Abstract
A histone H3 variant, H3T, is highly expressed in the testis, suggesting that it may play an important role in the chromatin reorganization required for meiosis and/or spermatogenesis. In the present study, we found that the nucleosome containing human H3T is significantly unstable both in vitro and in vivo, as compared to the conventional nucleosome containing H3.1. The crystal structure of the H3T nucleosome revealed structural differences in the H3T regions on both ends of the central α2 helix, as compared to those of H3.1. The H3T-specific residues (Met71 and Val111) are the source of the structural differences observed between H3T and H3.1. A mutational analysis revealed that these residues are responsible for the reduced stability of the H3T-containing nucleosome. These physical and structural properties of the H3T-containing nucleosome may provide the basis of chromatin reorganization during spermatogenesis.
During spermatogenesis, dramatic chromatin reorganization occurs, and most histones are eventually replaced by protamines (1). Several histone variants are highly expressed in the testis and are considered to be incorporated into the chromatin in the early stage of spermatogenesis.
In humans, about 4% of the haploid genome in the sperm is reportedly retained in nucleosomes, some containing the testis-specific histone H2B, hTSH2B/TH2B (2). Interestingly, the nucleosomes retained in the sperm are significantly enriched in loci that contain developmentally important genes. In addition, histone modifications, such as acetylation and methylation, are likely to occur after the incorporation of the histone variants during spermatogenesis (1). These observations suggest that nucleosomes containing testis-specific histone variants, with or without chemical modifications, may function as epigenetic markers in the sperm chromatin.
H3T is a variant of histone H3 that is robustly expressed in the human testis (3–5). We previously reported that H3T, like the conventional H3.1, can be assembled into nucleosomes with H2A, H2B, and H4 (H3T nucleosome) (6). A histone chaperone, Nap2, with 3-fold higher expression in the testis than in other somatic tissues (7), was found to be a more efficient chaperone for H3T nucleosome assembly than the ubiquitously expressed histone chaperone, Nap1 (6). Therefore, H3T may be assembled into the chromatin by a specific chaperone-mediated pathway in the testis. Comprehensive proteome analyses of nuclear extracts from HeLa cells suggested that H3T also exists in somatic cells (8, 9). However, the nucleosomes containing H3T probably comprise only a small proportion of the bulk chromatin in somatic cells, because the amount of H3T in HeLa cells is extremely low. Therefore, H3T may have a limited function in somatic cells that is currently unknown.
In the present study, we found that the H3T nucleosome is significantly unstable, as compared to the conventional H3.1 nucleosome, both in vitro and in vivo. The crystal structure of the H3T nucleosome was determined at 2.7 Å resolution, revealing that, although the overall structure was similar to that of the conventional H3.1 nucleosome, structural differences were observed at both ends of the central α2 helix of H3T and H3.1. The unique physical and structural characteristics of the H3T nucleosome were attributed to the Val111 and Met71 residues that are specific to H3T.
Results
H3T Nucleosome Is Less Stable than the Conventional Nucleosome.
The nucleosome containing human H3T was reconstituted by a salt-dialysis method, using human histones H3T, H2A, H2B, and H4, and a 146 base-pair DNA. To prepare a structurally homogeneous nucleosome, the reconstituted H3T nucleosome was incubated for 2 h at 55 °C to disrupt the inappropriate histone–DNA interactions (Fig. 1A) and was purified from the free DNA by gel electrophoresis (Fig. 1B). Conventional H3.1 nucleosomes were also prepared with the same procedure (Fig. 1B). Histone compositions of the H3T and H3.1 nucleosomes prepared in this procedure were confirmed by SDS-PAGE (Fig. 1C). We next compared the stabilities of the H3T and H3.1 nucleosomes by examining the gel migration distances of the nucleosomes exposed to different NaCl concentrations. Exposure to 0.4 M NaCl had no apparent effect on the migration distances of both the H3.1 and H3T nucleosomes (Fig. 1D, lanes 1 and 5, respectively). This observation indicates that the nucleosomes were intact at this salt concentration. The H3.1 nucleosome appeared stable even when exposed to 0.8 M NaCl (Fig. 1D, lane 4), and only a small fraction of the nucleosome migrated slower. These nucleosomes are probably multimers formed by enforced hydrophobic interactions from higher salt concentrations. By contrast, the band corresponding to the intact nucleosome was nearly absent for the H3T nucleosome that was exposed to 0.6 M NaCl (Fig. 1D, lane 6). Instead, multiple, nonnucleosomal bands, containing only H2A/H2B (Fig. S1), were detected (Fig. 1D, lanes 6–8). These results indicate that the H3T nucleosome is less stable than the H3.1 nucleosome under high salt concentrations.
Fig. 1.
