Abstract
E2F transcription factors regulate a variety of cellular processes but their role in angiogenesis is not clear. We find that many genes involved in angiogenesis like FLT-1, KDR and angiopoietin 2 have potential E2F1 binding sites in their promoter. ChIP assays showed that E2F1 can associate with these promoters and the recruitment of E2F1 was enhanced upon VEGF stimulation with concomitant dissociation of Rb leading to the transcriptional activation of these promoters. Transient transfection experiments showed that these promoters were induced by E2F1 and repressed by Rb while depletion of E2F1 decreased their expression. The increased binding of E2F1 to these promoters upon VEGF stimulation correlated with acetylation of histones and E2F1; this required VEGFR function, as seen in ChIP-re-ChIP experiments. This suggests the existence of a positive feedback loop regulating E2F1 acetylation and VEGFR expression. Acetylation associated with VEGF signaling appears to be predominantly mediated by PCAF and depletion of histone acetyl transferases disrupted the formation of angiogenic tubules. These results suggest a novel role for E2F1 and acetylation in the angiogenic process.
Keywords: FLT-1, KDR, cell cycle, endothelial cells, angiogenesis
Introduction
E2F family of transcription factors plays a major role in cell cycle control by regulating a group of genes involved in cell cycle progression and DNA replication. The transcriptional activity of E2Fs is regulated at many levels, but mainly through the association with the Rb family proteins (1–3). E2Fs 1–3 transactivate key cell cycle genes including cyclins, replication factors, and enzymes involved in nucleic acid synthesis (4, 5). E2F activity is interconnected through complexes with any of the nine E2Fs, two DP binding proteins (DP1 and DP2) and three pocket proteins (Rb, p130, p107) (5, 6). E2F4 and E2F5 are poor transcriptional activators and function as passive repressors by recruiting pocket proteins to the E2F regulated promoters (2, 3, 7). E2Fs 6 to 8 lack transactivation and pocket protein binding domains; they actively repress transcription independent of pocket proteins (6, 8–10).
Beyond the cell cycle, E2Fs have been implicated in the regulation of apoptosis, development, and differentiation (11, 12). Although the role of E2Fs and Rb in cell proliferation is well established, their involvement in the regulation of other processes that contribute to tumor growth like angiogenesis and invasion is not well characterized. Previous studies from our lab have shown that metallothionein 1G (MT1G) promoter is E2F responsive and VEGF induces this promoter by enhancing the binding of E2Fs (13). This suggested that E2Fs might be affecting the expression of genes involved in other aspects of tumor growth and progression, like angiogenesis.
To assess whether E2F contributes to VEGF mediated angiogenesis, we examined the promoters of VEGF receptors, FLT-1 and KDR, as well as Angiopoeitin 2, a regulator of angiogenesis, for the presence of E2F binding sites. Here we provide the evidence that the transcriptional activity of FLT-1, KDR and ANGPT2 are regulated by the E2F family of transcription factors. Depletion of E2F1 reduced the expression of these genes and prevented VEGF-induced angiogenic tubule formation in matrigel. Further, VEGF stimulation led to the association of E2F1 with these promoters, coinciding with a dissociation of Rb, leading to their transcriptional activation. Here we demonstrate that VEGF induces the recruitment of acetyl transferases like CBP, p300 and PCAF on FLT-1 and KDR promoters; there was also increased acetylation of the promoter region as well as E2F1, enhancing its recruitment to these promoters. These results suggest that the Rb-E2F pathway contributes to the expression of VEGF receptors facilitating angiogenesis and might promote the growth and progression of tumors in response to aberrant signaling events.
Materials and Methods
Cell lines and reagents
Human primary aortic endothelial cells (HAEC), Human umbilical vein endothelial cells (HUVEC) and Human microvascular endothelial cells from lungs (HMEC-L) were obtained from Clonetics, USA and cultured in EBM-2 supplemented with growth factors (EGM-2 bullet kit, Lonza). A549 cells were cultured in F12K medium supplemented with 10 % serum (CellGro, USA). For VEGF stimulation, HAECs, HUVECs and HMEC-Ls were rendered quiescent by growing in EBM2 without the supplements for 24 hours and stimulated by VEGF (100ng/ml) for 24 hours.
Transient transfections and Luciferase assays
A549 cells and HUVECs were transfected by calcium phosphate mediated transfection according to standard protocols (Sambrook and Russell, 2001). Cotransfection with 1μg of pRL construct containing Renilla reniformis luciferase gene was used as normalizing control. Total DNA per well was adjusted to an equal level by adding the empty vector PGL3 or salmon sperm DNA. Luciferase assays were done by using Dual Luciferase Assay System (Promega). Relative luciferase activity was defined as the mean value of the firefly luciferase/Renilla luciferase ratios obtained from three independent experiments.
ChIP assays
ChIP assays were carried out as described previously (14). HAEC, HUVECS and HMEC-L cells were serum starved for 24 hours and treated with VEGF for 24 hours and ChIP lysates were prepared. Immunoprecipitations were conducted using antibodies to E2F1 to 5, Rb, p300, CBP, PCAF (Santa Cruz Biotechnology) and anti-acetylated histone H3 monoclonal antibody (Upstate Biotechnology). Rabbit anti-mouse secondary antibody (Pierce) was used as the control. c-Fos promoter was used as a negative control to test the specificity of all ChIP assays.
