Abstract
Hypoxia-inducible factors (HIFs) are a family of evolutionary conserved alpha-beta heterodimeric transcription factors that induce a wide range of genes in response to low oxygen tension. Molecular mechanisms that mediate oxygen-dependent HIF regulation operate at the level of the alpha subunit, controlling protein stability, subcellular localization, and transcriptional coactivator recruitment. We have conducted an unbiased genome-wide RNA interference (RNAi) screen in Drosophila cells aimed to the identification of genes required for HIF activity. After 3 rounds of selection, 30 genes emerged as critical HIF regulators in hypoxia, most of which had not been previously associated with HIF biology. The list of genes includes components of chromatin remodeling complexes, transcription elongation factors, and translational regulators. One remarkable hit was the argonaute 1 (ago1) gene, a central element of the microRNA (miRNA) translational silencing machinery. Further studies confirmed the physiological role of the miRNA machinery in HIF–dependent transcription. This study reveals the occurrence of novel mechanisms of HIF regulation, which might contribute to developing novel strategies for therapeutic intervention of HIF–related pathologies, including heart attack, cancer, and stroke.
Author Summary
Adaptation of cells to low oxygen (hypoxia) is a physiological response related to important diseases, including heart attacks, stroke, cancer, and diabetes. The mechanisms that mediate adaptation to hypoxia in humans are almost identical to those operating in diverse animal species, including mice, worms, and insects. The master regulator of cellular responses to hypoxia is a transcription factor named HIF, which induces a set of genes that mediate adaptation to oxygen starvation. Although it is known that regulation of HIF occurs mainly at the level of protein degradation and transcriptional coactivator recruitment, a comprehensive screen for HIF regulators has not been performed before. In this work, we have conducted an RNAi-based screen of the genome of the fruit fly Drosophila melanogaster, searching for genes that are required for HIF activity. This screen carried out in a cell culture system led to the definition of 30 critical regulators of HIF, most of which have not been associated with hypoxia biology before. The hits of the screen included components of chromatin remodeling complexes, transcription elongation factors, and translational regulators. Our results open the possibility of performing detailed studies on HIF regulation that may lead to novel therapeutic strategies for important human diseases.
Introduction
The cellular response to low oxygen tension (hypoxia) involves changes in gene expression that mediate adaptation to this condition. The hypoxic response is primarily mediated by a family of highly conserved transcription factors named Hypoxia Inducible Factors (HIFs) [1]. HIFs are α/β heterodimers, in which the common β subunit is constitutive and α subunits are negatively regulated by O2 through several concurrent mechanisms that include oxygen-dependent proteasomal degradation [2], blockage of transcriptional co-activator recruitment [3], [4] and subcellular localization [5], [6]. HIFα proteolysis requires polyubiquitination, which in turn depends on the hydroxylation of two key prolyl residues localized in the so-called oxygen-dependent degradation domain (ODDD) [7], [8]. Hydroxylation is mediated by specific HIF prolyl-4-hydroxylases, named PHDs that utilize dioxygen as a co-substrate, and hence, are considered bonafide cellular oxygen sensors [9], [10].
The machinery that mediates the transcriptional response to hypoxia is conserved in Drosophila melanogaster [11], being Sima and Tango the fly orthologues of HIFα and HIFβ [12] respectively, and Fatiga, the single Drosophila PHD [13]. As in mammalian cells, Sima is stable in hypoxia but rapidly degraded in normoxic conditions; its degradation requires Fatiga-dependent hydroxylation of a specific prolyl residue localized in the Sima ODDD [12], [14]. The fatiga gene is in turn transcriptionally activated by HIF, defining a negative feedback loop [12], [15].
HIF plays a crucial role in several human pathologies, including coronary heart disease, stroke and cancer [16], [17], and thus, considerable effort has been devoted to the characterization of the cellular response to hypoxia, and to the identification of HIF regulators that may contribute to developing novel strategies for therapeutic intervention. Various small molecule screens searching for HIF regulators have been conducted using high-throughput approaches (see [18] for a review). Although these strategies have been instrumental for manipulating HIF-dependent transcription, they have resulted less informative for the identification of the molecular targets involved.
In this work, we have carried out a genome-wide RNAi screen in Drosophila Schneider (S2) cells, aimed to the identification of genes required for HIF activity in hypoxic conditions. We have identified 30 regulators of the HIF response, including some previously reported genes, such as members of the phosphoinositide 3-kinase (PI3K) and Target of Rapamycin (TOR) signaling pathways [19], subunits of the COP9 signalosome complex [20], [21], and components of the Brahma chromatin-remodeling complex [22]. Among the genes identified as novel regulators of HIF-dependent transcription, we found the chromatin modifying elements Reptin and Pontin, several transcriptional and translational regulators, and the miRNA pathway component Argonaute 1. Further analysis confirmed an absolute requirement of core components of the miRNA machinery for the hypoxic response, both in cell culture and in vivo, suggesting a physiological role of miRNAs in HIF activity.
