Abstract
The application of systems biology tools holds promise for rational industrial microbial strain development. Here, we characterize a Zymomonas mobilis mutant (AcR) demonstrating sodium acetate tolerance that has potential importance in biofuel development. The genome changes associated with AcR are determined using microarray comparative genome sequencing (CGS) and 454-pyrosequencing. Sanger sequencing analysis is employed to validate genomic differences and to investigate CGS and 454-pyrosequencing limitations. Transcriptomics, genetic data and growth studies indicate that over-expression of the sodium-proton antiporter gene nhaA confers the elevated AcR sodium acetate tolerance phenotype. nhaA over-expression mostly confers enhanced sodium (Na+) tolerance and not acetate (Ac-) tolerance, unless both ions are present in sufficient quantities. NaAc is more inhibitory than potassium and ammonium acetate for Z. mobilis and the combination of elevated Na+ and Ac- ions exerts a synergistic inhibitory effect for strain ZM4. A structural model for the NhaA sodium-proton antiporter is constructed to provide mechanistic insights. We demonstrate that Saccharomyces cerevisiae sodium-proton antiporter genes also contribute to sodium acetate, potassium acetate, and ammonium acetate tolerances. The present combination of classical and systems biology tools is a paradigm for accelerated industrial strain improvement and combines benefits of few a priori assumptions with detailed, rapid, mechanistic studies.
Keywords: ethanol, inhibitor, microarray, sequencing, systems biology
Recent high oil prices, concerns over energy security, and environmental goals have reawakened interest in producing alternative fuels via large-scale industrial fermentation. Improvement in methods producing ethanol from cellulosic biomass is crucial in this goal. A key barrier to convert cellulosic biomass to ethanol is the recalcitrance of biomass to hydrolysis (1 and 2). Severe biomass pretreatments are required to release the C-5 and C-6 sugars, which, along with by-products of fermentation can also create inhibitors to the production of ethanol (see recent reviews (3 and 4)). A range of inhibitory chemicals are generated by pretreatment regimes including sugar degradation products such as furfural and hydroxymethylfurfural (HMF); weak acids such as acetic, formic, and levulinic acids; and lignin degradation products such as the substituted phenolics vanillin and lignin monomers. High product yields, titers, and rapid product formation rates are considered prerequisites for industrial biofuel production (5). High substrate loadings are likely to be required in order to achieve an economically viable process, which in turn, is likely to lead to concomitant increases in inhibitor concentrations in fermentations and higher titers of the toxic, final product (5).
The development and deployment of robust ethanologenic microorganisms resistant to industrially relevant inhibitors and with high-yield ethanol production will be a critical component in the successful production of fuel ethanol in industrial-scale quantities. However, little progress has been made toward understanding the genetic basis of tolerance to ethanol (5) and to date there have been only a few examples of metabolic engineering using systems biology tools for bioprocess development (6). The revolutionary influence of genomics upon the field of microbiology and the application of DNA microarrays have been reviewed previously (7 and 8).
Yeast strains are the most frequently used microorganisms for industrial fuel production (9). However, engineered bacteria such as Escherichia coli, Zymomonas mobilis, and others are also being developed and deployed in search of superior production economics (2 and 10). Z. mobilis are Gram-negative facultative anaerobic bacteria with desirable industrial biocatalyst characteristics, such as high-specific productivity, high ethanol yield, and ethanol tolerance (12% v/v) (10–12). The genome sequence of Z. mobilis strain ZM4 has been determined (13) and its annotation improved by our group recently (14). Wild-type strains are able to ferment only a limited range of C-6 sugars, although this limitation has been overcome through the development of recombinant strains able to ferment both hexose and pentose sugars such as xylose and arabinose so that major components of cellulosic biomass can be utilized (15 and 16). Other recombinant strains have been engineered to ferment xylose, arabinose, and other substrates with high yields, but low tolerance to acetic acid and decreased ethanol tolerance have been reported (10, 17–19).
Acetic acid is an important inhibitor produced by the deacetylation of hemicelluloses during biomass pretreatment. At pH 5.0, 36% of acetic acid is in the uncharged and undissociated form and is able to permeate the bacterial plasma membrane leading to uncoupling and anion accumulation causing cytoplasmic acidification (20). The importance of acetic acid comes from the significant ratio of the concentration of acetate produced relative to fermentable sugars (21) and this ratio is higher in material from hardwoods (20). Synergistic or additive inhibitory effects are also likely with other hydrolysate inhibitors or metabolic by-products generated during the fermentation (3). Acetate removal processes have been described but they are energy or chemical-intensive and a full cost analysis has not been reported (21). The acetate tolerant Z. mobilis mutant (AcR) was generated by chemical mutagenesis and selection in continuous culture with a progressively increasing concentration of sodium acetate in the medium feed (22). AcR is capable of efficient ethanol production in the presence of 20 g/L NaAc, while the parent ZM4 is inhibited significantly above 12 g/L NaAc under the same conditions (22). Acetic acid was shown to be strongly inhibitory to wild-type derived strain ZM4(pZB5) on xylose medium and intracellular deenergization and acidification appeared to be the major inhibition mechanisms (23). A recombinant strain able to utilize both xylose and glucose with increased acetate resistance was generated by transforming plasmid pZB5 into the AcR and had improved fermentation characteristics in the presence of 12 g/L NaAc (24).
