Abstract
The t(5;12)(q33;p13) translocation associated with chronic myelomonocytic leukemia (CMML) generates a TEL/PDGFβR fusion gene. Here, we used a murine bone marrow transplant (BMT) assay to test the transforming properties of TEL/PDGFβR in vivo. TEL/PDGFβR, introduced into whole bone marrow by retroviral transduction, caused a rapidly fatal myeloproliferative disease that closely recapitulated human CMML. TEL/PDGFβR transplanted mice developed leukocytosis with Gr-1+ granulocytes, splenomegaly, evidence of extramedullary hematopoiesis, and bone marrow fibrosis, but no lymphoproliferative disease. We assayed mutant forms of the TEL/PDGFβR fusion protein — including 8 tyrosine to phenylalanine substitutions at phosphorylated PDGFβR sites to which various SH2 domain–containing signaling intermediates bind — for ability to transform hematopoietic cells. All of the phenylalanine (F-) mutants tested conferred IL-3-independence to a cultured murine hematopoietic cell line, but, in the BMT assay, different F-mutants displayed distinct transforming properties. In transplanted animals, tyrosines 579/581 proved critical for the development of myeloproliferative phenotype. F-mutants with these residues mutated showed no sign of myeloproliferation but instead developed T-cell lymphomas. In summary, TEL/PDGFβR is necessary and sufficient to induce a myeloproliferative disease in a murine BMT model, and PDGFβR residues Y579/581 are required for this phenotype.
Introduction
TEL/PDGFβR is a fusion protein that is expressed as a consequence of the t(5;12) (q33;p13) chromosomal translocation in patients with chronic myelomonocytic leukemia (CMML) (1), a disease characterized by splenomegaly, abnormal myelopoiesis, myelofibrosis, and a predisposition to acute myeloid leukemia (AML) (2). As a result of the t(5;12), the NH2-terminal ligand–binding domain of the receptor tyrosine kinase PDGFβR is replaced by the NH2-terminus of the transcription factor TEL, which contains the self-association domain, pointed (PNT; Figure 1a) (1). TEL/PDGFβR forms oligomers mediated by the PNT domain and is a constitutively active tyrosine kinase (3). The ability of TEL/PDGFβR to confer factor independence to Ba/F3 cells is dependent on the PNT domain of TEL and on the kinase activity of the PDGFβR portion of the fusion (3).
Figure 1.
Diagram of TEL/PDGFβR primary structure and retroviral constructs. (a) TEL/PDGFβR fuses the ets-family transcription factor TEL on chromosome 12p13 to the receptor tyrosine kinase PDGFβR protein on chromosome 5q33. The fusion does not include the DNA binding domain of TEL or the ligand binding domain of PDGFβR but retains the PNT oligomerization domain of TEL and the tyrosine kinase domain of PDGFβR. The transmembrane region of PDGFβR (tm) is also retained. (b) Schematic of TEL/PDGFβR and negative control retroviral constructs used in BMTs. Domains are labeled (pgk, phosphoglycerate kinase promoter; neo, neomycin resistance; IRES, internal ribosomal entry site; GFP, green fluorescent protein).
TEL/PDGFβR is one of a growing list of chimeric oncoproteins that are dysregulated protein tyrosine kinases. The native PDGFβR is a receptor tyrosine kinase, and ligand binding to PDGFβR results in dimerization and autophosphorylation (4, 5). Phosphotyrosine residues of the activated receptor bind to specific SH2 domain proteins including SRC, PI3 kinase (PI3K), PLCγ, GRB2, SHP-2, and STAT5 that couple the receptor to downstream signal transduction pathways (5). Tyrosine to phenylalanine mutants of PDGFβR that are incapable of binding specific signaling molecules have been used to demonstrate the roles of specific signal transduction pathways after activation by PDGF. In this way, it has been determined that either PI3K or PLCγ is required to stimulate mitogenesis in HepG2 cells (6).
Here we report that the TEL/PDGFβR fusion gene is necessary and sufficient to cause a myeloproliferative disease in a murine bone marrow transplant (BMT) assay that closely resembles the human disease CMML and is characterized by hepatosplenomegaly, extramedullary hematopoiesis, myelofibrosis, and a neutrophilic leukocytosis. Furthermore, we have constructed a series of tyrosine to phenylalanine mutants of TEL/PDGFβR to examine the role of specific signal transduction pathways in transformation by TEL/PDGFβR and assessed the effects of these mutations on the development of CMML in the BMT assay.
Methods
Plasmid constructs.
The 2.4-kb TEL/PDGFβR cDNA was subcloned into the multicloning site of the MSCVneoEB retroviral vector containing a modified murine Moloney leukemia virus LTR and a neomycin resistance cassette (murine stem cell virus [MSCV], kindly provided by R. Hawley, Red Cross, Rockville, Maryland, USA) (7). TEL/PDGFβR tyrosine to phenylalanine mutants were generated by subcloning F2, F5, and F7 wild-type PDGFβR mutants (provided by A. Kaslauskas, Boston, Massachusetts, USA) into the TEL/PDGFβR background. Vectors expressing TEL/PDGFβR and related mutants cotranscriptionally with green fluorescent protein (GFP) using an internal ribosomal entry site (IRES) expression cassette were created by subcloning TEL/PDGFβR and mutants into the MSCV2.2IRESGFP retroviral vector (provided by W. Pear, University of Pennsylvania, Philadelphia, Pennsylvania, USA).
Production of retrovirus and determination of viral titers.
