The authors report on the genetic characterization of a deletion in the feline CRX gene in the Rdy cat, defining a new large-animal model for Leber congenital amaurosis, retinitis pigmentosa, and cone–rod dystrophy.
Abstract
Purpose.
To elucidate the gene defect in a pedigree of cats segregating for autosomal dominant rod–cone dysplasia (Rdy), a retinopathy characterized extensively from a clinical perspective. Disease expression in Rdy cats is comparable to that in young patients with congenital blindness (Leber congenital amaurosis [LCA] or retinitis pigmentosa [RP]).
Methods.
A pedigree segregating for Rdy was generated and phenotyped by clinical ophthalmic examination methods including ophthalmoscopy and full-field flash electroretinography. Short tandem repeat loci tightly linked to candidate genes for autosomal dominant retinitis pigmentosa in humans were genotyped in the pedigree.
Results.
Significant linkage was established to the candidate gene CRX (LOD = 5.56, θ = 0) on cat chromosome E2. A single base pair deletion was identified in exon 4 (n.546delC) in affected individuals but not in unaffected littermates. This mutation generates a frame shift in the transcript, introducing a premature stop codon truncating the putative CRX peptide, which would eliminate the critical transcriptional activation region. Clinical observations corroborate previously reported clinical reports about Rdy. Results show that the cone photoreceptor system was more severely affected than the rods in the early disease process.
Conclusions.
A putative mutation causative of the Rdy phenotype has been described as a single base pair deletion in exon 4 of the CRX gene, thus identifying the first animal model for CRX-linked disease that closely resembles the human disease. As such, it will provide valuable insights into the mechanisms underlying these diseases and their variable presentation, as well as providing a suitable model for testing therapies for these diseases.
Inherited retinal disorders are genetically heterogeneous in humans and include well over 180 mapped disease-causing loci (http://www.sph.uth.tmc.edu/retnet/ provided in the public domain by the University of Texas Houston Health Science Center, Houston, TX). Three major distinctive phenotypes have been described: retinitis pigmentosa (RP), cone–rod dystrophy (CoRD), and Leber congenital amaurosis (LCA). LCA is the most severe of the three retinal dystrophies, leading to an early onset of visual loss mainly based on rod and variable cone photoreceptor dysfunction at or near birth, nystagmus, and absent or barely recordable ERG responses.1 The progression of RP in humans has been characterized primarily as a dysfunction of the rod photoreceptors, with later involvement of the cones, gradual loss of peripheral vision leading to tunnel vision, and finally a loss of central vision.2 Last, the progression of CoRD is characterized by a primary loss of cone function with variable degrees of reduced rod function. With the latter disease, there is usually an early loss of central vision followed by peripheral vision. The clinical symptoms of these retinal degenerative disorders are widely heterogeneous. Currently, there are 18 known human genes and one genomic region that are linked to autosomal dominant retinitis pigmentosa (http://www.sph.uth.tmc.edu/retnet/). Supplementary Table S1 lists genes that we considered to be candidates for Rdy (all Supplementary Tables are available at http://www.iovs.org/cgi/content/full/51/6/2852/DC1). Of note, mutations in several of these genes have also been reported as causative of autosomal dominant CoRD/LCA and/or autosomal recessive LCA/RP.
An early-onset rod–cone dysplasia (Rdy) was first described in a domestic cat of the Abyssinian breed in the United Kingdom.3 Out-crossings with unrelated domestic shorthaired cats demonstrated an autosomal dominant form of inheritance for the disorder.3 Affected kittens displayed dilated pupils and sluggish pupillary light reflexes at 2 weeks of age and an intermittent rotatory nystagmus developed by 4 to 6 weeks of age. Funduscopic changes were observed between 8 and 12 weeks of age, with the central fundus being affected before the peripheral parts. Preliminary ERG studies showed nonrecordable responses by 8 to 12 weeks of age.4 Further studies, however, including intravitreal recordings at 4.5 weeks of age, demonstrated recordable scotopic ERGs with prolonged a- and b-wave implicit times. The ERG was a-wave dominated5 with barely discernable b-waves. Photopic responses were nonrecordable. These studies, together with light- and electron microscopic findings,4,6 indicated abnormal photoreceptor development observed first at 22 days of age, with older individuals displaying more advanced photoreceptor degeneration and thinning of the neural retina. It was concluded that Rdy-affected cats display retarded development of the photoreceptor cells, followed by degeneration when these cells initiate functional differentiation.6
A breeding colony was established and maintained over a 20-year period, providing for thorough characterization of the pathologic features and preliminary candidate gene analysis,5–8 which excluded RDS,9 PDE6G, and ROM1 and partially excluded rhodopsin (RHO) as causative of Rdy.10 Recently, an affected breeding pair (siblings) was donated to one of the authors (KN) and was moved to the University of Missouri (MU) for further characterization of the phenotype and elucidation of the genetic defect. The clinical and laboratory findings performed at MU and a short description of mutation detection have been reported.11
The development of genomic mapping resources in the domestic cat, including genetic and radiation hybrid maps12–15 and the recent 1.9X whole-genome sequence of the domestic cat and interactive web browser16,17 have provided the critical resources necessary for mapping and characterizing mutations causative of hereditary diseases18,19 and of general phenotypic interest.12,20–23 Recently, we reported the causative mutation in the CEP290 gene for late-onset retinal degeneration in the Abyssinian cat (rdAc),24 a model of human LCA25 that was first described clinically in the cat by Narfström.26 In this article, we report on the mapping and mutational analysis for Rdy, identifying another new animal model for human retinal disease.
Methods
Animals
Two Rdy-affected siblings, members of the pedigree first established by Barnett and Curtis3 were crossed with five unrelated outbred American shorthaired cats (not a recognized cat breed) producing a pedigree of 19 offspring, including 9 normal and 10 affected progeny (Fig. 1). Blood and/or tissue samples were obtained from all individuals included in the study. The progeny were phenotyped by routine clinical ophthalmic examinations, according to a University of Missouri-approved Animal Care and Use Protocol (no. 3689). Further, all experiments performed adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
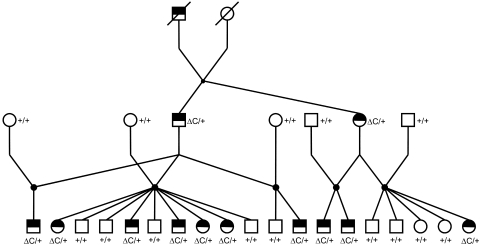
Figure 1.