Strain AcR was generated while many systems biology tools were being developed or had yet to be deployed, and the molecular mechanism of AcR sodium acetate tolerance had not been elucidated until the present study. The application of recombinant DNA technology is used for strain development purposes, although rational metabolic engineering approaches require a priori knowledge or assumptions (25). The systematic combination of classical strain development with comparative genome resequencing and functional genomics is an emerging science that has few successful examples to date, but offers great promise to provide unprecedented insights into microbial physiology and important bioprocessing traits (6). The objective of the present study was to determine mutant loci selected in the AcR strain as we hypothesized one or more of the mutations conferred sodium acetate tolerance. We identify and confirm important mutations in the AcR strain through the systematic combination of comparative genome resequencing, functional genomics, and genetics and extend our studies to test similar Saccharomyces cerevisiae genes. We use computational modeling for mechanistic insights and discuss technological limitations of resequencing approaches.
Results and Discussion
Identification of Genomic Changes in AcR Mutant Strain.
The combination of microarray-based CGS and next-generation 454 pyroresequencing was used to identify the genome-wide genetic changes associated with the Z. mobilis acetate tolerant strain AcR compared to the ZM4 parental strain. The CGS results from AcR identified a 1,461-bp (∼1.5 kb) region of deleted DNA and thirty-eight putative AcR SNPs, 26 of which were within coding regions and 12 within intergenic regions (Table S1). The 1.5-kb AcR deletion was confirmed using the PCR, agarose gel electrophoresis, and Sanger sequencing analysis (Table S2). We generated ∼42-megabases (Mb) of pyrosequencing data that resulted in ∼21-fold genome coverage for ZM4 and ∼36-Mb of sequence data or ∼17-fold coverage for AcR. The pyroresequencing datasets included shotgun and paired-end sequencing reads for ZM4 and AcR, which have been deposited in the National Center for Biotechnology Information (NCBI) short-read archive database (Study SRP000908 (14) and SRP001379, respectively).
The GSMapper application in the 454 GS FLX software package 1.1.03 (454 Life Sciences) was used to map the pyrosequencing reads onto the Z. mobilis ZM4 reference genome (GenBank accession: AE008692) (13). This identified 200 and 219 high confidence differences (HCDiffs) for strains ZM4 and AcR respectively (Table S3). An HCDiff reflects differences between the pyroresequencing data and the reference genome sequence, and is defined as having at least one read aligned to each DNA strand possessing the reported difference, with at least five bases on both sides of the difference and few other isolated sequence differences in the read. A comparison of HCDiffs between ZM4 and AcR pyrosequencing data revealed that 183 were identical in both strains (Table S3), i.e., unlikely to contribute to the acetate tolerance phenotype. The discrepancies between our experimental data and the Z. mobilis ZM4 reference genome led to a collaborative effort to provide a comprehensive update to the ZM4 genome annotation (14).
When the 454 pyroresequencing data were compared to the CGS results, 27 SNPs were identified that had matching HCDiffs in both ZM4 and AcR 454 pyroresequencing data and were unlikely candidates for playing a role in the AcR tolerance phenotype (Table S2). The confirmed SNPs contained two that were unique to AcR, with one synonymous SNP (i.e., no change at the amino acid level) found within ZMO1184 encoding a hypothetical protein and a nonsynonymous SNP (i.e., with an amino acid change) in kup (ZMO1209) encoding a putative potassium transporter (Table S3). Therefore, the combined comparative genomic analyses and Sanger sequencing confirmation identified that the only differences between AcR and ZM4 were the 1.5-kb deletion region that truncated ZMO0117 and DNA upstream of ZMO0119, and two SNPs that affected ZMO1184 and ZMO1209.
Transcriptomic Profiling Identifies Differentially Expressed AcR Genes.
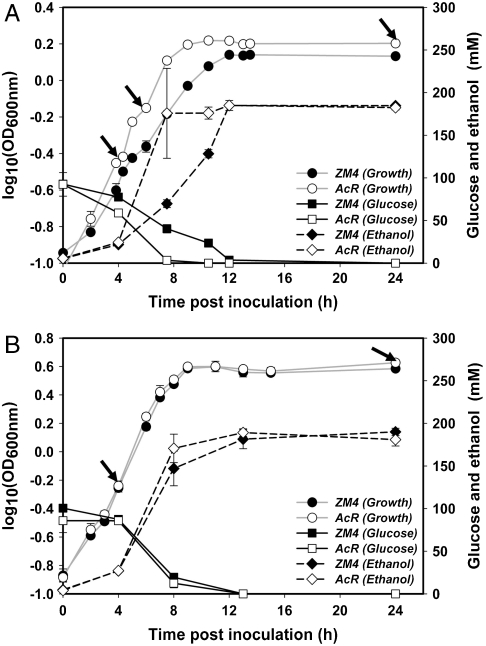
A microarray study was conducted to investigate differentially expressed genes in AcR and ZM4 and to correlate gene transcriptomic profiles with genotype data (GEO accession number GSE18106). An ANOVA was conducted using JMP Genomics (SAS Institute Inc.) to identify significant differences in exponential and stationary phase transcriptomic profiles for ZM4 and AcR growing either in the presence of NaAc (146 mM or 12 g/L NaAc, pH 5.0) or NaCl (146 mM or 8.6 g/L NaCl, pH 5.0) (Fig. 1 A and B, respectively). The quality of the expression data was assessed using JMP Genomics and validated by real-time quantitative (qRT-PCR) analysis (Fig. S1). High levels of concordance (R2 = 0.82–0.98) were observed between microarray and qRT-PCR data for 12 genes assayed under five conditions indicating that the microarray results were representative of the transcriptomic profiles (Fig. S1).
Fig. 1.