Results/Discussion
Genome-wide RNAi screen for HIF regulators
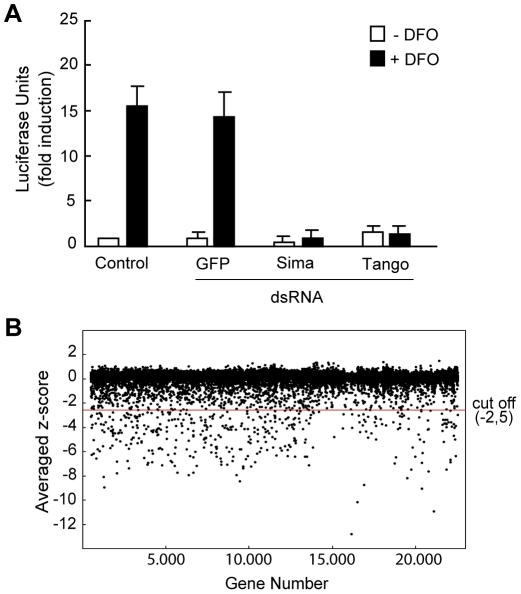
The genomic screen was carried out in Drosophila S2 cells bearing a stably-transfected hypoxia inducible reporter, in which a HIF-Responsive-Element (HRE) derived from the murine lactate dehydrogenase-A (ldh-A) enhancer drives the expression of firefly luciferase (Figure S1A; [15]). The HRE-Luc reporter was strongly induced upon exposure of the cells to hypoxia or to the iron chelating agent desferrioxamine (DFO), a compound that mimics the effect of hypoxia (Figure S1B) [15]. RNAi pilot experiments demonstrated that induction of the HRE-Luc reporter was dependent on Sima and Tango (Figure 1A) [15], and therefore, served as a reliable assay for testing the genomic double stranded RNA (dsRNA) library of the RNAi Screening Center (DRSC; http://flyrnai.org) that corresponds to more than 90% of the Drosophila transcriptome [23].
Figure 1. Primary screen for genes required for HIF–dependent transcription.
(A) S2-HRE-Luc cells treated with dsRNA against gfp (negative control), sima or tango were exposed or not to DFO. Luciferase induction by DFO was abrogated in cells depleted from sima or tango. (B) Scatter plot of the average Z-score (see Materials and Methods) of the whole set of data of the primary screen. dsRNAs which reduced reporter gene expression with a Z-score of less than −2.5 (cut-off line) were selected as positive hits of the primary screen for further analysis.
The screen was divided in 3 sequential phases (Table S1; see also Materials and Methods): I) a primary screen carried out in cells exposed to DFO, using a first-generation genomic library (DRSC 1.0 library) [23]; II) a secondary screen in which the genes that scored as positives in the primary screen were re-tested in cells also exposed to DFO, using a second generation library (DRSC Validation library) [24], [25], and normalizing the results with a constitutive transcriptional reporter (see below); and finally, III) a tertiary screen in which genes that scored as positives in the two previous phases were tested in hypoxia (1% O2).
I) The results of the primary screen were highly reproducible with Z score values (see Materials and Methods) showing a correlation coefficient of 0.6 between duplications (Figure S1C). A few dsRNAs rendered less reproducible results (i.e. the duplicates were more divergent), but nevertheless, were included in the secondary screen to avoid loosing potentially relevant hits. As shown in Figure 1B, approximately 97% of the dsRNAs rendered Z score values of around zero, indicating that, as expected, the majority of them do not affect HIF-dependent transcription. The screen was carried out in cells exposed to DFO and therefore, set up for the identification of positive regulators only. Thus, a substantial number of genes rendered negative Z score values (putative activators) but no genes with significant positive Z-score values (putative inhibitors) were obtained. We decided to define a Z score cut-off value of −2.5 for a gene to be considered a hit of the screen (Figure 1B) and, based on this criterion, 603 genes were initially selected for further analysis (Table S2). Noteworthy, both sima and tango -the Drosophila HIF-alpha and beta subunits respectively- scored as positives in this primary selection, with Z scores of −6.4 and −4.1 respectively, suggesting that the screen is reliable and has the potential to identify novel genes required for HRE-dependent transcription. Next, in order to eliminate genes that presumably interfere with basic cellular functions and prevent cell viability, the 603 hits were filtered against the results of a RNAi genome-wide screen for genes required for cell viability, previously carried out in the same cellular system with the same library [23]; 311 genes fell in the “cell viability” category, so they were not pursued further. Open reading frames that have been predicted but never demonstrated (the “Sanger collection”: 67 genes) were also eliminated from the analysis. Thus, after filtration, the number of positive genes from the primary screen was reduced to 225 (Table S3).
II) For the secondary screen, we developed a stably transfected cell line, which contained, along with the HRE-firefly luciferase reporter, a constitutive actin-Renilla-luciferase element, which was used to normalize the results (see Materials and Methods). This phase of the screen was carried out with a second-generation library (DRSC Validation Library; http://flyrnai.org), which was designed to eliminate false positives that arise from off-target effects of the original library [24], [25]; this new library includes more than one dsRNA for most genes (Table S4). Like in the primary screen, DFO was used as the hypoxic-mimetic agent (Table S1).
At the secondary screen, those genes that provoked a reduction of HRE-Luc reporter activity of more than 50% with at least one of the two dsRNAs were considered as hits. On this basis, 66 genes scored as positives, and based on the strength of the effect, this set of genes was further classified into two categories: Group A) Genes that rendered -with at least one of the corresponding dsRNAs- over 75% inhibition of HRE-luciferase activity (23 genes), and group B) Genes that provoked an inhibition of 50–75% of the activity -with at least one of the corresponding dsRNAs- (43 genes) (Table S4). As expected, sima and tango were among the hits of the secondary screen with approximately 96% inhibition.
III) Finally, we carried out a tertiary screen, in which genes that scored as positives in the secondary screen were tested in cells exposed to hypoxia (1% O2). All 23 genes that scored in group A (strong inhibition) were included in the tertiary screen, along with a selected set of genes from group B (12 genes) that are functionally related to those from group A. Thus, a total number of 35 genes were analyzed in hypoxia (Table 1). In this final screen 30 genes, including sima and tango, scored as positives with at least one of the two dsRNAs provoking more than 50% inhibition of HRE reporter activity (Table 1). Genes already known to be required for the HRE response, such as elements from the PI3K/TOR pathway [19] and the COP9 signalosome complex [20], [21], as well as genes that were not previously linked to HIF (see below), were among the hits in this final phase of the screen (Table 1).
