Abstract
Fatty-acyl CoA reductases (FAR) convert fatty acids into fatty alcohols in pro- and eukaryotic organisms. In the Lepidoptera, members of the FAR gene family serve in the biosynthesis of sex pheromones involved in mate communication. We used a group of closely related species, the small ermine moths (Lepidoptera: Yponomeutidae) as a model to investigate the role of FARs in the biosynthesis of complex pheromone blends. Homology-based molecular cloning in three Yponomeuta species led to the identification of multiple putative FAR transcripts homologous to FAR genes from the Bombyx mori genome. The expression of one transcript was restricted to the female pheromone-gland tissue, suggesting a role in pheromone biosynthesis, and the encoded protein belonged to a recently identified Lepidoptera-specific pgFAR gene subfamily. The Yponomeuta evonymellus pgFAR mRNA was up-regulated in sexually mature females and exhibited a 24-h cyclic fluctuation pattern peaking in the pheromone production period. Heterologous expression confirmed that the Yponomeuta pgFAR orthologs in all three species investigated [Y. evonymellus (L.), Yponomeuta padellus (L.), and Yponomeuta rorellus (Hübner)] encode a functional FAR with a broad substrate range that efficiently promoted accumulation of primary alcohols in recombinant yeast supplied with a series of biologically relevant C14- or C16-acyl precursors. Taken together, our data evidence that a single alcohol-producing pgFAR played a critical function in the production of the multicomponent pheromones of yponomeutids and support the hypothesis of moth pheromone-biosynthetic FARs belonging to a FAR gene subfamily unique to Lepidoptera.
Keywords: biosynthetic enzyme, chemical communication, gene family evolution, Lepidoptera, Yponomeuta
Long-chain fatty-alcohol molecules serve essential biological roles in a vast majority of living organisms (1–4). In particular, in most of the Lepidoptera, primary fatty alcohols and their aldehyde or acetate derivatives constitute the essential chemistry of mate attraction (5, 6). Most female moths synthesize their pheromone components in a specialized gland (Fig. 1) through de novo synthesis of palmitate or stearate followed by discrete enzymatic conversions, including selective two-carbon chain-shortening and Δ11 or other fatty-acyl CoA desaturation reactions (7–9). Fatty acids with specific chain lengths and appropriate number and position of double bonds are subsequently reduced by fatty-acyl CoA reductases (FAR) before subsequent oxidation and acetylation (8, 10, 11).
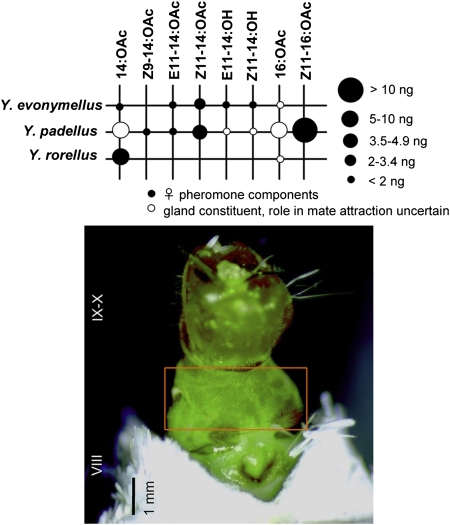
Fig. 1.
Sex-pheromone components of small ermine moths (Lepidoptera: Yponomeutidae) (Upper). The size of the dots is proportional to relative amounts found in an individual female PG (modified after ref. 20). (Lower) Ventral view of a Yponomeuta rorellus female extruding its terminal abdominal segments containing the sex pheromone-producing tissue located between abdominal segments XIII and IX (orange box).
FARs catalyze the NADPH-dependent reduction of fatty acyl-CoA precursors into fatty alcohols on the cytosolic side of the peroxisomal membrane in a two-step reaction that does not release intermediate aldehyde forms (12). They have an essential role in regulating the final steps of sex pheromone biosynthesis in certain moth species. In the European and Asian corn borers, Ostrinia nubilalis and Ostrinia furnacalis, in vivo labeling studies demonstrated that the selectivity of the reductase system could modulate ratios among final pheromone components by exclusive conversion of specific acid moieties into their corresponding alcohols (13, 14). In most species, pheromone production is also regulated by neurohormonal factors that in several cases have been shown to control activation of the reductase system (11, 15, 16). Despite the presumed central role of FARs in fatty-alcohol pheromone synthesis in moths, very few molecular and functional insights are available, except from the silkworm Bombyx mori (16) and the European corn borer O. nubilalis and its congener Ostrinia scapulalis (17, 18). Whereas the pheromone blends of B. mori and Ostrinia spp. are respectively composed of the single (E,Z)-10,12-hexadecadien-1-ol or of isomeric ratios of (Z)- and (E)-11-tetradecenyl acetate (Z/E11-14:OAc), many moth species produce multicomponent blends made of mixtures of C12, C14, and C16 fatty-acid-derived pheromone components (10, 19). The functional bases of the reduction step in the biosynthesis of complex pheromone blends remain largely unexplored, and in particular, the number of FAR genes implicated and their substrate affinities are yet to be determined.
In the present study, we used a molecular and functional approach to unravel the reduction step implicated in pheromone biosynthesis in three West-Paleartic species of small ermine moths, namely Yponomeuta evonymellus (L.), Yponomeuta padellus (L.), and Yponomeuta rorellus (Hübner) (Lepidoptera: Yponomeutidae). All of the nine yponomeutids present in Europe use pheromone blends made of chemically and structurally related C14- and C16-fatty alcohols and their derivatives (20). In six sympatric species, including Y. evonymellus and Y. padellus, the two primary components are Z11-14:OAc and E11-14:OAc, and complete reproductive isolation is ensured by the use of additional pheromone components (Fig. 1) (21). Y. rorellus is peculiar in that it uses only saturated (14:OAc) but no unsaturated components as pheromone (22). Screening of pheromone-gland FAR genes and heterologous expression allowed us to demonstrate that the reduction step of the long-chain C14- and C16-acyl pheromone precursors found in small ermine moths is accounted for by a single pheromone gland-specific FAR. Our results support that fatty alcohol precursors of complex blends used by most Lepidopteran species are likely to be biosynthesized by broad-range pheromone biosynthetic FARs belonging to a gene subfamily unique to the Lepidoptera.
