Abstract
Chimeric genes, which form through the genomic fusion of two protein-coding genes, are a significant source of evolutionary novelty in Drosophila melanogaster. However, the propensity of chimeric genes to produce adaptive phenotypic changes is not fully understood. Here, we describe the chimeric gene Quetzalcoatl (Qtzl; CG31864), which formed in the recent past and swept to fixation in D. melanogaster. Qtzl arose through a duplication on chromosome 2L that united a portion of the mitochondrially targeted peptide CG12264 with a segment of the polycomb gene escl. The 3′ segment of the gene, which is derived from escl, is inherited out of frame, producing a unique peptide sequence. Nucleotide diversity is drastically reduced and site frequency spectra are significantly skewed surrounding the duplicated region, a finding consistent with a selective sweep on the duplicate region containing Qtzl. Qtzl has an expression profile that largely resembles that of escl, with expression in early pupae, adult females, and male testes. However, expression patterns appear to have been decoupled from both parental genes during later embryonic development and in head tissues of adult males, indicating that Qtzl has developed a distinct regulatory profile through the rearrangement of different 5′ and 3′ regulatory domains. Furthermore, misexpression of Qtzl suppresses defects in the formation of the neuromuscular junction in larvae, demonstrating that Qtzl can produce phenotypic effects in cells. Together, these results show that chimeric genes can produce structural and regulatory changes in a single mutational step and may be a major factor in adaptive evolution.
Keywords: adaptation, new genes, regulatory evolution, frameshifts
Chimeric genes form when complex genetic changes fuse portions of existing genes to produce a novel ORF. Such rearrangements can produce novel combinations of existing modular elements, contributing to the development of genes with novel functions (1). Chimeric genes appear to be common in the genomes of multicellular organisms, including humans (2–5). They are formed often in Drosophila melanogaster (5, 6), and there are several known examples of chimeric genes that have been stably incorporated into the genome (5). Although a handful of chimeric genes shows signatures of positive selection in Drosophila (7–12), there are very few with known functions, and the factors influencing the physiological and evolutionary impacts of chimeric genes are largely unknown.
We previously identified 14 chimeric genes in D. melanogaster, which present candidates for studies of adaptation and the development of novel functions (5). Of these 14 genes, eight have appeared within the past 1 million years and are specific to D. melanogaster. These young chimeric genes formed recently in D. melanogaster and are the most likely of the 14 to have contributed to lineage-specific evolutionary changes. Here, we describe the recent formation and apparent fixation of one of these new chimeric genes, which we have named Quetzalcoatl (Qtzl) after the chimeric feathered serpent from Mesoamerican mythology. We find that Qtzl formed through a duplication on chromosome 2L, quickly swept to fixation, and has sequence signatures of adaptive evolution.
Results
Qtzl was previously identified as part of a bioinformatic study of chimeric genes in Drosophila melanogaster (5). Qtzl is located on chromosome 2L and bears strong similarity to two neighboring genes, escl and CG12264.
Formation of Qtzl.
The chimeric gene Qtzl was formed from the tandem duplication of a 3,911-bp region on chromosome 2L (Fig. 1). As described previously, the chimeric gene was formed entirely through DNA-based sequence changes, with no evidence to suggest action of transposable elements (5). This duplication united 276 bp from the 5′ end of CG12264 with 96 bp of coding sequence from the 3′ end of polycomb gene escl. The breakpoint forming the chimeric gene falls within an exon, such that the 3′ portion of the gene is out of frame with respect to the parental gene escl, as confirmed by sequencing amplified mRNA. There are no known transcripts of Qtzl that can produce peptide sequences equivalent to the Escl peptide, except perhaps through rare frameshift errors during translation. The predicted protein from Qtzl is 123 aa in length and inherits a 5′ mitochondrial target peptide from its parental gene CG12264 along with some short conserved sequence (Fig. 1); it does not, however, contain any known enzyme active sites or binding domains from either parental gene.
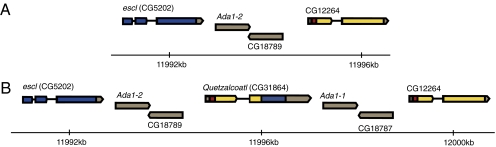
Fig. 1.