Instability of the H3T nucleosome. (A) H3T nucleosomes, reconstituted using 1.2 mg/mL total histones and 0.7 mg/mL DNA, were analyzed by nondenaturing 6% PAGE. Lane 1 indicates naked DNA. Lanes 2 and 3 indicate the H3T nucleosomes before and after a 55 °C incubation, respectively. DNA was visualized by ethidium bromide staining. Asterisks indicate bands corresponding to nonnucleosomal DNA–histone complexes. (B) The H3T and H3.1 nucleosomes were purified using a Prepcell apparatus, and were analyzed by nondenaturing 6% PAGE with ethidium bromide staining. (C) Histone compositions of the purified H3T and H3.1 nucleosomes were analyzed by 18% SDS-PAGE with Coomassie brilliant blue staining. (D) Salt titration. The nucleosomes were incubated in the presence of 0.4 M (lanes 1 and 5), 0.6 M (lanes 2 and 6), 0.7 M (lanes 3 and 7), and 0.8 M NaCl (lanes 4 and 8) at 42 °C for 2 h. The samples were analyzed by nondenaturing 6% PAGE with ethidium bromide staining. Lanes 1–4 and 5–8 indicate experiments with H3.1 and H3T nucleosomes, respectively. Bands corresponding to nucleosome monomers and nucleosome-nucleosome aggregates are indicated. Asterisks represent bands corresponding to nonnucleosomal DNA-histone complexes.
Weaker Association of H3T/H4 Tetramer to H2A/H2B Dimer.
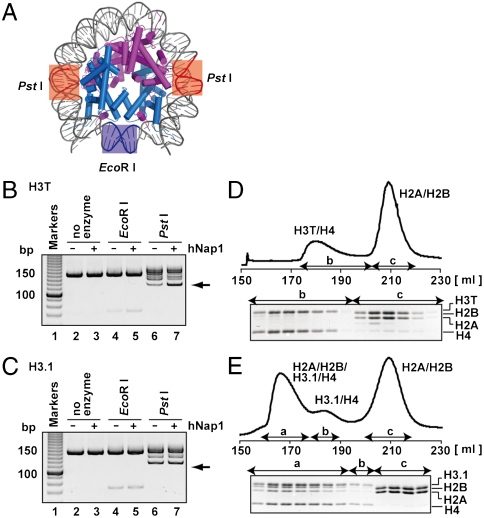
To investigate the stability of the H3T nucleosome at physiological ionic strengths, we examined the Nap1-mediated H2A/H2B disassembly from the H3T nucleosome. It is known that excess amount of the histone chaperone, human Nap1 (hNap1), promotes H2A/H2B disassembly from the nucleosome (10). The nucleosomal DNA used in this assay contains two PstI sites that are palindromically located in regions 13–18 bases away from both ends of the 146 base-pair DNA (Fig. 2A). These PstI sites are close to the binding sites of H2A/H2B within the nucleosome (Fig. 2A). The DNA also contains a single EcoRI site at the nucleosomal dyad, which is a binding site for H3/H4 (Fig. 2A). Therefore, if hNap1 disassembles H2A/H2B, the DNA is predicted to become more susceptible to digestion by PstI.
Fig. 2.
H2A/H2B associates weakly with H3T/H4. (A) The crystal structure of the H3.1 nucleosome determined in this study. Locations the PstI and EcoRI sites are indicated. The H2A/H2B and H3.1/H4 molecules are colored in purple and in dark blue, respectively. (B and C) H2A/H2B disassembly assay with hNap1. The nucleosomes were treated with PstI or EcoRI in the presence or absence of excess amount of hNap1 (6.5 μM). The resulting DNA fragments were extracted by Phenol/chloroform, and were analyzed by 10% PAGE with ethidium bromide staining. Arrows indicate the DNA fragment produced by complete PstI digestion. These results were confirmed to be reproduced in three independent experiments. (B) The H3T nucleosome. (C) The H3.1 nucleosome. (D and E) Interaction between H2A/H2B and H3T/H4 or H3.1/H4. H2A, H2B, H4, and H3T (D) or H3.1 (E) were incubated without DNA in the presence of 2 M NaCl. The samples were then subjected to HiLoad 26/60 Superdex 200 prep grade gel filtration column chromatography. Histone compositions of the peak fractions were analyzed by 18% SDS-PAGE with Coomassie brilliant blue staining. The peak fractions denoted as a, b, and c correspond to H2A/H2B/H3/H4 octamer, H3/H4 tetramer, and H2A/H2B dimer, respectively.
In the absence of hNap1, PstI partially digested the nucleosomal DNA of both H3T and H3.1 nucleosomes (Fig. 2 B and C, lane 6), whereas EcoRI only slightly digested the nucleosomal DNA (Fig. 2 B and C, lane 4). Therefore, the DNA segment located near the exit of the nucleosome is more susceptible to restriction nucleases than that located near the dyad. Interestingly, hNap1 substantially increased the PstI susceptibility of the DNA in the H3T nucleosome (Fig. 2B, compare lanes 6 and 7), probably by its H2A/H2B disassembly function. In contrast, this hNap1-dependent enhancement of the PstI susceptibility was not observed in the H3.1 nucleosome under the experimental conditions used in this study (Fig. 2C, compare lanes 6 and 7). These results suggested that H2A/H2B in the H3T nucleosome is less stably incorporated, as compared with those in the conventional H3.1 nucleosome. The instability of the H3T nucleosome may be primarily caused by a weaker association of the H2A/H2B dimer to the H3T/H4 tetramer. It should be noted that the DNA in the H3.1 nucleosome was more susceptible than that in the H3T nucleosome in the absence of hNap1, and that the PstI digestion patterns of the H3.1 and H3T nucleosomes were slightly different (Fig. 2 B and C). Differences in the DNA flexibility of the H3.1 and H3T nucleosomes were observed (Fig. S2B), which may be the reason for the distinct digestion patterns.