ChIP-Re-ChIP or Reverse ChIP
ChIP-re-ChIP assay was carried out as described by (15). The primary immunoprecipitation was done using E2F1 or acetylated lysine antibody (Cell Signaling). The immunoprecipitated complexes were eluted with re-ChIP buffer (15). The eluate from E2F1 IP was re-immunoprecipitated with acetylated lysine antibody and the eluate from this was precipitated by E2F1 antibody. The presence of the promoter sequences in the resulting re-ChIP immunoprecipitates were examined as described above for one-step ChIP.
Immunoprecipitation-Western Blots
To examine the role of VEGF receptors in VEGF induced acetylation of E2F1, immunoprecipitation was carried out with lysates from starved, VEGF stimulated or VEGF stimulated cells in the presence of KDR inhibitor SU1498 (Calbiochem) or KDR and FLT-1 inhibitor 676481 (Calbiochem) using acetylated lysine antibody (Cell Signalling) and western blotted with E2F1 monoclonal antibody (Santa Cruz). IP-Western was performed as described before (16).
si RNA transfections and real time PCR
Cells were grown on 60 mm dishes and transfected either with control siRNA (100 pmol) or different concentrations of E2F1 siRNA (50, 75 and 100 pmol) using oligofectamine (Invitrogen, CA). The cells were collected 24 hours after transfection and total RNA was isolated using the RNeasy kit (Qiagen). Levels of FLT-1 and KDR mRNA were analyzed by quantitative reverse transcription PCR using the Biorad icycler. Data were normalized using 18S rRNA as internal control and the fold change in the expression levels was determined using the nontarget siRNA as the control. All siRNAs were purchased from Santa Cruz Biotechnology.
Matrigel assays
Matrigel (B.D.) was used to promote the differentiation of HAECs and HUVECs into capillary tube-like structures. A total of 100 μl of Matrigel was added to 96 well plates, followed by incubation for 60 minutes at 37°C. SiRNA transfected HAECs (12,000cells/100 μl matrigel) were seeded on the gels and incubated overnight at 37°C. Capillary tube formation was assessed using a Leica DMIL phase contrast microscope (Weltzar).
Results
FLT-1, KDR and angiopoietin promoters have E2F1 binding sites
Our previous studies have shown that certain VEGF-responsive genes like MT1G have E2F1 binding sites in their promoters (13) raising the possibility that E2Fs might be regulating genes involved in the angiogenic process. To verify this possibility, we first examined the promoters of human FLT-1, KDR and ANGPT2 for the presence of potential E2F1 binding sites. A detailed analysis of the promoter region (1.2 kb) upstream of transcription start site (TSS) of FLT-1, KDR and ANGPT2 using MatInspector (Genomatix) revealed the presence of several putative E2F binding sites. Analysis of the FLT-1 promoter showed that there are eight potential E2F binding sites in the 1200 base pair region upstream of the TSS (Supplementary Data, Table 1); two additional E2F binding sites were present overlapping the TSS. Similar promoter analysis on a 1200 bp region of KDR and ANGPT2 promoters showed the presence of four and two E2F binding sites respectively. KDR promoter had one E2F binding site immediately downstream of TSS.
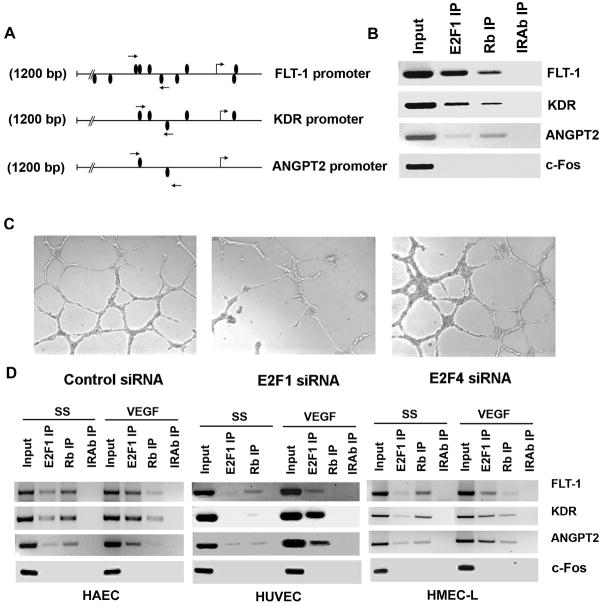
To experimentally verify whether E2F1 could bind to these promoters, we performed chromatin immunoprecipitation assays on asynchronously growing human aortic endothelial cells (HAECs) using primers spanning the putative E2F binding sites in these promoters. The primer pairs spanned four E2F binding sites located in FLT1 promoter and amplified a 203 bp fragment (indicated by arrows in Figure 1A). The primers designed for KDR promoter amplified a 182 bp region containing one putative E2F binding site whereas primer pairs for ANGPT2 promoter spanned two E2F binding site and amplified a 235 bp region. As shown in Figure 1B, there was a significant association of E2F1 to the promoter region of FLT-1, KDR and ANGPT2 in HAECs; like most E2F-regulated promoters, Rb protein could also be detected on these promoters. We had observed earlier that Rb was phosphorylated upon VEGF stimulation of HAECs and the association of Rb with these promoters was reduced upon VEGF stimulation, probably due to phosphorylation (17, 18). An unrelated promoter, c-Fos, used as the negative control could not be detected in the E2F1 and Rb immunoprecipitates; further, the promoters could not be detected in immunoprecipitations done with an irrelevant antibody, confirming the specificity of the assay. This experiment suggests that the E2F sites present on these three promoters are functional and can recruit E2F1 in vivo.
Figure 1.
