Abstract
Previous model studies of peptides and proteins have shown that protein–lipid interactions, primarily involving amino acid side chains near the membrane–water interface, modulate the position of transmembrane helices in bilayers. The present study examines whether such interfacial side chains stabilize the signaling states of a transmembrane signaling helix in a representative receptor, the aspartate receptor of bacterial chemotaxis. To examine the functional roles of signaling helix side chains at the periplasmic and cytoplasmic membrane–water interfaces, arginine and cysteine substitutions were scanned through these two interfacial regions. The chemical reactivities of the cysteine residues were first measured to determine the positions at which the helix crosses the membrane–water interface in both the periplasmic and cytoplasmic compartments. Subsequently, two antisymmetric in vitro activity measurements were carried out to determine the effect of each interfacial arginine or cysteine substitution on receptor signaling. Substitutions that stabilize the receptor on-state cause upregulation of receptor-coupled kinase activity and inhibition of methylation at receptor adaptation sites, while substitutions that stabilize the off-state have the opposite effects on these two activities. Notably, four substitutions at aromatic tryptophan and phenylalanine positions buried in the membrane near the membrane–water interface were found to stabilize the native on- or off-signaling state. The striking ability of these substitutions to drive the receptor toward a specific signaling state indicates that interfacial side chains are highly optimized to correctly position the native signaling helix in the membrane and to allow normal switching between the on- and off-signaling states. The analogous substitutions in model transmembrane helices are known to drive small piston-type displacements of the helix normal to the membrane. Thus, the simplest molecular interpretation of the present findings is that the signal-stabilizing substitutions drive piston displacements of the signaling helix, providing further support for the piston model for transmembrane signaling in bacterial chemoreceptors. More generally, the findings indicate that the interfacial phenylalanine, tryptophan, and arginine side chains widespread in the transmembrane α-helices of receptors, channels, and transporters can play important roles in modulating transitions between signaling and conformational states.
The chemoreceptors of Eschericia coli and Salmonella typhimurium are the best studied examples of a large class of transmembrane receptors that regulate cytoplasmic histidine kinases in bacterial taxis pathways (1–5). The structural unit of these receptors is a homodimer of two identical subunits, in which two α-helices from each subunit assemble to form a transmembrane four-helix bundle (Figure 1) (6, 7). Multiple homodimers can assemble to form trimers-of-dimers or larger oligomers, in which the individual dimers can signal cooperatively (8–12). Studies of chimeric receptors indicate that broad classes of bacterial chemoreceptors may share a conserved mechanism of transmembrane signaling (13–16). Moreover, multiple lines of evidence indicate that a specific transmembrane helix, termed the signaling helix, plays a central role in the transmission of conformational signals across the membrane (4, 17–19). Various mechanistic models propose that the conformational signal is initiated by a ligand-induced displacement of the signaling helix, with the proposed displacement described as a helix rotation or a helix tilt, or a piston-type sliding of the helix along its long axis (4, 17–21).
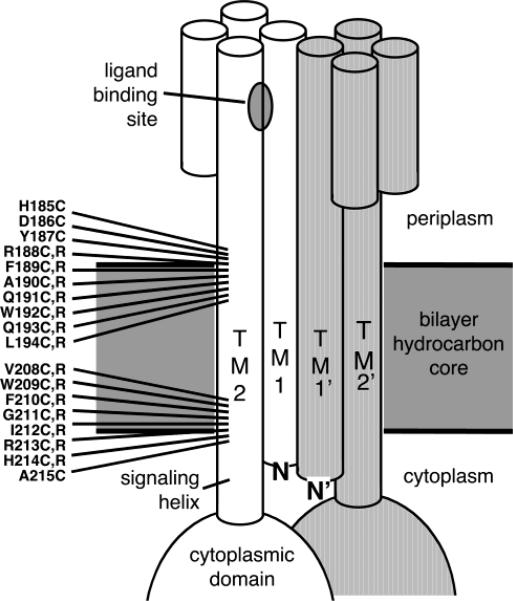
Figure 1.
Scanning approach used to map the membrane–water interfaces and search for signal-stabilizing interfacial mutations. Shown are the sites of cysteine and arginine substitutions selected for the present study, all located at interfacial positions on the signaling helix TM2 that carries signals across the membrane. The two symmetric subunits of the homodimer are shown in white and gray, respectively, and individual helices are depicted as cylinders.
Protein engineering approaches can be used to generate certain types of transmembrane helix displacements, which may be useful in determining the forces that hold helices in the correct position, as well as the type of helix displacements that occur during transitions between signaling states. Recent studies of model transmembrane peptides and proteins have revealed that amino acid side chains at the membrane–water interface have strong electrostatic and hydrophobic interactions with the surrounding lipids that control the precise positioning of transmembrane helices in membranes (22–24). As a result, side chain substitutions near the membrane–water interface can cause helix displacement. For example, the introduction of a charged side chain such as arginine into the bilayer hydrocarbon core gives rise to an electrostatic perturbation, which is relieved by displacing the entire helix toward the aqueous compartment closest to the arginine, thereby allowing the terminal guanidinium ion to snorkel into the polar region. The helix displacement ranges approximately from 1 Å (for arginine substitutions one to six residues from the end of the transmembrane helix) to 3 Å (for arginine substitutions 10 or 11 residues from the end of the helix) (22, 23). Alternatively, removal of a hydrophobic anchor such as phenylalanine near the end of a transmembrane helix can yield a helix translation of several angstroms toward the nearest aqueous compartment (24). In both cases, if the side chain substitution is placed near the cytoplasmic compartment, the helix will translate toward the cytoplasm. By contrast, a substitution near the extracellular compartment will yield helix translation in the opposite direction. Thus, the introduction of an arginine or the removal of a bulky hydrophobic can generate a transmembrane helix displacement up to several angstroms in a known direction.