Cat pedigree derived at MU, segregating for autosomal dominant rod-cone dysplasia (Rdy). White symbols: unaffected individuals; partially filled symbols: Rdy-affected individuals.
The cats were examined clinically at 6 to 8 weeks of age27–29 by indirect ophthalmoscopy and biomicroscopy after induction of mydriasis (1% tropicamide, Bausch and Lomb Inc., Tampa, FL). Funduscopic changes were documented by using a digital camera (model NM-100; Nidek Co. Ltd., Freemont, CA). Scotopic and photopic full-field flash electroretinograms (ERGs) were obtained in cats under general anesthesia (medetomidine, 0.15 mL/kg IM; Domitor, Pfizer Animal Health, Exton, PA) and ketamine, 5 mg/kg IM (Ketaset III, Fort Dodge Animal Health, Fort Dodge, IA) at 7 to 12 weeks of age. A portable ERG unit was used (HMsERG; RetVetCorp. Inc, Columbia, MO) with an automated protocol for evaluation of rod and cone function.30,31 In short, cats were dark adapted overnight, anesthetized, and prepared for the ERG in the dark (under dim red light). Either a short or a longer protocol was used, the latter protocol in five normal and six Rdy-affected cats, in which a scotopic intensity series was performed (light stimuli in steps from 0.1 mcd · s/m2 to a maximum of 25 cd · s/m2). After 10 minutes of light adaptation, a photopic intensity series was performed (10 mcd · s/m2 to 25 cd · s/m2).
Genotyping and Marker Development
Short tandem repeat (STR) loci were selected from the domestic cat genome browser, GARField (http://lgd.abcc.ncifcrf.gov/cgi-bin/gbrowse/cat/ Laboratory of Genomic Diversity, National Cancer Institute, Frederick, MD), tightly linked to 16 candidate genes and one genomic region previously reported to be mutated/linked to autosomal dominantly inherited retinitis pigmentosa in humans or other animal models (http://www.sph.uth.tmc.edu/retnet/) (Supplementary Table S1). Amplification of the STRs was performed by a touchdown PCR reaction protocol as described by Menotti-Raymond et al.32 PCR products were fluorescently labeled with M13-tailed primers, as described by Boutin-Ganache et al.33 Products were analyzed on a genetic analyzer (model 3100; Applied Biosystems, Inc., [ABI], Foster City, CA, with Genescan ver. 3.7 and Genotyper ver. 2.5 software; ABI).
Linkage Analysis
Linkage analysis computations were performed with Superlink (http://bioinfo.cs.technion.ac.il/superlink-online/ provided by Technion, The Israel Institute of Technology, Haifa, Israel).34,35 We modeled Rdy as a fully penetrant, autosomal dominant disease with disease allele frequency of 0.001. Marker allele frequencies were set as equal.
Genomic DNA Extraction and Amplification
Genomic DNA was extracted from whole blood (QIAampDNA Mini Kit; Qiagen, Valencia, CA). Genomic DNA from one sample was obtained from testes with the tissue protocol in the kit and the DNA was quantified (NanoDrop method; NanoDrop Technologies, Wilmington, DE). Genomic DNA was amplified with a touchdown procedure and sequenced as previously described.21
RNA Extraction and Generation of cDNAs
Retinal neuronal and pigment epithelial tissues harvested from Rdy-affected cats and retinal tissue from normal cats was stored at −80°C in RNA stabilizer (RNAlater; Ambion, Austin, TX). RNA was extracted with a PCR kit (RNAqueous-4; Ambion). Reverse transcription-PCR (RT-PCR) was performed (SuperScript III One-Step RT-PCR kit; Invitrogen, Carlsbad, CA), to generate amplified cDNA product. RT-PCR products were visualized on 2% agarose gels and sequenced as previously described.21 When multiple bands were present, individual bands were isolated by touching each band separately with a sterile toothpick and placing the toothpick in a microcentrifuge tube with 10 μL of water. Then, 1 μL of this solution was PCR amplified by a published touchdown procedure with PCR reaction conditions and cycling times.32 The PCR primers used for amplification of the Rdy cDNA are listed in Supplementary Table S2. Complimentary DNA (cDNA) sequences were aligned on computer (Sequencher ver. 4.8; Gene Codes Corp., Ann Arbor, MI).
Amino Acid Alignment of CRX
The predicted cat CRX amino acid sequence, as determined from genomic DNA and cDNA generated from extracted RNA was aligned to human and dog CRX sequences reported in GenBank with CLUSTALW.36
Population Genetic Survey of CRX Mutation in 19 Cat Breeds
The Rdy mutation was first characterized in a cat of the Abyssinian breed. We conducted a population genetic survey for the Rdy mutation to determine whether there was any incidence of the Rdy allele in Abyssinian and Somali (long-haired Abyssinian) cat populations. Abyssinian and Somali cats from Scandinavia (n = 69) collected for another study37 and 30 Abyssinian and Somali cats from North America were examined for the presence of the CRX Rdy allele. PCR and sequence analysis were performed on an approximate 280-bp fragment of exon 4 of the CRX gene in which the Rdy segregating risk allele (n.546delC) was identified. Primers listed in Supplementary Table S2 were used with touchdown PCR and DNA sequencing performed as described earlier. A second population survey was conducted to determine whether there was any incidence of the CRX Rdy mutation in 17 cat breeds collected for another study32 and in the LGD collection,38 including the American Curl (n = 1), American Shorthair (n = 2), American Wirehair (n = 5), Bengal (n = 16), Japanese Bobtail (n = 12), Cornish Rex (n = 20), Devon Rex (n = 17), Exotic (n = 7), Egyptian Mau (n = 8), Himalayan (n = 15), Maine Coon Cat (n = 13), Manx (n = 15), Ocicat (n = 6), Persian (n = 20), Scottish Fold (n = 16), Selkirk Rex (n = 15), and Sphynx (n = 9).