Results and Discussion
Cloning of Biosynthetic FAR Homolog Candidates.
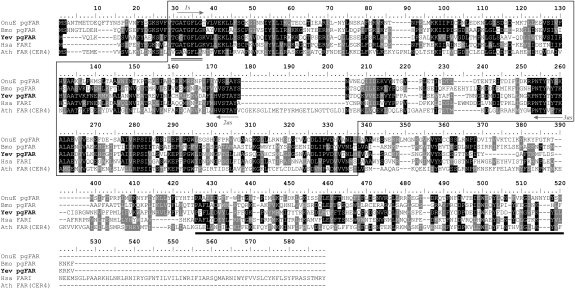
Consensus oligonucleotide primers were designed based on homologies between discrete N-termini motifs from various functional eukaryotic FARs (Fig. 2). Their use in PCR reactions in combination with Y. evonymellus pheromone gland (PG) cDNA as template led to the amplification of three distinct FARs from the PG transcriptome. The cloned full-length cDNA transcripts spanned respectively 1,988 bp, 2,259 bp, and 2,114 bp that encompassed ORFs of 1,734 bp, 1,350 bp, and 1,575 bp, which corresponded respectively to proteins with 578 amino acid residues (aa) (Yev-FARI), 450 aa (Yev-FARII), and 525 aa (Yev-FARIII). On average, the three predicted proteins presented 32 to 36% sequence homology in pairwise comparisons. GenBank searches in the nonredundant protein database revealed that the three encoded proteins from Y. evonymellus shared 26 to 35% sequence homology to the B. mori pgFAR (16), 21 to 26% to the Arabidopsis thaliana cuticular wax CER4 FAR (23), and 31 to 39% sequence homology to the Homo sapiens FAR1 implicated in the synthesis of the alcohol precursors of wax monoesters and ether lipids (24). The Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) enzyme nomenclature and sequence analysis in the GenBank nonredundant protein database indicated the encoding-genes to represent long-chain fatty acyl reductases (E.C.1.2.1.50). In addition, protein family domain architecture (Pfam) indicated that the transcripts shared characteristic domains of eukaryotic FARs, including a Rossmann-fold NAD(P)(+)-binding domain encompassed between amino acid positions 30 and 330 and a Sterile domain found in the C-terminal portion of the polypeptide chains (Fig. 2).
Fig. 2.
Alignment of the pheromone biosynthetic reductase from Yponomeuta evonymellus (Yev-pgFAR) and functional FAR proteins from eukaryotic organisms: OnuE, Ostrinia nubilalis (GenBank accession no. FJ807735); Bmo, Bombyx mori (GenBank accession no. BAC79426); Hsa, Homo sapiens (GenBank accession no. AAT42129); Ath, Arabidopsis thaliana (GenBank accession no. NP567936). Sequence alignments were computed in ClustalW (39) and edited in BOXSHADE (http://www.ch.embnet.org/index.html). Identical amino acid residues and conservative substitutions are shaded in black or gray, respectively. The FAR structural elements include a N-teminal Rossmann-fold NAD(P)(+)-binding domain (black box) and the NADH-binding motif (double underline) as well as a Sterile protein domain (thick black line). Yev-pgFAR was isolated using the 1s and 3as oligonucleotide primers (arrows).
Tissue Distribution and Temporal and Diurnal Analysis of Yev-pgFAR Gene Expression.
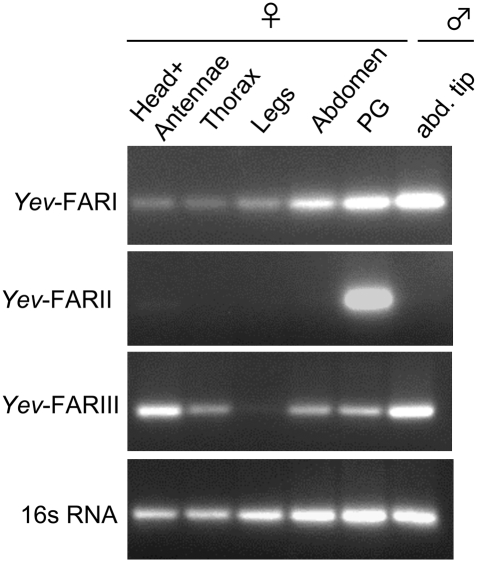
The production and release of moth pheromones is an elaborate task that takes place in the glandular cells constituting the PG of mature females (10). Monitoring of Y. evonymellus FARs by RT-PCR demonstrated that Yev-FARII exhibited a female PG-specific expression (Fig. 3), whereas negligible signal was observed in other tissues, which is similar to the expression pattern of pheromone biosynthetic Δ11-desaturase genes (i.e., refs. 25 and 26), thereby supporting Yev-FARII, hereafter referred to as Yev-pgFAR, as a candidate pheromone biosynthetic gene. In contrast, Yev-FARI and Yev-FARIII appeared broadly distributed and not female-specific (Fig. 3).
Fig. 3.
Tissue distribution of the small ermine moth Y. evonymellus FAR mRNAs monitored by reverse-transcriptase PCR. Amplicon sizes: FARI, 330 bp; FARII (Yev-pgFAR), 320 bp, FARIII, 333 bp; 16s RNA, 397 bp.