We then tested the interaction between H2A/H2B and H3T/H4 or H3.1/H4 in the absence of DNA. Gel filtration analysis in the presence of 2 M NaCl revealed that the H2A/H2B dimers did not tightly associate with the H3T/H4 tetramer (Fig. 2D). This result sharply contrasts with the fact that two H2A/H2B dimers stably associates with the conventional H3.1/H4 tetramer and forms a stable H2A/H2B/H3.1/H4 octamer (Fig. 2E) (11). Therefore, the weaker association between H2A/H2B and H3T/H4 may be responsible for the instability of the H3T nucleosome. A possible explanation for this difference could be the fact that the self-association of the H3T/H4 tetramer prevents the interaction between the H3T/H4 tetramer and the H2A/H2B dimer. A dynamic light scattering analysis revealed that the Stokes radius of the H3T/H4 tetramer in the presence of 2 M NaCl was approximately twice the size of that of the H3.1/H4 tetramer, which was judged to be monodisperse (Table S1), suggesting that the H3T/H4 tetramer aggregation competes with the H3T/H4–H2A/H2B interaction.
Rapid Exchange of H3T in Nucleosomes of Living Cells.
We next compared the mobility of H3T with H3.1, as GFP-fusion proteins in living cells, by fluorescence recovery after photobleaching (FRAP) (12). Because GFP-H3.1 was stably incorporated into nucleosomes in living cells, only a subtle recovery was observed, even at 20 min after photobleaching (Fig. 3 A and B). By contrast, the fluorescence of GFP-H3T recovered within several minutes (Fig. 3 A and B), suggesting that H3T in the nucleosome is more rapidly exchanged compared to the conventional H3.1 in living cells. GFP-H3T was detected in the mono- and oligo-nucleosomal fractions prepared by micrococcal nuclease digestion followed by sucrose gradient centrifugation (Fig. 3C), indicating that GFP-H3T was actually incorporated into chromatin. These in vivo results are consistent with the instability of the H3T nucleosome.
Fig. 3.
FRAP. HeLa cells expressing GFP-H3.1 or GFP-H3T were subjected to the FRAP analysis. (A) The mobility of GFP-H3.1 or GFP-H3T in living cells was analyzed by bleaching one-half of the nucleus. (B) The averages of the relative fluorescence intensity of bleached area were plotted with the standard deviations (n = 5). (C) GFP-H3T was incorporated into the HeLa cell chromatin. (Upper ) DNA fragments of mono-, di-, and trinucleosomes fractionated by sucrose gradient centrifugation were analyzed by agarose gel electrophoresis with ethidium bromide staining. (Right and Left) The nucleosome samples from the HeLa cells with and without GFP-H3T expression, respectively. The sucrose gradient fraction numbers are indicated at the top of each panel. Middle panel. Histone compositions of the purified nucleosomes were analyzed by 16% SDS-PAGE with Coomassie brilliant blue staining staining. (Lower) GFP-H3T was detected with anti-GFP monoclonal antibody.
Crystal Structure of the H3T Nucleosome.
To understand the structural basis of the instability observed in the H3T nucleosome, the crystal structure of the nucleosome core particle containing H3T was solved at 2.7 Å resolution (Table S2). The overall structure was essentially similar to that of the H3.1 nucleosome (13) (Fig. 4A). There are four amino acid differences between H3T (Val24, Met71, Ser98, and Val111) and the conventional H3.1 (Ala24, Val71, Ala98, and Ala111), and three of them (Met71, Ser98, and Val111) are located in the visible, histone-fold domain (Fig. 4B). The Val24 residue, which is located in the N-terminal tail outside of the histone-fold domain, was not visible in the present H3T nucleosome structure (Fig. 4 A and B). H3.1 and its variants contain a central helix (α2) with two shorter α-helices (α1 and α3) flanking both ends of α2. The Met71 (α1) and Val111 (α2) residues of H3T are located near the L1 and L2 loops, respectively, which connect the flanking helices (α1 and α3) to α2 (Fig. 4B). The Ser98 residue is located near the middle of the α2 helix (Fig. 4B).
Fig. 4.
Crystal structure of the H3T nucleosome. (A) Two views of the H3T-nucleosome structure are represented. The H3T molecules are shown in red. Locations of the Met71 and Val111 residues are indicated. (B) Structural differences between H3T and H3.1 in the nucleosomes. The H3T and H3.1 structures are superimposed, and the rmsd values for each residue pair is calculated and plotted. The secondary structure of H3T in the nucleosome is shown in the top of the panel. Arrows indicate the locations of the H3T-specific amino acid residues, Met71, Ser98, and Val111. (C and D) Comparison of the H3T structure (red) with the H3.1 structure (green). The side chains of the H3T-M71, H3.1-V71, H3T-V89, H3.1-V89, H3T-V111, H3.1-A111, H3T-R116, H3.1-R116, H3T-D123, and H3.1-D123 residues are represented by space-filling models. The H3T and H3.1 regions containing the amino acid residues 71 (C) and 111 (D) are shown. Arrows in C and D indicate the locations of H3T and H3.1 that are structurally different from each other.
