Abstract
To push the 100-plex envelope of suspension array technology, we have developed fully automated methods to acquire multi-spectral images of multiplexed quantum-dot (QD) encoded microspheres, to segment them in the images, to classify them based on their color code, and to quantify the multiplexed assays. Instead of coding microspheres with two colors and n levels, microspheres were coded with n colors and two levels (present or absent), thus transforming the classification problem from analog to digital. Images of multiplexed microspheres, sedimented at the bottom of microwells, were acquired through a tunable filter at the peak luminescence wavelength of each QD coding species in the system and the assay label wavelength. Another image of the light scattered from microspheres was captured in the excitation bandwidth that was utilized to localize microspheres in multispectral luminescence images. Objects in the acquired images are segmented and luminescence from each identified microsphere in each channel is recorded, based on which the ‘color code’ of each microsphere is determined by applying a mathematical model and a classification algorithm. Our image analysis procedures could identify and classify microspheres with more than 97% accuracy, and the assay CVs were under 20%. These proof-of-principle results demonstrate that highly multiplexed quantification of specific proteins is possible with this rapid, small-sample volume format.
Keywords: multiplexed assays, suspension arrays, microsphere-based arrays, quantum dots, microscopy, image analysis, imaging-based systems, imaging cytometry
INTRODUCTION
In response to emerging needs to perform highly parallel quantification of specific proteins, a number of rapid, low cost, small sample volume analytical methods have been developed (1,2). Planar protein arrays have been designed which can analyze many thousands of proteins in parallel (3,4). They are printed on glass slides and each array element is identified by its location within the array’s grid. However, planar arrays require immobilization of each protein on the surface with the same linking chemistry under common conditions, which limits ones ability to optimize epitope presentation and binding site density. Moreover, since each array spot is 100-200μm in diameter, these arrays are limited to less than 104 assays/cm2 that require relatively large sample volumes for highly multiplexed assays.
Microsphere-based suspension arrays were developed to overcome the disadvantages of planar arrays (5-7). Microspheres offer the potential of utilizing different surfaces so each protein can be coated under optimal buffer, pH, and salt concentration conditions. They also allow assays to be performed in suspension thereby accelerating kinetics, reducing sample volumes and allowing the reaction to proceed under homogeneous conditions. With suspension arrays, microspheres are coated in bulk, producing a large stock of immobilized proteins that can be tested for functionality prior to use in an array. Such arrays have been successfully used for screening and diagnostic applications such as cytokine measurements (8-10), kinase testing (11), cystic fibrosis screening (12), and proteomics and immunoassays (13-15).
With current suspension array technology, microspheres are encoded with fluorescent dyes where the concentrations of fluorophores identify each array element. With n levels of two spectrally distinct dyes, n2 unique analog codes can be generated. Most commonly used formats analyze microspheres in suspension with flow cytometry (16,17). Luminex (Austin, TX) has developed a Luminex 200™ suspension array system based on two spectrally distinct fluorophores with ten distinct levels thereby theoretically providing a 100-plex suspension array. However, flow cytometers typically can read only 3-4 fluorescent wavelengths that limits the number of codes which can be discriminated. Flow readers have additional drawbacks of short signal integration times, long read-out time per sample and inability to re-read a microsphere. In view of these disadvantages, imaging based screening systems have been developed that can analyze assayed microspheres immobilized (18) or sedimented (19) on planar surfaces. These systems have the capability to read an unlimited number of wavelengths in visible spectrum, thereby allowing for the generation and use of many more color codes if a high degree of multiplexing is required.
In view of aforementioned advantages, we developed an imaging-based, fully-automated, high-throughput screening (HTS) system based on QD-encoded microspheres. In this paper, we describe the image analysis procedures required to segment and classify microspheres, and quantify levels of specific proteins in the assay. Successful localization and classification of QD-encoded microspheres present in a multiplexed assay mixture demonstrated the capability of our system to be used in highly multiplexed ultra-HTS applications.
QDs are ideal labels for use in highly multiplexed microsphere-based assays. Their unique properties, which include common broad-band UV excitation, size-tunable luminescence wavelength, higher quantum yields, narrower emission bandwidths (~30nm), and higher photostability, make them preferable to fluorescent dyes for generating color codes. Nie and coworkers first employed QDs to encode microspheres in multiplexed assays (20), and others have recently extended their approach to other applications (21-23).
In all previous reports, analog coding schemes have been employed with n (>2) levels of m dyes to generate mn codes. Because fluorescent dyes have broad emission spectra, the number of dyes with well-separated emission spectra has been limited to two. Because only 3-4 levels per decade can be differentiated with acceptable accuracy, the number of levels has been limited to ten. Thus, the number of distinguishable color codes that can be generated with these schemes is limited to 102. By using multiple QD species with narrow emission spectra in conjunction with a tunable filter, the number of coding wavelengths can be large so that only two levels are sufficient to generate a large number of codes. By having only to determine if each color is present or absent, identification is thus converted from an analog to a digital process.
To demonstrate the feasibility of a 24 coding scheme, in our system microspheres were encoded with four QD species with only two levels (present, 1 or absent, 0), and target molecules were tagged with a fifth QD species (reporter label). Mixtures of multiplexed sedimented microspheres were then analyzed by reading luminescence through a tunable filter at the peak emission wavelength of each QD species. Microspheres were segmented, and localized by processing the scattered light image (SLI) that revealed their location regardless of their QD color code. Identification of the color code (classification) indicated the capture probe on each microsphere and the intensity of reporter QD was used to quantify the amount of captured target.