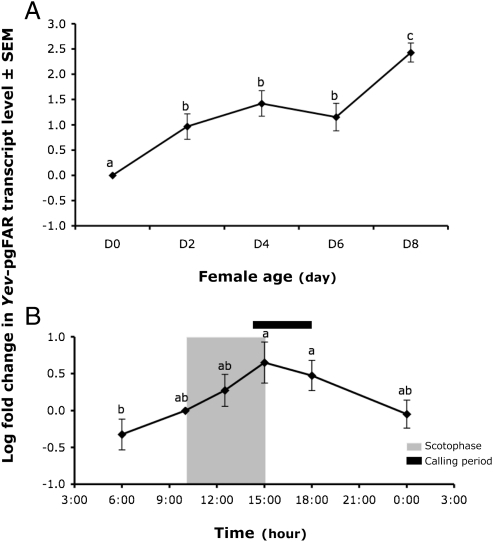
The production and emission of chemical cues mediating conspecific-male attraction in moths is also a tightly regulated process; the release of chemicals is concordant with female maturity and usually coincides with a peak in behavioral activity occurring during a specific time window (27, 28). In Yponomeuta spp. pheromone production is typically acquired 2 to 4 d after eclosion (21) and the diel rhythmicity in pheromone production and periods of calling behavior are synchronized to be maximal at dawn (28). Quantitative PCR was used to examine the changes in transcript level of the pheromone biosynthetic candidate Yev-pgFAR in adult Y. evonymellus females at different ages and over a 24-h period. Our data showed that transcription occurs in newly emerged immature females (0 d old) and continues up to at least 8 d after eclosion, indicating that the Yev-pgFAR gene is activated before maturity. However, the gene is up-regulated over ages with a significant increase in mRNA expression level from 2-d-old females (Fig. 4A). Whether the pgFAR-encoded gene-expression levels in Y. evonymellus could also be synchronized with the interval of pheromone production was examined over 24 h by sampling PGs of 5- to 6-d-old females at defined time-points during early-, mid-, and late-photophase and scotophase periods. Our data showed that the Yev-pgFAR gene expression fluctuates in accordance with the photoperiodic cues and with a peak of transcriptional activity at the onset of the photophase (Fig. 4B): that is, the observed period of behavioral activity and pheromonal emission in this species (21). In moths, the tight synchronization between behavioral, physiological, and biochemical cues is usually achieved by neuroendocrine mechanisms entrained by photoperiodic cues (29). Pheromone production can be stimulated by a neurohormone [pheromone biosynthesis activating neuropeptide (PBAN)] (reviewed in ref. 29) of which the circadian release elicits a pheromonotropic response up-regulating key pheromone biosynthetic enzymes including the reductase system (7, 11, 15, 30). In B. mori, a PBAN-mediated cascade has been demonstrated to activate the reductase enzyme responsible for Bombykol production (31, 32). Importantly, the Yev-pgFAR gene transcriptional up-regulation is concordant with the interval of pheromone emission in Y. evonymellus and indicated that activation of the reduction stage could contribute to the rhythmicity of pheromone production in this species, thereby supporting the pgFAR transcript to encode a key element of the reductase system in yponomeutids.
Fig. 4.
Temporal and diurnal expression patterns of Yev-pgFAR monitored using quantitative PCR. RNAs were extracted from the PG of adult females (A) x days after emergence (x = 0–8 d) and (B) from females (5–6 d old) at different time points over a 24-h period. The mean relative fold-change expression scores were calculated from raw cycle threshold (Ct) values, relative to the expression of the 16S RNA control gene and are calibrated to female day 0 (A) and time point 10.00 AM (B), respectively. Transcript levels were log2 transformed (±SEM, n = 18). Treatments with different letters are statistically different (ANOVA, Ryan-Einot-Gabriel-Weilsh test, P < 0.05). The functional Yev-pgFAR is up-regulated in mature females and its diurnal fluctuations are correlated with the rhythmicity of pheromone release in mature females.
Phylogenetic Analyses Indicate Yev-pgFAR to Be Orthologous to Pheromone-Specific Moth FARs.
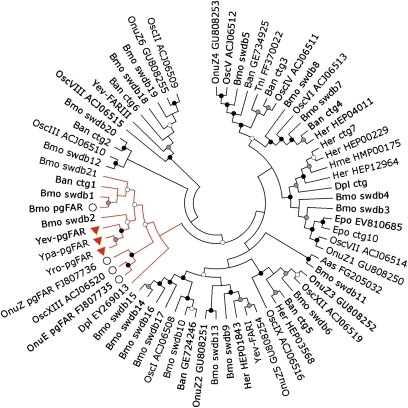
A phylogeny of the lepidopteran FAR repertoire was constructed using moth and butterfly FAR genes collected from GenBank, genomic, and EST databases. Twenty-one gene paralogs of the functional B. mori pgFAR gene were found in the silkworm genome database, of which many had homologs in O. nubilalis (17) and its congener O. scapulalis (18) or in various butterfly EST datasets (Fig. 5). Ten well-defined reductase groups were supported with nodes of bootstrap values over 90. The three Y. evonymellus FAR genes clustered in distinct clades and Yev-pgFAR grouped together with the pgFARs from B. mori and Ostrinia spp. (16–18), which we suggest to be a monophyletic group within the Lepidoptera. Consistent with its tissue distribution, as well as temporal and diurnal expression patterns, phylogenetic data supported the Yev-pgFAR as a candidate for pheromone biosynthesis. Therefore, we cloned its orthologs from Y. padellus and Y. rorellus, which similarly grouped into the pgFAR specific clade (Fig. 3) and exhibited a PG-specific expression pattern (Fig. S1). The three orthologs appeared extremely conserved in their ORFs at the nucleotide level and diverged by only a few amino acid residues (Yev-pgFAR/Yro-pgFAR, uncorrected P distance = 0.038; Yev-pgFAR/Ypa-pgFAR, uncorrected P distance = 0.043; and Yro/Ypa pgFAR, uncorrected P distance = 0.016). The Nei-Gojobori codon-based test indicated that overall all sequence pairs were evolving under purifying selection (dN/dS < 1; P = 0.003), suggesting that the genes have a function of essential importance.