Results
A pedigree was established that segregated for the Rdy phenotype with two affected siblings from the colony in which Rdy was first described3 (Fig. 1). Nineteen progeny were generated, including 10 Rdy-affected and 9 unaffected individuals, demonstrating and verifying the previously reported autosomal dominant mode of inheritance for Rdy.3 Clinically, affected cats were easily distinguished from normal littermates by demonstrating moderately dilated, sluggish pupillary light reflexes (PLRs) by the first scheduled time of examination, at 6 to 8 weeks of age. At this time, all affected cats also showed a slight but rapidly quivering nystagmus. Further, bilateral funduscopic changes were observed including a distinct mottling and discoloration of the area centralis, followed only a few weeks later by a generalized vascular attenuation (Fig. 2). Electroretinograms (ERGs) at 7 weeks of age showed extremely low-amplitude a- and b-wave responses, both with abnormally long implicit times in scotopic conditions (Fig. 3), whereas the responses in photopic conditions were nonrecordable (flat lines; data not shown). At 11 weeks of age in affected cats, both scotopic and photopic responses were nonrecordable. Further details of the ERG findings will be given in a separate publication.
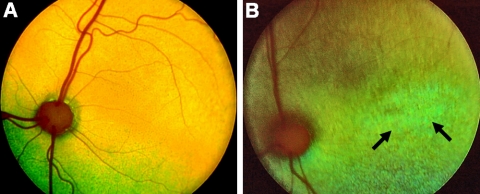
Figure 2.
Fundus photographs of a 12-week-old normal Abyssinian cat (A) and an affected mixed breed (Rdy) cat (B). The generalized grayish discoloration of the fundus in the affected cat is most notable in the area centralis, as is the generalized vascular attenuation. Arrows: marked changes in the area centralis of the affected cat.
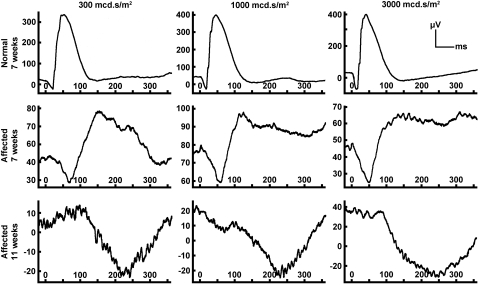
Figure 3.
Dark-adapted (scotopic) ERGs from a 7-week-old normal mixed-breed kitten showing responses to three different light intensities: 300, 1000, and 3000 mcd · s/m2. Similar recordings are shown for an Rdy-affected littermate at ages 7 and 11 weeks, obtained with identical procedures as for the normal kitten. Note the initial recordable ERG in the affected kitten at 7 weeks, with clear a- and b-wave recordings, both of low amplitude and increased implicit time. Within 4 weeks a- and b-waves from the same kitten were nonrecordable and were replaced by a late-onset negative waveform. Amplitude and implicit time calibrations are shown on the ordinate in microvolts and on the abscissa in milliseconds. Note that the amplitude calibration varies in the ordinate for most of the recordings.
A candidate gene approach was used to identify the gene defect causative of Rdy disease. Primers were designed for STRs tightly linked to 16 candidate genes and one genomic region (Supplementary Table S1) selected from the domestic cat GARField genome browser. Significant linkage was established with two markers, CRX-6 and - 7, tightly linked to the feline CRX gene on cat chromosome E2 (LOD = 5.6, θ = 0; LOD = 3.9, θ = 0.05, respectively; Table 1). Four additional STRs, adjacent to the CRX gene, also demonstrated significant linkage with Rdy (LOD 4.66–5.6; Table 1). LOD scores obtained for additional candidate genes were below the significance value, with the majority demonstrating scores of −2 or below (Supplementary Table S3). STRs positioned 5′ and 3′ of CRX display increasing θ values with increased distance from CRX (Table 1). Analyses of sequence from cDNA generated from retinal tissue and genomic DNA from affected and unaffected individuals demonstrated the presence of a 1-bp deletion in exon 4 of CRX (n.546delC), that co-segregated with Rdy (Figs. 1, 4). The deletion generates a frame shift with the subsequent introduction of a premature stop codon that would truncate 114 (38%) amino acids of the putative CRX peptide (Figs. 5, 6). All phenotypically normal cats (n = 14) were homozygous for the wild-type CRX sequence. (GenBank accession numbers for feline CRX sequences [GQ369523–GQ369525; http://www.ncbi.nlm.nih.gov/Genbank/ National Center for Biotechnology Information, Bethesda, MD.])
Table 1.
Linkage Mapping of the Domestic Cat Rdy Locus
| LOD | Theta | Cat Chrom. E2 Position (Mb)* | Dog Chrom. 1 Position (Mb) | Human Chrom. 19 Position (Mb) | |
|---|---|---|---|---|---|
| CRX-STR-1 | 1.37 | 0.1 | 4.41 | 106.60 | 58.60 |
| CRX-STR-2 | 5.30 | 0 | 8.47 | 109.92 | 54.66 |
| CRX-STR-3 | 4.66 | 0 | 9.12 | 110.47 | 53.94 |
| CRX-STR-4 | 5.57 | 0 | 9.84 | 111.10 | 53.18 |
| CRX-STR-5 | 5.63 | 0 | 9.87 | 111.12 | 53.06 |
| CRX Gene | — | — | 9.88 | 111.13 | 53.02 |
| CRX-STR-6 | 5.60 | 0 | 9.93 | 111.17 | 52.98 |
| CRX-STR-7 | 3.87 | 0.05 | 10.07 | 111.25 | 52.87 |
Markers are shown in genomic order along Fca chromosome E2 based on the cat genome browser GARfield.17 Columns 5 and 6 show the position of cat markers in the CFA build 2 genome assembly (http://www.ncbi.nlm.nih.gov/projects/mapview/stats/BuildStats.cgi?taxid=9615&build=2&ver=1) an the human (build 36) (http://www.ncbi.nlm.nih.gov/projects/mapview/stats/BuildStats.cgi?taxid=9606&build=36&ver=3) genome assembly by BLAT analysis to be orthologous to sequences flanking the feline markers. Peak LOD scores for linkage of polymorphic markers to the CRX locus and the estimated recombination fraction (θ) for each are in columns 2 and 3, respectively.
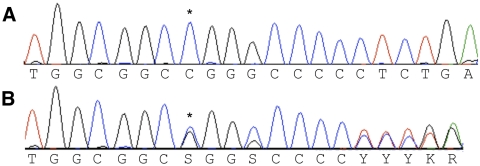
Figure 4.
(A) Genomic nucleotide sequence of the feline CRX gene, nucleotides 539-559 of the coding sequence, Rdy unaffected cat; (B) genomic sequence of homologous region in an Rdy-affected cat carrying one wild-type and one affected allele. Note the polymorphism (C/G) at the marked (*) position where the affected allele demonstrates a 1-bp deletion. Downstream of the deletion, the wild-type and affected alleles are 1 bp out of frame with one another, which can be clearly seen in the remaining sequence.