The present study begins by scanning arginine and cysteine substitutions through signaling helix positions at both the periplasmic and cytoplasmic membrane–water interfaces. The chemical reactivities of the cysteine residues reveal the precise locations at which the two interfaces cross this helix. In addition, the effect of each substitution on receptor activity is determined to identify arginine and cysteine substitutions that drive the receptor into the on- or off-signaling state. Most of these substitutions perturb receptor signaling in an uninteresting fashion, yielding nonnative signaling states. However, four interesting substitutions drive the receptor toward the native on- or off-state and thus may shed light on the native signaling mechanism. The locations and chemical nature of these four substitutions are identical to the substitutions previously observed to drive piston displacements of model transmembrane helices (22–24). Together, the results indicate that the native interfacial side chains are carefully optimized to correctly position the signaling helix within the membrane and to modulate native helix movements. Further, the findings are fully consistent with the predictions of the piston model for transmembrane signaling in bacterial chemoreceptors (4, 17–19). More generally, the optimization of interfacial side chains is likely to be a widespread feature of transmembrane helices in receptors, channels, and transporters, where these residues stabilize the native structure and help determine the energetics of transitions between different conformational and functional states.
MATERIALS AND METHODS
Materials
E. coli strain RP3808, kindly provided by Dr. John S. Parkinson, University of Utah (Salt Lake City, UT), was used for plasmid overexpression of the aspartate receptor since its genome lacks the major chemoreceptors including the aspartate receptor (Δ(cheA-cheZ)DE2209 tsr-1 leuB6 his-4 eda-50 rpsL136 [thi-1 Δ(gal-attl)DE99 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx-78]/mks/). The expression plasmid pSCF6, which carries the S. typhimurium aspartate receptor gene, has been previously described, as have the plasmids, strains, and protocols used to produce purified CheA, CheW, CheY, and CheR (25). The sulfhydryl-specific probe 5-iodoacetamido fluorescein (5-IAF)1 was obtained from Molecular Probes. Enzyme substrates S-adenosyl-l-[methyl-3H]-methionine and [γ-32P]-ATP were purchased from Amersham. Deoxyoligonucleotides were synthesized by GibcoBRL and Integrated DNA Technology. Kunkel mutagenesis reagents (T7 DNA polymerase, T4 DNA ligase, and deoxynucleotide triphosphates) were purchased from BioRad. Unless specifically noted, all other reagents were obtained from Sigma and were reagent grade.
Creation and Isolation of Mutant Aspartate Receptors
Arginine and cysteine substitutions were created by oligo-nucleotide-directed mutagenesis of the plasmid pSCF6 using the Kunkel method as described previously (25, 26). Modified plasmids were transformed into E. coli strain RP3808 and expressed, and the products were isolated in native cell membranes as previously described (27).
Chemical Reactivity Assays
Cysteine reactivity assays were performed as previously described using single-cysteine receptors (Cys185 through Cys190) in isolated cell membranes (28). Briefly, each single-cysteine receptor (5 μM receptor monomer) was incubated in reaction buffer (10 mM sodium phosphate, pH 6.5 with NaOH, 50 mM NaCl, 50 mM KCl, and 10 mM EDTA) containing the sulfhydryl-specific probe 5-IAF (300 μM final). The reaction was allowed to proceed for 5 min at 25 °C, at which time a 20 μL aliquot was removed and quenched with 1.25 μL of β-mercaptoethanol. The remaining 20 μL of the reaction was denatured with heat (95 °C) and SDS (0.6% w/v) and allowed to react for an additional 3 min before quenching. 5-IAF-tagged receptors were resolved on 10% Laemmli acrylamide gels (acrylamide-to-bisacrylamide ratio of 40:0.2) and illuminated on a UV light box. Fluorescent bands were visualized and quantitated with a digital camera (Alpha Innotec). After fluorescent detection, gels were stained with Coomassie and receptor bands were quantitated with a digital camera to normalize for variations in receptor amounts. Chemical reactivity was defined as the ratio of receptor alkylation in the folded versus the unfolded states.
In Vitro Activity Assays
The in vitro receptor-coupled kinase assay was performed using receptors in isolated cell membranes essentially as described with the following modifications (27). The receptor-coupled kinase reaction was initiated by the addition of [γ-32P] ATP to a reaction mixture containing receptor, CheW, CheA, and CheY. Stoichiometries were maintained such that receptor-regulated CheA autophosphorylation was the rate-determining step; specifically, each 10 μL of reaction mixture contained 40 pmol of receptor monomer, 20 pmol of CheW, 5 pmol of CheA, 100 pmol of CheY, and 1000 pmol of ATP in reaction buffer (50 mM Tris, pH 7.5 with HCl, 50 mM KCL, and 5 mM MgCl2). After 10 s, an aliquot was removed and quenched with 2× Laemmli sample buffer supplemented with 25 mM EDTA to prevent further phosphorylation and dephosphorylation reactions. The [32P]-phospho-CheY formed by receptor-regulated autophosphorylation of CheA followed by rapid phosphotransfer to CheY was resolved on a 15% Laemmli SDS–polyacrylamide gel (acrylamide-to-bisacryl-amide ratio of 40:1.25). Gels were dried, and the amount of [32P]-phospho-CheY was quantitated by phosphorimaging (Molecular Dynamics).
