Abstract
Background
Rare variants in cardiac ion channel genes are associated with sudden cardiac death (SCD) in rare primary arrhythmic syndromes; however, it is unknown whether common variation in these same genes may contribute to SCD risk at the population level.
Methods and Results
We examined the association between 147 single nucleotide polymorphisms (SNPs) (137 tag, 5 non-coding SNPs associated with QT interval duration and 5 nonsynonymous SNPs) in 5 cardiac ion channel genes, KCNQ1, KCNH2, SCN5A, KCNE1 and KCNE2 and sudden and/or arrhythmic death in a combined nested case-control analysis among 516 cases and 1522 matched controls of European ancestry enrolled in six prospective cohort studies. After accounting for multiple testing, two SNPs (rs2283222 located in intron 11 in KCNQ1 and rs11720524 located in intron 1 in SCN5A) remained significantly associated with sudden/arrhythmic death (FDR = 0.01 and 0.03 respectively). Each increasing copy of the major T allele of rs2283222 or the major C allele of rs1172052 was associated with an OR = 1.36 (95% CI 1.16-1.60, P=0.0002) and 1.30 (95% CI 1.12-1.51, P=0.0005) respectively. Control for cardiovascular risk factors and/or limiting the analysis to definite SCDs did not significantly alter these relationships.
Conclusion
In this combined analysis of 6 prospective cohort studies, two common intronic variants in KCNQ1 and SCN5A were associated with SCD in individuals of European ancestry. Further study in other populations and investigation into the functional abnormalities associated with non-coding variation in these genes may lead to important insights into predisposition to lethal arrhythmias.
Keywords: death, sudden, genetics, ion channels, epidemiology
Introduction
There are an estimated 250,000 to 400,000 sudden cardiac deaths (SCD) annually in the United States1, and over half are the initial manifestation of heart disease2. Although the majority of these deaths are associated with coronary artery disease3; the factors determining predisposition to arrhythmia in the presence of ischemia or infarction remain poorly understood. There is emerging evidence that a proportion of the familial contribution to SCD risk is distinct from that associated with other manifestations of atherosclerosis4-6. For example, individuals with a familial history of SCD are more likely to experience SCD6 and/or ventricular fibrillation during acute myocardial infarction5. These data suggest that there may be genetic factors that confer a predisposition to fatal ventricular arrhythmias even in the setting of atherosclerosis or other acquired disease states.
Cardiac ion channels play a critical role in cardiac electrophysiology7, and perturbation of their function can result in SCD by lethal ventricular arrhythmias in a wide range of conditions8. While many ion channels and other proteins are involved in the genesis and maintenance of normal cardiac rhythm, genetic analyses of rare arrhythmic disorders have definitively established a critical role for 5 particular ion channel genes in arrhythmia vulnerabilitys8, 9. These genes encode the cardiac sodium channel (SCN5A), and ion channels regulating the slow component (KCNQ1 and KCNE1) and the rapid component (KCNH2 and KCNE2) of the delayed rectifier potassium current (IKs and IKr respectively). While the primary arrhythmic disorders associated with mutations in these same genes are rare, there are data to suggest that common variants may result in subclinical alterations in ion channel function10-17 that may only become clinically manifest in the setting of other proarrhythmic factors, such as atherosclerosis or drugs.
In order to address the hypothesis that common variants in these genes might be associated with SCD within the general population, we assembled SCD cases from six NIH-funded prospective cohorts and utilized a prospective nested case-control design to test for associations between common genetic variation in coding and non-coding regions of these five ion channel genes and SCD among individuals of European ancestry.
Methods
Study Populations
The prospective cohorts included in the present investigation include the Physicians’ Health Study (PHS I and II), the Nurses’ Health Study (NHS), the Health Professionals Follow-up Study (HPFS), the Women’s Health Study (WHS), and the Women’s Antioxidant Cardiovascular Study (WACS). Together, these cohorts include a total of 40,878 men and 67,093 women with stored blood samples. The details of the cohorts along with the proportion of participants who donated blood samples at baseline are outlined in Supplemental Table 1. In brief, the NHS and HPFS are prospective observational cohort investigations, and the PHS I, WHS, and WACS studies were initially randomized trials of aspirin and/or vitamin supplements in which the intervention phases have ended. Prospective follow-up is ongoing in PHS I and WHS. The PHS II is an ongoing randomized trial of vitamin supplementation. Information about medical history, lifestyle choices, and incident disease is assessed either annually or biennially by self-administered questionnaires.
Endpoint Confirmation
The study end points include incident cases of sudden and/or arrhythmic cardiac death that occurred after return of the blood sample and before April 1, 2007. All cohorts employed similar methods to document endpoints as has been previously described in detail18. Among the individuals who donated blood samples at baseline, 540 sudden/arrhythmic deaths were confirmed, and 536 of these had DNA samples that passed our quality control standards. A cardiac death was considered a definite SCD if the death or cardiac arrest that precipitated death occurred within one hour of symptom onset as documented by medical records or next-of-kin reports (n=389, 72.6%) or had an autopsy consistent with SCD (i.e. acute coronary thrombosis or severe coronary artery disease without myocardial necrosis or other pathologic findings to explain death; n=23, 4.3%). Unwitnessed deaths or deaths that occurred during sleep were considered probable SCDs if the participant was documented to be symptom free when last observed within the preceding 24 hours, and circumstances suggested that the death could have been sudden (n= 92, 17.2%)19.
Deaths were also classified as arrhythmic or non-arrhythmic based on the definition of Hinkle and Thaler20. An arrhythmic death was defined as an abrupt spontaneous collapse of the circulation (pulse disappeared) without evidence of prior circulatory impairment (shock, congestive heart failure) or neurologic dysfunction (change in mental status, loss of consciousness, or seizure). Deaths before which the pulse gradually disappeared and/or those preceded by circulatory or neurologic impairment were considered non-arrhythmic deaths, and these deaths were excluded from the SCD endpoint even if the death occurred within one hour of symptom onset. Deaths which fulfilled the criteria for arrhythmic death, but were preceded by greater than one hour of symptoms (n= 32, 6.0%) were also included in the combined endpoint of sudden and/or arrhythmic cardiac death.