Figure 5.
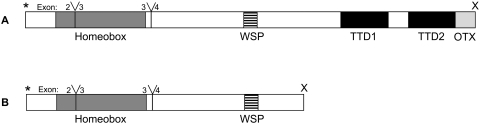
CRX protein structure in Felis catus. Comparison between the wild-type feline CRX protein (A) to the putative truncated CRX protein (B). Y, exon splice junctions; *start codon; X stop codons. Shaded boxes: protein domains, defined as the homeobox, the WSP domain, the transcriptional transactivation domains 1 and 2 (TTD1 and TTD2), and the OTX tail. Domains are drawn to scale.
Figure 6.
CLUSTAL W (1.83) multiple-sequence amino acid alignment of human CRX protein compared with the wild-type and Rdy feline CRX protein. Underscore: the conserved homeobox (DNA binding) domain; italic: the reported WSP domain; bold: the putative transcriptional activation domains. The amino acids after the second transcription domain demarcate the conserved OTX tail. X, the premature termination codon.
In addition to the single base pair deletion (n.546delC), we observed eight more SNPs in the CRX sequence, including one SNP in the 5′UTR, and seven synonymous substitutions in the coding region (Table 2), none of which segregated with the disease phenotype. These SNPs were identified based on sequence differences observed in the two affected and two unaffected individuals from which the entire coding sequence was determined. In addition, an approximately 280-bp fragment of exon 4, which included the CRX (n.546delC) SNP, was sequenced in all individuals of the pedigree and 280 individuals of a population genetic screen (see below).
Table 2.
Polymorphisms Observed in the Feline CRX Coding Region
| Polymorphism | Nucleotide Position* | Exon | Phenotype | Amino Acid Change | Effect | Rdy Cats (n = 12) Frequency (%) | Normal Cats† (n = 14) Frequency (%) | Breed Cats (n = 283) Frequency (%) |
|---|---|---|---|---|---|---|---|---|
| CC | −82 | 1 | N‡ and Rdy | 5′UTR | Unknown | NC§ | NC | NC |
| CT | 1 | N and Rdy | 5′UTR | Unknown | NC | NC | NC | |
| TT | 1 | BC‖ | 5′UTR | Unknown | NC | NC | NC | |
| CC | 468 | 4 | N, Rdy and BC | C = Ala | Synonymous | 91.7 | 21.4 | 42.2 |
| CG | 4 | N, Rdy and BC | G = Ala; C = Ala | Synonymous | 8.3 | 57.1 | 40.1 | |
| GG | 4 | N, BC | G = Ala | Synonymous | 0 | 21.4 | 17.7 | |
| TT | 492 | 4 | N, Rdy and BC | T = Ser | Synonymous | 100 | 100 | 99.3 |
| TC | 4 | BC | C = Ser; T = Ser | Synonymous | 0 | 0 | 0.7 | |
| CC | 4 | Not observed | C = Ser | Synonymous | 0 | 0 | 0 | |
| GG | 531 | 4 | N, Rdy and BC | G = Ala | Synonymous | 91.7 | 85.7 | 80.9 |
| GA | 4 | N, Rdy and BC | A = Ala; G = Ala | Synonymous | 8.3 | 14.3 | 9.2 | |
| AA | 4 | BC | A = Ala | Synonymous | 0 | 0 | 9.9 | |
| CC | 535 | 4 | N, Rdy and BC | C = Leu | Synonymous | 100 | 100 | 99.6 |
| CT | 4 | BC | T = Leu; C = Leu | Synonymous | 0 | 0 | 0.4 | |
| TT | 4 | Not observed | T = Leu | Synonymous | 0 | 0 | 0 | |
| GG | 543 | 4 | N, Rdy and BC | G = Ala | Synonymous | 100 | 100 | 99.3 |
| GT | 4 | BC | T = Ala; G = Ala | Synonymous | 0 | 0 | 0.7 | |
| TT | 4 | Not observed | T = Ala | Synonymous | 0 | 0 | 0 | |
| ΔCΔC | 546 | 4 | Not observed | Δ1 bp PTC/D1 bp PTC¶ | Trunc. protein 185 aa | 0 | 0 | 0 |
| CΔC | 4 | Rdy | A182; Δ1 bp PTC | Wild-type and trunc. prot. | 100 | 0 | 0 | |
| CC | 556 | 4 | N, Rdy and BC | A182 | Wild-type prot. | 0 | 100 | 100 |
| CC | 4 | N, Rdy and BC | C = Leu | Synonymous | 100 | 100 | 97.9 | |
| CT | 4 | BC | T = Leu; C = Leu | Synonymous | 0 | 0 | 1.4 | |
| TT | 4 | BC | T = Leu | Synonymous | 0 | 0 | 0.7 | |
| CC | 564 | 4 | N, Rdy and BC | C = Ser | Synonymous | 91.7 | 85.7 | 80.9 |
| CT | 4 | N, Rdy and BC | T = Ser; C = Ser | Synonymous | 8.3 | 14.3 | 9.2 | |
| TT | 4 | Not observed | T = Ser | Synonymous | 0 | 0 | 9.9 |
Number indicates the nucleotide position from the ATG start position in the CRX DNA.
Normal cats are mixed breed individuals in the Rdy pedigree.
N represents normal (unaffected) cats in the Rdy pedigree.
NC not enough samples were sequenced in this area to calculate a frequency.
BC represents cat(s) from the breed study.
PTC, premature termination codon at the 186th codon.
The complete feline CRX peptide is 299 amino acids in length, similar to that reported in human (Fig. 6) and dog.39 The human and canine homologues of CRX are located on chromosome 19, 53.0 Mb, and chromosome 1, 111.1 Mb, respectively. The feline CRX gene demonstrates a high degree of nucleotide sequence homology to the orthologous mammalian CRX genes, exhibiting 91%, 89%, 86%, 85%, and 90% nucleotide homology to the dog, human, rat, mouse, and cow sequences, respectively (Supplementary Table S4). The predicted feline CRX peptide demonstrates 96%, 94%, 92%, 93%, and 94% homology to the dog, human, rat, mouse, and cow CRX peptides, respectively (Supplementary Table S4). An alignment of the full-length human and feline CRX proteins and the putative feline truncated CRX peptide is presented in Figure 6. The feline CRX peptide exhibits common motifs demonstrated in the human, mouse, and dog, including the homeodomain, WSP domain, transcriptional activating domains, and OTX tail (Figs. 5, 6). Truncation of the putative CRX peptide would eliminate the OTX tail and the two transactivational domains that have been demonstrated to be critical in transcriptional activation.40,41
We observed no incidence of the Rdy mutation in a sample set of Abyssinian/Somali cats (n = 85), including 69 Scandinavian and 16 North American Abyssinian/Somali cats, the breed in which the Rdy phenotype was first described.3 An approximate 280-bp fragment of exon 4, which included the CRX (n.546delC) SNP was sequenced in these breeds. We additionally observed no incidence of the Rdy risk allele in a population survey of 267 individuals from North America representing 17 additional cat breeds (Supplementary Table S5).