Table 1. List of genes that scored as positives at the tertiary screen.
| Grouping criteria | Drosophila gene | Human homologue | Known or inferred function | inhibition (%) amplicon 1 | inhibition (%) amplicon 2 |
| HIF | Sima | HIF alpha | Alpha subunit of HIF | 92,3+/−6,4 | 89,3+/−5,4 |
| Tango | HiF beta/Arnt | Beta subunit of HIF | 59,0+/−1,1 | - | |
| Protein translation | CG9769 | eIF3f | Subunit-f of eukaryotic translation initiation factor 3 complex | 91,9+/−3,7 | - |
| Tango7 | eIF3m | Subunit-m of eukaryotic translation initiation factor 3 complex | 87,4+/−2,8 | 61,7+/−25,7 | |
| Trip1 | eIF3i | Subunit-i of eukaryotic translation initiation factor 3 complex | 87,5+/−3,4 | 84,7+/−11,8 | |
| CG8636 | eIF3g | Subunit-g of eukaryotic translation initiation factor 3 complex | 85,1+/−10,2 | - | |
| Pixie | RLI | Assembly of translation initiation complexes | 74,5+/−7,9 | 77,7+/−14,2 | |
| CG4849 | eEF2 | Putative translation elongation factor (downstream of TOR/S6K) | 83,3+/−2,1 | 42,0+/−11 | |
| Ef2b | eEF2 | Translation elongation factor (downstream of TOR/S6K) | 73,9+/−16,1 | 67,7+/−12 | |
| PI3K/TOR signaling | dTOR | TOR | Target of Rapamycin kinase | 89,5+/−5,5 | - |
| dRaptor | Raptor | Component of TORC1 complex | 89,3+/−2,3 | 66,9+/−3,1 | |
| dRheb | Rheb | GTPase required for TOR activity | 82,1+/−11,9 | 54,6+/−31,2 | |
| dPDK1 | PDK1 | 3-phosphoinositide-dependent protein kinase 1 | 56,2+/−8,3 | 51,5+/−24,5 | |
| Chromatin remodelling | Brahma | Brahma | ATPase component of the SWI/SNF complex | 83,9+/−11,6 | 84,8+/−0,6 |
| Bap155/Moira | BAF170 | Component of the SWI/SNF complex | 77,9+/−5,3 | 77,5+/−2,3 | |
| Snr1 | Ini1 | Component of the SWI/SNF complex | 38,0+/−15,3 | 28,2+/−11,4 | |
| Bap60 | BAF60a | Component of the SWI/SNF complex | 34,2+/−2,3 | 16,1+/−33.3 | |
| Dalao | BAF57 | Component of the SWI/SNF complex | 35,3+/−7,9 | - | |
| Reptin | Reptin | AAA+ ATPase component of various complexes | 80,3+/−0,4 | 74,6+/−8,7 | |
| Pontin | Pontin | AAA+ ATPase component of various complexes | 75,3+/−4,6 | 67,3+/−17,5 | |
| mRNA processing | CG14641 | RBM22 | RNA binding motif protein 22 -Putative pre mRNA splicing factor | 85,2+/−7,5 | 64,3+/−16,8 |
| Prp8 | Prp8 | RNAse H component of the spliceosome catalytic core | 72,5+/−20,5 | 71,1+/−2,3 | |
| Clipper | CPSF-30K | Subunit of Cleavage and Polyadenylation Specificity Factor | 63,0+/−10,6 | - | |
| Symplekin | Symplekin | Protein associated to mRNA polyadenylation complex | 58,8+/−1,0 | 46,2+/−16,9 | |
| Peanuts | - | ATP dependent RNA helicase involved in RNA splicing | 31,2+/−41,3 | - | |
| microRNA | Argonaute 1 | Ago proteins | Component of the miRNA pathway | 88,4+/−8,1 | 69,7+/−9,7 |
| Signalosome | CSN3 | COPS3 | COP9 complex subunit 3 | 60,3+/−2,6 | 30,5+/−3,7 |
| CSN6 | COPS6 | COP9 complex subunit 6 | 41,6+/−7,0 | - | |
| Miscellanea | Spt6 | Spt6 | Transcription elongation factor involved in heat shock response | 88,9+/−2,7 | 75,2+/−0,1 |
| CG2446 | - | Unknown | 84,9+/−5,3 | - | |
| TER94 | p97 | ER chaperone involved in ERAD | 78,7+/−0,2 | 75,3+/−1,8 | |
| Cryptocephal | ATF4 | Transcription factor involved in stress responses | 70,7+/−2,8 | - | |
| MBD-R2 | PHF20 | Unknown function - DNA interacting protein | 69,9+/−8,9 | 31,9+/−18,9 | |
| CG7065 | - | Unknown | 64,8+/−19,6 | - | |
| NSL1 | - | tRNA aminoacylation | 63,0+/−12 | 62,8+/−11,7 |
Cells were exposed to hypoxia, and dsRNAs corresponding to genes that provoked strong reduction of the response to DFO in the secondary screen (“Group A” genes) as well as some selected genes that rendered milder reduction of the response to DFO (“Group B” genes) were tested for their capacity to interfere with HRE-Luc reporter induction. Depicted genes are grouped according to their molecular function. Normalized luciferase activity (firefly to renilla luciferase activity ratio) for each well was calculated and expressed as the percentage of inhibition respect to hypoxic control cells treated with dsRNA against GFP. One or two amplicons (dsRNAs) were used for each gene. Amplicon identity is depicted in Table S4; their sequence can be found in http://flyrnai.org.
Four genes of the PI3K and TOR pathways -PDK1, TOR, Rheb and Raptor- were among the positive hits. Although it is still a matter of some controversy, several studies suggested that activation of PI3K/TOR pathway is required for HIF-dependent transcription [19], [26]. The fact that four elements belonging to these pathways were in the final list of hits strongly supports the notion that they are critically required for HIF activity.