Fig. 5.
Phylogeny of Lepidopteran FARs. The Neighbor-joining algorithm analysis was computed using MEGA (v. 4.0) and the JTT model for amino acids with 1,500 bootstrap replicates (40). Nodes with bootstrap comprised between 50 and 70% are marked with open circles. Nodes with bootstrap support between 71 and 90% or over 90% are marked with gray or black circles, respectively. Lepidopteran FAR sequences were retrieved from GenBank and EST databases using BLASTP and TBLASTN searches and the B. mori pgFAR as query (16). The predicted paralogs of B. mori pgFAR were retrieved from the Silkworm Genome Database (accession numbers available in Table S1) (41). TBLASTN searches were made against the clustered ESTs database on ButterflyBase (42) and EST sequences were manually assembled into contigs (ctg) and translated using ExPASy (43). Predicted protein sequences were aligned in MAFFTv6 (44) followed by manual inspection. The clade in red contains the functional pgFARs from Y. evonymellus (Yev-pgFAR), Y. padellus (Ypa-pgFAR), and Y. rorellus (Yro-pgFAR) (upside-down red triangles). Other FAR gene members implied in pheromone biosynthesis in Lepidoptera are labeled with large open circles. The abbreviated species names correspond to (i) butterflies species: Ban, Bicyclus anynana; Dpl, Danaus plexippus; Her, Helioconius erato; Hme, Helioconius melpomene; Pxu, Papilio xuthus and (ii) moth species: Aas, Antheraea assama; Amy, Antheraea mylitta; Bmo, Bombyx mori; Epo, Epiphyas postvittana; Pin, Plodia interpunctella; Tni, Trichoplusia ni; Onu, Ostrinia nubilalis, and Osc, Ostrinia scapulalis.
Yeast Expression Demonstrated That the Yponomeutid pgFARs Are Multisubstrate Reductases.
Molecular characterization of Yev-pgFAR and its orthologs evidenced the genes as candidate biosynthetic FARs. To investigate their reductase activity and substrate specificity, we expressed the Yev-pgFAR, Ypa-pgFAR, or Yro-pgFAR ORFs placed under the control of a GAL promoter in the InvSc1 strain of the yeast Saccharomyces cerevisiae. To assess the reliability of tissue distribution patterns and phylogenetic studies in predicting the functionality of pheromone biosynthetic lepidopteran FARs, Yev-FARI or Yev-FARIII were included as controls. Yeast cells were incubated for 24 h in the presence of galactose and equal concentrations of individually supplied FAME precursors (17, 33) before extraction with n-hexane and GC-MS analyses. Yeast cells supplemented with fatty-acyl moieties and expressing Yev-pgFAR did catalyze the formation of primary fatty alcohols, whereas yeast cells transformed with the pYES2.1 expression vector alone did not convert any acyl substrate (Fig. 6 and Fig. S2). As predicted, Yev-FARI and Yev-FARIII failed to reduce any biosynthetic fatty acyl precursors, and thus we concluded that they were inactive in pheromone production.
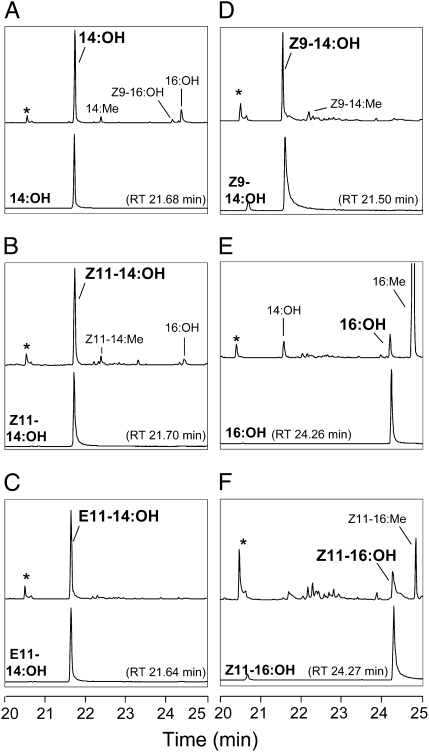
Fig. 6.
GC-MS analyses of fatty alcohol extracts from galactose-induced yeast transformed with pYES2.1-Yev-pgFAR in presence of 0.5 mM FAME substrates. The upper chromatogram traces represent the total ion currents (TICs) of fatty alcohol products from yeast cells transformed with pYES2.1-Yev-pgFAR and supplemented with (A) 14:Me, (B) Z11-14:Me, (C) E11-14:Me, (D) Z9-14:Me, (E) 16:Me, and (F) Z11-16:Me. The lower traces represent the TIC of the corresponding alcohol reference compounds (RT = retention time). Asterisks (*) indicate the internal standard (150 ng Z11-13:OH). The y axes represent the relative abundance.
