
0.941 |
0 |
153 |
0 |
0.762 |
0 |
153 |
0 |
0.976 |
0 |
153 |
0 |
1.019 |
0 |
153 |
0 |
0.889 |
0 |
153 |
0 |
0.938 |
0 |
153 |
0 |
0.961 |
0 |
154 |
0 |
0.999 |
0 |
154 |
0 |
0.880 |
0 |
154 |
0 |
0.971 |
0 |
154 |
0 |
0.924 |
0 |
154 |
0 |
0.939 |
0 |
154 |
0 |
0.966 |
0 |
154 |
0 |
0.981 |
0 |
154 |
0 |
0.974 |
0 |
154 |
0 |
0.965 |
0 |
154 |
0 |
0.938 |
0 |
154 |
0 |
0.903 |
0 |
154 |
0 |
1.094 |
0 |
154 |
0 |
0.867 |
0 |
154 |
0 |
1.007 |
0 |
54 |
750 |
0.768 |
0 |
54 |
750 |
0.731 |
0 |
54 |
750 |
0.716 |
0 |
54 |
750 |
0.693 |
1 |
54 |
750 |
0.769 |
0 |
54 |
750 |
0.731 |
0 |
54 |
750 |
0.791 |
0 |
54 |
750 |
0.765 |
1 |
54 |
750 |
0.611 |
1 |
54 |
750 |
0.745 |
0 |
59 |
750 |
0.747 |
0 |
59 |
750 |
0.755 |
0 |
59 |
750 |
0.600 |
0 |
59 |
750 |
0.796 |
0 |
59 |
750 |
0.719 |
0 |
59 |
750 |
0.458 |
0 |
59 |
750 |
0.599 |
1 |
59 |
750 |
0.635 |
0 |
59 |
750 |
0.775 |
1 |
59 |
750 |
0.660 |
0 |
59 |
750 |
0.736 |
0 |
59 |
750 |
0.745 |
0 |
59 |
750 |
0.732 |
0 |
59 |
750 |
0.653 |
1 |
62 |
750 |
0.869 |
0 |
62 |
750 |
0.834 |
1 |
62 |
750 |
0.892 |
0 |
62 |
750 |
0.542 |
0 |
62 |
750 |
0.940 |
0 |
62 |
750 |
0.814 |
1 |
62 |
750 |
0.872 |
0 |
62 |
750 |
0.772 |
0 |
62 |
750 |
0.847 |
0 |
62 |
750 |
0.946 |
0 |
62 |
750 |
0.742 |
0 |
67 |
750 |
0.959 |
0 |
67 |
750 |
0.931 |
0 |
67 |
750 |
0.967 |
0 |
67 |
750 |
0.851 |
0 |
67 |
750 |
1.061 |
0 |
67 |
750 |
0.866 |
0 |
67 |
750 |
0.838 |
0 |
67 |
750 |
0.911 |
0 |
67 |
750 |
0.885 |
0 |
67 |
750 |
1.041 |
0 |
67 |
750 |
0.940 |
0 |
67 |
750 |
0.951 |
0 |
70 |
750 |
0.836 |
0 |
70 |
750 |
0.897 |
0 |
70 |
750 |
0.738 |
0 |
70 |
750 |
0.793 |
1 |
70 |
750 |
0.764 |
0 |
70 |
750 |
0.759 |
0 |
70 |
750 |
0.872 |
0 |
70 |
750 |
0.802 |
0 |
70 |
750 |
0.857 |
0 |
70 |
750 |
0.875 |
0 |
70 |
750 |
0.830 |
0 |
70 |
750 |
0.778 |
0 |
75 |
750 |
0.860 |
0 |
75 |
750 |
0.871 |
0 |
75 |
750 |
0.794 |
1 |
75 |
750 |
0.879 |
0 |
75 |
750 |
0.674 |
1 |
75 |
750 |
0.817 |
0 |
75 |
750 |
0.913 |
0 |
75 |
750 |
0.846 |
0 |
75 |
750 |
0.931 |
1 |
75 |
750 |
0.953 |
0 |
75 |
750 |
0.809 |
0 |
75 |
750 |
0.918 |
0 |
75 |
750 |
0.850 |
0 |
78 |
750 |
0.940 |
0 |
78 |
750 |
0.871 |
0 |
78 |
750 |
0.834 |
0 |
78 |
750 |
0.963 |
0 |
78 |
750 |
0.945 |
0 |
78 |
750 |
0.966 |
0 |
78 |
750 |
0.911 |
0 |
78 |
750 |
0.934 |
0 |
78 |
750 |
1.018 |
0 |
78 |
750 |
0.948 |
0 |
78 |
750 |
0.948 |
0 |
78 |
750 |
0.947 |
0 |
78 |
750 |
0.963 |
0 |
78 |
750 |
0.837 |
0 |
78 |
750 |
0.855 |
0 |
86 |
750 |
0.815 |
0 |
86 |
750 |
0.961 |
0 |
86 |
750 |
0.920 |
0 |
86 |
750 |
0.963 |
0 |
86 |
750 |
0.812 |
0 |
86 |
750 |
0.960 |
0 |
86 |
750 |
0.982 |
0 |
86 |
750 |
0.966 |
0 |
86 |
750 |
0.874 |
0 |
86 |
750 |
0.854 |
0 |
86 |
750 |
0.900 |
0 |
91 |
750 |
