Abstract
Loss-of-function Aspergillus nidulans CclA, a Bre2 ortholog involved in histone 3 lysine 4 methylation, activated the expression of cryptic secondary metabolite (SM) clusters in A. nidulans. One novel cluster generated monodictyphenone, emodin and emodin derivatives while a second encoded two anti-osteoporosis polyketides, F9775A and F9775B. Modification of the chromatin landscape in fungal SM clusters allows for a simple technological means to express silent fungal secondary metabolite gene clusters.
Aspergilli are ubiquitous filamentous fungi whose members include human and plant pathogens and industrial fungi with tremendous medical, agricultural and biotechnological importance. Although demonstrating synteny along large tracks of their sequenced genomes, members of this genus vary remarkably in their secondary metabolome, possibly a reflection of a chemical arsenal important in niche securement1, 2. The sheer numbers of unique secondary metabolite (SM) genes highlight the genus as a potentially rich source of bioactive metabolites for medicinal and pharmaceutical use. Gene wealth, however, has not translated well into compound production, in part due to an inability to find conditions promoting expression of SM gene clusters.
Some progress has been achieved in activating SM gene cluster expression using the model organism Aspergillus nidulans. Genome sequence analysis of A. nidulans reveals dozens of putative SM gene clusters including the well-studied penicillin (1) and sterigmatocystin (2) clusters3. Yet the expression of most SM clusters and their concomitant products remain veiled. Two approaches for activating otherwise silent clusters were recently described. One strategy, utilizing the knowledge that many SM clusters contain a pathway specific transcription factor, fused an inducible promoter to a cluster transcription factor leading to the production of hybrid polyketide-nonribosomal peptide metabolites, the cytotoxic aspyridones A (3) and B (4)4. A second approach, based on genomic mining of microarrays generated from mutants of the global regulator of secondary metabolism LaeA5, 6, 7, led to the identification of the anti-tumor compound terrequinone A (5)8. Efforts to uncover the regulatory role of LaeA revealed that some subtelomeric SM clusters were located in heterochromatic regions of the genome where suppression was relieved by deletion of a key histone deacetylase9. The importance of histone modifications in SM clusters was further reflected in the initiation and spread of histone H4 acetylation concurrent with transcriptional activation of the subtelomeric A. parasiticus aflatoxin (6) gene cluster10.
A consideration of the accruing evidence linking chromatin modifications with SM cluster regulation led us to examine the hypothesis that additional chromatin modifying proteins were important in SM cluster regulation. In particular, we examined a member of the COMPASS (complex associated with Set1) complex for possible regulatory roles in SM silencing. The COMPASS complex is a conserved eukaryotic transcriptional effector both facilitating and repressing chromatin-mediated processes through methylation of lysine 4 of histone 3 (H3K4)11, 12. While H3K4me2 and H3K4me3 are found predominantly on active loci 12, the COMPASS complex also regulates homothallic mating silencing, ribosomal DNA silencing, telomere length, and subtelomeric gene expression in yeast 13-15.
A critical member of the COMPASS complex is the SPRY domain protein designated Bre2 in Saccharomyces cerevisiae11. Analysis of the A. nidulans genome revealed a putative ortholog, here named CclA. Extracts of cclA deletants (cclAΔ, Supplementary Methods), deficient in H3K4 di- and trimethylation (Fig. 1 and Supplementary Fig. 1), presented an altered chemical landscape as depicted by thin layer chromatography (Supplementary Fig. 2). Previous work has shown the major SM produced by A. nidulans is the polyketide sterigmatocystin (2, ST). To reduce ST and ST precursor backgrounds, stcJ encoding a fatty acid synthase required for ST production16, was also deleted, generating a double stcJΔ, cclAΔ mutant. HPLC profiles of stcJΔ showed the production of two known metabolites of A. nidulans austinol (7) and dehydroaustinol (8) and the absence of ST (2). Analysis of the stcJΔ, cclAΔ double mutant yielded at least six additional aromatic compounds. Full one- and two- dimensional NMR analysis revealed the compounds as monodictyphenone (9), emodin (10) and four other emodin analogs (11 – 14) (Fig. 1a and Fig. 1b).
