Abstract
Research in aging laboratory animals has characterized physiological and cellular alterations in medial temporal lobe structures, particularly the hippocampus, that are central to age-related memory deficits. The current study compares molecular alterations across hippocampal subregions in a rat model that closely mirrors individual differences in neurocognitive features of aging humans, including both impaired memory and preserved function. Using mRNA profiling of the CA1, CA3 and dentate gyrus subregions, we have distinguished between genes and pathways related to chronological age and those associated with impaired or preserved cognitive outcomes in healthy aged Long Evans rats. The CA3 profile exhibited the most prominent gene expression differences related to cognitive status and of the three subregions, best distinguished preserved from impaired function among the aged animals. Within this profile differential expression of synaptic plasticity and neurodegenerative disease-related genes suggests recruitment of adaptive mechanisms to maintain function and structural integrity in aged unimpaired rats that does not occur in aged impaired animals.
1. Introduction
Cognitive impairment occurs in a substantial segment of the elderly population. The use of animal models has established that disability at older ages, such as memory impairment, can have a functional basis in brain systems with largely preserved neuronal circuits. In the hippocampus, for example, the numbers of principle neurons in its major subdivisions in the rat brain do not differ as a function of age or cognitive status (Rapp and Gallagher, 1996). Connectional integrity is also remarkably preserved with limited reductions that are highly circuit specific. For example, in aged rats with impaired performance in spatial tasks, connections from Layer II neurons of the entorhinal cortex are diminished (approximately 20% decreased) in the dentate gyrus and CA3 region, while connections in the CA1 region, both from Schaffer collaterals and Layer III of entorhinal cortex are preserved (Smith et al., 2000; Geinisman et al., 2004). Against a background of largely preserved structure, other research has provided ample evidence for functional alterations in the hippocampus that are closely tied to age-related cognitive decline (Wilson et al., 2006). Here too, however, the effects of aging differ across hippocampal subregions (Wilson et al., 2005; Burke and Barnes, 2006).
Prior aging studies have examined the global mRNA expression profiles in the hippocampus in its entirety (Rowe et al., 2007; Pawlowski et al., 2009) or only in one specific subregion (Blalock et al., 2003; Verbitsky et al., 2004; Burger et al., 2007; Burger et al., 2008; Kadish et al., 2009) in the rodent brain. These studies have generally confirmed an abundance of molecular alterations that occur as a function of age in the hippocampal system and also identified markers associated with the presence and severity of cognitive impairment independent of chronological age. Notably no published studies have examined expression patterns in the CA3 subregion despite strong evidence of a role for this subfield in information encoding and age-related memory deficits (Nakazawa et al., 2003; Leutgeb et al., 2004; Vazdarjanova and Guzowski, 2004; Wilson et al., 2005; Leutgeb et al., 2007; Kesner et al., 2008). The current study examines the profile of gene expression in a well-characterized, Long Evans rat model of neurocognitive aging across the three major subregions of the hippocampal formation. Aged Long Evans rats are a potentially informative model for this purpose because subjects from a single cohort perform with a range of ability in hippocampal-dependent memory tasks (Gallagher et al., 1993; Robitsek et al., 2008). In the study presented here, we expected that the expression pattern in each hippocampal subregion would distinguish impaired from intact memory performance in the aging population and, additionally, that comparison of the regional profiles could be informative as to factors affecting specific circuits within this system.
Here we report that large-scale changes in the CA3 subregion are closely coupled to cognitive status. Aged rats with impaired performance in a hippocampal-dependent behavioral task exhibit a molecular profile that segregates these subjects from both young adults and aged cohorts with preserved cognition. Moreover the CA3 molecular profile segregates expression patterns of aged rats with good cognitive outcomes from both impaired aged subjects and young rats. Overall, the results point to the CA3 region as an important component of the neurocognitive network that determines aging outcomes.
2. Methods
2.1 Subjects
Aged, male Long-Evans rats were obtained at 8–9 mo of age from Charles River Laboratories (Raleigh, NC) and housed in a vivarium at The Johns Hopkins University until 24–26 mo of age for the present experiments. Young rats at 6 months of age were tested alongside the aged rats. All rats were individually housed at 25°C and maintained on a 12 hr light/dark cycle. Food and water were provided ad libitum. All rats included in the study were determined to be healthy as confirmed by pathogen-free status throughout the experiments, screening for disability, as well as by necropsies at the time of sacrifice. All procedures were approved by the institutional animal care and use committee in accordance with the National Institutes of Health directive.
2.2 Behavioral characterization
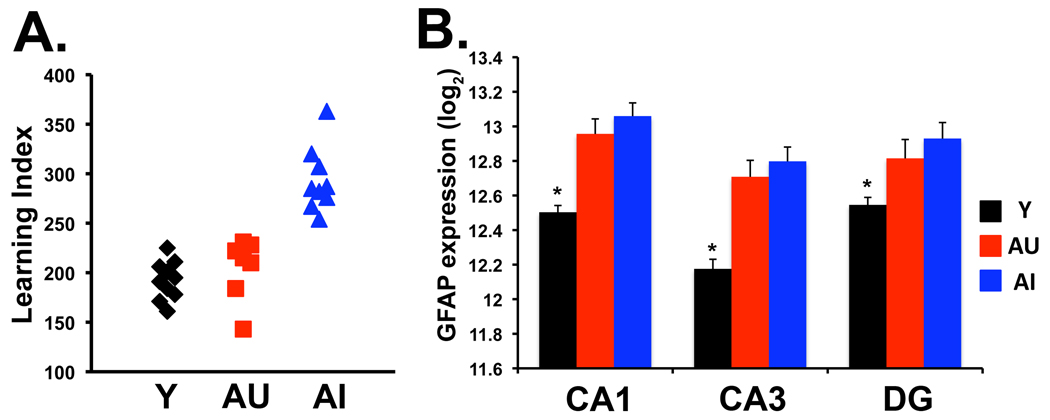
Behavioral assessment of memory function in a Morris water maze task was conducted as previously described (Gallagher et al., 1993). The primary measure used in the spatial learning task was proximity to the escape platform location, a sensitive method for behavioral analysis in this task (Maei et al., 2009). A learning index was generated from the proximity of the rat to the escape platform during probe trials interpolated throughout training and was used to define impairment in the rats. Lower scores reflect better performance as they indicate a search closer to the platform location. Aged rats were categorized based on the normative range of young performance established across years of testing within this paradigm. Those performing as well as young were designated aged unimpaired (AU) whereas those performing worse than young were considered aged impaired (AI). Cue training (visible escape platform) occurred on the last day of training to test for sensorimotor and motivational factors independent of spatial learning. In order to complete dissections at a standard timepoint after behavioral characterization, animals were included in this study from 4 different behavioral testing runs over a 3-month span. Each run included aged and young subjects. Animals were selected for inclusion in this study to represent a complete range of performance as indicated by learning index (Figure 1A).
Figure 1.
A. Morris water maze performance for all aged and young rats used in microarray study. Learning index scores were derived from proximity measures during probe trials interpolated throughout training as in Gallagher et al (1993). Lower scores indicate better performance. The graph illustrates that aged unimpaired rats perform within the range of normal young rats. B. Microarray assessment of glial fibrillary acidic protein (GFAP) shows increased expression with age in all three subregions consistent with previously published data using different analysis methods (Smith et al., 2001). Log2 expression values are graphed; *, SAM p<0.0001 relative to all aged rats. Y, young; AU, Aged unimpaired; AI, Aged impaired.
2.3 Microarray hybridization and analysis
All rats (16 aged, 10 young) were sacrificed at two weeks after completion of behavioral testing by rapid decapitation and brains removed from the skull. The two-week time frame was chosen to maximize the likelihood of detecting stable, baseline mRNA profile differences and minimizing the effects of learning or swimming induced differences. The CA1, CA3, and DG were microdissected from 400 micron transverse sections of the hippocampus along its entire longitudinal extent. Tissue was snap frozen and stored at −80C until all samples were collected. All subsequent procedures were performed on a subregional basis with equal inclusion (as best possible) of Y, AU and AI subjects per batch when processing required multiple batches. Total RNA was extracted by homogenization in TRIzol reagent (Invitrogen) followed by application to Qiagen RNeasy columns. RNA samples (7AU, 8 AI and 9 Y for each region) were sent to the Johns Hopkins Microarray core facility for cRNA labeling and hybridization to Affymetrix rat 230 2.0 microarrays using standard Affymetrix recommended procedures. All subjects were the same for the three regions except one Y and one AI were substituted in the CA3 subregion due to insufficient RNA quantity after extraction. All quality control, normalization, differential expression, and exploratory analysis of microarray data were performed by using the open-source R statistical language (www.r-project.org). The quality of microarray data was assessed on many levels, resulting in the omission of 5 of the 72 hybridizations from the analysis. The gcRMA package in Bioconductor (www.bioconductor.org) (Irizarry et al., 2003) was used to normalize microarray data. In the CA1 data a batch effect resulting from microarray processing procedures was removed individually from each probe set signal by simple subtraction of the mean difference of each batch to the grand mean. Significance analysis in microarrays (SAM) d-statistics (Tusher et al., 2001) were combined with an empirically-derived low-intensity cut-off to assess differential expression across comparison groups of animals. A false detection rate (FDR) was calculated by comparing the observed differential expression statistics to those expected by chance (estimated by permuting the group labels of the data many times and recalculating differential expression statistics). For comparisons that included all subjects, i.e. age and age impaired comparisons, a 5% FDR was imposed, but this was increased to 10% for comparison between the aged subgroups due to the reduced number of subjects. A principal component analysis (PCA) was used to visualize global differences between arrays for each subfield. Each array is plotted as a single point on a on a 2 dimensional graph. PCA attempts to express the maximal amount of variance across all genes in the first principal component (PC1). The second principal component is orthogonal to PC1 within the high dimensional gene expression space. The PCA is intrinsically blind to group identity and samples are labeled by cognitive status and age after the analysis.
