Abstract
Recently, much progress has been made in elucidating the chemistry and metabolism of retinoids and carotenoids as well as the structures of processing proteins related to vision. Because carotenoids and their retinoid metabolites are isoprenoids, only a limited number of chemical transformations are possible and just a few of these naturally occur. Although there is an intriguing evolutionary conservation of the key components involved in chromophore production and recycling, these genes also have adapted to the specific requirements of both insect and vertebrate vision. These ancestral footprints in animal genomes bear witness to the common origin of the chemistry of vision and will further stimulate research across evolutionary boundaries.
Keywords: vision, phototransduction, carotenoids, retinoids, vitamin A, retinoid cycle, visual cycle, retina, retinal pigment epithelium, inherited retinal disease
A enzymatic pathway of chromophore trans-to-cis isomerization is intrinsic to animal vision
A cycle of cis-to-trans isomerization of the visual chromophore is an intrinsic part of animal vision 1. Phototransduction, the process by which energy from light is translated into a photoreceptor's electrical response, has long been at the forefront of sensory transduction and cell signaling studies in general. The elucidation of the biochemical steps involved in photoreceptor excitation, together with seminal work on hormone-stimulated adenylate cyclase, led to the discovery and characterization of G protein signaling 2. Cascades whereby heptahelical transmembrane receptors, such as rhodopsin, catalytically activate heterotrimeric G proteins are involved in a broad range of physiological processes throughout the body, where they respond to a wide variety of chemical messengers including hormones, neurotransmitters, odorants, and food ingredients. Visual pigments comprise one class of G protein-coupled receptors (GPCRs) and consist of an integral transmembrane protein (opsin) and a covalently bound retinylidene chromophore that mediates phototransduction 2.
Visual GPCR signaling is unique with respect to its dependence on a diet-derived chromophore (retinal or 2-dehydro-retinal in vertebrates; retinal and 3-hydroxy-retinal in insects). The chromophore is naturally generated by oxidative cleavage of carotenoids (C40) to retinoids (C20). Then the retinoid cleavage product must be metabolically converted to the respective 11-cis-retinal derivative in either the same carotenoid cleavage reaction or a separate reaction. The 11-cis-stereoisomer binds by a Schiff-base linkage to a membrane-embedded Lys residue in opsin to form functional visual pigments 3. The phototransduction cascade in vertebrate rods is well understood and widely cited as the textbook example of G protein signaling. Absorption of light triggers an 11-cis to all-trans isomerization of the chromophore that converts rhodopsin into an activated state termed Meta II. Meta II is catalytically active and binds transducin (Gt), a photoreceptor-specific G protein, thereby initiating a signal-amplifying cascade involving cGMP that results in plasma membrane hyperpolarization. In contrast to vertebrate signaling, rhodopsin activation and binding of Gt in invertebrates such as Drosophila melanogaster, initiates phosphoinositide signaling, culminating in the opening of transient receptor potential (TRP) channels and depolarization of the photoreceptor cell membrane 4.
In vertebrates, activated rhodopsin and cone visual pigments decay into the opsin and the all-trans-retinal photoproduct. Subsequent regeneration of the cis-chromophore depends on an enzymatic pathway known as the visual (retinoid) cycle. In rod photoreceptors, this pathway operates between rod outer segments (ROS) and the adjacent retinal pigmented epithelium (RPE) 5. Although outnumbered more than 20:1 by rod photoreceptors, cone photoreceptors in the human eye mediate daylight vision and are critical for visual acuity and color discrimination 6. Cones operate under bright light, which saturates rods; however rods still consume 11-cis-retinal. This scenario might require an additional cone-specific regeneration pathway to avoid competition for 11-cis-retinal between rod and cone visual pigments. By contrast, an enzymatic visual cycle is believed to be absent in invertebrates such as Drosophila. Here activated rhodopsin thought to be thermostable, and can be reisomerized back to rhodopsin by absorption of another incoming photon 7.
All animals endowed with the ability to detect light through visual pigments must have evolved pathways in which dietary precursors for chromophore, such as carotenoids and retinoids, are first absorbed in the gut, and then transported, metabolized and stored within the body to establish and sustain vision (Box 1). Not surprisingly, mutations in genes encoding the involved components have emerged as important causes for not only blinding diseases, but also fatal diseases such as the Matthew-Wood syndrome, which is characterized by pleiotropic, multisystem malformations including cardiac deformities and ocular defects. Carotenoids and their retinoid metabolites are isoprenoid compounds, which physiologically undergo only a limited number of possible chemical transformations (Fig. 1). Recent research has revealed that enzymes catalyzing these different transformations are closely related between animal classes, including humans, findings that suggest a monophyletic origin of these pathways. In particular, biochemical and structural analyses of the involved proteins have provided atomic details of the mechanisms for trans-to-cis isomerization of retinoids. This review will summarize the advanced state of knowledge about pathways for visual chromophore production, and will place a special focus on the molecular and biochemical basis of trans-to-cis retinoid isomerization, the fundamental chemical reaction which underlies vision.
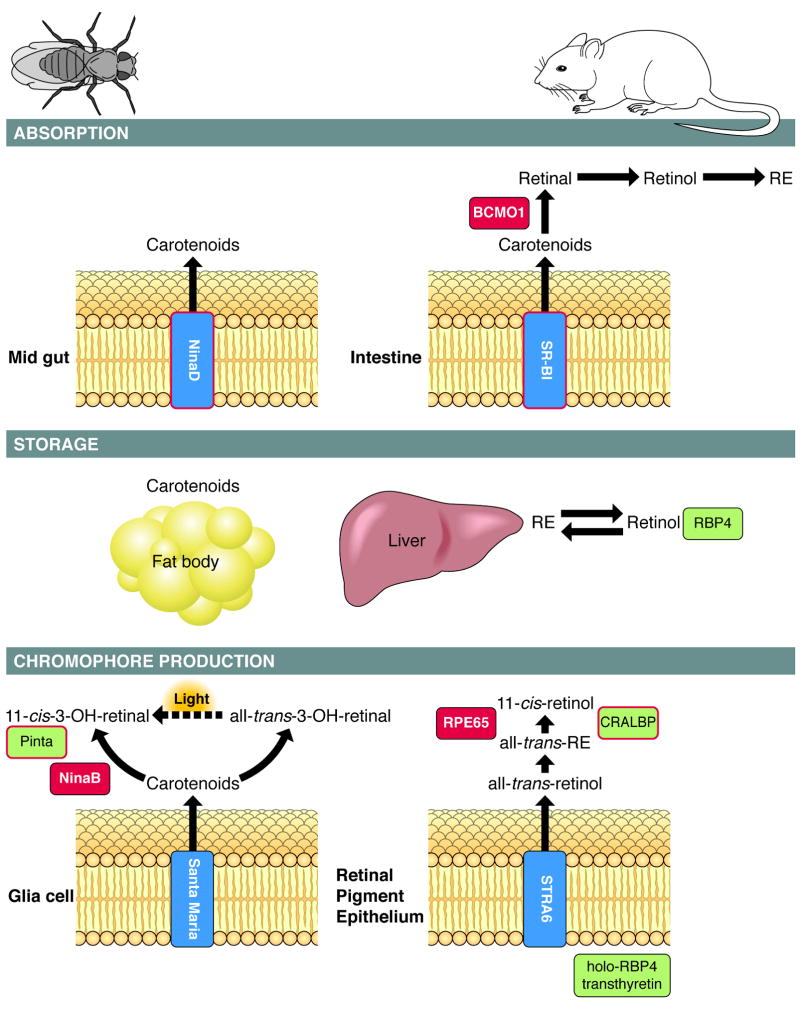
Box 1. [SC3] Related proteins are involved in carotenoid/retinoid metabolism in insects and mammals.