TEL/PDGFβR and related MSCV retroviral constructs were used to make replication-incompetent retroviral supernatant by transient transfection of 293T cells. Bone marrow cells from normal mice pretreated with 5-fluorouracil was incubated with high-titer retroviral supernatant and intravenously injected into lethally irradiated syngeneic mice. The TEL/PDGFβR cDNA described by Golub et al. (1) was cloned into the MSCVneoEB retroviral vector (6) to create MSCVneoT/P. The plasmid containing packaging sequences, pIK6.1MCV.ecopac.UTd (Ecopac), was obtained from M. Finer (Cell Genesys, Redwood City, California, USA). 293T cells were transfected with MSCVneoT/P and Ecopac using the SuperPhect (Pharmacia, Uppsala, Sweden) calcium phosphate method per the manufacturer’s instructions. All supernatants were collected 48 hours after transfection and passed through a 0.45-μm filter before freezing at –80°C. Supernatant was tested by infecting 3T3 cells, and viral titer was estimated by quantitating neomycin-resistant colonies as described (8).
Retrovirally mediated gene transfer and BMT.
On day –8 of each BMT procedure, 6- to 8-week-old BALB/c mice were pretreated with 150 mg/kg of 5-fluorouracil administered by intraperitoneal injection. On day –2, bone marrow was harvested from these donor mice and red blood cells were lysed by incubation for 5 minutes on ice with an ammonium chloride lysis solution (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA [pH 7.4]). The remaining mononuclear marrow cells were then resuspended in RPMI medium containing 1 mL viral supernatant, 20% FBS, 6 ng/mL recombinant mouse IL-3 (Genzyme, Cambridge, Massachusetts, USA), 100 ng/mL recombinant mouse stem cell factor (Genzyme), and 10 ng/mL recombinant mouse IL-6 (PeproTech, Rocky Hill, New Jersey, USA). The cells were plated onto a 10-cm tissue culture plate treated with the fibronectin fragment CH-296 (RetroNectin PanVera, Madison, Wisconsin, USA) per the manufacturer’s instructions. At 24 hours, an additional 1 mL of viral supernatant was added; cells were recovered by gentle scraping at 48 hours after plating, counted, and resuspended in Hanks’ buffered saline. A total of 5 × 105 cells were injected by tail vein into syngeneic mice that had been lethally irradiated with 900 cGy in 2 fractions. A total of 144 mice were transplanted in 3 separate experiments that all yielded equivalent results. Sublethally irradiated syngeneic mice used for secondary transplants were subjected to a single dose of 450 cGy.
Assessment of mice.
Animals were monitored 3 times per week for the development of disease by inspection and palpation of the spleen and femoral lymph nodes. Peripheral blood was obtained by retroorbital phlebotomy using heparinized Natelson blood collecting tubes (Fisher Scientific, Pittsburgh, Pennsylvania, USA) after adequate methoxyflurane (Mallinckrodt Veterinary, Mundelein, Illinois, USA) anesthesia. Mice with disease evidenced by abnormal white blood cell counts, splenomegaly, lymphadenopathy, or shortness of breath were sacrificed by CO2 asphyxiation followed by cervical dislocation. Bone marrow cells were isolated by flushing femurs and tibias with PBS. Single-cell suspensions were prepared by passing spleen and lymph node tumor tissue through nylon mesh (Falcon, Lincoln Park, New Jersey, USA) wetted with PBS.
Expression analysis.
Single-cell suspensions of tissues were prepared as just described. Total cellular RNA was isolated by resuspending 107 cells in 1 mL of TRIzol (Life Technologies, Gaithersburg, Maryland, USA) and processing according to the manufacturer’s instructions. Total RNA (4 μg) from each sample was reverse transcribed using AMV RT (Life Technologies) for 1 hour at 42°C. To assess TEL/PDGFβR message in transplanted mice, 1 mL of cDNA solution was subjected to 40 cycles of PCR (94°C for 60 seconds, 53°C for 60 seconds, and 72°C for 60 seconds) using primers spanning the translocation breakpoint: TEL Bam-196, 5′- GCG GAT CCC AAT TTA CTG GAG CAG-3′ and PDGFR-1979R 5′-GCG TCC CAG CAC AAG CTG GT-3′. Wild-type murine TEL message was used as a cDNA control subjecting the same cDNA solutions to 40 cycles of PCR using primers MTEL-1184, 5′-CCA GAG CCC TGC GCC ACT AC-3′ and MTEL-1487R, 5′-TCC CTG CCA TTT CTG GGT CG-3′.
Ba/F3 cell growth rates.
Polyclonal Ba/F3 cell lines stably expressing TEL/PDGFβR were made by retroviral infection using 1 × 106 cells, polybrene (American Bioanalytical, Natick, Massachusetts, USA), and supernatants prepared as already described here. Cells were selected in RPMI media containing 10% FBS, 1 ng/mL IL-3, and 1 mg/mL G418 (Sigma Chemical Co., St. Louis, Missouri, USA) for 7 days. Expression of TEL/PDGFβR and related mutants was confirmed by Western blot analysis using anti-tail PDGFβR antibody (PharMingen, San Diego, California, USA). Cells containing TEL/PDGFβR and related mutants were washed twice in media without IL-3 and allowed to grow to confluence. Cells containing vector alone and kinase-inactive TEL/PDGFβR were maintained in media containing IL-3. Cells were then plated at a concentration of 104 per mL, and cells excluding Trypan blue (Sigma Chemical Co.) were counted daily for 1 week.
Proviral integration and T-cell receptor rearrangements.
Genomic DNA was isolated from assorted tissues as described elsewhere (9). A total of 10 μg of each genomic DNA sample was digested with either XhoI (cuts twice in provirus) or EcoRI (cuts once) and subjected to agarose gel electrophoresis and transferred to nylon membranes (Hybond N+; Amersham, Arlington Heights, Illinois, USA) using alkaline transfer as described previously (10). Provirus was detected by Southern blotting using a 760-bp EagI-BamHI fragment of the neomycin gene as a probe random labeled using 32Pγ dCTP (Boehringer Mannheim, Indianapolis, Indiana, USA). Hybridization was performed at 65°C for 16 hours, and membranes were washed with 2 × SSC, 0.1% SDS, for 20 minutes at room temperature and with 1 × SSC, 0.1% SDS, at 65°C for 20 minutes and were exposed to photographic film (BioMax; Eastman Kodak, Rochester, New York, USA) at –80°C overnight. T-cell receptor β (TCR-β) chain gene rearrangements were detected using a 2-kb EcoRI fragment of Jβ2B plasmid (provided by B. Rich, Brigham and Women’s Hospital, Boston, Massachusetts, USA), 10 μg of genomic tumor DNA digested with HindIII, and an identical Southern blot protocol.