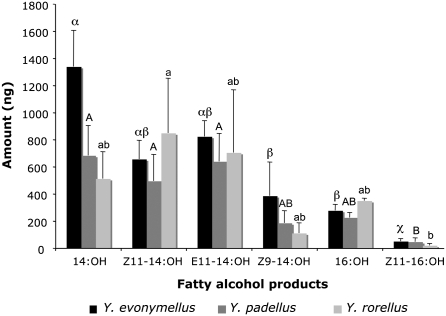
In a series of assays using structurally similar saturated and unsaturated C14 and C16 substrates, Yev-pgFAR converted the supplied pheromone precursors with an overall preference for C14 substrates (Figs. 6 and 7). Yev-pgFAR concomitantly reduced the 14:acyl and 16:acyl naturally occurring in the InvSc1 yeast strain (Fig. 6), and some Z9-16:acyl (Fig. S2). The Ypa-pgFAR and Yro-pgFAR orthologs were similarly expressed, and the resulting amounts of the various fatty alcohols produced were compared between and within species. All characterized Yponomeuta pgFARs exhibited an overall identical reduction activity on the supplemented substrates (two-way ANOVA, test for interaction between species and substrate; P = 0.27) (Fig. 7). Similarly to observations from expression studies of Arabidopsis fatty-alcohol reductases (34), the produced fatty alcohols were found both in culture medium and yeast cells, with both fractions having an identical composition (Figs. S3 and S4). There was no correlation between the yeast/medium ratio and the incubation time (Spearman's rank correlation; P ≥ 0.09 for all alcohol products considered individually) and the yeast/medium ratio did not differ significantly between the different fatty alcohol products (Kruskal-Wallis test; all time-points considered; P = 0.12) (Fig. S3). All together, the yeast fatty alcohol production thus reflects the enzymes’ total production and functionality.
Fig. 7.
Fatty alcohol production of Yev-pgFAR, Ypa-pgFAR, and Yro-pgFAR orthologous genes characterized from Y. evonymellus, Y. padellus, and Y. rorellus, respectively, and expressed in a heterologous S. cerevisiae yeast strain supplemented with various fatty-acyl precursors as shown in Fig. 6. Bars indicate the SEMs for each pgFAR (n = 3 per substrate tested). Greek, uppercase, and lowercase letters correspond to Yev-pgFAR, Ypa-pgFAR, and Yro-pgFAR, respectively. Same letters within each series indicate mean values that are not statistically different [ANOVA, Tukey's HSD test (P < 0.05)]. Overall, the three orthologous encoded genes display an analogous biochemical activity, with a preference for the reduction of C14 substrates versus C16 substrates.
We further tested two rare C14 pheromone precursors (Z- and E12-14:Me, respectively) that are not found in glands of Yponomeuta species. We found that the pgFAR enzyme was consistently able to reduce these two exotic substrates (Fig. S2), pheromone precursors of the Asian corn borer (ACB) O. furnacalis. These data stress that the reductase system of some moth species is inclined toward reducing a broad set of fatty acyls and support the theory that the structural composition of pheromone blends greatly depends on precursors made available by upstream biosynthetic enzymes (i.e., β-oxidases and desaturases). Hence, to produce its unique blend composed of the Z- and E12–14:OAc (35), the ACB uses a unique Δ14-desaturase absent in yponomeutids and that acts upon palmitic acid to produce Z- and E14–16:acyl (followed by β-oxidation, reduction, and acetylation) (36). Assuming that the ACB ancestor had a reductase enzyme as broad as the yponomeutid pgFAR, these unique pheromone components could have been produced directly following the recruitment of the new functional type of desaturase.
In a second set of experiments, we tested the pgFAR substrate chain-length preference by supplementing mixtures of even carbon-saturated substrates ranging from C8 to C24. The pgFAR orthologs were able to reduce the C14 and C16 acyls in a similar ratio as when supplementing precursors individually (Fig. 7), plus minor amounts of the C12 acyl (6% of the corresponding saturated C14 alcohol production) but failed to reduce any saturated fatty acyl substrates shorter than C12 (C8 and C10) or longer than C16 (C18 to C24). We rationalize that the production of 12:OH may further indicate the ability of the pgFAR orthologs to reduce other C12 acyl substrates, including the precursor of Z9-12:OAc used as pheromone component in Yponomeuta malinellus (reviewed in ref. 20).
The different yponomeutid pgFAR orthologs thus possess a similar biochemical activity, which implies that the function evolved before radiation within the Yponomeuta genus and that the ancestral enzyme was exquisitely nonspecific. Overall, the Yponomeuta pgFAR enzymes selectively recognize the substrate length of potential Yponomeuta spp. pheromone precursors but accept a broad range of substrates, being restricted neither to the isomeric nature of the substrates (Z or E) nor to the location of the double-bond position in C14 substrates because Δ9-, Δ11-, or Δ12-14:acyls were equally reduced (Fig. 6 and Fig. S2). Evolutionary changes have nevertheless occurred upstream in the biosynthetic machinery and the in vivo pgFAR enzyme activity differs according to the pool of fatty-acyl precursors available for pheromone production in the distinct Yponomeuta species. For example, the deactivation of the Δ11-desaturase in Y. rorellus (22) prevents the production of any Δ11-unsaturated alcohols in this species. Because of its essential role in pheromone biosynthesis, the Y. rorellus pgFAR gene is evolving under selective constraints, which has most likely prevented its functional divergence from its sister species pgFARs. In Y. padellus, the low reductive activity on the Z11-16:acyl is compensated by a large pool of the same precursor in the insect gland. When supplementing a mixture of the E11-14, Z11-14, and Z11-16:acyls in a ratio corresponding to the relative abundance of each precursor in the gland (1:1:100) (20), the final yeast fatty-alcohol ratio corresponded to a 1:1:5 (Fig. S4) similarly to the pheromone blend (Fig. 1). Although we cannot exclude that another FAR displaying a substrate-specificity for the Z11-16:acyl may also exist, these findings support that providing specific pools of precursors the reduction step in Y. padellus can be accounted for by a single enzyme. The in vitro specificity of the pgFARs thus reflects to a great degree the specificity in insecta, but the specific pheromone blends of yponomeutids result from the concerted action of a specific set of biosynthetic genes. For example, the combined action of a Δ11-desaturase and the Ypa-pgFAR explains to a great part the Z11-16:OAc ratio in this species. In Y. padellus and Y. evonymellus, both the E11-14 and Z11-14:OH and acetates are produced, and Y. padellus produces the Z9-14:OAc, which emphasizes that the specificity of the—yet uncharacterized—acetyl transferase system certainly plays a role in regulating the alcohols/acetates ratios in these species.