One subunit of the eIF3 translation initiation complex, eIF3e/Int6, was previously shown to contribute to mammalian HIF-2α degradation [27]. In this screen, 4 additional subunits of this complex scored as positives as well (Table 1), implying that eIF3 complex involvement in HIF regulation might be broader than previously anticipated. The eIF3 complex is a scaffold for protein translation initiation composed of 12–13 polypeptides [28], and noteworthy, some of eIF3 subunits are associated to specific cellular events, such as oncogenic transformation [29] and apoptosis [30]. This work has now revealed that additional eIF3 subunits are required for HIF-dependent transcription. Several genes involved in chromatin remodeling, including 5 genes from the Brahma (also known as SWI/SNF) complex, and two from unrelated complexes -pontin and reptin- were also hits of the screen (Table 1). One previous report suggested a role of the SWI/SNF in the response to hypoxia [22], and a central role of chromatin remodeling in HIF-dependent gene expression is increasingly evident [31]. Therefore, the current screen, along with previous reports, strengthens the notion that an array of chromatin remodeling factors contribute to HIF-dependent transcriptional responses to hypoxia. Drosophila Pontin and Reptin are closely related members of the highly conserved AAA+ family of DNA helicases, which, besides participating in chromatin remodeling, are involved in responses to DNA double-strand breaks and transcriptional regulation mediated by β-catenin, E2F1 or c-Myc [32] [33]. The precise role of Pontin and Reptin in HIF-dependent responses needs to be investigated in detail.
A transcription elongation factor, Spt6, which had not been linked before to HIF regulation, was also identified in the screen (Table 1). Spt6 is known to co-localize with the phosphorylated (active) form of RNA polymerase II in areas of active transcription, particularly during induction of stress-related genes [34]. Spt6 is recruited to heat-shock (HS) dependent promoters upon the HS stimulus; recruitment occurs within 2 minutes after the HS and depends on the Heat Shock Factor (HSF) [35], [36]. Our results therefore expand the notion that Spt6 is a component of transcriptional responses to stress, including now the cellular response to hypoxia.
The Drosophila ATF4 homologue cryptocephal was another remarkable hit of the screen (Table 1). ATF4 is a bZIP transcription factor expressed at high levels in hypoxic areas of human cervix, brain, breast and skin tumors [37], and considered a central component of cellular responses to different types of stress, including the unfolded protein response (UPR), amino acid deprivation, oxidative stress and hypoxia. In hypoxia, PERK, an endoplasmic reticulum (ER) transmembrane protein kinase, is activated, leading to general inhibition of protein synthesis, thereby allowing upregulated translation of selective proteins including ATF4. As a result, ATF4 induces the expression of genes in response to hypoxia, but remarkably, this response is HIF-independent [19], [38]. Our data now suggest that ATF4 is required for HIF activity, adding a new layer of complexity to the mechanisms involved in the cellular response to hypoxia.
Argonaute 1 and the miRNA machinery are necessary for the transcriptional response to hypoxia
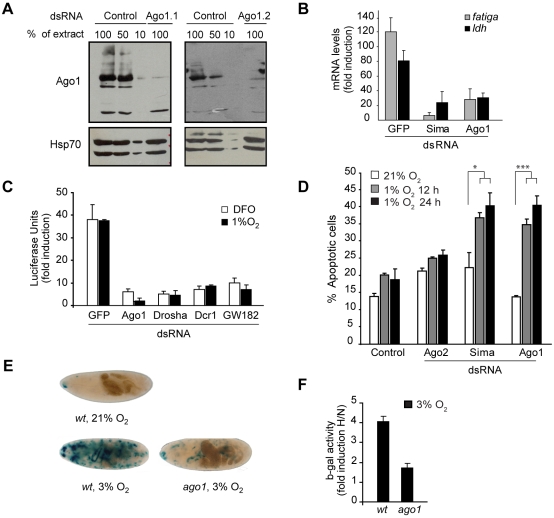
Argonaute 1 (Ago1), a central component of the microRNA silencing machinery [39] also scored as positive in the screen (Table 1). Given that little is known about the participation of the miRNA machinery in HIF regulation, we sought to further characterize Ago1 involvement in this process. We began by checking that dsRNA treatments were effective in reducing Ago1 protein levels (Figure 2A), and consistent with this, we confirmed that the function of the miRNA machinery was impaired (Figure S2A). To determine if inhibition of HRE-Luc reporter activity after Ago1 depletion reflects the behavior of endogenous hypoxia-inducible genes, we examined transcript levels of two well-established Sima downstream targets: ldh and PHD/fatiga [15]. The two transcripts were strongly upregulated in cells exposed to hypoxia, and this induction was dramatically reduced in cells treated with ago1 dsRNA (Figure 2B). In order to assess if Ago1 is required in the hypoxic response as part of the miRNA pathway, we silenced other components of the miRNA machinery. dsRNAs for dicer-1, drosha or gw182 strongly reduced luciferase reporter induction in cells exposed to DFO or hypoxia (Figure 2C), suggesting that the miRNA pathway is required for the transcriptional response to hypoxia. Other genes related to ago1, which have no reported function in the miRNA pathway, were also evaluated: Depletion of argonaute2, piwi or dicer-2, did not affect the HRE-response in S2 cells (Figure S2B). It is well known that HIF play a crucial role in the adaptive response to hypoxia by controlling the expression of genes that eventually promote cell survival. Thus, we studied if Ago1 does indeed contribute to cell viability in hypoxia. As depicted in Figure 2D, cells treated with ago1 dsRNA and exposed to hypoxia enter apoptosis at a higher proportion than untreated cultures, or cells treated with control ago2 dsRNA, suggesting a physiological requirement of Ago1 in the response to hypoxia. We next sought to test whether Ago1 is required for the HRE response in vivo. We analyzed the expression of the hypoxia-responsive transgenic reporter LDH-LacZ [12] in ago1k08121 mutant embryos. As previously reported, in wild type embryos LDH-LacZ is silent in normoxia and induced in hypoxia in a characteristic expression pattern that corresponds to some of the developing tracheal branches [12] (Figure 2E). In contrast, in ago1k08121 homozygous embryos, induction of the LDH-LacZ reporter in hypoxia was much weaker, indicating that Ago1 contributes to HIF/Sima dependent transcription in vivo (Figure 2E and 2F).