Structure of a segment of chromosome 2L before (A) and after (B) the formation of Qtzl. Qtzl was formed when approximately 4 kb of sequence encompassing portions of escl and CG12264 experienced a tandem duplication, such that the duplicated 5′ end of CG12264 was united with the duplicated 3′ end of escl. In the diagram, gray areas correspond to 5′ and 3′ UTRs and black lines represent introns. Genes are shaded in blue and yellow to show relationships to parental genes. Full transcript structure and coding sequence boundaries are not depicted for the two pairs of duplicate genes in the region, Ada1-1/Ada1-2 and CG18789/CG18787. The chimeric gene inherits a mitochondrial target peptide (shown in red) from CG12264, a gene that encodes a mitochondrial peptide involved in iron modification of proteins and tRNAs. The 3′ end of Qtzl is inherited out of frame. If translated, the mRNA from Qtzl could encode a peptide of 123 aa.
Parental Gene Functions.
Parental gene CG12264 encodes an aminotransferase, a member of the AATI superfamily. The CG12264 protein shares 78% amino acid similarity with the Saccharomyces cerevisiae protein Nfs1 (13). Nfs1 is an essential gene responsible for the modification of sulfur residues on cystine-bearing mitochondrial peptides and for proper posttranscriptional modification of nuclear and mitochondrial tRNAs (14). Nfs1 protein in yeast is active in the mitochondrial matrix, but also appears to be targeted to the cell nucleus (14). CG12264 is the only full-length orthologue of Nfs1 in D. melanogaster. The protein retains both the mitochondrial target peptide and the nuclear localization signal found in yeast, as well as the known active site. Therefore, CG12264 has likely retained many of the functions of Nfs1 and may be an essential gene in D. melanogaster.
Escl is a polycomb gene containing seven repeated WD domains, which fold into a propeller-type structure and serve as a scaffold during the assembly of a D. melanogaster histone modification complex (15, 16). It has a single full-length paralogue, esc, which shows 63% amino acid identity and 78% similarity with escl (15). Esc is a polycomb gene, which trimethylates histones to regulate homeotic genes during early development (16). In vitro assays have shown that Esc and Escl proteins bind similar targets, suggesting that they have similar cellular roles (15, 16), and escl has been shown to substitute for most esc functions during development (15). As the peptide sequence that Qtzl inherited from escl is in a different frame, its phenotypic effects are unlikely to reflect any known function of esc or escl.
Probable Fixation in D. melanogaster.
In the D. melanogaster reference genome, we found no nucleotide differences between the coding sequence of Qtzl and its parental genes, suggesting a recent origin (5). We amplified and sequenced DNA from 35 strains of D. melanogaster, all of which carry Qtzl, placing the lower bound at a frequency of 0.920 (95% one-sided CI). That this gene could be at high frequency despite appearing so recently suggests that its fixation was through nonneutral processes. One allele carries a modified version of Qtzl that would still encode a valid peptide sequence, although its current functional status is uncertain. This strain appears to have derived from WT sequence very recently and is consistent with a selective sweep in the past, regardless of its current function.
Reduced Diversity and Skewed Site Frequency Spectra.
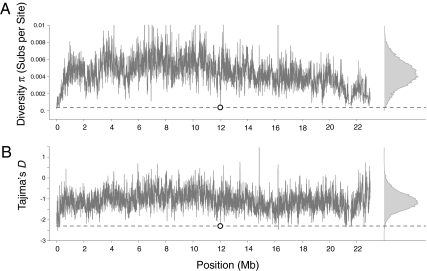
We calculated π, θW, and Tajima’s D (17) for sliding windows of 10 kb at 1-kb intervals across chromosome 2L. We find a region centered about the chimeric gene Qtzl from 11,990 kb to 12,000 kb, which displays reduced diversity and skewed site frequency spectra (Fig. 2). Such reductions in π, θW, and Tajima’s D are consistent with a selective sweep occurring at Qtzl. Diversity measures are drastically reduced in a region of approximately 100 kb around the gene (Fig. 3), and Tajima’s D is significantly reduced in the surrounding region. The window containing Qtzl has the 10th lowest measure of diversity and the 35th lowest measure of Tajima’s D among 22,033 windows. A one-sided percentile rank test yields a P value of 4.5 × 10−4 for diversity and a P value of 1.59 × 10−3 for Tajima’s D (Table S1). As we began our study with 14 chimeric genes (5), a Bonferroni adjustment for both tests yields a corrected P value of 0.0064 for diversity and a corrected P value of 0.02 for Tajima’s D (Table S1).