0.943 |
0 |
91 |
750 |
0.898 |
0 |
91 |
750 |
0.895 |
0 |
91 |
750 |
0.938 |
0 |
91 |
750 |
0.870 |
0 |
91 |
750 |
0.878 |
0 |
91 |
750 |
0.849 |
0 |
91 |
750 |
0.802 |
0 |
94 |
750 |
1.014 |
0 |
94 |
750 |
1.043 |
0 |
94 |
750 |
0.967 |
0 |
94 |
750 |
0.967 |
0 |
94 |
750 |
0.936 |
0 |
94 |
750 |
0.996 |
1 |
94 |
750 |
0.909 |
0 |
94 |
750 |
0.962 |
0 |
94 |
750 |
0.714 |
0 |
94 |
750 |
0.979 |
0 |
94 |
750 |
0.921 |
0 |
99 |
750 |
1.010 |
0 |
99 |
750 |
1.053 |
0 |
99 |
750 |
1.006 |
0 |
99 |
750 |
0.961 |
0 |
99 |
750 |
1.043 |
0 |
99 |
750 |
0.984 |
0 |
99 |
750 |
1.034 |
0 |
99 |
750 |
1.031 |
0 |
99 |
750 |
0.894 |
0 |
99 |
750 |
1.004 |
0 |
102 |
750 |
0.970 |
0 |
102 |
750 |
0.952 |
0 |
102 |
750 |
0.932 |
0 |
102 |
750 |
0.954 |
0 |
102 |
750 |
1.030 |
0 |
102 |
750 |
0.979 |
0 |
102 |
750 |
0.890 |
0 |
102 |
750 |
1.100 |
0 |
102 |
750 |
0.965 |
0 |
102 |
750 |
1.110 |
0 |
102 |
750 |
0.965 |
0 |
102 |
750 |
1.010 |
0 |
102 |
750 |
1.080 |
1 |
102 |
750 |
1.020 |
0 |
108 |
750 |
0.812 |
0 |
108 |
750 |
0.813 |
0 |
108 |
750 |
0.833 |
0 |
108 |
750 |
0.739 |
0 |
108 |
750 |
0.819 |
0 |
108 |
750 |
0.874 |
0 |
108 |
750 |
0.800 |
0 |
108 |
750 |
0.728 |
0 |
108 |
750 |
0.774 |
1 |
108 |
750 |
0.877 |
0 |
116 |
750 |
0.564 |
0 |
116 |
750 |
0.835 |
0 |
116 |
750 |
0.722 |
0 |
116 |
750 |
0.778 |
0 |
116 |
750 |
0.810 |
0 |
116 |
750 |
0.816 |
0 |
116 |
750 |
0.838 |
0 |
116 |
750 |
0.691 |
0 |
116 |
750 |
0.695 |
0 |
116 |
750 |
0.903 |
0 |
116 |
750 |
0.731 |
0 |
116 |
750 |
0.811 |
0 |
116 |
750 |
0.770 |
0 |
116 |
750 |
0.791 |
0 |
121 |
750 |
0.928 |
0 |
121 |
750 |
0.879 |
0 |
121 |
750 |
0.906 |
1 |
121 |
750 |
0.841 |
0 |
121 |
750 |
0.906 |
0 |
121 |
750 |
0.899 |
0 |
121 |
750 |
0.888 |
0 |
121 |
750 |
0.865 |
0 |
121 |
750 |
0.914 |
0 |
121 |
750 |
0.859 |
0 |
121 |
750 |
0.927 |
0 |
121 |
750 |
0.905 |
0 |
124 |
750 |
0.936 |
0 |
124 |
750 |
0.930 |
0 |
124 |
750 |
0.845 |
0 |
124 |
750 |
0.757 |
0 |
124 |
750 |
0.868 |
0 |
124 |
750 |
0.759 |
0 |
124 |
750 |
0.931 |
0 |
124 |
750 |
0.920 |
0 |
124 |
750 |
0.921 |
1 |
124 |
750 |
0.848 |
0 |
129 |
750 |
1.016 |
0 |
129 |
750 |
0.988 |
0 |
129 |
750 |
0.900 |
0 |
129 |
750 |
0.820 |
0 |
129 |
750 |
0.949 |
0 |
129 |
750 |
0.826 |
1 |
129 |
750 |
0.828 |
0 |
129 |
750 |
1.003 |
0 |
129 |
750 |
0.924 |
0 |
129 |
750 |
0.879 |
0 |
129 |
750 |
0.913 |
0 |
132 |
750 |
0.835 |
1 |
132 |
750 |
0.876 |
0 |
132 |
750 |
0.894 |
0 |
132 |
750 |
0.841 |
0 |
132 |
750 |
0.853 |
0 |
132 |
750 |
0.733 |
0 |
132 |
750 |
0.860 |
0 |
132 |
750 |
0.814 |
0 |
132 |
750 |
0.835 |
0 |
132 |
750 |
0.995 |
1 |
132 |
750 |
0.864 |
0 |
132 |
750 |
0.870 |
0 |
132 |
750 |
0.802 |
1 |
137 |
750 |
0.875 |
0 |
137 |
750 |
0.925 |
0 |