The in vitro receptor methylation assay was carried out on receptors in isolated cell membranes as previously described with the following modifications (25, 29). Receptor membranes were mixed in buffer (50 mM potassium phosphate, pH 7.0 with NaOH, and 0.5 mM EDTA) containing 1 μL of a CheR-containing cytosolic extract (15–20 mg/mL total protein). Samples were then allowed to equilibrate for at least 30 min at 30 °C to allow the receptor–CheR complex to form. Methylation was initiated by the addition of a 1:1 mixture of S-adenosyl-l-methionine (SAM) and S-adenosyl-l-[methyl-3H]-methionine ([3H]-SAM) to yield a final total SAM concentration of 0.1 mM. The reaction was allowed to proceed for 20, 40, and 60 s and then stopped by spotting aliquots on a filter paper and immersing in a 10% w/v trichloroacetic acid (TCA) bath with subsequent stirring for 10 min. The TCA wash was repeated twice, followed by a pair of 2 min methanol washes to remove residual TCA. Filter papers were placed in eppendorf tubes, 100 μL of 1 M NaOH was added to the filter papers, and each tube was floated in a vial containing scintillation cocktail (Ecoscint H; National Diagnostics). Liberated [3H]-methanol partitioned into the scintillation cocktail during a >18 h incubation at 37 °C then was quantitated by scintillation counting.
Standard Deviation
Error ranges represent the standard deviation of the mean for n > 3.
RESULTS
Locations of Membrane–Water Interfaces
To examine the role of interfacial side chains in aspartate receptor signaling, it is first necessary to know which signaling helix positions are located in the membrane and which positions are exposed to aqueous solvent. Previous work developed a chemical reactivity scan method to identify the position where the signaling helix crosses the cytoplasmic lipid–water interface (28), and the same type of scan was employed herein to locate the periplasmic lipid–water interface. Briefly, six cysteine substitutions were introduced by site-directed mutagenesis at signaling helix positions 185–190 (Figure 1). Since the native receptor lacks cysteine, each engineered receptor subunit contained a single cysteine residue. The resulting mutant receptors were overexpressed in E. coli, and native cell membranes containing a given mutant protein were isolated. Subsequently, the chemical reactivity of each cysteine sulfhydryl was determined in the membrane-bound receptor by reaction with the anionic probe 5-iodoacetamidofluorescein (5-IAF), which specifically alkylates the cysteine sulfanion to generate a fluorescent product quantitated by electrophoresis and digitization of the fluorescent gel image. Reaction conditions were optimized to produce rapid labeling of cysteine residues exposed to aqueous solvent or the polar lipid headgroup phase but slow labeling of cysteine residues exposed to the apolar bilayer hydrocarbon core where the sulfhydryl pKa is substantially elevated and the anionic probe is present at low concentration.
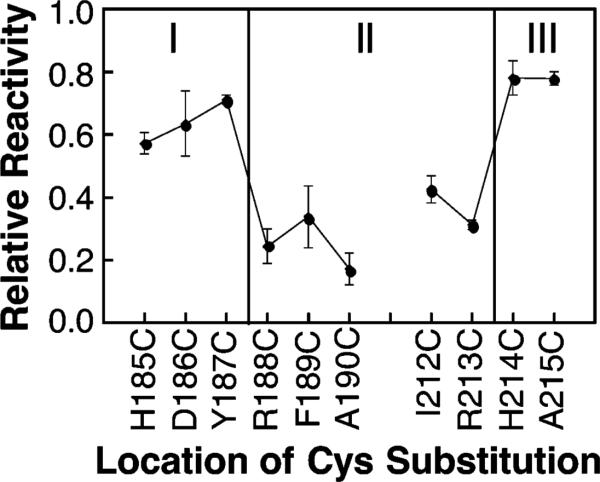
Figure 2 summarizes the chemical reactivities measured for cysteine residues at the periplasmic and cytoplasmic interfaces (present results and ref 28, respectively). The results indicate that positions 188–213 of the signaling helix span the bilayer hydrocarbon core, where all positions tested exhibit low chemical reactivities with 5-IAF. By contrast, positions outside this region exhibit significantly higher reactivities with 5-IAF. Notably, in the native receptor, both interfacial positions 188 and 213 are occupied by the cationic residue arginine. The arginine side chain is often found near the ends of transmembrane helices, where its apolar methylenes are buried in the bilayer hydrocarbon core while its cationic guanidinium group snorkels into the polar headgroup region (23). Evidence for such snorkeling is provided by the low chemical reactivities of Cys188 and Cys213, which indicate that the β-carbon atoms of side chains at these positions are indeed buried in the bilayer hydrocarbon core.
Figure 2.
Relative chemical reactivities of the cysteine substitutions used to map the membrane–water interface onto the surface of the signaling helix. Each indicated mutant receptor possessing a single cysteine per subunit was isolated in native E. coli membranes and reacted with the aqueous alkylating agent 5-iodoacetamidofluorescein (5-IAF) under fixed conditions as described in Materials and Methods. The initial reaction rate was measured in triplicate and normalized to the reaction rate of a fully exposed cysteine residue as previously described for the chemical reactivity scan of the cytoplasmic interface (28). The reactivities of solvent- and headgroup-exposed cysteine residues are significantly higher than those exposed to the bilayer hydrocarbon core (see text).
Signaling States of Engineered Receptors
To examine the role of each interfacial position in transmembrane signaling, seven consecutive signaling helix positions were targeted for arginine and cysteine substitutions at both the periplasmic and cytoplasmic membrane–water interfaces (positions 188–194 and 208–214, respectively, as illustrated in Figure 1). Each arginine or cysteine substitution was created by site-directed mutagenesis, and then the resulting mutant receptors were expressed in E. coli strain RP3808 lacking the soluble chemotaxis proteins and the major chemoreceptors. Subsequently, each overexpressed receptor was isolated in native cell membranes. Finally, the signaling state of each receptor was determined by activity measurements in two in vitro assays selected to positively identify both the on- and off-signaling states (25, 27, 29). The receptor on-state is defined as the signaling state of the apo receptor, which stimulates CheA autophosphorylation in the in vitro kinase assay and inhibits methyl esterification of receptor adaptation sites by the CheR enzyme in the in vitro methylation assay. By contrast, the receptor off-state inhibits CheA autophosphorylation and activates receptor methylation. The use of these two antisymmetric assays ensures that each state will yield a positive response in one assay and a negative response in the other, which facilitates the accurate identification of substitutions that stabilize the native on- or off-state. Table 1, section A, summarizes the activities of each arginine mutant in the two assays, both in the apo and attractant-occupied states, while Table 1, section B, summarizes the activities of each cysteine mutant.