Z. mobilis ZM4 and AcR fermentations for transcriptomics studies. (A) Growth, glucose consumption, and ethanol production of ZM4 and AcR in RM with 146 mM (12 g/L) NaAc; and (B) with 146 mM (8.55 g/L) NaCl. Samples were collected for transcriptomic profiling at time points indicated by black arrows from pH 5.0, controlled anaerobic fermentations. The averages of two replicate fermentors for each strain and standard deviations are presented.
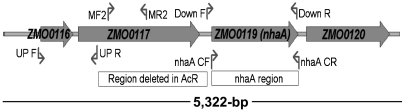
In contrast to the other genes assayed, microarray analysis revealed nhaA expression was significantly greater (> 16-fold) in strain AcR compared to ZM4 under all the conditions tested (Table 1, Fig. S2). The 1,461-bp region of AcR DNA deleted included a 1,363-bp region of ZMO0117, which left a 275-bp 5′ fragment (Fig. 2). The 160-bp region upstream of ZM4 nhaA was also truncated and left only 98-bp of nhaA upstream region unchanged in AcR (Fig. 2). Our expression data showed that a consistently lower AcR ZMO0117 signal was detected in each condition compared to that of ZM4 (Table 1, Fig. S2). These findings lead us to hypothesize that the AcR deletion increases nhaA expression and conferred NaAc tolerance in the strain.
Table 1.
Summary of expression data for nhaA and adjacent genes
| NaAc condition | NaCl condition | |||
| Gene* | Exp† | StPh† | Exp† | StPh† |
| ZMO0117 | −2.1 (8.8)‡ | −2.2 (7.2) | −2.2 (5.9) | −2.0 (5.0) |
| ZMO0119 | 4.6 (34.4) | 4.3 (31.7) | 4.4 (27.8) | 4.4 (27.9) |
| ZMO0120 | 1.7 (20.2) | 1.0 (20.7) | 1.8 (17.8) | 1.0 (9.5) |
*ZMO0117 encodes hydroxylamine reductase, ZMO0119 encodes Na+/H+ antiporter NhaA, and ZMO0120 encodes dihydroorotate dehydrogenase. ZMO0118 was deleted and ZMO0119 (encoding Na+/H+ antiporter NhaA) extended in a recent genome reannotation11.
†Expression ratio values (log 2) for AcR against ZM4 (AcR/ZM4) in either exponential (Exp) or stationary (StPh) phase comparisons.
‡ANOVA significance values are represented as a - log 10 (p-value) and shown in brackets.
Fig. 2.
Primer positions and regions of DNA deleted, disrupted, and used in expression and mutant construction studies. ZMO0116, ZMO0117, ZMO0119, and ZMO0120 indicate Z. mobilis ZM4 genes. The open box labeled “Region deleted in AcR” is present in ZM4, but deleted in the AcR mutant. The open box labeled “nhaA region” represents the region of DNA cloned for nhaA overexpression (p42-0119). nhaA_CF and nhaA_CR indicate positions of primers used to generate p42-0119. UP_F, UP_R, Down_F, and Down_R indicate the positions of primers used to construct of the deletion in ZM4 that mimics the AcR deletion. Primers MF2 and MR2 were used to amplify an internal region of ZMO0117 and create insertional mutant ZM4IM0117.
nhaA Genetic Studies.
To test the hypothesis that the deletion in AcR resulted in higher nhaA expression and conferred the reported NaAc tolerance (22), we recreated the 1,461-bp AcR deletion in strain ZM4 by marker exchange and designated it strain ZM4DM0117 (Table S4 and Materials and Methods). To test the influence of ZMO0117 on NaAc tolerance, we also constructed a ZMO0117 insertion mutant strain ZM4IM0117 (Table S4 and Materials and Methods). In addition, to test the correlation between nhaA overexpression and NaAc tolerance, we generated a plasmid for nhaA overexpression, designated p42-0119, and introduced it into wild-type ZM4 background through conjugation and selection (Table S4 and Materials and Methods). The ZMO0118 gene was combined with ZMO0119 in our recent update to the ZM4 genome (14) and the regions of DNA deleted, disrupted, and used in overexpression studies are presented (Fig. 2).
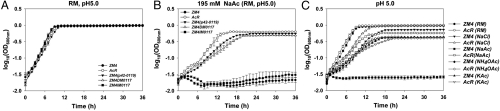
The newly-generated strains grew similarly under anaerobic conditions in rich medium (RM) broth not amended with NaAc, although a slightly lower growth rate was observed in wild-type ZM4 strains with p42-0119, indicative of increased metabolic burden on the strain (Table S5 and Fig. 3A). In the presence of 195 mM (or 16 g/L) NaAc at pH 5.0, ZM4 wild type and the ZM4IM0117 were unable to grow, while the positive control strain AcR grew well (Table S5 and Fig. 3B). The expression of nhaA in ZM4 via plasmid p42-0119 permitted strain ZM4 to grow under these selective conditions, reaching three-fourths of the AcR growth rate (Table S5 and Fig. 3B). The final cell density (OD600 nm) of ZM4 (p42-0119) was only 13% less than that of AcR (Table S5 and Fig. 3B). The facsimile of the 1,461-bp deletion in strain ZM4 (ZM4DM0117) also permitted growth in RM with 195 mM NaAc at pH 5 achieving more than half of the growth rate and three-fourths of the final cell density of AcR strain (Table S5 and Fig. 3B). The insertional mutant (ZM4IM0117) growth data indicated ZM0117 was not responsible for NaAc tolerance (Table S5 and Fig. 3). Despite the ZM4 NaAc tolerance being augmented substantively by either additional copies of nhaA provided via plasmid DNA or by recreating the deleted DNA region (ZM4DM0117), the AcR phenotype was not emulated completely (Table S5 and Fig. 3B). Possible reasons for this include that the region of DNA deleted in AcR may contain a repressor binding site(s) and its removal could relieve repression from the nhaA promoter, and/or that the elimination of a potential ZMO0117 transcriptional terminator may lead to a fusion transcript being expressed from the ZMO0117 promoter.