137 |
750 |
0.813 |
0 |
137 |
750 |
0.819 |
0 |
137 |
750 |
0.961 |
0 |
137 |
750 |
0.957 |
0 |
137 |
750 |
0.903 |
0 |
137 |
750 |
0.895 |
0 |
137 |
750 |
0.876 |
0 |
137 |
750 |
0.943 |
0 |
137 |
750 |
0.882 |
0 |
137 |
750 |
0.831 |
0 |
137 |
750 |
0.911 |
0 |
137 |
750 |
0.857 |
0 |
137 |
750 |
0.912 |
0 |
140 |
750 |
0.874 |
0 |
140 |
750 |
0.867 |
0 |
140 |
750 |
0.815 |
0 |
140 |
750 |
0.832 |
0 |
140 |
750 |
0.875 |
0 |
140 |
750 |
0.876 |
0 |
140 |
750 |
0.820 |
0 |
140 |
750 |
0.857 |
0 |
140 |
750 |
0.861 |
0 |
140 |
750 |
0.858 |
0 |
140 |
750 |
0.878 |
0 |
140 |
750 |
0.836 |
0 |
140 |
750 |
0.828 |
0 |
140 |
750 |
0.903 |
0 |
140 |
750 |
0.874 |
0 |
145 |
750 |
0.957 |
1 |
145 |
750 |
1.074 |
0 |
145 |
750 |
0.993 |
0 |
145 |
750 |
0.854 |
0 |
145 |
750 |
1.043 |
1 |
145 |
750 |
0.912 |
0 |
145 |
750 |
0.952 |
0 |
148 |
750 |
1.008 |
0 |
148 |
750 |
1.048 |
0 |
148 |
750 |
0.911 |
0 |
148 |
750 |
0.998 |
0 |
148 |
750 |
0.851 |
0 |
148 |
750 |
0.868 |
1 |
148 |
750 |
0.842 |
0 |
148 |
750 |
0.744 |
0 |
148 |
750 |
1.037 |
0 |
148 |
750 |
1.027 |
0 |
148 |
750 |
1.015 |
0 |
148 |
750 |
1.033 |
0 |
152 |
750 |
1.045 |
0 |
152 |
750 |
1.026 |
0 |
152 |
750 |
1.116 |
0 |
152 |
750 |
0.794 |
1 |
152 |
750 |
1.038 |
0 |
152 |
750 |
1.105 |
0 |
152 |
750 |
0.939 |
0 |
155 |
750 |
0.809 |
0 |
155 |
750 |
0.914 |
0 |
155 |
750 |
0.772 |
1 |
155 |
750 |
0.837 |
0 |
155 |
750 |
0.960 |
0 |
155 |
750 |
0.913 |
0 |
155 |
750 |
0.883 |
0 |
155 |
750 |
0.892 |
0 |
155 |
750 |
0.831 |
0 |
55 |
1500 |
0.673 |
1 |
55 |
1500 |
0.750 |
1 |
55 |
1500 |
0.680 |
0 |
55 |
1500 |
0.501 |
0 |
55 |
1500 |
0.712 |
1 |
55 |
1500 |
0.662 |
1 |
55 |
1500 |
0.631 |
1 |
55 |
1500 |
0.647 |
1 |
55 |
1500 |
0.708 |
1 |
55 |
1500 |
0.657 |
1 |
58 |
1500 |
0.670 |
1 |
58 |
1500 |
0.716 |
0 |
58 |
1500 |
0.749 |
0 |
58 |
1500 |
0.672 |
1 |
58 |
1500 |
0.676 |
1 |
58 |
1500 |
0.682 |
1 |
58 |
1500 |
0.735 |
1 |
58 |
1500 |
0.717 |
1 |
58 |
1500 |
0.727 |
0 |
58 |
1500 |
0.679 |
0 |
58 |
1500 |
0.674 |
0 |
63 |
1500 |
0.673 |
0 |
63 |
1500 |
0.644 |
0 |
63 |
1500 |
0.605 |
0 |
63 |
1500 |
0.627 |
0 |
63 |
1500 |
0.457 |
1 |
63 |
1500 |
0.656 |
1 |
63 |
1500 |
0.665 |
0 |
63 |
1500 |
0.748 |
0 |
63 |
1500 |
0.755 |
1 |
63 |
1500 |
0.743 |
1 |
63 |
1500 |
0.744 |
1 |
63 |
1500 |
0.645 |
0 |
66 |
1500 |
0.574 |
0 |
66 |
1500 |
0.619 |
1 |
66 |
1500 |
0.607 |
1 |
66 |
1500 |
0.637 |
1 |
66 |
1500 |
0.573 |
1 |
66 |
1500 |
0.600 |
1 |
66 |
1500 |
0.623 |
1 |
66 |
1500 |
0.554 |
1 |
66 |
1500 |
0.658 |
1 |
66 |
1500 |
0.624 |
0 |
66 |
1500 |
0.582 |
1 |
66 |
1500 |
0.526 |
1 |
66 |
1500 |
0.467 |
1 |
71 |
1500 |
0.722 |
1 |
71 |
1500 |
0.419 |
1 |
71 |
1500 |
0.704 |
1 |
71 |
1500 |
0.718 |
1 |
71 |
1500 |