MATERIALS AND METHODS
Microspheres
Polystyrene microspheres (diameter~5μm) were encoded with four QD species (lot#316 from Crystalplex Corporation, Pittsburgh, PA) with emission peaks centered at 525nm, 575nm, 620nm and 675nm in a binary manner wherein each of the QD species could either be present, 1, or absent, 0. Each QD species is represented by QDxxx where ‘xxx’ represents its emission wavelength in nm. Color codes of the microsphere populations are specified as ‘x1 x2 x3 x4’, where xi denotes the level (1 or 0) of ith QD species in the microspheres in the order QD525, QD575, QD620 and QD675. For example, 1001 code indicates presence of QD525 and QD675, and absence of QD575 and QD620 in the microsphere.
Samples of all the classes of microspheres except 0000 were supplied as 1ml DI H2O suspension (with 0.01% Sodium Azide), with 5mg/ml concentration. The 0000 microspheres were supplied as 1ml DI H2O suspension (with 0.1% Sodium Azide), with 250μg/ml concentration. Reactive group on each microsphere surface was COOH.
Streptavidin coating and reporter label (QD497) tagging
We covalently linked SAv to six classes (0000, 0001, 0010, 0100, 1000 and 1001) of QD-encoded microspheres with EDC coupling reaction (24). For each class of microspheres, we started with 100μl of 0.5% w/v microspheres and finally achieved a volume of 50μl of SAv conjugated microspheres at a concentration of 1% w/v. We further tagged a fraction of SAv coated 0000 and 0001 microspheres with QD497 by incubating 10μl of the suspension containing microspheres from each class with 4μl of 14.8μM solution containing biotinylated QD497 (Evident Technologies, Troy, NY) using Na-P (pH~5.8) binding buffer (total volume = 100μl). After incubation, we centrifuged the suspension, removed the supernatant, and resuspended the ‘labeled’ microspheres in 20μl DI H2O at 0.5% w/v concentration.
Assay system
For our model biotin-streptavidin (SAv) assay system, we mixed the suspension of QD497 labeled 0001 microspheres, *0001 (‘*’ marks the presence of reporter label QD497) in equal proportions with microsphere suspensions for four other classes (0010, 0100, 1000 and 1001) and diluted the mixture with DI H2O to achieve a final concentration of 0.02% w/v. We filled a microwell (9mm diameter; 8mm deep) with coverslip bottom with 45μl of this suspension mixture and allowed the microspheres to settle for one hour.
In our assay system, we selected 0001 microspheres for tagging because spillover luminescence from QD497 into 525nm channel due to spectral overlap could make them potentially be mistaken as 1001 microspheres. Accordingly, performance of our analysis would be judged mainly on the basis of correct classification of *0001 and 1001 microspheres from the mixture.
Calibrator sets
To train our image analysis procedures, we prepared eight calibrator sets of microspheres (SAv coated only: 0000, 0001, 0010, 0100, 1000 and 1001; QD497 labeled: *0000 and *0001) at a concentration of 0.02% w/v each. An aliquot of 45μl from each set was pipetted into distinct microwells, and the microspheres were allowed to settle for one hour. These calibrator sets were utilized to estimate relative concentrations of different QD species in any given microsphere based on its intensities in different channels based on a mathematical model.
Standard curve
A 10-point standard curve for our SAv-biotin assay system was generated based on which the reporter signal in the assay could be correlated to the amount of analyte (QD497-tagged biotin) present in the sample. Towards this purpose, we first performed serial dilutions of biotinylated QD497 solution (stock concentrations: 14.8μM-0.029μM; dilution factor: 2). Next, we mixed 4μl of each biotinylated QD497 dilution (final concentration: 592nM-1.16nM) with 10μl of the 1% w/v suspension of SAv coated 0001 microspheres (same class was tagged in our assay system) and 86μl Na-P (pH~5.8) binding buffer (total volume = 100μl), in duplicates. After incubation, we centrifuged the suspension, removed the supernatant, and resuspended the ‘labeled’ microspheres in 500μl DI H2O at 0.02% w/v concentration. An aliquot of 45μl from each set was pipetted into distinct microwells, and the microspheres were allowed to settle for one hour.
Imaging system
We used an automated epifluorescence microscope equipped with a tunable filter for acquiring images of sedimented microspheres. The excitation optics consisted of an AttoArc 100-W mercury arc lamp (Carl Zeiss Microimaging, Inc., Thornwood, NY) in combination with a band pass high-Q interference filter (Chroma Technology, Brattleboro, VT) with allowable range of 360nm-460nm and a low pass dichroic mirror with the cutoff wavelength at ~460nm. The emission optics included a Zeiss Fluar 40×/1.30 NA oil objective, a VariSpec™ liquid crystal tunable band pass filter (LCTF) (Cambridge Research & Instrumentation Inc., Woburn, MA) and the QImaging (Burnaby, BC, Canada) Retiga 1350 EX cooled charge-coupled device (CCD) digital camera that is equipped with a 1392×1040 chip with a 6.45μm pixel size and on-chip binning up to 4×4. The operating range of wavelengths for the LCTF was 400nm-720nm and the allowable bandwidth was ~20nm. A demagnification factor of 0.72 was associated with the relay lenses in the LCTF. The CCD had a 12-bit A/D converter, low dark current, and over 50% quantum efficiency in the visible range. Other specifications of our automated imaging system and the workstation interfaced with the microscope have been discussed in ref. 18.
Image acquisition
Images of microspheres sedimented onto coverslip bottoms of microwells were brought into focus with an auto-focusing program that maximized a contrast-based function (18). We acquired images at five wavelengths corresponding to the emission peaks of reporter QDs (QD497) and coding QDs (QD525, QD575, QD620, and QD675). In addition, we also acquired scattered light image (SLI) at 455nm, which used backscattered light to locate the microspheres in the field-of-view (FOV). The wavelength for acquiring SLI was empirically selected based on lower exposure time required for image acquisition and higher signal to background ratio (data not shown).