Fig. 3.
NhaA plays a role in Z. mobilis sodium acetate and sodium chloride tolerance. The growth differences of different strains were monitored by Bioscreen C (Growth Curves USA) under anaerobic conditions in RM, pH 5.0 (A); RM with 195 mM NaAc, pH 5.0 (B); 195 mM NaCl, NaAc, NH4OAc, and KAc at pH 5.0 (C). Strains included in this study were: ZM4: Z. mobilis ZM4 wild-type; AcR: previously described ZM4 acetate tolerant mutant; ZM4 (p42-0119): ZM4 containing p42-0119 for nhaA (ZMO0119) expression; ZM4IM0117: ZM4 ZMO0117 insertional mutant; ZM4DM0117: deletion mutant affecting ZMO0117 and part of upstream nhaA (Fig. 2; Table S4). This experiment has been repeated at least three times with similar results. Duplicates were used for each condition.
To investigate the role of nhaA on different forms of acetate, we grew ZM4 and AcR strains in the presence of the same molar concentrations (195 mM) of sodium chloride (NaCl), sodium acetate (NaAc), potassium acetate (KAc), or ammonium acetate (NH4OAc). Both the sodium and acetate ions had a toxic effect on Z. mobilis growth, with decreases in both growth rate and final cell density (Table S5 and Fig. 3C). The acetate ion was more toxic than the sodium ion: Z. mobilis grew more rapidly in the presence of 195 mM NaCl and the final cell density was higher compared to growth in the presence of same molar concentration of NH4OAc or KAc (Table S5). NaAc was more inhibitory than the same molar concentration (195 mM) of KAc or NH4OAc for ZM4 and the combination of elevated Na+ and Ac- ions appeared to exert a synergistic inhibitory effect for strain ZM4 with the growth of Z. mobilis totally inhibited (Table S5 and Fig. 3C).
The AcR strain was selected for sodium acetate tolerance, but also had enhanced tolerance to NaCl, but not NH4OAc or KAc compared to the wild-type ZM4 (Table S5 and Fig. 3C). Strain ZM4DM0117 and ZM4 harboring the nhaA-expressing plasmid p42-0119 similarly had enhanced NaCl tolerance that did not extend to NH4OAc or KAc (Table S5 and Fig. 3C). The increased tolerance to NaAc for these strains may therefore be due mostly to an increased sodium ion tolerance arising from the overexpression of Na+/H+ antiporter gene nhaA. The strains were also tested for tolerance to other classes of pretreatment inhibitors such as vanillin, furfural, or HMF and advantages were not observed (Fig. S3). These data indicate that NhaA mostly confers enhanced specific Na+ tolerance and not Ac- tolerance unless both ions are present in sufficient quantities (Fig. 3C).
Z. mobilis NhaA Structural Studies.
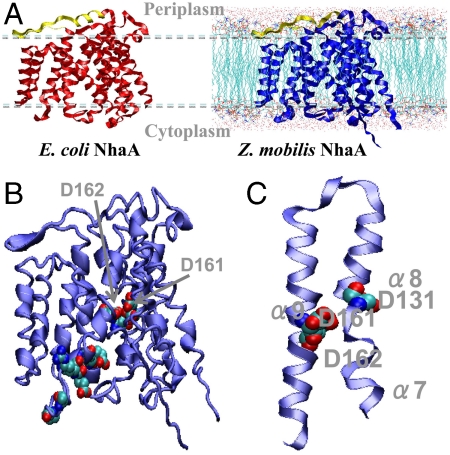
To gain further insights into the Na+/H+ antiporter tolerance mechanism, the Z. mobilis NhaA molecular structure was predicted. In the present case, it was possible to construct an atomic-detail structural model of the Z. mobilis NhaA protein, by using homology with the existing crystal structure of the E. coli NhaA (PDB id: 1ZCD, (26)). The amino acid sequence of Z. mobilis NhaA exhibits a 95% similarity score when compared to the E. coli NhaA, as obtained with the 3Dcoffee (27 and 28) multiple-sequence sequence-structure alignment tool. Hence, a Z. mobilis NhaA homology model could be constructed that is highly likely to be accurate. The model (Fig. 4A) was generated using the Swiss-model program (29) with the E. coli structure as the template (26). The Tmbase program (30) was applied to predict the orientations of the transmembrane helices. All modeling results indicate that the Z. mobilis Na+/H+ antiporter is structurally very similar to that of E. coli. The main region of low homology (and thus the region least likely to be structurally similar) was found to be located away from the ion transport region and in the periplasmic loop, which consists of a coil region and an antiparallel β-sheet and comprises residues Glu36 to Asn56 in the model of Z. mobilis NhaA. As in the E. coli NhaA crystal structure, the Z. mobilis NhaA structural model possesses a clear pH transduction pathway (Fig. 4B). The ion binding site of ZM4 NhaA consists of Asp161 and Asp162. Most of the charged or polar residues are also conserved in the “pH sensor” region of the funnel entrance (31). Similar to E. coli NhaA, ZM4 NhaA seems to have a convertible transmembrane segment (TMS IV, including helices α7 and α8, Fig. 4) and the structure of this depends upon the protonation state of residue Asp131. When Asp131 is protonated (at acidic pH), the helix α7 is closely parallel to α8 (Fig. 4C), while when Asp131 is deprotonated (at higher pH), α7 is crossed with α8 (32). Overall, the template fold is preserved in the homology model.