Figure 2. Argonaute 1 (Ago1) and the miRNA machinery are necessary for adaptation to hypoxia.
(A) Western blot showing Ago1 strong reduction in cells treated with dsRNA against ago1 during 4 days. Two different dsRNAs, ago 1.1 and ago 1.2 were used with identical results. Extracts from control cells were loaded at different amounts. Remaining Ago1 protein levels were 10% relative to controls after 4 days of RNAi treatment. Hsp70 was used as a loading control. (B) mRNA levels of two different HIF target genes, fatiga and ldh, were analyzed by real time PCR in cells exposed to hypoxia (1% O2) for 16 hours in comparison to those of cells maintained in normoxia. sima or ago1 dsRNAs largely prevented hypoxic induction of ldh and fatiga transcripts. (C) S2-HRE-Luc cells were treated with dsRNA against gfp, ago1, dicer-1, drosha or gw182 and then exposed to DFO or 1% O2. Whereas the gfp dsRNA had no effect on luciferase induction, silencing of any of the other genes strongly reduced luciferase induction by DFO or hypoxia. Data are represented as fold induction respect to control cells treated with dsRNA against gfp, and maintained in normoxia. (D) Analysis of the proportion of cells in apoptosis revealed that cells treated with ago1 dsRNA were as sensitive to hypoxia as cells treated with sima dsRNA, whereas untreated cells or cells treated with ago2 dsRNA were remarkably more resistant to low oxygen. After exposure to hypoxia, cells were stained with propidium iodide (PI) and Hoescht, and observed under a fluorescence microscope. The proportion of dying cells (PI positive) was determined using the CellProfiler cell image analysis software (Chi2 test *p<0.05; ***p<0.001). (E–F) Transgenic embryos bearing the hypoxia inducible reporter LDH-lacZ were exposed to hypoxia (3% O2) during 4 hours, and reporter gene activity was analyzed by X-gal staining (E) or quantitative β-galactosidase assays (F). The transgenic reporter is silent in normoxic wild type individuals, and strongly induced upon exposure to hypoxia (E). In ago1k08121 homozygous mutants the expression of the reporter in hypoxia is clearly reduced (E–F; p<0.01). N = Normoxia; H = Hypoxia.
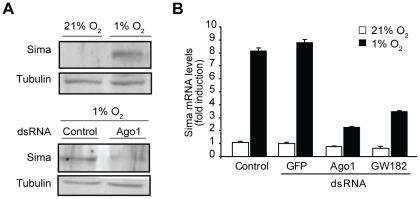
We have recently shown that oxygen-dependent subcellular localization is an important mechanism of Sima regulation: Sima shuttles continuously between the nucleus and cytoplasm, and nuclear export is inhibited in hypoxia [6], [40]. To get insights into how Ago1 depletion affects the transcriptional response to hypoxia, we studied Sima subcellular localization, and found no differences between Ago1 mutant embryos and wild type controls (Figure S3). Next we sought to study if Sima protein accumulation in hypoxic cells is affected upon Ago1 depletion. As depicted in Figure 3A, hypoxic induction of Sima protein is clearly reduced in S2 cells treated with ago1 dsRNA. The next step was to analyze sima transcript levels. Real time PCR analysis revealed a striking upregulation of sima mRNA levels upon exposure of the cells to hypoxia, and that ago1 RNAi treatment inhibited this induction (Figure 3B). These results indicate that HIF transcriptional induction or mRNA stabilization plays a role in the Drosophila hypoxic response, and suggests that the miRNA machinery participates in this regulation.
Figure 3. Hypoxic accumulation of Sima protein and mRNA is prevented in cells treated with ago1 dsRNA.
(A) Anti-Sima western blot analysis reveals that hypoxic accumulation of Sima is reduced in ago1 RNAi treated cells (24 h at 1% O2). (B) Real time PCR revealed that sima mRNA is strongly induced in cells exposed to hypoxia, and this induction is largely prevented in cells treated with ago1 or GW182 dsRNA.
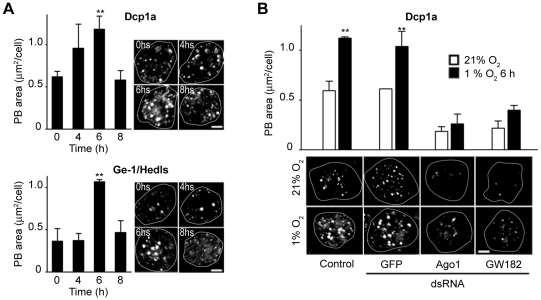
The above results prompted us to analyze possible changes in the miRNA machinery in hypoxia: a well known effect of miRNA dependent translational silencing is the accumulation of Processing Bodies (PBs), which are cytosolic foci that contain untranslated mRNAs and proteins, as well as small RNAs involved in translational silencing [41]. Thus, we explored whether exposure to hypoxia stimulates accumulation of PBs. As shown in Figure 4A and 4B, a clear increase of PBs was observed after exposing the cells to hypoxia, as revealed by anti-DCP1 or anti-Hedls antibody staining [42]. The accumulation of PBs in hypoxia was transient, reaching a peak 6 hours after transferring the cells to 1% O2 (Figure 4A). To explore if this effect is related to the miRNA pathway, we analyzed PB formation in cells depleted of Ago1 or GW182 and exposed to hypoxia. As shown in Figure 4B, both basal PB levels and induction of PBs by hypoxia were strongly reduced in these cells. It is unclear whether PB accumulation per se is required for HIF-dependent transcription or if alternatively, PB accumulation only reflects the activity of the miRNA machinery in the hypoxic response.
Figure 4. PBs accumulate in cells exposed to hypoxia in an Ago1- and GW182-dependent manner.