Selection of Controls
Using risk-set sampling21, we randomly selected up to three controls for each case matched on study cohort, sex, age (+/− 1 year), ethnicity, smoking status (current, never, past), time and date of blood sampling, fasting status, and presence or absence of cardiovascular disease (MI, angina, CABG, or stroke) prior to death. Since very few cases among other ethnicities (n= 20) occurred in these predominantly European-derived cohorts and to reduce the risk of population stratification, analyses were limited to self-described whites (n=516).
Linkage Disequilibrium Characterization and SNP Selection
Given recent data highlighting the importance of non-coding regions in genetic association studies22 and on ion channel function14, we tested for associations between SNPs in both coding and non-coding regions of the candidate genes, including 5kb upstream and downstream using a tag SNP strategy. We used genotypes available from the International HapMap CEU (release 22) sample of northern and western European ancestry (120 independent chromosomes), and the program Tagger23 was used to select tag SNPs that capture SNPs with minor allele frequency ≥5% in the reference pedigrees at a minimum r2 of 0.8. A total of 137 tag SNPs were selected and successfully genotyped. We also genotyped 5 additional non-coding variants shown to be associated with QT interval duration in a recent genome-wide association study16 as well as 5 single nonsynonymous SNPs with reported frequencies over 1 percent in Caucasians known either to be associated with intermediate markers, such as QT interval, or documented to have functional significance in cellular models11, 12, 15, 24, 25. These SNPs captured the majority of common variation at the 5 gene loci, with mean r2 to the target SNPs with minor allele frequency ≥5% in HapMap CEU of 0.87 (KCNQ1), 0.90 (KCNH2), 0.95 (SCN5A), 0.77 (KCNE1), and 0.86 (KCNE2). The SNPs captured a large proportion of the SNPs at each locus at r2 > 0.50 (0.85, 0.96, 1.0, 0.92, 1.0, respectively)
Finemapping of associated SNPs
For tag SNPs found to be associated with sudden/arrhythmic death, we attempted to further refine the signals of association by additionally genotyping partially correlated SNPs. Specifically, we examined all SNPs within 50kb of the associated variant (sentinel SNP) in HapMap CEU (release 24) and identified variants (target SNPs) with r2 > 0.20 to the sentinel SNP. We then selected a parsimonious subset of finemapping SNPs using Tagger such that every target SNP was captured by pairwise correlation r2 > 0.80 to any one of the finemapping SNPs.
Genotyping and Quality Control
Genomic DNA was extracted from the buffy coat fraction of centrifuged blood using Qiagen Autopure kits (Valencia, CA) in NHS, HPFS, and WACS and from whole blood in PHS I. In WHS and PHS I, DNA was extracted using the MagNA Pure LC instrument with the MagNA Pure LC DNA isolation kit (Roche Applied Science, Penzberg, Germany). All assays were conducted without knowledge of case status, and samples were labeled by study code only. Matched case-control pairs were handled identically and assayed in the same analytical run. All cohort DNA samples were genotyped together in the same experiment. For all SNPs except rs1805123 in KCNH2, genotyping was performed on the Sequenom platform (San Diego, CA) which resolves allele-specific single-base extension products using mass spectrometry (MALDI-TOF). DNA samples with successful genotyping on fewer than 60% of SNPs selected were excluded from analysis. Genotypes for SNPs included in the analysis passed our quality control thresholds (call rate ≥95%, Hardy-Weinberg equilibrium p>0.01 in controls). Blinded replicate quality control samples were included and genotyped with 99.82 percent agreement. Genotyping for rs1805123 was performed using a TaqMan 5-nuclease allelic discrimination assay with standard protocols and blinded replicate quality control samples were genotyped with 100% agreement.
Statistical analysis
Means or proportions for baseline cardiac risk factors were calculated for cases and controls. The significance of risk factor associations were tested with the Chi-square statistic for categorical variables and with the Student’s t-test for continuous variables. For each cohort, we investigated deviation from Hardy-Weinberg equilibrium among controls using a χ2 goodness-of-fit test. We analyzed the association between each SNP and the risk of sudden and/or arrhythmic cardiac death using conditional logistic regression analysis. With risk-set analysis, the odds ratio derived from the logistic regression directly estimates the hazard ratio, and thus, the rate ratio or relative risk26.
In the primary meta-analysis, the conditional odds ratios for each of the individual tag SNPs were estimated for each cohort separately under an additive model of inheritance. For the exonic SNPs, we assessed genetic effects under all three modes of inheritance: additive, dominant, and recessive, since prior studies reported testing of different genetic models for the nonsynonymous SNPs. Fixed effect meta-analyses using inverse variance weights were conducted based on the summary conditional logistic regression results for each cohort27, and PROC MIXED in SAS was used for estimation of the summary effects. Tests for heterogeneity of the genetic effect across sites were conducted using the Q-statistic28. To adjust for multiple comparisons, the false discovery rate (FDR)29 was computed for each SNP based upon the number of SNPs tested for each candidate gene, and an FDR <0.05 was utilized as the threshold for statistical significance for tag SNPs without a pre-specified hypothesis.
We then further adjusted these conditional logistic regression models for additional factors. Of the matching variables, age and smoking were not perfectly matched, and therefore these variables were also entered into the conditional logistic regression models to avoid any potential for residual confounding. The first multivariable model adjusted for age, and the second multivariable model further adjusted for standard cardiac risk factors including body-mass index; history of diabetes, hypertension, hyperlipidemia, and smoking. The third additionally adjusted for family history of myocardial infarction, alcohol intake, physical activity, and aspirin use (randomized aspirin treatment in WHS, PHS I, and reported aspirin use in the other studies). The randomized vitamin therapies in the trials had no effect on any cardiovascular outcome, and therefore, we do not adjust for their use in the model.