Fig. 2.
(A) Diversity π, measured as substitutions per site, and (B) Tajima’s D. Both plots are for sliding windows of 10 kb across chromosome 2L. The position of the chimeric gene Qtzl is shown by the open circle and dashed line. Density plots of each metric are juxtaposed to the right of each graph. Both π and D are significantly reduced in the 10-kb window surrounding Qtzl.
Fig. 3.
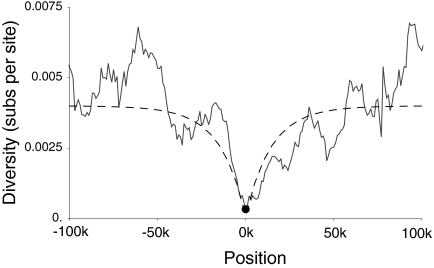
Local diversity π, measured as substitutions per site, surrounding Qtzl (solid line) fitted with the expectation after a selective sweep (dashed line). The fitted curve describes a selective sweep with s of 0.0098, which occurred 15,000 y ago.
Additional analysis provides further support for the occurrence of a selective sweep. Haplotype patterns correspond well with plots of diversity (Fig. S1), with a single high-frequency haplotype at Qtzl. Extended haplotype homozygosity (EHH) for the region is 1.0, with a slow decay of EHH across approximately 20 kb to the left and 40 kb to the right of the gene (Fig. S1). Additionally, Qtzl shows a coalescent effective population size (Ne) of 6.76 × 104 individuals and a time to the most recent common ancestor (TMRCA) of 2.04 × 104 y (Fig. S2 and Table S1). These estimates differ significantly from the chromosomal average of Ne of 1.85 × 106 individuals and TMRCA of 3.05 × 105 y (Fig. S2 and Table S1). The associated estimate of the relative rate of nonsynonymous to synonymous changes, ω for Qtzl is elevated above the chromosomal average but is not significantly greater than 1.0 (Table S1).
The region surrounding Qtzl shows the strongest reduction in diversity among the nonheterochromatic regions of chromosome 2L in the sample of strains studied. This alone would suggest that the selective advantage associated with Qtzl is substantial. In general, more recent selective sweeps show a deeper reduction in genetic diversity, whereas stronger selective sweeps show a wider reduction in diversity (18). Fitting observed patterns of genetic diversity surrounding Qtzl to theoretical expectations suggests that the selective advantage s conferred by Qtzl was 0.0098 and that the swept allele fixed 15,000 y ago (Fig. 3).
Demographic effects such as population expansion are insufficient to explain reduced diversity in this region, as these factors would affect large portions of the genome in a fairly uniform fashion. Similarly, previous searches have found no evidence for significantly reduced recombination that could influence these results (19, 20), although abnormalities within exceptionally narrow windows could be difficult to detect. This region shows a marked departure from the pattern displayed across the majority of chromosome 2L, and the reduced diversity is likely the product of a selective sweep. The excess of singleton sites, long haplotypes, and deep, wide valleys in diversity are incompatible with models of adaptation arising from standing variation (21, 22). In addition to the D. melanogaster samples, we have assayed six Drosophila sechellia and 13 Drosophila simulans fly lines, and have failed to amplify the chimeric gene, suggesting that each of the other two species has only a single copy of the region. The chimeric gene is not found in the reference genome for any other Drosophila species. These data suggest that Qtzl emerged recently in the D. melanogaster lineage and rapidly increased to fixation.
No Differentiation Among Duplicate Copies.
The region that duplicated to form Qtzl also created two pairs of duplicate genes: CG18787/CG18789 and Ada1-1/Ada1-2 (Fig. 1). CG18787 and CG18789 have no known function, but are suggested to bind nucleic acids based on computational comparisons of protein domains (Flybase). Ada1-1 and Ada1-2 are homologous to the yeast gene Ada1, a core protein in the yeast SAGA complex, which is known to influence transcription through histone acetylation (23). To determine whether duplicate genes in this region have differentiated, we mapped all available reads for 15 Raleigh strains of D. melanogaster to a single-copy template of the region. If duplicate sites in this region have not differentiated between the two copies, mappings should have a single sequence, whereas differentiation of duplicate copies will be manifest in two different types of reads mapping to any single site. No single site with two types of reads could be found in all 15 strains examined. These results suggest that the adaptive mutation that swept to fixation did not involve differentiation of protein or regulatory sequences in the duplicate genes Ada1-1 and Ada1-2 or CG18787 and CG18789. Similarly, no single nucleotide change differentiating escl or CG12264 from Qtzl can account for the sweep to fixation.