(A) S2R+ cells were maintained in normoxia or exposed to hypoxia (1% O2) for different time periods, then fixed and stained with an anti-DCP1 or anti-Hedls antibodies, two PBs specific markers. The PB area per cell was determined, revealing that PBs accumulate in a transient manner in cells exposed to hypoxia, peaking at 6 h after the onset of the hypoxic treatment, and decreasing at 8 h (one-way ANOVA and Dunnett multiple comparison post-Test, **p<0.01). (B) ago1 or GW182 dsRNA treatment affect PB basal levels and prevent PB accumulation upon exposure of the cells to 1% O2 for 6 h. (one-way Anova and SNK multiple comparisons post-test, p<0.01).
Taken together, our results suggest that the miRNA pathway plays a physiological role in cellular responses to hypoxia. Why does Ago1 depletion prevent sima mRNA induction? Although the identity of the target molecules that are controlled by the miRNA machinery is unknown, we can speculate that such unknown regulators directly or indirectly prevent sima transcriptional induction or alternatively contribute to sima messenger degradation (Figure 5). In mammalian cells, HIFα mRNAs are induced by NF-κB, so that NF-κB regulation plays an important role in the response to hypoxia [43]. It is not known if in Drosophila an NF-κB protein is required for sima transcriptional induction. If this was the case, it should be investigated if an inhibitor of the NF-κB pathway (i.e. IκB/Cactus) is subjected to miRNA-dependent translational regulation during adaptation to hypoxia.
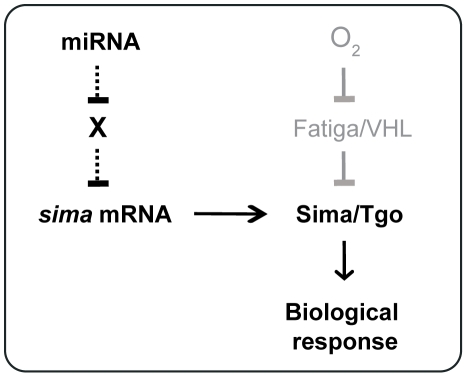
Figure 5. Model for sima regulation by the miRNA machinery.
An unknown (“X”) factor that directly or indirectly inhibits sima transcription is silenced by the miRNA machinery. When cells are depleted from Ago1, the factor X accumulates thereby preventing sima transcriptional induction in hypoxia.
Most major HIF regulators including PHDs, the von Hippel-Lindau tumour suppressor protein (VHL) [2] and factor inhibiting HIF (FIH) [4] are all inhibitors of the hypoxic response. The screen we have carried out here was instead focused on positive regulators of HIF. One remarkable feature of the results we have obtained is that most of the hits belong to just a few multiprotein complexes or signaling pathways. These include the PI3K/TOR pathway (translational regulation), eIF3 and eEF2 complexes (translational regulation), the COP9 signalosome (protein degradation/translational regulation), and the Brahma/SWI/SNF complex (chromatin remodeling). Noteworthy, besides genes belonging to these complexes, other hits of the screen are also linked to translational control (Ago1) or chromatin remodeling (Reptin, Pontin). Thus, one central conclusion of the results of this screen is that translational control and chromatin remodeling are two important mechanisms of HIF regulation, whose characterization in detail will broaden our understanding of HIF regulation and the cellular response to hypoxia.
Materials and Methods
Vectors, reporters, and cell culture
The reporter plasmids HRE- firefly luciferase (HRE-Luc) and act-renilla luciferase were previously described [15], [44]. The miRNA reporter pAC-miR-12 and CG10011-luc were a gift from E. Izaurralde [45]. pBLAST (Blasticidine resistance) and pPUR (Puromycin resistance) vectors were used to generate S2 stable cell lines. Drosophila Schneider's lines S2 or S2R+ cells were maintained at 25°C in Schneider or M3 medium (Sigma), supplemented with 10% fetal bovine serum (Gibco), 50 units/ml penicillin and 50 µg/ml streptomycin in 25 or 75 cm2 T-flasks (Greiner). Cells were grown in 12, 24, 96 or 384-well plates (Greiner), during 3 days and treated with 100 µM of DFO (Sigma) or exposed to hypoxia in a Forma Scientific 3131 incubator.
Synthesis of dsRNA and RNAi treatments
For dsRNAs not obtained from the Drosophila RNAi Screening Center (DRSC), fragments of the genes were amplified by PCR from cDNA or genomic DNA using T7-tailed oligonucleotides as primers. dsRNA synthesis was carried out with a T7 Megascript kit (Ambion) following manufacturer's instructions. The “bathing” method was utilized to introduce dsRNAs into S2 or S2R+ cells as previously described [46].
RNAi Screens
For screening experiments Drosophila S2 cells were maintained at 25°C in Schneider's medium. The primary screen was carried out at the Drosophila RNAi Screening Center (DRSC), the secondary and tertiary screens were performed in our laboratory with dsRNAs obtained from the DRSC. Primer and amplicon information can be found at http://flyrnai.org.
Primary screen
Two sets of 58 384-well screening plates (Costar) containing approximately 0.2 µg of dsRNA per well were provided by the DRSC (DRSC 1.0 library). Sima or GFP were used as positive and negative controls, respectively. Three days after plating, the cells were stimulated with DFO (100 µM) for 20 h and then firefly luciferase activity was determined using the SteadyGlo reagent (Promega). The Z-score value for each well was calculated as the luciferase activity of the well minus the average of the luciferase activity of the whole plate divided by the standard deviation of the plate.
Secondary screen
The secondary screen was carried out in 96-well plates. Firefly and Renilla luciferase activities were determined using the DualGlo reagent (Promega) in a Veritas Luminometer. Normalized luciferase activity (firefly to renilla luciferase activity ratio) for each well was calculated as a percentage of the control wells treated with GFP dsRNA.