Statistical analysis was performed using SAS statistical software (SAS Institute Inc, Cary, NC), Version 9.1.
Results
A total of 516 cases (188 women and 328 men) of sudden and/or arrhythmic cardiac deaths occurred among subjects with European ancestry in the six cohorts over an average follow-up of 13.0 years. The clinical characteristics of the 516 cases and 1522 controls are displayed by study cohort in Table 1. The mean age of the cases was 64.2 years old and 201 (39.0%) reported a history of cardiovascular disease (CVD) prior to the SCD. In pooled analyses, cases were more likely to report a history of diabetes, hypertension, and a higher body mass index (p<0.005 for all comparisons). Cases did not differ significantly with respect to a history of hypercholesterolemia, family history of MI and/or aspirin use from the controls. Of the total study group, 509 cases and 1507 matched controls were successfully genotyped according to our pre-specified criteria. The genotyping call rates for the 142 SNPs ranged from 96.1% to 100%. There was no difference in the average call rate between cases and controls (97.2 vs 97.1%).
Table 1.
Cohort Specific* and Pooled Prevalence of Cardiac Risk Factors at the Time of the Blood Draw according to Case and Control Status.
| Cohort* | Case/ Control |
n | Age† (SD) years |
Body mass index (SD) kg/m2 |
History of prior CVD† N (%) |
Current smoking† N (%) |
Diabetes N (%) |
Hypertension N (%) |
High cholesterol N (%) |
Family History of MI N (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| HPFS | case | 120 | 68.0 (7.7) | 26.5 (3.8) | 50 (41.7 %) | 10 (8.9%) | 18 (15.0%) | 64 (53.3%) | 62 (51.7%) | 18 (15.0%) |
| HPFS | control | 366 | 67.7 (7.4) | 25.7 (3.5) | 150 (41.0 %) | 36 (10.1%) | 24 (6.6%) | 131 (35.8%) | 163 (44.5 %) | 50 (13.7 %) |
|
| ||||||||||
| PHS I | case | 133 | 59.8 (8.9) | 25.4 (3.0) | 34 (25.6 %) | 16 (12.0%) | 14(10.5%) | 63 (47.7%) | 14 (11.4%) | 20 (15.0%) |
| PHS I | control | 391 | 59.6 (8.8) | 24.6 (3.0) | 94 (24.0 %) | 46 (11.8%) | 16 (4.1 %) | 97 (25.0 %) | 45 (12.5 %) | 38 (9.7 %) |
|
| ||||||||||
| PHS II | case | 75 | 73.0 (9.0) | 26.0 (4.1) | 34 (45.3 %) | 3 (4.0%) | 13 (17.3%) | 46 (61.3%) | 30 (40.5%) | 13 (17.3%) |
| PHS II | control | 223 | 72.9 (8.8) | 25.4 (2.9) | 100 (44.8 %) | 8(3.6 %) | 15 (6.7 %) | 115 (51.6%) | 100 (44.8%) | 27 (12.1 %) |
|
| ||||||||||
| NHS | case | 113 | 60.8 (6.1) | 27.2 (5.4) | 48 (42.5 %) | 30 (26.6%) | 25 (22.1%) | 69 (61.1%) | 66 (58.4%) | 30 (26.6%) |
| NHS | control | 327 | 60.7 (6.1) | 26.5 (5.1) | 134 (41.0 %) | 67 (20.5%) | 31 (9.5 %) | 149 (45.6 %) | 188 (57.5%) | 65 (19.9%) |
|
| ||||||||||
| WACS | case | 37 | 66.4 (7.1) | 29.5 (7.1) | 28 (75.7 %) | 9 (24.3%) | 11 (29.7%) | 30 (81.1%) | 24 (64.9%) | 10 (27.0%) |
| WACS | control | 109 | 66.1 (6.8) | 28.9 (6.0) | 83 (76.2 %) | 27 (24.8%) | 19 (17.4 %) | 82 (75.2 %) | 83 (76.2 %) | 39 (35.8 %) |
|
| ||||||||||
| WHS | case | 38 | 58.8 (8.5) | 26.7 (4.6) | 7 (18.4 %) | 10 (26.3%) | 4 (10.5%) | 20 (52.6%) | 12 (31.6%) | 3 (7.9%) |
| WHS | control | 106 | 58.5 (8.3) | 26.9 (5.6) | 17 (16.0%) | 30 (28.3%) | 6 (5.7 %) | 38 (35.9 %) | 41 (38.7 %) | 15 (14.2 %) |
|
| ||||||||||
| Total | case | 516 | 64.2 (9.4) | 26.5 (4.5) | 201 (39.0 %) | 78 (15.3%) | 85 (16.5 %) | 292 (56.7 %) | 208 (41.2 %) | 94 (18.2 %) |
| Total | control | 1522 | 64.1 (9.2) | 25.9 (4.3) | 578 (38.0 %) | 214 (14.1%) | 111 (7.3 %) | 612 (40.3 %) | 620 (41.6 %) | 234 (15.4 %) |
PHS I, PHSII, HPFS participants are all men. NHS, WHS, and WACS participants are all men
Matching Factor
Association between Ion Channel Tag SNPs and SCD