We acquired images of each FOV in the following order of wavelengths: 497nm, 525nm, 455nm, 575nm, 620nm and 675nm. SLI at 455nm was acquired in the middle of the sequence to minimize the effects of lateral displacement of microspheres (if any) due to local convection currents.
We acquired all the images with 2×2 binning in 12-bit format and then converted them to 16-bit before storing as .tif files. Images had a final resolution of 516×676 pixels with the final pixel width at the sample of 0.45μm.
Image analysis
Image analysis was significantly simplified by using SLI for identifying zones that contained individual microspheres in a FOV. Zones of interest could thus be defined from a single image, and only luminescence data need to be extracted from images at other wavelengths. Having zones larger than microsphere dimensions allows room for greater lateral displacement of microspheres in sparser images. Aptly, for denser images, where microspheres zones were smaller, lateral movement was observed to be small.
Image analysis consisted of following sequence of steps: preprocessing, zone identification, thresholding, area filtering, and signal extraction.
Preprocessing images
As a first step, we corrected luminescent images in all five channels for uneven illumination by utilizing ‘pseudo-flood’ images (25) corresponding to each wavelength (flattening correction). Pseudo-flood images were images of densely packed microspheres containing only single QD species. We also corrected SLI with a psuedo-flood image constructed from dense images of blank (0000) microspheres, acquired at 455nm. The microsphere suspensions utilized for this purpose were at 0.2% w/v concentration. We divided intensities in each image by intensities in the corresponding pseudo-flood image pixel-by-pixel, and obtained an image with essentially uniform illumination across the FOV.
Microsphere zone identification
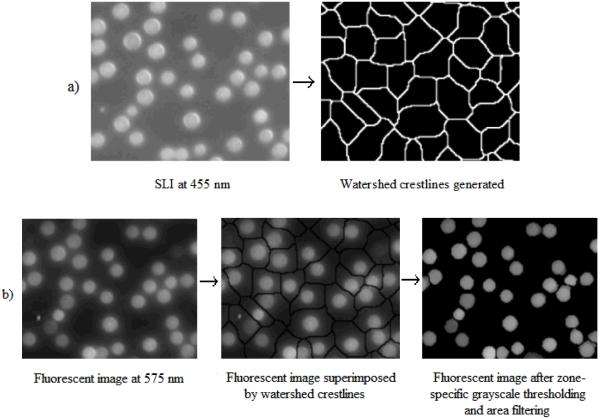
Our fully-automated watershed-based segmentation algorithm (25) was applied to the SLI to define watershed crestlines in the image. These crestlines were then superimposed on the luminescent images to form zones that presumably contained single microspheres (Fig. 1a).
Figure 1.
a) Microsphere zones defined by watershed crestlines by applying watershed-based segmentation algorithm to SLI at 455nm; b) identification and localization of microspheres in a fluorescent image (at 575nm) by superimposing watershed crestlines identified in a), followed by zone-specific grayscale thresholding and area filtering
Grayscale thresholding
After identifying microsphere zones, we applied Otsu’s thresholding (26) to each identified zone in the luminescent images. This zone-specific grayscale thresholding removed the background from each zone leaving only the foreground pixels, which presumably belonged to the microsphere in that zone.
Area filtering
After picking foreground objects by zone-specific grayscale thresholding, we area-filtered the segmented image to remove isolated small clusters of pixels, which did not belong to microspheres. These objects could either be small contaminants or background pixels erroneously assigned to the foreground in the thresholding step. We removed such clusters by setting a lower limit on the size of the permissible clusters as 10% of the average size of the microspheres in the system
Fig. 1b illustrates localization of microspheres in a typical luminescent image based on microspheres zones defined in Fig. 1a.
Intensity estimation
After localizing microspheres in the luminescent images, we recorded the intensity of each microsphere in each image as the signal averaged across the interconnected pixels constituting the identified object (normalization across different sizes). This mean intensity was further divided by the exposure time required for image acquisition to yield intensity per unit area (pixel) per unit exposure time (IPAT) in each channel (normalization across different exposure times). Expectedly, we observed that the CVs in IPAT values were lower than CVs in either total intensity values or mean intensity values across the objects identified in an image (data not shown).
We identified and localized the microspheres in the multispectral images, acquired for the assay mixture, for each one of the calibrator sets 0001, *0001, 0010, 0100, 1000, 1001, 0000 and *0000, and for generation of the standard curve, and characterized the signal from all of them by calculating IPAT values at all five wavelengths (497nm, 525nm, 575nm, 620nm and 675nm).
Estimation of relative number of QDs, r
Since IPAT values for objects in an image acquired at the emission peak (primary channel) of one QD species would have contributions from off-peak wavelengths (secondary channels) of other QD species that overlap with band pass, we first estimated the relative concentrations of each QD species present in each microsphere with respect to the average concentration of the respective QD species in the microspheres with only that species present, based on a mathematical model. Unlike IPAT values, the relative concentration of a QD species is not influenced by other QD species that may or may not be present in the microsphere.