Discussion
The CRX gene is a member of the OTD/OTX homeodomain protein family that is requisite for mammalian eye development and that transcribes one of a network of photoreceptor transcription factors acting to control photoreceptor gene expression, development, and maintenance.42,43 It has been demonstrated that CRX interacts with numerous photoreceptor-specific transcriptional regulators including NRL and NR2E3, QRX, and general factors in the SP family of transcription factors, as well as several chromatin-remodeling factors,44–47 (for review, see Hennig43). Peng and Chen48 theorize that CRX may act to alter the configuration of chromatin of photoreceptor genes by recruiting histone acetyl-transferases that act to enhance chromatin configurations for transcription. CRX is also expressed in the pineal gland and functions as a transcription factor for pineal-specific genes.49 CRX-knockout mice (Crx−/Crx−) are viable and fertile but lack photoreceptor outer segments, show no detectable cones, and have markedly decreased rod ERG responses.50 In addition, CRX-knockout mice demonstrate alterations in circadian rhythm cycles.50
Mutations in the CRX gene have been characterized in several human retinal diseases exhibiting a wide range of clinical phenotypes.51 First described relative to the role of CRX in autosomal dominant CoRD, mutations in the gene have since been described that are also causative of autosomal dominant RP and autosomal dominant and recessive LCA.52–55 Functional studies have not elucidated a clear relationship between the nature and position of a mutation with the severity or onset of the disease's effects (Supplementary Table S6).41
A 1-bp deletion in exon 4 of the feline CRX gene introduces a frame shift and a premature stop codon immediately downstream. The putative feline CRX peptide would contain conserved CRX motifs, including an intact homeobox, demonstrated previously as responsible for the DNA-binding and nuclear localization of the CRX protein41,56 and WSP domain (Figs. 5, 6). However, the putative feline CRX peptide would lack the two domains critical for transcriptional transactivation. Chen et al.41 have used deletion and heterologous promoter constructs to demonstrate that the C-terminal fraction of the CRX peptide (amino acids 200-284) is the critical region of the peptide involved with transcriptional activation. This region is precisely the one that is deleted in the putative feline CRX peptide (Figs. 5, 6). All previously reported frameshift mutations in the human CRX gene, other than Pro9 (1-bp ins), have also been observed in the last exon, including two truncations (Tyr191[1-bp del]; Ala196 [4-bp del]; Ala196[1-bp ins]51,57,58), which would generate putative CRX peptides that are 6 and 11 amino acids longer than the truncated feline product. Individuals with these frameshift mutations also exhibit congenital retinal blindness (Supplementary Table S6). The prospect that the allele we define is not the causative SNP is an ongoing issue in disease gene identification by linkage and by association analysis. The putative disease SNP could track the causative SNP by linkage disequilibrium (LD). However, we believe that we have provided compelling reasons to demonstrate that n.546delC is the causative mutation. We cannot formally exclude that there is another noncoding SNP traveling in complete LD with our candidate SNP that may also contribute to the phenotype.
Nonsense-mediated decay (NMD) is a mechanism first reported in yeast that leads to degradation of mRNA with premature termination codons (PTCs) and is theorized to be a mechanism that prevents the accumulation of deleterious truncated protein products.59 In humans, a dominant form of retinitis pigmentosa is reported to be a consequence of haploinsufficiency of the product of nonsense-mediated mRNA decay in the PRPF31 gene.60 As Figure 4b demonstrates, Rdy-affected individuals demonstrated a mixture of two cDNA products downstream of the CRX nucleotide position 545. This finding suggests that the deleterious CRX allele is not subject to substantial NMD and that nonsense mediated decay is unlikely to be the mechanism responsible for the disease of Rdy. In addition, a relatively benign phenotype is observed in the heterozygous Crx/null (Crx/−) mouse model, which although haploinsufficient for the CRX peptide, develops photoreceptor outer segments and slightly reduced or no reduction in ERGs relative to wild-type mice.50
Therefore, it is anticipated that a mutant CRX protein could be present in Rdy individuals from translation of the truncated mRNA, but with diminished or nonexistent transactivational activity.40,41 As the DNA recognition homeodomain is still present in the putative mutant protein, CRX interaction with cis-regulatory elements and other transcription factors, such as NRL61 could occur, essentially competing with binding and activity of the wild-type CRX transcription factor.62 It will be important to assess whether the truncated CRX peptide is present in Rdy-affected individuals.
The clinical investigations performed in the present study demonstrated that in the Rdy cat, photopic retinal function was nonrecordable at a time point when there was still some abnormal scotopic retinal function. With these findings, it appears that cone function is more severely affected than rod function, at least at the time points studied, and that it is likely that the disease can be categorized as a CoRD. Further studies are in progress that include light and electron microscopy and immunohistochemistry, for structural evaluation of rod and cone photoreceptors in young affected kittens. The early funduscopic changes appear to corroborate the functional studies, in that marked pathologic changes were first noted in the area centralis, where the concentration of cones in the feline retina reaches a peak, and can therefore in some ways be compared to the human macula. All other clinical and laboratory results from the present study corroborated previous reports relative to the Rdy model.4–6,8
In humans affected by mutations in the CRX gene, there is an association with retinopathies that share phenotypic features but vary in disease severity. The disease involves central vision loss as an early symptom. It can be manifested in infancy as nystagmus or in adulthood as an acuity disturbance and reduced visual sensitivity, especially in dim lighting conditions. In some patients, central vision is lost entirely, with only peripheral islands of functional retina retained. Thus, it is apparent that the clinical disease in Rdy cats shares some similarities with the human counterpart.