However, duplication may have affected the dosage of these neighboring genes. Whole transcriptome sequencing data for D. melanogaster and D. sechellia (24) indicates that the singleton orthologues of Ada1-1 and CG18787 have significantly higher expression (Welch two-sample t test, P < 10−15; SI Materials and Methods) than the total of these duplicate genes in D. melanogaster. However, these results are at odds with microarray expression data of whole adult female flies, which shows that the total dosage of the genes CG18787 and CG18789 equals that of the singleton orthologue in D. simulans (25). Hence, the dosage effects of this duplication cannot be determined readily, even for this single life stage. Yeast Ada1 is an important component of the SAGA histone acetylation complex in yeast (23), and deletions are known to have massive impacts on SAGA complex formation and gene expression. If expression in D. melanogaster is indeed substantially lower than in the ancestral state, such changes in the expression of the Ada1 orthologues in D. melanogaster seem more likely to have negative than positive selective effects.
Expression Patterns.
We used RT-PCR assays as well as modENCODE transcriptome sequencing data (26) to assess the expression profiles of the chimeric gene Qtzl and its corresponding parental genes (Table 1 and Fig. S3). The parental gene escl was found in only the testes of adult male flies, in early pupae, in developing embryos from 0 to 2 h, and in whole adult females but not in female head tissues. The parental gene CG12264 is transcribed at high levels in every tissue and life stage examined, a result consistent with its putative role as an essential enzyme in the mitochondria and nucleus. Expression of Qtzl appears to be strongest at the stages when the parental gene escl is also expressed. Transcripts of Qtzl amplify from head and testes of adult males, but we found no evidence of the mRNA in the male carcass (excluding the head and testes). Transcripts could not be amplified from the heads of adult females, a result that has been confirmed by data from modENCODE. We find Qtzl at high levels in adult females, early pupae, and embryos 0 to 2 h after fertilization. Yet, expression of Qtzl has been clearly decoupled from that of escl in two key ways: it is found at moderate to high levels in middle to late embryonic development when escl is absent, and it is also present at low levels in the head tissues of adult males. As Qtzl carries no mutations that can differentiate it from its parental genes, all differences in regulatory profiles can only be caused by chimeric gene formation, and not subsequent divergence. Hence, some regulatory change based on the new 3′ end of Qtzl has had a critical influence over expression, and the combination of modular regulatory elements in the 5′ and 3′ ends was essential in the creation of the distinct regulatory profile of Qtzl. The observed expression differences could be produced through elements determining both transcription and mRNA stability, and it is uncertain which of these might define the regulation of Qtzl.
Table 1.
Expression patterns of chimeric and parental genes
| Stage or tissue | CG12264 | Qtzl | escl |
| Male carcass | + | – | – |
| Testes | + | + | + |
| Male head | + | + | – |
| Early embryos | + | + | + |
| Late embryos | + | + | – |
| Larvae | + | – | – |
| Pre-pupae | + | + | + |
| Pupae | + | – | – |
| Adult whole males | + | – | – |
| Adult whole females | + | + | + |
+, Present; –, absent.
Mutant Phenotype.