2.4 Differential Expression of Functional Gene Groups
Functionally related gene groups for each subregion were defined by annotations within the Gene Ontology (GO), the Kyoto Encyclopedia of Genes and Genomes (KEGG), the PFAM database (protein sequence motifs), the National Center for Biotechnology Information (NCBI, including genes sharing particular transcription factor binding sites, and protein-protein interactions), as well as functional gene groups assembled by the Broad Institute at MIT (http://www.broad.mit.edu/gsea/downloads.jsp) and the Stanford Microarray group (http://www-stat.stanford.edu/~tibs/GSA/). This categorization method allows a single gene to be a member of more than one group and thus results overlapping and partially redundant functional groups. Groups were limited to those with greater than 5 and less than 600 members resulting in 4302, 4497 and 4356 partially overlapping groups for the CA3, CA1 and dentate gyrus respectively. These groups were then queried for an over-representation of increased or decreased genes. The assessment of differential expression in defined groups was carried out by implementation of the Wilcox rank sum test in the GeneSetTest function in the limma package in Bioconductor (Smyth, 2004). Listed p-values are not corrected for multiple comparisons and should be interpreted accordingly.
2.5 In Situ Hybridization
A biologically independent set of behaviorally characterized young and aged rats was used to confirm microarray findings with in situ hybridization histochemistry and to provide anatomical resolution. Probe templates were synthesized de novo by PCR from whole hippocampal RNA and were cloned in pGem7zf+ plasmid or modified to contain SP6 and T7 RNA polymerase binding sites by PCR. Thirty-micrometer sections from paraformaldehyde fixed brains were taken through the hippocampus and hybridized with 35S-UTP-labeled probe generated from the templates. Hybridized sections were exposed in a phosphorimager cassette and quantified using Imagequant.
2.6 Immunohistochemistry
A biologically independent set of behaviorally characterized young and aged rats was used to confirm kcnd2 microarray findings with immunohistochemistry and to provide anatomical resolution. Forty-micrometer histological sections were labeled with a commercially available Neuromab, monoclonal antibody against Kv4.2 (Antibodies Inc., Davis, CA) in an immunoperoxidase protocol. Protein expression in CA3 was quantified using ImageJ.
2.7 Ingenuity pathways analysis
Significant gene regulations within the AI and AU animal groups were analyzed using Ingenuity Pathway Analysis (IPA version 6.0, Ingenuity inc, Redwood city, CA 94063, USA). For each of the sub-fields; CA1, CA3 and DG genes were selected which were regulated more than 1.2 fold between AI and AU individuals and were associated with a p < 0.01. 166 (DG), 211 (CA3), and 68 (CA1) genes were mapped to pathways using these criteria. Additional methodological details for all procedures can be found in supplemental text.
3. Results
3.1 mRNA expression analysis of hippocampal subregions
Gene expression profiles were generated for each of the three major hippocampal subregions from a model of neurocognitive aging in which aged Long Evans rats exhibit individual differences on a hidden platform water maze task that is sensitive to spatial memory impairment. Ranging from preserved performance to markedly impaired spatial memory, approximately half of the subjects in this study population perform on par with young animals while the rest perform more poorly. For this research, all subjects were behaviorally characterized on the standardized hidden platform watermaze with aged subjects selected for inclusion representing the full range of individual differences based on the behavioral learning index score (Gallagher et al., 1993). Aged rats falling within the range of young (Y) performance were designated aged unimpaired (AU) while those which performed worse than young were considered aged impaired (AI) (Fig. 1A). As expected, aged rats differed from Y during training trials (Fig. S1) but no difference was observed among the groups during cue training to a visible platform (data not shown). Using a comprehensive rat expression array (Affymetrix rat 230 2.0) microarray analysis was conducted on dissected CA1, CA3 and dentate gyrus (DG) subregions of the aged and young rats. For each subregion, mRNA from 7 AU, 8 AI and 9 Y rats was assessed independently resulting in the initial processing of 72 microarrays. Extensive quality control analysis of all data identified 5 arrays that did not meet minimum standards and were excluded from further analyses. The resulting Ns were 9Y, 6AU, 8AI for CA1; 9Y, 6AU, 8AI for CA3 and 8Y, 6AU, 7AI for dentate gyrus. For each subregion, an empirically defined low expression cut-off was applied to gcRMA derived values to remove probesets targeting extremely low or absent mRNAs which may lead to inaccurate results. All raw microarray data has been made available in the public GEO database: http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE14726.
3.2 Individual and global expression patterns associated with chronological age are broadly evident across subregions and reflect known “aging” molecular phenotypes
A substantial body of research has established specific genes and pathways that are modulated with chronological age. Although not the main focus of this research, our experimental design provided an opportunity to assess gene expression patterns associated with chronological age for the purpose of dataset validation by comparison to established aging phenotypes and to distinguish “age changes” from those related to cognitive performance. Thus, prior to further unsupervised analyses, datasets were queried for changes in select genes or gene groups from prior research with empirically defined gene expression changes associated with chronological age. gcRMA data acquired from the microarrays were analyzed for expression changes using significance analysis in microarray (SAM; Tusher et al., 2001). Chronological age changes were identified by comparing mRNA expression levels from all aged rats (AU and AI) to those of young rats. For each probeset, SAM generates a d-statistic and p-value used to determine if a significant group difference is evident. Increased GFAP mRNA in the entire hippocampus is observed in our study population and widely reported elsewhere (Nichols et al., 1993; Laping et al., 1994; Sugaya et al., 1996; Smith et al., 2001). SAM analysis of the current microarray data revealed increased expression of this gene in aged subjects in all three subregional comparisons, consistent with the published data (Fig 1B). Similarly, an age-related increase in prodynorphin mRNA was detected in the DG (p<0.0002) consistent with the age-related increase in hippocampal dynorphin mRNA and protein as previously determined in this model (Jiang et al., 1989). DG prodynorphin expression far exceeds levels elsewhere in the hippocampus thus it is not unexpected that in CA1 and CA3, only 1 of the 2 prodynorphin probesets exceeded the low expression cutoff in those regions. For validation of genes with decreased expression, in situ hybridization was performed in a separate set of behaviorally characterized aged and young rats for two genes, lipoprotein lipase (LPL) and the gene encoding the α5 subunit of the GABAA receptor (Gabra5). CA3-specific diminished expression was confirmed for both (p<0.03 for LPL and p<0.01 for Gabra5).
We have also found that our observed microarray age changes recapitulate findings from aging studies of other investigators. This point is best illustrated by a comparison to a meta-analysis of microarray studies from a variety of tissues and species (de Magalhaes et al., 2009) where high correspondence (>65% of matched genes) was found between lists of genes consistently increased or decreased with age in that study and the CA1 and CA3 datasets described here. Moreover, pair-wise comparisons between our subregional datasets of differential expression statistics for individual genes (SAM d-statistic) found rather strong correlations (Table 1, top row), suggesting a measure of similarity in the identities and direction of gene changes associated with chronological age across subregions. We also performed functional groups analysis of our expression data using multiple publicly available databases to define gene groups. Gene groups were assessed for overrepresentation of increased or decreased genes, the most highly significant of which are listed in Table S1 (online supplementary data). Many of these groups correspond well to functional groups found by others. For example, gene groups for lysosomal constituents and immune function were increased with age in all three subregions (Wilcox rank sum, p<0.001) consistent with whole hippocampal datasets that find similar changes (Blalock et al., 2003; Rowe et al., 2007). Together the above described comparisons of chronological age-dependent expression changes serve to corroborate the reliability of our subregional datasets supporting the assertion that any observed differences in molecular profiles, as described further below for changes specifically associated with cognitive outcomes, are not confounded by experimental factors affecting sensitivity or variability across our subregional datasets.
Table 1.
Subregional pair-wise Pearson correlations of individual gene SAM d-statistics for each statistical comparison.
| Group Comparison |
CA3 v CA1 | CA3 v DG | CA1 v DG |
|---|---|---|---|
| AI+AU v Y | 0.56 | 0.35 | 0.48 |
| AI v AU+Y | 0.37 | 0.23 | 0.38 |
| AI v AU | 0.19 | 0.06 | 0.09 |
3.3 Global assessment of gene expression profiles indicates distinct subfield characteristics
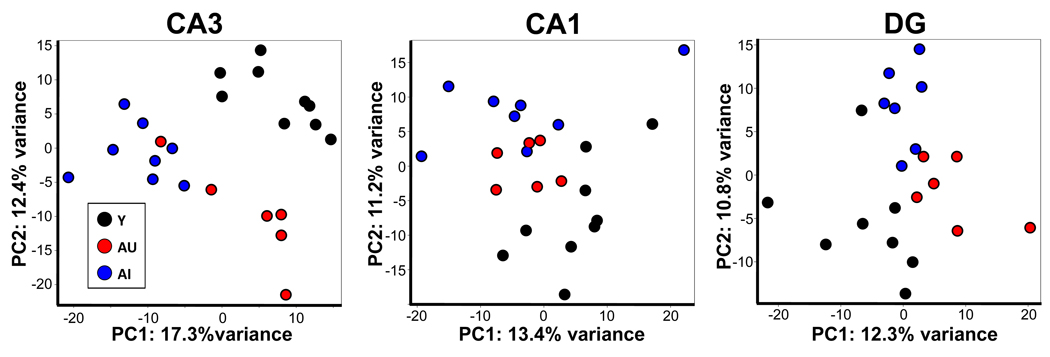
For a global assessment of the gene profiles, we employed a principal component analysis (PCA) to characterize relationships between individual subjects and subject groups within a given subregion (Fig. 2). The PCA visualization attempts to express variance across all genes in two-dimensional space. Each point in the plot represents a single array, and the distance between points illustrates the overall similarity of expression intensities with more proximal points representing arrays that have more similarity. In all three regions, subjects of a particular cognitive status and age tended to cluster together indicating that the expression profiles reflected features of age and behavioral phenotypes. However the relative relationships among those groups differs between subfields and CA3 prominently distinguishes between the three groups of animals. Within the CA3 plot, the horizontal axis (the first principal component, PC1) largely segregates the AI rats from both the Y and AU, separating out the subjects with impaired behavioral performance irrespective of chronological age. Additionally, five of the six AU subjects form a group distinct from both AI and Y clusters, based primarily on the vertical axis (PC2), indicating unique mRNA expression patterns for these animals. The AU subject outside this group, which had the poorest behavioral score of all AU rats, segregated with the AI subjects, consistent with a behavioral characterization of borderline performance. The consistent segregation of subjects relative to aging and cognitive groups illustrates the sensitivity of the molecular analysis to individual gene expression differences related to cognitive status across subjects and hippocampal subregions.