Histology.
Murine tissues were fixed for 24 hours in 10% neutral buffered formalin and embedded in paraffin. Femurs were subjected to an additional decalcification step in RDO (Apex Engineering Products, Plainfield, Illinois, USA) for 4 hours before processing. Sections (3 μm) were deparaffinized and stained with hematoxylin and eosin (H&E).
Flow cytometry.
Spleen cell suspensions were processed for flow cytometry as described previously (11). Antibodies used in the current analysis were biotin conjugates of anti–Gr-1 and anti-CD3 (PharMingen), phycoerythrin conjugates of anti-B220, anti–Thy-1.2, and anti-CD8α (PharMingen), and allophycocyanin-conjugated anti-CD4 (Caltag, South San Francisco, California, USA). Binding of the biotinylated primary antibodies to cells was detected using streptavidin labeled with allophycocyanin (Caltag). Acquisition and data analysis were performed on a FACStar running CellQuest 3.1 software (Becton Dickinson Immunocytometry Systems, San Jose, California, USA).
Results
TEL/PDGFβR causes a fatal myeloproliferative syndrome in mice.
The in vivo transforming properties of TEL/PDGFβR were tested in a murine BMT assay. Briefly, TEL/PDGFβR was introduced into primary bone marrow cells by retroviral transduction and transplanted into lethally irradiated syngeneic recipient mice. Controls included 2 nontransforming TEL/PDGFβR mutants, K634R and TELΔPNT/PDGFβR mutants (3). The K634R mutant contains a single amino acid substitution in the catalytic domain of the PDGFβR portion of the fusion that abrogates tyrosine kinase activity and transformation of Ba/F3 cells by TEL/PDGFβR. Deletion of the TEL PNT domain prevents TEL/PDGFβR from forming activated oligomers, and this mutant is similarly unable to transform Ba/F3 cells to factor independence (3).
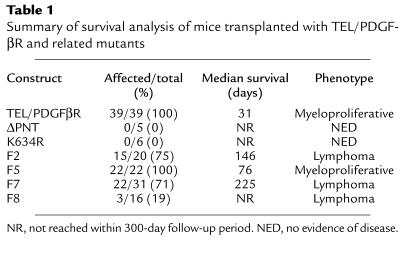
Fifty mice were transplanted with TEL/PDGFβR (39 mice), kinase-inactive K634R (6 mice), or ΔPNT mutants (controls, 5 mice) (Table 1). All (39/39; 100%) TEL/PDGFβR transplanted mice developed peripheral leukocytosis and splenomegaly and died within 65 days of transplantation (median survival = 31 days). Nine of these mice were transplanted with a TEL/PDGFβR retroviral construct containing, instead of a neomycin resistance cassette, a cassette allowing for expression of GFP from an IRES (Figure 1b). These mice developed an identical myeloproliferative disease with a similar latency (data not shown). In contrast, 0 of 11 (0%) mice transplanted with the kinase-inactive K634R TEL/PDGFβR mutant or the TELΔPNT/PDGFβR mutant developed disease, and they remain alive and free of disease at 1 year after transplant (Table 1). TEL/PDGFβR transplanted mice had peripheral blood leukocyte counts as high as 5.1 × 108 per mL (range: 4.9 × 107 to 5.1 × 108; normal: 2.0 × 106 to 12.0 × 106) consisting of myeloid lineage cells at all stages of maturation (Figure 2a), analogous to CMML in humans. There was a marked expansion of maturing granulocytes and precursors within normal sites of hematopoiesis, the bone marrow and splenic red pulp, occasionally accompanied by splenic infarction (Figure 2b). The marrow also demonstrated increased collagen deposition, based on silver impregnation stains (data not shown). Extramedullary hematopoiesis was also present at abnormal sites, including the hepatic and pulmonary parenchyma and lymph nodes (Figure 2c and data not shown). There was no evidence of lymphoproliferative disease in TEL/PDGFβR transplanted mice.
Table 1.
Summary of survival analysis of mice transplanted with TEL/PDGFβR and related mutants
Figure 2.
Histopathology of TEL/PDGFβR myeloproliferative disease. (a) Wright-Giemsa–stained peripheral blood smear demonstrates marked leukocytosis composed predominantly of myeloid cells in various states of maturation (×100). (b) Formalin fixed spleen tissue stained with H&E shows effacement of normal splenic architecture as well as a large area of infarction (×10). Higher-power views of the spleen demonstrate effacement by myeloid precursors (data not shown). (c) Formalin fixed lung tissue stained with H&E shows congestion of pulmonary vasculature with granulocytes (×40).
Expression of TEL/PDGFβR.