Figure 1.
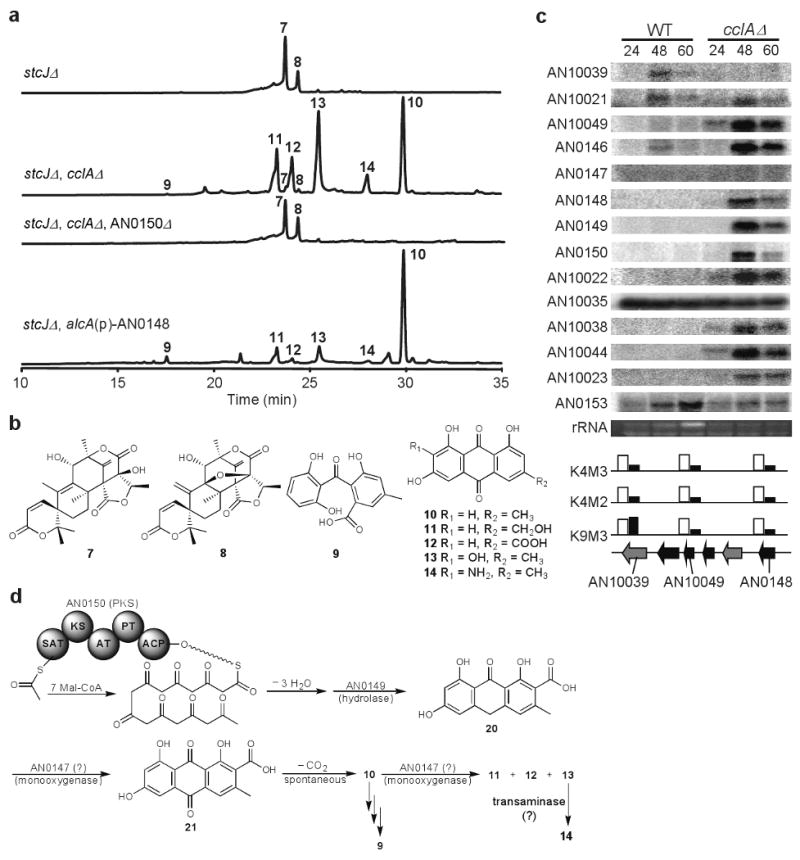
Identification of monodictyphenone and its gene cluster.
(a) HPLC profiles (UV 254 nm) of A. nidulans strains. Ethyl acetate extract of stcJΔ mutant, stcJΔ, cclAΔ double mutant, stcJΔ, cclAΔ, AN0150Δ triple mutant, and stcJΔ, alcA(p)-AN0148 mutant under inducing conditions.
(b) Chemical structures of compounds 7 – 14 isolated from A. nidulans.
(c) Top: Gene cluster expression in cclAΔ. mRNAs from liquid shake cultures grown for 24, 48 and 60 hr were examined for expression in wild type and cclAΔ strains. Bottom: Histone 3 lysine 4 trimethylation (K4M3), dimethylation (K4M2) and histone 3 lysine 9 trimethylation (K9M3) of 3 genes in wild type (white bars) and cclAΔ (black bars) at 48 hr correlating with mRNA in top. Raw data is presented in Supplementary Fig. 1.
(d) Proposed biosynthetic pathway of monodictyphenone (9) and emodin analogs (10 – 14).
Monodictyphenone (9) has been previously isolated from a marine fungus Monodictys putredinis17 as well as an engineered strain of A. nidulans18. This strain of A. nidulans expressed the Glarea lozoyensis polyketide synthase gene encoding for 6-methylsalicylic acid (15) and the authors could not determine whether the monodictyphenone (9) produced in addition to 6-methylsalicylic acid (15) was due to the heterologous gene or expression of an endogenous A. nidulans PKS218. Our data clearly shows that monodictyphenone (9) is a product of A. nidulans and not derived from the heterologously expressed gene. Monodictyphenone (9), a metabolite with antimicrobial properties, shares structural similarity to a known A. terreus metabolite sulochrin (16) which is derived from the anthraquinone emodin (10)19. Emodin (10), not known until now to be produced by A. nidulans, is an active anthraquinone constituent demonstrating anti-mutagenic, anti-cancer, vasorelaxant, immunosuppressive anti-inflammation and anti-apoptosis activities20.