Figure 2.
Global comparisons of gene expression profiles between the three hippocampal subregions. Principal component analysis of CA3 (left), CA1 (middle) and dentate gyrus (right). PCA compares expression intensities of all probes for each of the microarrays of a given subregion. 2-dimensional depiction of this analysis is accomplished by plotting the relative position of each array along the 2 vectors representing the greatest variance. The percentage of variance along each axis is noted on the graphs. Total variance accounted for by PC1 and PC2 combined is as follows: CA3, 29.7%; CA1, 24.6%; DG, 23.1%. Each dot represents a single array: Y, black; AU, red; AI, blue.
While each of the first two principal components in the CA3 analysis more clearly segregates one of the three subject groups from the other two, CA1 expression patterns appear to form a graded continuum with the AU subjects intermediate between Y and AI on both principle components. This analysis demonstrates that in CA3, expression differences that distinguish Y and AU subjects are largely independent of those that distinguish Y and AI subjects, while in CA1 it is primarily the same expression differences of differing magnitude that separate the subjects groups. Hence, Y, AI and AU expression differences in CA3 are qualitatively different, while in CA1 they are more qualitatively similar, but quantitatively different. Examination of additional principal components (3 and 4) along with PC1 and 2 in each subregion revealed that the first two principal components in the CA3 analysis were the only to display this unique group separation (data not shown). In addition, an algorithmically distinct global analysis, multi-dimensional scaling (MDS), revealed patterns similar to those observed in the PCA, particularly for CA1 and CA3 subregions (Fig. S2A). Thus the unique pattern of arrays in these analyses suggests fundamentally different expression characteristics between subregions with regard to cognitive status in aging.
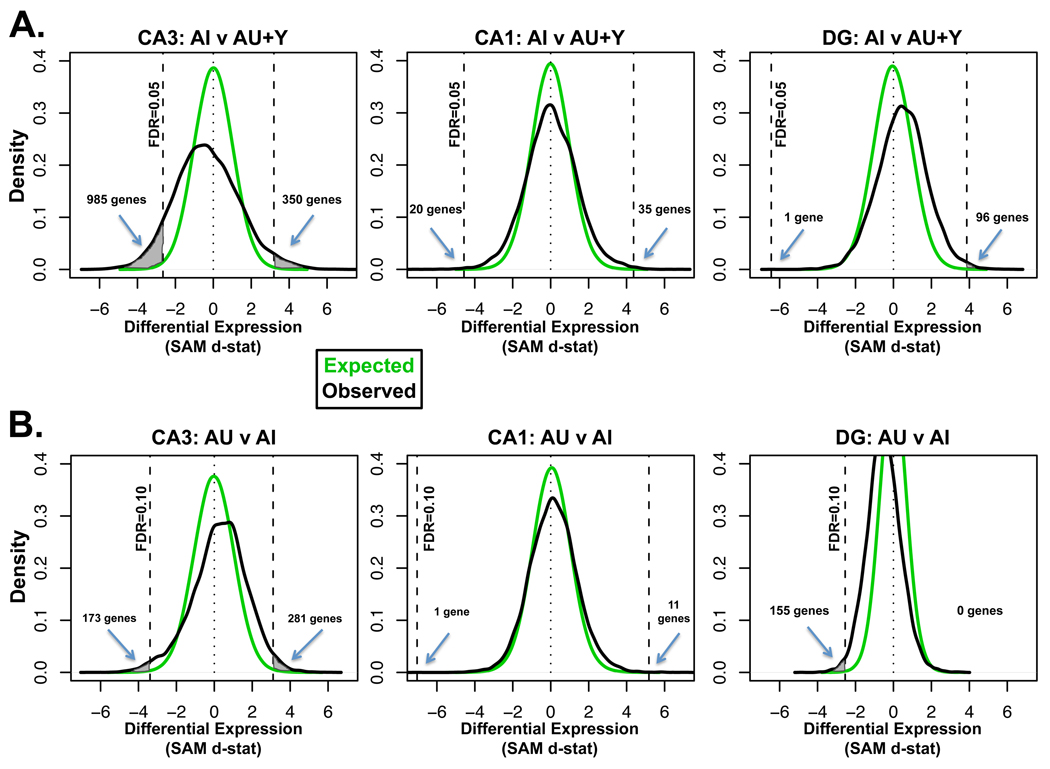
Unique subregional expression characteristics were also revealed in differential expression analyses targeting age and cognitive phenotypes. In addition to the assessment of chronological age changes described above, two comparisons were performed to capture genes changes associated with impairment [AI compared to the combined group of young and AU (AI v Y+AU)] and cognitive status within the aged cohort (AU v AI). CA3 was found to have the greatest number of significantly differentially expressed probesets with p<0.05 (Table S2). To visualize these comparisons graphically, estimates of observed d-statistic distributions were plotted (Fig 3A, B and S2B, black line) and compared to a distribution expected by chance (green line). Implementing a false detection rate (FDR) cut-off of 5% for the impairment comparison (AI v AU+Y, Fig 3A) and 10% for cognitive status within aged group (AU v AI, Fig 3B) spotlights the subregional differences and the prominence of CA3 changes. A distinctly different analysis of the gene array data, MAS5.0 coupled with student t-test, similarly found more CA3 expression differences irrespective of the p-value cut-off employed (Table S2 and data not shown). Pair-wise subregional correlations of SAM d-statistics for individual probesets exhibited less similarity between subregions in the AI comparison and, particularly, in the AI v AU comparison than for the chronological age comparison (Table 1) suggesting genes differing with cognitive performance exhibit more subregional specificity. While analyses from all subregions detected expression differences related to both cognitive status and age, CA3 clearly exhibited more expression differences and the most robust changes associated with cognitive function. Because of these data, along with minimal existing data on this subregion, we focused the remainder of the manuscript on the CA3 analysis although data from other subregions is noted when appropriate.
Figure 3.
Graphical visualization of SAM d-statistic distributions illustrates significant CA3 expression changes associated with cognition. The density of observed d-statistic distributions (black lines) were plotted for A. the aged impaired comparison (AI v AU+Y) and B. comparison between aged subjects (AI v AU) for each subregion. The distribution expected by chance (green line) was used to determine the false detection rate (FDR; see methods for details). Dotted vertical lines to the left and right demarcate the point beyond which the false detection rate is 5% for A. and 10% for B. Differentially expressed probe sets exceeding this FDR are represented by the shaded space between the green and black lines to the outside of the dotted lines and the numbers of genes meeting the FDR are noted on each graph. Numbers on the x-axis to the right (positive d-stat) indicate increased expression and numbers to the left (negative d-stat) indicate decreased expression in AI subjects (A.) or AU subjects (B.).
3.4 CA3 subregion exhibits a prominent profile associated with impairment in aged subjects
As seen in Fig 3A gene expression changes associated with impaired spatial learning performance were far greater in the CA3 subregion than in either of the others. The 5% false detection rate cut-off revealed more than 1300 probe sets differentially expressed in CA3 but many fewer in both CA1 and DG. A functional groups analysis was performed on the CA3 dataset to better understand pathways and gene classes altered with impairment (Table S3). Consistent with the earlier analysis for genes associated with chronological age, many highly significant groups such as those associated with lysosome, immune function or mRNA translation were increased in this analysis and, notably, changes in these groups were not specific for the CA3 subregion (Table S3). The overlap between analyses suggests that some pathways differentially expressed with chronological age are altered more substantially in impaired subjects than in the aged unimpaired subjects.