To identify the finer structural differences between the H3T and H3.1 nucleosomes, we next compared the structures of the H3T and H3.1 histones. Although several H3 structures are available for comparison, differences in crystallization conditions and crystal packing may obscure the fine structural differences. To minimize these possibilities, the H3.1 nucleosome was prepared and crystallized under identical procedures as those of the H3T nucleosome, and its structure was determined at a similar resolution (2.5 Å) (Fig. S2 and Table S2). All residues in the crystal structures, including the H3T-specific residues, were well structured, as judged from their B factors (Table S3). The H3T and H3.1 histones were then superimposed, and the rmsd for each residue pair was calculated and plotted against each other. The largest deviations were found near both ends of the central α2 helix (Fig. 4B). At the N terminus of the H3T α2 helix, the van der Waals radii of the Met71 residue side chain come in close contact with that of the Val89 residue (Fig. 4C, Left). This close contact is absent in the corresponding location of the H3.1 structure (Fig. 4C, Right). At the other end of the α2 helix, the side chain of the Val111 residue contacts the side chain of the Asp123 residue, which forms a salt bridge with Arg116 (Fig. 4D, Left). This van der Waals interaction between 111 and 123 residues is not observed in the H3.1 structure (Fig. 4D, Right). This difference between H3T and H3.1 appears important for the structural differences observed near the C terminus of the α2 helix (Fig. 4 B and D). All other locations of H3T, including the region around the H3T-specific Ser98 residue, were essentially identical in the main chain structure to those of H3.1 (Fig. 4B). Therefore, the Met71 and Val111 residues in H3T appear to be important for the formation of the specific structure of the H3T nucleosome and may contribute to the reduced stability of the H3T nucleosome.
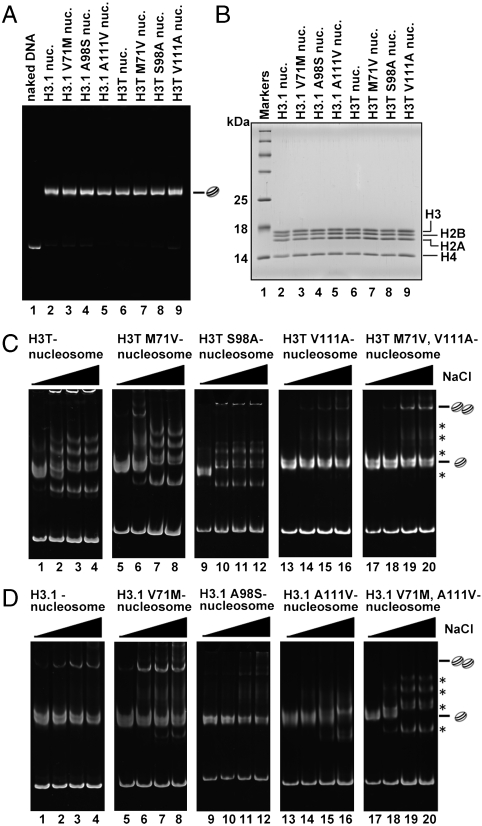
Contribution of the V71M and A111V Substitutions in the H3T Nucleosome Stability.
In the crystal structure of the H3T nucleosome, we found that Met71 and Val111 are the key residues affecting the H3T-specific conformation (Fig. 4). This structural property of the H3T nucleosome may be responsible for its reduced stability. Therefore, we then sought to determine whether the nucleosome stability is affected by substituting H3T-specific amino acids for the H3.1 types, by creating the H3T-M71V, -S98A, and -V111A mutants. All of the mutants were competent in nucleosome formation (Fig. 5A), and the histone compositions of these nucleosomes were confirmed (Fig. 5B). The nucleosome stability, as judged by the resistance to high NaCl concentrations, was enhanced drastically by the H3T-V111A mutation (Fig. 5C, lanes 13–16) and moderately by the H3T-M71V mutation (Fig. 5C, lanes 5–8). By contrast, the nucleosome stability was not affected by the S98A mutation (Fig. 5C, lanes 9–12). An H3T-M71V/V111A double mutant exhibited essentially similar stability to the H3.1 nucleosome (Fig. 5C, lanes 17–20). Consistent results were obtained by reciprocal experiments using H3.1 mutants, in which Val71, Ala98, and Ala111 were replaced by the corresponding H3T amino acids. The nucleosome stability was decreased drastically and moderately by H3.1-A111V and H3.1-V71M mutations, respectively (Fig. 5D, lanes 13–16 and lanes 5–8). The H3.1-V71M/A111V double mutant, like the H3T nucleosome, was very unstable (Fig. 5D, lanes 17–20). As expected, the H3.1-A98S mutation did not affect the stability of the H3.1 nucleosome (Fig. 5D, lanes 9–12). Consistently, the H2A/H2B disassembly assay revealed that the H3T-V111A and H3.1-A111V mutations suppressed and enhanced, respectively, the H2A/H2B disassembly by hNap1 (Fig. S3). These results indicate that the reduced stability of the H3T nucleosome is attributable mainly to the V111A substitution and partly to the M71V substitution.