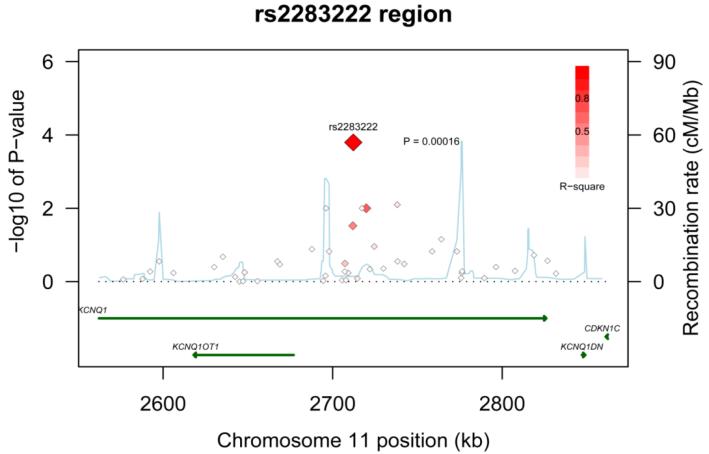
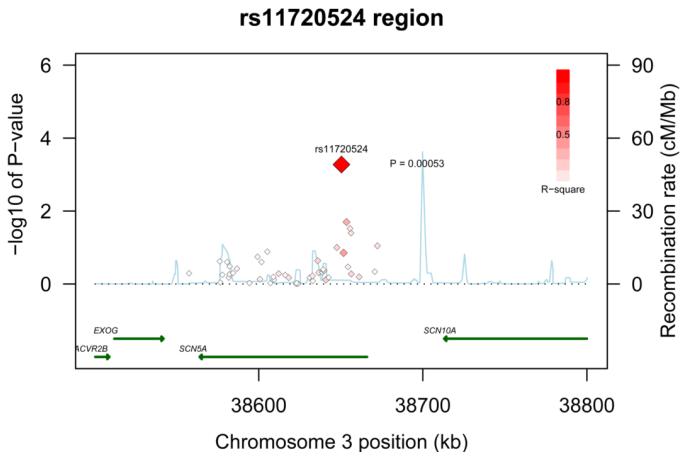
The full results from the primary fixed-effects meta-analysis for each of the 137 ion channel tag SNPs and the 5 intronic SNPs known to be associated with QT interval are outlined in the supplement (Supplementary Table 2). Fifteen of the 142 SNPs were nominally significant (P < 0.05) under an additive genetic model of inheritance in three of the genes. These included 4 out of 9 tested in KCNE1, 7 out of 68 in KCNQ1, and 4 out of 47 in SCN5A. None of the intronic SNPs known to be associated with QT interval or the tag SNPs in KCNE2 and KCNH2 reached this level of significance. To account for multiple tests within each gene, we computed the false discovery rate, and two tag SNPs (rs2283222 in KCNQ1 and rs11720524 in SCN5A) remained strong candidates for sudden/arrhythmic death (FDR = 0.01 and 0.03 respectively). SNP rs2283222 is located in intron 11 in KCNQ1 (Figure 1) and rs11720524 is located in intron 1 of SCN5A near the promoter region (Figure 2). Another SNP in KCNE1, rs11701049, was of borderline significance (FDR =0.051).
Figure 1. Regional association plot across KCNQ1 locus.
Statistical significance of associated SNPs at each locus are shown on the −log10(p) scale as a function of chromosomal position (NCBI Build 36). The primary associated SNP at the locus (rs2283222) is shown as a large red diamond. The correlation of the primary SNP to other SNPs (smaller diamonds) at the locus is shown on a scale from minimal (white) to maximal (bright red). Estimated recombination rates from HapMap and RefSeq annotations are shown.
Figure 2. Regional association plot across SCN5A locus.
Statistical significance of associated SNPs at each locus are shown on the −log10(p) scale as a function of chromosomal position (NCBI Build 36). The primary associated SNP at the locus (rs11720524) is shown as a large red diamond. The correlation of the primary SNP to other SNPs (smaller diamonds) at the locus is shown on a scale from minimal (white) to maximal (bright red). Estimated recombination rates from HapMap and RefSeq annotations are shown.
In an attempt to further refine the signals of association at KCNQ1 and SCN5A, we genotyped an additional 10 SNPs with r2 > 0.20 to rs2283222 (KCNQ1) or rs11720524 (SCN5A), respectively, and one SNP at each locus failed. None of the 8 passing finemapping SNPs were more strongly associated with sudden/arrhythmic death than the two sentinel SNPs (supplementary table 3).
Table 2 displays the individual cohort specific associations for the “at risk” allele for the two sentinel SNPs that remained associated with sudden/arrhythmic death after accounting for multiple testing. Allele frequencies did not differ among the control groups (P=0.33 for rs2283222 and P=0.84 for rs11720524), and the pooled at-risk or major allele frequency was 0.67 for the T allele in rs2283222 and 0.60 for the C allele in rs11720524. The associations of both SNPs with sudden/arrhythmic death were largely consistent across the six studies (Table 2), and the test for heterogeneity of the odds ratios was non-significant (P= 0.65 for rs2283222 and P=0.58 for rs11720524).
Table 2. Cohort Specific Associations between rs2283222 in KCNQ1 and rs11720524 in SCN5A and Sudden Cardiac Death.
Shown are the individual cohort specific and combined meta-analysis odds ratios (95% CI) for increasing copy of the “at risk” T allele of rs2283222 in KCNQ1 and C allele of rs11720524 in SCN5A from the conditional logistic regression models under an additive model of inheritance.