A schematic overview of pathways involved in chromophore production in Drosophila and mammals. The process can be divided into three major events: absorption of precursors in the gut, transport and storage in the body, and uptake into cells that produce chromophore. In Drosophila, with carotenoid function restricted to vision, molecular players were identified by genetic dissection and screening of blind rhodopsin-deficient mutants. Absorption of dietary carotenoids essentially depends on NinaD 11 which is expressed in the midgut 12. This cytoplasmatic trans-membrane protein facilitates carotenoid uptake from micelles into cells 71. Upon absorption, non-hydroxylated carotenoids such as β,β-carotene are hydroxylated to zeaxanthin for storage 72. These carotenoids are mobilized in a NinaD-dependent manner 71 and taken up into neuronal and glial cells of the optic lobes. Absorption of carotenoids into both cell types depends on the NinaD-related scavenger receptor, Santa Maria 12. Both cell types also express NinaB, which converts carotenoids into retinoids 12, 73. NinaB is a carotenoid isomeroxygenase that combines the activities of mammalian BCMO1 and RPE65 to produce 11-cis and all-trans-3-hydroxy-retinal 10. The all-trans-stereoisomer cleavage product is light-dependently converted to chromophore 10, 13.
Studies in knockout mice and human intestinal cells showed that the NinaD-related scavenger receptor class B type 2 (SR-BI) mediates intestinal carotenoid absorption in mammals 15, 74. Upon absorption, carotenoids such as β, β-carotene are oxidatively cleaved to retinal by the action of BCMO1 75, 76. SR-BI and BCMO1 activity are regulated by negative feedback at the transcriptional level to avoid excess vitamin A production. The primary retinaldehyde cleavage product is then successively converted to retinol and REs by the action of retinol dehydrogenases (RDHs) and LRAT 77. Such REs, together with REs formed from preformed dietary vitamin A, are packed into chylomicrons, secreted into the circulation, and taken up by target tissues (e.g. liver) for storage in an LRAT-dependent manner 27, 78. Retinoids are secreted from storage compartments in the form of all-trans-retinol bound to RBP4 17, 79. From this complex, all-trans-retinol uptake into the RPE is accomplished by the cytoplasmatic membrane protein STRA6 as evidenced by analyses in cell cultures 18, 19. Accumulation of all-trans-retinol is driven by its esterification via LRAT 19. The resulting REs serve as substrates for the retinoid isomerase RPE65 that catalyses 11-cis-retinol formation. In both mammals and Drosophila, related retinoid-binding proteins, respectively termed CRALBP and Pinta, act downstream of these enzymes to supply photoreceptors with chromophore 13, 80.
Box 1, Figure I. Key components (highlighted in blue) in the pathways for chromophore production are well conserved between flies and mammals.
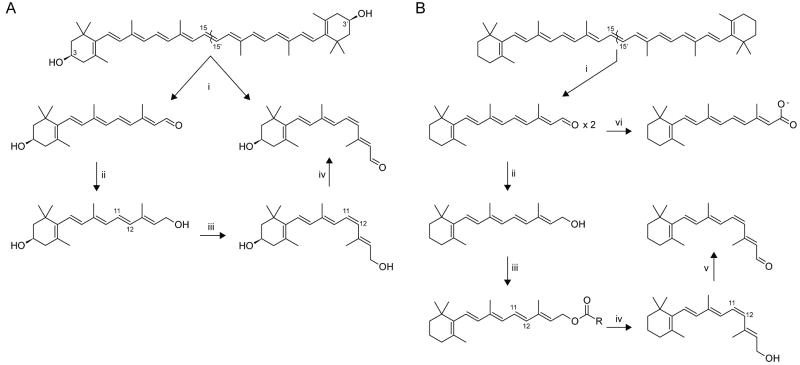
Figure 1. Key enzymatic steps in carotenoid/retinoid metabolism in insects and mammals.
A comparison of the chemical transformations of carotenoids and their retinoid metabolites in the pathways for chromophore production in different animal classes. These include oxidative cleavage of double bonds, oxidation of alcohols to aldehydes and aldehydes to acids and aldehyde reduction to alcohols, esterification of alcohols, hydroxylation of carbons in ionone ring structures, and trans-to-cis isomerization of carbon-carbon double bonds. A) In insects, [i] carotenoids such as zeaxanthin are converted to one molecule of 11-cis and one molecule of all-trans-3-hydroxy-retinal in an isomerooxygenase reaction. [ii] all-trans-3-hydroxy-Retinal is converted to all-trans-3-hydroxy-retinol. [iii] all-trans-3-hydroxy-Retinol is light-dependently isomerized to 11-cis-3-hydroxy-retinol. [iv] 11-cis-3-hydroxy-Retinol is oxidized to 11-cis-3-hydroxy-retinal. B) In mammals, [i] β,β-carotene is symmetrically cleaved to two molecules of all-trans-retinal. [ii] all-trans-Retinal is reduced to all-trans-retinol (vitamin A). [iii] all-trans-Retinol is converted to retinyl esters for storage or [iv] formation of 11-cis-retinol. [v] 11-cis-Retinol is oxidized to 11-cis-retinal. [vi] all-trans-Retinal can be also oxidized to retinoic acid.
Absorption, metabolism and transport of carotenoids and retinoids
Two fundamental processes in chromophore metabolism defied molecular analysis for a long time: the conversion of the parent C40 carotenoid precursor into C20 retinoids and the all-trans to 11-cis isomerization and cleavage involved in continuous chromophore renewal. The molecular basis of retinoid production was resolved by cloning carotenoid oxygenases from different animals, including humans. Interestingly, based on their primary amino acid sequences, carotenoid-oxygenases belong to the same enzyme family as retinoid isomerases8, 9. Moreover, insect enzyme family members can combine both activities in a single polypeptide 10.
Chromophore metabolism begins with the absorption of precursors in the gut. In insects such as Drosophila with vitamin A function restricted to vision, carotenoids are absorbed intact by the gut, transported in lipoproteins within the hemolymph and then absorbed by neuronal and glial cells of the optic lobes in close proximity to photoreceptor cells. Absorption in the gut as well as into target cells is protein-mediated 11, 12. Carotenoids are converted to cis- and trans-chromophore by the action of a carotenoid isomerooxygenase 10 (Fig. 1A). A second light-dependent pathway carries out the cis-isomerization of the trans-chromophore 13. The cis-chromophore then binds the opsin and promotes rhodopsin maturation.
In contrast to insects, mammals efficiently use both dietary preformed vitamin A (mainly retinyl esters; REs) and provitamin A carotenoids (mainly β,β-carotene) for chromophore production (Box 1) 14. Dietary REs are hydrolyzed to retinol in the intestine and retinol diffuses into the enterocytes in a concentration-dependent manner 15. By contrast, carotenoid uptake is a protein-mediated and regulated process 15. Absorbed β,β-carotene is converted into two molecules of all-trans-retinal by β,β-carotene 15,15′-monooxygenase (BCMO1) (Fig. 1B). The primary cleavage product is reduced to all-trans-retinol and then esterified by acyl transferase enzymes to form REs. These REs along with REs formed from absorbed retinol are packaged into lipoprotein particles called chylomicrons and transported to target tissues. Most REs in chylomicrons are taken up by the liver and stored as esters in hepatic stellate cells (also called Ito cells, lipocytes or fat-storing cells) 16. The remaining REs in chylomicrons are taken up by target tissues that include adipose tissue, heart, muscle, lungs, reproductive organs and bone marrow, but not the eyes. REs from body stores are re-secreted into the circulation as all-trans-retinol bound to serum retinol-binding protein (RBP4) 17. Holo-RBP4 serves as the major transport mode for vitamin A and its blood concentrations remain well controlled even in the absence of dietary retinoids. This retinol is taken up by target tissues for the production of biologically active derivatives such as retinoic acid and visual chromophore. Retinoic acid, a hormone-like compound binds to retinoic acid receptors that act as transcription factors. The uptake of retinol from holo-RBP4 is a protein-mediated process that depends on a transmembrane receptor encoded by STRA6 (stimulated by retinoic acid 6 gene)18, which mediates the bidirectional flow of all-trans-retinol between RBP4 and target cells 19. This uptake is driven by retinol esterification with fatty acids 19. REs are converted to 11-cis-retinol by RPE-specific 65-kDa retinoid isomerase (RPE65) and subsequent oxidation to the visual chromophore 20-22. Despite differences in the pathways for chromophore production between insects and mammals, several key protein components are closely related (Box 1).