Fig. 5.
Mutational analysis of H3T and H3.1 nucleosomes. (A) Nucleosomes containing H3T and H3.1 mutants were purified using a Prepcell apparatus and were analyzed by nondenaturing 6% PAGE with ethidium bromide staining. (B) Histone compositions of the purified nucleosomes containing H3T and H3.1 mutants were analyzed by 18% SDS-PAGE with Coomassie brilliant blue staining. (C and D) Salt titration. Nucleosomes were incubated in the presence of 0.4 M (lanes 1, 5, 9, 13, and 17), 0.6 M (lanes 2, 6, 10, 14, and 18), 0.7 M (lanes 3, 7, 11, 15, and 19), and 0.8 M NaCl (lanes 4, 8, 12, 16, and 20) at 42 °C for 2 h. The samples were analyzed by nondenaturing 6% PAGE with ethidium bromide staining. Bands corresponding to nucleosome monomers and nucleosome–nucleosome aggregates are indicated. Asterisks represent bands corresponding to nonnucleosomal DNA-histone complexes. (C) The stability of H3T mutants. Lanes: 1–4, H3T; 5–8, H3T-M71V; 9–12, H3T-S98A; 13–16, H3T-V111A; and 17–20, H3T-M71V/V111A nucleosomes. (D) The stability of H3.1 mutants. Lanes: 1–4, H3.1; 5–8, H3.1-V71M; 9–12, H3.1-A98S; 13–16, H3.1-A111V; and 17–20, H3.1-V71M/A111V.
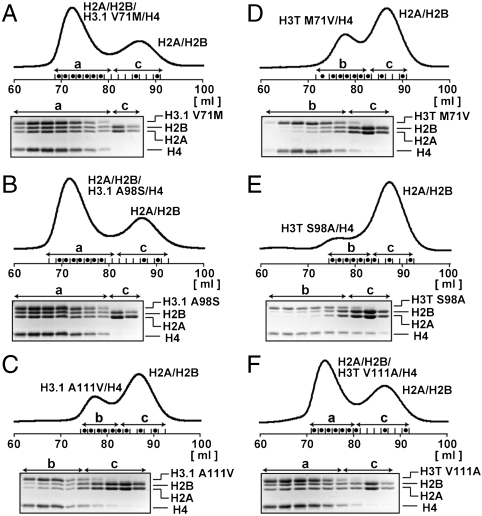
To confirm whether the Val111 residue is responsible for the weaker H2A/H2B association to H3T/H4, we performed gel filtration analyses with these H3 mutants. As shown in Fig. 6C, H3.1/H4 containing the H3.1-A111V mutant was significantly defective in the H2A/H2B binding in the absence of DNA, like H3T/H4 (Fig. 2D). The H3.1-V71M and H3.1-A98S mutations did not significantly affect the H2A/H2B binding of H3.1/H4 (Fig. 6 A and B). Reciprocal results were obtained with H3/H4 containing H3T-M71V, H3T-S98A, and H3T-V111A (Fig. 6 D–F). These biochemical results are consistent with the differences observed in the crystal structure, and therefore, the V111A substitution in H3T may be mainly responsible for the specific structure and function of the H3T nucleosome.
Fig. 6.
Mutational analyses of interactions of H2A/H2B with H3T/H4 or H3.1/H4. Gel filtration analyses were performed as described in Fig. 2 D and E, except that a HiLoad 16/60 Superdex 200 prep grade column was used. Fractions indicated with dots are analyzed by 18% SDS-PAGE with Coomassie brilliant blue staining. The peak fractions denoted as a, b, and c correspond to H2A/H2B/H3/H4 octamer, H3/H4 tetramer, and H2A/H2B dimer, respectively. (A) H3.1-V71M. (B) H3.1-A98S. (C) H3.1-A111V. (D) H3T-M71V. (E) H3T-S98A. (F) H3T-V111A.
Discussion
In the present study, we found that a prominent property of the H3T nucleosome is its instability. The crystal structure of the H3T nucleosome revealed that the H3T-specific amino acid residues, Met71 and Val111, corresponding to Val71 and Ala111 in H3.1, are responsible for the structural differences observed between H3T and H3.1. Consistently, mutational analyses revealed that the reduced stability of the H3T nucleosome is caused mainly by Val111 and partly by Met71. Therefore, we conclude that the Met71 and Val111 residues of H3T are essential for the H3T-specific structure and function.
Our structural and biochemical analyses revealed that the single amino acid substitution of H3, at position 111, significantly affected the nucleosome stability, probably by weakening the association of the H3T/H4 tetramer with H2A/H2B dimers. Whereas the Val111 of H3T does not directly interact with H2A/H2B and DNA, a mutation at Arg116 (to His) close to Val111 reportedly destabilized the nucleosome (14, 15). This H3-R116H mutation in Saccharomyces cerevisiae has been identified as a Sin mutation that alleviates the requirement for the nucleosome-remodeling factor, Swi/Snf, which activates transcription (14, 16). A structural study revealed that the H3-Arg116 residue may not directly interact with the DNA backbone and H2A/H2B, but instead forms a salt bridge with H3-Asp123 (17). This salt bridge may be very important to arrange the residues appropriately around position 116 for hydrogen bond formation with the DNA backbone and H2A/H2B (18). Therefore, this Sin mutation at Arg116 of H3 may destabilize the nucleosome by allosterically reducing the histone-DNA and/or histone-histone interactions. In the present study, we found that the Val111 residue of H3T sterically affected the side chain orientation of the Arg116 and Asp123 residues within the H3T monomer. The H3T-Val111 residue may destabilize nucleosomes by a similar mechanism to that proposed for the H3-R116H mutation. Consistent with this idea, a histone H3-A111G mutant exhibited the Swi/Snf-independent phenotype in S. cerevisiae (19).