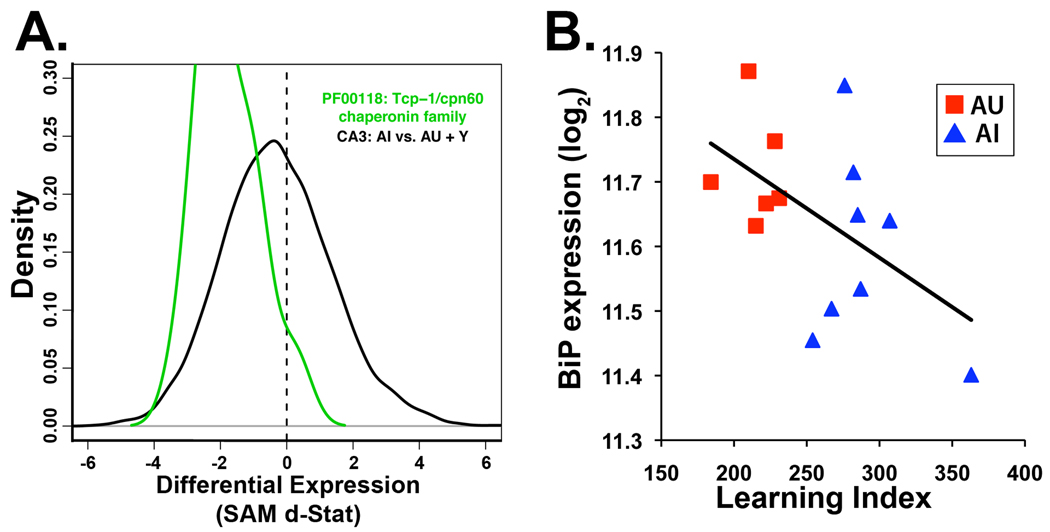
Additional significant functional group changes were more limited to the CA3 dataset. Decreased expression in AI rats was found for chaperone/protein folding genes, such as members of chaperonin containing Tcp1 complex (CCT; Fig 4A). In situ hybridization follow-up of Tcp1, a prominent member of this complex, confirmed decreased expression in AI rats in the CA3 subregion but not in CA1 (Table 2, Fig S3). Further investigation into the status of other chaperones found decreased expression of the endoplasmic reticulum chaperone Hspa5/BiP in the CA3 of AI subjects via both microarray and in situ hybridization (Table 2). For this gene expression levels correlated with learning ability among the aged animals in the study such that lower expression was found in rats with poorer performance on the watermaze (Fig 4B). In situ hybridization further uncovered decreased expression of an additional endoplasmic reticulum chaperone, calreticulin, in AI rats (data not shown), suggesting a consistent decrease in ER chaperone capacity. In contrast to Tcp1, decreased expression of ER chaperones was not strictly limited to the CA3, but rather showed lowered expression in both CA3 and CA1 subregions in the follow-up analysis (Table 2). As expected, in situ hybridization images confirmed strong expression of all three genes in the pyramidal cell layers of both CA3 and CA1 (Fig S3). Consistent with those findings, Hspa5/BiP and Calr were recently found to be decreased in aged impaired mice in a whole hippocampal microarray study (Pawlowski et al., 2009). Nonetheless, our data demonstrate that the CA3 subregion exhibits a broader range of chaperone decline in AI subjects with potentially far reaching effects on protein function and stability, including both transmembrane and cytoplasmic constituents.
Figure 4.
Chaperone expression is consistently decreased in CA3 of AI rats. A. CA3 d-statistic distribution of the TCP1/cpn-60 chaperonin family (green line) relative to the observed distribution from the AI comparison (AI v AU+Y, black line). Expression of genes in this group is shifted to the left indicating decreased expression (Wilcox rank sum p=0.00015). B. Individual expression data of Hspa5/BiP plotted against learning index for aged subjects (AU, Red; AI, blue). Higher learning index indicates poorer performance in watermaze testing and correlates with lower BiP expression in the aged rats (black line, Pearson correlation across all aged subjects; r = −0.51).
Table 2.
Microarray (left) and in situ hybridization (right) raw values for 2 validated genes, Tcp1 and Hspa5/BiP.
| Microarraya | In situ hybridizationa | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Young | Aged Unimpaired |
Aged Impaired |
Young | Aged Unimpaired |
Aged Impaired |
||||
| Tcp1 | CA3 | 1660 ± 44 | 1647 ± 32 | 1551 ± 36 | ** | 187.2 ±1.4 | 181.5 ± 8.5 | 169.6 ± 4.8 | * |
| CA1 | 1908 ± 55 | 1835 ± 18 | 1878 ± 45 | 131.0 ± 1.3 | 136.9 ± 6.7 | 130.0 ± 3.0 | |||
|
Hspa5/ BiP |
CA3 | 3707 ± 164 | 3373 ± 85 | 3104 ± 115 | *** | 472.1±6.9 | 446.1± 10.5 | 416.0±12.5 | * |
| CA1 | 4455 ± 188 | 4386 ± 84 | 3751 ± 122 | *** | 354.4±10.0 | 332.2 ± 3.6 | 327.0 ± 9.9 | # | |
Statistical analysis compared values for aged impaired subjects to the combined group of young and aged unimpaired.
SAM p<0.01
SAMp<0.05
ANOVA p<0.05
ANOVA p=0.07.
Expression pattern differences between CA3 and CA1 are more pronounced when considering mitochondrial (GO: 0005739) and oxidative phosphorylation (Kegg: 00190) gene groups in aged impaired subjects. Both subregions show significant gene expression shifts in AI rats compared with Y and AU, but in opposite directions. Mitochondrion and oxidative phosphorylation groups are increased in the impaired subjects in CA1 (WRS p=6.21e-9 and p=0.0027 respectively) but decreased in CA3 (WRS p=0.0024 and p=0.00018 respectively, Table S2). Prior studies have also noted alterations in pathways regulating energy metabolism (Baskerville et al., 2008; Kadish et al., 2009), however our data reinforce the notion that aging, at least in cognitively impaired subjects, places differential demands on the individual hippocampal subfields.
3.5 CA3 expression profiles distinguish aged unimpaired rats from both aged impaired and young
In vitro studies of the hippocampus have provided evidence for a mechanistic shift in synaptic plasticity in aged unimpaired rats as compared to young rats (Lee et al., 2005; Boric et al., 2008). The gene profiles, albeit under basal conditions, similarly distinguish aged unimpaired from young as well as from aged impaired, as revealed by the PCA (Fig 2). This was particularly evident in the CA3 analysis and further suggests that CA3-specific physiological remodeling occurring with age may contribute to the preservation of cognitive function.
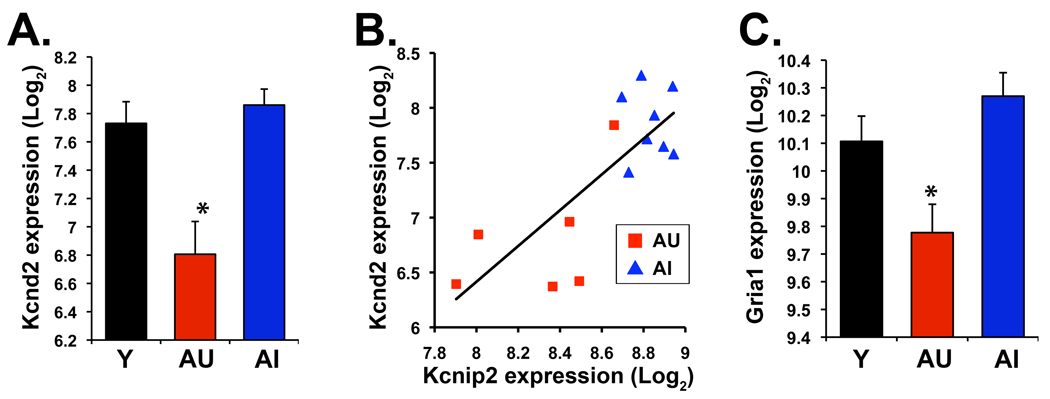
In order to identify the expression characteristics of AU subjects several comparisons were integrated to form a CA3 AU selective list (Table 3 and Table 4). We required that genes be significantly differentially expressed (SAM p<0.05) in AU v AI and AU v Y comparisons and not changed generally with age. This last criterion was defined as no expression difference in the AI v Y comparison (requiring SAM p>0.3) except when the direction of the AI change was in the opposite direction from the AU change relative to young. Of the 289 probesets, 172 (60%) showed decreased expression in AU subjects relative to both AI and Y (Table 3). Genes related to synaptic transmission were among those with decreased expression including the potassium channel, Kcnd2 (Fig. 5A), and its interacting protein, Kcnip2 (Table 3). Kcnd2 encodes the A-type potassium channel Kv4.2 which negatively regulates dendritic backpropagating action potentials (Lauver et al., 2006). Kcnip2 and a second gene decreased in AU rats, Dpp6 (AU v AI p=0.022), regulate KV4.2 cell surface expression and channel function (Birnbaum et al., 2004). These genes exhibit a coordinate decrease specifically in AU rats, illustrated by the Kcnd2 and Kcnip2 correlation (Fig. 5B), likely resulting in downregulation of a molecular complex that modulates plasticity at distal dendrites. Additionally, expression levels of Kcnd2 and Kcnip2 each correlate significantly with behavioral scores among the aged rats (Pearson r = 0.75 and 0.67, respectively), further supporting a possible role for these genes in mechanisms contributing to maintenance of cognitive capacity in aging.
Table 3.