Visual chromophore recycling
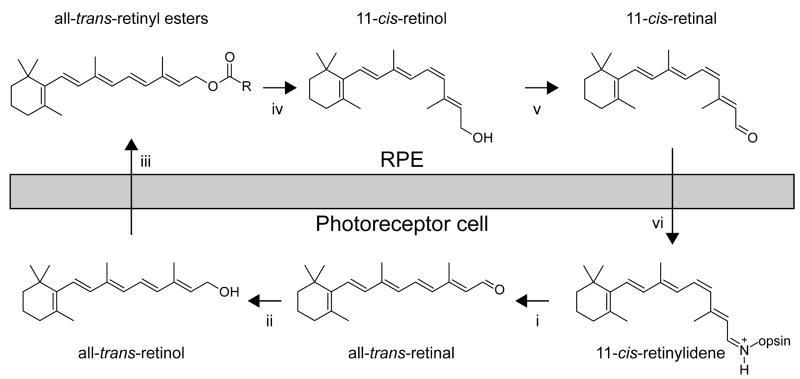
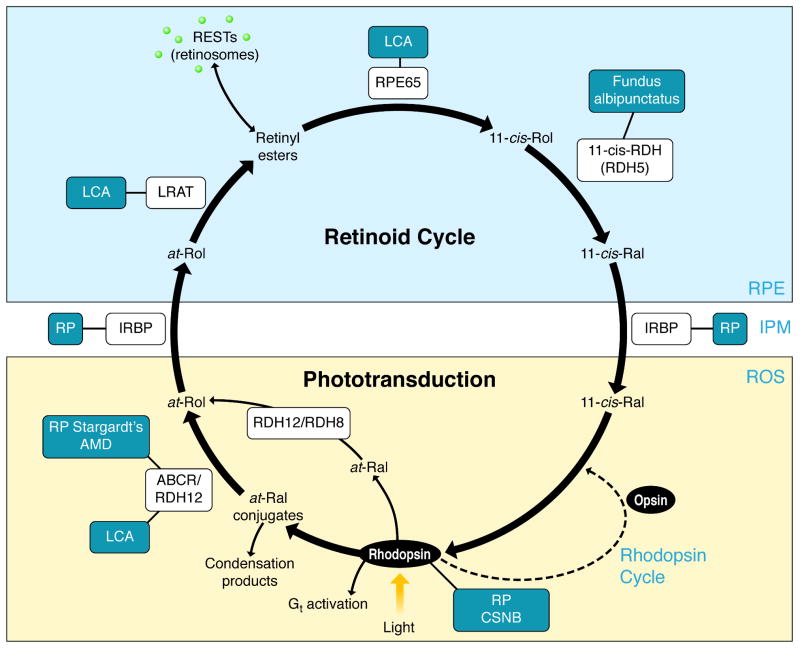
To sustain vision, all-trans-retinal released from light-activated rhodopsin must be continuously isomerized back to its 11-cis isomer. This process occurs via a sequence of enzymatic reactions called the retinoid or visual cycle that occur in rod and cone photoreceptor outer segments (OS) and the RPE (Fig. 2). The first step in the retinoid cycle involves reduction of all-trans-retinal to all-trans-retinol catalyzed by retinol dehydrogenases (RDHs). In mouse photoreceptors, two enzymes that belong to the short-chain dehydrogenase/reductase (SRD) family and utilize NADPH as a cofactor, RDH8 in photoreceptor OS and RDH12 in photoreceptor inner segments, are primarily responsible for catalyzing this reaction 23. However, the redundancy of retinal reductase activity observed in mice suggests that photoreceptors contain additional functional RDHs aside from RDH12 and RDH8 24. All-trans-retinol formed in OS is then transported to the RPE where it is esterified. This process is facilitated by two retinoid-binding proteins: interphotoreceptor retinoid-binding protein (IRBP) that binds retinoids in the extracellular space and cellular retinol-binding protein-1 (CRBP1) located within RPE cells 25, 26. The major ester synthase in RPE is lecithin:retinol acyl transferase (LRAT) 27. Due to their high hydrophobicity, all-trans-retinyl esters constitute a stable storage form of vitamin A within internal membranes and oil droplet-like structures called retinosomes 28. This pool of all-trans-retinyl esters serves a substrate for RPE65 which catalyses the endothermic transformation of all-trans-retinoid to its 11-cis conformation 20, 21. The product of this isomerization reaction, 11-cis-retinol, is subsequently oxidized in the final catalytic step of the retinoid cycle to form 11-cis-retinal. Enzymatic activities of SDRs such as RDH5, RDH10 and RDH11 are primarily responsible for this reaction 29 but additional 11-cis-RDHs might participate within the RPE 24. Newly synthesized 11-cis-retinal is protected by binding to cellular retinaldehyde-binding protein (CRALBP) which mediates its transport to the apical plasma membrane of the RPE. Following transport across the interphotoreceptor matrix that in part can involve IRBP, 11-cis-retinal enters the photoreceptor OS where it couples to opsin, thereby completing the cycle 30.
Figure 2. The visual cycle regenerates 11-cis-RAL.
In rod cells, 11-cis-retinal couples to a protein opsin, forming rhodopsin. Absorption of a photon of light by rhodopsin causes photoisomerization of 11-cis-RAL to all-trans-RAL leading to release of all-trans-RAL from the chromophore-binding pocket of opsin [i]. [ii] All-trans-RAL is reduced to all-trans-retinol in a reversible reaction catalyzed by an NADPH-dependent all-trans-RDH. [iii] All-trans-ROL diffuses into the RPE where it is esterified in a reaction catalyzed by LRAT. [iv] There all-trans-RE is the substrate for RPE65 that converts it to 11-cis-ROL, which is further oxidized back to 11-cis-RAL by RDH5, RDH11 and other RDHs [v]. [vi] 11-cis-RAL formed in the RPE diffuses back into the ROS and COS, where it completes the cycle by recombining with opsins to form rhodopsin and cone pigments.
Rhodopsin regeneration requires 11-cis-retinal supplied from RPE, but cones are not exclusively dependent on RPE65-mediated isomerization 31. Biochemical studies in cone-dominant ground-squirrels and chickens 32 as well as genetic studies in zebrafish (Danio rerio) support the existence of a separate “cone visual cycle” 33, 34. In the alternative, cone-specific visual cycle, all-trans-retinol released from cone OSs is taken up by Müller cells where, in contrast to the RPE, it is directly isomerized to the 11-cis configuration and subsequently esterified to 11-cis-retinyl esters by acyl-CoA:retinol acyltransferase (ARAT) 32, 34-36. 11-cis-Retinyl esters can be mobilized by 11-cis-retinyl ester hydrolase (REH) to yield 11-cis-retinol which then binds CRALBP and is transported back to cone photoreceptors. Finally, NADP+/NADPH-dependent 11-cis-RDH activity found exclusively in cone photoreceptors expedites the regeneration of visual chromophore from 11-cis-retinol 32. Ultimate confirmation of this pathway will require identification of the respective genes.
In contrast to vertebrates, the bleached chromophore in activated rhodopsin of invertebrates is re-isomerized back to the ground state by another incoming photon. Nevertheless, insects also require a light-independent pathway for all-trans to 11-cis isomerization of chromophore. The 11-cis-stereoisomer is not only important for phototransduction, but also for targeting insect visual pigments to photoreceptor membranes via the secretory pathway 38.[SC1] This cis-chromophore-dependent ‘targeting’ is also essential for cone pigment maturation and cone photoreceptor survival 39.
Structural insights into retinoid metabolism and transport
In recent years, substantial progress has been made in determining high-resolution structures of a few key components of the retinoid cycle. In addition to constituting a fundamental contribution to our understanding of the chemistry of vision, such information will help delineate the consequences of amino acid substitutions in these enzymes that are associated with impaired ocular vitamin A metabolism in several blinding disease.