Monodictyphenone (9) and emodin (10) and its derivatives (11 – 14), share a similar aromatic polyketide structure suggesting that a single non-reduced polyketide synthase (PKS) is involved in their biosynthesis. We selected ten of the twelve non-reduced polyketide synthases (NR-PKS) in A. nidulans for disruption. The two known NR-PKSs not targeted were the ST PKS and the wA PKS (required for production of a spore pigment). Metabolite analysis of the ten PKS mutant strains identified a single PKS (AN0150, 65.5% identical to the A. terreus emodin anthrone polyketide synthase gene) responsible for production of all compounds 9 – 14 (Fig. 1b).
AN0150 is located 0.5 Mb from the right telomere of 5 Mb chromosome VIII and is surrounded by several genes with high homologies to genes found in the ST (2) and aflatoxin (6) clusters (Supplementary Table 1 and Supplementary Fig. 3). One of these genes, AN0148, showed similarity to AflR, a Zn2Cys6 binuclear transcription factor required for expression of enzymatic genes in the ST (2) cluster21. Replacement of the promoter region of AN0148 with the alcA inducible promoter (Supplementary Table 2 and 3) allowed induction of compounds 9 – 14 (Supplementary Table 4 and Supplementary Fig. 4) and created an HPLC profile similar to the cclAΔ, stcJΔ double mutant (Fig. 1a).
We next determined if increased production of compounds 9 – 14 was reflected in gene expression in cclAΔ. Figure 1c shows up-regulated gene expression in cclAΔ from AN10021 through AN10023, two exception being AN0147 and AN10035 that were expressed equally well in the control strain. An examination of histone H3 methylation and acetylation levels in cclAΔ by chromatin immunoprecipitation of two cluster genes (AN0148 and AN10049) and one flanking gene (AN10039) indicated a strong reduction of H3K4me2 and H3K4me3 in all three genes confirming the role of the putative COMPASS complex member CclA in lysine 4 methylation of H3 (Fig. 1c and Supplementary Fig. 1). Interestingly, reduced levels of H3K4me2/3 also resulted in low levels of H3K9me2/3, a chromatin mark associated with gene silencing and heterochromatin formation, in the two genes belonging to the cluster, but not in the flanking, non-expressed gene in cclAΔ. Thus, strongly reduced levels of H3K4me2/3 as well as H3K9me2/3 at the 5′ end of cluster genes are required for de-repression during secondary metabolism. We hypothesize that this cluster of genes encodes enzymes or regulatory proteins required for monodictyphenone (9) and emodin (10) production and name them mdpA – mdpL. AN10039 and AN0153 may represent boundaries of this gene cluster allowing us to propose a likely pathway (Fig. 1d and Supplementary Fig. 3).
Finally we asked if CclA regulation extended to other clusters. Interestingly, two anti-osteoporosis yellow polyketides, F9775B (17) and F9775A (18), isolated from Paecilomyces carneus22 could also be detected from the cclAΔ strain grown on YAG solid medium after acidified extraction (Figs. 2a and 2b and Supplementary Table 5). Disruption of the NR-PKS AN7909 resulted in the loss of F9775B (17) and F9775A (18) (Fig. 2a). AN7909 is located in a cluster of genes 0.21 Mb from the left telomere of the 1.44 Mb chromosome II (Supplementary Table 1). A comparison of gene expression between cclAΔ and a wild type control in this region confirmed CclA regulation of the F9775 cluster (Fig. 2c). AN7909 and at least some of the contiguous genes (AN7910-AN7915) are predicted to be necessary for F9775 biosynthesis.
Figure 2.