Fig. 4.
Z. mobilis NhaA structural predictions. (A) E. coli NhaA X-ray structure (left, red, (26)) and Z. mobilis NhaA model (right, blue). The periplasmic loops that are parallel to the membrane (light blue, dashed lines) are shown in yellow. The helix (α1, Ser35 to Asp41) of the loop in E. coli NhaA is modeled as a coil (Glu36 to Lys40) in Z. mobilis NhaA; (B) the active site and “pH sensor” in Z. mobilis NhaA. The protein is shown as a ribbon and the active site and pH sensor residues are shown as spheres; and (C) the protonation state of Asp131 may interconvert the topology of the transmembrane segment it is located in (transmembrane segment IV, including helices α7 and α8) between two different states (33).
Cytoplasm acidification by uncoupling and anion accumulation is one of the inhibitory effects of acetic acid on Z. mobilis (20). Weak acids such as acetic acid inhibit S. cerevisiae by ATP depletion, toxic anion accumulation, and inhibition of aromatic amino acid uptake, as reviewed previously (4). The E. coli NhaA antiporter exports a sodium ion from the cytoplasm while importing two protons (32) and if Z. mobilis were to function in a similar manner to the E. coli NhaA protein, the expulsion of sodium ions would lead to further cytoplasm acidification. Methanosarcina barkeri has a Na+/H+ antiporter that operates in the opposite direction to the E. coli NhaA, i.e., it imports a sodium ion into the cytoplasm while exporting two protons (33). Although the structural results (Fig. 4) are consistent with the Z. mobilis NhaA protein operating like the E. coli NhaA rather than the M. barkeri system, this would lead to two protons accumulating in the cytoplasm for each sodium ion that was exported. Conceivably Z. mobilis may then have another mechanism to control for cytoplasmic proton accumulation that we did not detect in the current study.
NaAc Tolerance Mechanism Testing in Other Microorganisms.
E. coli possesses three Na+/H+ antiporter genes, with NhaA being the archetype and most well studied Na+/H+ antiporter (32). E. coli provided with Z. mobilis NhaA (p42-0119) did not have enhanced tolerance to NaAc (Fig. S4), likely due to E. coli functional redundancy and regulation. Wu et al. (34) transferred the E. coli nhaA gene into rice (Oryza sativa L. ssp. japonica) and detected high expression in the transgenic rice, resulting in enhanced salt and drought tolerance (34). The Schizosaccharomyces pombe Na+/H+ plasma membrane transporter gene SOD2 has been shown to be involved in salt tolerance (35) and expression of SOD2 in a S. cerevisiae strain lacking any Na+-ATPase activity restores the ability of these cells to export sodium and greatly increases their resistance to both Na+ and Li+ in the medium (36). NHA1, a homolog of SOD2, was cloned from S. cerevisiae and found to contribute to Na+ extrusion (37).
The discovery that the ZM4 nhaA gene confers Z. mobilis NaAc tolerance and the existence of NhaA homologues in S. cerevisiae led us to investigate whether this target gene has a similar role in other industrial strains such as S. cerevisiae. S. cerevisiae has three Na+/H+ antiporters, a cytoplasmic Na+/H+ antiporter NHA1 (YLR138W), a vacuolar Na+/H+ exchanger VNX1 (YNL321W), and an endosomal Na+/H+ exchanger NHX1 (YDR456W). We assayed three S. cerevisiae Na+/H+ antiporter gene deletion mutants and a NHA1 over-expression strain (Table S4) in the presence of NaAc, NH4OAc, or KAc and found that, especially at early stages, each Na+/H+ antiporter mutant was more sensitive to acetate than the wild-type control strain (Fig. S5), and that yeast tolerance could be further enhanced by expression of plasmid encoded yeast Na+/H+ antiporter genes (Fig. S5). These yeast data are consistent with earlier reports that these yeast systems can function as monovalent cation/H+ antiporters (38 and 39), and Na+/H+ antiporter genes in S. cerevisiae are involved in acetate tolerance (Fig. S5).
Resequencing Limitations.
Thirty-eight putative SNPs and a 1,461-bp deletion region were identified by CGS in strain AcR (Table S1). Sanger sequencing analysis validated that 76% of the putative AcR SNP position changes had been identified correctly. However, both the correct position and nucleotide base call were confirmed by Sanger sequence analysis for only 61% of the AcR SNPs identified by CGS (Table S2). Although a number of SNPs and the deletion were predicted correctly, these data (Table S2) were also consistent with an earlier report that CGS has difficulties in identifying nucleotide differences of more than one bp or insertions/deletions(40).
Here, next-generation resequencing more accurately and rapidly assessed genomic differences between ZM4 and AcR strains than CGS, which is conduced in two phases (Table S1, Table S2, Table S3). CGS readily identified the 1,461-bp region of DNA that was deleted in AcR, as did the pyrosequencing approach. The addition of paired-end pyrosequencing data to shotgun libraries generated 12 and 11 contigs during mapping analysis for ZM4 and AcR, respectively. The 1,461-bp deletion region was at the boundary of one of the pyrosequencing gaps. The combination of pyroresequencing and CGS allowed a rapid and independent assessment of SNPs and gaps against the ZM4 reference genome.