| Cohort | Case/ control |
rs2283222 (KCNQ1) | rs11720524 (SCN5A) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Frequency of T- Allele |
OR per T-Allele (95% CI) |
P-value | n | Frequency of C- Allele |
OR per C-Allele (95% CI) |
P-value | ||
| HPFS | case | 116 | 0.720 | 1.24 (0.90-1.72) |
0.18 | 116 | 0.599 | 1.08 (0.80-1.46) |
0.62 |
| HPFS | control | 340 | 0.671 | 339 | 0.586 | ||||
|
| |||||||||
| PHS I | case | 130 | 0.735 | 1.27 (0.92-1.75) |
0.15 | 132 | 0.686 | 1.40 (1.04-1.87) |
0.03 |
| PHS I | control | 378 | 0.689 | 385 | 0.608 | ||||
|
| |||||||||
| PHS II | case | 72 | 0.771 | 1.58 (1.02–2.44) |
0.04 | 73 | 0.623 | 1.12 (0.76-1.64) |
0.56 |
| PHS II | control | 208 | 0.676 | 212 | 0.594 | ||||
|
| |||||||||
| NHS | case | 109 | 0.734 | 1.57 (1.12-2.21) |
0.01 | 108 | 0.690 | 1.56 (1.12-2.17) |
0.008 |
| NHS | control | 320 | 0.636 | 317 | 0.585 | ||||
|
| |||||||||
| WACS | case | 37 | 0.770 | 1.69 (0.92-3.11) |
0.09 | 36 | 0.653 | 1.34 (0.77-2.35) |
0.30 |
| WACS | control | 109 | 0.665 | 106 | 0.590 | ||||
|
| |||||||||
| WHS | case | 38 | 0.697 | 0.99 (0.56 -1.73) |
0.96 | 38 | 0.724 | 1.53 (0.86-2.75) |
0.15 |
| WHS | control | 103 | 0.704 | 106 | 0.632 | ||||
|
| |||||||||
| Meta- analysis |
case | 502 | 0.736 | 1.36 (1.16-1.60) |
0.0002 | 503 | 0.658 | 1.30 (1.12-1.51) |
0.0005 |
| Meta- analysis |
control | 1458 | 0.670 | 1465 | 0.596 | ||||
Further control for age and other cardiovascular and lifestyle risk factors (Table 3) did not materially affect these associations. In the full multivariable model, the OR for sudden/arrhythmic death was 1.39 per T-allele copy (95% CI; 1.16 to 1.66, P=0.0003) for rs2283222 and 1.34 (95% CI; 1.13 to 1.58, P=0.0007) per C-allele copy for rs11720524. Results for both SNPs were also not materially altered in sensitivity analyses excluding possible SCDs (Table 3). To demonstrate the independent associations for each SNP, we included both SNPs in the fully adjusted multivariable model, and found that both SNPs remained strongly associated with sudden/arrhythmic death (OR=1.38/T-allele copy, 95%CI: 0.1.16-1.64, P= 0.0002 for rs2283222 and OR=1.36/C-allele copy, 95%CI, 1.16 – 1.60, P= 0.0002 for rs11720524).
Table 3. Multivariable and Sensitivity Analyses.
Association of sudden cardiac death with two ion channel variants. Shown are the meta-analysis odds ratios (95% CI) with increasing levels of risk factor adjustment for increasing copy of the T-allele of rs2283222 in KCNQ1 and the C-allele of rs11720524 in SCN5A.
| Genetic Variant | OR(95% CI) for All SCD (Primary Analysis) |
P-value | OR(95% CI) for Definite SCD* (N= 425) |
P-value |
|---|---|---|---|---|
| rs2283222 | ||||
| Age-adjusted | 1.36 (1.15-1.59) | 0.0002 | 1.33 (1.11-1.59) | 0.002 |
| Multivariate Model 1† | 1.37 (1.16-1.63) | 0.0003 | 1.34 (1.11-1.63) | 0.003 |
| Multivariate Model 2‡ | 1.39 (1.16-1.66) | 0.0003 | 1.36 (1.11-1.67) | 0.003 |
| rs11720524 | ||||
| Age-adjusted | 1.31 (1.12-1.52) | 0.0005 | 1.32 (1.12-1.56) | 0.001 |
| Multivariable Model 1† | 1.33 (1.13-1.55) | 0.0004 | 1.35 (1.13-1.62) | 0.0009 |
| Multivariate Model 2‡ | 1.34 (1.13-1.58) | 0.0007 | 1.35 (1.12-1.64) | 0.002 |
Sensitivity Analysis: Excluding probable sudden cardiac death cases (i.e. Unwitnessed deaths or deaths that occurred during sleep where the participant was documented to be symptom free when last observed within the preceding 24 hours)
Multivariable Model 1: Controlled simultaneously for age, smoking status (current, past, never), BMI (continuous), history of diabetes, hypertension, and high Cholesterol.
Multivariable Model 2: Controlled for variables listed above in multivariable Model 2 and family history of myocardial infarction, alcohol intake (<weekly, weekly, daily, 2 or more per day); physical activity (at least once per week) and aspirin (> or = 11 days/month
Association between Non-Synonymous SNPs and SCD
In addition to the linkage disequilibrium-based approach relating common genetic variation in the five candidate genes, we directly tested 5 nonsynonymous SNPs. Table 4 lists the results obtained in the primary fixed-effects meta-analysis for these SNPs under dominant, additive and recessive genetic modes of inheritance. None of these SNPs achieved a nominal level of significance in any of the genetic models or after multivariable adjustment (data not shown).
Table 4. Association between Ion Channel Amino Acid Polymorphisms and Sudden Cardiac Death.
Primary fixed-effects meta-analysis odds ratios and 95 percent confidence intervals for the minor allele according to additive, dominant and recessive modes of inheritance
| Gene | Amino Acid Change |
SNP ID | Genomic Context |
Allele (A/a) |
MAF | Additive | Dominant | Recessive | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | ||||||
| KCNE1 | D85N | rs1805128 | 34743550 | G/a | 0.02 | 1.18 | (0.60-2.31) | 0.63 | 1.18 | (0.60-2.31) | 0.63 | - | - | - |
| KCNE1 | S38G | rs1805127 | 34743691 | C/t | 0.35 | 1.02 | (0.88-1.19) | 0.77 | 1.14 | (0.93-1.41) | 0.20 | 0.82 | (0.59-1.12) | 0.21 |
| KCNE2 | T8A | rs2234916 | 34664669 | A/g | 0.01 | 1.12 | (0.31-4.03) | 0.87 | 1.12 | (0.31-4.03) | 0.87 | - | - | - |
| KCNH2 | K897T | rs1805123 | 150083182 | A/c | 0.23 | 0.91 | (0.76-1.10) | 0.32 | 0.89 | (0.72-1.10) | 0.28 | 1.02 | (0.59-1.77) | 0.93 |
| SCN5A | H558R | rs1805124 | 38620424 | A/g | 0.23 | 1.08 | (0.91-1.27) | 0.39 | 1.14 | (0.92-1.40) | 0.23 | 0.94 | (0.61-1.46) | 0.79 |
Discussion
In this combined nested case-control analysis from six prospective cohorts, two common intronic variants in KCNQ1 and SCN5A were significantly associated with sudden and/or arrhythmic death in individuals of European ancestry after adjustment for multiple testing. The at-risk alleles are common, with population frequencies of 67% for the T allele at rs2283222 in KCNQ1 and 60% for the C allele at rs11720524 in SCN5A. The associations between each allele and sudden and/or arrhythmic death were independent of the other and were not mediated by established CHD risk factors. In the full multivariable models including both variants, each of the at risk alleles was associated with a 39 to 41% higher odds ratio of sudden/arrhythmic death. None of the five exonic SNPs tested were associated with sudden/arrhythmic death in the combined sample.