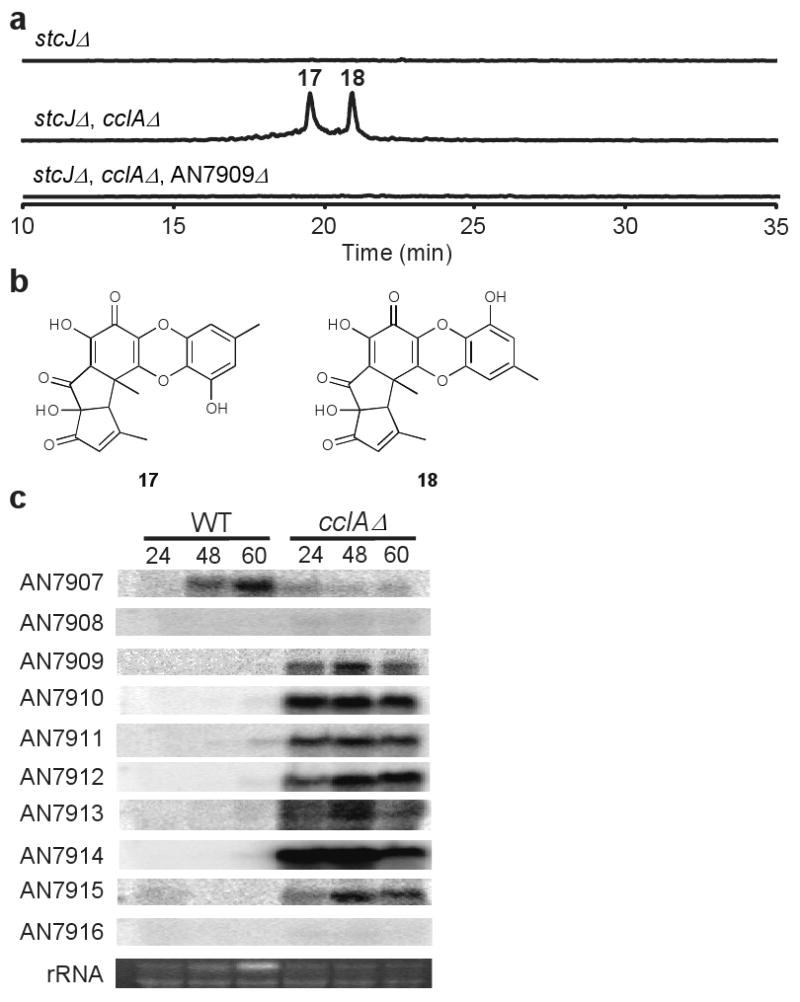
Identification of F9775 and its gene cluster.
(a) EIC profiles at m/z 395 (negative mode) of A. nidulans strains. Ethyl acetate extract after acidification of stcJΔ mutant, stcJΔ, cclAΔ double mutant, and stcJΔ, cclAΔ, AN7909Δ triple mutant analyzed by LCMS.
(b) Chemical structures of F9775B (17) and F9775A (18).
(c) Gene cluster expression in cclAΔ. mRNAs from liquid shake cultures grown for 24, 48 and 60 hr were examined for expression in wild type and cclAΔ strains. Genes AN7909 – AN7915 are predicted to be required for F9775 biosynthesis.
Sequencing of Aspergilli genomes1 and those of several other ascomycete genera23, 24 has exposed a wealth of secondary metabolite genes, conveniently arranged in clusters, thought to aid the fungus in competing successfully with other organisms in its natural habitat2. A literature survey of 1500 fungal metabolites isolated and characterized between 1993 and 2001, showed that more than half of these molecules had antibacterial, antifungal or antitumour activity25. A few of these metabolites have translated into highly profitable pharmaceuticals including antibiotics, cholesterol-lowering agents, tumor inhibitors and immunosuppressants for transplant operations.
A major impediment in identifying these metabolites is finding the conditions in which they are produced. SM clusters are frequently silent and efforts to elicit expression in wild type strains have often proved futile. Recent technological advances, such as over expressing pathway specific transcription factors4, does not work to activate all SM clusters – nor do all clusters (e.g. F9775) contain transcription factors. We present here a revolutionary approach to hunting and harvesting fungal metabolites by exploiting the concept of a chromatin landscape composed of silent natural product islands that can be conveniently manipulated to active chromatin states.