0.695 |
0 |
71 |
1500 |
0.867 |
1 |
71 |
1500 |
0.800 |
1 |
71 |
1500 |
0.827 |
0 |
71 |
1500 |
0.756 |
0 |
74 |
1500 |
0.677 |
1 |
74 |
1500 |
0.749 |
0 |
74 |
1500 |
0.686 |
1 |
74 |
1500 |
0.788 |
0 |
74 |
1500 |
0.719 |
0 |
74 |
1500 |
0.652 |
1 |
74 |
1500 |
0.771 |
0 |
74 |
1500 |
0.740 |
1 |
74 |
1500 |
0.570 |
1 |
74 |
1500 |
0.778 |
0 |
74 |
1500 |
0.809 |
0 |
74 |
1500 |
0.658 |
1 |
74 |
1500 |
0.758 |
1 |
79 |
1500 |
1.154 |
0 |
82 |
1500 |
0.760 |
0 |
82 |
1500 |
0.786 |
1 |
82 |
1500 |
0.828 |
0 |
82 |
1500 |
0.791 |
0 |
82 |
1500 |
0.644 |
1 |
82 |
1500 |
0.815 |
0 |
82 |
1500 |
0.730 |
0 |
82 |
1500 |
0.872 |
0 |
82 |
1500 |
0.943 |
0 |
82 |
1500 |
0.833 |
0 |
82 |
1500 |
0.809 |
1 |
87 |
1500 |
0.925 |
0 |
87 |
1500 |
0.866 |
0 |
87 |
1500 |
0.789 |
1 |
87 |
1500 |
0.857 |
0 |
87 |
1500 |
0.904 |
1 |
87 |
1500 |
0.793 |
1 |
87 |
1500 |
0.800 |
1 |
87 |
1500 |
0.822 |
1 |
87 |
1500 |
0.789 |
1 |
87 |
1500 |
0.711 |
1 |
87 |
1500 |
0.703 |
1 |
90 |
1500 |
0.654 |
0 |
90 |
1500 |
0.707 |
0 |
90 |
1500 |
0.674 |
0 |
90 |
1500 |
0.629 |
0 |
90 |
1500 |
0.772 |
0 |
90 |
1500 |
0.784 |
0 |
90 |
1500 |
0.861 |
0 |
90 |
1500 |
0.700 |
0 |
90 |
1500 |
0.763 |
1 |
90 |
1500 |
0.834 |
0 |
90 |
1500 |
0.645 |
1 |
90 |
1500 |
0.716 |
0 |
90 |
1500 |
0.712 |
0 |
90 |
1500 |
0.714 |
0 |
95 |
1500 |
0.942 |
0 |
95 |
1500 |
0.871 |
0 |
95 |
1500 |
0.973 |
0 |
95 |
1500 |
0.893 |
0 |
95 |
1500 |
0.788 |
0 |
95 |
1500 |
0.835 |
0 |
95 |
1500 |
0.917 |
0 |
95 |
1500 |
0.821 |
1 |
95 |
1500 |
0.870 |
0 |
95 |
1500 |
0.816 |
0 |
95 |
1500 |
0.848 |
0 |
95 |
1500 |
0.951 |
0 |
95 |
1500 |
0.874 |
0 |
98 |
1500 |
0.881 |
0 |
98 |
1500 |
0.804 |
0 |
98 |
1500 |
0.795 |
1 |
98 |
1500 |
0.733 |
1 |
98 |
1500 |
0.656 |
1 |
98 |
1500 |
0.850 |
1 |
98 |
1500 |
0.737 |
1 |
98 |
1500 |
0.851 |
0 |
98 |
1500 |
0.883 |
1 |
98 |
1500 |
0.814 |
1 |
98 |
1500 |
0.715 |
1 |
98 |
1500 |
0.728 |
1 |
98 |
1500 |
0.831 |
0 |
98 |
1500 |
0.792 |
0 |
98 |
1500 |
0.711 |
1 |
103 |
1500 |
0.895 |
0 |
103 |
1500 |
0.931 |
1 |
103 |
1500 |
0.916 |
1 |
103 |
1500 |
0.848 |
0 |
103 |
1500 |
0.654 |
0 |
103 |
1500 |
0.613 |
1 |
103 |
1500 |
0.842 |
0 |
103 |
1500 |
0.669 |
1 |
103 |
1500 |
0.831 |
0 |
103 |
1500 |
0.788 |
1 |
103 |
1500 |
0.835 |
0 |
105 |
1500 |
0.877 |
0 |
105 |
1500 |
0.954 |
0 |
105 |
1500 |
0.886 |
0 |
105 |
1500 |
0.895 |
0 |
105 |
1500 |
0.840 |
0 |
105 |
1500 |
0.723 |
0 |
105 |
1500 |
0.797 |
0 |
105 |
1500 |
0.836 |
0 |
105 |
1500 |
0.726 |
0 |
105 |
1500 |
0.727 |
1 |
105 |
1500 |
0.837 |
0 |
105 |
1500 |
0.715 |
0 |
112 |
1500 |
0.810 |
0 |
112 |
1500 |
0.812 |
0 |
112 |
1500 |
0.782 |
1 |
112 |
1500 |
0.757 |
0 |
112 |
1500 |
0.749 |
0 |
112 |
1500 |
0.779 |
0 |
112 |
1500 |