Rare mutations in these two ion channel genes have been associated with rare Mendelian arrhythmic disorders, most notably the long QT syndrome. KCNQ1 encodes the alpha subunit of the slow component of the delayed rectifier potassium current and mutations in this gene account for the majority of long QT syndrome cases7, 9. Mutations in SCN5A, which encodes the cardiac sodium channel, are a less common cause of long QT syndrome, but are also associated with multiple other discrete phenotypes with a propensity for ventricular arrhythmias, most notably the Brugada syndrome8, 9. Recently, mutations and rare variants with functional effects in SCN5A and KCNQ1 were found in 6.5% of SIDS victims30, and rare variants in SCN5A were found in 10% of a sample of SCD cases among women within one of these populations31.
To our knowledge, this is the first study linking common variation in these ion channel genes to sudden arrhythmic death within the general population. Common variants in introns and exons of these genes have previously been documented to modify ion channel function in cellular electrophysiology studies10, 14 and to be associated with conduction intervals13 and repolarization phenotypes16, 17 on the electrocardiogram. As has been previously documented for intermediate traits such as QT interval32 and other complex phenotypes22, common variation in intronic DNA sequence exhibited stronger associations with sudden/arrhythmic death than known common exonic missense SNPs. Presumably these intronic variants exert their influence through gene expression, and although neither SNP is in strong LD with a known coding variant, further work will be required to exclude coding variants as the source of these association signals. Future studies on the genetic basis of arrhythmic risk will need to include functional analyses of non-coding variants as well as those that result in amino acid changes.
Although, we found that sequence variants at the KCNQ1 and SCN5A loci are associated with sudden/arrhythmic death risk, it is not known whether these specific SNPs are the functional variants or are in LD with a functional allele. Our fine-mapping did not find a more strongly associated correlated allele; however, deep resequencing would be required to fully define the set of all candidate alleles, and much larger sample sizes would be required to distinguish among all correlated alleles at the locus. The SNPs are also not in LD with other common intronic variants in these same genes recently found to be associated with QT interval16, 17, and five non-coding variants in KCNQ, KCNH2 and SCN5A that influence QT interval16 were not associated with sudden/arrhythmic death in these cohorts.
From a pathophysiologic standpoint, these data suggest that common variation in genes that directly influence cardiac electrophysiology may influence SCD risk at the population level. Recently, variation at a genetic locus found to influence QT interval, the nitric oxide synthase 1 adaptor protein (NOS1AP) gene, was also found to be associated with SCD in two population-based studies33. These genetic markers, once validated in other populations, may eventually be useful for SCD risk prediction in combination with markers for other pathways contributing to SCD risk such as atherosclerosis and/or autonomic instability. Regardless of such population-level application, further study of the functional consequences of these variants on cardiac electrophysiology may lead to important advances in our understanding of the mechanisms underlying SCD and could ultimately lead to novel therapeutic approaches.
Strengths of the present analysis include the large well-characterized cohorts, the combined large number of rigorously confirmed sudden and/or arrhythmic cardiac deaths, and the prospective design that allowed us to match on follow-up time at risk reducing survival bias. The study also has several limitations that deserve consideration. First, the low frequency at which SCD occurs in the general population and the inherent difficulties in documenting the circumstances surrounding out of hospital deaths in free living populations limits the number of rigorously phenotyped SCD cases even in large cohort studies. As a result, we needed to pool cases from independent cohorts to achieve adequate statistical power. Although the associations between both of the SNPs and sudden/arrhythmic death were relatively consistent across the six studies, this does not substitute for an independent replication study of comparable size and phenotype, which would be needed to establish certainty regarding the observed associations.
Second, although CHD underlies the majority of SCDs3, other pathologies account for a sizeable minority, especially among women34. Therefore, associations may differ depending on presence and type of underlying structural heart disease. The absence of autopsy data in this population based study precludes our examination of this important question. Also, we did not have power to evaluate with certainty other plausible interactions by age, sex, and cardiovascular disease status as well as gene-gene interactions. Similarly, there were variants of borderline significance that may have reached statistical significance in a larger sample size. Third, our study sample is of European ancestry, and the relevance of the 2 variants described to populations of different ancestry is unknown. Fourth, baseline electrocardiograms were not collected in these cohorts, and therefore, we did not have data on potential electrocardiographic intermediate markers, such as QRS duration and QT interval. Finally, the selective composition of the cohorts, U.S. health professionals, may limit the generalizability of the findings to other populations with differing prevalence of CVD and/or CVD risk factors.