CA3 genes significantly decreased in AU subjects relative to both Y and AI.
| Decreased Expressiona | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Symbolb |
AU v Y SAMp |
Gene Symbol |
AU v Y SAMp |
Gene Symbol |
AU v Y SAMp |
Gene Symbol |
AU v Y SAMp |
Gene Symbol |
AU v Y SAMp |
| Abhd8 | 0.0235 | Cs* | 0.0028 | Igf2r | 0.0074 | Osbpl1a | 0.0193 | Sidt2 | 0.0001 |
| Adora1 | 0.0313 | Csnk1d* | 0.0230 | Ina | 0.0344 | Pacsin1 | 0.0128 | Slc17a7 | 0.0054 |
| Alg2 | 0.0069 | Ctbp2 | 0.0295 | Jun | 0.0009 | Parvb | 0.0319 | Slc24a2 | 0.0417 |
| Ap3d1 | 0.0232 | Dab2 | 0.0002 | Kcnd2* | 0.0036 | Pde4dip | 0.0035 | Slc25a44 | 0.0452 |
| Apeg3 | 0.0003 | Dag1 | 0.0023 | Kcnip2 | 0.0054 | Pdia3 | 0.0285 | Slc2a3 | 0.0448 |
| Aplp2 | 0.0404 | Dbc1 | 0.0430 | Kif2a | 0.0161 | Pfkm | 0.0365 | Slco3a1 | 0.0132 |
| App | 0.0307 | Dnajc5 | 0.0297 | Kif5c | 0.0455 | Pigt | 0.0026 | Slitrk3 | 0.0419 |
| Arf3 | 0.0478 | Dnd1 | 0.0220 | Klc1 | 0.0111 | Pik3r2 | 0.0249 | Slu7 | 0.0040 |
| Arhgef11 | 0.0127 | Dos | 0.0234 | Klhl22 | 0.0051 | Pnn | 0.0151 | Smap2 | 0.0185 |
| Arl2bp | 0.0273 | Ehd3 | 0.0110 | Lingo1 | 0.0468 | Ppp1r14b | 0.0015 | Smarca4 | 0.0228 |
| Atcay | 0.0379 | Eif2c2 | 0.0108 | Lrp11* | 0.0112 | Ppp2r1a | 0.0459 | Snurf | 0.0479 |
| Atp1a1 | 0.0294 | Eif4a1 | 0.0109 | Lrrc59 | 0.0118 | Ppp2r4 | 0.0304 | Srp72 | 0.0010 |
| Atp1a3 | 0.0062 | Emg1 | 0.0042 | Lsamp | 0.0063 | Prg-2 | 0.0304 | Srpk2 | 0.0241 |
| Atp2a2 | 0.0052 | Enc1 | 0.0140 | Marcks* | 0.0001 | Prkar1a | 0.0135 | Ssbp3 | 0.0451 |
| Atp2b3 | 0.0342 | Epha5 | 0.0040 | Marcksl1 | 0.0016 | Rab14 | 0.0349 | Stmn4 | 0.0009 |
| B3gat1 | 0.0123 | Faim2 | 0.0069 | Mgll | 0.0408 | Rab5b | 0.0250 | Stx1a | 0.0070 |
| Bcat1 | 0.0080 | Fcho1 | 0.0064 | Mphosph8 | 0.0280 | Rapgefl1 | 0.0400 | Syt4 | 0.0407 |
| Bcl11b* | 0.0074 | Fgfr1 | 0.0037 | Mpp3 | 0.0478 | Rasl10b | 0.0259 | Tcf25 | 0.0091 |
| Blcap | 0.0246 | Fts | 0.0033 | Mpp6 | 0.0012 | Rdx | 0.0199 | Thra | 0.0160 |
| Calm3 | 0.0178 | Gabarapl1 | 0.0130 | Mrpl18 | 0.0383 | Runx1t1 | 0.0099 | Tmem38a | 0.0241 |
| Car11* | 0.0039 | Gnao1 | 0.0479 | Mtap2 | 0.0058 | Ryr2 | 0.0085 | Tpm3* | 0.0053 |
| Ccdc55 | 0.0076 | Gpr85 | 0.0127 | Mtap6 | 0.0273 | Sbf1* | 0.0129 | Traf7 | 0.0042 |
| Ccnl2 | 0.0110 | Gria1 | 0.0233 | Ncam1 | 0.0158 | Sec14l2 | 0.0169 | Tspan2 | 0.0133 |
| Cdk7 | 0.0331 | Herc1 | 0.0452 | Ncor1 | 0.0388 | Sel1l | 0.0231 | Ttc13 | 0.0312 |
| Centa1 | 0.0017 | Hnrnpa2b1 | 0.0118 | Neu1 | 0.0035 | Sept3 | 0.0215 | Unc13a | 0.0446 |
| Centg3* | 0.0186 | Hpcal4 | 0.0230 | Nfix | 0.0346 | Sept8 | 0.0020 | Usp7* | 0.0138 |
| Cplx2 | 0.0399 | Hsp90aa1 | 0.0070 | Nisch | 0.0017 | Sf3b2 | 0.0409 | Vamp1 | 0.0208 |
| Crim1 | 0.0318 | Hsp90ab1 | 0.0110 | Oaz2 | 0.0101 | Sfrs11 | 0.0473 | Zmiz1 | 0.0041 |
| Crym | 0.0213 | Ifnar1 | 0.0105 | Odz2 | 0.0381 | ||||
Genes listed are decreased in AU relative to both Y and AI.
b Only annotated (named) genes are listed; those marked with a * were represented by multiple probesets.
Table 4.
CA3 genes significantly increased in AU subjects relative to both Y and AI.
| Increased Expressiona | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Symbolb |
AU v Y SAMp |
Gene Symbol |
AU v Y SAMp |
Gene Symbol |
AU v Y SAMp |
Gene Symbol |
AU v Y SAMp |
Gene Symbol |
AU v Y SAMp |
| Acsl3 | 0.0003 | Eif1a* | 0.0126 | Helz | 0.0484 | Paip1 | 0.0007 | Sfrs15 | 0.0104 |
| Amigo2 | 0.0032 | Elovl4 | 0.0137 | Hip1 | 0.0441 | Pcmtd2 | 0.0021 | Stx8 | 0.0216 |
| Ank | 0.0001 | Emx2 | 0.0495 | Hnrpll | 0.0420 | Pdcl | 0.0203 | Syne1 | 0.0027 |
| Ank2 | 0.0498 | Ep300 | 0.0025 | Idi1 | 0.0243 | Pi4kb | 0.0064 | Tbl1x | 0.0041 |
| Apbb2 | 0.0034 | Exosc4 | 0.0051 | Ints5 | 0.0277 | Ppig | 0.0038 | Tm9sf2 | 0.0350 |
| Arhgef3 | 0.0153 | Fam49b | 0.0198 | Lactb2 | 0.0161 | Ppm1d | 0.0008 | Tmem126a | 0.0140 |
| Bcs1l | 0.0355 | Fam76a | 0.0169 | Larp1* | 0.0412 | Psme3 | 0.0144 | Tmem131 | 0.0113 |
| Btbd14a | 0.0035 | Fam98b | 0.0121 | Lrfn5 | 0.0492 | Ptpn3 | 0.0401 | Trappc5 | 0.0382 |
| Cdc37l1 | 0.0242 | Fbxl14 | 0.0058 | Magt1 | 0.0106 | Pum2 | 0.0053 | Utp15 | 0.0043 |
| Cdo1 | 0.0050 | Fbxo22 | 0.0236 | Mcm4 | 0.0068 | Rab2b | 0.0034 | Uxs1 | 0.0025 |
| Cebpz | 0.0263 | Fbxo44 | 0.0222 | Med13l | 0.0139 | Rbm6 | 0.0393 | Vps4b | 0.0467 |
| Cfl2 | 0.0414 | Fndc3a | 0.0070 | Mgat2 | 0.0060 | Rlf | 0.0202 | Wdfy3 | 0.0477 |
| Cpeb4 | 0.0234 | Foxn3 | 0.0039 | Mll1 | 0.0426 | Rnf111 | 0.0044 | Wdr37 | 0.0244 |
| Cpne8 | 0.0135 | Fxr1 | 0.0076 | Mrps11 | 0.0219 | Rpp14 | 0.0454 | Wnt2 | 0.0107 |
| Cpsf3 | 0.0432 | Gclm | 0.0428 | Msrb2 | 0.0211 | Sc5d* | 0.0008 | Xpr1 | 0.0227 |
| Cpsf5 | 0.0069 | Ggnbp2 | 0.0434 | Nebl | 0.0019 | Scaf1 | 0.0066 | Yme1l1 | 0.0065 |
| Cpsf6 | 0.0469 | Gjb6 | 0.0471 | Nek6 | 0.0221 | Scd1 | 0.0240 | Zbtb4 | 0.0320 |
| Crebl2 | 0.0402 | Gosr1 | 0.0049 | Nfia | 0.0028 | Sec31a | 0.0352 | Zc3h7b | 0.0290 |
| Ctbs | 0.0206 | Gpatch1 | 0.0331 | Ntrk2 | 0.0125 | Senp6 | 0.0472 | Zfp329 | 0.0257 |
| Cul3 | 0.0163 | Gtdc1 | 0.0047 | Nup153 | 0.0057 | Setd3 | 0.0018 | Znf124 | 0.0415 |
| Dnajc21 | 0.0471 | Hccs | 0.0131 | Nxt2 | 0.0402 | ||||
Genes listed are increased in AU relative to both Y and AI.
b Only annotated (named) genes are listed; those marked with a * were represented by multiple probesets.
Figure 5.
CA3 microarray expression data for Kcnd2 and Gria1. A. Graph of Kcnd2 mean expression data illustrates decreased expression in AU rats compared to both Y and AI (*, SAMp=0.0021, AU v AI; SAMp=0.0036, AU v Y). B. Expression intensities for Kcnd2 and Kcnip2 are intercorrelated in the aged subjects (black line; Pearson correlation across all aged subjects, r= 0.78). C. Gria1 expression is similarly decreased in AU subjects relative to both AI and Y (*, SAMp=0.0021, AU v AI; SAMp=0.023, AU v Y). Error bars indicate SEM.