Tertiary screen in hypoxia
Fifty-nine dsRNAs from the DRSC Validation library were used to cover the 35 selected genes. S2-HRE-Luc cells were incubated with dsRNAs in 96-well plates as described above and then exposed to hypoxia (1% O2) in a Forma Scientific 3131 incubator for 20 hours. Firefly and Renilla luciferase activities were determined and normalized as above.
Real-time PCR
Total RNAs from cells exposed to different treatments were isolated using the Trizol reagent (Invitrogen). One to 5 µg of total RNA were used as a template for cDNA synthesis, using the SuperScript III First Strand Synthesis System for RT-PCR (Invitrogen). Quantitative real time PCR was performed in the MX3005P real time PCR instrument (Stratagene, La Jolla, CA) with Syber Green, the hot start Platinum Taq DNA polymerase (Invitrogen) and the ROX reference dye (Invitrogen). Primers for amplifying 100–300 bp of each PCR product were used. PCR reactions were carried out for 5 min at 95°C followed by 35 cycles of three-step PCR for 30 seconds at 95°C, 1 min at 60°C, and 1 min at 72°C. Each sample was analyzed in triplicate. The data were normalized by subtracting the difference of the CT values between the target gene of interest (Tgene) and that of tubulin mRNA, thereby obtaining a ΔCT (Tgene CT −Tubulin CT). Relative expression (fold induction) was calculated as 2−(SΔC T −CΔC T ) where SΔCT−CΔCT is the difference between the sample ΔCT (treated cells) and the control ΔCT (RNAi GFP cells). Both target gene and tubulin reactions approached 100% efficiency as determined by standard curves. PCR products were analyzed on agarose gels to check that a single band was amplified.
Fly stocks
Flies used were yw, ldh- LacZ [12] and yw, ago1k08121/CyO.
β-galactosidase activity
Wild type or ago1k08121 embryos were exposed to 3% or 21% O2 for 4 h, homogenized in lysis buffer (50 mM Tris HCl [pH 7.8], 2 mM EDTA, 10% glycerol, 2 mM DTT, 1% Triton X-100, 1 mM PMSF) and centrifuged at 2,500×g for 3 min at 4°C. Enzymatic reactions were carried out by incubating 20 to 50 µg of protein extract in 180 µl buffer, containing 80 mM Na3PO4 (pH 7.3), 102 mM β-mercaptoethanol, 9 mM MgCl2, and 4 mM CPRG (Chlorophenol Red β-d-galactopyranoside; Roche Diagnostics) at 37°C, and absorbance at 574 nm was recorded at 10, 30, 60, 120, and 180 min time points; color development was linear throughout this time period. Endogenous background was subtracted using a heat-inactivated sample.
Immunofluorescence
For PB staining either a mouse monoclonal anti-DCP1 antibody (Abnova) was utilized at a 1∶1000 dilution were used or a rabbit anti-HEDLS antiserum (Bethyl) was used at 1∶500 dilution. Images were acquired in LSM510 Meta confocal microscopes (Carl Zeiss), using a Plan-Apochromat 63×/1.4 oil objective. Equipment adjustment was assessed by using 1µm Focal Check fluorescent microspheres (Molecular Probes). Pictures were exported to Adobe Photoshop software for cropping. Neither filters nor gamma-adjustments were applied. PB number and size in µm2 were determined with the “Analyze Particles” tool of the Image J software (NIH) in randomly selected micrographs.
Supporting Information
HRE-luciferase reporter induction in cells exposed to hypoxia or DFO. (A) Schematic representation of the HIF-responsive firefly luciferase reporter element used in this study (HRE-Luc). A dimerized regulatory sequence derived from the murine lactate dehydrogenase enhancer was cloned upstream of a firefly luciferase gene in a pGL3 plasmid bearing a fly hsp70 minimal promoter. Each 51 bp sequence contains two HIF responsive elements (HREs) and one cyclic AMP responsive element (CRE). (B) S2-HRE-luc cells were seeded in 96-wells tissue culture plates (1×104 cells per well), grown for 3 days, and stimulated with DFO (100 µM), or exposed to hypoxia (1% O2) for 20 hours. Strong induction of luciferase activity was observed in cells stimulated with DFO or hypoxia. Results are expressed as fold induction of luciferase activity respect to normoxic untreated cells. (C) Scatter plot of the duplicate results (Z-scores; see Materials and Methods) of the primary screen, showing the overall reproducibility of the data.
(0.03 MB PDF)
miRNAs and the response to hypoxia. (A) Upper panel, schematic representation of the miRNA reporter CG10011-luc; the miR-12 miRNA binds to the 3′ UTR of the luciferase mRNA, thereby inhibiting translation. Over-expression of miR-12 is therefore expected to provoke strong inhibition of translation. Lower panel, S2 cells were co-transfected with the CG10011-luc reporter and the pAC-miR-12 over-expression plasmid, or with an empty vector (pAC) as a control, and exposed to ago1 or gfp dsRNA treatments during 4 days. miR-12 over-expression inhibits 80% of luciferase expression in the control cells treated with gfp dsRNA, whereas in cells depleted from ago1 (ago1.1 or ago1.2 dsRNAs) miR-12 over-expression failed to inhibit luciferase expression to a large extent. (B) S2-HRE-luc cells were treated with dsRNA against gfp (control), sima, ago1, ago2, piwi, or dicer-2, grown during 4-8 days, and stimulated with DFO (100 µM). Cells depleted from ago1 or sima showed strong reduction of reporter activity, whereas cells depleted from ago2, dicer-2, or piwi exhibited normal induction of the reporter upon DFO exposure.
(0.01 MB PDF)
Regulation of Sima subcellular localization is not affected in Ago1 homozygous mutant embryos. We have analyzed Sima subcellular localization in en-Gal4/UAS-sima transgenic embryos carrying a homozygous mutation in the Ago1 locus (ago1k0208), and compared with Sima localization in en-Gal4/UAS-sima wild type individuals. The analysis was carried out as we reported previously (Dekanty et al., 2005) [15]. Three categories of Sima subcellular localization were defined for quantitative purposes: “Nuclear” (black color), “Ubiquitous” (grey) and “Cytoplasmic” (white). The Ago1 mutation does not impinge on Sima subcellular localization neither in normoxia nor in hypoxia.