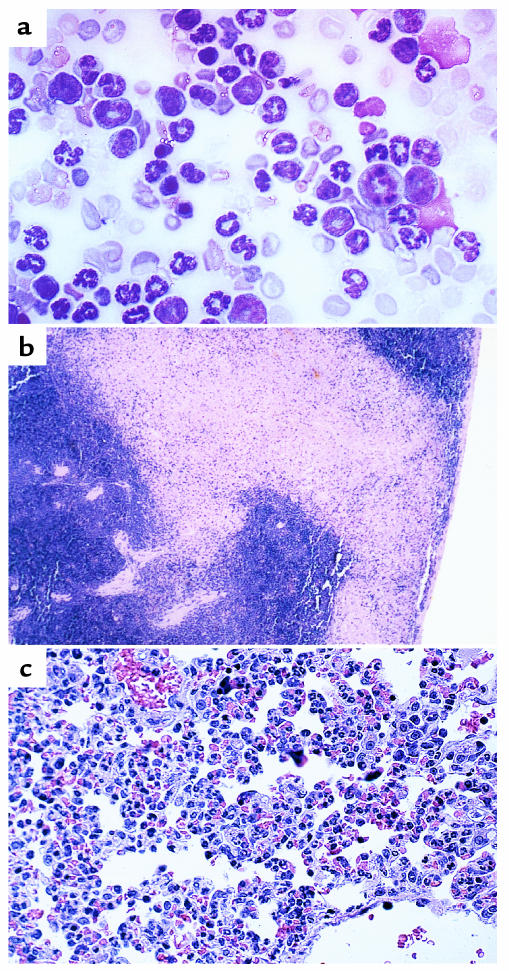
Expression was confirmed in affected tissues of transplanted animals by RT-PCR and by GFP expression from appropriate vectors. In RT-PCR experiments (Figure 3a), RNA integrity was confirmed by amplification of wild-type TEL RNA, with RT-negative and water controls. Because GFP is expressed from a single transcript containing TEL/PDGFβR and an IRES/GFP cassette, GFP expression in intact cells, as assessed by fluorescent microscopy is an informative surrogate for expression of TEL/PDGFβR. As shown in Figure 3b, there were high levels of GFP expression in hematopoietic cells.
Figure 3.
Expression of TEL/PDGFβR and proviral integration in CMML mice. (a) RT-PCR of RNA isolated from spleen and bone marrow of transplanted and control animals. Reactions containing RT and RT-negative controls are indicated by plus and minus symbols, respectively. In the top panel, primers were chosen complementary to human TEL and PDGFβR sequences spanning the breakpoint of the fusion. In the lower panel, primers to endogenous murine TEL were used to control for cDNA quality. Although the TEL control in the T/P BMT marrow sample is faint, the TEL/PDGFβR transcript is easily seen. (b) Unstained peripheral blood from TEL/PDGFβR-IRES-GFP BMT mouse. Image was taken using a scanning laser confocal microscope. (c) Southern blot detection of provirus in tissues of TEL/PDGFβR mice (M, bone marrow; S, spleen; N, lymph node; B, blood; T, thymus). XbaI cuts in the viral LTR, so digestion results in a band of expected size in tissues that contain provirus. EcoRI cuts only once in the provirus, resulting in bands of different sizes corresponding to different genomic integration sites. The smearing indicates the polyclonal nature of these cells.
Clonality of myeloproliferative disease.
Southern blotting of genomic DNA isolated from transplanted animals using a neomycin probe confirmed proviral integration in all affected tissues, but not in unaffected or control tissues (Figure 3c). Provirus was present in bone marrow, spleen, and blood, consistent with flow cytometry and histopathological findings, whereas there was no evidence for proviral integration in lymph node or thymus (Figure 3c). Clonality of the abnormal cells was assessed using a restriction endonuclease (EcoRI) that cleaves once within the provirus. Each independent viral integration event thus yielded a unique band. The tumors in mice transplanted with TEL/PDGFβR were oligo- or polyclonal, as evidenced by multiple bands in these lanes (Figure 3c).
Transplantability.
Cells from diseased mice were transplanted into secondary recipients. Two experiments using 1 × 106 spleen cells (4 mice) or marrow cells (4 mice) from mice with TEL/PDGFβR disease failed to cause disease in sublethally irradiated secondary recipients. In a third experiment, 8 lethally irradiated mice were injected with 5 × 106 spleen cells each, and 8 mice were injected with 1 × 106 bone marrow cells each. One of 8 mice from the spleen group developed a myeloproliferative disease with splenomegaly and peripheral leukocytosis (data not shown).
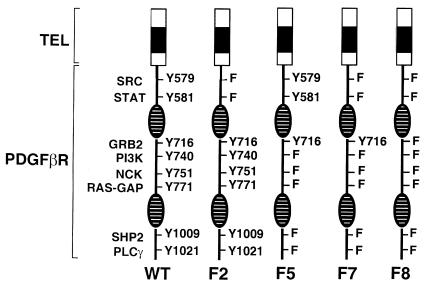
Signal transduction pathways associated with PDGFβR activation are not required for the transformation of Ba/F3 cells by TEL/PDGFβR. The native PDGFβR requires specific tyrosine residues for signaling through distinct transduction pathways (5). To determine the tyrosine residues required for the development of the myeloproliferative phenotype, we analyzed a series of TEL/PDGFβR mutants containing tyrosine to phenylalanine point mutations designed to abrogate the binding of specific SH2 adapter signaling molecules (Figure 4). Control experiments showed that these mutations had no effect on PDGFβR kinase activity in the context of the TEL/PDGFβR fusion (D.W. Sternberg and D.G. Gilliland, unpublished observation). Ba/F3 cell lines expressing each of these mutants were generated and assessed for their ability to grow in the absence of IL-3. Each of the F-mutants conferred IL-3–independent growth to Ba/F3 cells (Figure 5). Furthermore, stable Ba/F3 cell lines expressing the various F-mutants had identical growth rates when deprived of IL-3.
Figure 4.
Tyrosine to phenylalanine signaling mutants of TEL/PDGFβR. The NH2-terminal TEL portion of the fusions, indicated by a shaded box, remains unchanged in the wild-type and mutants. Phosphotyrosines in the PDGFβR portion of the fusion are indicated by “Y” and are numbered according to the wild-type PDGFβR sequence. Mutants were generated with tyrosine to phenylalanine (F) substitutions at specific residues as indicated. The split kinase domain of PDGFβR is indicated by striped ovals.
Figure 5.