Another critical component of excitatory neurotransmission and synaptic plasticity, Gria1, encodes the GluR1 subunit of the AMPA subtype of glutamate receptors, and exhibited decreased expression in the CA3 AU selective gene list (Table 3 and Fig. 5C). Further inspection of the microarray data showed that a gene for a second AMPA receptor subunit, Gria2, likewise showed significantly decreased expression in AU compared to AI and Y (data not shown) and AU v AI functional group analysis confirmed a decrease in the ionotropic glutamate receptor signaling pathway (GO:0035235, WRSp=7.93e-5). Along with the potassium channel findings, these data illustrate that molecules central to synaptic plasticity are downregulated in aged animals with maintained cognition as compared to those with impaired function. This differential expression is consistent with electrophysiological evidence of higher activity, e.g. firing rates, of CA3 neurons in AI rats (Wilson et al. 2005) and shifts in mechanisms of successful memory encoding in AU subjects.
3.6 Decreased protein levels of voltage-gated potassium channel, Kv4.2, in the CA3 subregion characterizes aged rats with preserved cognition
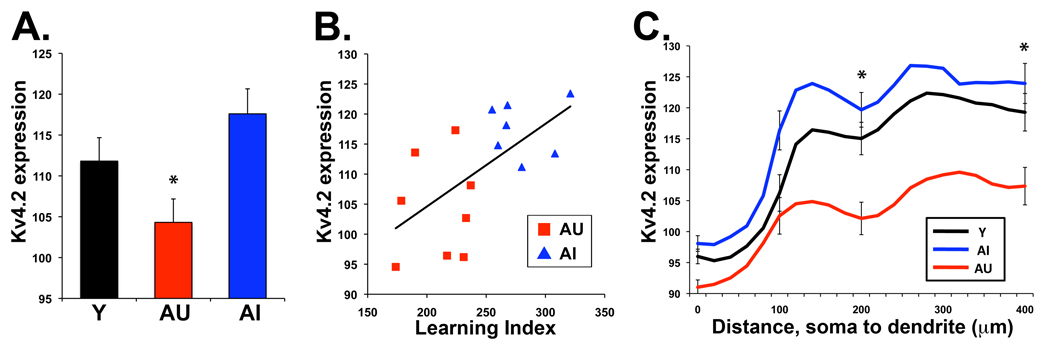
To determine if these findings extend to the protein level, immunohistochemical analysis of Kv4.2, the protein product of the Kcnd2 gene, revealed a significant reduction in protein expression in AU rats compared to Y and AI in CA3 (p < 0.05; Fig 6A and Fig S4) consistent with the decreased Kcnd2 gene expression observed among AU rats in the microarray. Additionally, Kv4.2 protein expression levels significantly correlated with learning ability among the aged animals in the study such that lower expression was found in rats with preserved performance on the watermaze (Pearson r = 0.63; Fig 6B).
Figure 6.
Kv4.2 protein expression is decreased in the CA3 of AU rats. A. Graph of Kv4.2 mean CA3 expression data illustrates decreased expression in AU rats compared to both Y and AI (ANOVA p = 0.016; *, pcon < 0.05, AU v Y and AI). B. Individual CA3 protein expression data of Kv4.2 plotted against learning index score for aged subjects (AU, Red; AI, blue). Lower learning index indicates better performance in watermaze testing and correlates with lower Kv4.2 expression in the aged rats (black line, Pearson correlation across all aged subjects; r=0.63). C. Kv4.2 protein expression in CA3b plotted from pyramidal soma to perforant path input within stratum lacunosum-moleculare. Plot of Kv4.2 mean expression reveals decreased protein expression in CA3b stratum radiatum and stratum lacunosum-moleculare in AU rats compared to both Y and AI (*, pcon < 0.05, AU v Y+AI). Kv4.2 expression levels depicted in gray scale intensity units from ImageJ. Error bars indicate SEM.
In normal young rats Kv4.2 protein exhibits a distinct pattern with regard to subcellular localization. Relatively low levels are found at the cell body with increasing amounts along the dendrites such that the distal dendrites have the greatest amount (Birnbaum et al., 2004). Examination of Kv4.2 expression from pyramidal cell body to distal apical dendrite in the stratum lacunosum-moleculare in CA3 revealed a linear trend in the relationship between Kv4.2 expression and distance on the dendrite from the soma (p < .001). Furthermore an interaction was found between animal group and protein expression at distances out on the dendrite, which was driven by the AU group (p = 0.021; Fig 6C). A significant reduction of Kv4.2 protein expression in AU rats compared to Y and AI was found at distal dendritic regions nearing the perforant path input from the entorhinal cortex, within both the CA3 stratum radiatum and stratum lacunosum-moleculare (Fig 6C).
3.7 Decreased expression of potassium channel conductance and Alzheimer's disease genes in the CA3 subregion characterizes aged rats with preserved cognition
Molecular pathway analysis is an additional method to determine relationships among genes differentially expressed as a function of neurocognitive aging outcomes. As with the other comparisons, CA3 has the most robust differential expression between AU and AI subjects (Fig 3B). In order to capture interactions amongst the most salient genes, an Ingenuity pathways analysis was performed for each subregion with the subset of genes from the AU v AI comparison that met a p-value <0.01 and at least a 1.2 fold change in expression (Table 5). Several pathways containing 20 or more genes meeting these criteria were found for CA3 but fewer for CA1 and DG. In CA3, two of the most involved pathways identified were related to neurodegenerative disease (APP signaling) and neuronal activity (potassium signaling). Twenty-three genes in the APP signaling pathway showed modulated mRNA expression in the CA3 subregion, of which 19 were downregulated in AU rats relative to AI. Likewise in the potassium signaling pathway, a large majority of the differentially expressed genes were decreased in AU relative to AI. Both Kcnd2 and Kchip2 were included in this pathway.
Table 5.
Ingenuity pathways found for genes significantly differentially expressed in the AU v AI comparison for each subregion.
| Subregion | Pathway | # Regulated genesa |
|---|---|---|
| CA3 | K+ signaling | 24 |
| Calcium signaling/vesicular transport and release |
23 | |
| APP signaling/Neurological disease |
23 | |
| Map kinase signaling | 22 | |
| PKC/p38 signaling | 21 | |
| CA1 | Erk/Akt/MapK3 signaling | 20 |
| DG | Cell morphology | 24 |
| Nervous system function | 20 | |
Only pathways with 20 or more genes listed.
4. Discussion
Recent studies have demonstrated differential contributions of hippocampal subregions to information encoding and retrieval functions in memory (Brun et al., 2002; Nakazawa et al., 2003; Leutgeb et al., 2004; Leutgeb et al., 2007). In particular, neurons in the CA3/dentate gyrus regions produce distinct representations of inputs with common elements, a process referred to as pattern separation (Leutgeb et al., 2007). A corresponding function has also been observed in human CA3/DG in functional neuroimaging (Bakker et al., 2008). This function is impaired in aging, as indicated by in vivo recordings in behaving rats and, more recently, by functional neuroimaging in man (M.A. Yassa, M. Albert, M Gallagher, and C.E.L. Stark, unpublished observations). In the CA3 region specifically, neurons exhibit reduced plasticity when AI rats are exposed to novel spatial environments and also exhibit elevated activity. Increased activity is an ongoing condition affecting principal neurons in the CA3 region that is not observed in the in vivo recordings from CA1 neurons (Wilson et al., 2005). The current study expands on this background using microarrays to identify regionally specific pathways that characterize impairment in aged subjects as well those that are modulated to preserve cognition with age. The notion that aged unimpaired subjects differ from young adults is supported by evidence for shifts in molecular and cellular mechanisms that support plasticity in those animals (Lee et al., 2005; Boric et al., 2008). The expression profiles garnered here confirm differential contribution of the three major hippocampal subregions to neurocognitive aging and highlight the importance of specific changes in the CA3 subregion.
PCA and MDS analysis enable 2-dimensional visualization of complex comparisons between probe sets on individual arrays. Both of these methods when applied to the current datasets reveal distinctive patterns for CA1 and CA3. While the CA3 profile distinctively segregated AU subjects from Y and AI, the three groups formed a continuum in CA1. This strongly suggests that aging trajectories in the CA3 contribute to differential cognitive outcomes in aging. The separation of AU and Y profiles, most notably, suggests that preservation of cognitive ability with age is not merely due to maintenance of young physiology but, instead, may require adaptive changes. Data from human imaging studies of young and old subjects with equivalent memory integrity show different connectivity profiles with other brain regions between young and old supporting the contention that successful cognitive performance in aging occurs differently than in young (Della-Maggiore et al., 2000; Grady et al., 2003).
Identification of molecular pathways in CA3 that are differentially expressed between the age unimpaired and aged impaired rats support a model in which excessive neuronal excitability in this subregion, as observed in the case of cognitive impairment (Wilson et al., 2005), leads to further degradation of synaptic and cellular function. Moreover, the robust profile in the CA3 region that distinguishes cognitive outcomes in this model may provide a context through which aging confers risk or resistance to neurodegenerative conditions. Ingenuity pathways analysis highlighted molecular groups/pathways associated with synaptic plasticity and neurodegenerative disease processes in the comparison of subgroups of aged rats with differing cognitive outcomes. Expression changes reflect increased synaptic transmission/disease components in aged impaired subjects relative to aged unimpaired. These data are further consistent with electrophysiological recording analysis in rodents and functional neuroimaging in humans, both of which find increased hippocampal activity in elderly subjects with impaired memory. In AI rats, in vivo recordings reveal high firing rates in CA3 place cells concomitant with a reduced capacity to encode features of a new environment (Wilson et al., 2005). Firing rates of the CA3 pyramidal neurons are increased both in basal activity and when information processing occurs. Functional neuroimaging in aged humans has indicated increased hippocampal activation in subjects with mild cognitive impairment relative to age-matched normal controls under conditions that tax pattern separation in a memory task. These subjects are at significantly increased risk for progressing to Alzheimer’s disease (AD) and elevated hippocampal activation in adults with mild cognitive impairment predicts subsequent cognitive decline (Dickerson et al., 2004; Miller et al., 2008). Similarly increased activation is found in cognitively normal individuals carrying an ApoE4 allele (Bookheimer et al., 2000), who also exhibit localized structural changes confined to the CA3/DG (Mueller et al., 2008). Together these data suggest a potential link between increased hippocampal excitation and neurodegenerative disease progression.