Although pyrosequencing analysis of Z. mobilis ZM4 and AcR is a powerful approach for strain characterization we also identified systematic issues with certain differences reported as HCDiffs by the GSMapper software. The frequency of “mapped reads” and “other reads” varied widely among the 200 and 219 HCDiffs for AcR and ZM4 compared to the ZM4 reference genome sequence (Table S3). A previous study established a criterion by which mapping alignments having < 80% identity to the reference genome were ignored (41). In the present study, Sanger sequencing data were used to investigate reported AcR and ZM4 HCDiffs that had a range of different identity values (Table S3). We found a false positive rate of 100% among HCDiffs reported by the GSMapper software that had frequency variation (or identity) values of < 50% by Sanger sequencing analysis (Table S3). When these differences were included in the analysis an overall false positive rate of > 15% was found. In contrast, better results were obtained if we used our previously reported more stringent criteria (14), with each pyrosequencing difference tested being validated by analysis of Sanger data. When we examined pyrosequencing SNPs with variation values between 50% and 80% by Sanger data analysis we found one out of nine was reported correctly for AcR and the seven ZM4 pyroresequencing SNPs in this category were incorrect (Table S3).
We also identified one false negative in our pyrosequencing data during our Sanger sequence analysis, and noted occasions where the GSMapper software represented identical differences in AcR and ZM4 in different formats (Table S3). We expect developments in chemistry and bioinformatics will improve the accuracy and reporting of SNP detection in the future, but suggest our insights related to more stringent reporting of pyrosequencing differences (14) will be immediately useful for other microbial resequencing projects. In addition, the large number of SNPs shared in ZM4 and AcR compared to the reference genome showed the value in resequencing both strains in this study and for providing adequate controls when designing strategies for strain selection and resequencing in general.
Conclusions
The present study shows the potential of systems biology tools and genetics for the rapid identification and characterization of process-relevant traits. The expression profiles generated in these studies are the most comprehensive for Z. mobilis to date and will likely serve as useful reference data for future systems biology studies. By demonstrating that Z. mobilis nhaA overexpression confers the AcR tolerance phenotype and that most of the advantage conferred is against the sodium ion, our data reinforce the idea that one obtains what one selects for during adaptive evolution experiments. The present work also demonstrates S. cerevisiae Na+/H+ antiporter gene overexpression enhances its tolerance to acetate with three different counter ions. Our study also provides a caveat in using reference genome sequences for SNP identification and insights into technological limitations. We have affirmed the notion that near-term pathway engineering approaches benefit from a combinatorial approach (2). The combined approach of employing the advantages of classical selection, which lack mechanistic a priori assumptions, with systems biology tools is a paradigm for industrial strain characterization and development.
Materials and Methods
Strains and Culture Conditions.
Bacterial strains and plasmids used in this study are listed in Table S4. E. coli strains were cultured using LB medium ZM4 was obtained from the American Type Culture Collection (ATCC 31821). AcR is the ZM4 acetate tolerant strain described previously (22). ZM4 and AcR were cultured in RM at 30 °C. S. cerevisiae wild-type, deletion mutant YKO strains and GST-fusion ORF overexpression strains were obtained through Open Biosystems. S. cerevisiae strains were cultured in minimum complete medium purchased from Teknova Inc. Growth was monitored turbidometrically by measuring optical density at the wavelength of 600 nanometer (nm) intermittently with a Bioscreen C (Growth Curves USA). Each experiment has been repeated at least three times. Replicates were used for each condition.
DNA Manipulations, CGS, and 454 Genomic Pyroresequencing.
Genomic DNA from Z. mobilis was isolated using a Wizard Genomic DNA purification kit (Promega). The QIAprep Spin Miniprep and HiSpeed Plasmid Midi kits (Qiagen) were used for plasmid isolation. PCR, restriction enzyme digestion, ligation, cloning, and DNA manipulations followed standard molecular biology approaches (42). Genomic DNA of ZM4 and AcR was sent to NimbleGen facility for CGS service. Pyrosequencing using the Roche 454 GS FLX System (454 Life Sciences) was carried out using both shotgun and paired-end DNA library preparation methods. The GSMapper application in the 454 GS FLX software package 1.1.03 (454 Life Sciences) was used to map the reads generated from GS FLX onto the ZM4 reference genome (GenBank accession: AE008692) and the GSAssembler application was used to assemble the sequence reads into contigs, which were then compared with ZM4 genome sequence (13).
HPLC.
HPLC analysis was used for the measurements of the concentration of glucose, acetate, and ethanol in 0.2 μm-filtered samples taken at different time points (Fig. 1) and analyzed as described previously (43).
Microarray Analysis.
Transcriptomic profiles of exponential and stationary phase cells were analyzed for the ZM4 and AcR in the presence of 146 mM (or 12 g/L) NaAc or 146 mM (or 8.55 g/L) NaCl and samples were harvested at different time points (Fig. 1). Duplicate batch fermentations were conducted in approximately 2.5-L RM medium in 7.5-L BioFlo110 bioreactors (New Brunswick Scientific) and RNA was isolated as described previously (43). ds-cDNA was generated using an Invitrogen ds-cDNA synthesis kit (Invitrogen) and sent to NimbleGen for labeling, hybridization, and scanning following company's protocols. Statistical analysis was done with JMP Genomics 4.0 software (SAS Institute) as described previously (43).