(A) A schematic representation of FLT-1 promoter, KDR promoter and Angiopoietin 2 promoter showing the potential E2F binding sites. Position of primers used for ChIP assay spanning the putative E2F binding sites are shown as arrows. (B) E2F1 and Rb associates with FLT-1, KDR and ANGPT2 promoters in vivo. ChIP assays were carried out on asynchronously growing HAECs. Input indicates an aliquot of total DNA. Antibodies used for immunoprecipitation are indicated above the lanes. (C) Depletion of E2F1 by transfecting HAECs with siRNA targeted to E2F1 abrogates angiogenic tubule formation while non-targeting siRNA and E2F4 siRNA did not affect the formation of angiogenic tubules. HAECs were transfected with the indicated siRNAs and plated on matrigel using standard protocols and observed under light microscopy. (D) VEGF induces recruitment of E2F1 on FLT-1, KDR and ANGPT2 promoters. ChIP assay was carried out on serum starved and VEGF stimulated HAEC, HUVEC and HMEC-L cells. In serum starved cells, a significant amount of Rb was found to be associated with these promoters. VEGF stimulation induced the binding of E2F1 to the promoter region while dissociating Rb in all the three cell types tested.
Since we found that E2F1 is associated with FLT1, KDR and ANGPT2 promoters, we next examined whether depletion of E2F1 by siRNA had any effect on angiogenic tubule formation in matrigel. To this end, HAECs were transfected with E2F1-siRNA (50 pmol) and 24 hours after transfection cells were plated in matrigel and grown for 18 hours. HAECs transfected with E2F1 siRNA showed significantly reduced angiogenic tubule formation in matrigel while non-targeting control siRNA showed well formed angiogenic tubules suggesting that E2F1 contributes to angiogenic tubule formation (Figure 1C). Transfecting HAECs with E2F4 siRNA did not show any effect on the tubule formation suggesting a role for E2F1 in regulating angiogenesis. Taken together, it can be imagined that E2F1 can regulate the angiogenic process by modulating the expression of such genes involved in angiogenesis.
VEGF induces the binding of E2F1 and dissociation of Rb from FLT1, KDR and ANGPT2 promoters
ChIP assays were performed to test whether VEGF induced the binding of E2F1 to these promoters. ChIP lysates were prepared from serum starved as well as VEGF stimulated HAECs and immunoprecipitated with an antibody to E2F1, Rb or a control antibody. As can be seen from the Figure 1D, there was only minimal amount of E2F1 associated with FLT-1, KDR and ANGPT2 promoters in serum starved cells while VEGF induced recruitment of E2F1 in these promoters. A significant amount of Rb could be found in association with these promoters in quiescent cells; Rb dissociated from these promoters upon VEGF stimulation. The promoter fragments were not present in immunoprecipitates obtained with the control antibody. Similar results were obtained from ChIP assays conducted on other endothelial cells like human microvascular endothelial cells from the lung (HMEC-L) and human umbilical cord vein endothelial cells (HUVECs). It was found that VEGF induced the binding of E2F1 to all the three promoters in HUVECs as well as HMEC-Ls (Figure 1D) suggesting that E2F1 can bind to the promoter region of FLT-1, KDR and ANGPT2 genes in a signal-dependent manner.
FLT-1, KDR and angiopoietin promoters are E2F1 and Rb responsive
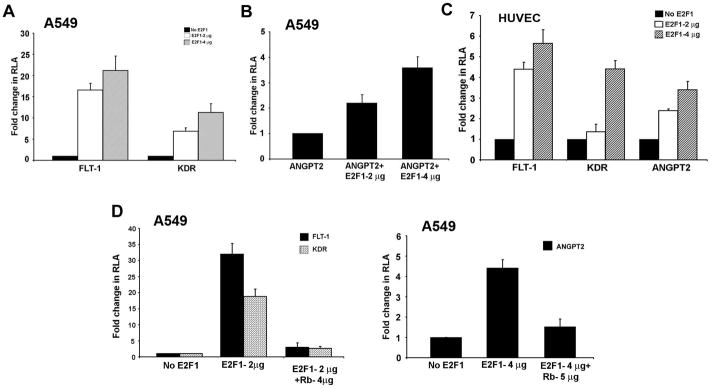
Since E2F1 could bind to the promoters in a VEGF dependent fashion, we hypothesized that the expression of FLT-1, KDR and ANGPT2 could be E2F1 regulated. In order to test the functionality of the E2F binding sites, A549 cells (Figure 2A and 2B) or HUVECs (Figure 2C) were transiently transfected with luciferase reporter constructs driven by FLT-1, KDR or ANGPT2 promoters in the presence or absence of an E2F1 expression vector. There was a significant increase in the expression of FLT-1 promoter when co-transfected with E2F1 expression construct in A549 and HUVEC cells. Similarly, KDR promoter activity was also remarkably induced by E2F1 in both the cell lines (Figure 2C). There was a four-fold increase in the transcriptional activity of ANGPT2 promoter in presence of 4 μg of E2F1 in both A549 and HUVECs (Figure 2B and 2C). We next examined whether Rb could repress E2F1 mediated transcription of FLT-1, KDR and ANGPT2 promoters. Transient transfection of A549 and HUVECs with a Rb construct showed a significant inhibition of E2F1 mediated transcription of FLT-1 and KDR promoters (Figure 2D, left panel) as well as the ANGPT2 promoter (Figure 2D, right panel). These results suggest that the E2F1-Rb pathway might be involved in the regulation of FLT1, KDR and ANGPT2 expression.
Figure 2.
FLT-1, KDR and ANGPT2 promoters are E2F1 and Rb responsive. Transient transfection experiments carried out in A549 (A and B) and HUVEC (C) cells showed that E2F1 could induce FLT-1, KDR and ANGPT2 promoters. Two and four μg of E2F1 could induce a significant amount of luciferase activity. Transfection with 4 or 5 μg of Rb construct expressing the large pocket domain of Rb showed a significant inhibition of E2F1 mediated transcription from these promoters (D, left and right panels).