Aged unimpaired rats exhibit decreased synaptic transmission mRNA signatures relative not only to aged impaired rats but also relative to young in CA3. Genes found to be downregulated in this group include both those that mediate excitatory function (e.g. Gria1) and those normally contributing to inhibitory mechanisms (Kcnd2). As noted previously, Kcnd2 encodes a somatodendritic potassium channel known as Kv4.2 that regulates back-propagation of action potentials (Lauver et al., 2006). In correspondence with the downregulation of the kcnd2 gene, a decrease in protein expression among our aged unimpaired rats was confirmed for Kv4.2 in an immunohistochemical analysis. Reduction of this gene product, together with its binding partners, could result in greater dendritic depolarization, thereby promoting synaptic plasticity. Thus the complement of changes in synaptic transmission components in AU rats could result in altered plasticity mechanisms rather than an overall reduction in plasticity per se. In that context, it is notable that synaptic integrity at the entorhinal connections onto the most distal segments of CA3 dendrites is specifically reduced in aged rats with cognitive deficits and the downregulation in Kv4.2 protein expression relative to AI rats was localized to the entorhinal input at the perforant path (Smith et al., 2000). The hypotheses that adaptive plasticity mechanisms are recruited to maintain cognitive function in aging has not been tested directly for the CA3 region, but evidence for altered plasticity mechanisms in CA1 support the possibility of such an adaptation in CA3.
In addition to the global PCA, SAM d-statistic distributions support CA3 as a significant contributor to differing cognitive outcomes in aged subjects. The numbers of genes differentially expressed in CA3 that meet the FDR exceed the other two regions substantially in all comparisons. When assessing gene profiles associated with chronological age and age-associated impairment, expression changes correlated to some degree between subregions. Analysis of this overlap across subregions suggests that correlated genes generally fall into categories associated with homeostatic processes important for normal cellular function such as lysosomal function and translation. However, CA3 consistently exhibited more robust differential expression. Thus CA3 may be more sensitive to age-dependent neuronal dysfunction, representing in some cases prominent molecular alterations that also occur to a lesser extent in the other subregions.
Changes in chaperones in aged impaired rats exemplify this relationship between CA1 and CA3. Chaperones provide an essential cellular function, folding and trafficking many types of proteins so that they obtain their proper structure and localization. In the work presented here we demonstrated that both cytosolic and endoplasmic reticulum (ER) chaperones were decreased in CA3 but only endoplasmic reticulum chaperones were likewise decreased in CA1. BiP along with additional ER chaperones are essential in folding membrane bound and secreted proteins as well as in mediating a cellular stress response known as the unfolded protein response (UPR) (Schroder and Kaufman, 2005). Many necessary molecular components of the synapse must traffic through ER and be correctly folded by the resident ER machinery. In addition, proteins associated with neurodegenerative disease such as APP and presenilin 1 interact with the components of the UPR. Evidence suggests these interactions can modulate APP cleavage as well as UPR activation (Katayama et al., 1999; Katayama et al., 2001; Kudo et al., 2006). Tcp1 assembles with several other CCT proteins to form the chaperonin TriC/CCT complex whose major substrates are the structural proteins actin and tubulin, essential components of synapse structure (Dunn et al., 2001). In addition, TriC/CCT complex has been shown to modulate protein aggregation in a cellular neurodegenerative disease model (Kitamura et al., 2006; Tam et al., 2006). Thus reduction of mRNA for both of these chaperone pathways in CA3 increases the likelihood not only of impaired synapse function but also of potential for neurodegenerative cellular phenotypes associated with advancing age.
Although the numbers of differentially expressed genes in the dentate gyrus were roughly comparable to those of CA1, we kept the discussion of this subregion to a minimum as this profile was considerably more difficult to relate to known physiological age-and impairment-related modifications and showed less correspondence to the CA3 profile than CA1. Previous studies in this model have identified substantially decreased neurogenesis (Bizon and Gallagher, 2003) and reduced synaptic innervation from the entorhinal cortex in the dentate (Smith et al., 2000). Although large-scale evidence of such phenomena was not apparent in the microarray profile, gene expression changes consistent with earlier findings were found in the profiles, e.g. an age and AI-related increase in dynorphin mRNA (Jiang et al., 1989). This difficulty in identifying coherent phenotypic signatures may result from the inherent complexity of the neuronal subtypes within dentate gyrus. The DG tissue recovered from our dissection method included the pyramidal cell layer that extends between the blades of the DG (sometimes referred to as CA4) and hilar neurons in addition to dentate granule cells. The heterogeneity of this tissue may obscure signals generated by a single neuronal subtype. A more targeted approach, such as laser capture microdissection of the granule cell layer might produce a more robust and informative dataset.
This broad molecular analysis provides the first such study to directly compare across hippocampal subfields in a well-characterized model of neurocognitive aging. The data support a previously unidentified role for robust changes in CA3 that are closely coupled to the wide spectrum of cognitive outcomes in aged individuals. CA3 expression profiles identified molecular pathways that likely contribute to dysfunctional neuronal activity and information encoding capacity found in AI rats and present evidence that adaptive changes occur in rats that maintain neuronal function and cognitive abilities with age.
Supplementary Material
Acknowledgements
The authors would like to thank the JHMI Microarray core facility for array processing, Weidong Hu, Ming Teng Koh and Joanne Lee for technical assistance, Giovanni Parmigiani for bioinformatics consultation. This work was supported by National Institute on Aging/National Institutes of Health Grant P01AG09973, and a Freedom to Discover Award to MG from the Bristol Myers Squibb Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: The authors M.G. and R.P.H. are inventors on a provisional patent filed by Johns Hopkins University containing portions of the data described in this manuscript. Licensing agreements based on this patent are in negotiation.
References
- Bakker A, Kirwan CB, Miller M, Stark CE. Pattern separation in the human hippocampal CA3 and dentate gyrus. Science. 2008;319:1640–1642. doi: 10.1126/science.1152882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville KA, Kent C, Personett D, Lai WR, Park PJ, Coleman P, McKinney M. Aging elevates metabolic gene expression in brain cholinergic neurons. Neurobiol Aging. 2008;29:1874–1893. doi: 10.1016/j.neurobiolaging.2007.04.024. [DOI] [PubMed] [Google Scholar]
- Birnbaum SG, Varga AW, Yuan LL, Anderson AE, Sweatt JD, Schrader LA. Structure and function of Kv4-family transient potassium channels. Physiol Rev. 2004;84:803–833. doi: 10.1152/physrev.00039.2003. [DOI] [PubMed] [Google Scholar]
- Bizon JL, Gallagher M. Production of new cells in the rat dentate gyrus over the lifespan: relation to cognitive decline. Eur J Neurosci. 2003;18:215–219. doi: 10.1046/j.1460-9568.2003.02733.x. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Chen KC, Sharrow K, Herman JP, Porter NM, Foster TC, Landfield PW. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci. 2003;23:3807–3819. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookheimer SY, Strojwas MH, Cohen MS, Saunders AM, Pericak-Vance MA, Mazziotta JC, Small GW. Patterns of brain activation in people at risk for Alzheimer's disease. N Engl J Med. 2000;343:450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boric K, Munoz P, Gallagher M, Kirkwood A. Potential adaptive function for altered long-term potentiation mechanisms in aging hippocampus. J Neurosci. 2008;28:8034–8039. doi: 10.1523/JNEUROSCI.2036-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun VH, Otnass MK, Molden S, Steffenach HA, Witter MP, Moser MB, Moser EI. Place cells and place recognition maintained by direct entorhinal-hippocampal circuitry. Science. 2002;296:2243–2246. doi: 10.1126/science.1071089. [DOI] [PubMed] [Google Scholar]