The Drosophila Gene Search Project has produced multiple P-element insertions that drive overexpression of nearby genes (27). The sequence surrounding one such P-element construct was mapped to the parental gene CG12264 using D. melanogaster genome annotation release 4.1.1 (2005). However, with more extensive sequence from the surrounding region, we have shown that the insertion instead maps to Qtzl (SI Materials and Methods and Fig. S4). The overexpression construct inserted into the 5′ end of the gene approximately 48 bp upstream from the start of the putative peptide sequence. Dominant-negative mutants in an unrelated gene, Nsf2, normally display an overgrowth of neurons at the neuromuscular junction (28, 29). Laviolette et al. (28) assayed the Qtzl overexpression line in these Nsf2 mutants and discovered that overexpression is sufficient to partially rescue the neural overgrowth. Hence the chimeric gene Qtzl can affect neural development in some genetic contexts, although the mechanism by which it corrects this overgrowth phenotype is unknown. The transgenic insertion is also effectively a disruption mutant, as it falls between the natural promoter and the start codon of the gene; we have not been able to amplify Qtzl mRNA from homozygous flies. Flies homozygous for the disruption insertion are viable, suggesting that Qtzl is not essential, as is expected for a recently created gene. However, no offspring have been isolated from crosses of homozygous flies, indicating a possible sterility phenotype for Qtzl, consistent with expression in the testes. The stock has been kept with a balancer for several generations and this sterility could also be a result of the accumulation of factors at other loci.
One sequence of Qtzl from a Malawi strain lacks 104 bp beginning just after the reported transcription start and includes the annotated start codon. This deletion point corresponds exactly to the insertion point of the transgenic P-element construct described earlier. The sequence also displays one indel, which removed 25 bp downstream from the intron, replacing it with a new 17-bp sequence (Fig. S4). The remaining portion of the allele shows no point mutations compared with the reference strain even at synonymous or intronic sites, suggesting that the allele was only recently derived from a functional WT sequence. For this transcript, the longest putative translation would still include the frameshifted sequence from escl as well as some small portion from CG12264. The mitochondrial target peptide, however, would be lost.
We have isolated RNA from early pupae from this strain and were able to amplify a portion of the mRNA using gene specific primers. RNA levels appear to be high, resulting in expression that appears to be equivalent to the WT allele. An alternative set of primers, designed to amplify a longer segment of the mRNA did not produce product, further confirming the deletion. Additionally, we have amplified the transcript from the testes of adult males. No amplification was observed from heads of adult males, possibly suggesting a role for the 5′ deleted region in head-specific expression. Still, WT mRNA levels in male heads appear to be low, and even slight knock-downs could limit our ability to detect the transcript. Flies appear to develop normally, based on cursory examination. It may be that this shortened peptide sequence, if translated, is entirely functional and can perform the same adaptive activities as WT alleles of Qtzl or that Qtzl acts primarily as a noncoding RNA that is not perturbed by these deletions. Alternatively, this allele may be entirely dysfunctional, in which case it could be destined for loss, assuming continued selective pressure. Nevertheless we find it surprising that this strain underwent multiple complex mutations but still retained the ability to encode a putative peptide containing the latter portion of Qtzl.
Discussion
We describe a unique chimeric gene, Qtzl, located on chromosome 2L that was involved in a selective sweep. The region surrounding this gene displays a clear reduction in diversity, skewed site frequency spectra, and extended high-frequency haplotypes. Expression analysis indicates that Qtzl developed a unique regulatory profile through the shuffling of modular regulatory elements and molecular analysis of an overexpression line suggests that Qtzl can produce phenotypic effects in neurons.
Frameshifts and Evolution.
Gilbert first suggested that introns might be beneficial for their ability to facilitate exon shuffling while maintaining reading frame and thus would promote formation of new proteins (30). Yet, we know that chimeric gene formation commonly occurs within exons, although Qtzl was the only gene among the 14 examined that exhibited a frameshift (5). Based on current available transcript models, there is no available start codon that could possibly result in the faithful translation of peptide sequence similar to that of parental gene escl. Adaptive benefit solely from duplication of the 5′ end of CG12264 seems especially unlikely, as this segment lacks any known active sites or binding domains. CG12264 also exhibits strong constitutive expression, which may “drown out” any regulatory benefit of a partial duplication. Furthermore, we have isolated a single allele from a Malawi strain that has undergone a series of complex mutations but preserves transcription of the sequence inherited from escl. Hence, the frameshifted section inherited from escl is likely to be instrumental in any adaptive effects of Qtzl. One previously identified chimeric gene, Sdic, is known to have recruited former noncoding DNA to form new peptide sequence (9, 31). However, the use of out-of-frame sequence to form a novel peptide is clearly unusual. In many cases, RNAs produced from frameshifted genes with premature stop codons are degraded through nonsense-mediated decay (32). Yet, we have found that Qtzl mRNAs are still found at high levels in select tissues and life stages based on whole transcriptome sequencing (Table 1 and Fig. S3).