Visual cycle enzymes
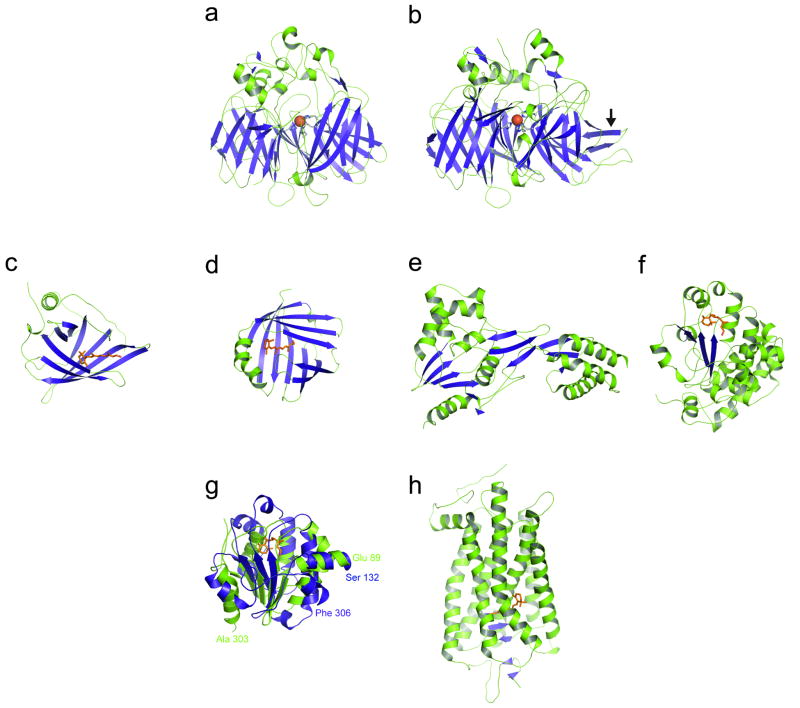
After transport to the RPE, the task of converting all-trans-retinol to 11-cis-retinal is performed by a series of microsomal enzymes. As these enzymes are membrane-bound and require detergents for effective solubilization, their study by X-ray crystallography has proceeded much more slowly than that of water-soluble retinoid-binding proteins. A structure for a water-soluble homolog of RPE65 from Synechocystis, sp. PCC 6803 apocarotenoid oxygenase (ACO), which belongs to the carotenoid cleavage oxygenase (CCO) family, was determined in 2005 40. This structure revealed that the general fold adopted by CCOs is a 7-bladed β-propeller and that the required non-heme ferrous iron cofactor is coordinated by four absolutely conserved His residues (Fig. 3A).
Figure 3. Crystal structures of proteins involved in the visual cycle, retinoid transport and phototransduction.
A variety of protein folds are utilized in nature to bind retinoids for metabolism, transport and signal transduction. In panels A-F and H, β strands and α helices are colored blue and green, respectively. A) Apocarotenoid oxygenase (ACO) from Synechocystis (PDB ID: 2BIW). B) 65 kDa retinal pigment epithelium-specific protein (RPE65, retinoid isomerase) from Bos taurus (PDB ID: 3FSN). The arrow indicates an insertion found in vertebrate members of the carotenoid cleavage enzyme family but not in cyanobacterial members. Despite the overall similar architecture of A) and B), the proteins catalyze fundamentally different reactions and have only about 22% sequence identity. C) Human serum retinol-binding protein (PDB ID: 1RBP). D) Cellular retinol-binding protein from Rattus norvegicus (PDB ID: 1CRP). E) Module two of Xenopus laevis interphotoreceptor retinoid-binding protein (PDB ID: 1J7X). F) Human cellular retinaldehyde-binding protein (PDB ID: 3HY5). Proteins that preferentially bind all-trans-retinol (C and D) have retinoid binding sites composed exclusively of β strands whereas those proteins that bind 11-cis-retinal (E and F) are composed of a mixture of α helices and β sheets. G) Structural superpositioning of the C-terminal domain of CRALBP and the B domain of IRBP module 2 reveals similar chain folds. Superimposed structures of the C-terminal domain of CRALBP consisting of residues 132-306 (in blue) and the B domain of IRBP module two consisting of residues 89-169, 194-240 and 275-303 (in green) are shown. The bound 11-cis-retinal ligand in the CRALBP structure is shown as orange sticks. Both domains exhibit asymmetric αβα sandwich folds that superimpose with an RMSD of 3.5 Å over 107 matched Cα positions. This observation might indicate that the 11-cis-retinoid-binding site of IRBP resides in the B domain. The superposition was performed with the DALI server (http://ekhidna.biocenter.helsinki.fi/dali_server/). H) Ground-state rhodopsin from Bos taurus (PDB ID: 1U19). The retinylidene binding site is composed entirely of α helices.
The structure determined for native RPE65 from Bos taurus revealed a monotopic mode of membrane insertion for this protein 41 (Fig. 3B). The major hydrophobic patch on the protein surrounds the entrance of a tunnel that leads to the active site of the enzyme defined by the iron cofactor. Unlike ACO, there is only one tunnel through which substrates and products can travel. This observation suggests that retinoid substrates enter the active site from the membrane and after metabolism the products are released back into the membrane where they can diffuse to another membrane-bound enzyme, RDH5, for further processing. Thus, it seems likely that after delivery of all-trans-retinol to LRAT by CRBP, all steps of retinoid processing that lead to the production of 11-cis-retinal occur in endoplasmic reticulum membranes without involvement of retinoid-binding proteins.
Retinal and retinol-binding proteins
Retinoid-binding proteins are essential regulators of retinoid transport throughout the body. This functional class of proteins comprises four structurally distinct members: RBP 42, CRBP 43, IRBP 44 CRALBP 45. RBP and CRBP, single-domain proteins that adopt cup-shaped, anti-parallel β-barrel folds that form their ligand-binding sites, belong to the lipocalin and intracellular lipid-binding proteins, respectively, and both preferentially bind all-trans-retinol (Fig. 3C,D). Retinol molecules in both proteins are found buried deep within the β-barrel in a hydrophobic cavity; however, these ligands are oriented oppositely in the two proteins with the β-ionone ring found near the cavity entrance in CRBP but positioned at the cavity base in RBP 46.
IRBP is a soluble lipoglycoprotein produced by photoreceptor neurons and secreted into the interphotoreceptor matrix where it is thought to be involved in the transport of retinoids to and from the photoreceptor and RPE cell layers. It is the largest known retinoid-binding protein, with a molecular mass of ∼130 kDa, and it folds into 4 separate modules that share considerable homology. IRBP possesses three or four retinoid-binding sites per molecule rather than the single-binding site observed for other retinoid-binding proteins. An electron microscopic study revealed that IRBP has a flexible, rod-shaped structure approximately 23 nm in length and 4 nm in width 47. A higher percentage of particles in a bent conformation were observed when these proteins were saturated with retinoid ligands, indicating that they can undergo major conformational changes. No complete structure has yet been reported for full-length IRBP; however, a high resolution crystal structure of a functional module of Xenopus laevis IRBP was solved 44 (Fig. 3E). This module is composed of two domains (A and B) linked by a lipophilic hinge. Interestingly, the full module is highly homologous to a photosystem II-processing protease, D1P, whereas domain B superimposes reasonably well with members of the enoyl-CoA hydratase/isomerase superfamily 44. The module possesses two hydrophobic cavities postulated to be ligand-binding sites: one site resides in the hinge region between domains A and B, whereas the second is found within domain B.