It appears that the disease mechanism in felines affected with the CRX mutation involves abnormal photoreceptor differentiation and development, with an earlier involvement of cones than of rods. Humans affected by mutations in the CRX gene all manifest maculopathy and show either equal loss of rod and cone function or greater cone dysfunction than rod dysfunction, as demonstrated by ERG. Cats with the CRX (n.546delC) mutation have an early area centralis involvement and a CoRD with blindness at the time of normal feline retinal functional maturation,6 which occurs at approximately 7 weeks of age.63 The Rdy cat may thus provide a valuable animal model for treatment strategies of early-onset primary photoreceptor disease.
In recent years, considerable progress and advancements have been made in gene therapy intervention for retinal degenerative disease.64 Improved vectors provide cell-specific targeting64 and cell-specific promoters.65 rAAV vectors, which have shown efficacy in long-term trials in the RPE65 Briard dog model,66,67 have advanced to human clinical trials with promising results.68–70
Large animal models are important in assessment of treatment modalities, particularly when gene therapy is considered, providing a perspective that cannot be gained from rodent models.71 They provide background genetic heterogeneity similar to that in humans and ultimately facilitate critical long-term studies. The availability of a large-animal model for a dominantly inherited eye disorder is particularly valuable. Gene-related therapies pose a particular challenge with regard to dominant disorders, because (1) the one functional gene may result in a haploinsufficiency of product, and (2) a truncated or aberrant protein product may be causative of the disease. Novel approaches are being explored in rodent models, including RNAi, to target host transcript while providing wild-type transcripts resistant to RNAi silencing.72–74 A second large-animal autosomal dominant disorder, the T4R opsin gene mutation in the English mastiff affects glycosylation patterns of rhodopsin and poses challenges unique from the Rdy model.75,76 The Rdy model will provide a valuable large-animal model with which to explore gene and therapeutic interventions that have potential relevance, not only in CRX-related disorders, but in exploring treatment modalities for conditions in which truncated or aberrant protein products could be causative of the disease.
Supplementary Material
Acknowledgments
The authors thank Leilani Castaner for technical support with the clinical parts of the project.
Footnotes
Supported by the University of Iowa Foundation, an unrestricted grant from Research to Prevent Blindness, and federal funds from the National Cancer Institute, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Disclosure: M. Menotti-Raymond, None; K.H. Deckman, None; V. David, None; J. Myrkalo, None; S.J. O'Brien, None; K. Narfström
References
- 1.den Hollander AI, Roepman R, Koenekoop RK, et al. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res 2008;27:391–419 [DOI] [PubMed] [Google Scholar]
- 2.Daiger SP, Bowne SJ, Sullivan LS. Perspective on genes and mutations causing retinitis pigmentosa. Arch Ophthalmol 2007;125:151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett KC, Curtis R. Autosomal dominant progressive retinal atrophy in Abyssinian cats. J Hered 1985;76:168–170 [DOI] [PubMed] [Google Scholar]
- 4.Curtis R, Barnett KC, Leon A. An early-onset retinal dystrophy with dominant inheritance in the Abyssinian cat: clinical and pathological findings. Invest Ophthalmol Vis Sci 1987;28:131–139 [PubMed] [Google Scholar]
- 5.Leon A, Hussain AA, Curtis R. Autosomal dominant rod-cone dysplasia in the Rdy cat. 2. Electrophysiological findings. Exp Eye Res 1991;53:489–502 [DOI] [PubMed] [Google Scholar]
- 6.Leon A, Curtis R. Autosomal dominant rod-cone dysplasia in the Rdy cat. 1. Light and electron microscopic findings. Exp Eye Res 1990;51:361–381 [DOI] [PubMed] [Google Scholar]
- 7.Holmes NG, Curtis R. Changes in a photoreceptor polypeptide correlating with an early-onset retinal dystrophy in the cat. Mol Cell Biochem 1991;107:111–117 [DOI] [PubMed] [Google Scholar]
- 8.Chong NH, Alexander RA, Barnett KC, et al. An immunohistochemical study of an autosomal dominant feline rod/cone dysplasia (Rdy cats). Exp Eye Res 1999;68:51–57 [DOI] [PubMed] [Google Scholar]
- 9.Gorin MB, Snyder S, To A, et al. The cat RDS transcript: candidate gene analysis and phylogenetic sequence analysis. Mamm Genome 1993;4:544–548 [DOI] [PubMed] [Google Scholar]
- 10.Gould DJ, Sargan DR. Autosomal dominant retinal dystrophy (Rdy) in Abyssinian cats: exclusion of PDE6G and ROM1 and likely exclusion of rhodopsin as candidate genes. Anim Genet 2002;33:436–440 [DOI] [PubMed] [Google Scholar]
- 11.Narfström K, David V, Deckman KH, et al. Mutation discovered in a feline model for early-onset severe retinal blinding disease. Invest Ophthalmol Vis Sci 2009:50:ARVO Abstract 3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt-Küntzel A, Nelson G, David VA, et al. Linkage map and the sex-linked orange locus-mapping of orange, multiple origins, and epistasis over non-agouti. Genetics 2009;4:181:1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menotti-Raymond M, David VA, Schäffer AA, et al. An autosomal genetic linkage map of the domestic cat, Felis silvestris catus. Genomics 2009;93:305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy WJ, Davis B, David VA, et al. A 1.5-Mb-resolution radiation hybrid map of the cat genome and comparative analysis with the canine and human genomes. Genomics 2007;89:189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis BW, Raudsepp T, Pearks Wilkerson A, et al. A high-resolution cat radiation hybrid and integrated FISH mapping resource for phylogenomic studies across Felidae. Genomics 2009;93:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pontius JU, Mullikin JC, Smith DR, et al. Initial sequence and comparative analysis of the cat genome. Genome Res 2007;17:1675–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pontius JU, O'Brien SJ. Genome Annotation Resource Fields–GARField: a genome browser for Felis catus. J Hered 2007;98:386–389 [DOI] [PubMed] [Google Scholar]