Still, this duplicated region may have produced adaptive changes that do not depend on the translation products of Qtzl. The mRNA of Qtzl is polyadenylated and shows a properly spliced intron. These two factors suggest that the RNA is processed properly and that the peptide from Qtzl is likely to be translated. Furthermore, we can find no antisense targets for Qtzl that could imply action as a regulatory RNA. However, the phenotypic effects associated with the overexpression mutant may be a result of some form of functional or regulatory action of the RNA transcript. Similarly, unknown regulatory effects from the duplication, independent of Qtzl cannot be entirely discounted. However, certain possibilities common to duplicate gene evolution, such as differentiation of duplicate copies or nucleotide substitutions after chimeric gene formation, cannot explain the reduction in diversity. Clearly, Qtzl is the most distinct structure to appear after duplication. This obvious novelty coupled with its position at the center of the selective sweep, as well as its demonstrated effects on neural growth, are suggestive of a role for Qtzl in adaptation.
Chimeric Genes and Regulatory Evolution.
Chimeric genes often form through local rearrangements of DNA placing the newly formed chimeric gene between its parental genes (5). Chimeric genes carry coding sequence and adjacent DNA from their parental genes, such that any functional noncoding elements are typically faithfully inherited. Here we have found Qtzl emerged with a unique regulatory profile through the shuffling of 5′ and 3′ regulatory elements. Such regulatory divergence is likely to influence the ultimate function and phenotypic effects of the gene. Changes in gene expression are known to be instrumental for phenotypic changes in multicellular organisms (33). Multicellular genomes contain particularly intricate gene regulatory structure, with significant effects from insulators, enhancers, UTRs, chromatin modeling signals, posttranslational modification sites, and various cellular targeting peptides. Genomes that house such a complex architecture of regulatory elements may provide an exceptionally dynamic mutational landscape within which chimeric genes can shuffle components to develop novel expression.
Role of Chimeric Genes in Adaptation.
Large numbers of chimeric genes have been discovered in Caenorhabditis elegans (2) and in plants (3) and humans (4, 34, 35), where they are known for their detrimental effects in human tumor formation (36). However, the role of chimeric genes in adaptation is drawn primarily from studies of the genus Drosophila, from which we know of four other chimeric genes that have been involved in adaptive changes (7–12). Our results describing the origins and fixation of Qtzl add to a growing body of work that suggests chimeric genes are major players in adaptation and the development of new traits.
Chimeric genes may be especially well poised to act as a source of adaptive changes. When a novel chimeric gene appears, it is initially different from each of its parental genes, and may be more likely than a duplicate gene to develop a novel function. Although the products of some chimeric genes will have entirely defective protein-folding patterns, others may shuffle portions of protein sequences to occasionally produce entirely novel folds that cannot be reached through point mutations alone (37, 38). These leaps across the fitness landscape may be more likely than smaller changes to provide for adaptation under selective pressure. Indeed, mutations of large effect appear to play a substantial role in early adaptation whereas mutations of smaller effect may fine-tune novel functions afterward (39). The total possible set of protein sequences is extremely vast; evolution has, until this point, been able to explore only a small subset of this space. It seems likely that advantageous protein sequences exist that cannot be reached by simple mutational pathways. The formation of chimeric genes presents a complex mutational pathway that, although rare, may be an indispensable mechanism as it can reach remote and innovative functions. Thus, if mutation substantially limits the available adaptive possibilities (40), chimeric gene formation may indeed serve as a crucial force that can drive evolutionary change.
Materials and Methods
Identification of Chimeric Genes.
We previously identified a core set of chimeric genes through a bioinformatic study of the D. melanogaster genome (5). This work identified 14 chimeric genes that warranted further study. Here we focus on the chimeric gene CG31864, which we have named Qtzl. This chimeric gene was formed by uniting a copy of the 5′ end of CG12264 with a copy of the 3′ end of escl (CG5202).
Sequencing and DNA Polymorphism Analysis.