In the present study, we showed that the H3T nucleosome structurally and biochemically differs from the conventional H3.1 nucleosome. The histone H3T variant is highly expressed in the testis (5). Therefore, H3T is anticipated to have a specific function in chromatin reorganization during meiosis and/or the postmeiotic maturation of male germ cells. In germ cells, drastic chromatin reorganization occurs by histone replacement with histone variants, and the histones are eventually replaced by protamines during sperminogenesis (1, 20). The reduced stability of the H3T nucleosome may be favorable to promote this global transition of the chromosome architecture during meiotic and/or postmeiotic events. Intriguingly, a mouse testis-specific H2A variant, H2AL2, reportedly formed a specific nucleosome that was distinct from the conventional nucleosome. The H2AL2 nucleosome was quite susceptible to nucleases, suggesting that these nucleosomes have different structural properties, as compared to those of the conventional nucleosome (21). A possible human counterpart of H2AL2, H2A.Bbd, which is highly expressed in the testis, was also suggested to form a specific nucleosome structure with reduced stability (22–24). Furthermore, nucleosomes containing a testis-specific H2B variant, hTSH2B/TH2B, were reportedly unstable, as compared to the conventional nucleosome (25). Therefore, the instability of these nucleosomes formed by the testis-specific histone variants may be a common property required for chromatin reorganization during spermatogenesis.
H3T may also be a constituent of sperm chromatin, in which about 4% of the human sperm genome retains nucleosomes (2). These sperm nucleosomes are suggested to have a specific epigenetic function, because they are significantly enriched around developmentally important genes, such as the imprinted gene clusters, microRNA clusters, and HOX gene clusters. Testis-specific H2B variants, hTSH2B/TH2B and H2BFWT, were found in the sperm nuclei (2, 26). H2BFWT was stably incorporated into nucleosomes in vitro and in vivo (27) and was suggested to have telomere-associated function (26). Whereas it remains to be revealed whether sperm chromatin indeed retains H3T nucleosomes, the robust expression of H3T in the testis suggests its presence in the sperm. The specific biochemical and structural properties of the H3T nucleosome, like those containing the H2B variants, could also play an important epigenetic role in the regulation of genes containing nucleosomes in the sperm.
Materials and Methods
Purification of Human Histones.
Human H2A, H2B, H3.1, H3T, and H4 were overexpressed in Escherichia coli cells, and were purified by a method according to previous papers (6, 28) with modifications. Details are described in SI Materials and Methods.
Preparation of the H3T and H3.1 Nucleosomes.
Details are described in SI Materials and Methods. The 146 base-pair DNA (13, 17) was prepared as described previously (11). Purified H2A/H2B (1.7 mg), H3T/H4 (1.7 mg) or H3.1/H4 (1.7 mg), and the DNA (2 mg) were mixed in a solution containing 2 M KCl. The H3T and H3.1 nucleosomes were reconstituted by the salt-dialysis method and were purified from the free DNA and histones by nondenaturing polyacrylamide gel electrophoresis, using a Prepcell apparatus (Bio-Rad).
Salt Resistance Assay.
The nucleosomes (240 ng/μL) were incubated in the presence of 0.4, 0.6, 0.7, and 0.8 M NaCl at 42 °C for 2 h. After incubation, NaCl concentration of the samples were adjusted to 0.4 M, and the samples were analyzed by nondenaturing 6% PAGE.
Crystallization and Structure Determination.
Details are described in SI Materials and Methods. Briefly, crystals of the purified H3T and H3.1 nucleosomes were obtained by the hanging drop method. The H3T-nucleosome crystals belonged to the orthorhombic space group P212121, with unit cell constants of a = 105.5 Å, b = 109.5 Å, and c = 181.1 Å, and contained one nucleosome per asymmetric unit. The H3.1-nucleosome crystals also belonged to the same space group, with unit cell constants of a = 105.8 Å, b = 109.5 Å, and c = 180.9 Å, and contained one nucleosome per asymmetric unit. High-resolution diffraction data were obtained using the synchrotron radiation source at the beamline BL41XU station of SPring-8, Harima, Japan.
The structures of the H3T and H3.1 nucleosomes were initially solved to 2.7 and 2.5 Å resolutions, respectively, by the molecular replacement method, using the MOLREP program (29) and the human nucleosome structure (Protein Data Bank ID code 2CV5) as a guide (13). For H3T nucleosome, the Ramachandran plot of the final structure showed 93.3% of the residues in the most favorable regions and no residues in the disallowed region. For H3.1 nucleosome, the Ramachandran plot of the final structure showed 94.9% of the residues in the most favorable regions and no residues in the disallowed region. Summary of the data collection and refinement statistics is provided in Tables S2 and S3. All structure figures were created using the PyMOL program (30). The atomic coordinates of the H3T nucleosome and the H3.1 nucleosome have been deposited, with the RCSB ID codes, 3A6N and 3AFA, respectively.
hNap1-Mediated H2A/H2B Disassembly Assay.