E2F1 depletion leads to downregulation of FLT-1 and KDR gene expression
To confirm the role of E2F1 in the transcriptional activation of angiogenic promoters, we transfected HAEC, HUVEC and HMEC-Ls with different concentrations of E2F1 siRNA as well as non-targeting control siRNA. E2F1 siRNA treatment in these cells effectively reduced the level of E2F1 in a dose dependent manner as shown in western blot analysis (Figure 3). Depletion of E2F1 by siRNA transfection resulted in the downregulation of FLT-1 and KDR expression in realtime PCR experiments. There was a 50% decrease in the expression of FLT-1 mRNA and a 45% decrease in KDR expression levels with E2F1 siRNA (75 pmol) in HAEC cells compared to control siRNA treated cells (Figure 3A). A similar pattern of downregulation of FLT1 and KDR was observed in HMEC-Ls and HUVECs confirming that E2F1 indeed is involved in the transcriptional activation of these pro-angiogenic genes (Figure 3B and 3C).
Figure 3.
Depletion of E2F1 leads to downregulation of FLT-1, KDR and ANGPT2. Depletion of E2F1 by transiently transfecting 50 pmol, 75 pmol or 100 pmol E2F1 si RNA reduced the expression of FLT1 and KDR genes in HAECs (A), HMEC-Ls (B) and HUVECs (C) as seen by Real-Time PCR. Western blot showing depletion of E2F1 after siRNA transfection is also shown. Real-time PCR data showing the involvement of E2F1 in regulating VEGF induced expression of FLT-1 and KDR (D). VEGF stimulation induced expression of FLT-1 and KDR in control siRNA transfected cells while E2F1 siRNA treated cells did not show an induction of FLT-1 and KDR gene expression.
Since VEGF stimulation leads to recruitment of E2F1 to FLT-1 and KDR promoters, we further examined whether E2F1 is involved in VEGF induced expression of FLT-1 and KDR genes. As shown in Figure 3D, VEGF stimulation induced expression of FLT-1 and KDR in control siRNA treated cells while E2F1 siRNA treated cells did not respond to VEGF treatment suggesting that E2F1 facilitates the VEGF induced expression of FLT-1 and KDR.
VEGF stimulation enhances histone acetylation of FLT-1, KDR and ANGPT2 promoters
Histone acetylation has been implicated in altering nucleosome structure facilitating transcriptional activation (19). We examined whether VEGF stimulation leads to acetylation of histones on FLT-1, KDR and ANGPT2 promoters. ChIP assays were carried out using antibodies against the histone acetyl transferase p300, on lysates from serum-starved or VEGF stimulated HUVECs. The results revealed that there was only a very low amount of p300 associated with the promoter region of FLT1 and ANGPT2 in serum starved cells, suggesting that the promoters are not acetylated and are inactive (Figure 4A). Further, there was HDAC1 associated with these two promoters, supporting this hypothesis. At the same time, there was a certain amount of histone H3 acetylation and p300 binding observed on the KDR promoter; KDR promoter also showed the presence of more HDAC1 compared to the other promoters. There was a significant increase in the acetylation of histones on all the three promoters with VEGF stimulation and this correlated with increased association of p300. HDAC1 was completely dissociated from the promoters upon VEGF stimulation. This shows that VEGF stimulation leads to the recruitment of p300 to these promoters, facilitating histone acetylation and transcriptional induction.
Figure 4.

(A) VEGF stimulation induces recruitment of p300 and dissociation of HDAC-1 in angiogenic promoters. ChIP assay showing the recruitment of acetylated Histone H3 and p300 on FLT-1, KDR and ANGPT2 promoters with VEGF induction. In quiescent cells these promoters are occupied by HDAC-1 while with VEGF stimulation, HDAC-1 dissociates from the promoter with concomitant recruitment of acetylated histone H3 and p300 suggesting promoter activation. (B) VEGF induced recruitment of CBP, p300 and PCAF on FLT-1 and KDR promoters. In serum starved cells promoter occupancy of acetyl transferases CBP, p300 and PCAF is minimal while with VEGF stimulation, there is a significant increase in the association of these acetyl transferases in the FLT-1 and KDR promoters. (C) Matrigel assay demonstrating the role of CBP, p300 and PCAF in angiogenic tubule formation. HUVECs transfected with PCAF siRNA showed significantly reduced angiogenic tubule formation in matrigel while non-targeting control siRNA showed well formed angiogenic tubules.