Construction of ZMO0119 Expression Plasmid p42-0119 and Z. mobilis Mutant.
The construction of the entry vector and expression clone of target gene ZMO0119 was carried out as described previously (44). ZMO0119 was amplified by PCR using ZM4 genomic DNA as the template and primers nhaA_CF and nhaA_CR (Table S4). The plasmid p42-0119 sequence was confirmed to be correct by sequencing analysis.
The ZMO0117 and ZMO0119 genes, as well as the primer positions used in mutant construction and construction of the nhaA overexpression vector are shown (Fig. 2). To create the ZMO0117 insertion mutant a 529-bp internal region of the gene was amplified by PCR, purified, and cloned into pCR2.1-TOPO™ (Invitrogen). The insert DNA sequence was confirmed and then ligated into pKnock-Km plasmid. The presence and integrity of pKm-0117 in E. coli WM3064 was verified by sequence analysis. pJK100 was used to recreate the 1,461-bp AcR deletion in ZM4 as described previously (45). Primer pairs UP_F/UP_R and Down_F/Down_R were used to amplify 805 and 1,050-bp regions of DNA that flanked either up or downstream regions of DNA from the region of interest as shown (Fig. 2 and Table S4). The deletion plasmid construct was named pJK_hcp_nhaA. It was verified by PCR and sequencing and transformed in E. coli WM3064. Ex-conjugants were selected by plating on RM agar plates containing 20 μg/mL tetracycline for p42-0119 plasmid ex-conjugants or 200 μg/mL kanamycin for both pKm-0117 and pJK_hcp_nhaA plasmid ex-conjugants at 30 °C. PCR and sequencing were used to confirm the presence of the correct plasmid or mutant constructs.
Supplementary Material
Acknowledgments.
The authors thank Peter Rogers for generously providing strain AcR; M.K. Kerley, M. Rodriguez Jr., and L. Dice for technical assistance with Sanger sequencing, HPLC, and qRT-PCR, respectively. We also thank T. Phelps, B. Davison, and X. Cheng (ORNL) and J. Wiegel (University of Georgia, Athens) for insightful discussions and gratefully acknowledge D. Graham (ORNL) for critical review during manuscript preparation. This work is sponsored by the Laboratory Directed Research and Development Program of ORNL. The BioEnergy Science Center is a Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental in the Department Of Energy Office of Science. This manuscript has been authored by UT-Battelle, LLC, under Contract No. DE-AC05-00OR22725 with the Department of Energy.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this article have been deposited in the National Center for Biotechnology Information (NCBI) short-read archive database (Study SRP001379) GEO (accession number GSE18106).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914506107/-/DCSupplemental.
References
- 1.Himmel ME, et al. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 2.Alper H, Stephanopoulos G. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat Rev Microbiol. 2009;7:715–723. doi: 10.1038/nrmicro2186. [DOI] [PubMed] [Google Scholar]
- 3.Mills T, Sandoval N, Gill R. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnology for Biofuels. 2009;2:26. doi: 10.1186/1754-6834-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almeida JRM, et al. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biot. 2007;82:340–349. [Google Scholar]
- 5.Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science. 2007;315:801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- 6.Park JH, Lee SY, Kim TY, Kim HU. Application of systems biology for bioprocess development. Trends Biotechnol. 2008;26:404–412. doi: 10.1016/j.tibtech.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Medini D, et al. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6:419–430. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 8.Gresham D, Dunham MJ, Botstein D. Comparing whole genomes using DNA microarrays. Nat Rev Genet. 2008;9:291–302. doi: 10.1038/nrg2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn-Hagerdal B, Galbe M, Gorwa-Grauslund MF, Liden G, Zacchi G. Bio-ethanol—the fuel of tomorrow from the residues of today. Trends Biotechnol. 2006;24:549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Dien BS, Cotta MA, Jeffries TW. Bacteria engineered for fuel ethanol production: current status. Appl Microbiol Biotechnol. 2003;63:258–266. doi: 10.1007/s00253-003-1444-y. [DOI] [PubMed] [Google Scholar]
- 11.Panesar PS, Marwaha SS, Kennedy JF. Zymomonas mobilis: an alternative ethanol producer. J Chem Technol Biot. 2006;81:623–635. [Google Scholar]
- 12.Rogers PL, Jeon YJ, Lee KJ, Lawford HG. Biofuels. Vol 108. Berlin: Springer-Verlag Berlin; 2007. Zymomonas mobilis for fuel ethanol and higher value products; pp. 263–288. (Advances in Biochemical Engineering / Biotechnology). [DOI] [PubMed] [Google Scholar]
- 13.Seo JS, et al. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat Biotechnol. 2005;23:63–68. doi: 10.1038/nbt1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S, et al. Improved genome annotation for Zymomonas mobilis. Nat Biotechnol. 2009;27:893–894. doi: 10.1038/nbt1009-893. [DOI] [PubMed] [Google Scholar]
- 15.Deanda K, Zhang M, Eddy C, Picataggio S. Development of an arabinose-fermenting Zymomonas mobilis strain by metabolic pathway engineering. Appl Environ Microbiol. 1996;62:4465–4470. doi: 10.1128/aem.62.12.4465-4470.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Eddy C, Deanda K, Finkestein M, Picataggio S. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science. 1995;267:240–243. doi: 10.1126/science.267.5195.240. [DOI] [PubMed] [Google Scholar]