Table 1.
Effects of Arginine and Cysteine Substitutions on Receptor Kinase and Methylation Activitiesa
| A. Arginine Substitutions | ||||
|---|---|---|---|---|
| kinase activity |
methylation activity |
|||
| position | (–)Asp | (+)Asp | (–)Asp | (+)Asp |
| R188 (WT) | 1.00 ± 0.03 | 0.00 ± 0.00 | 1.00 ± 0.13 | 2.63 ± 0.08 |
| F189R | 2.05 ± 0.45 | 0.05 ± 0.03 | 0.29 ± 0.05 | 0.40 ± 0.05 |
| A190R | 0.77 ± 0.12 | 0.07 ± 0.03 | 0.09 ± 0.02 | 0.03 ± 0.00 |
| Q191R | 0.59 ± 0.18 | 0.04 ± 0.03 | 0.32 ± 0.02 | 0.92 ± 0.09 |
| W192R | 3.55 ± 0.77 | 0.00 ± 0.01 | 0.28 ± 0.04 | 0.58 ± 0.04 |
| Q193R | 0.00 ± 0.02 | 0.03 ± 0.04 | 0.01 ± 0.06 | 0.08 ± 0.04 |
| L194R | 0.03 ± 0.02 | 0.00 ± 0.01 | 0.20 ± 0.04 | 0.31 ± 0.06 |
| V208R | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.09 ± 0.07 | 0.10 ± 0.03 |
| W209R | 0.03 ± 0.05 | 0.03 ± 0.03 | 2.37 ± 0.22 | 2.57 ± 0.20 |
| F210R | b | b | b | b |
| G211R | 0.34 ± 0.05 | 0.01 ± 0.20 | 0.61 ± 0.10 | 0.95 ± 0.03 |
| I212R | b | b | b | b |
| R213 (WT) | 1.00 ± 0.03 | 0.00 ± 0.00 | 1.00 ± 0.13 | 2.63 ± 0.08 |
| H214R | 0.63 ± 0.24 | 0.04 ± 0.02 | 0.24 ± 0.06 | 0.05 ± 0.03 |
| B. Cysteine Subsitutions | ||||
|---|---|---|---|---|
| kinase activity |
methylation activity |
|||
| position | (–)Asp | (+)Asp | (–)Asp | (+)Asp |
| R188 (WT) | 1.00 ± 0.03 | 0.00 ± 0.00 | 1.00 ± 0.13 | 2.63 ± 0.08 |
| R188C | 0.38 ± 0.03 | 0.05 ± 0.01 | 0.92 ± 0.13 | 1.80 ± 0.16 |
| F189C | 3.90 ± 0.46 | 0.06 ± 0.02 | 0.43 ± 0.01 | 1.42 ± 0.15 |
| A190C | 0.22 ± 0.02 | 0.03 ± 0.00 | 0.34 ± 0.03 | 0.77 ± 0.03 |
| Q191C | 0.46 ± 0.08 | 0.03 ± 0.02 | 0.58 ± 0.09 | 1.18 ± 0.04 |
| W192C | 0.44 ± 0.16 | 0.02 ± 0.02 | 0.68 ± 0.06 | 1.22 ± 0.17 |
| Q193C | 0.02 ± 0.01 | 0.01 ± 0.01 | 1.09 ± 0.15 | 1.39 ± 0.08 |
| L194C | 0.01 ± 0.01 | 0.02 ± 0.02 | 0.14 ± 0.02 | 0.01 ± 0.11 |
| V208C | 1.74 ± 0.39 | 0.05 ± 0.03 | 0.40 ± 0.02 | 1.35 ± 0.19 |
| W209C | 0.03 ± 0.02 | 0.04 ± 0.02 | 0.24 ± 0.05 | 0.37 ± 0.10 |
| F210C | 1.22 ± 0.37 | 0.02 ± 0.02 | 0.25 ± 0.07 | 0.97 ± 0.12 |
| G211C | 0.45 ± 0.09 | 0.01 ± 0.01 | 0.12 ± 0.01 | 0.33 ± 0.03 |
| I212C | 0.07 ± 0.03 | 0.04 ± 0.16 | 0.53 ± 0.18 | 1.05 ± 0.07 |
| R213C | 0.34 ± 0.01 | 0.00 ± 0.00 | 0.19 ± 0.04 | 0.62 ± 0.12 |
| H214C | 0.23 ± 0.04 | 0.03 ± 0.03 | 0.26 ± 0.01 | 1.06 ± 0.09 |
Shown are the activities of (A) arginine-substituted and (B) cysteine-substituted receptors in two antisymmetric functional assays, which together identify receptors in the on- or off-state. In both assays, activities are measured in triplicate and expressed relative to the wild-type apo receptor. The kinase assay reconstitutes receptor-containing membranes with the purified, soluble pathway components CheA, CheW, and CheY to form the receptor-kinase signaling complex. Following addition of γ-[32P]-ATP to trigger the kinase reaction, the initial rate of [32P]-phospho-CheY formation at 25 °C is measured under conditions where the autophosphorylation of CheA is rate-determining, both in the absence and presence of the attractant l-aspartate (Asp) (see Materials and Methods and ref 27). The methylation assay reconstitutes receptor-containing membranes with the adaptation enzyme CheR. Following the addition of [3H]-S-adenosyl methionine, the initial rate of receptor methyl esterification is measured at 30 °C (see text and ref 25). The receptor on-state is recognized by its activation of the cytoplasmic kinase CheA and inhibition of the cytoplasmic methylation enzyme CheR, while the off-state is recognized by its inhibition of kinase activity and stimulation of methylation activity.