- 18.Fyfe JC, Menotti-Raymond M, David VA, et al. An approximately 140-kb deletion associated with feline spinal muscular atrophy implies an essential LIX1 function for motor neuron survival. Genome Res 2006;16:1084–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young AE, Biller DS, Herrgesell EJ, et al. Feline polycystic kidney disease is linked to the PKD1 region. Mamm Genome 2005;16:59–65 [DOI] [PubMed] [Google Scholar]
- 20.Schmidt-Küntzel A, Eizirik E, O'Brien SJ, et al. Tyrosinase and tyrosinase related protein 1 alleles specify domestic cat coat color phenotypes of the albino and brown loci. J Hered 2005;96:289–301 [DOI] [PubMed] [Google Scholar]
- 21.Ishida Y, David VA, Eizirik E, et al. A homozygous single-base deletion in MLPH causes the dilute coat color phenotype in the domestic cat. Genomics 2006;88:698–705 [DOI] [PubMed] [Google Scholar]
- 22.Eizirik E, Yuhki N, Johnson WE, et al. Molecular genetics and evolution of melanism in the cat family. Curr Biol 2003;13:448–453 [DOI] [PubMed] [Google Scholar]
- 23.Kehler JS, David VA, Schäffer AA, et al. Four independent mutations in the feline fibroblast growth factor 5 gene determine the long-haired phenotype in domestic cats. J Hered 2007;98:555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menotti-Raymond M, David VA, Schäffer AA, et al. Mutation in CEP290 discovered for cat model of human retinal degeneration. J Hered 2007;98:211–220 [DOI] [PubMed] [Google Scholar]
- 25.den Hollander AI, Koenekoop RK, Yzer S, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet 2006;79:556–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narfström K. Progressive retinal atrophy in the Abyssinian cat: clinical characteristics. Invest Ophthalmol Vis Sci 1985;26:193–200 [PubMed] [Google Scholar]
- 27.Hyman JA, Vaegan, Lei B, et al. Electrophysiologic differentiation of homozygous and heterozygous Abyssinian-crossbred cats with late-onset hereditary retinal degeneration. Am J Vet Res 2005;66:1914–1921 [DOI] [PubMed] [Google Scholar]
- 28.Vaegan, Narfström K. Electroretinographic diagnosis of feline hereditary rod cone degeneration is most efficient when amax to scotopic Imax is the only measure used. Doc Ophthalmol 2008;117:1–12 [DOI] [PubMed] [Google Scholar]
- 29.Vaegan, Narfström K. Optimal discrimination of an Abyssinian cat recessive retinal degeneration: a short electroretinogram protocol is more efficient than a long one. Clin Exp Ophthalmol 2004;32:619–625 [DOI] [PubMed] [Google Scholar]
- 30.Katz ML, Coates JR, Cooper JJ, et al. Retinal pathology in a canine model of late infantile neuronal ceroid lipofuscinosis. Invest Ophthalmol Vis Sci 2008;49:2686–2695 [DOI] [PubMed] [Google Scholar]
- 31.Narfström K, Ekesten B, Rosolen SG, et al. Guidelines for clinical electroretinography in the dog. Doc Ophthalmol 2002;105:83–92 [DOI] [PubMed] [Google Scholar]
- 32.Menotti-Raymond MA, David VA, Wachter LL, et al. An STR forensic typing system for genetic individualization of domestic cat (Felis catus) samples. J Forensic Sci 2005;50:1061–1070 [PubMed] [Google Scholar]
- 33.Boutin-Ganache I, Raposo M, Raymond M, et al. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. BioTechniques 2001;31:24–26, 28 [PubMed] [Google Scholar]
- 34.Fishelson M, Geiger D. Exact genetic linkage computations for general pedigrees. Bioinformatics 2002;18(suppl 1):S189–S198 [DOI] [PubMed] [Google Scholar]
- 35.Fishelson M, Geiger D. Optimizing exact genetic linkage computations. J Comput Biol 2004;11:263–275 [DOI] [PubMed] [Google Scholar]
- 36.Chenna R, Sugawara H, Koike T, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 2003;31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narfström K, David V, Jarret O, et al. Retinal degeneration in the Abyssinian cat (rdAc); correlation between genotype and phenotype and rdAc allele frequency in two continents. Vet Ophthalmol 2009;12(5):285–291 [DOI] [PubMed] [Google Scholar]
- 38.O'Brien SJ, Menotti-Raymond M, Murphy WJ, et al. The Feline Genome Project. Ann Rev Genet 2002:36:657–686 [DOI] [PubMed] [Google Scholar]
- 39.Akhmedov NB, Baldwin VJ, Zangerl B, et al. Cloning and characterization of the canine photoreceptor specific cone-rod homeobox (CRX) gene and evaluation as a candidate for early onset photoreceptor diseases in the dog. Mol Vis 2002;8:79–84 [PubMed] [Google Scholar]
- 40.Chau KY, Chen S, Zack DJ, et al. Functional domains of the cone-rod homeobox (CRX) transcription factor. J Biol Chem 2000;275:37264–37270 [DOI] [PubMed] [Google Scholar]
- 41.Chen S, Wang QL, Xu S, et al. Functional analysis of cone-rod homeobox (CRX) mutations associated with retinal dystrophy. Hum Mol Genet 2002;11:873–884 [DOI] [PubMed] [Google Scholar]
- 42.Hsiau TH-C, Diaconu C, Myers CA, et al. The Cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS one 2007;7:e643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res 2008;1192:114–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mears AJ, Kondo M, Swain PK, et al. Nrl is required for rod photoreceptor development. Nat Genet 2001;29:447–452 [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Xu S, Rivolta C, et al. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J Biol Chem 2002;277:43288–43300 [DOI] [PubMed] [Google Scholar]
- 46.Lerner LE, Gribanova YE, Ji M, et al. Nrl and Sp nuclear proteins mediate transcription of rod-specific cGMP-phosphodiesterase beta-subunit gene: involvement of multiple response elements. J Biol Chem 2001;276:34999–35007 [DOI] [PubMed] [Google Scholar]
- 47.Lerner LE, Peng GH, Gribanova YE, et al. Sp4 is expressed in retinal neurons, activates transcription of photoreceptor-specific genes, and synergizes with Crx. J Biol Chem 2005;280:20642–20650 [DOI] [PubMed] [Google Scholar]
- 48.Peng GH, Chen S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum Mol Genet 2007;16:2433–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X, Chen S, Wang Q, et al. A pineal regulatory element (PIRE) mediates transactivation by the pineal/retina-specific transcription factor CRX. Proc Natl Acad Sci. U S A 1998;95:1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furukawa T, Morrow EM, Li T, et al. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet 1999;23:466–470 [DOI] [PubMed] [Google Scholar]