- 17.Lawford HG, Rousseau JD. Improving fermentation performance of recombinant Zymomonas in acetic acid-containing media. Appl Biochem Biotechnol. 1998;70:161–172. doi: 10.1007/BF02920133. [DOI] [PubMed] [Google Scholar]
- 18.Lawford HG, Rousseau JD, Tolan JS. Comparative ethanol productivities of different Zymomonas recombinants fermenting oat hull hydrolysate. Appl Biochem Biotechnol. 2001;91:133–146. doi: 10.1385/abab:91-93:1-9:133. [DOI] [PubMed] [Google Scholar]
- 19.Ranatunga TD, Jervis J, Helm RF, McMillan JD, Hatzis C. Identification of inhibitory components toxic toward Zymomonas mobilis CP4(pZB5) xylose fermentation. Appl Biochem Biotechnol. 1997;67:185–198. [Google Scholar]
- 20.Lawford HG, Rousseau JD. The effect of acetic acid on fuel ethanol-production by Zymomonas. Appl Biochem Biotechnol. 1993;39:687–699. [Google Scholar]
- 21.McMillan JD. Enzymatic conversion of biomass for fuels production. Vol 566. Washington, DC: Ed Himmel; 1994. Conversion of hemicellulose hydrolyzates to ethanol; pp. 411–437. (ACS Symposium Series). [Google Scholar]
- 22.Joachimstahl E, Haggett KD, Jang JH, Rogers PL. A mutant of Zymomonas mobilis ZM4 capable of ethanol production from glucose in the presence of high acetate concentrations. Biotechnol Lett. 1998;20:137–142. [Google Scholar]
- 23.Kim IS, Barrow KD, Rogers PL. Nuclear magnetic resonance studies of acetic acid inhibition of rec Zymomonas mobilis ZM4(pZB5) Appl Biochem Biotechnol. 2000;84-6:357–370. doi: 10.1385/abab:84-86:1-9:357. [DOI] [PubMed] [Google Scholar]
- 24.Jeon YJ, Svenson CJ, Joachimsthal EL, Rogers PL. Kinetic analysis of ethanol production by an acetate-resistant strain of recombinant Zymomonas mobilis. Biotechnol Lett. 2002;24:819–824. [Google Scholar]
- 25.Bailey JE. Toward a science of metabolic engineering. Science. 1991;252:1668–1675. doi: 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- 26.Hunte C, et al. Structure of a Na + /H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 27.O’Sullivan O, Suhre K, Abergel C, Higgins DG, Notredame C. 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J Mol Biol. 2004;340:385–395. doi: 10.1016/j.jmb.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 28.Armougom F, et al. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann K, Stoffel W. TM base—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166–170. [Google Scholar]
- 31.Padan E, Kozachkov L, Herz K, Rimon A. NhaA crystal structure: functional-structural insights. J Exp Biol. 2009;212:1593–1603. doi: 10.1242/jeb.026708. [DOI] [PubMed] [Google Scholar]
- 32.Arkin IT, et al. Mechanism of Na+/H+ antiporting. Science. 2007;317:799–803. doi: 10.1126/science.1142824. [DOI] [PubMed] [Google Scholar]
- 33.Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 34.Wu LQ, et al. Over-expression of the bacterial nhaA gene in rice enhances salt and drought tolerance. Plant Sci. 2005;168:297–302. [Google Scholar]
- 35.Jia ZP, Mccullough N, Martel R, Hemmingsen S, Young PG. Gene amplification at a locus encoding a putative Na+/H+ antiporter confers sodium and lithium tolerance in fission yeast. EMBO J. 1992;11:1631–1640. doi: 10.1002/j.1460-2075.1992.tb05209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahnenberger KM, Jia ZP, Young PG. Functional expression of the Schizosaccharomyces pombe Na+/H+ antiporter gene, sod2, in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5031–5036. doi: 10.1073/pnas.93.10.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prior C, Potier S, Souciet JL, Sychrova H. Characterization of the NHA1 gene encoding a Na+/H+-antiporter of the yeastSaccharomyces cerevisiae. FEBS Lett. 1996;387:89–93. doi: 10.1016/0014-5793(96)00470-x. [DOI] [PubMed] [Google Scholar]
- 38.Banuelos MA, Sychrova H, Bleykasten-Grosshans C, Souciet JL, Potier S. The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology. 1998;144:2749–2758. doi: 10.1099/00221287-144-10-2749. [DOI] [PubMed] [Google Scholar]
- 39.Cagnac O, Leterrier M, Yeager M, Blumwald E. Identification and characterization of Vnx1p, a novel type of vacuolar monovalent cation/H+ antiporter of Saccharomyces cerevisiae. J Biol Chem. 2007;282:24284–24293. doi: 10.1074/jbc.M703116200. [DOI] [PubMed] [Google Scholar]
- 40.Herring CD, Palsson BO. An evaluation of comparative genome sequencing (CGS) by comparing two previously-sequenced bacterial genomes. BMC Genomics. 2007;8:274. doi: 10.1186/1471-2164-8-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brockman W, et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch EF, Manatis T, editors. Molecular cloning, a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Yang S, et al. Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentation. BMC Genomics. 2009;10:34. doi: 10.1186/1471-2164-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelletier DA, et al. A general system for studying protein-protein interactions in gram-negative bacteria. J Proteome Res. 2008;7:3319–3328. doi: 10.1021/pr8001832. [DOI] [PubMed] [Google Scholar]
- 45.Denef VJ, et al. Genetic and genomic insights into the role of benzoate catabolic pathway redundancy in Burkholderia xenovorans LB400. Appl Environ Microbiol. 2006;72:585–595. doi: 10.1128/AEM.72.1.585-595.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.