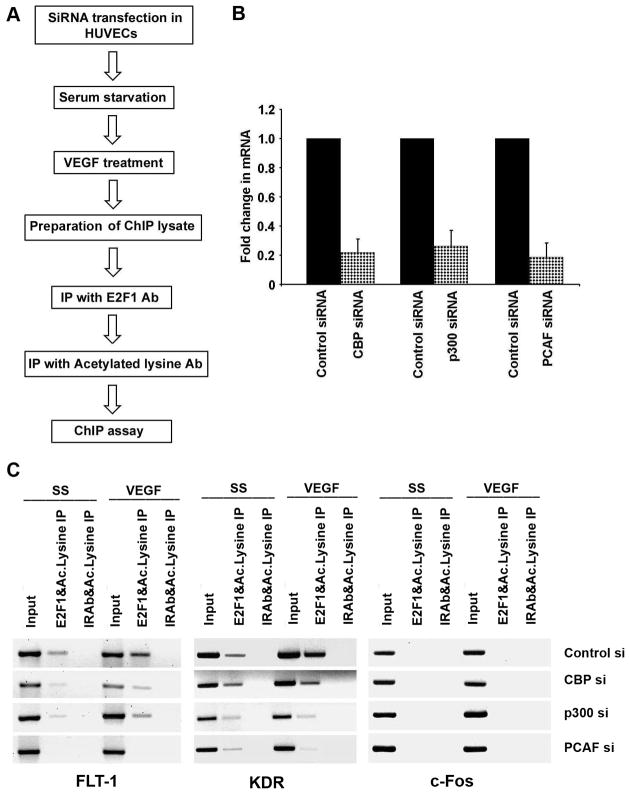
Since the acetyl transferases CBP, p300 and PCAF are known to acetylate histones leading to transcriptional activation, we examined whether these acetyl transferases are involved in acetylation of FLT-1 and KDR promoter region in response to VEGF stimulation. To verify this, we performed ChIP assays on quiescent and VEGF induced HUVEC cells using CBP, p300 and PCAF antibodies. The ChIP assay revealed an enhanced recruitment of all the three acetyl transferases on KDR and FLT-1 promoters upon VEGF stimulation (Figure 4B). The level of PCAF on these promoters was greater compared to the levels of CBP and p300, suggesting that PCAF might be playing a major role in acetylating E2F1.
We next examined the role of CBP, p300 and PCAF in angiogenic tubule formation. To this end, we depleted the levels of CBP, p300 and PCAF in HUVECs using siRNAs and 24 hours after transfection cells were plated in matrigel and grown for 18 hours. HUVECs transfected with PCAF siRNA showed significantly reduced angiogenic tubule formation in matrigel while those transfected with a non-targeting control siRNA showed well formed angiogenic tubules suggesting that PCAF might be the major acetyl transferase contributing to angiogenic tubule formation (Figure 4C). Transfecting HUVECs with CBP and P300 siRNA also showed effect on the tubule formation suggesting the involvement of all the three acetyl transferases in regulating angiogenesis. Figure 6B shows the efficiency of siRNA transfection as shown by Real-time PCR for CBP, p300 and PCAF.
Figure 6.
E2F1 acetylation is mediated mainly by PCAF. (A) Schematic of the workflow of the experiment. (B) Efficiency of siRNA transfection shown by Real-time PCR for CBP, p300 and PCAF. (C) ChIP-re-ChIP showing the occupancy of acetylated E2F1 on angiogenic promoters with VEGF stimulation in control, CBP, P300 and PCAF siRNA treated cells. Antibodies used for the first and second immunoprecipitation are indicated above the lanes. Control siRNA treated cells showed enhanced occupancy of acetylated E2F1 on FLT-1 and KDR promoters with VEGF stimulation while PCAF siRNA treated cells did not show increase in acetylated E2F1 on these promoters.
VEGF stimulation leads to binding of E2F1 to angiogenic promoters
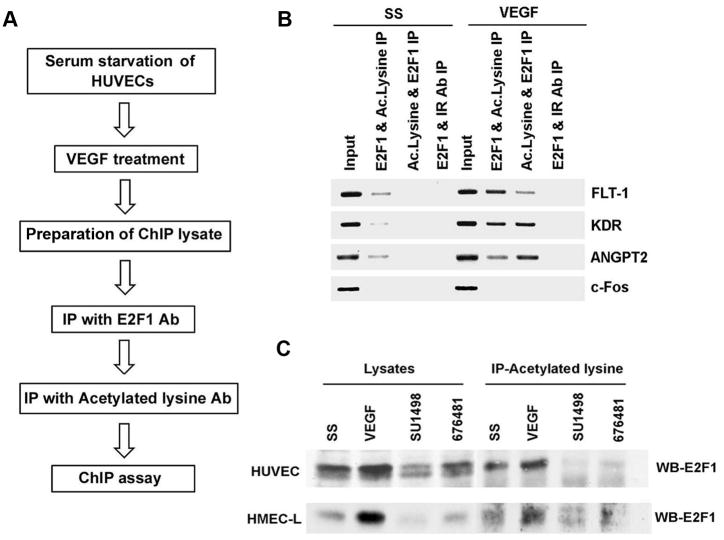
Acetylation of E2F1 has been shown to occur in response to DNA damage and enhances its DNA binding activity (20). This acetylation is brought about mainly by p300/PCAF protein and leads to increased stabilization and accumulation of E2F1 (21). Our previous studies showed that stimulation of HAECs with VEGF led to an increase in the acetylation of E2F1 (18). We further examined whether VEGF induced increase in E2F1 acetylation facilitates its interaction with angiogenic promoters. Towards this purpose we conducted ChIP-re-ChIP on these promoters using E2F1 and acetylated lysine antibodies (Figure 5A). ChIP assay lysates were prepared from quiescent and VEGF stimulated HUVECs and immunoprecipitated with E2F1 antibody to bring down all the promoter elements bound by E2F1. To determine whether the E2F1 associated with these promoters as well as the histones are acetylated, the chromatin complexes were eluted and re-immunoprecipitated with antibodies to acetylated lysine. Immunoprecipitation in the reverse fashion using acetylated lysine for the primary IP followed by E2F1 IP was also done to confirm the results. ChIP assays were done from the second immunoprecipitated DNA to amplify promoter fragments of angiogenic genes. The results from ChIP- re-ChIP revealed that there is increased acetylation of E2F1 as well as histones associated with the angiogenic promoters when stimulated with VEGF (Figure 5B). As control, IP was performed using E2F1 antibody followed by a second IP using anti-rabbit secondary antibody. An unrelated promoter c-Fos was also analyzed to confirm the specificity of the experiment.