We hypothesized that silencing of SM clusters could be reversed by removal of genes important in the establishment of a repressive chromatin configuration. We tested this hypothesis by removing from the A. nidulans genome the ortholog of S. cerevisiae Bre2, which was shown to be required in yeast to silence a marker gene inserted near the telomere15. Loss-of-function CclA strains allowed for expression of at least two silent A. nidulans gene clusters, one yielding the active anthraquinone constituents monodictyphenone (9) and emodins (10 – 14), and another the anti-osteoporosis polyketides F9775A (18) and F9775B (17). The critical role for chromatin level regulation by histone methylation of fungal SM cluster genes raises the possibility that epigenetic mechanisms based on histone posttranslational modifications are a general mechanism to silence fungal SM gene clusters.
Supplementary Material
Acknowledgments
This research was funded in part by NSF grant MCB-0236393 to N.P.K., NIH grant GM084077 to N.P.K., B.R.O. and C.C.C.W and GM031837 to B.R.O. Work in Vienna was supported by grant P19731-B11 from the Austrian Science Fund (FWF) to J.S.
Footnotes
Author Contributions: J.W.B. contributed to design and execution of experiments and to writing. Y.-M.C., J.F.S., H.-C.L. were involved in metabolite analysis of Aspergillus strains by LC/MS and in the isolation and structural determination of monodictyphenone, emodins, and F9755A/B by 1D and 2D NMR. Y. R.-D. conducted the ChIP analysis. K.W. was involved in obtaining LC/MS data. E.S. and A.D.D. were involved in generating Aspergillus mutant strains. B.R.O., N.P.K., J.S. and C.C.C.W. contributed to design and writing.
Competing Interests Statement: There are no competing interests.
References
- 1.Nierman WC, et al. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 2.Rohlfs M, Albert M, Keller NP, Kempken F. Biol Lett. 2007;3:523–525. doi: 10.1098/rsbl.2007.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keller NP, Turner G, Bennett JW. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann S, et al. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 5.Bok JW, Keller NP. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayram O, et al. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 7.Perrin RM, et al. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bok JW, et al. Chem Biol. 2006;13:31–37. doi: 10.1016/j.chembiol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Shwab EK, et al. Eukaryot Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roze LV, Arthur AE, Hong SY, Chanda A, Linz JE. Mol Microbiol. 2007;66:713–726. doi: 10.1111/j.1365-2958.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- 11.Mueller JE, Canze M, Bryk M. Genetics. 2006;173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sims RJ, 3rd, Rienberg D. Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 13.Wood A, Schneider J, Shilatifard A. Biochem Cell Biol. 2005;83:460–467. doi: 10.1139/o05-116. [DOI] [PubMed] [Google Scholar]
- 14.Ottaviani A, Gilson E, Magdinier F. Biochimie. 2008;90:93–107. doi: 10.1016/j.biochi.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Schneider J, et al. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Brown DW, Adams TH, Keller NP. Proc Natl Acad Sci USA. 1996;93:14873–14877. doi: 10.1073/pnas.93.25.14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krick A, et al. J Nat Prod. 2007;70:353–360. doi: 10.1021/np060505o. [DOI] [PubMed] [Google Scholar]
- 18.Lu P, et al. Mol Genet Genomics. 2005;273:207–216. doi: 10.1007/s00438-005-1132-y. [DOI] [PubMed] [Google Scholar]
- 19.Couch RD, Gaucher GM. J Biotechnol. 2004;108:171–178. doi: 10.1016/j.jbiotec.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y, et al. Life Sci. 2007;81:1332–1338. doi: 10.1016/j.lfs.2007.08.040. [DOI] [PubMed] [Google Scholar]
- 21.Fernandes M, Keller NP, Adams TH. Mol Microbiol. 1998;28:1355–1365. doi: 10.1046/j.1365-2958.1998.00907.x. [DOI] [PubMed] [Google Scholar]
- 22.Sato S, Morishita T, Hosoya T, Ishikawa Y. In: Koho JKT, editor. Japan: 1999. 11. [Google Scholar]
- 23.Dean RA, et al. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 24.Cuomo CA, et al. Science. 2007;317:1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- 25.Pelaez F. Biological activities of fungal metabolites. In: An Z, editor. Handbook of Industrial Mycology. Marcel Dekker; 2005. pp. 49–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