Receptors that failed to accumulate in cell membranes, presumably because membrane insertion or protein stability is disrupted.
All 28 substitutions tested yield significant perturbations of receptor function in one or both activity assays, indicating that the native interfacial side chains play essential roles. Two arginine substitutions (F210R, I212R) prevent accumulation of receptor in the membrane during expression (Table 1A), suggesting that these substitutions significantly decrease receptor membrane insertion or stability. Twenty-two other arginine and cysteine substitutions yield significant activity losses in one or both assays (Table 1, sections A and B), indicating that these substitutions alter protein-lipid or protein–protein interactions and thereby perturb receptor function. Cysteine substitutions are generally less perturbing than arginine ones, particularly in the methylation assay (Table 1, sections A and B). For example, the least perturbed mutant is V208C, which yields activities within 2-fold of wild type in both assays, while V208R is completely inactive. Thus, overall, receptor function is highly sensitive to 24 substitutions near the membrane–water interface, and argi-nine substitutions are typically more perturbing than cysteine ones.
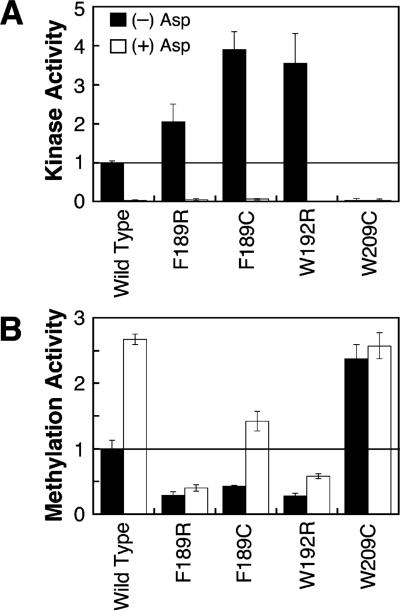
Notably, the remaining four substitutions perturb the receptor in a fashion that stabilizes one of the two native signaling states, as illustrated in Table 1, sections A and B, and Figure 3. Three substitutions, F189R, F189C, and W192R, superactivate the kinase by a factor of 2- to 4-fold (relative to the wild-type receptor) but inhibit receptor methylation 1.5- to 6-fold, as expected for mutations that stabilize the native on-state. For these three substitutions, aspartate binding still inactivates the kinase, but receptor methylation is only partially stimulated by aspartate, indicating that attractant binding can still drive the receptor partially, but not completely, into the off-state. Together these findings indicate that the F189R, F189C, and W192R substitutions, termed on-state stabilizers, drive the receptor equilibrium toward the on-state but do not irreversibly trap the receptor in this state. One substitution, W209R, inhibits the kinase over 10-fold and yields the maximum receptor methylation activity. Moreover, aspartate binding has little or no effect on this modified receptor. These findings indicate that the W209R substitution, termed a lock-off mutation, strongly locks the receptor in the native off-state even in the absence of attractant to yield essentially the same signaling behavior as the attractant-occupied, wild-type receptor.
Figure 3.
Use of in vitro activity assays to identify substitutions that stabilize the on- or off-signaling state. Receptors containing arginine or cysteine substitutions were isolated in native E. coli membranes, and their kinase and methylation activities were measured in triplicate as detailed in Table 1. The two plots summarize the kinase and methylation activities of four mutant receptors exhibiting on- or off-state stabilization, where activities are expressed relative to the wild-type apo receptor. The F189C, F189R, and W192R mutations stabilize the on-state since they superactivate the kinase relative to wild-type protein while receptor methylation is inhibited. The W209R mutation locks the off-state since it inhibits kinase activity but yields maximal receptor methylation activity even when saturated with aspartate.
DISCUSSION
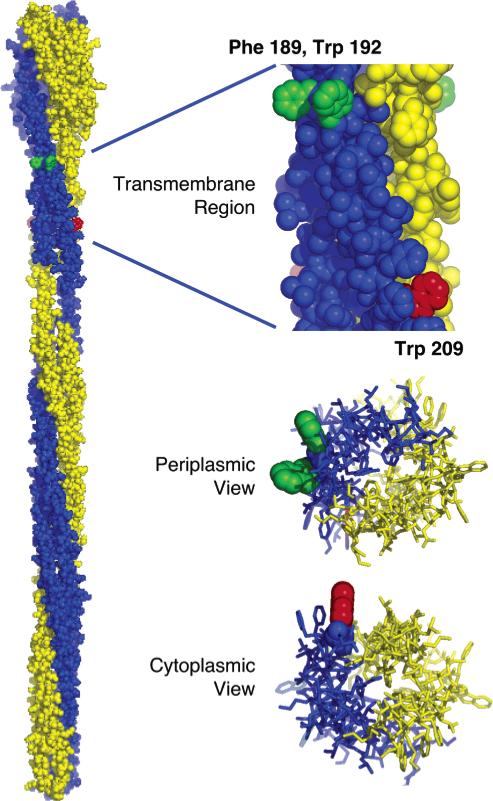
The results map out the locations of the periplasmic and cytoplasmic membrane–water interfaces on the transmembrane signaling helix of the aspartate receptor and also identify four amino acid substitutions near these interfaces that drive the receptor into its on- or off-signaling state. The locations of these four signal-stabilizing substitutions relative to the membrane–water interface have important implications for the mechanism of transmembrane signaling. All four substitutions are located at positions exposed to the bilayer hydrocarbon core but near the headgroup region (Figure 4). These positions do not have stable contacts with adjacent transmembrane helices in the same or nearby receptor dimers, since they point away from the central dimer axis and are located far from the cytoplasmic region where interdimer contacts occur within the trimer-of-dimers (10, 30). Furthermore, the larger diameter of the periplasmic domain, which is 18 Å broader than the membrane-spanning four-helix bundle, prevents contacts between the transmembrane helices of adjacent dimers (Figure 4). Together these points indicate that these four residues stabilize the on- or off-signaling state via perturbations of native protein–lipid interactions.