Figure 5.
VEGF stimulation leads to binding of acetylated E2F1 to angiogenic promoters and VEGF induced E2F1 acetylation is mediated by VEGF receptors. (A) Flow chart showing the steps involved in ChIP-re-ChIP. (B) ChIP-Re-ChIP showing the enhanced occupancy of acetylated E2F1 on angiogenic promoters with VEGF stimulation. Antibodies used for the first immunoprecipitation and second immunoprecipitation are indicated above the lanes. (C) Immunoprecipitation-Western blot analysis demonstrating the role of KDR and FLT-1 in mediating VEGF induced acetylation of E2F1. KDR and FLT1 inhibitors could block E2F1 acetylation significantly indicating the possibility of a positive feedback loop in regulating FLT-1 and KDR expression by E2F1.
VEGF induced acetylation requires signaling through the VEGF Receptors
Since we observed that E2F1 binding is enhanced and correlated with induction of acetylation in response to VEGF stimulation in endothelial cells, we examined the role of FLT-1 and KDR function in mediating VEGF induced acetylation using KDR inhibitor SU1498 and KDR and FLT-1 inhibitor 676481 (Calbiochem). HUVECs and HMEC-Ls were serum starved and subsequently stimulated with VEGF in the presence or absence of the inhibitors. Acetylation of E2F1 was determined by IP-Western blot analysis using antibodies to E2F1 and acetylated lysine. The results from the IP-Western blots revealed that KDR and FLT1 inhibitors could block E2F1 acetylation significantly (Figure 5C). Inhibition of E2F1 acetylation by VEGF-R inhibitors suggests that these receptors might be involved in downstream signaling leading to the acetylation of E2F1. This indicates the possibility of a positive feedback loop in regulating FLT-1 and KDR expression by E2F1.
VEGF induced acetylation is mediated mainly by PCAF
We next explored the involvement of histone acetyl transferases in VEGF induced acetylation. Towards this purpose we knocked down CBP, p300 and pCAF using siRNAs (100 pmols) and subsequently serum starved and induced with VEGF for 24 hours. ChIP-re-ChIP assay was performed on quiescent as well as VEGF stimulated HUVECs (Figure 6A). First immunoprecipitation reaction was performed using E2F1 antibody followed by subsequent immunoprecipitation with acetylated lysine antibody to determine the association of acetylated E2F1 on KDR and FLT-1 promoters. Immunoprecipitation using an irrelevant antibody (anti-rabbit secondary antibody) followed by a second immunoprecipitation with acetylated lysine antibody was used as control. Increased amounts of E2F1 was found associated with KDR and FLT-1 promoter upon VEGF stimulation in control siRNA treated cells (Figure 6C). Transient transfection with PCAF siRNA resulted in significantly lower amount of E2F1 in these promoters whereas knocking down CBP and p300 did not have much effect on the recruitment of E2F1. Collectively, these results suggest that PCAF may be the major acetyl transferase involved in acetylating E2F1 as well as the histones on these promoters. Real-time PCR was performed to assess the efficiency of siRNA transfection (Figure 6B). Taken together, we conclude that acetylation of E2F1 as well as the histones in the promoter regions plays a role in regulating the angiogenic process by modulating the expression of FLT-1, KDR and ANGPT2 genes.
Discussion
Angiogenesis plays a role in several diseases including cancer where angiogenic pathway is elevated during tumor growth, progression and metastasis (22, 23). Vascular endothelial growth factor, exerts its biological effects through specific interactions with VEGF receptors. VEGFR1 (FLT-1) and VEGFR2 (KDR) are found on the surface of endothelial cells, hematopoietic stem cells, leukocytes, osteoblasts and also on many tumor cells (24, 25). Knock out studies targeting FLT-1 have implicated it as a critical mediator of both developmental and physiological angiogenesis (26). It has been reported that FLT-1 acts as a decoy receptor that inhibits KDR mediated angiogenesis (27, 28); however, there is evidence that FLT-1 may also cooperatively activate angiogenesis with KDR in a ligand and cell context dependent manner. FLT-1 is also expressed as a soluble truncated form which lacks the intracellular receptor tyrosine kinase domains and it binds to VEGF and inhibits angiogenesis (29). Recent reports suggests that FLT-1 is expressed in several pancreatic and colorectal cancer cell lines where its activation led to EMT which is linked to increased invasion and migration (30, 31).
KDR is considered as the principal receptor of VEGF dependent angiogenic signals with an important role in normal vessel growth and hematopoiesis (32, 33). The angiopoietin/Tie-2 family plays an important role in regulating vessel stability. ANGPT2 shows context dependent proangiogenic and antiangiogenic activities (34). ANGPT2 knock out studies revealed that ANGPT2 is not needed for embryonic vascular development, but is required for postnatal angiogenic remodelling (35). Recent studies show that in the presence of VEGF, ANGPT2 promotes endothelial proliferation and migration and stimulates sprouting of new blood vessels while ANGPT2 promotes endothelial cell death and vessel regression if the activity of endogenous VEGF is inhibited (36).
Our studies show for the first time that FLT-1, KDR and ANGPT2 genes are VEGF inducible and that these promoters are E2F responsive. Previous studies had shown that expression of FLT-1 and KDR was regulated by Sp proteins (25, 37). It is also known that E2F1 and Sp1 proteins physically and functionally interact and show functional synergism in promoters having binding site for both(38).