Growth rates of Ba/F3 cells transformed with TEL/PDGFβR F-mutants. Ba/F3 cell lines expressing TEL/PDGFβR, TEL/PDGFβR mutants, or vector alone control were plated in media without IL-3 at the indicated concentration. Cells excluding Trypan blue were counted daily. Similar results were obtained from 3 replicate experiments. A representative example is plotted here. Cells expressing TEL/PDGFβR and all F-mutants continued to proliferate in the absence of IL-3 at the same rate. Small differences in growth rates were not consistently reproduced in repeat experiments.
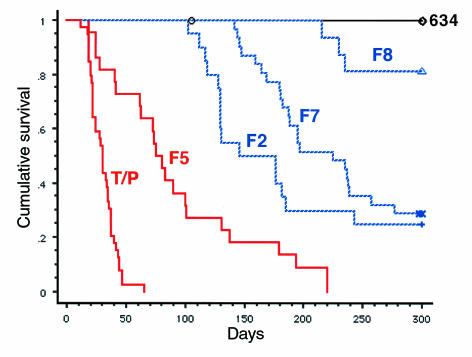
Expression of TEL/PDGFβR tyrosine to phenylalanine mutants in primary hematopoietic cells results in distinct phenotypes in mice. To confirm the findings with the TEL/PDGFβR F-mutants in primary hematopoietic cells, murine BMT was performed using viral supernatants of equivalent titer, with the F2 (20 mice), F5 (22 mice), F7 (31 mice), and F8 (16 mice) TEL/PDGFβR tyrosine to phenylalanine mutants (Table 1). In contrast with data obtained in Ba/F3 cells, the TEL/PDGFβR mutants showed distinct differences in the murine BMT assay. The F5 mutant, which lacks the SH2 binding sites required for PI3 kinase and PLCγ in the context of the native receptor (11) and in the context of TEL/PDGFβR (D.W. Sternberg and D.G. Gilliland, unpublished observation), caused a myeloproliferative phenotype similar to wild-type TEL/PDGFβR with elevated white blood cells counts and splenomegaly, but with a prolonged median survival (76 days versus 31 days, respectively; Figure 6). The most striking difference observed was in mice transplanted with the F2, F7, and F8 mutants that maintained normal white blood counts (data not shown) and did not develop myeloproliferative disease (Figure 7). Initially, these animals appeared to be unaffected by the transplanted tyrosine kinase fusions. However, after a latency of several months, these mice developed mediastinal tumors that expressed high levels of the mutant TEL/PDGFβR as assessed by Western blot (Figure 7, and data not shown). The median survival of the F2 and F7 were 146 and 225 days, respectively, whereas the median survival of the F8 mice was not reached by 300 days after transplant (Figure 6).
Figure 6.
Kaplan-Meier survival analysis of mice transplanted with TEL/PDGFβR and F-series mutants. The wild-type TEL/PDGFβR (MSCVneoT/P or MSCV-T/P-IRES-GFP) caused a rapidly fatal myeloproliferative syndrome with a median survival of 31 days (39/39 mice affected, 100%). Mice transplanted with a kinase-inactive point mutant of TEL/PDGFβR (MSCVneoK634R) remain free of disease 1 year after transplant (0/6 mice affected, 0%). F5 mice developed myeloproliferative disease with a longer median survival of 76 days (22/22 mice affected, 100%). F2, F7, and F8 mice did not develop myeloproliferation, but after several months of latency they developed lethal mediastinal lymphomas. Median survivals of F2 and F7 mice were 146 and 225 days, respectively (15/20 mice, 75%; 22/31 mice, 71%), whereas the median survival of mice transplanted with the F8 mutant was not achieved by 300 days (3/16 mice affected, 19%). Red color indicates myeloproliferative phenotype; blue indicates lymphoma phenotype.
Figure 7.
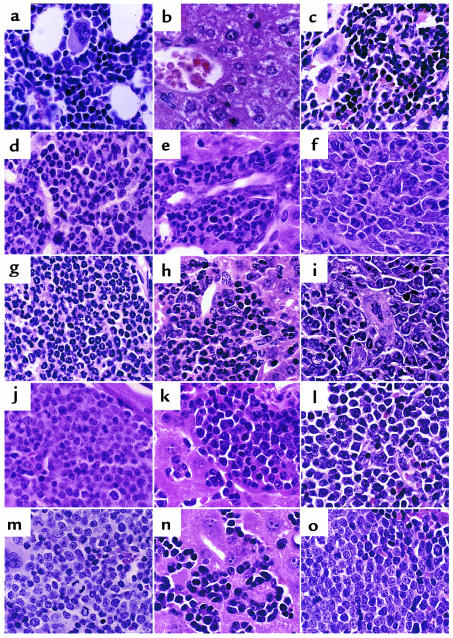
Histopathology of mice transplanted with TEL/PDGFβR and related mutants. High-power (×640) views of H&E-stained bone marrow (a, d, g, j, m), liver (b, e, h, k, n), and spleen (c, f, i, l, o) of transplanted mice. (a–c) Negative control mouse transplanted with the ΔPNT mutant of TEL/PDGFβR demonstrates normal tissues after transplant. (a) Intramedullary adipocytes and a normal mixture of maturing myeloid and erythroid precursors and scattered megakaryocytes are seen in the bone marrow. (b) A central venule surrounded by normal hepatocytes. Splenic red pulp (c) demonstrates a mixture of maturing erythroid and myeloid elements similar to that seen in the marrow. TEL/PDGFβR (d–f) and F5 mutant (g–i) tissues both demonstrate marked expansion of myeloid precursors (identified as cells with folded or ringlike nuclei) within bone marrow and splenic red pulp, as well as marked extramedullary hematopoiesis comprising mainly myeloid precursors within the liver. In contrast, F2 (j–l), F7 (m–o), and F8 (not shown) tissues show a monomorphous population of lymphoblasts that efface the marrow and splenic red pulp and infiltrate liver sinusoids.