Mathematical model for calculating relative concentration of QDs
The primary channel IPAT signal for a microsphere having multiple QD species can be broken down into three components: a) background, b) signal from QDs with emission peak in primary channel, and c) signal due to spillover from QDs with emission peaks not in primary channel. Accordingly, in an assay system with n QD species and with n detection channels corresponding to their emission peaks at which images are acquired, if the concentration of QDs with peak emission in channel j in a given microsphere is denoted by qj (j∈ [1, n]), the IPAT signal detected in any detection channel i (Si) can be represented mathematically by the following linear model:
or,
| (1) |
where i ∈ [1, n] denotes the primary (detection) channel, j∈ [1, n] denotes the emission peak channel and index of corresponding QD species, b is the background signal, q represents the concentration of QDs, αii denotes the primary channel coefficient (when j = i indicating that the emission peak lies in the primary channel), and αij denotes the secondary channel coefficient (when j ≠ i) representing the spillover of emission from QDs with emission peak in channel j into the primary channel i. The concentration of QDs of a given species in a microsphere can be expressed relative to their average concentration in a set of standard microspheres containing QDs of only that particular species, leading to:
| (2) |
where is the average concentration of jth species (j∈ [1, n]) of QDs in the standard set with only jth species present and is the ratio of concentrations of QDs of jth species in a given microsphere to its average concentration in the standard set with only jth species present. As a corollary, for standard set with only ith species present, mean value of rj across the microspheres in the set equals to 1 for the QD species that is present (when j = i) and 0 for all other QD species that are absent (when j ≠ i). If we denote the mean IPAT signal in any channel j (∈ [1, n]) from the standard microsphere containing only ith species as and substitute terms in Eq. (2) with Aji -bi (= Cij), we get
| (3) |
Thus, the signal vector S from an arbitrary microsphere with relative numbers of QDs can be written in the matrix format as:
| (4) |
where r = [ri]n×1 is a column vector containing relative concentrations of each of the n QD species with respect to the standard set of microspheres with only corresponding QD species present, [Cij]n×n = [Aji -bi]n×n, and b = [bi]n×1. Eq. (4) is a general form for estimating the signals in all detection channels from microspheres coded with any arbitrary combination of any number of QD species. Conversely, by measuring signals in individual detection channels, we can calculate the relative concentrations for different QD species in an arbitrary class of microspheres using,
| (5) |
Estimation of C and b from calibrator sets
The calibration matrix, C, and the background vector, b in Eq. (5) were estimated from calibrator sets that contained either no QDs or only single QD species. For the proof-of-principle case, our system consisted of 5 QD species (n = 5; 4 coding, 1 reporter) with the subscripts i, j ∈ [1,5] representing the emission wavelengths 497nm, 525nm, 575nm, 620nm and 675nm, in order. Hence, S = [Si]5×1 represents a vector formed by the IPAT values at five wavelengths for the microsphere under consideration, b = [bi]5×1 represents the average background at five wavelengths calculated as mean IPAT values for 0000 microspheres. We computed elements [Aji]5×5 by calculating mean IPAT values of microspheres from images of individual calibrator sets *0000, 1000, 0100, 0010 and 0001, in all five detection channels. Calibration matrix C = [Cij]5×5 = [Aji - bi]5×5 represents the mean IPAT values (Aji) in channel i (primary channel), for the microspheres with only jth QD species present minus the background (bi) in channel i (primary channel) (i, j ∈ [1,5]). Each element in the column vector r (= [ri] 5×1) represents the ratio of the concentration of ith QD species in the microsphere under consideration to the average concentration of same QD species in the microspheres with only that species present.
Estimation of r for microspheres in calibrator sets and assay mixture
After localizing microspheres in multispectral images of the assay mixture, their individual color codes were determined based on the comparison of their individually calculated r vectors with the r vectors of microspheres in calibrator sets. We used Eq. (5) to calculate r for microspheres from calibrator sets from individual classes *0000, 0001, 0010, 0100, 1000, 1001 and *0001, and for the microspheres in the assay mixture.
Classification
In order to classify microspheres in the assay mixture containing classes *0001, 0010, 0100, 1000 and 1001, we applied quadratic discriminant analysis (QDA) based on r values for coding QDs (rQD525, rQD575, rQD620 and rQD675). QDA classifies a set of observations in n-dimensional space into predefined classes based on the corresponding calibrator sets (27). It utilizes a multivariate quadratic discriminant function and assigns an observation to the class for which the function has highest value.
In order to judge the performance of this 4-channel classification for our system, we analyzed and compared rQD497 distributions for microspheres identified as *0001 and 1001 from the assay mixture. The efficiency of our assay system and associated analysis depends on the relative accuracy of 4-channel classification in correctly identifying microsphere color codes. Since all 0001 microspheres in our assay mixture were tagged with QD497, a measure of the efficiency was defined as the fraction of identified 0001 microspheres that were identified to be tagged with QD497. Only those microspheres in the assay mixture that have rQD497 greater than a critical threshold value would be declared as ‘tagged’. The method of setting this threshold value in the current study is based on rQD497 distributions for microspheres in *0001 and 0001 calibrator sets and is explained further in the next section. Thus, mathematically,
| (6) |
RESULTS
Computing b and C
Instead of calculating b and C directly, we performed bootstrapping analysis to estimate elements of these matrices and associated variability. For estimating b, we randomly picked 1,000 microspheres from the image of 0000 microspheres and averaged the IPAT values across all microspheres in each of the five channels. We performed this calculation 100 times and computed elements of b as mean values with associated standard deviation as:
For computing the calibrator matrix C, every time we randomly picked 1,000 microspheres from 0000 class and subtracted their mean IPAT values in each channel from mean IPAT values in the corresponding channel for 1,000 microspheres randomly picked from class with only one QD species present. We performed this calculation 100 times and computed elements of C as mean values with associated standard deviation as:
Elements of b and C showed little variability across different sets of 1,000 microspheres. CVs for elements in b were all less than 1%. For the primary channels along matrix diagonal in C, CVs were 0.18%, 1.01%, 1.09%, 0.90% and 0.81%. For most of the secondary channels (non-diagonal elements) in C too, CVs were all under 5%. Most of these CVs were well under the corresponding standard deviations divided by (standard error). Thus, we inferred that 1,000 microspheres from each calibrator set were sufficient to generate reliable estimates of b and C.