In the Z and E strains of O. nubilalis, pheromone specificity is controlled by one gene-encoding strain-specific reductase alleles that resulted in the evolution of substrate-specific reductases (17), which convert both Z- and E11 to 14:acyl precursor moieties in ratios corresponding to the Z and E strains pheromone blend composition (13). In contrast and similarly to the Yponomeuta genus, many moth species produce complex multicomponent mating signals resulting from the concerted action and inherent specificity of more than one biosynthetic gene product (8). Taken together, our findings demonstrate that in Yponomeuta spp. a single biosynthetic pgFAR can catalyze all terminal-reduction modifications taking place in pheromone biosynthesis and support that extant reductase systems in moths might have evolved under different evolutionary constraints, notably as a result of the variable complexity in the female-emitted blends.
Materials and Methods
Insects.
Yponomeuta evonymellus and Y. padellus pupae were collected from Prunus padus L. and from Crataegus sp. or Sorbus aucuparia L., respectively (Lund, Sweden). Y. rorellus pupae were collected from Salix sp. (Leiden, The Netherlands). Males and females were separated before eclosion and maintained in rearing chambers at 22 ± 1 °C in a 19-h:5-h light:dark photoperiod. Newly emerged females were separated daily before the scotophase and considered to be 0 d old.
cDNA Cloning.
PGs were excised from 15 virgin females on the day of eclosion and snap-frozen in liquid nitrogen. Total RNA isolation and DNase purification were performed using the RNeasy Isolation kit (Qiagen) and first strand cDNA was synthesized from 1 μg total RNA and a Stratascript reverse transcriptase (Stratagene). Oligonucleotide primers were designed based on the N terminus of eukaryotic FARs: Forward: 5′-GGHGCNACBGGNTTYHTDG-3′ (1s), Reverse: 5′-TMRTADGCHGTNGANACRTRA-3′ (2as) or 5′-TTBGTVWRNRYRTABGTRTTNG-3′ (3as) (Fig. 2). PCR thermal cycling conditions consisted of 95 °C for 5 min, 35 cycles at 95 °C for 30 s, 55 °C for 45s, 72 °C for 90s, and 72 °C for 10 min. Specific 420-bp (primers 1s-2as) or 560-bp (primers 1s-3as) amplification products were cloned into the pCR2.1-TOPO TA cloning vector system (Invitrogen) and resultant plasmids were sequenced using the Big Dye Terminator cycle sequencing kit v1.1 followed by analysis on a capillary ABI 3100 sequencer instrument (Applied Biosystems). The 5′- and 3′- cDNA ends were obtained using the SMART RACE Kit (Clontech) and gene-specific primers (GSPs) (Table S2). DNA sequences were analyzed using BioEdit followed by BLAST searches (37).
RT-PCR.
Total RNAs were isolated from head, thorax, legs, abdomen, and PG tissues of 15 female individuals as well as from 15 male abdominal tips on day 0 postemergence. RT-PCR reactions were carried out using the SuperScript III One-Step RT-PCR System with Platinum Taq (Invitrogen) in a 25-μL reaction mix containing 50 ng total RNA and 0.8 μM of each GSP (Table S2). PCR conditions were as follows: 55 °C for 30 min, 94 °C for 2 min, 30 cycles of 94 °C for 15s, 55 °C for 30s, 68 °C for 45s, and 68 °C for 2 min. PCR products were separated on a 2% agarose gel.
Functional Assays.
GSPs were designed (Table S2) to amplify all FAR candidate ORFs using the corresponding species PG cDNA as template in combination with the Advantage2 PCR system (Clontech). Each FAR ORF was cloned in the pYES2.1 expression vector (Invitrogen) downstream the GAL1 promoter before sequencing. The pYES2.1 plasmid alone and the distinct FAR constructs were transformed into the InvSc1 strain of the yeast S. cerevisiae (Invitrogen) and propagated on SC-U plates containing 2% glucose. Individual prototroph colonies were inoculated in 5 mL SC-U medium and incubated for 48 h at 30 °C and 300 rpm (Innova 42, New Brunswick Scientific), diluted to an OD600 = 0.4 to a final volume of 20 mL SC-U 2% galactose and 0.1% glucose in 250-mL flasks, and incubated for 24 h at 30 °C and 200 rpm. For precursor assays, yeast cultures were diluted to 1:10 in 2 mL SC-U 2% galactose, 1% tergitol (Nonidet P-40, Sigma), and 0.5-mM alcohol-free precursors in the form of methyl-esters (18, 33). The Z11-14:Me, E11-14:Me, Z12-14:Me, E12-14:Me, and Z11-16:Me were synthesized from their corresponding free acids, as described in ref. 17. The Z9-14:Me, 14:Me, Z9-16:Me, 16:Me, and mixtures 20% w:w of saturated C8 to C16 and C16 to C24 FAMEs were purchased from Larodan. All FAMEs were dissolved in 95% ethanol at a stock concentration of 0.02 M. After incubation for 24 h at 30 °C and 300 rpm, cells were collected by centrifugation at 2,000 × g (Labofuge 200, Heraeus Instruments) and washed in sterile water. Cell pellets were extracted with 1-mL n-hexane (16–18) spiked with 150 ng Z11-13:OH as an internal standard followed by shaking at 200 cycles/min (Vibramax 100, Heidolph) for 60 min. Samples were stored at −20 °C until gas chromatography analyses (SI Materials and Methods). For statistical analyses, the amounts (ng) of fatty-alcohol products were log transformed [log10 (amount+1)] and subjected to a two-way ANOVA (SPSS 16.0) with species and product as variables. The amounts were subsequently treated for each species individually using a one-way ANOVA and the Tukey's honestly significant difference procedure.