In summary, common variants in the genes encoding the slow component of the cardiac potassium channel (KCNQ1) and the alpha subunit of the cardiac sodium channel (SCN5A) are associated with significantly increased risks of sudden and/or arrhythmic cardiac death in this combined population of European-derived men and women. These findings highlight the important role these two cardiac ion channel channels may play in propensity toward fatal ventricular arrhythmias not only in the rare primary arrhythmic disorders, but also in more common forms of sudden cardiac death. Therefore, further investigation into the functional abnormalities associated with non-coding variation in these genes may lead to important insights regarding mechanisms underlying ventricular arrhythmias in the general population. Ultimately, if these findings are replicated in other populations, variants in these genes in combination with other validated genetic, biologic and environmental SCD risk markers may advance risk prediction for this lethal and poorly predicted disease.
Supplementary Material
Acknowledgement
We are indebted to the participants in the Harvard Cohort Studies and their families for their outstanding commitment and cooperation, and to Julie Pester, Lisa Dunn, Barbara Egan, Helena Judge Ellis, Joanne Smith, and Olga Veysman for their expert assistance. We thank Gabriel Crawford for his genotyping of the SCD samples, and Patrick Ellinor for his assistance with the genotyping for rs1805123.
Sources of Funding: This project was supported by grants from the National Heart, Lung, and Blood Institute (HL068070 to Dr. Albert) and the Doris Duke Foundation (Clinical Innovation Award to Drs. Albert and MacRae). Additional support for the DNA extraction in the Women’s Health Study came from the Donald W. Reynolds Foundation. Dr. Albert is also supported by an Established Investigator Award from the American Heart Association. Dr. Newton-Cheh was supported by an award from the NIH (K23HL080025), a Doris Duke Charitable Foundation Clinical Scientist Development Award, and a Burroughs Wellcome Fund Career Award for Medical Scientists. The cohort studies were supported by grants: HL-26490, HL-34595, HL-34594, HL-35464, HL-043851, HL-46959, HL-080467 from the National Heart, Lung, and Blood Institute and CA-34944, CA 40360, CA-47988, CA55075, CA-87969, CA 97193 from the National Cancer Institute.
Footnotes
Disclosures: The authors have no potential financial relationships to disclose other than the grants above.
References
- 1.Zheng ZJ, Croft JB, Giles WH, Mensah GA. Sudden cardiac death in the United States, 1989 to 1998. Circulation. 2001;104:2158–2163. doi: 10.1161/hc4301.098254. [DOI] [PubMed] [Google Scholar]
- 2.Cupples LA, Gagnon DR, Kannel WB. Long- and short-term risk of sudden coronary death. Circulation. 1992;85:I11–18. [PubMed] [Google Scholar]
- 3.Huikuri HV, Castellanos A, Myerburg RJ. Sudden death due to cardiac arrhythmias. N Engl J Med. 2001;345:1473–1482. doi: 10.1056/NEJMra000650. [DOI] [PubMed] [Google Scholar]
- 4.Jouven X, Desnos M, Guerot C, Ducimetiere P. Predicting sudden death in the population: the Paris Prospective Study I. Circulation. 1999;99:1978–1983. doi: 10.1161/01.cir.99.15.1978. [DOI] [PubMed] [Google Scholar]
- 5.Dekker LR, Bezzina CR, Henriques JP, Tanck MW, Koch KT, Alings MW, Arnold AE, de Boer MJ, Gorgels AP, Michels HR, Verkerk A, Verheugt FW, Zijlstra F, Wilde AA. Familial sudden death is an important risk factor for primary ventricular fibrillation: a case-control study in acute myocardial infarction patients. Circulation. 2006;114:1140–1145. doi: 10.1161/CIRCULATIONAHA.105.606145. [DOI] [PubMed] [Google Scholar]
- 6.Kaikkonen KS, Kortelainen ML, Linna E, Huikuri HV. Family history and the risk of sudden cardiac death as a manifestation of an acute coronary event. Circulation. 2006;114:1462–1467. doi: 10.1161/CIRCULATIONAHA.106.624593. [DOI] [PubMed] [Google Scholar]
- 7.Ackerman MJ, Clapham DE. Ion channels--basic science and clinical disease. N Engl J Med. 1997;336:1575–1586. doi: 10.1056/NEJM199705293362207. [DOI] [PubMed] [Google Scholar]
- 8.Shah M, Akar FG, Tomaselli GF. Molecular basis of arrhythmias. Circulation. 2005;112:2517–2529. doi: 10.1161/CIRCULATIONAHA.104.494476. [DOI] [PubMed] [Google Scholar]
- 9.Roberts R. Genomics and cardiac arrhythmias. J Am Coll Cardiol. 2006;47:9–21. doi: 10.1016/j.jacc.2005.08.059. [DOI] [PubMed] [Google Scholar]
- 10.Makielski JC, Ye B, Valdivia CR, Pagel MD, Pu J, Tester DJ, Ackerman MJ. A ubiquitous splice variant and a common polymorphism affect heterologous expression of recombinant human SCN5A heart sodium channels. Circ Res. 2003;93:821–828. doi: 10.1161/01.RES.0000096652.14509.96. [DOI] [PubMed] [Google Scholar]
- 11.Anson BD, Ackerman MJ, Tester DJ, Will ML, Delisle BP, Anderson CL, January CT. Molecular and functional characterization of common polymorphisms in HERG (KCNH2) potassium channels. Am J Physiol Heart Circ Physiol. 2004;286:H2434–2441. doi: 10.1152/ajpheart.00891.2003. [DOI] [PubMed] [Google Scholar]