Computing r
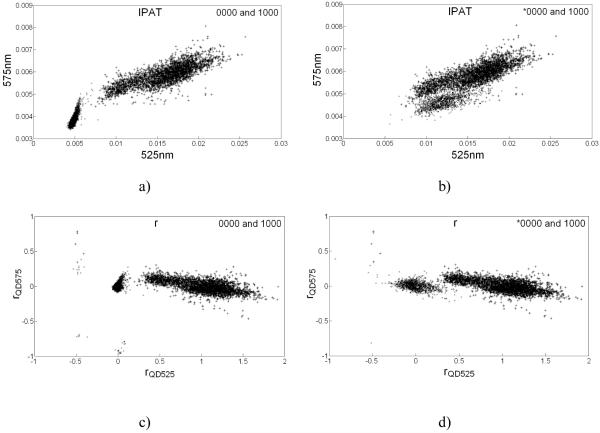
We calculated r for microspheres in calibrator sets and in assay mixture using Eq. (5) based on b and C calculated from IPAT values extracted from the multispectral luminescent images. The necessity for transformation of IPAT signals into r is evident from the scatter plot between IPAT values in channels 525nm and 575nm wherein *0000 and 1000 clusters overlapped and hence were difficult to distinguish (Fig. 2b). However, in the scatter plot in r space (rQD575 vs. rQD525) they formed distinct clusters and hence were relatively easier to demarcate (Fig. 2d). Scatter plots for 0000 and 1000 also show the location of untagged 0000 microspheres with respect to 1000 microspheres in r and IPAT spaces (Figs. 2a and 2c). It was conceived that spectral overlap of emission from reporter label (QD497) enhanced the signal at 525nm and shifted *0000 cluster towards right to overlap with 1000 cluster in IPAT space. Since ratios (r) are not influenced by the presence or absence of additional QD species unlike IPAT values, *0000 and 1000 clusters in r space were distinct with their centroids near (0,0) and (1,0), respectively (Fig. 2d), similar to 0000 and 0001 clusters in r space (Fig. 2c). Thus, we proposed using ratios of coding QD species, instead of IPAT values, for classifying microspheres.
Figure 2.
Scatter plots explaining the rationale for utilizing r values instead of IPAT values to perform classification on assayed microspheres: a) and b) show scatter plots between IPAT values observed in 525nm and 575nm detection channels for classes 0000-1000 and *0000-1000, respectively; c) and d) represent rQD525 vs. rQD575 scatter plots for classes 0000-1000 and *0000-1000, respectively. Labeling with QD497 (*) brings the 0000 cluster lie very close to and overlap with 1000 cluster in scatter plots in IPAT space lowering the classification efficiency; however in r space *0000 and 1000 clusters remain well demarcated and classification efficiency is high. 0000 and *0000 clusters are marked with ‘×’, and 1000 clusters are marked with ‘+’.
Classification of assayed microspheres in the mixture was based on rQD525, rQD575, rQD620 and rQD675 distributions for calibrator sets.
Verification of QDA
Although QDA assumes the data to be multivariate normal, it is relatively robust to this requirement. However, an empirical tolerance test should be performed to verify its applicability. QDA’s robustness was ascertained by its ability to classify a subset of microspheres from five calibrator sets (1000, 0100, 0010, 0001 and 1001) accurately. Towards this, we picked 1,000 data points (microspheres) from each individual calibrator set to form the training set that was utilized to classify the remaining data points from all classes. We compiled remaining data points from all classes into a single matrix and randomized the order while keeping the track of their original classes. This matrix had 8,490 rows (corresponding to 8,490 microspheres) and 4 columns (rQD525, rQD575, rQD620 and rQD675). We classified these microspheres using QDA based on the initially picked 5,000 data points (1,000 from each class) in the training set. We compared the identified classes of these 8,490 microspheres with their actual classes and constructed the classification table (see Table 1). Overall accuracy of 4-channel classification was 98.68%. High accuracy in classifying microspheres proved QDA to be a rational method for classifying microspheres from the unknown assay mixture.
Table 1. Classification of microspheres using QDA based on training set derived from 1,000 microspheres from each class selected randomly from calibrator sets. Row labels and column labels depict actual classes and identified classes, respectively.
| CLASS | 0001 | 1000 | 0100 | 0010 | 1001 | Number of microspheres under consideration |
|---|---|---|---|---|---|---|
| 0001 | 98.03 | 0.79 | 0.13 | 0.92 | 0.13 | 762 |
| 1000 | 0.00 | 97.72 | 0.05 | 0.00 | 2.23 | 2,104 |
| 0100 | 0.17 | 0.08 | 99.71 | 0.04 | 0.00 | 2,404 |
| 0010 | 0.12 | 0.54 | 0.27 | 99.04 | 0.04 | 2,602 |
| 1001 | 0.32 | 2.10 | 0.32 | 0.00 | 97.25 | 618 |
| TOTAL = 8,490 | ||||||
For classifying microspheres in the assay mixture, we utilized all 13,490 microspheres in the calibrator sets from all five classes (1000, 0100, 0010, 0001 and 1001). Table 2 shows centroids and associated standard deviations in r values for individual clusters of calibrator sets. From Table 2, we observed that all five clusters were well separated from each other in 4-dimensions and thus would allow for an efficient classification.