The recent high-resolution structure determination of human CRALBP with bound 11-cis-retinal constitutes a major breakthrough in our understanding of 11-cis-retinal transport 45 (Fig. 3F). CRALBP belongs to the CRAL_TRIO family of proteins that typically bind hydrophobic ligands and contain a highly basic patch of amino acid residues thought to mediate membrane binding. 11-cis-Retinal is bound deep within the protein in a horseshoe-shaped cavity. In contrast to both RBP4 and CRBP, the center of the retinoid molecule is nearest to the cavity opening. The aldehyde functional group forms hydrogen bonds with Y180 and E202 and a π stacking interaction with F161. The conformation of the retinoid polyene chain is markedly different from that observed in ground-state rhodopsin. In rhodopsin, the 11-12 cis double bond is twisted such that conversion to the trans configuration is favored when the molecule absorbs light. By contrast, the 11-12 cis double bond of the retinal ligand bound to CRALBP is in a nearly perfect cis configuration, which presumably makes photoisomerization of the retinal 11-12 cis double bond much less favorable when retinal is bound to CRALBP. Considerable insight into the molecular pathogenesis of Bothnia dystrophy, an autosomal recessive disease characterized by early-onset night blindness and macular degeneration, was achieved by determining the structure of CRALBP containing the disease-associated R234W substitution 45. R234 is located in the basic cleft of the protein thought to be involved in membrane binding. The structure revealed that a series of amino acid side chains undergo a domino-like rearrangement that alter the retinoid-binding cavity. This conformational change decreases the volume of the cavity and creates a more snug fit for the retinoid ligand. This structural observation is consistent with biochemical data that demonstrated that the mutant protein binds ligand more tightly, making it somewhat less susceptible to cis-to-trans photoisomerization 45, 48. It is conceivable that similar conformational changes that ultimately reduce the affinity of CRALBP for retinoid ligand could occur in response to interactions of the basic patch with negatively charged phospholipid head groups which promotes dissociation of the retinoid ligand 49. Comparative analysis of the B domain of the second IRBP module and the C-terminal domain of CRALBP reveals that these domains are structurally related, with both assuming asymmetric αβα sandwich folds. Indeed, a superposition of the IRBP B module and CRALBP with the DALI server 50 resulted in an root mean square deviation (RMSD) of 3.5 Å over 107 matched Cα atoms (Fig. 3G). Similarity of the IRBP B domain to the retinoid-binding domain of CRALBP supports a hypothesis that this domain contains a retinoid-binding site. 44
Photochemical and chemical retinoid isomerization
Retinoids contain conjugated double bonds and readily undergo light-induced geometrical isomerization. The structures of both vertebrate and invertebrate rhodopsin have been determined, along with further structural refinements and determinations of their photointermediate structures (Fig. 3H)(reviewed in 51-53). In light perception, 11-cis-retinylidene is photochemically converted to all-trans-retinylidene and photons provide the energy required to transiently “break” one of the double bonds (Fig. 4A). To ensure continuity of vision, “in the dark” enzymatic regeneration of light-sensitive chromophore must occur. From a chemical and thermodynamic point of view, this reaction can be accomplished only by lowering the bond order. Then rotation and reaction of the resulting carbocation with water yields the product in a proper cis-configuration. This process is frequently called isomerohydrolase activity, a term introduced many years ago 54, because the putative substrate is believed to be all-trans-retinyl palmitate (or other ester-containing long-chain fatty acids) while the products are 11-cis-retinol and free fatty acid. The name, however, implies isomerization and hydrolysis by water. Because no chemical mechanism theoretically exists to explain such a reaction, we instead prefer the name retinoid isomerase.
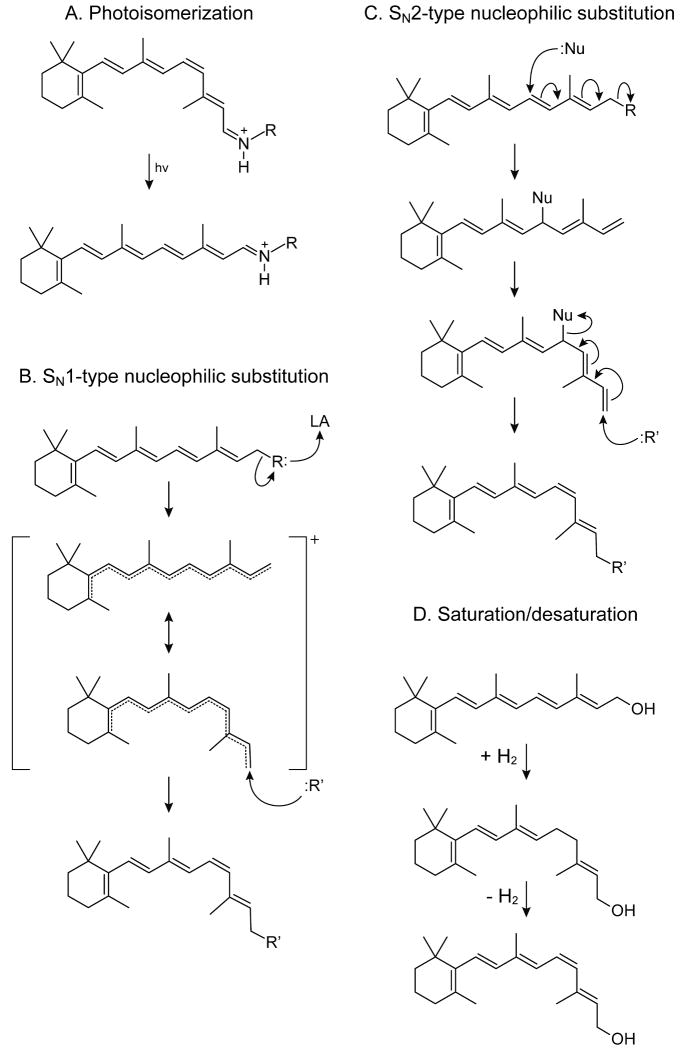
Figure 4. Observed and hypothetical mechanisms by which retinoid isomerization occurs.
A) Photoisomerization. Here energy from visible light temporarily reduces the π bond order through generation of anti-bonding orbitals of the polyene chain that allow rotation about the σ bonds. This mechanism is found throughout nature for both trans to cis and cis to trans isomerization reactions. B) Putative unimolecular nucleophilic substitution. In this mechanism, dissociation of a leaving group that may be promoted by a Lewis acid (LA) creates a retinyl carbocation with a lowered π bond order. Consequently, the activation energy for geometric isomerization is reduced. C) Putative bimolecular nucleophilic substitution. Here substitution of an active site nucleophile (Nu:) for the terminal retinoid R group and the consequent rearrangement of double bonds results in an enzyme-retinoid covalent intermediate with a single bond connecting the retinyl C11 and C12 atoms. After low energy rotation, a strong nucleophile, such as hydroxide, attacks the retinyl C15 atom, which rearranges the double bonds locking the C11-C12 in a cis configuration and leading to expulsion of the enzyme-linked nucleophile. D) Saturation/desaturation. Here, reduction of a π bond by addition of two hydrogen atoms allows free rotation of the C11-C12 sigma bond. Subsequent removal of H2 locks C11-C12 in the cis configuration.
Although crystal structures of ACO and RPE65 provide a structural framework for the isomerization reaction, the precise mechanism of isomerization awaits future structural and biochemical studies. From an enzymatic and chemical perspective, several options are possible. Insights into this reaction mechanism have been gained primarily by using O18 labeled retinyl esters, bulk labeled water, and selected stereospecific reactions 41, 55-58.
In our opinion, the most likely mechanism involves an SN1-type nucleophilic substitution (Fig. 4B) 57, 59. A general Lewis acid could promote an alkyl-cleavage of the ester group (between the alcohol oxygen and carbon), and a simple nucleophilic attack of hydroxide or water followed by deprotonation on the C15 position would yield the alcohol group of retinol. This reaction is possible because conjugation of double bonds throughout the substrate stabilizes the carbocation 57. Indeed, the retinyl carbocation is one of the most stable found in nature 60. The RPE65 active site contains a strong electron density that cannot be attributed to protein atoms but instead is suggestive of a bound fatty acid molecule 41. Based on this observation, we proposed that RPE65-catalyzed isomerization of all-trans-retinyl esters proceeds via an SN1-like nucleophilic substitution mechanism (Fig. 4B) with the iron cofactor acting as a Lewis acid. Once the carbocation is formed, a conformation resembling that of 11-cis-retinol is induced by the enzyme. Finally, addition of a nucleophile rather than hydrolysis completes the reaction 57(. CRALBP and/or 11-cis-RDH pull out the alcohol product for the subsequent oxidation reaction. Assuming some tolerance in the active site, this mechanism is also compatible with catalytic formation of 13-cis-retinal, a product that can be obtained in vitro by changing retinoid-binding protein specificity 57, 61-63. Predictably, positively charged retinylamine is a potent inhibitor of this isomerization, both in vitro and in vivo 59, 64.
Over 20 years ago R. Rando 65 proposed an alternative mechanism (Fig. 4C) postulating that a nucleophile adds specifically to carbon 11, and hence the 11-12 cis isomer could be formed, although rotation around C13-C14 without rotation around C11-C12, would also produce 13-cis-retinol. Addition of water to C15 would complete the reaction. As delineated previously, there are numerous problems with this mechanism 65. Moreover, the RPE65 structure fails to reveal a suitable nucleophilic group, presumably a Cys residue thiol group, in its active site 40, 41.