It is intriguing that in serum starved cells there was a significant amount of Rb and HDAC associated with these promoters as seen in ChIP assays, which dissociated upon VEGF stimulation; this is reminiscent of the changes taking place on proliferative promoters during cell cycle progression. Indeed, we had observed that VEGF stimulation of quiescent HAECs lead to Rb phosphorylation (17, 18). Rb could significantly repress E2F mediated transcription of these promoters. It is well established that Rb represses gene expression by recruiting HDACs (39) and this appears to be true for FLT-1, KDR and ANGPT2 promoters. VEGF stimulation also led to the enhanced recruitment of histone acetyl transferases, suggesting transcriptional activation. There are recent evidences supporting the role of Rb in the regulation of proangiogenic factor expression at the transcriptional level (40). For example, over expression of cyclin D1 induces FGFR1 promoter in mouse fibroblasts in an Rb-E2F dependent fashion (41, 42). Another study suggested that human FGFR1 expression is also regulated by E2F1 through two non-consensus E2F1 responsive sequences (43). Recently, a novel variant of human PDGFRα transcript identified in a subset of human cell lines was found to be regulated by E2F-1 (44). Taken together, these findings demonstrate a new role of E2Fs targeting factors that can modulate angiogenesis.
Although E2F activity is regulated principally by association with different regulator proteins, posttranslational modifications such as acetylation and phosphorylation have been suggested as possible mechanisms of regulation of E2F activity (21, 45). The p300/CBP associated factor PCAF and also p300/CBP itself was observed to acetylate E2F1, and resulted in stimulating the functions of non-Rb bound form of E2F1. Acetylation of E2F1 by PCAF increases its DNA binding ability, activation potential and protein half-life. Although the major targets of CBP, p300 and PCAF are histones, these acetylases also modify a number of transcription factors (46–48). There are many recent reports of acetylation sites on proteins involved in diverse biological processes (49). p73 is an E2F1 target gene in DNA damage response, and acetylation of E2F1 was required to be recruited on p73 promoter (50). Our results show time that VEGF stimulation leads to recruitment of E2F1 on FLT-1 and KDR promoters and this correlates with enhanced acetylation of the histones as well as E2F1. VEGF induced acetylation is mediated through its receptors, suggesting the possibility of a positive feedback loop in E2F1 activity.
While it is known that E2Fs function in a wide range of biological processes, its role in regulating the expression of molecules involved in angiogenesis had not been well explored. Our findings indicate that E2F and Rb might be contributing to tumor growth by regulating the transcription of genes involved in angiogenesis as well, in addition to their established roles in cell proliferation.
Supplementary Material
Acknowledgments
Assistance of the Microscopy and Molecular Biology Core Facilities at the Moffitt Cancer Center is greatly appreciated. This study was supported by the grant CA63136 from the NCI to SPC. The authors have no conflicts of interest to declare.
References
- 1.Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–61. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 2.Nevins JR. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–93. [PubMed] [Google Scholar]
- 3.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–62. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 4.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 5.Slansky JE, Farnham PJ. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 6.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 7.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 8.Trimarchi JM, Fairchild B, Verona R, Moberg K, Andon N, Lees JA. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci U S A. 1998;95:2850–5. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen J, Cloos P, Toftegaard U, et al. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 2005;33:5458–70. doi: 10.1093/nar/gki855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Logan N, Graham A, Zhao X, et al. E2F-8: an E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene. 2005;24:5000–4. doi: 10.1038/sj.onc.1208703. [DOI] [PubMed] [Google Scholar]
- 11.Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−)mice. Nat Genet. 1998;18:360–4. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 12.Field SJ, Tsai FY, Kuo F, et al. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–61. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 13.Joshi B, Ordonez-Ercan D, Dasgupta P, Chellappan S. Induction of human metallothionein 1G promoter by VEGF and heavy metals: differential involvement of E2F and metal transcription factors. Oncogene. 2005;24:2204–17. doi: 10.1038/sj.onc.1208206. [DOI] [PubMed] [Google Scholar]
- 14.Pillai S, Chellappan SP. ChIP on chip assays: genome-wide analysis of transcription factor binding and histone modifications. Methods Mol Biol. 2009;523:341–66. doi: 10.1007/978-1-59745-190-1_23. [DOI] [PubMed] [Google Scholar]
- 15.Reid G, Hubner MR, Metivier R, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 16.Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278:47853–61. doi: 10.1074/jbc.M305171200. [DOI] [PubMed] [Google Scholar]