We identified all polymorphisms on chromosome 2L in the DPGP D. melanogaster Solexa Assemblies (release 0.5) of 39 sequenced strains of D. melanogaster from Raleigh, NC, and six strains from Malawi (http://www.dpgp.org/; accessed June 2009). This dataset provides aligned sequence for all uniquely mapping reads in all 45 genomes. We calculated π, θW, and Tajima’s D (17) for 10-kb windows sliding along chromosome 2L at 1-kb intervals, using only sites that had mapped reads for all 45 strains. We considered only windows with more than 1 kb of sites with full coverage. Significance was assessed by using a percentile rank test, correcting for multiple testing. Because duplicate regions do not map uniquely, sequence for the chimeric gene was not available from the DPGP 50 genomes release. We therefore collected approximately 20 adult males from each of 10 strains from Malawi, 12 strains from Raleigh, NC, and 12 strains from a worldwide collection of D. melanogaster. Chimeric gene sequences for each strain were amplified and sequenced (SI Materials and Methods, Dataset S1).
We used BEAST coalescent software (41) to date the age of the sweep in the region of Qtzl in D. melanogaster. We extracted fourfold synonymous sequences for each of 16 genes immediately surrounding the chimera as well as the chimera itself, and the calculated coalescent effective population size (Ne) and time to TMRCA for each gene. These single-gene estimates of Ne and TMRCA were compared with the average estimate across 100 randomly selected coding sequences from nonheterochromatic regions on chromosome 2L. We also estimated and compared ω, the ratio of rates of nonsynonymous to synonymous substitutions in the population.
Expression Patterns.
We dissected 10 adult male flies from the D. melanogaster reference strain, y1cn1sp1bw1, and isolated mRNA from heads, testes, and carcasses. We then performed RT-PCR reactions using gene specific primer pairs internal to the coding sequence to amplify each chimeric and parental gene. Additionally, we amplified Qtzl mRNA sequence using primers designed to amplify a long segment of the transcript, including start and stop codons. We gel-purified and sequenced products to confirm the FlyBase mRNA annotation. Primer sequences are provided in Table S2. Additional expression data for various life stages was gathered from modENCODE transcriptome sequencing data (26).
Overexpression Mutant.
We obtained a misexpression line containing a Gene Search Vector insertion from the Kyoto Stock Center (stock no. 201–346). We isolated genomic DNA from approximately 20 flies using the Wizard Genomic DNA purification kit (Promega). We amplified the surrounding sequence using a P-element specific forward primer and gene-specific reverse primers. An approximately 500 bp product was isolated priming off of Qtzl and gel purified using a Qiagen kit and sequenced. No amplification was observed priming off of CG12264.
Supplementary Material
Acknowledgments
We thank Timothy Sackton, Michael B. Eisen (University of California, Berkeley), C. R. Young, Kyle M. Brown, and Philipp Messer for helpful discussions. In addition, we thank Alexis S. Harrison for her help in naming our chimeric gene. Daven Presgraves (University of Rochester), Michael Eisen, and Naoyuki Fuse (Kyoto University) provided D. melanogaster stocks and Sarah B. Kingan (University of Rochester) donated stocks of D. simulans and D. sechellia. John Wakeley provided access to nodes on the Harvard Odyssey cluster supported by the Faculty of Arts and Sciences Research Computing Group. This work was supported by National Institutes of Health grants GM065169 and GM084236.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006503107/-/DCSupplemental.