The nucleosomes (0.8 μM) were incubated with hNap1 (6.5 μM), and were treated with PstI (30 unit) or EcoRI (36 unit) in 10 μL of 50 mM Tris·HCl buffer (pH 7.5), containing 10 mM MgCl2 and 100 mM NaCl. After a 120 min incubation at 37 °C, the DNA was extracted with phenol/chloroform. The DNA fragments were then analyzed by 10% PAGE in 0.5 × TBE buffer (45 mM Tris base, 45 mM Boric acid, and 1 mM EDTA) (21 V/cm for 1 h) and ethidium bromide staining.
Superdex 200 Gel Filtration Chromatography.
Freeze-dried H3T (2.1 mg), H3.1 (2.1 mg), or H3 mutants (2.1 mg) was mixed with H2A (1.9 mg), H2B (1.9 mg), and H4 (1.5 mg) in 5 mL of 20 mM Tris·HCl buffer (pH 7.5), containing 7 M guanidine hydrochloride, and 20 mM 2-mercaptoethanol. The samples were dialyzed against 10 mM Tris·HCl buffer (pH 7.5), containing 2 M NaCl, and 2 mM 2-mercaptoethanol, and were analyzed by Superdex 200 gel filtration chromatography (GE Healthcare). The peak fractions were analyzed by 18% SDS-PAGE.
Fluorescence Recovery After Photobleaching.
HeLa cells stably expressing GFP-H3.1 or GFP-H3T were grown on a glass-bottom dish (Mat-tek). FRAP was performed using a confocal microscope (FV-1000; Olympus) with a 60× UPlanSApo N.A. = 1.35 lens, as described previously (12). Three confocal images of a field containing 4–10 nuclei were collected (800 × 800 pixels, zoom 3, scan speed 2 μs/pixel, pinhole 800 μm, Kalman filtration for four scans, LP505 emission filter, and 0.1% transmission of 488-nm Ar laser). One-half of each nucleus was bleached using 75% transmission of 488 nm and 100% of 514 nm (two iterations), and images were collected using the original setting every 1 min. The fluorescence intensity of the bleached area was measured using Image J 1.39u. After subtracting the background, the intensity was normalized to the initial intensity before bleaching.
Analysis of the GFP-H3T Incorporation into Chromatin in HeLa Cells.
Details are described in SI Materials and Methods. Nucleosomes were prepared essentially according to previous paper (31) with slight modifications. Briefly, HeLa cells, in which GFP-H3T was exogenously expressed, were collected and resuspended in 1 mL ice-cold 10 mM Hepes-NaOH (pH 7.4), 15 mM NaCl, and 1.5 mM MgCl2 containing 1% Triton X100 and protease inhibitor cocktail (Nacalai Tesque). After disrupting the cells using Dounce homogenizer (tight pestle; five times), nuclei were collected and treated with micrococcal nuclease (2 × 106 Gel units/mL; New England Biolabs) at 30 °C for 1 h (mixing by inverting every 15 min). After adding 10 mM EDTA (pH 8.0) and centrifugation (10,000 × g; 10 min; 4 °C), the pellet was suspended in 540 μL 10 mM EDTA (pH 8.0). After the addition of 5 M NaCl solution (final 0.3 M), the sample was centrifuged (20,000 × g; 10 min; 4 °C), and the supernatant was collected. The supernatant was then incubated at 55 °C for 2 h to denature nonnucleosomal proteins. After the centrifugation (10,000 × g; 5 min; 4 °C) to remove denatured proteins, the supernatant (nucleosome sample) was collected. The nucleosome sample (0.6 mL) was fractionated in a 12 mL sucrose gradient (10–30%) by centrifugation (209,541 × g; 21 h; 4 °C) using a Beckman SW41Ti rotor. Fractions (0.6 mL each) were collected from the top.
For DNA analysis, fractions were mixed with SDS (0.2%) and analyzed by 2% agarose gel electrophoresis in 1 × TAE (40 mM Tris base, 20 mM acetic acid, and 1 mM EDTA) (13.3 V/cm for 1 h) followed by ethidium bromide staining. For protein analysis, fractions containing mono-, di-, and trinucleosomes were separated by 16% SDS-PAGE, and either stained with Coomassie brilliant blue or transferred to Hybond-P PVDF membrane (GE Healthcare) using a semidry blotting system (BIO CRAFT). The GFP-H3T signals were detected with anti-GFP antibody (Nacalai Tesque), peroxidase-conjugated anti-mouse Ig, and ECL Western Blotting Detection Reagents (GE Healthcare) using a LAS-4000 (Fujifilm).
Supplementary Material
Acknowledgments.