- 51.Sohocki MM, Sullivan LS, Mintz-Hittner HA, et al. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am J Hum Genet 1998;63:1307–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S, Wang QL, Nie Z, et al. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 1997;19:1017–1030 [DOI] [PubMed] [Google Scholar]
- 53.Freund CL, Gregory-Evans CY, Furukawa T, et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 1997;91:543–553 [DOI] [PubMed] [Google Scholar]
- 54.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 1997;91:531–541 [DOI] [PubMed] [Google Scholar]
- 55.Swaroop A, Wang QL, Wu W, et al. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum Mol Genet 1999;8:299–305 [DOI] [PubMed] [Google Scholar]
- 56.Fei Y, Hughes TE. Nuclear trafficking of photoreceptor protein crx: the targeting sequence and pathologic implications. Invest Ophthalmol Vis Sci 2000;41:2849–2856 [PubMed] [Google Scholar]
- 57.Rivolta C, Peck NE, Fulton AB, et al. Novel frameshift mutations in CRX associated with Leber congenital amaurosis. Hum Mutat 2001;18:550–551 [DOI] [PubMed] [Google Scholar]
- 58.Swain PK, Chen S, Wang QL, et al. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron 1997;19:1329–1336 [DOI] [PubMed] [Google Scholar]
- 59.Wen J, Brogna S. Nonsense-mediated mRNA decay. Biochem Soc Trans 2008;36:514–516 [DOI] [PubMed] [Google Scholar]
- 60.Rio Frio T, Wade NM, Ransijn A, et al. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J Clin Invest 2008;118:1519–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitton KP, Swain PK, Chen S, et al. The leucine zipper of NRL interacts with the CRX homeodomain: a possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem 2000;275:29794–29799 [DOI] [PubMed] [Google Scholar]
- 62.Kitiratschky VB, Nagy D, Zabel T, et al. Cone and cone-rod dystrophy segregating in the same pedigree due to the same novel CRX gene mutation. Br J Ophthalmol 2008;92:1086–1091 [DOI] [PubMed] [Google Scholar]
- 63.Hamasaki DI, Maguire GW. Physiological development of the kitten's retina: an ERG study. Vision Res 1985;25:1537–1543 [DOI] [PubMed] [Google Scholar]
- 64.Colella P, Cotugno G, Auricchio A. Ocular gene therapy: current progress and future prospects. Trends Mol Med 2009;15:23–31 [DOI] [PubMed] [Google Scholar]
- 65.Alexander JJ, Umino Y, Everhart D, et al. Restoration of cone vision in a mouse model of achromatopsia. Nat Med 2007;13:685–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Acland GM, Aguirre GD, Ray J, et al. Gene therapy restores vision in a canine model of childhood blindness. Nat Genet 2001;28:92–95 [DOI] [PubMed] [Google Scholar]
- 67.Narfström K, Vaegan, Katz M, et al. Assessment of structure and function over a 3-year period after gene transfer in RPE65−/− dogs. Doc Ophthalmol 2005;111:39–48 [DOI] [PubMed] [Google Scholar]
- 68.Bainbridge JW, Smith AJ, Barker SS, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med 200822;358:2231–2239 [DOI] [PubMed] [Google Scholar]
- 69.Cideciyan AV, Aleman TS, Boye SL, et al. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc Natl Acad Sci U S A 2008;105:15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maguire AM, Simonelli F, Pierce EA, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med 2008;358:2240–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casal M, Haskins M. Large animal models and gene therapy. Eur J Hum Genet 2006;14:266–272 [DOI] [PubMed] [Google Scholar]
- 72.Tam LC, Kiang AS, Kennan A, et al. Therapeutic benefit derived from RNAi-mediated ablation of IMPDH1 transcripts in a murine model of autosomal dominant retinitis pigmentosa (RP10). Hum Mol Genet 2008;17:2084–2100 [DOI] [PubMed] [Google Scholar]
- 73.O'Reilly M, Millington-Ward S, Palfi A, et al. A transgenic mouse model for gene therapy of rhodopsin-linked Retinitis Pigmentosa. Vision Res 2008;48:386–391 [DOI] [PubMed] [Google Scholar]
- 74.Chadderton N, Millington-Ward S, Palfi A, et al. Improved retinal function in a mouse model of dominant retinitis pigmentosa following AAV-delivered gene therapy. Mol Ther 2009;17:593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kijas JW, Cideciyan AV, Aleman TS, et al. Naturally occurring rhodopsin mutation in the dog causes retinal dysfunction and degeneration mimicking human dominant retinitis pigmentosa. Proc Natl Acad Sci U S A 2002;99:6328–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu L, Jang GF, Jastrzebska B, et al. A naturally occurring mutation of the opsin gene (T4R) in dogs affects glycosylation and stability of the G protein-coupled receptor. J Biol Chem 2004;279:53828–53839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silva E, Yang JM, Li Y, et al. A CRX null mutation is associated with both Leber congenital amaurosis and a normal ocular phenotype. Invest Ophthalmol Vis Sci 2000;41(8):2076–2079 [PubMed] [Google Scholar]
- 78.Dharmaraj SR, Silva ER, Pina AL, et al. Mutational analysis and clinical correlation in Leber congenital amaurosis. Ophthalmic Genet 2000;21(3):135–150 [PubMed] [Google Scholar]
- 79.Tzekov RT, Sohocki MM, Daiger SP, et al. Visual phenotype in patients with Arg41Gln and ala196+1bp mutations in the CRX gene. Ophthalmic Genet 2000;21(2):89–99 [PubMed] [Google Scholar]
- 80.Sankila EM, Joensuu TH, Hamalainen RH, et al. A CRX mutation in a Finnish family with dominant cone-rod retinal dystrophy. Hum Mutat 2000;16(1):94 [DOI] [PubMed] [Google Scholar]
- 81.Sohocki MM, Daiger SP, Bowne SJ, et al. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum Mutat 2001;17(1):42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tzekov RT, Liu Y, Sohocki MM, et al. Autosomal dominant retinal degeneration and bone loss in patients with a 12-bp deletion in the CRX gene. Invest Ophthalmol Vis Sci 2001;42(6):1319–1327 [PMC free article] [PubMed] [Google Scholar]
- 83.Jacobson SG, Cideciyan AV, Huang Y, et al. Retinal degenerations with truncation mutations in the cone-rod homeobox (CRX) gene. Invest Ophthalmol Vis Sci 1998;39:2417–2426 [PubMed] [Google Scholar]
- 84.Freund CL, Wang QL, Chen S, et al. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet 1998;18(4):311–312 [DOI] [PubMed] [Google Scholar]
- 85.Perrault I, Hanein S, Gerber S, et al. Evidence of autosomal dominant Leber congenital amaurosis (LCA) underlain by a CRX heterozygous null allele. J Med Genet 2003;40(7):e90 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.