References
- 1.Patthy L. Modular assembly of genes and the evolution of new functions. Genetica. 2003;118:217–231. [PubMed] [Google Scholar]
- 2.Katju V, Lynch M. On the formation of novel genes by duplication in the Caenorhabditis elegans genome. Mol Biol Evol. 2006;23:1056–1067. doi: 10.1093/molbev/msj114. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers RL, Bedford T, Hartl DL. Formation and longevity of chimeric and duplicate genes in Drosophila melanogaster. Genetics. 2009;181:313–322. doi: 10.1534/genetics.108.091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, et al. On the origin of new genes in Drosophila. Genome Res. 2008;18:1446–1455. doi: 10.1101/gr.076588.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long M, Langley CH. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- 8.Wang W, Zhang J, Alvarez C, Llopart A, Long M. The origin of the Jingwei gene and the complex modular structure of its parental gene, yellow emperor, in Drosophila melanogaster. Mol Biol Evol. 2000;17:1294–1301. doi: 10.1093/oxfordjournals.molbev.a026413. [DOI] [PubMed] [Google Scholar]
- 9.Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature. 1998;396:572–575. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- 10.Jones CD, Begun DJ. Parallel evolution of chimeric fusion genes. Proc Natl Acad Sci USA. 2005;102:11373–11378. doi: 10.1073/pnas.0503528102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones CD, Custer AW, Begun DJ. Origin and evolution of a chimeric fusion gene in Drosophila subobscura, D. madeirensis and D. guanche. Genetics. 2005;170:207–219. doi: 10.1534/genetics.104.037283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shih HJ, Jones CD. Patterns of amino acid evolution in the Drosophila ananassae chimeric gene, siren, parallel those of other Adh-derived chimeras. Genetics. 2008;180:1261–1263. doi: 10.1534/genetics.108.090068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Nakai Y, et al. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J Biol Chem. 2004;279:12363–12368. doi: 10.1074/jbc.M312448200. [DOI] [PubMed] [Google Scholar]
- 15.Kurzhals RL, Tie F, Stratton CA, Harte PJ. Drosophila ESC-like can substitute for ESC and becomes required for Polycomb silencing if ESC is absent. Dev Biol. 2008;313:293–306. doi: 10.1016/j.ydbio.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tie F, Stratton CA, Kurzhals RL, Harte PJ. The N terminus of Drosophila ESC binds directly to histone H3 and is required for E(Z)-dependent trimethylation of H3 lysine 27. Mol Cell Biol. 2007;27:2014–2026. doi: 10.1128/MCB.01822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan NL, Hudson RR, Langley CH. The “hitchhiking effect” revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hey J, Kliman RM. Interactions between natural selection, recombination and gene density in the genes of Drosophila. Genetics. 2002;160:595–608. doi: 10.1093/genetics/160.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh ND, Arndt PF, Petrov DA. Genomic heterogeneity of background substitutional patterns in Drosophila melanogaster. Genetics. 2005;169:709–722. doi: 10.1534/genetics.104.032250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–2323. [PubMed] [Google Scholar]
- 22.Barrett RD, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Wu PY, Winston F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McManus CJ, et al. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 2010 doi: 10.1101/gr.102491.109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ranz JM, Namgyal K, Gibson G, Hartl DL. Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res. 2004;14:373–379. doi: 10.1101/gr.2019804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Celniker SE, et al. modENCODE Consortium Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aigaki T, Ohsako T, Toba G, Seong K, Matsuo T. The gene search system: its application to functional genomics in Drosophila melanogaster. J Neurogenet. 2001;15:169–178. doi: 10.3109/01677060109167374. [DOI] [PubMed] [Google Scholar]
- 28.Laviolette MJ, Nunes P, Peyre JB, Aigaki T, Stewart BA. A genetic screen for suppressors of Drosophila NSF2 neuromuscular junction overgrowth. Genetics. 2005;170:779–792. doi: 10.1534/genetics.104.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart BA, et al. Dominant-negative NSF2 disrupts the structure and function of Drosophila neuromuscular synapses. J Neurobiol. 2002;51:261–271. doi: 10.1002/neu.10059. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 31.Ranz JM, Ponce AR, Hartl DL, Nurminsky D. Origin and evolution of a new gene expressed in the Drosophila sperm axoneme. Genetica. 2003;118:233–244. [PubMed] [Google Scholar]
- 32.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: What defines a substrate? Curr Opin Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 34.Babushok DV, et al. A novel testis ubiquitin-binding protein gene arose by exon shuffling in hominoids. Genome Res. 2007;17:1129–1138. doi: 10.1101/gr.6252107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y, et al. Evolution and expression of chimeric POTE-actin genes in the human genome. Proc Natl Acad Sci USA. 2006;103:17885–17890. doi: 10.1073/pnas.0608344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 37.Giver L, Arnold FH. Combinatorial protein design by in vitro recombination. Curr Opin Chem Biol. 1998;2:335–338. doi: 10.1016/s1367-5931(98)80006-9. [DOI] [PubMed] [Google Scholar]
- 38.Cui Y, Wong WH, Bornberg-Bauer E, Chan HS. Recombinatoric exploration of novel folded structures: a heteropolymer-based model of protein evolutionary landscapes. Proc Natl Acad Sci USA. 2002;99:809–814. doi: 10.1073/pnas.022240299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orr HA. The genetic theory of adaptation: A brief history. Nat Rev Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- 40.Nei M. The new mutation theory of phenotypic evolution. Proc Natl Acad Sci USA. 2007;104:12235–12242. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.