TEL/PDGFβR mutants lacking PDGFβR juxtamembrane tyrosines 579/581 develop clonal T-cell lymphomas.
The mice transplanted with F2, F7, and F8 mutants developed a characteristic clinical syndrome consisting of labored breathing and exophthalmos after a latency of at least 14 weeks. Necropsy showed mediastinal masses that were frequently associated with pleural effusions (data not shown). Microscopic analysis revealed that these tumors invaded the chest wall, tracked along the bronchi, encased the lungs, and replaced the spleen and bone marrow (Figure 7, and data not shown).
Flow cytometry of cells isolated from the spleens of affected animals demonstrated an abnormal population with a B220–, CD3lo, Thy-1+, and CD4/CD8 double-positive immunophenotype, consistent with an immature T-cell phenotype (Figure 8). An identical immunophenotype was seen in mediastinal tumors and in bone marrow (data not shown).
Figure 8.
Flow cytometric analysis of spleen cells from mice transplanted with TEL/PDGFβR and F-mutants. Cells isolated from the spleens of affected animals were analyzed by dual-color flow cytometry using antibodies indicated. Wild-type TEL/PDGFβR, T/P (WT), and F5 spleens contain an abnormal population of Gr-1–positive cells, consistent with mature granulocytes. Most but not all of these cells coexpress EGFP. The F2, F7, and F8 samples do not contain significant numbers of Gr-1–positive cells but instead contain an abnormal population with the immunophenotype, CD3lo, B200–, Thy-1+, CD4+, CD8+, consistent with immature T lymphocytes (lymphoblasts). The F7 samples were generated using the MSCVneo vector backbone and do not express GFP.
Proviral integration and gene rearrangement studies.
Mediastinal tumors from mice transplanted with the F2, F7, and F8 mice contained proviral DNA as assessed by Southern blot (Figure 9a). However, in contrast to the pattern of oligo- or polyclonality seen in animals with myeloproliferative disease, these clonality analyses revealed either a single or a double band, indicating that these were clonal tumors. This conclusion was confirmed by the gene rearrangement studies described below.
Figure 9.
Assessment of proviral integration and TCR-β gene rearrangements. (a) As in Figure 3c, Southern blot analysis of tumors from transplanted animals demonstrates the presence of provirus. A human TEL probe that did not cross-hybridize with the endogenous murine TEL was used as a virus-specific probe. Neomycin vectors yield a 5-kb band (filled arrowhead), and GFP-containing vectors yield a 4-kb band (open arrowhead). (b) Genomic DNA isolated from F2, F7, and F8 mediastinal tumors subjected to Southern blot analysis using a TCR-β probe. Rearrangement of the TCR-β locus in each case proves the T-cell identity of these tumors. Clonality is demonstrated by the presence of 2 unique TCR-β alleles per case and is seen in the majority of tumors. The tumor from mouse 13.17 shows 4 bands, suggesting a biclonal tumor in this case. Contamination of tumor samples with stromal cells results in the presence of the unrearranged germline band (arrowhead).
The T-cell origin of these tumors was confirmed by Southern blot analysis of the TCR-β gene (Figure 9b). In all cases tested, the tumors demonstrated clonal rearrangement of the TCR-β locus. In 8 of 9 mice analyzed, there were 2 bands of equal intensity corresponding to rearrangement of both TCR-β alleles, consistent with monoclonal derivation. In one case, there were 4 bands, indicating the presence of at least 2 independent T-cell clones. The immunoglobulin heavy chain gene locus was not rearranged in these tumors (data not shown). The T-cell lymphomas associated with the F2, F7, and F8 mutants were readily transplantable into sublethally irradiated secondary recipients with as few as 104 cells and also gave rise to mediastinal T-cell lymphomas (data not shown).
Discussion
We have demonstrated that expression of TEL/PDGFβR in unfractionated bone marrow results in a myeloproliferative disease with features of the human disease CMML. Features of CMML that are recapitulated in the murine BMT model include leukocytosis with normal maturation and differentiation, splenomegaly, bone marrow fibrosis, and extramedullary hematopoiesis in multiple organs including spleen and liver. It has also recently been reported that expression of TEL/PDGFβR under the control of the CD11b promoter in transgenic mice can cause a myeloproliferative phenotype (12).
The rapid onset of disease after BMT, and the polyclonal nature of the disease, favor the hypothesis that TEL/PDGFβR is both necessary and sufficient to cause this myeloproliferative phenotype. We cannot exclude the possibility that retroviral integration itself contributes to the myeloproliferative phenotype. However, this event would have to be both common enough to yield polyclonal disease and yet have no effect in animals transplanted with retrovirus containing inactive mutants of TEL/PDGFβR, an unlikely possibility.
TEL/PDGFβR is exclusively associated with CMML in humans (13, 14). Nonetheless, it was unexpected that TEL/PDGFβR would cause a myeloproliferative phenotype in the murine BMT model without histopathological evidence of lymphoid or other lineage involvement. First, the introduction of TEL/PDGFβR into primary hematopoietic cells via retroviral gene transfer allows for expression in multiple hematopoietic lineages, including lymphoid cells (15). Use of this murine BMT technique with the BCR/ABL fusion (16) or the TEL/JAK2 (17) tyrosine kinase fusion produces phenotypes consisting of tumors derived from multiple hematopoietic lineages. Second, we have previously reported that transgenic mice expressing TEL/PDGFβR under the control of the immunoglobulin promoter-enhancer EμPμ develop B-cell lymphoblastic lymphomas, demonstrating that TEL/PDGFβR is capable of causing lymphoid tumors (11). However, transformation of lymphoid cells in EμPμ-TEL/PDGFβR transgenic mice requires a relatively long latency of 2–4 months, and these tumors are clonal, indicating that additional genetic events are required for transformation of lymphoid cells (10). The lack of involvement of the lymphoid lineage after BMT is therefore probably because longer latency lymphoid disease in transplanted mice is masked by the development of a rapidly fatal myeloproliferative disease. Collectively, these data demonstrate the importance of the target cell in transformation by oncogenic fusion proteins, as recently reviewed by Westervelt and Ley (18).