The structure[SC2] determination of ACO and RPE65 revealed that the iron centers of these enzymes are extremely similar. Additionally, both proteins require the ferrous form of iron for their activity 40, 41, 66. Therefore, on the basis of evolutionary arguments, ACO and RPE65 might be hypothesized to have similar catalytic mechanisms. However, the isomerization reaction does not involve a change in redox state of the retinoid, and the oxygen atom found in the 11-cis-retinol product is known to be derived from water 41. Although a role for molecular oxygen in the isomerization cannot be definitively ruled out, there is currently no experimental data to support this idea.
Formally, another possibility exists in the biology of retinoids and carotenoids. Carotenoid isomerase was identified through elegant work on tomato ripening from yellow to red (lycopene isomerization), 67. Enzymes of this family catalyze saturation-rotation-desaturation reactions 9. The mammalian homolog, RetSat can catalyze only half of the reaction, that is conversion of retinol to 13,14-dihydroretinol (Fig. 4D). Because these enzymes utilize FAD as a cofactor that manifests a characteristic primary sequence signature, this mechanism can be excluded for RPE65 or ACO.
Concluding remarks and future perspectives
Understanding the oxidative cleavage of carotenoids, isomerization of retinol, and retinoic acid-mediated gene regulation is at the forefront of modern biological chemistry. Mutations in humans and homologous knockout animal models demonstrate the physiological importance of these pathways. Indeed, even obesity and type 2 diabetes are linked to retinoid metabolism. Moreover, this knowledge has greatly facilitated the identification of disease-causing mutations visual cycle genes (Box 2). Defects in nearly every component of this cycle cause human inherited retinal dystrophies that can be divided into two etiologic groups: one involves impaired synthesis of visual chromophores whereas the other manifests accumulation of cytotoxic products derived from all-trans-retinal.
Recently, we elucidated the pathway for de novo chromophore production in insects. Interestingly, both carotenoid oxygenase and retinoid isomerase activities are combined in a single protein, NinaB 10. Likewise, the crystal structure of native bovine RPE65 provided a breakthrough in understanding the structural basis of trans-to-cis isomerization 41. Together, these contributions provide a platform for determining the mechanistic underpinnings of the enzymes that are associated with impaired chromophore metabolism and blindness in humans. Moreover, recent analysis in flies revealed that a chromophore regeneration pathway also is intrinsic to non-vertebrate species 68 These results establish Drosophila as an animal model for further study of chromophore regeneration pathways.
Despite these advances, several questions remain open including the mechanistic and structural basis for retinoid trans-to-cis isomerization and the pathways of chromophore production for cone pigments and the non-visual light-sensory protein, melanopsin. Additionally, the pathway for chromophore production in non-vertebrate species remains poorly understood.
Gene therapy in animal models of blinding diseases such as retinitis pigmentosa has successfully replaced enzymes involved in chromophore regeneration and restored vision 69. Dystrophies resulting from impaired chromophore synthesis also respond to supplementation with a readily available chromophore analog precursor (9-cis-retinyl acetate), and those derived from accumulation of toxic retinoid derivatives can be treated by inhibiting the visual cycle or limiting the supply of vitamin A to the eyes via pharmacological intervention 59, 70. Recent progress in both areas provides hope that many inherited retinal diseases will soon be treatable by pharmaceutical remedies. Only progress in understanding the basic chemistry of vision can guarantee that clinical studies will progress in parallel to benefit afflicted patients.
Box 2. [SC4] Retinoids and degenerative retinal diseases.
Several molecular mechanisms associated with disordered retinoid metabolism contribute to diverse retinopathies 81. Elucidation of these mechanisms has been greatly advanced by the availability of both natural and laboratory-generated animal models for human retinopathies. Animal models featuring anomalies in the retinoid cycle not only illustrate the importance of chromophore regeneration, they also provide an approach to elucidating mechanisms involved in human retinal dysfunction and disease. For example, lack of vitamin A transport from the liver to the eye, either because of global vitamin A deprivation or a genetic lack of RBP4, affects visual performance. Two sisters were identified with compound heterozygous missense mutations (resulting in Ile41Asn and Gly75Asp) in RBP4. Both lacked detectable serum RBP, had one sixth of the normal retinol level, and normal retinyl esters, but only mild clinical visual symptoms (night blindness and a modest retinal dystrophy 82). The lack of any other visual disorder provides strong evidence for an alternative tissue source of vitamin A in these siblings, most likely retinyl esters from chylomicron remnants 83. Similarly RBP4-/- mice maintained on a vitamin A-sufficient diet evidenced normal vision even though their blood retinol levels remained low. RBP-/- mice can acquire hepatic retinol stores, but these cannot be mobilized. Thus, their vitamin A status is extremely tenuous and dependent on a sustained vitamin A intake 79. Other examples include enzymes involved in the retinoid cycle. Lecithin:retinol acyl transferase is a key enzyme involved in formation of retinyl esters, and consequently the propensity of retinyl esters to aggregate allows retinoid storage in the liver and in the RPE. Lack of retinoids in Lrat-/- mice results in slowly progressive death of rods 27 attributed to continuous activation of visual phototransduction by unliganded opsin 84-86. Disordered vectorial transport of cone visual pigments lacking bound-chromophore also leads to very rapid cone degeneration 39. It is possible that this effect is exaggerated in mice, because the drop in cone numbers is greater than in humans 70. Obviously, either ocular retinoid deficiency or inadequate retinoid isomerization and production of 11-cis-retinal, can severely affect visual performance. These disorders, e.g. inactivating mutations in LRAT, are associated with Leber congenital amaurosis (LCA) 87. Paradoxically, an abnormally high flux of retinoids through the retinoid cycle can also induce retinopathies in humans, such as those related to ATP-binding cassette transporter ABCA4 mutations 88 and mouse models 89, 90. In addition, other disabling mutations in genes encoding proteins of the retinoid cycle can cause a spectrum of retinal diseases affecting vision (Fig. I) 91.
Acknowledgments
We thank Dr. Leslie T. Webster, for critical comments on the manuscript.
This research was supported in part by grants EY008061 (KP), EY08123 (J.vL), and a core grant P30 EY11373 from the National Institutes of Health, and Foundation Fighting Blindness. PDK was supported by the Visual Sciences Training Program grant T32EY007157 from the NEI.
Glossary
- Isoprenoid
A lipophilic compound that contains polymeric isoprene (2-methyl-1,3-butadiene).
- Müller cell
A support glial cell located in the neuroretina that houses enzymes needed to regenerate light-sensitive retinoid chromophores for cone photoreceptors.
- ROS
Rod Outer Segment. Part of a rod photoreceptor cell pointed toward the back of the eye and adjoining the RPE that contains the light-absorbing material rhodopsin.
- RPE
Retinal pigment epithelium. The monolayer of pigmented cells between the neurosensory retina and the choroid that nourishes and provides visual chromophore (11-cis-retinal) to photoreceptor cells.
- RPE65
Retinal pigment epithelium-specific 65 kDa protein that catalyzes conversion of all-trans-retinyl esters to 11-cis-retinol.