- 17.Dasgupta P, Sun J, Wang S, et al. Disruption of the Rb--Raf-1 interaction inhibits tumor growth and angiogenesis. Mol Cell Biol. 2004;24:9527–41. doi: 10.1128/MCB.24.21.9527-9541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinkade R, Dasgupta P, Carie A, et al. A small molecule disruptor of Rb/Raf-1 interaction inhibits cell proliferation, angiogenesis, and growth of human tumor xenografts in nude mice. Cancer Res. 2008;68:3810–8. doi: 10.1158/0008-5472.CAN-07-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell. 2006;23:289–96. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Trouche D, Cook A, Kouzarides T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996;24:4139–45. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–71. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beecken WD, Engl T, Ringel EM, et al. An endogenous inhibitor of angiogenesis derived from a transitional cell carcinoma: clipped beta2-glycoprotein-I. Ann Surg Oncol. 2006;13:1241–51. doi: 10.1245/s10434-006-9009-9. [DOI] [PubMed] [Google Scholar]
- 23.Zetter BR. Angiogenesis and tumor metastasis. Annu Rev Med. 1998;49:407–24. doi: 10.1146/annurev.med.49.1.407. [DOI] [PubMed] [Google Scholar]
- 24.Bellamy WT. Vascular endothelial growth factor as a target opportunity in hematological malignancies. Curr Opin Oncol. 2002;14:649–56. doi: 10.1097/00001622-200211000-00010. [DOI] [PubMed] [Google Scholar]
- 25.Abdelrahim M, Baker CH, Abbruzzese JL, et al. Regulation of vascular endothelial growth factor receptor-1 expression by specificity proteins 1, 3, and 4 in pancreatic cancer cells. Cancer Res. 2007;67:3286–94. doi: 10.1158/0008-5472.CAN-06-3831. [DOI] [PubMed] [Google Scholar]
- 26.Fong GH, Zhang L, Bryce DM, Peng J. Increased hemangioblast commitment, not vascular disorganization, is the primary defect in flt-1 knock-out mice. Development. 1999;126:3015–25. doi: 10.1242/dev.126.13.3015. [DOI] [PubMed] [Google Scholar]
- 27.Park JE, Chen HH, Winer J, Houck KA, Ferrara N. Placenta growth factor. Potentiation of vascular endothelial growth factor bioactivity, in vitro and in vivo, and high affinity binding to Flt-1 but not to Flk-1/KDR. J Biol Chem. 1994;269:25646–54. [PubMed] [Google Scholar]
- 28.Rahimi N, Dayanir V, Lashkari K. Receptor chimeras indicate that the vascular endothelial growth factor receptor-1 (VEGFR-1) modulates mitogenic activity of VEGFR-2 in endothelial cells. J Biol Chem. 2000;275:16986–92. doi: 10.1074/jbc.M000528200. [DOI] [PubMed] [Google Scholar]
- 29.Goldman CK, Kendall RL, Cabrera G, et al. Paracrine expression of a native soluble vascular endothelial growth factor receptor inhibits tumor growth, metastasis, and mortality rate. Proc Natl Acad Sci U S A. 1998;95:8795–800. doi: 10.1073/pnas.95.15.8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wey JS, Fan F, Gray MJ, et al. Vascular endothelial growth factor receptor-1 promotes migration and invasion in pancreatic carcinoma cell lines. Cancer. 2005;104:427–38. doi: 10.1002/cncr.21145. [DOI] [PubMed] [Google Scholar]
- 31.Fan F, Wey JS, McCarty MF, et al. Expression and function of vascular endothelial growth factor receptor-1 on human colorectal cancer cells. Oncogene. 2005;24:2647–53. doi: 10.1038/sj.onc.1208246. [DOI] [PubMed] [Google Scholar]
- 32.Ferrara N. Role of vascular endothelial growth factor in physiologic and pathologic angiogenesis: therapeutic implications. Semin Oncol. 2002;29:10–4. doi: 10.1053/sonc.2002.37264. [DOI] [PubMed] [Google Scholar]
- 33.Shalaby F, Rossant J, Yamaguchi TP, et al. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–6. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 34.Maisonpierre PC, Suri C, Jones PF, et al. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 35.Gale NW, Thurston G, Hackett SF, et al. Angiopoietin-2 is required for postnatal angiogenesis and lymphatic patterning, and only the latter role is rescued by Angiopoietin-1. Dev Cell. 2002;3:411–23. doi: 10.1016/s1534-5807(02)00217-4. [DOI] [PubMed] [Google Scholar]
- 36.Lobov IB, Brooks PC, Lang RA. Angiopoietin-2 displays VEGF-dependent modulation of capillary structure and endothelial cell survival in vivo. Proc Natl Acad Sci U S A. 2002;99:11205–10. doi: 10.1073/pnas.172161899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higgins KJ, Abdelrahim M, Liu S, Yoon K, Safe S. Regulation of vascular endothelial growth factor receptor-2 expression in pancreatic cancer cells by Sp proteins. Biochem Biophys Res Commun. 2006;345:292–301. doi: 10.1016/j.bbrc.2006.04.111. [DOI] [PubMed] [Google Scholar]
- 38.Lin SY, Black AR, Kostic D, Pajovic S, Hoover CN, Azizkhan JC. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol Cell Biol. 1996;16:1668–75. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–73. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 40.Gabellini C, Del Bufalo D, Zupi G. Involvement of RB gene family in tumor angiogenesis. Oncogene. 2006;25:5326–32. doi: 10.1038/sj.onc.1209631. [DOI] [PubMed] [Google Scholar]
- 41.Tashiro E, Minato Y, Maruki H, Asagiri M, Imoto M. Regulation of FGF receptor-2 expression by transcription factor E2F-1. Oncogene. 2003;22:5630–5. doi: 10.1038/sj.onc.1206636. [DOI] [PubMed] [Google Scholar]
- 42.Tashiro E, Maruki H, Minato Y, Doki Y, Weinstein IB, Imoto M. Overexpression of cyclin D1 contributes to malignancy by up-regulation of fibroblast growth factor receptor 1 via the pRB/E2F pathway. Cancer Res. 2003;63:424–31. [PubMed] [Google Scholar]
- 43.Kanai M, Tashiro E, Maruki H, Minato Y, Imoto M. Transcriptional regulation of human fibroblast growth factor receptor 1 by E2F-1. Gene. 2009;438:49–56. doi: 10.1016/j.gene.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Minato Y, Tashiro E, Kanai M, Nihei Y, Kodama Y, Imoto M. Transcriptional regulation of a new variant of human platelet-derived growth factor receptor alpha transcript by E2F-1. Gene. 2007;403:89–97. doi: 10.1016/j.gene.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 45.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–44. [PMC free article] [PubMed] [Google Scholar]
- 46.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 47.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–8. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci U S A. 1998;95:9855–60. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 50.Pediconi N, Ianari A, Costanzo A, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003;5:552–8. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.