Two additional chromosomal translocations associated with CMML, t(5;7)(q33;q11.2) and t(5;10)(q33;q11.2), have recently been cloned and also result in the fusion of the PDGFβR gene to the HIP1 and H4 genes, respectively (19, 20). TEL, HIP1, and H4 are unrelated proteins that each contribute a dimerization/oligomerization motif to the respective PDGFβR fusion. Taken together, these observations suggest that PDGFβR tyrosine kinase activation can be subserved by a spectrum of fusion partners using different dimerization motifs and that a critical event in the pathogenesis of CMML is constitutive activation of the PDGFβR kinase domain and its downstream targets.
The rapid onset of disease in transplanted mice enabled us to use the murine BMT as an assay to test the role of individual downstream signal transduction pathways in the development of CMML. In the Ba/F3 cell context, each of the tyrosine to phenylalanine mutants was capable of conferring IL-3–independent growth, and the growth rates of stable cell lines expressing each F-mutant were identical when the cell lines were grown in the absence of IL-3. However, there was a dramatic difference in phenotype of TEL/PDGFβR and the F5 mutants that conferred a myeloproliferative disease phenotype, and the F2, F7, and F8 mutants, which conferred a T-cell lymphoblastic lymphoma phenotype. Although the F8 mutant lacks the major SH2 interaction sites in the context of the native receptor, the F8 mutant remains constitutively phosphorylated on tyrosine (D.S. Sternberg and D.G. Gilliland, unpublished observation). Thus, other SH2 interaction sites may be functioning in this mutant. The difference in phenotype cannot be explained by differences in viral supernatant titers, which were equivalent (data not shown), or by differences in PDGFβR kinase activity in the context of the mutants, which was also equivalent. These findings suggest that mutational analysis of oncogenic proteins in hematopoietic cell lines must be interpreted with caution, and data should be corroborated in transformation assays of primary hematopoietic cells.
Our data indicate that the juxtamembrane tyrosine residues of the wild-type PDGFβR, 579/581, are critical for the development of myeloid disease in the BMT assay. The 579/581 tyrosines of the wild-type PDGFβR are required for the binding and activation of SRC family kinases (21), as well as activation of STAT family members by PDGFβR (22, 23). Activated SRC exerts its downstream effects in part through activation of MYC (24). TEL/PDGFβR has been shown to activate MYC (25), and dominant negative MYC inhibits growth of TEL/PDGFβR transformed cells (25). Thus, activation of MYC by SRC family members may play an important role in transformation of myeloid cells by TEL/PDGFβR.
STAT5 activation may also play an important role in transformation of hematopoietic cells and is known to be activated by a spectrum of tyrosine kinase fusions associated with human leukemias, including BCR/ABL (26), TEL/ABL (27), TEL/JAK2 (17), HIP1/PDGFβR (28), and TEL/PDGFβR (D. W. Sternberg and D. G. Gilliland, unpublished observation). Furthermore, STAT5 is required for TEL/JAK2-mediated myeloproliferation in a murine BMT model (29). Thus, activation of STAT5 by residues Y579/581 may also be important in transformation of myeloid cells by TEL/PDGFβR.
Mutations affecting residues besides 579/581 incrementally prolonged the latency of lymphoid disease (Figure 6 and Table 1). The graded decrement in oncogenicity cannot be attributed to variations in PDGFβR kinase activity or to differences in viral supernatant titer. However, a similar mechanism of action for native PDGFβR function, invoking signal strength, has recently been proposed. Fambrough et al. used oligonucleotide microarrays to examine the immediate early genes induced by PDGFβR and a similar series of tyrosine to phenylalanine mutants as those used in this report in the context of the native PDGFβR (30). The overall levels of expression of these target genes were diminished with progressive tyrosine to phenylalanine substitutions, and it was suggested that the various signaling pathways activated by PDGFβR converge upon a common set of downstream targets. The data from the murine BMT assay of TEL/PDGFβR tyrosine to phenylalanine mutants would be consistent with this hypothesis.
Acknowledgments
The authors thank M. Ryan for secretarial and administrative assistance; E. Lander and K. Weilbaecher for critical review of the manuscript; and members of the Gilliland lab for valuable discussion. This research was supported in part by a grant from the Leukemia Society of America (5461-98 to M.H. Tomasson) and grants from the National Institutes of Health (K08 CA81197-01 to M.H. Tomasson; P01CA66996-01 and P010K50654 to D.G. Gilliland; R01 CA57593 to R.A.Van Etten; and K08 HL03310 to R.L. Ilaria.) D.G. Gilliland is an Associate Investigator in the Howard Hughes Medical Institute.
References
- 1.Golub TR, Barker GF, Lovett M, Gilliland DG. Fusion of PDGF receptor β to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 2.Lichtman, M.A., and Brennan, J.K. 1983. Dyshemopoietic (preleukemic) disorders. In Hematology. 3rd edition. W.J. Williams, E. Beutler, A.J. Erslev, and M.A. Lichtman, editors. McGraw-Hill. New York, NY. 175–184.