0.686 |
1 |
112 |
1500 |
0.760 |
1 |
112 |
1500 |
0.795 |
0 |
112 |
1500 |
0.785 |
0 |
112 |
1500 |
0.708 |
0 |
112 |
1500 |
0.660 |
0 |
112 |
1500 |
0.644 |
0 |
117 |
1500 |
0.785 |
0 |
117 |
1500 |
0.762 |
0 |
117 |
1500 |
0.744 |
0 |
117 |
1500 |
0.709 |
0 |
117 |
1500 |
0.718 |
0 |
117 |
1500 |
0.785 |
0 |
117 |
1500 |
0.820 |
0 |
117 |
1500 |
0.674 |
0 |
117 |
1500 |
0.811 |
0 |
117 |
1500 |
0.845 |
0 |
117 |
1500 |
0.693 |
0 |
117 |
1500 |
0.743 |
0 |
117 |
1500 |
0.723 |
0 |
125 |
1500 |
0.705 |
1 |
125 |
1500 |
0.607 |
1 |
125 |
1500 |
0.709 |
1 |
125 |
1500 |
0.715 |
0 |
125 |
1500 |
0.713 |
1 |
125 |
1500 |
0.765 |
1 |
125 |
1500 |
0.723 |
1 |
128 |
1500 |
0.820 |
0 |
128 |
1500 |
0.909 |
0 |
128 |
1500 |
0.925 |
0 |
128 |
1500 |
0.884 |
0 |
128 |
1500 |
0.788 |
1 |
128 |
1500 |
0.822 |
0 |
128 |
1500 |
0.816 |
0 |
128 |
1500 |
0.789 |
1 |
136 |
1500 |
0.877 |
0 |
136 |
1500 |
1.000 |
0 |
136 |
1500 |
0.817 |
1 |
141 |
1500 |
0.808 |
0 |
141 |
1500 |
0.879 |
0 |
141 |
1500 |
0.816 |
0 |
141 |
1500 |
0.939 |
0 |
141 |
1500 |
0.824 |
0 |
141 |
1500 |
0.922 |
0 |
141 |
1500 |
0.947 |
0 |
144 |
1500 |
0.792 |
0 |
144 |
1500 |
0.868 |
0 |
144 |
1500 |
0.526 |
1 |
144 |
1500 |
0.816 |
0 |
144 |
1500 |
0.770 |
1 |
144 |
1500 |
0.760 |
1 |
144 |
1500 |
0.771 |
1 |
144 |
1500 |
0.854 |
0 |
144 |
1500 |
0.921 |
0 |
144 |
1500 |
0.840 |
0 |
149 |
1500 |
0.844 |
0 |
149 |
1500 |
0.957 |
0 |
149 |
1500 |
0.929 |
0 |
149 |
1500 |
0.809 |
0 |
149 |
1500 |
0.883 |
0 |
149 |
1500 |
0.856 |
0 |
149 |
1500 |
0.755 |
0 |
149 |
1500 |
0.693 |
0 |
149 |
1500 |
0.914 |
0 |
149 |
1500 |
0.905 |
0 |
149 |
1500 |
0.860 |
0 |
149 |
1500 |
0.828 |
0 |
56 |
3000 |
0.678 |
1 |
56 |
3000 |
0.765 |
0 |
56 |
3000 |
0.518 |
1 |
56 |
3000 |
0.629 |
1 |
56 |
3000 |
0.763 |
0 |
56 |
3000 |
0.658 |
1 |
56 |
3000 |
0.732 |
0 |
57 |
3000 |
0.550 |
1 |
57 |
3000 |
0.633 |
1 |
57 |
3000 |
0.498 |
1 |
57 |
3000 |
0.633 |
1 |
57 |
3000 |
0.619 |
0 |
57 |
3000 |
0.521 |
1 |
57 |
3000 |
0.507 |
1 |
57 |
3000 |
0.696 |
1 |
57 |
3000 |
0.599 |
1 |
57 |
3000 |
0.605 |
1 |
64 |
3000 |
0.718 |
0 |
64 |
3000 |
0.615 |
0 |
64 |
3000 |
0.634 |
0 |
64 |
3000 |
0.514 |
0 |
64 |
3000 |
0.716 |
1 |
64 |
3000 |
0.708 |
0 |
64 |
3000 |
0.668 |
1 |
64 |
3000 |
0.693 |
0 |
64 |
3000 |
0.627 |
1 |
64 |
3000 |
0.623 |
0 |
64 |
3000 |
0.734 |
1 |
64 |
3000 |
0.599 |
0 |
65 |
3000 |
0.702 |
1 |
65 |
3000 |
0.678 |
1 |
65 |
3000 |
0.733 |
1 |
65 |
3000 |
0.656 |
1 |
65 |
3000 |
0.647 |
1 |
65 |
3000 |
0.719 |
1 |
65 |
3000 |
0.644 |
1 |
65 |
3000 |
0.628 |
1 |
65 |
3000 |
0.653 |
1 |
65 |
3000 |
0.597 |
1 |
65 |
3000 |
0.531 |
1 |
72 |
3000 |
0.624 |
1 |
72 |
3000 |
0.827 |
1 |
72 |
3000 |
0.846 |
1 |
72 |
3000 |
0.778 |
1 |