Figure 4.
Model of the full-length receptor structure illustrating the two identical subunits of the homodimer (yellow and blue) (10), built using information provided by crystallographic and chemical studies (4, 7, 25, 32, 33, 38, 39). The expanded views show the transmembrane four-helix bundle as seen from the center of the bilayer (top), the periplasm (middle), and the cytoplasm (bottom). Positions on the signaling helix where side chain substitutions stabilize the receptor on-state (Phe189 and Trp192) or off-state (Trp209) are indicated in green and red, respectively. Notably, the native side chains at these positions project into the surrounding lipids. For simplicity, positions of signal-stabilizing substitutions are highlighted for only one of the two identical subunits. Note that the residue numbers of the model structure (10), which are based on the serine receptor primary sequence, have been replaced with the corresponding residue numbers for the aspartate receptor of the present study. Moreover, the illustrated Phe189 was modeled in place of Gln at the homologous serine receptor position.
Notably, the three on-state stabilizers (F189R, F189C, W192R) are located near the periplasmic membrane–water interface (Figure 4). Two of these mutations, F189R and F189C, remove a large apolar side chain, Phe189, which is proposed to help anchor the helix in the hydrocarbon core of the lipid bilayer. Although the arginine side chain of F189R has considerable hydrophobic character, its greater flexibility prevents it from being an effective anchor. Loss of such a hydrophobic anchor would allow the entire helix to slide toward the periplasm (Figure 5). While the magnitude of this helix displacement is difficult to predict, the removal of two phenylalanine residues near the end of the transmembrane helix of the M13 major coat protein triggers a helix displacement of several angstroms (24); thus the loss of a single phenylalanine likely triggers a smaller displacement. The third on-state stabilizer is W192R, which places a charged arginine side chain at a native tryptophan position one full helical turn deeper in the bilayer core than the interfacial Arg188 residue. Studies of model transmembrane helices suggest that a tryptophan residue is most energetically stable at a position where its side chain amide can interact with the backbone carbonyls of lipid residues, whereas an arginine residue is most stable at a position where its side chain guanidinium can form a salt bridge to a lipid headgroup phosphate (22, 23). Significantly, for an arginine side chain to reach the headgroup phosphates, it must be located at least one residue closer to the aqueous compartment than a tryptophan residue interacting with the lipid backbone (22, 23). It follows that the W192R substitution buries the arginine side chain in the membrane one position too deeply to interact with the headgroup phosphates, providing a driving force for helix displacement toward the periplasm (Figure 5). The resulting displacement would have a magnitude as large as one residue (1.5 Å). Consistent with this picture, placement of the arginine side chain one position closer to the periplasm in the Q191R mutant yields kinase and methylation activities more similar to wild-type protein, while placement of arginine one position farther from the periplasm in the Q193R mutant virtually eliminates both activities suggesting a highly perturbing piston displacement that is significantly larger than that in wild-type protein.
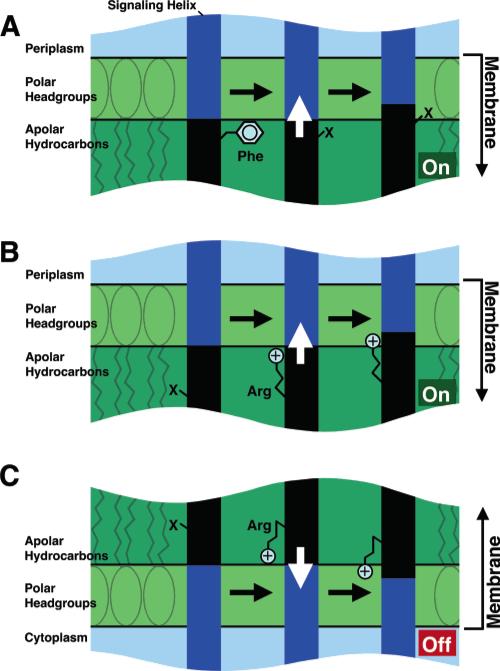
Figure 5.
Proposed effects of each signal-stabilizing substitution on the position of the transmembrane signaling helix. For simplicity, the other three transmembrane helices of the receptor dimer are omitted. The blue and black areas indicate regions of the signaling helix initially exposed to polar and hydrophobic environments, respectively. (A) Loss of the anchoring hydrophobic Phe189 near the periplasmic membrane–water interface allows displacement of the signaling helix toward the periplasm, as proposed for the F189C and F189R substitutions. (B) Similarly, gain of a buried positive charge via the W192R substitution drives the signaling helix toward the periplasm, where the arginine guanidinium moiety is proposed to interact with headgroup phosphates. (C) Gain of a buried positive charge via the W209R substitution near the cytoplasmic membrane–water interface drives the signaling helix toward the cytoplasm, where the arginine guanidinium moiety is proposed to interact with headgroup phosphates. The functional effect of each helix displacement is to turn the associated cytoplasmic CheA kinase on (green) or off (red).
By contrast, the lone lock-off substitution is located at the opposite side of the membrane near the cytoplasmic membrane–water interface, again at a position exposed to the hydrophobic core of the bilayer (Figure 4). Like the symmetric W192R substitution, W209R places a cationic arginine side chain at a native tryptophan position one full helical turn deeper in the hydrocarbon core than the interfacial Arg213. Since W209R is closest to the cytoplasmic compartment, it drives a helix displacement toward the cytoplasm with a magnitude as large as one residue in a standard R-helix (1.5 Å, Figure 5). However, W209R exhibits significantly more signal-locking character than the symmetric W192R substitution, indicating that the W209R perturbation is less easily reversed by ligand binding. It follows that mechanical forces within the receptor molecule may energetically favor cytoplasmic signaling helix displacements more than periplasmic displacements. Alternatively, the local electrostatic environments of arginine could be different at the 192 and 209 positions due to nearby lipid or protein charges, yielding an electrostatic force favoring cytoplasmic displacements. Notably, the native tryptophan residues at positions 192 and 209 are perfectly conserved in the major chemoreceptors for aspartate and serine from both E. coli and S. typhimurium, further highlighting their importance.