- 12.Bezzina CR, Verkerk AO, Busjahn A, Jeron A, Erdmann J, Koopmann TT, Bhuiyan ZA, Wilders R, Mannens MM, Tan HL, Luft FC, Schunkert H, Wilde AA. A common polymorphism in KCNH2 (HERG) hastens cardiac repolarization. Cardiovasc Res. 2003;59:27–36. doi: 10.1016/s0008-6363(03)00342-0. [DOI] [PubMed] [Google Scholar]
- 13.Bezzina CR, Shimizu W, Yang P, Koopmann TT, Tanck MW, Miyamoto Y, Kamakura S, Roden DM, Wilde AA. Common sodium channel promoter haplotype in asian subjects underlies variability in cardiac conduction. Circulation. 2006;113:338–344. doi: 10.1161/CIRCULATIONAHA.105.580811. [DOI] [PubMed] [Google Scholar]
- 14.Yang P, Koopmann TT, Pfeufer A, Jalilzadeh S, Schulze-Bahr E, Kaab S, Wilde AA, Roden DM, Bezzina CR. Polymorphisms in the cardiac sodium channel promoter displaying variant in vitro expression activity. Eur J Hum Genet. 2008;16:350–357. doi: 10.1038/sj.ejhg.5201952. [DOI] [PubMed] [Google Scholar]
- 15.Friedlander Y, Vatta M, Sotoodehnia N, Sinnreich R, Li H, Manor O, Towbin JA, Siscovick DS, Kark JD. Possible association of the human KCNE1 (minK) gene and QT interval in healthy subjects: evidence from association and linkage analyses in Israeli families. Ann Hum Genet. 2005;69:645–656. doi: 10.1046/j.1529-8817.2005.00182.x. [DOI] [PubMed] [Google Scholar]
- 16.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA, Sotoodehnia N, Smith NL, Rotter JI, Kors JA, Witteman JC, Hofman A, Heckbert SR, O’Donnell CJ, Uitterlinden AG, Psaty BM, Lumley T, Larson MG, Stricker BH. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, Ehret GB, Orru M, Pattaro C, Kottgen A, Perz S, Usala G, Barbalic M, Li M, Putz B, Scuteri A, Prineas RJ, Sinner MF, Gieger C, Najjar SS, Kao WH, Muhleisen TW, Dei M, Happle C, Mohlenkamp S, Crisponi L, Erbel R, Jockel KH, Naitza S, Steinbeck G, Marroni F, Hicks AA, Lakatta E, Muller-Myhsok B, Pramstaller PP, Wichmann HE, Schlessinger D, Boerwinkle E, Meitinger T, Uda M, Coresh J, Kaab S, Abecasis GR, Chakravarti A. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albert CM, Chae CU, Grodstein F, Rose LM, Rexrode KM, Ruskin JN, Stampfer MJ, Manson JE. Prospective study of sudden cardiac death among women in the United States. Circulation. 2003;107:2096–2101. doi: 10.1161/01.CIR.0000065223.21530.11. [DOI] [PubMed] [Google Scholar]
- 19.Chugh SS, Jui J, Gunson K, Stecker EC, John BT, Thompson B, Ilias N, Vickers C, Dogra V, Daya M, Kron J, Zheng ZJ, Mensah G, McAnulty J. Current burden of sudden cardiac death: multiple source surveillance versus retrospective death certificate-based review in a large U.S. community. J Am Coll Cardiol. 2004;44:1268–1275. doi: 10.1016/j.jacc.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 20.Hinkle LE, Jr., Thaler HT. Clinical classification of cardiac deaths. Circulation. 1982;65:457–464. doi: 10.1161/01.cir.65.3.457. [DOI] [PubMed] [Google Scholar]
- 21.Prentice RL, Breslow NE. Retrospective studies and failure time models. Biometrika. 1978;65:153–158. [Google Scholar]
- 22.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 24.Tan BH, Valdivia CR, Rok BA, Ye B, Ruwaldt KM, Tester DJ, Ackerman MJ, Makielski JC. Common human SCN5A polymorphisms have altered electrophysiology when expressed in Q1077 splice variants. Heart Rhythm. 2005;2:741–747. doi: 10.1016/j.hrthm.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 25.Sesti F, Abbott GW, Wei J, Murray KT, Saksena S, Schwartz PJ, Priori SG, Roden DM, George AL, Jr., Goldstein SA. A common polymorphism associated with antibiotic-induced cardiac arrhythmia. Proc Natl Acad Sci U S A. 2000;97:10613–10618. doi: 10.1073/pnas.180223197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prentice RL, Breslow NE. Retrospective studies and failure time models. Biometrika. 1998;65:153–158. [Google Scholar]
- 27.Whitehead A. Meta-Analysis of Controlled Clinical Trials. Wiley; New York: 2002. [Google Scholar]
- 28.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JRSSB. 1995;57:125–133. [Google Scholar]
- 30.Arnestad MCL, Rognum TO, Insolia R, Pedrazzini M, Ferrandi C, Vege A, Wang DW, Rhodes TE, George AL, Schwartz PJ. Prevalence of long-QT syndrome gene variants in sudden death syndrome. Circulation. 2007;115:361–376. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]
- 31.Albert CM, Nam EG, Rimm EB, Jin HW, Hajjar RJ, Hunter DJ, MacRae CA, Ellinor PT. Cardiac sodium channel gene variants and sudden cardiac death in women. Circulation. 2008;117:16–23. doi: 10.1161/CIRCULATIONAHA.107.736330. [DOI] [PubMed] [Google Scholar]
- 32.Rubart M, Zipes DP. Genes and cardiac repolarization: the challenge ahead. Circulation. 2005;112:1242–1244. doi: 10.1161/CIRCULATIONAHA.105.563015. [DOI] [PubMed] [Google Scholar]
- 33.Kao WH, Arking DE, Post W, Rea TD, Sotoodehnia N, Prineas RJ, Bishe B, Doan BQ, Boerwinkle E, Psaty BM, Tomaselli GF, Coresh J, Siscovick DS, Marban E, Spooner PM, Burke GL, Chakravarti A. Genetic variations in nitric oxide synthase 1 adaptor protein are associated with sudden cardiac death in US white community-based populations. Circulation. 2009;119:940–951. doi: 10.1161/CIRCULATIONAHA.108.791723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albert CM, McGovern BA, Newell JB, Ruskin JN. Sex differences in cardiac arrest survivors. Circulation. 1996;93:1170–1176. doi: 10.1161/01.cir.93.6.1170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.