- STRA6
Stimulated by retinoic acid gene 6 that encodes a cell-surface receptor specific for holo serum retinol-binding protein. STRA6 mediates cellular uptake of all-trans-retinol by facilitating its transport across the plasma membrane.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wald G. Molecular basis of visual excitation. Science. 1968;162:230–239. doi: 10.1126/science.162.3850.230. [DOI] [PubMed] [Google Scholar]
- 2.Bitensky MW, et al. The mechanism of activation of light-activated phosphodiesterase and evidence for homology with hormone-activated adenylate cyclase. Adv Cyclic Nucleotide Protein Phosphorylation Res. 1984;17:227–237. [PubMed] [Google Scholar]
- 3.Filipek S, et al. G protein-coupled receptor rhodopsin: A Prospectus. Annu Rev Physiol. 2003;65:851–879. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yau KW, Hardie RC. Phototransduction motifs and variations. Cell. 2009;139:246–264. doi: 10.1016/j.cell.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBee JK, et al. Confronting complexity: the interlink of phototransduction and retinoid metabolism in the vertebrate retina. Progress in retinal and eye research. 2001;20:469–529. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- 6.Mustafi D, et al. Structure of cone photoreceptors. Progress in retinal and eye research. 2009;28:289–302. doi: 10.1016/j.preteyeres.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamdorf K, et al., editors. Biochemistry and Physiology of Visual Pigments. Springer; 1973. [Google Scholar]
- 8.von Lintig J, Vogt K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J Biol Chem. 2000;275:11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- 9.Moise AR, et al. Related enzymes solve evolutionarily recurrent problems in the metabolism of carotenoids. Trends Plant Sci. 2005;10:178–186. doi: 10.1016/j.tplants.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Oberhauser V, et al. NinaB combines carotenoid oxygenase and retinoid isomerase activity in a single polypeptide. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19000–19005. doi: 10.1073/pnas.0807805105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiefer C, et al. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10581–10586. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T, et al. Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. The Journal of cell biology. 2007;177:305–316. doi: 10.1083/jcb.200610081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voolstra O, et al. NinaB is essential for Drosophila vision but induces retinal degeneration in opsin-deficient photoreceptors. J Biol Chem. 2009;285:2130–2139. doi: 10.1074/jbc.M109.056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paik J, et al. Vitamin A: overlapping delivery pathways to tissues from the circulation. The Journal of nutrition. 2004;134:276S–280S. doi: 10.1093/jn/134.1.276S. [DOI] [PubMed] [Google Scholar]
- 15.During A, Harrison EH. Mechanisms of provitamin A (carotenoid) and vitamin A (retinol) transport into and out of intestinal Caco-2 cells. Journal of lipid research. 2007;48:2283–2294. doi: 10.1194/jlr.M700263-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Blaner WS, et al. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochimica et biophysica acta. 2009;1791:467–473. doi: 10.1016/j.bbalip.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blomhoff R, et al. Transport and storage of vitamin A. Science. 1990;250:399–404. doi: 10.1126/science.2218545. [DOI] [PubMed] [Google Scholar]
- 18.Kawaguchi R, et al. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 19.Isken A, et al. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell metabolism. 2008;7:258–268. doi: 10.1016/j.cmet.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin M, et al. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122:449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Redmond TM, et al. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13658–13663. doi: 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moiseyev G, et al. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12413–12418. doi: 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda A, et al. Role of photoreceptor-specific retinol dehydrogenase in the retinoid cycle in vivo. J Biol Chem. 2005;280:18822–18832. doi: 10.1074/jbc.M501757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda A, et al. Redundant and unique roles of retinol dehydrogenases in the mouse retina. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19565–19570. doi: 10.1073/pnas.0707477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redmond TM, et al. Isolation and characterization of monkey interphotoreceptor retinoid-binding protein, a unique extracellular matrix component of the retina. Biochemistry. 1985;24:787–793. doi: 10.1021/bi00324a038. [DOI] [PubMed] [Google Scholar]
- 26.Edwards RB, Adler AJ. Exchange of retinol between IRBP and CRBP. Experimental eye research. 1994;59:161–170. doi: 10.1006/exer.1994.1094. [DOI] [PubMed] [Google Scholar]
- 27.Batten ML, et al. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem. 2004;279:10422–10432. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imanishi Y, et al. Noninvasive two-photon imaging reveals retinyl ester storage structures in the eye. The Journal of cell biology. 2004;164:373–383. doi: 10.1083/jcb.200311079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haeseleer F, et al. Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J Biol Chem. 2002;277:45537–45546. doi: 10.1074/jbc.M208882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saari JC, Bredberg DL. Photochemistry and stereoselectivity of cellular retinaldehyde-binding protein from bovine retina. J Biol Chem. 1987;262:7618–7622. [PubMed] [Google Scholar]
- 31.Jones GJ, et al. Retinoid requirements for recovery of sensitivity after visual-pigment bleaching in isolated photoreceptors. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:9606–9610. doi: 10.1073/pnas.86.23.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mata NL, et al. Isomerization and oxidation of vitamin a in cone-dominant retinas: a novel pathway for visual-pigment regeneration in daylight. Neuron. 2002;36:69–80. doi: 10.1016/s0896-6273(02)00912-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schonthaler HB, et al. Evidence for RPE65-independent vision in the cone-dominated zebrafish retina. The European journal of neuroscience. 2007;26:1940–1949. doi: 10.1111/j.1460-9568.2007.05801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleisch VC, et al. Subfunctionalization of a retinoid-binding protein provides evidence for two parallel visual cycles in the cone-dominant zebrafish retina. J Neurosci. 2008;28:8208–8216. doi: 10.1523/JNEUROSCI.2367-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mata NL, et al. Chicken Retinas Contain a Retinoid Isomerase Activity that Catalyzes the Direct Conversion of all-trans-Retinol to 11-cis-Retinol. Biochemistry. 2005;44:11715–11721. doi: 10.1021/bi050942m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das SR, et al. Muller cells of chicken retina synthesize 11-cis-retinol. The Biochemical journal. 1992;285(Pt 3):907–913. doi: 10.1042/bj2850907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samardzija M, et al. R91W mutation in Rpe65 leads to milder early-onset retinal dystrophy due to the generation of low levels of 11-cis-retinal. Human molecular genetics. 2008;17:281–292. doi: 10.1093/hmg/ddm304. [DOI] [PubMed] [Google Scholar]
- 38.Ozaki K, et al. Maturation of major Drosophila rhodopsin, ninaE, requires chromophore 3-hydroxyretinal. Neuron. 1993;10:1113–1119. doi: 10.1016/0896-6273(93)90059-z. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, et al. Trafficking of membrane-associated proteins to cone photoreceptor outer segments requires the chromophore 11-cis-retinal. J Neurosci. 2008;28:4008–4014. doi: 10.1523/JNEUROSCI.0317-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kloer DP, et al. The structure of a retinal-forming carotenoid oxygenase. Science. 2005;308:267–269. doi: 10.1126/science.1108965. [DOI] [PubMed] [Google Scholar]
- 41.Kiser PD, et al. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17325–17330. doi: 10.1073/pnas.0906600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zanotti G, et al. Crystal structure of the trigonal form of human plasma retinol-binding protein at 2.5 A resolution. Journal of molecular biology. 1993;230:613–624. doi: 10.1006/jmbi.1993.1173. [DOI] [PubMed] [Google Scholar]
- 43.Kleywegt GJ, et al. Crystal structures of cellular retinoic acid binding proteins I and II in complex with all-trans-retinoic acid and a synthetic retinoid. Structure. 1994;2:1241–1258. doi: 10.1016/s0969-2126(94)00125-1. [DOI] [PubMed] [Google Scholar]
- 44.Loew A, Gonzalez-Fernandez F. Crystal structure of the functional unit of interphotoreceptor retinoid binding protein. Structure (Camb) 2002;10:43–49. doi: 10.1016/s0969-2126(01)00698-0. [DOI] [PubMed] [Google Scholar]
- 45.He X, et al. Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18545–18550. doi: 10.1073/pnas.0907454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newcomer ME, Ong DE. Plasma retinol binding protein: structure and function of the prototypic lipocalin. Biochimica et biophysica acta. 2000;1482:57–64. doi: 10.1016/s0167-4838(00)00150-3. [DOI] [PubMed] [Google Scholar]
- 47.Adler AJ, et al. Size and shape of bovine interphotoreceptor retinoid-binding protein by electron microscopy and hydrodynamic analysis. J Biol Chem. 1987;262:13198–13203. [PubMed] [Google Scholar]