Quantitative PCR Analysis.
Two biological cDNA samples were obtained from fifteen Y. evonymellus female PGs dissected at the onset of the photophase at 0, 2, 4, 6, and 8 d after emergence (expression over age) and from PGs of 5- to 6-d-old females dissected every sixth hour during the photophase and every 2.5 h during the scotophase, which corresponded to early-, mid-, and late-periods, respectively (diurnal expression). Triplicated 25-μL quantitative PCR reactions were run on an Mx3000P v4.01 (Stratagene) using 10 ng of PG cDNA as template with 200 nM GSPs (Table S2) designed in the AlleleID software (PREMIER, Biosoft International), 50 nM Rox Dye, and the Platinum SYBR green qPCR SuperMix-UDG (Invitrogen). The amplification efficiency (E) of each primer pair was initially assessed from serial dilutions of cDNAs, and the raw cycle threshold values (Ct) were averaged from triplicated reactions. Quantitative PCR products from initial runs were checked on 2% agarose gels and sequenced to assess primer-specificity. The 16S RNA was used as normalizer gene, and all relative expression levels were calculated in the Mx3000 program (Stratagene) using the (1+E)−ΔΔCt algorithm (38). Fluorescence background baselines and amplification thresholds were calculated automatically. Log fold-change in expression level were calculated against a calibrator sample and subjected to a one-way ANOVA analysis. The groups were compared using the Ryan-Einot-Gabriel-Weilsh procedure (SPSS 16.0).
Supplementary Material
Acknowledgments
We thank S. Menken (Institute of Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam) and R. Koi (Institute of Biology, Leiden, The Netherlands) for providing Yponomeuta rorellus pupae; E. Hedenström (Department of Natural Sciences, Engineering and Mathematics, Sundsvall, Sweden) and his group for synthesizing FAME precursors; T. Johansson (Department of Biology, Lund University, Lund, Sweden) for useful discussions; and anonymous reviewers for valuable comments. This study was supported by the Crafoord Foundation and the Swedish Research Council (Vetenskapsrådet).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition:The sequences reported in this paper have been deposited in the GenBank database: Y. evonymellus (accession nos. GQ907231–GQ907233), Y. rorellus (accession no. GQ907234), and Y. padellus (accession no. GQ907235).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000823107/-/DCSupplemental.
References
- 1.Gilbert LI, Chino H. Transport of lipids in insects. J Lipid Res. 1974;15:439–456. [PubMed] [Google Scholar]
- 2.Gilby AR. Transpiration, temperature and lipids in insect cuticle. Advanced Insect Physiology. 1980;15:1–33. [Google Scholar]
- 3.Blomquist GJ, Nelson DR, de Renobales M. Chemistry, biochemistry, and physiology of insect cuticular lipids. Arch Insect Biochem Physiol. 1987;6:227–265. [Google Scholar]
- 4.Buckner JS. In: Insect Lipids: Chemistry, Biochemistry, and Biology. Stanley-Samuelson DW, Nelson DR, editors. Lincoln: University of Nebraska Press; 1993. pp. 227–270. [Google Scholar]
- 5.Roelofs WL. Chemistry of sex attraction. Proc Natl Acad Sci USA. 1995;92:44–49. doi: 10.1073/pnas.92.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardé RT, Haynes KF. In: Advances in Insect Chemical Ecology. Cardé RT, Millar JG, editors. Cambridge, United Kingdom: Cambridge University Press; 2004. pp. 283–332. [Google Scholar]
- 7.Tillman JA, Seybold SJ, Jurenka RA, Blomquist GJ. Insect pheromones—An overview of biosynthesis and endocrine regulation. Insect Biochem Mol Biol. 1999;29:481–514. doi: 10.1016/s0965-1748(99)00016-8. [DOI] [PubMed] [Google Scholar]
- 8.Blomquist GJ, Jurenka RA, Schal C, Tittiger C. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill S, editors. San Fransisco: Elsevier Academic; 2005. pp. 705–751. [Google Scholar]
- 9.Roelofs WL, Rooney AP. Molecular genetics and evolution of pheromone biosynthesis in Lepidoptera. Proc Natl Acad Sci USA. 2003;100:9179–9184. doi: 10.1073/pnas.1233767100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjostad LB, Wolf W, Roelofs WL. In: Pheromone Biochemistry. Blomquist GJ, Prestwich GD, editors. New York: Academic Press; 1987. pp. 77–120. [Google Scholar]
- 11.Jurenka RA. In: Insect Pheromone Biochemistry and Molecular Biology. Vogt G, Blomquist R, editors. New York: Academic Press; 2003. pp. 53–80. [Google Scholar]
- 12.Wanders RJA, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 13.Zhu JW, Zhao C-H, Lu F, Bengtsson M, Löfstedt C. Reductase specificity and the ratio regulation of E/Z isomers in pheromone biosynthesis of the european corn borer, Ostrinia nubilalis (Lepidoptera: Pyralidae) Insect Biochem Mol Biol. 1996;26:171–176. [Google Scholar]
- 14.Zhao C-H, Fan J, Bengtsson M, Löfstedt C. Substrate specificity of acetyltransferase and reductase enzyme systems used in pheromone biosynthesis by asian corn borer, Ostrinia furnacalis. J Chem Ecol. 1995;21:1495–1510. doi: 10.1007/BF02035148. [DOI] [PubMed] [Google Scholar]
- 15.Rafaeli A. Pheromone biosynthesis activating neuropeptide (PBAN): regulatory role and mode of action. Gen Comp Endocrinol. 2009;162:69–78. doi: 10.1016/j.ygcen.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Moto K, et al. Pheromone gland-specific fatty-acyl reductase of the silkmoth, Bombyx mori. Proc Natl Acad Sci USA. 2003;100:9156–9161. doi: 10.1073/pnas.1531993100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lassance J-M, et al. Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature. 2010 doi: 10.1038/nature09058. 10.1038/nature09058. [DOI] [PubMed] [Google Scholar]
- 18.Antony B, et al. Pheromone-gland-specific fatty-acyl reductase in the adzuki bean borer, Ostrinia scapulalis (Lepidoptera: Crambidae) Insect Biochem Mol Biol. 2009;39:90–95. doi: 10.1016/j.ibmb.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 19.El-Sayed AM. The Pherobase: Database of insect pheromones and semiochemicals. 2010. [Accessed May 21, 2010]. http://www.pherobase.com.