- 3.Carroll M, Tomasson M, Barker GF, Golub TR, Gilliland DG. The TEL-PDGFβR fusion protein dimerizes and transforms hematopoietic cells through activation of PDGFβR dependent signaling pathways. Proc Natl Acad Sci USA. 1996;93:14845–14850. doi: 10.1073/pnas.93.25.14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claesson-Welsh L. Platelet-derived growth factor receptor signals. J Biol Chem. 1994;269:32023–32026. [PubMed] [Google Scholar]
- 5.Heldin C-H, Östman A, Rönnstrand L. Signal transduction via platelet-derived growth factor receptors. Biochim Biophys Acta. 1998;1378:79–113. doi: 10.1016/s0304-419x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 6.Valius M, Kazlauskas A. Phospholipase C-gamma1 and phosphatidylinositol 3 kinase are the downstream mediators of the PDGF receptor’s mitogenic signal. Cell. 1993;73:321–334. doi: 10.1016/0092-8674(93)90232-f. [DOI] [PubMed] [Google Scholar]
- 7.Hawley RG, Fong AZ, Burns BF, Hawley TS. Transplantable myeloproliferative disease induced in mice by an interleukin 6 retrovirus. J Exp Med. 1992;176:1149–1163. doi: 10.1084/jem.176.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller A, Rosman G. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 9.Davis RW. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65:404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- 10.Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY. 9.31–9.57.
- 11.Tomasson MH, et al. TEL/PDGFβR induces hematologic malignancies in mice that respond to a specific tyrosine kinase inhibitor. Blood. 1999;93:1707–1714. [PubMed] [Google Scholar]
- 12.Ritchie K, et al. The tel-PDGFRbeta fusion gene produces a chronic myeloproliferative syndrome in transgenic mice. Leukemia. 1999;13:1790–1803. doi: 10.1038/sj.leu.2401494. [DOI] [PubMed] [Google Scholar]
- 13.Lerza R, Castello G, Sessarego M, Cavallini D, Pannacciulli I. Myelodysplastic syndrome associated with increased bone marrow fibrosis and translocation (5;12)(q33;p12-13) Br J Haematol. 1992;82:476–477. doi: 10.1111/j.1365-2141.1992.tb06450.x. [DOI] [PubMed] [Google Scholar]
- 14.Wessels JW, et al. t(5;12)(q31;p12): a clinical entity with features of both myeloid leukemia and chronic myelomonocytic leukemia. Cancer Genet Cytogenet. 1993;65:7–11. doi: 10.1016/0165-4608(93)90051-m. [DOI] [PubMed] [Google Scholar]
- 15.Hawley RG, Lieu FH, Fong AZ, Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 16.Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the p210 bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 17.Schwaller J, et al. Transformation of hematopoietic cell lines to growth-factor independence and induction of a fatal myeloid- and lymphoproliferative disease in mice by retrovirally transduced TEL/JAK2 fusion gene. EMBO J. 1998;17:5321–5333. doi: 10.1093/emboj/17.18.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westervelt P, Ley TJ. Seed vs. soil: the importance of the target cell for transgenic models of human leukemias. Blood. 1999;93:2143–2148. [PubMed] [Google Scholar]
- 19.Ross TS, Bernard OA, Berger R, Gilliland DG. Fusion of Huntingtin interacting protein 1 to platelet-derived growth factor β receptor (PDGFβR) in chronic myelomonocytic leukemia with t(5;7)(q33;q11.2) Blood. 1998;91:4419–4426. [PubMed] [Google Scholar]
- 20.Anastasiadou E, et al. H4(10S170), a gene frequently rearranged in papillary thyroid carcinoma, is fused to the platelet-derived growth factor β receptor (PDGFβR) gene in atypical chronic myeloid leukemia with t(5;10)(q33;q22) Blood. 1999;94(Suppl. 1):51a. doi: 10.1182/blood.v97.12.3910. (Abstr.) [DOI] [PubMed] [Google Scholar]
- 21.Mori S, et al. Identification of two juxtamembrane autophosphorylation sites in the PDGF beta-receptor: involvement in the interaction with Src family tyrosine kinases. EMBO J. 1993;12:2257–2264. doi: 10.1002/j.1460-2075.1993.tb05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valgeirsdottir S, Paukku K, Silvennoinen O, Heldin C-H, Claeson-Welsh L. Activation of STAT5 by platelet-derived growth factor (PDGF) is dependent on phosphorylation sites in PDGF β-receptor juxtamembrane and kinase insert domains. Oncogene. 1998;16:505–515. doi: 10.1038/sj.onc.1201555. [DOI] [PubMed] [Google Scholar]
- 23.Sachsenmaier C, Sadowski HB, Cooper JA. STAT activation by the PDGF receptor requires juxtamembrane phosphorylation sites but not Src tyrosine kinase activation. Oncogene. 1999;18:3583–3592. doi: 10.1038/sj.onc.1202694. [DOI] [PubMed] [Google Scholar]
- 24.Barone MV, Courtneidge SA. Myc but not Fos rescue of PDGF signalling block caused by kinase-inactive Src. Nature. 1995;378:509–512. doi: 10.1038/378509a0. [DOI] [PubMed] [Google Scholar]
- 25.Bourgeade M-F, Défachelles A-S, Cayre YE. Myc is essential for transformation by TEL/platelet-derived growth factor receptor β (PDGFRβ) Blood. 1998;91:3333–3339. [PubMed] [Google Scholar]
- 26.Carlesso N, Frank D, Griffin J. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J Exp Med. 1996;183:811–820. doi: 10.1084/jem.183.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okuda K, Gollub TR, Gilliland DG, Griffin J. p210BCR/ABL, p190BCR/ABL, and TEL/ABL activate similar signal transduction pathways in hematopoietic cell lines. Oncogene. 1996;13:1147–1152. [PubMed] [Google Scholar]
- 28.Ross TS, Gilliland DG. Transforming properties of the Huntingtin interacting protein 1/platelet-derived growth factor β receptor fusion protein. J Biol Chem. 1999;274:22328–22336. doi: 10.1074/jbc.274.32.22328. [DOI] [PubMed] [Google Scholar]
- 29.Schwaller J, et al. Stat5a/b, but not Stat1, is essential for induction of a lethal myelo-and lymphoproliferative disease in mice by the TEL/JAK2 fusion protein associated with human leukemia. Blood. 1999;94(Suppl. 1):380a. (Abstr.) [Google Scholar]
- 30.Fambrough D, McClure K, Kazlauskas A, Lander ES. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]