72 |
3000 |
0.764 |
0 |
72 |
3000 |
0.838 |
1 |
72 |
3000 |
0.841 |
1 |
72 |
3000 |
0.919 |
1 |
72 |
3000 |
0.852 |
0 |
72 |
3000 |
0.833 |
0 |
73 |
3000 |
0.665 |
1 |
73 |
3000 |
0.665 |
1 |
73 |
3000 |
0.563 |
0 |
73 |
3000 |
0.649 |
1 |
73 |
3000 |
0.617 |
1 |
73 |
3000 |
0.617 |
1 |
73 |
3000 |
0.550 |
1 |
73 |
3000 |
0.621 |
1 |
73 |
3000 |
0.607 |
1 |
73 |
3000 |
0.599 |
1 |
80 |
3000 |
1.025 |
1 |
80 |
3000 |
0.490 |
1 |
80 |
3000 |
1.004 |
1 |
80 |
3000 |
0.968 |
0 |
81 |
3000 |
0.687 |
1 |
81 |
3000 |
0.593 |
1 |
81 |
3000 |
0.631 |
1 |
81 |
3000 |
0.713 |
1 |
81 |
3000 |
0.747 |
1 |
81 |
3000 |
0.685 |
1 |
81 |
3000 |
0.791 |
1 |
89 |
3000 |
0.660 |
0 |
89 |
3000 |
0.640 |
0 |
89 |
3000 |
0.756 |
0 |
89 |
3000 |
0.727 |
1 |
96 |
3000 |
0.619 |
1 |
96 |
3000 |
0.601 |
1 |
96 |
3000 |
0.665 |
1 |
96 |
3000 |
0.572 |
1 |
96 |
3000 |
0.472 |
1 |
96 |
3000 |
0.561 |
1 |
96 |
3000 |
0.637 |
1 |
96 |
3000 |
0.627 |
1 |
96 |
3000 |
0.621 |
1 |
96 |
3000 |
0.637 |
1 |
96 |
3000 |
0.596 |
1 |
96 |
3000 |
0.591 |
1 |
97 |
3000 |
0.686 |
1 |
97 |
3000 |
0.685 |
1 |
97 |
3000 |
0.464 |
1 |
97 |
3000 |
0.649 |
1 |
97 |
3000 |
0.595 |
1 |
97 |
3000 |
0.523 |
1 |
97 |
3000 |
0.640 |
1 |
97 |
3000 |
0.618 |
1 |
97 |
3000 |
0.566 |
1 |
97 |
3000 |
0.539 |
1 |
97 |
3000 |
0.606 |
1 |
97 |
3000 |
0.585 |
1 |
104 |
3000 |
0.684 |
1 |
104 |
3000 |
0.748 |
1 |
104 |
3000 |
0.701 |
1 |
104 |
3000 |
0.484 |
0 |
104 |
3000 |
0.779 |
1 |
104 |
3000 |
0.734 |
1 |
104 |
3000 |
0.777 |
1 |
104 |
3000 |
0.893 |
1 |
104 |
3000 |
0.812 |
0 |
104 |
3000 |
0.852 |
1 |
104 |
3000 |
0.770 |
1 |
104 |
3000 |
0.734 |
1 |
104 |
3000 |
0.733 |
0 |
104 |
3000 |
0.751 |
1 |
110 |
3000 |
0.686 |
1 |
110 |
3000 |
0.762 |
0 |
110 |
3000 |
0.798 |
0 |
110 |
3000 |
0.804 |
0 |
110 |
3000 |
0.779 |
0 |
110 |
3000 |
0.731 |
0 |
110 |
3000 |
0.830 |
1 |
110 |
3000 |
0.644 |
1 |
110 |
3000 |
0.764 |
0 |
110 |
3000 |
0.750 |
1 |
110 |
3000 |
0.721 |
1 |
119 |
3000 |
0.669 |
0 |
119 |
3000 |
0.716 |
0 |
119 |
3000 |
0.717 |
0 |
119 |
3000 |
0.629 |
0 |
119 |
3000 |
0.690 |
0 |
119 |
3000 |
0.755 |
1 |
119 |
3000 |
0.663 |
0 |
119 |
3000 |
0.618 |
1 |
119 |
3000 |
0.717 |
1 |
119 |
3000 |
0.690 |
1 |
119 |
3000 |
0.675 |
0 |
119 |
3000 |
0.677 |
0 |
126 |
3000 |
0.727 |
0 |
126 |
3000 |
0.677 |
0 |
126 |
3000 |
0.623 |
1 |
126 |
3000 |
0.618 |
1 |
126 |
3000 |
0.641 |
1 |
126 |
3000 |
0.592 |
0 |
126 |
3000 |
0.768 |
1 |
126 |
3000 |
0.816 |
0 |
126 |
3000 |
0.498 |
0 |
126 |
3000 |
0.644 |
1 |
126 |
3000 |
0.743 |
0 |
126 |
3000 |
0.662 |
0 |
127 |
3000 |
0.713 |
0 |
127 |
3000 |
0.411 |
1 |
127 |
3000 |
0.768 |
0 |
127 |
3000 |
0.665 |
0 |
127 |
3000 |
0.711 |
0 |
127 |
3000 |
0.719 |
0 |
127 |
3000 |
0.710 |
0 |