Table 2. Centroids and associated standard deviations in r values for individual clusters of calibrator sets.
| rQD525 | rQD575 | rQD620 | rQD675 | |
|---|---|---|---|---|
| 0001 | 0.00±0.07 | 0.00±0.13 | 0.00±0.13 | 1.00±0.40 |
| 1000 | 1.00±0.31 | 0.00±0.11 | 0.00±0.12 | 0.00±0.30 |
| 0100 | 0.00±0.06 | 1.00±0.32 | 0.00±0.15 | 0.00±0.33 |
| 0010 | 0.00±0.05 | 0.00±0.07 | 1.00±0.36 | 0.00±0.46 |
| 1001 | 0.72±0.19 | −0.06±0.07 | −0.01±0.12 | 0.94±0.61 |
Setting rQD497 threshold to distinguish tagged from untagged microspheres
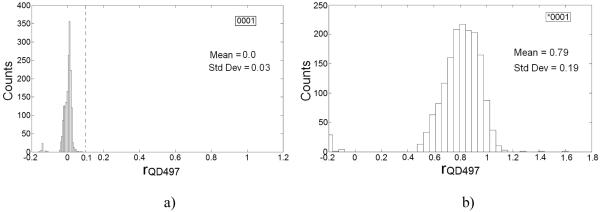
In the assay mixture, we distinguished microspheres tagged with QD497 label from those which were untagged based on rQD497 distributions for *0001 and 0001 calibrator sets (Fig. 3). Untagged microspheres did not have rQD497 values greater than 0.1 (Fig. 3a); accordingly, we set rQD497 = 0.1 as the upper limit for untagged microspheres. Only a small percentage (~2.6%) of microspheres in *0001 calibrator set exhibited rQD497 values lower than 0.1 (Fig. 3b). All microspheres with rQD497>0.1 were classified as tagged microspheres.
Figure 3.
rQD497 distributions for a) 0001 and, b) *0001 calibrator sets. rQD497 distribution for 0001 was used to set 0.1 as upper threshold (marked by --- in histogram for 0001) on rQD497 for untagged microspheres.
Assay performance
We implemented QDA on 1572 microspheres in the assay mixture (consisting of *0001, 0010, 0100, 1000 and 1001 microspheres) based on calibrator sets for 0001, 1000, 0100, 0010 and 0001 microspheres for 4-channel classification.
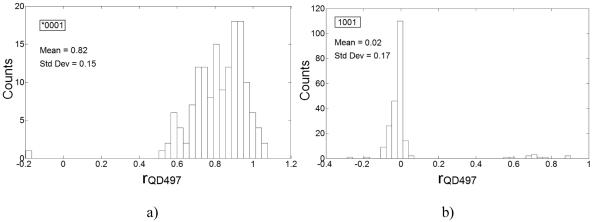
We observed that 99.33% of 149 microspheres from the class identified as 0001 by QDA, had rQD497>0.1 and thus were correctly identified as tagged microspheres. The mean and the associated CV in rQD497 values were 0.82 and 18.3%, respectively. Also, 209 out of 220 i.e. 95% of the microspheres from class 1001 had rQD497<0.1 and had mean value of 0.02 and thus, were correctly classified as untagged microspheres. We observed that both *0001 and 1001 populations have distinct rQD497 distributions and could be distinguished with high accuracy (Fig. 4). We also observed that mean and standard deviation in rQD497 values for identified *0001 class were similar to that for *0001 calibrator set underscoring the ability of our analysis to quantify the reporter tag accurately. For other untagged microspheres in the mixture classified as 1000, 0100 and 0010, we fittingly observed that 97.71%, 99.48% and 97.19% of them, respectively had rQD497<0.1.
Figure 4.
rQD497 distributions for microspheres from classes a) *0001 and, b) 1001, as identified by our classification algorithm, in the assay mixture. Mean rQD497 values for *0001 and 1001 microspheres were close to the expected values and thus, underscored the capability of our 4-channel classification to correctly classify microspheres and to correctly quantify the capture target on them. Also, rQD497 values have low variability emphasizing accuracy of our analysis procedures.
A successful distinction between tagged and untagged microspheres from the assay mixture demonstrated that tagged microspheres could be identified solely on the basis of intensity information at wavelengths corresponding to the emission peaks of QDs that code the microspheres. This is desirable for any assay system as intensity in the reporter label channel is directly correlated with the concentration of analyte in the target sample mixture, which is not known a priori and thus, cannot be utilized for classification. We also demonstrated that 1,000 microspheres from each calibrator set were sufficient enough to generate reliable estimates of b and C, which were used to calculate r for microspheres.
Extending our study further to sixteen classes of microspheres, generated by absence or presence of four QD species, we could successfully classify 97.79% of 10,502 microspheres, based on 1,000 microspheres from each class as training sets and utilizing ratios for four coding QD species (data not shown).
Standard Curve
We acquired multispectral images of the microwells meant for generating the standard curve, and identified and localized the microspheres in them. Based on the mathematical model described above, we computed r vector and extracted rQD497 values for all the localized microspheres. For each duplicate per dilution, we calculated mean rQD497 value across all microspheres and then averaged the means for both duplicates to get an average estimate of rQD497 at that dilution (rQD497avg). Next, rQD497avg values were plotted on y-axis against the concentration of biotinylated QD497 on x-axis and fitted with four-parameter logistic model using SoftMax Pro v. 5.2 software (Molecular Devices, Sunnyvale, CA) to generate a standard curve (Fig. 5). The rQD497 values attained in the assay can be correlated to the concentrations of QD497-tagged biotin using this standard curve. Based on rQD497 = 0.1 as upper threshold for untagged microspheres, we observed that the lowest limit of detection for our SAv-Biotin assay system is ~4.63nM. Similar curves correlating r values for reporter label to the concentration for each analyte tested can be generated for a multiplexed system to quantify specific analyte levels in test samples.
Figure 5.