We thank Dr. K. Luger (Colorado State University) for providing the plasmid to prepare the 146 base-pair DNA used in this study. We also thank Drs. N. Fujikawa and S. Yokoyama (RIKEN) for technical assistance on nucleosome preparation, Dr. T. Fukagawa (National Institute of Genetics) for general discussion, and Dr. S.-Y. Park (Yokohama City University) for assistance on dynamic light scattering experiments. We are also grateful to the beamline scientists, Drs. Y. Kawano and N. Shimizu for their assistance in data collection at the BL41XU beamline of SPring-8. This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology, Japan. H. Kurumizaka is a research fellow in the Waseda Research Institute for Science and Engineering.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank, www.pdb.org (RCSB ID codes 3A6N and 3AFA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003064107/-/DCSupplemental.
References
- 1.Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem. 2004;271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 2.Hammoud SS, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franklin SG, Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977;266:273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- 4.Albig W, Ebentheuer J, Klobeck G, Kunz J, Doenecke D. A solitary human H3 histone gene on chromosome 1. Hum Genet. 1996;97:486–491. doi: 10.1007/BF02267072. [DOI] [PubMed] [Google Scholar]
- 5.Witt O, Albig W, Doenecke D. Testis-specific expression of a novel human H3 histone gene. Exp Cell Res. 1996;229:301–306. doi: 10.1006/excr.1996.0375. [DOI] [PubMed] [Google Scholar]
- 6.Tachiwana H, Osakabe A, Kimura H, Kurumizaka H. Nucleosome formation with the testis-specific histone H3 variant, H3t, by human nucleosome assembly proteins in vitro. Nucleic Acids Res. 2008;36:2208–2218. doi: 10.1093/nar/gkn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu RJ, Lee MP, Johnson LA, Feinberg AP. A novel human homologue of yeast nucleosome assembly protein, 65 kb centromeric to the p57KIP2 gene, is biallelically expressed in fetal and adult tissues. Hum Mol Genet. 1996;5:1743–1748. doi: 10.1093/hmg/5.11.1743. [DOI] [PubMed] [Google Scholar]
- 8.Andersen JS, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 9.Govin J, Caron C, Rousseaux S, Khochbin S. Testis-specific histone H3 expression in somatic cells. Trends Biochem Sci. 2005;30:357–359. doi: 10.1016/j.tibs.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Okuwaki M, Kato K, Nagata K. Functional chracterization of human nucleosome assembly protein 1-like proteins as histone chaperones. Genes Cells. 2010;15:13–27. doi: 10.1111/j.1365-2443.2009.01361.x. [DOI] [PubMed] [Google Scholar]
- 11.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 12.Kimura H, et al. A novel histone exchange factor, protein phosphatase 2Cgamma, mediates the exchange and dephosphorylation of H2A-H2B. J Cell Biol. 2006;175:389–400. doi: 10.1083/jcb.200608001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsunaka Y, Kajimura N, Tate S, Morikawa K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005;33:3424–3434. doi: 10.1093/nar/gki663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kruger W, et al. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 15.Kurumizaka H, Wolffe AP. Sin mutations of histone H3: Influence on nucleosome core structure and function. Mol Cell Biol. 1997;17:6953–6969. doi: 10.1128/mcb.17.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wechser MA, Kladde MP, Alfieri JA, Peterson CL. Effects of Sin- versions of histone H4 on yeast chromatin structure and function. EMBO J. 1997;16:2086–2095. doi: 10.1093/emboj/16.8.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 18.Muthurajan UM, et al. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Q, Yu C, Morse RH. Dispersed mutations in histone H3 that affect transcriptional repression and chromatin structure of the CHA1 promoter in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:1649–1660. doi: 10.1128/EC.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ooi SL, Henikoff S. Germline histone dynamics and epigenetics. Curr Opin Cell Biol. 2007;19:257–265. doi: 10.1016/j.ceb.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Syed SH, et al. The incorporation of the novel histone variant H2AL2 confers unusual structural and functional properties of the nucleosome. Nucleic Acids Res. 2009;37:4684–4695. doi: 10.1093/nar/gkp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao Y, et al. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 2004;23:3314–3324. doi: 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautier T, et al. Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep. 2004;5:715–720. doi: 10.1038/sj.embor.7400182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doyen CM, et al. Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome. EMBO J. 2006;25:4234–4244. doi: 10.1038/sj.emboj.7601310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li A, et al. Characterization of nucleosomes consisting of the human testis/sperm-specific histone H2B variant (hTSH2B) Biochemistry. 2005;44:2529–2535. doi: 10.1021/bi048061n. [DOI] [PubMed] [Google Scholar]
- 26.Churikov D, et al. Novel human testis-specific histone H2B encoded by the interrupted gene on the X chromosome. Genomics. 2004;84:745–756. doi: 10.1016/j.ygeno.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Boulard M, et al. The NH2 tail of the novel histone variant H2BFWT exhibits properties distinct from conventional H2B with respect to the assembly of mitotic chromosomes. Mol Cell Biol. 2006;26:1518–1526. doi: 10.1128/MCB.26.4.1518-1526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka Y, et al. Expression and purification of recombinant human histones. Methods. 2004;33:3–11. doi: 10.1016/j.ymeth.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project. Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific LLC; 2008. www.pymol.org. [Google Scholar]
- 31.Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.