Overall, the present results reveal that the native signaling helix side chains located near membrane–water interfaces are highly optimized to allow normal switching between the on- and off-signaling states. Single substitutions at these positions can disrupt signaling in uninteresting ways or can stabilize a physiologically important signaling state. Interestingly, the native side chains at the signal-stabilizing positions (Phe189, Trp192, Trp209) are aromatic residues commonly found at interfacial positions in transmembrane helices (23, 24). Such aromatics possess specific interactions with surrounding lipids that correctly position helices relative to the membrane surface. It follows that the present results support chemoreceptor signaling models in which the trans-membrane helix TM2 plays an essential role in transmitting signals across the bilayer (4), further justifying its identification as the signaling helix.
The findings disfavor models proposing that the ligand-induced signaling helix displacement is dominated by helix rotational or tilting movements. The four signal-stabilizing substitutions do not appear to provide a significant driving force for rotation of the signaling helix about its long axis, since these substitutions are all exposed to the lipid hydro-carbon core, which is homogeneous for small rotations. (Multiple previous lines of evidence have indicated that the signaling helix displacement is small (4)). Furthermore, three of the four substitutions incorporate arginine residues with a long, flexible side chain not well-suited for driving such rotations. In principle, the signal-stabilizing mutations could generate tilting movements of the signaling helix by driving only one end of the helix toward an aqueous compartment (23). However, simple tilting models with the tilt axis located inside the membrane cannot explain the opposite signaling effects of substitutions on opposite sides of the membrane. Moreover, helix packing interactions would generally damp tilting motions triggered by perturbations on the helix face oriented toward lipid, which would push one end of the signaling helix into the other helices of the dimer.
Multiple lines of evidence from previous studies have supported a piston model in which ligand binding drives a small (1–2 Å) piston-type displacement of the signaling helix along the helix axis. The approaches providing this evidence have ranged from high-resolution structural studies of the ligand-induced conformational change in the isolated periplasmic domain to disulfide trapping and spectroscopic studies of the membrane-bound receptor to analysis of disulfide formation rates in vivo (4, 17–19, 31). In the piston model, displacements of the signaling helix toward the periplasm or cytoplasm yield kinase activation or inhibition, respectively. Such a piston mechanism provides mechanically efficient, long-range information transfer along the virtually incompressible axis of the signaling helix connecting the periplasmic and cytoplasmic domains (4, 17). The present findings provide further support for the piston model. In addition, as previously demonstrated by disulfide bonds between TM1 and TM2 that lock the receptor in the on- and off- signaling states (32), the present results indicate that the asymmetric displacement of a single signaling helix within the homodimer is not essential for transmembrane signaling. While aspartate binding does trigger an asymmetric signaling helix displacement due to negative cooperativity between the two symmetric aspartate binding sites of the homodimer (33–35), this asymmetry is not required since disulfide bonds (32) and interfacial substitutions that perturb both signaling helices of the homodimer in a symmetric fashion are able to induce transitions between the on- and off-signaling states. The fact that signaling can occur when both signaling helices are displaced in the same direction can account for the ability of two attractants such as aspartate and the maltose binding protein to generate additive signals, which are proposed to be carried through the two different signaling helices of the homodimer (36, 37). Finally, new evidence suggests that the piston displacement of the signaling helix may trigger cooperative signals in nearby dimers (8–12).
The same piston mechanism described for the aspartate chemoreceptor is believed to be widely utilized by other members of the large prokaryotic chemoreceptor family (4). More generally, the transmembrane helices of many receptors, channels, and transport proteins possess conserved phenylalanine, tryptophan, and arginine side chains near the membrane–water interface. Often these membrane-spanning helices are involved in conformational transitions between different functional states. Such functionally important helices are proposed to occupy highly optimized locations in the bilayer, as observed for the asparate receptor signaling helix, at least in part due to strong interactions between interfacial side chains and surrounding lipids. It follows that modification of interfacial side chains or the lipid bilayer may enable stabilization of specific functional states in a wide range of transmembrane proteins.
ACKNOWLEDGMENT
The authors gratefully acknowledge helpful comments by Profs. Sandy Parkinson (University of Utah), Jerry Hazel-bauer (University of Missouri), and other members of the Falke group.
Footnotes
Support provided by NIH Grant R01 GM-40731 (to J.J.F.) and by NIH Grant T32 GM-65103 (to A.S.M.).
Abbreviations: 5-IAF, 5-iodoacetamido fluorescein; [3H]-SAM, S-adenosyl-l-[methyl-3H] methionine; TCA, trichloroacetic acid.