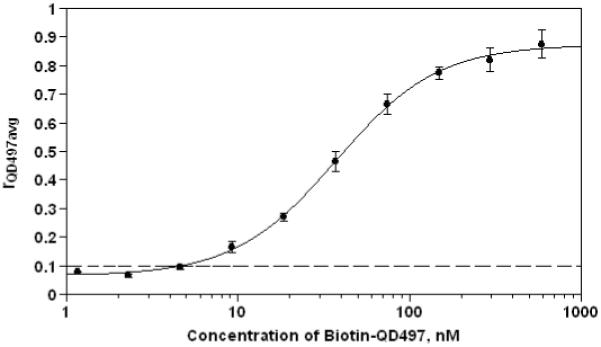
Standard curve (model fitted as four-parameter logistic using SoftMax Pro v. 5.2) for QD497-tagged biotin was generated via microsphere-based immunoassay. Data was generated from standard dilutions at concentrations 592, 296, 148, 74, 37, 18.5, 9.25, 4.63, 2.31 and 1.16nM using duplicates for each concentration. Error bars denote the standard deviation between duplicates. Based on rQD497 = 0.1 as upper threshold for untagged microspheres (see Figure 3), marked by ‘---’, we observed that the lowest limit of detection for our SAv-Biotin assay system is ~ 4.63nM. For the generation of curve, microspheres from class 0001 were used for tagging with biotin-QD497.
DISCUSSION
Binary coding is much more powerful than having multiple levels of colors as only the presence of a color needs to be ascertained. This scheme increases the tolerance levels for the variability in the QD concentrations inside microspheres, which is important in view of the variability associated with the encoding process. The presence or absence of multiple colors identifies the code of each microsphere present in the mix, and thereby indicates the capture reagent present on it. Results reported here have strongly demonstrated that this system is capable of performing multiplexed assays on 24 digitally encoded microspheres and quantifying the targets with acceptable precision. The segmentation algorithm can accurately identify and locate microspheres in images. Utilization of IPAT values made our analysis insensitive to the variability in the sizes of microspheres in the system, their lateral movement due to local convection currents and different exposure times used for image acquisition. Furthermore, the suggested mathematical model, developed for quantifying the relative concentrations of different QD species in microspheres (r), was reasonably accurate as the identified ratios were well within the expected range. Classification using QDA efficiently categorized tagged microspheres based on concentration ratios of encoding QD species. Thus, by characterizing the calibrator sets of microspheres beforehand, the proposed system could be used repeatedly for performing multiple assays. Use of ratios is a novel approach for classifying microspheres as opposed to the established use of intensities in multiple channels, such as in Luminex systems (Luminex, Inc., Austin, TX). Ratios-based approach removes the spectral overlap component and makes the classification problem straightforward. Thus, it is more useful than the intensity-based scheme wherein the number of dyes used is limited mainly to avoid significant spectral overlap into secondary detection channels which could make the classification problem difficult. Furthermore, the generation of a standard curve depicting dose-dependent increase in the r values of the reporter tag with the concentration of the target analyte substantiated the claim of our system to be able to quantify differential levels of protein in the assay.
Though, we have demonstrated the results for a single assay, our analysis procedures can readily be extended to evaluate multiple assays simultaneously. Our system is well integrated and fully automated that makes it applicable to much higher levels of multiplexing. In an extended study, with 4 QD coding species, we have performed sixteen assays at a time (data not shown). Increasing the number of QD species that encode microspheres would exponentially increase the number of assays that could be performed simultaneously. For instance, by using 10 species of QDs for encoding microspheres with their emission peaks spanning the visible spectrum (400nm-700nm) and still well separated (>30nm), we can increase the number of codes to 210 i.e. more than a thousand. By halving the distance between emission peaks, we can further increase the number of QD species to 20 and number of codes and possible assays to more than a million. Since microsphere clusters from different classes were well separated in 4-color space, addition of more QD species for coding microspheres without having the clusters overlap is a distinct possibility. Thus, our fully-automated HTS system in conjunction with our image analysis procedures has the potential of breaking the 100-plex barrier of suspension arrays posed by Luminex, Inc. (Austin, TX) and can be used effectively for performing thousands (and even a million) of assays concurrently in ultra-high throughput format.
ACKNOWLEDGMENT
We thank Crystalplex Corporation (Pittsburgh, PA) for generously providing us multicolored QD-encoded microspheres.
Footnotes
This research was supported by National Institute for Biomedical Imaging and Bioengineering grant R01-EB001418.
Contributor Information
Abhishek Mathur, Bioassay and Biological Characterization, Amgen, Inc., 1 Amgen Center Dr, Thousand Oaks, CA 91320.