- 48.McBee JK, et al. Isomerization of 11-cis-retinoids to all-trans-retinoids in vitro and in vivo. J Biol Chem. 2001;276:48483–48493. doi: 10.1074/jbc.M105840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saari JC, et al. Release of 11-cis-retinal from cellular retinaldehyde-binding protein by acidic lipids. Molecular vision. 2009;15:844–854. [PMC free article] [PubMed] [Google Scholar]
- 50.Holm L, et al. Searching protein structure databases with DaliLite v.3. Bioinformatics (Oxford, England) 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lodowski DT, et al. Comparative analysis of GPCR crystal structures. Photochemistry and photobiology. 2009;85:425–430. doi: 10.1111/j.1751-1097.2008.00516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palczewski K. G protein-coupled receptor rhodopsin. Annual review of biochemistry. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hofmann KP, et al. A G protein-coupled receptor at work: the rhodopsin model. Trends in biochemical sciences. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 54.Rando RR. Membrane phospholipids as an energy source in the operation of the visual cycle. Biochemistry. 1991;30:595–602. doi: 10.1021/bi00217a001. [DOI] [PubMed] [Google Scholar]
- 55.Deigner PS, et al. Membranes as the energy source in the endergonic transformation of vitamin A to 11-cis-retinol. Science. 1989;244:968–971. doi: 10.1126/science.2727688. [DOI] [PubMed] [Google Scholar]
- 56.Jang GF, et al. Stereoisomeric specificity of the retinoid cycle in the vertebrate retina. J Biol Chem. 2000;275:28128–28138. doi: 10.1074/jbc.M004488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McBee JK, et al. Isomerization of all-trans-retinol to cis-retinols in bovine retinal pigment epithelial cells: dependence on the specificity of retinoid-binding proteins. Biochemistry. 2000;39:11370–11380. doi: 10.1021/bi001061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jang GF, et al. Characterization of a dehydrogenase activity responsible for oxidation of 11-cis-retinol in the retinal pigment epithelium of mice with a disrupted RDH5 gene. A model for the human hereditary disease fundus albipunctatus. J Biol Chem. 2001;276:32456–32465. doi: 10.1074/jbc.M104949200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Golczak M, et al. Positively charged retinoids are potent and selective inhibitors of the trans-cis isomerization in the retinoid (visual) cycle. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8162–8167. doi: 10.1073/pnas.0503318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gurzadyan GG, et al. Photoionization versus photoheterolysis of all-trans-retinol. The effects of solvent and laser radiation intensity. Phys Chem Chem Phys. 2007;9:288–298. doi: 10.1039/b609165m. [DOI] [PubMed] [Google Scholar]
- 61.Stecher H, et al. Isomerization of all-trans-9- and 13-desmethylretinol by retinal pigment epithelial cells. Biochemistry. 1999;38:13542–13550. doi: 10.1021/bi9913294. [DOI] [PubMed] [Google Scholar]
- 62.Stecher H, et al. Preferential release of 11-cis-retinol from retinal pigment epithelial cells in the presence of cellular retinaldehyde-binding protein. J Biol Chem. 1999;274:8577–8585. doi: 10.1074/jbc.274.13.8577. [DOI] [PubMed] [Google Scholar]
- 63.Redmond TM, et al. RPE65, visual cycle retinol isomerase, is not inherently 11-cis specific: Support for a carbocation mechanism of retinol isomerization. J Biol Chem. 2009 doi: 10.1074/jbc.M109.027458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Golczak M, et al. Lecithin:retinol acyltransferase is responsible for amidation of retinylamine, a potent inhibitor of the retinoid cycle. J Biol Chem. 2005;280:42263–42273. doi: 10.1074/jbc.M509351200. [DOI] [PubMed] [Google Scholar]
- 65.Kuksa V, et al. Retinoid cycle in the vertebrate retina: experimental approaches and mechanisms of isomerization. Vision research. 2003;43:2959–2981. doi: 10.1016/s0042-6989(03)00482-6. [DOI] [PubMed] [Google Scholar]
- 66.Moiseyev G, et al. RPE65 is an iron(II)-dependent isomerohydrolase in the retinoid visual cycle. J Biol Chem. 2006;281:2835–2840. doi: 10.1074/jbc.M508903200. [DOI] [PubMed] [Google Scholar]
- 67.Isaacson T, et al. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. The Plant cell. 2002;14:333–342. doi: 10.1105/tpc.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, et al. Requirement for an Enzymatic Visual Cycle in Drosophila. Curr Biol. 2009 doi: 10.1016/j.cub.2009.1012.1022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maguire AM, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. The New England journal of medicine. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maeda T, et al. Loss of cone photoreceptors caused by chromophore depletion is partially prevented by the artificial chromophore pro-drug, 9-cis-retinyl acetate. Human molecular genetics. 2009;18:2277–2287. doi: 10.1093/hmg/ddp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voolstra O, et al. The Drosophila class B scavenger receptor NinaD-I is a cell surface receptor mediating carotenoid transport for visual chromophore synthesis. Biochemistry. 2006;45:13429–13437. doi: 10.1021/bi060701u. [DOI] [PubMed] [Google Scholar]
- 72.Giovannucci DR, Stephenson RS. Identification and distribution of dietary precursors of the Drosophila visual pigment chromophore: analysis of carotenoids in wild type and ninaD mutants by HPLC. Vision research. 1999;39:219–229. doi: 10.1016/s0042-6989(98)00184-9. [DOI] [PubMed] [Google Scholar]
- 73.von Lintig J, et al. Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation invivo. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1130–1135. doi: 10.1073/pnas.031576398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Bennekum A, et al. Class B scavenger receptor-mediated intestinal absorption of dietary beta-carotene and cholesterol. Biochemistry. 2005;44:4517–4525. doi: 10.1021/bi0484320. [DOI] [PubMed] [Google Scholar]
- 75.Hessel S, et al. CMO1 deficiency abolishes vitamin A production from beta-carotene and alters lipid metabolism in mice. J Biol Chem. 2007;282:33553–33561. doi: 10.1074/jbc.M706763200. [DOI] [PubMed] [Google Scholar]
- 76.Fierce Y, et al. In vitro and in vivo characterization of retinoid synthesis from beta-carotene. Archives of biochemistry and biophysics. 2008;472:126–138. doi: 10.1016/j.abb.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lobo GP, et al. ISX is a retinoic acid sensitive gatekeeper that controls intestinal β,β-carotene absorption and vitamin A production. Faseb J. 2010 doi: 10.1016/j.cub.2009.1012.1022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O'Byrne SM, et al. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT) J Biol Chem. 2005;280:35647–35657. doi: 10.1074/jbc.M507924200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quadro L, et al. Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. The EMBO journal. 1999;18:4633–4644. doi: 10.1093/emboj/18.17.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saari JC, et al. Visual cycle impairment in cellular retinaldehyde binding protein (CRALBP) knockout mice results in delayed dark adaptation. Neuron. 2001;29:739–748. doi: 10.1016/s0896-6273(01)00248-3. [DOI] [PubMed] [Google Scholar]
- 81.Travis GH, et al. Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. Annu Rev Pharmacol Toxicol. 2007;47:469–512. doi: 10.1146/annurev.pharmtox.47.120505.105225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Biesalski HK, et al. Biochemical but not clinical vitamin A deficiency results from mutations in the gene for retinol binding protein. The American journal of clinical nutrition. 1999;69:931–936. doi: 10.1093/ajcn/69.5.931. [DOI] [PubMed] [Google Scholar]
- 83.Seeliger MW, et al. Phenotype in retinol deficiency due to a hereditary defect in retinol binding protein synthesis. Investigative ophthalmology & visual science. 1999;40:3–11. [PubMed] [Google Scholar]
- 84.Fan J, et al. Opsin activation of transduction in the rods of dark-reared Rpe65 knockout mice. J Physiol. 2005;568:83–95. doi: 10.1113/jphysiol.2005.091942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Palczewski K, et al. Rod outer segment retinol dehydrogenase: substrate specificity and role in phototransduction. Biochemistry. 1994;33:13741–13750. doi: 10.1021/bi00250a027. [DOI] [PubMed] [Google Scholar]
- 86.Jager S, et al. Opsin/all-trans-retinal complex activates transducin by different mechanisms than photolyzed rhodopsin. Biochemistry. 1996;35:2901–2908. doi: 10.1021/bi9524068. [DOI] [PubMed] [Google Scholar]
- 87.den Hollander AI, et al. Leber congenital amaurosis: genes, proteins and disease mechanisms. Progress in retinal and eye research. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 88.Allikmets R, et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nature genetics. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 89.Maeda A, et al. Retinol dehydrogenase (RDH12) protects photoreceptors from light-induced degeneration in mice. J Biol Chem. 2006;281:37697–37704. doi: 10.1074/jbc.M608375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wenzel A, et al. The Rpe65 Leu450Met variation increases retinal resistance against light-induced degeneration by slowing rhodopsin regeneration. J Neurosci. 2001;21:53–58. doi: 10.1523/JNEUROSCI.21-01-00053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thompson DA, Gal A. Vitamin A metabolism in the retinal pigment epithelium: genes, mutations, and diseases. Progress in retinal and eye research. 2003;22:683–703. doi: 10.1016/s1350-9462(03)00051-x. [DOI] [PubMed] [Google Scholar]