- 20.Löfstedt C, Herrebout W, Menken S. Sex pheromones and their potential role in the evolution of sex reproductive isolation in small ermine moths (Yponomeutidae) Chemoecology. 1991;2:20–28. [Google Scholar]
- 21.Löfstedt C, Van der Pers JNC. Sex pheromones and reproductive isolation in four European small ermine moths. J Chem Ecol. 1985;11:649–666. doi: 10.1007/BF00988574. [DOI] [PubMed] [Google Scholar]
- 22.Löfstedt C, Herrebout W, Du J-W. Evolution of the ermine moth pheromone tetradecenyl acetate. Nature. 1986;323:621–623. [Google Scholar]
- 23.Rowland O, et al. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006;142:866–877. doi: 10.1104/pp.106.086785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng JB, Russell DW. Mammalian wax biosynthesis. I. Identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions. J Biol Chem. 2004;279:37789–37797. doi: 10.1074/jbc.M406225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenfield C-L, You KM, Marsella-Herrick P, Roelofs WL, Knipple DC. Structural and functional conservation and divergence among acyl-CoA desaturases of two noctuid species, the corn earworm, Helicoverpa zea, and the cabbage looper, Trichoplusia ni. Insect Biochem Mol Biol. 2001;31:949–964. doi: 10.1016/s0965-1748(01)00043-1. [DOI] [PubMed] [Google Scholar]
- 26.Moto K, et al. Involvement of a bifunctional fatty-acyl desaturase in the biosynthesis of the silkmoth, Bombyx mori, sex pheromone. Proc Natl Acad Sci USA. 2004;101:8631–8636. doi: 10.1073/pnas.0402056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyatt TD. Pheromones and Animal Behaviour. Communication by Smell and Taste. Cambridge, United Kingdom: Cambridge University Press; 2004. p. 391. [Google Scholar]
- 28.Löfstedt C, Herrebout W. Sex pheromones of three small ermine moths found on the European spindle tree. Entomol Exp Appl. 1988;46:29–38. [Google Scholar]
- 29.Rafaeli A. Neuroendocrine control of pheromone biosynthesis in moths. Int Rev Cytol. 2002;213:49–91. doi: 10.1016/s0074-7696(02)13012-9. [DOI] [PubMed] [Google Scholar]
- 30.Jurenka RA. Signal transduction in the stimulation of sex pheromone biosynthesis in moths. Arch Insect Biochem Physiol. 1996;33:245–258. [Google Scholar]
- 31.Fónagy A, et al. Pheromone-producing cells in the silkmoth, Bombyx mori: Identification and their morphological changes in response to pheromonotropic stimuli. J Insect Physiol. 2000;46:735–744. doi: 10.1016/s0022-1910(99)00162-6. [DOI] [PubMed] [Google Scholar]
- 32.Ohnishi A, Hull JJ, Matsumoto S. Targeted disruption of genes in the Bombyx mori sex pheromone biosynthetic pathway. Proc Natl Acad Sci USA. 2006;103:4398–4403. doi: 10.1073/pnas.0511270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, Jiao H, Murray NC, O'Connor M, Roelofs W. Gene characterized for membrane desaturase that produces (E)-11 isomers of mono- and diunsaturated fatty acids. Proc Natl Acad Sci USA. 2001;99:620–624. doi: 10.1073/pnas.221601498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doan TTP, et al. Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J Plant Physiol. 2009;166:787–796. doi: 10.1016/j.jplph.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Z-Q, et al. Sex pheromone components isolated from China corn borer, O. furnacalis Guernée (Lepidoptera:Pyralidae) (E)- and (Z)-12-tetradecnyl acetates. J Chem Ecol. 1981;7:841–851. doi: 10.1007/BF00992382. [DOI] [PubMed] [Google Scholar]
- 36.Roelofs WL, et al. Evolution of moth sex pheromones via ancestral genes. Proc Natl Acad Sci USA. 2002;99:13621–13626. doi: 10.1073/pnas.152445399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, et al. SilkDB: A knowledgebase for silkworm biology and genomics. Nucleic Acids Res. 2005;33(Database issue):D399–D402. doi: 10.1093/nar/gki116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papanicolaou A, Gebauer-Jung S, Blaxter ML, Owen McMillan W, Jiggins CD. ButterflyBase: A platform for lepidopteran genomics. Nucleic Acids Res. 2008;36(Database issue):D582–D587. doi: 10.1093/nar/gkm853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gasteiger E, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.