David M. Kelso, Department of Biomedical Engineering, Northwestern University, 2145 Sheridan Rd, Evanston, IL 60208
REFERENCES
- 1.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biology. 2001;2(2):1–13. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacBeath G. Protein microarrays and proteomics. Nature Genetics. 2002;32(Suppl 2):526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 3.MacBeath G, Schreiber SL. Printing proteins as microarrays for high throughput function determination. Science. 2000;289(5485):1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 4.Templin MF, Stoll D, Schrenk M, Traub PC, Vohringer CF, Joos TO. Protein microarray technology. Trends in Biotechnology. 2002;20(4):160–166. doi: 10.1016/s0167-7799(01)01910-2. [DOI] [PubMed] [Google Scholar]
- 5.Meza MB. Bead-based HTS applications in drug discovery. Drug Discovery Today: HTS supplement. 2000;1(1):38–41. [Google Scholar]
- 6.Yingyongnarongkul BE, How SE, Diaz-Mochon JJ, Muzerelle M, Bradley M. Parallel and multiplexed bead-based assays and encoding strategies. Combinatorial Chemistry and High Throughput Screening. 2003;6:577–587. doi: 10.2174/138620703771981179. [DOI] [PubMed] [Google Scholar]
- 7.Soini JT, Waris ME, Hanninen PE. Detection methods of microsphere based single-step bioaffinity and in vitro diagnostic assays. Journal of Pharmaceutical and Biomedical Analysis. 2004;34:753–760. doi: 10.1016/S0731-7085(03)00562-4. [DOI] [PubMed] [Google Scholar]
- 8.McNeill A, Kastrup J, Bäckström BT. A simplified cytokine immunoassay using magnetic polymer particles. Scandinavian Journal of Immunology. 2004;60:287–291. doi: 10.1111/j.0300-9475.2004.01484.x. [DOI] [PubMed] [Google Scholar]
- 9.Kellar KL, Kalwar RR, Dubois KA, Crouse D, Chafin WD, Kane BE. Multiplexed fluorescent bead-based immunoassays for quantitation of human cytokines in serum and culture supernatants. Cytometry. 2001;45(1):27–36. doi: 10.1002/1097-0320(20010901)45:1<27::aid-cyto1141>3.0.co;2-i. Erratum in: Cytometry 2003;53A(2):112. [DOI] [PubMed] [Google Scholar]
- 10.de Jager W, te Velthuis H, Prakken BJ, Kuis W, Rijkers GT. Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells. Clinical and Diagnostic Laboratory Immunology. 2003;10(1):133–139. doi: 10.1128/CDLI.10.1.133-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turek-Etienne TC, Kober TP, Stafford JM, Bryant RW. Development of a fluorescence polarization AKT serine/threonine kinase assay using an immobilized metal ion affinity-based technology. Assay and Drug Development Technologies. 2003;1(4):545–553. doi: 10.1089/154065803322302808. [DOI] [PubMed] [Google Scholar]
- 12.Zhang XM, Wang XT, Yue H, Leung SW, Thibodeau PH, Thomas PJ, Guggino SE. Organic solutes rescue the functional defect in ΔF508 cystic fibrosis transmembrane conductance regulator. Journal of Biological Chemistry. 2003;278(51):51232–51242. doi: 10.1074/jbc.M309076200. [DOI] [PubMed] [Google Scholar]
- 13.Liu MY, Xydakis AM, Hoogeveen RC, Jones PH, Smith EO, Nelson KW, Ballantyne CM. Multiplexed analysis of biomarkers related to obesity and the metabolic syndrome in human plasma, using the Luminex-100 system. Clinical Chemistry. 2005;51:1102–1109. doi: 10.1373/clinchem.2004.047084. [DOI] [PubMed] [Google Scholar]
- 14.Faucher S, Martel A, Sherring A, Ding T, Malloch L, Kim JE, Bergeron M, Sandstrom P, Mandy FF. Protein bead array for the detection of HIV-1 antibodies from fresh plasma and dried-blood-spot specimens. Clinical Chemistry. 2004;50:1250–1253. doi: 10.1373/clinchem.2004.032995. [DOI] [PubMed] [Google Scholar]
- 15.Tewari PC, Williams JS. Analytical characteristics of seminal fluid PSA differ from those of serum PSA. Clinical Chemistry. 1998;44:191–193. [PubMed] [Google Scholar]
- 16.Taylor JD, Briley D, Nguyen Q, Long K, Iannone MA, Li M-S, Ye F, Afshari A, Lai E, Wagner M, Chen J, Weiner MP. Flow cytometric platform for high-throughput single nucleotide polymorphism analysis. BioTechniques. 2001;30:661–669. doi: 10.2144/01303dd04. [DOI] [PubMed] [Google Scholar]
- 17.Nolan JP, Sklar LA. Suspension array technology: Evolution of the flat-array paradigm. Trends in Biotechnology. 2002;20:9–12. doi: 10.1016/s0167-7799(01)01844-3. [DOI] [PubMed] [Google Scholar]
- 18.Stevens PW, Kelso DM. Imaging and analysis of immobilized particle arrays. Analytical Chemistry. 2003;75(5):1147–1154. doi: 10.1021/ac0205816. [DOI] [PubMed] [Google Scholar]
- 19.Swartzman EE, Miraglia SJ, Mellentin-Michelotti J, Evangelista L, Yuan P-M. A homogeneous and multiplexed immunoassay for high-throughput screening using fluorometric microvolume assay technology. Analytical Biochemistry. 1999;271:143–151. doi: 10.1006/abio.1999.4128. [DOI] [PubMed] [Google Scholar]
- 20.Han J, Gao X, Su JZ, Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nature Biotechnology. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 21.Goldman ER, Clapp AR, Anderson GP, Uyeda HT, Mauro JM, Medintz IL, Mattoussi H. Multiplexed toxin analysis using four colors of quantum dot fluororeagents. Analytical Chemistry. 2004;76(3):684–688. doi: 10.1021/ac035083r. [DOI] [PubMed] [Google Scholar]
- 22.Xu H, Sha MY, Wong EY, Uphoff J, Xu Y, Treadway JA, Truong A, O’Brien E, Asquith S, Stubbins M, Spurr NK, Lai EH, Mahoney W. Multiplexed SNP genotyping using the Qbead™ system: a quantum dot-encoded microsphere-based assay. Nucleic Acids Research. 2003;31(8):e43, 1–10. doi: 10.1093/nar/gng043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldman ER, Uyeda HT, Hayhurst A, Mattoussi H. Luminescent biocompatible quantum dots: A tool for immunosorbent assay design. Methods in Molecular Biology. 2007;374:207–27. doi: 10.1385/1-59745-369-2:207. [DOI] [PubMed] [Google Scholar]
- 24.Hermanson GT. Bioconjugate Techniques. Academic Press, Inc.; San Diego, CA: 1996. [Google Scholar]
- 25.Mathur A, Kelso DM. Segmentation of microspheres in ultrahigh density multiplexed microsphere-based assays. Proceedings of SPIE. 2006;6064:60640B1–60640B10. [Google Scholar]
- 26.Otsu N. A threshold selection method from graylevel histogram. IEEE Transactions on Systems, Man, and Cybernetics. 1978;SMC-8:62–66. [Google Scholar]
- 27.Rencher AC. Methods of multivariate analysis. Wiley-Interscience; New York, NY: 2002. [Google Scholar]