- Burger C, Lopez MC, Baker HV, Mandel RJ, Muzyczka N. Genome-wide analysis of aging and learning-related genes in the hippocampal dentate gyrus. Neurobiol Learn Mem. 2008;89:379–396. doi: 10.1016/j.nlm.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger C, Cecilia Lopez M, Feller JA, Baker HV, Muzyczka N, Mandel RJ. Changes in transcription within the CA1 field of the hippocampus are associated with age-related spatial learning impairments. Neurobiology of Learning and Memory. 2007;87:21. doi: 10.1016/j.nlm.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Burke SN, Barnes CA. Neural plasticity in the ageing brain. Nat Rev Neurosci. 2006;7:30–40. doi: 10.1038/nrn1809. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Maggiore V, Sekuler AB, Grady CL, Bennett PJ, Sekuler R, McIntosh AR. Corticolimbic interactions associated with performance on a short-term memory task are modified by age. J Neurosci. 2000;20:8410–8416. doi: 10.1523/JNEUROSCI.20-22-08410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Salat DH, Bates JF, Atiya M, Killiany RJ, Greve DN, Dale AM, Stern CE, Blacker D, Albert MS, Sperling RA. Medial temporal lobe function and structure in mild cognitive impairment. Ann Neurol. 2004;56:27–35. doi: 10.1002/ana.20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AY, Melville MW, Frydman J. Review: cellular substrates of the eukaryotic chaperonin TRiC/CCT. J Struct Biol. 2001;135:176–184. doi: 10.1006/jsbi.2001.4380. [DOI] [PubMed] [Google Scholar]
- Gallagher M, Burwell R, Burchinal M. Severity of spatial learning impairment in aging: development of a learning index for performance in the Morris water maze. Behav Neurosci. 1993;107:618–626. doi: 10.1037//0735-7044.107.4.618. [DOI] [PubMed] [Google Scholar]
- Geinisman Y, Ganeshina O, Yoshida R, Berry RW, Disterhoft JF, Gallagher M. Aging, spatial learning, and total synapse number in the rat CA1 stratum radiatum. Neurobiol Aging. 2004;25:407–416. doi: 10.1016/j.neurobiolaging.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Grady CL, McIntosh AR, Craik FI. Age-related differences in the functional connectivity of the hippocampus during memory encoding. Hippocampus. 2003;13:572–586. doi: 10.1002/hipo.10114. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jiang HK, Owyang VV, Hong JS, Gallagher M. Elevated dynorphin in the hippocampal formation of aged rats: relation to cognitive impairment on a spatial learning task. Proc Natl Acad Sci U S A. 1989;86:2948–2951. doi: 10.1073/pnas.86.8.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadish I, Thibault O, Blalock EM, Chen KC, Gant JC, Porter NM, Landfield PW. Hippocampal and cognitive aging across the lifespan: a bioenergetic shift precedes and increased cholesterol trafficking parallels memory impairment. J Neurosci. 2009;29:1805–1816. doi: 10.1523/JNEUROSCI.4599-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Imaizumi K, Honda A, Yoneda T, Kudo T, Takeda M, Mori K, Rozmahel R, Fraser P, George-Hyslop PS, Tohyama M. Disturbed activation of endoplasmic reticulum stress transducers by familial Alzheimer's disease-linked presenilin-1 mutations. J Biol Chem. 2001;276:43446–43454. doi: 10.1074/jbc.M104096200. [DOI] [PubMed] [Google Scholar]
- Katayama T, Imaizumi K, Sato N, Miyoshi K, Kudo T, Hitomi J, Morihara T, Yoneda T, Gomi F, Mori Y, Nakano Y, Takeda J, Tsuda T, Itoyama Y, Murayama O, Takashima A, St George-Hyslop P, Takeda M, Tohyama M. Presenilin-1 mutations downregulate the signalling pathway of the unfolded-protein response. Nat Cell Biol. 1999;1:479–485. doi: 10.1038/70265. [DOI] [PubMed] [Google Scholar]
- Kesner RP, Hunsaker MR, Warthen MW. The CA3 subregion of the hippocampus is critical for episodic memory processing by means of relational encoding in rats. Behav Neurosci. 2008;122:1217–1225. doi: 10.1037/a0013592. [DOI] [PubMed] [Google Scholar]
- Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- Kudo T, Okumura M, Imaizumi K, Araki W, Morihara T, Tanimukai H, Kamagata E, Tabuchi N, Kimura R, Kanayama D, Fukumori A, Tagami S, Okochi M, Kubo M, Tanii H, Tohyama M, Tabira T, Takeda M. Altered localization of amyloid precursor protein under endoplasmic reticulum stress. Biochem Biophys Res Commun. 2006;344:525–530. doi: 10.1016/j.bbrc.2006.03.173. [DOI] [PubMed] [Google Scholar]
- Laping NJ, Teter B, Anderson CP, Osterburg HH, O'Callaghan JP, Johnson SA, Finch CE. Age-related increases in glial fibrillary acidic protein do not show proportionate changes in transcription rates or DNA methylation in the cerebral cortex and hippocampus of male rats. J Neurosci Res. 1994;39:710–717. doi: 10.1002/jnr.490390612. [DOI] [PubMed] [Google Scholar]
- Lauver A, Yuan LL, Jeromin A, Nadin BM, Rodriguez JJ, Davies HA, Stewart MG, Wu GY, Pfaffinger PJ. Manipulating Kv4.2 identifies a specific component of hippocampal pyramidal neuron A-current that depends upon Kv4.2 expression. J Neurochem. 2006;99:1207–1223. doi: 10.1111/j.1471-4159.2006.04185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Min SS, Gallagher M, Kirkwood A. NMDA receptor-independent long-term depression correlates with successful aging in rats. Nat Neurosci. 2005;8:1657–1659. doi: 10.1038/nn1586. [DOI] [PubMed] [Google Scholar]
- Leutgeb JK, Leutgeb S, Moser M-B, Moser EI. Pattern Separation in the Dentate Gyrus and CA3 of the Hippocampus. Science. 2007;315:961–966. doi: 10.1126/science.1135801. [DOI] [PubMed] [Google Scholar]
- Leutgeb S, Leutgeb JK, Treves A, Moser M-B, Moser EI. Distinct Ensemble Codes in Hippocampal Areas CA3 and CA1. Science. 2004;305:1295–1298. doi: 10.1126/science.1100265. [DOI] [PubMed] [Google Scholar]
- Maei HR, Zaslavsky K, Teixeira CM, Frankland PW. What is the Most Sensitive Measure of Water Maze Probe Test Performance? Front Integr Neurosci. 2009;3:4. doi: 10.3389/neuro.07.004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SL, Fenstermacher E, Bates J, Blacker D, Sperling RA, Dickerson BC. Hippocampal activation in adults with mild cognitive impairment predicts subsequent cognitive decline. J Neurol Neurosurg Psychiatry. 2008;79:630–635. doi: 10.1136/jnnp.2007.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Schuff N, Raptentsetsang S, Elman J, Weiner MW. Selective effect of Apo e4 on CA3 and dentate in normal aging and Alzheimer's disease using high resolution MRI at 4 T. Neuroimage. 2008;42:42–48. doi: 10.1016/j.neuroimage.2008.04.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa K, Sun LD, Quirk MC, Rondi-Reig L, Wilson MA, Tonegawa S. Hippocampal CA3 NMDA receptors are crucial for memory acquisition of one-time experience. Neuron. 2003;38:305–315. doi: 10.1016/s0896-6273(03)00165-x. [DOI] [PubMed] [Google Scholar]
- Nichols NR, Day JR, Laping NJ, Johnson SA, Finch CE. GFAP mRNA increases with age in rat and human brain. Neurobiol Aging. 1993;14:421–429. doi: 10.1016/0197-4580(93)90100-p. [DOI] [PubMed] [Google Scholar]
- Pawlowski TL, Bellush LL, Wright AW, Walker JP, Colvin RA, Huentelman MJ. Hippocampal gene expression changes during age-related cognitive decline. Brain Res. 2009;1256:101–110. doi: 10.1016/j.brainres.2008.12.039. [DOI] [PubMed] [Google Scholar]
- Rapp PR, Gallagher M. Preserved neuron number in the hippocampus of aged rats with spatial learning deficits. Proc Natl Acad Sci U S A. 1996;93:9926–9930. doi: 10.1073/pnas.93.18.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitsek RJ, Fortin NJ, Koh MT, Gallagher M, Eichenbaum H. Cognitive aging: a common decline of episodic recollection and spatial memory in rats. J Neurosci. 2008;28:8945–8954. doi: 10.1523/JNEUROSCI.1893-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe WB, Blalock EM, Chen K-C, Kadish I, Wang D, Barrett JE, Thibault O, Porter NM, Rose GM, Landfield PW. Hippocampal Expression Analyses Reveal Selective Association of Immediate-Early, Neuroenergetic, and Myelinogenic Pathways with Cognitive Impairment in Aged Rats. J Neurosci. 2007;27:3098–3110. doi: 10.1523/JNEUROSCI.4163-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Smith DR, Hoyt EC, Gallagher M, Schwabe RF, Lund PK. Effect of age and cognitive status on basal level AP-1 activity in rat hippocampus. Neurobiol Aging. 2001;22:773–786. doi: 10.1016/s0197-4580(01)00240-8. [DOI] [PubMed] [Google Scholar]
- Smith TD, Adams MM, Gallagher M, Morrison JH, Rapp PR. Circuit-specific alterations in hippocampal synaptophysin immunoreactivity predict spatial learning impairment in aged rats. J Neurosci. 2000;20:6587–6593. doi: 10.1523/JNEUROSCI.20-17-06587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Sugaya K, Chouinard M, Greene R, Robbins M, Personett D, Kent C, Gallagher M, McKinney M. Molecular indices of neuronal and glial plasticity in the hippocampal formation in a rodent model of age-induced spatial learning impairment. J Neurosci. 1996;16:3427–3443. doi: 10.1523/JNEUROSCI.16-10-03427.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, Guzowski JF. Differences in hippocampal neuronal population responses to modifications of an environmental context: evidence for distinct, yet complementary, functions of CA3 and CA1 ensembles. J Neurosci. 2004;24:6489–6496. doi: 10.1523/JNEUROSCI.0350-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbitsky M, Yonan AL, Malleret G, Kandel ER, Gilliam TC, Pavlidis P. Altered hippocampal transcript profile accompanies an age-related spatial memory deficit in mice. Learn Mem. 2004;11:253–260. doi: 10.1101/lm.68204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Gallagher M, Eichenbaum H, Tanila H. Neurocognitive aging: prior memories hinder new hippocampal encoding. Trends Neurosci. 2006;29:662–670. doi: 10.1016/j.tins.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Ikonen S, Gallagher M, Eichenbaum H, Tanila H. Age-associated alterations of hippocampal place cells are subregion specific. J Neurosci. 2005;25:6877–6886. doi: 10.1523/JNEUROSCI.1744-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.