REFERENCES
- 1.Zhulin IB, Nikolskaya AN, Galperin MY. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 2003;185:285–94. doi: 10.1128/JB.185.1.285-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 2002;277:9625–8. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- 3.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 2001;26:257–65. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armitage JP. Bacterial tactic responses. Adv. Microb. Physiol. 1999;41:229–89. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- 6.Milligan DL, Koshland DE., Jr. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 1988;263:6268–75. [PubMed] [Google Scholar]
- 7.Pakula AA, Simon MI. Determination of transmembrane protein structure by disulfide cross-linking: the Escherichia coli Tar receptor. Proc. Natl. Acad. Sci. U.S.A. 1992;89:4144–8. doi: 10.1073/pnas.89.9.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bornhorst JA, Falke JJ. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry. 2000;39:9486–93. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7060–5. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–92. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 11.Lybarger SR, Maddock JR. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol. 2001;183:3261–7. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. U.S.A. 2002;99:123–7. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rampersaud A, Utsumi R, Delgado J, Forst SA, Inouye M. Ca2(+)-enhanced phosphorylation of a chimeric protein kinase involved with bacterial signal transduction. J. Biol. Chem. 1991;266:7633–7. [PubMed] [Google Scholar]
- 14.Krikos A, Conley MP, Boyd A, Berg HC, Simon MI. Chimeric chemosensory transducers of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1985;82:1326–30. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumgartner JW, Kim C, Brissette RE, Inouye M, Park C, Hazelbauer GL. Transmembrane signaling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J. Bacteriol. 1994;176:1157–63. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung KH, Spudich EN, Trivedi VD, Spudich JL. An archaeal photosignal-transducing module mediates phototaxis in Escherichia coli. J. Bacteriol. 2001;183:6365–71. doi: 10.1128/JB.183.21.6365-6371.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. U.S.A. 1996;93:2545–50. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughson AG, Hazelbauer GL. Detecting the conformational change of transmembrane signaling in a bacterial chemoreceptor by measuring effects on disulfide cross-linking in vivo. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11546–51. doi: 10.1073/pnas.93.21.11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ottemann KM, Xiao W, Shin YK, Koshland DE., Jr. A piston model for transmembrane signaling of the aspartate receptor [see comments] Science. 1999;285:1751–4. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 20.Cochran AG, Kim PS. Imitation of Escherichia coli aspartate receptor signaling in engineered dimers of the cytoplasmic domain. Science. 1996;271:1113–6. doi: 10.1126/science.271.5252.1113. [DOI] [PubMed] [Google Scholar]
- 21.Maruyama IN, Mikawa YG, Maruyama HI. A model for transmembrane signaling by the aspartate receptor based on random-cassette mutagenesis and site-directed disulfide cross-linking. J. Mol. Biol. 1995;253:530–46. doi: 10.1006/jmbi.1995.0571. [DOI] [PubMed] [Google Scholar]
- 22.Monne M, Nilsson I, Johansson M, Elmhed N, von Heijne G. Positively and negatively charged residues have different effects on the position in the membrane of a model transmembrane helix. J. Mol. Biol. 1998;284:1177–83. doi: 10.1006/jmbi.1998.2218. [DOI] [PubMed] [Google Scholar]
- 23.Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 2000;25:429–34. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 24.Meijer AB, Spruijt RB, Wolfs CJ, Hemminga MA. Membrane-anchoring interactions of M13 major coat protein. Biochemistry. 2001;40:8815–20. doi: 10.1021/bi002956s. [DOI] [PubMed] [Google Scholar]
- 25.Chervitz SA, Lin CM, Falke JJ. Transmembrane signaling by the aspartate receptor: engineered disulfides reveal static regions of the subunit interface. Biochemistry. 1995;34:9722–33. doi: 10.1021/bi00030a010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkel TA, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–39. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 27.Bornhorst JA, Falke JJ. Quantitative analysis of aspartate receptor signaling complex reveals that the homogeneous two-state model is inadequate: development of a heterogeneous two-state model. J. Mol. Biol. 2003;326:1597–614. doi: 10.1016/s0022-2836(03)00026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998;37:10746–56. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chelsky D, Gutterson NI, Koshland DE., Jr. A diffusion assay for detection and quantitation of methyl-esterified proteins on polyacrylamide gels. Anal. Biochem. 1984;141:143–8. doi: 10.1016/0003-2697(84)90437-8. [DOI] [PubMed] [Google Scholar]
- 30.Mehan RS, White NC, Falke JJ. Mapping out regions on the surface of the aspartate receptor that are essential for kinase activation. Biochemistry. 2003;42:2952–9. doi: 10.1021/bi027127g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isaac B, Gallagher GJ, Balazs YS, Thompson LK. Site-directed rotational resonance solid-state NMR distance measurements probe structure and mechanism in the transmembrane domain of the serine bacterial chemoreceptor. Biochemistry. 2002;41:3025–36. doi: 10.1021/bi015759h. [DOI] [PubMed] [Google Scholar]
- 32.Chervitz SA, Falke JJ. Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J. Biol. Chem. 1995;270:24043–53. doi: 10.1074/jbc.270.41.24043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr., Kim SH. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–7. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 34.Lin LN, Li J, Brandts JF, Weis RM. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry. 1994;33:6564–70. doi: 10.1021/bi00187a025. [DOI] [PubMed] [Google Scholar]
- 35.Biemann HP, Koshland DE., Jr. Aspartate receptors of Escherichia coli and Salmonella typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochemistry. 1994;33:629–34. doi: 10.1021/bi00169a002. [DOI] [PubMed] [Google Scholar]
- 36.Mowbray SL, Koshland DE., Jr. Additive and independent responses in a single receptor: aspartate and maltose stimuli on the tar protein. Cell. 1987;50:171–80. doi: 10.1016/0092-8674(87)90213-3. [DOI] [PubMed] [Google Scholar]
- 37.Gardina PJ, Bormans AF, Hawkins MA, Meeker JW, Manson MD. Maltose-binding protein interacts simultaneously and asymmetrically with both subunits of the Tar chemoreceptor. Mol. Microbiol. 1997;23:1181–91. doi: 10.1046/j.1365-2958.1997.3001661.x. [DOI] [PubMed] [Google Scholar]
- 38.Bass RB, Falke JJ. The aspartate receptor cytoplasmic domain: in situ chemical analysis of structure, mechanism and dynamics. Struct. Fold. Des. 1999;7:829–40. doi: 10.1016/s0969-2126(99)80106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee GF, Burrows GG, Lebert MR, Dutton DP, Hazelbauer GL. Deducing the organization of a transmembrane domain by disulfide cross- linking. The bacterial chemoreceptor Trg. J. Biol. Chem. 1994;269:29920–7. [PubMed] [Google Scholar]