127 |
3000 |
0.722 |
0 |
127 |
3000 |
0.603 |
1 |
127 |
3000 |
0.662 |
0 |
127 |
3000 |
0.649 |
1 |
127 |
3000 |
0.614 |
0 |
134 |
3000 |
0.402 |
1 |
134 |
3000 |
0.507 |
1 |
134 |
3000 |
0.451 |
1 |
134 |
3000 |
0.517 |
1 |
134 |
3000 |
0.475 |
1 |
134 |
3000 |
0.576 |
1 |
134 |
3000 |
0.651 |
0 |
134 |
3000 |
0.618 |
1 |
134 |
3000 |
0.539 |
1 |
134 |
3000 |
0.705 |
1 |
135 |
3000 |
0.687 |
0 |
135 |
3000 |
0.847 |
0 |
135 |
3000 |
0.817 |
0 |
135 |
3000 |
0.842 |
0 |
135 |
3000 |
0.699 |
0 |
135 |
3000 |
0.838 |
0 |
135 |
3000 |
0.801 |
0 |
135 |
3000 |
0.635 |
0 |
135 |
3000 |
0.761 |
0 |
142 |
3000 |
0.696 |
0 |
142 |
3000 |
0.573 |
1 |
142 |
3000 |
0.656 |
1 |
142 |
3000 |
0.688 |
1 |
142 |
3000 |
0.689 |
1 |
142 |
3000 |
0.587 |
0 |
142 |
3000 |
0.608 |
0 |
142 |
3000 |
0.643 |
1 |
142 |
3000 |
0.618 |
0 |
143 |
3000 |
1.198 |
0 |
143 |
3000 |
0.758 |
0 |
143 |
3000 |
0.929 |
0 |
143 |
3000 |
0.930 |
0 |
143 |
3000 |
1.005 |
1 |
143 |
3000 |
0.917 |
1 |
143 |
3000 |
0.910 |
0 |
150 |
3000 |
0.733 |
0 |
150 |
3000 |
0.731 |
1 |
150 |
3000 |
0.810 |
1 |
150 |
3000 |
0.934 |
0 |
150 |
3000 |
0.785 |
0 |
150 |
3000 |
0.799 |
0 |
150 |
3000 |
0.742 |
0 |
150 |
3000 |
0.921 |
0 |
150 |
3000 |
0.957 |
0 |
150 |
3000 |
0.894 |
0 |
150 |
3000 |
0.870 |
0 |
150 |
3000 |
0.791 |
1 |
151 |
3000 |
0.883 |
0 |
151 |
3000 |
0.900 |
0 |
151 |
3000 |
0.756 |
1 |
151 |
3000 |
0.776 |
0 |
151 |
3000 |
0.857 |
0 |
151 |
3000 |
0.769 |
0 |
151 |
3000 |
0.890 |
1 |
156 |
3000 |
0.811 |
1 |
156 |
3000 |
1.002 |
0 |
156 |
3000 |
0.877 |
0 |
156 |
3000 |
0.790 |
0 |
156 |
3000 |
0.902 |
0 |
156 |
3000 |
0.864 |
0 |
156 |
3000 |
0.856 |
0 |
156 |
3000 |
0.797 |
0 |
156 |
3000 |
0.839 |
0 |
156 |
3000 |
0.867 |
0 |
156 |
3000 |
0.724 |
0 |
156 |
3000 |
0.829 |
0 |
; | |||
run; | |||
%inc ‘C:\ml.mac’; | |||
options nocenter mprint; | |||
%ml (data=EG_data, | |||
bin=Fetal_Mal,norm=Fetal_Wt, | |||
x=Dose, | |||
id=Litter_ID, | |||
method=ISAMP, | |||
qpoints=200, maxit=100, | |||
tech=NRRIDG, it_his=yes); | |||
Acknowledgments
The authors are grateful for the support provided by grants ROI-CA69222 and ROI-AI60373 from the National Institutes of Health.
Footnotes
Supplementary Materials: Web Appendix derivations of equations (10) and (11) for Xij = 0, as well as the EG data analyzed in Section 4 and an SAS macro implementing the ML method are available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
References
- Aerts M, Geys H, Molenberghs G, Ryan LM. Topics in Modelling of Clustered Data. Boca Raton, Florida: Chapman and Hall; 2002. [Google Scholar]
- Catalano P, Ryan LM. Bivariate latent variable models for clustered discrete and continuous outcomes. Journal of the American Statistical Association. 1992;87:651–658. [Google Scholar]
- Cleveland WS. Robust locally weighted regression and smoothing scatterplots. Journal of the American Statistical Association. 1979;74:829–836. [Google Scholar]
- Cox DR, Wermuth N. Response models for mixed binary and quantitative variables. Biometrika. 1992;79:441–461. [Google Scholar]
- Cox DR, Wermuth N. Multivariate Dependencies: Models, Analysis and Interpretation. London: Chapman and Hall; 1994. [Google Scholar]
- Dunson DB, Chen Z, Harry J. A Bayesian approach for joint modeling of cluster size and subunit-specific outcomes. Biometrics. 2003;59:521–530. doi: 10.1111/1541-0420.00062. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice GM, Laird NM. Regression models for a bivariate discrete and continuous outcome with clustering. Journal of the American Statistical Association. 1995;90:845–852. [Google Scholar]
- Geys H, Molenberghs G, Ryan LM. Pseudolikelihood modeling of multivariate outcomes in developmental toxicology. Journal of the American Statistical Association. 1999;94:734–745. [Google Scholar]
- Gueorguieva R, Agresti A. A correlated probit model for joint modeling of clustered binary and continuous responses. Journal of the American Statistical Association. 2001;96:1102–1112. [Google Scholar]
- Hougaard P. Analysis of Multivariate Survival Data. New York: Springer; 2000. [Google Scholar]
- Krzanowski WJ. Principles of Multivariate Analysis. Oxford: Clarendon Press; 1988. [Google Scholar]
- Lehmann EL, Casella G. Theory of Point Estimation. 2nd edition. New York: Springer; 1998. [Google Scholar]
- Little RJA, Schluchter MD. Maximum likelihood estimation for mixed continuous and categorical data with missing values. Biometrika. 1985;72:497–512. [Google Scholar]
- Price CJ, Kimmel CA, Tyl RW, Marr MC. The developmental toxicity of ethylene glycol in rats and mice. Toxicological Applications in Pharmacology. 1985;81:113–127. doi: 10.1016/0041-008x(85)90126-7. [DOI] [PubMed] [Google Scholar]
- Regan MM, Catalano PJ. Likelihood models for clustered binary and continuous outcomes: Applications to developmental toxicology. Biometrics. 1999;55:760–768. doi: 10.1111/j.0006-341x.1999.00760.x. [DOI] [PubMed] [Google Scholar]
- Royston P, Altman DG. Regression using fractional polynomials of continuous covariates: Parsimonious parametric modelling. Applied Statistics. 1994;43:429–467. [Google Scholar]
- Schafer JL. Analysis of Incomplete Multivariate Data. London: Chapman and Hall; 1997. [Google Scholar]
- Wang Z, Louis TA. Matching conditional and marginal shapes in binary mixed-effects models using a bridge distribution function. Biometrika. 2003;90:765–775. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