(0.01 MB PDF)
Summary of the three phases of the overall screen for genes required for HIF activity.
(0.04 MB PDF)
Results of the primary screen carried out in cells exposed to DFO are shown. The screen was performed in duplicate; genes in which at least one of the two Z scores values was below −2.5 are depicted in the table. Under this criterion, 603 genes scored as positives in this initial phase of the screen.
(0.43 MB PDF)
The data obtained at the primary screen (Table S2) were filtered against the results of a cell viability screen previously carried out at the DRSC (Boutros et al. 2004) [23]. Sequences from the “Sanger collection” were also eliminated from the study; the 225 genes that remained as positive hits of the primary screen are depicted.
(0.27 MB PDF)
The secondary screen was also carried out in cells exposed to DFO. A second-generation library (DRSC 2.0 library) was used, in which most genes are represented by more than one dsRNA. Normalized luciferase activity (firefly/renilla luciferase activity ratio) for each well was calculated and expressed as a percentage of the inhibition respect to control cells treated with dsRNA against GFP that were exposed to DFO. The screen was carried out in duplicate and the mean percentage of inhibition is depicted.
(0.26 MB PDF)
Acknowledgments
We wish to thank the Drosophila RNAi screening center at Harvard Medical School, where the primary screen was conducted, Elisa Izaurralde for reagents, the Bloomington stock center for fly stocks, and the Wappner lab for discussions.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by a PICT 2007 ANPCyT grant, the HHMI grant 55005973, and the Wellcome Trust grant WT087675MA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 3.Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 4.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kallio PJ, Okamoto K, O'Brien S, Carrero P, Makino Y, et al. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. Embo J. 1998;17:6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero NM, Irisarri M, Roth P, Cauerhff A, Samakovlis C, et al. Regulation of the Drosophila hypoxia-inducible factor alpha Sima by CRM1-dependent nuclear export. Mol Cell Biol. 2008;28:3410–3423. doi: 10.1128/MCB.01027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 8.Ivan M, Kondo K, Yang H, Kim W, Valiando J, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 9.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, et al. C. elegans EGL-9 and Mammalian Homologs Define a Family of Dioxygenases that Regulate HIF by Prolyl Hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 10.Bruick RK, McKnight SL. A Conserved Family of Prolyl-4-Hydroxylases That Modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 11.Romero NM, Dekanty A, Wappner P. Cellular and developmental adaptations to hypoxia: a Drosophila perspective. Methods Enzymol. 2007;435:123–144. doi: 10.1016/S0076-6879(07)35007-6. [DOI] [PubMed] [Google Scholar]
- 12.Lavista-Llanos S, Centanin L, Irisarri M, Russo DM, Gleadle JM, et al. Control of the Hypoxic Response in Drosophila melanogaster by the Basic Helix-Loop-Helix PAS Protein Similar. Mol Cell Biol. 2002;22:6842–6853. doi: 10.1128/MCB.22.19.6842-6853.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centanin L, Ratcliffe PJ, Wappner P. Reversion of lethality and growth defects in Fatiga oxygen-sensor mutant flies by loss of Hypoxia-Inducible Factor-alpha/Sima. EMBO Rep. 2005;6:1070–1075. doi: 10.1038/sj.embor.7400528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arquier N, Vigne P, Duplan E, Hsu T, Therond PP, et al. Analysis of the hypoxia-sensing pathway in Drosophila melanogaster. Biochem J. 2006;393:471–480. doi: 10.1042/BJ20050675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekanty A, Lavista-Llanos S, Irisarri M, Oldham S, Wappner P. The insulin-PI3K/TOR pathway induces a HIF-dependent transcriptional response in Drosophila by promoting nuclear localization of HIF-(alpha}/Sima. J Cell Sci. 2005;118:5431–5441. doi: 10.1242/jcs.02648. [DOI] [PubMed] [Google Scholar]
- 16.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–685. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melillo G. Hypoxia-inducible factor 1 inhibitors. Methods Enzymol. 2007;435:385–402. doi: 10.1016/S0076-6879(07)35020-9. [DOI] [PubMed] [Google Scholar]
- 19.Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- 20.Miyauchi Y, Kato M, Tokunaga F, Iwai K. The COP9/signalosome increases the efficiency of von Hippel-Lindau protein ubiquitin ligase-mediated hypoxia-inducible factor-alpha ubiquitination. J Biol Chem. 2008;283:16622–16631. doi: 10.1074/jbc.M710599200. [DOI] [PubMed] [Google Scholar]
- 21.Bemis L, Chan DA, Finkielstein CV, Qi L, Sutphin PD, et al. Distinct aerobic and hypoxic mechanisms of HIF-alpha regulation by CSN5. Genes Dev. 2004;18:739–744. doi: 10.1101/gad.1180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang F, Zhang R, Beischlag TV, Muchardt C, Yaniv M, et al. Roles of Brahma and Brahma/SWI2-related gene 1 in hypoxic induction of the erythropoietin gene. J Biol Chem. 2004;279:46733–46741. doi: 10.1074/jbc.M409002200. [DOI] [PubMed] [Google Scholar]
- 23.Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 24.Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, et al. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 25.Flockhart I, Booker M, Kiger A, Boutros M, Armknecht S, et al. FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 2006;34:D489–494. doi: 10.1093/nar/gkj114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, et al. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–785. doi: 10.1038/nature05029. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Uchida K, Endler A, Shibasaki F. Mammalian tumor suppressor Int6 specifically targets hypoxia inducible factor 2 alpha for degradation by hypoxia- and pVHL-independent regulation. J Biol Chem. 2007;282:12707–12716. doi: 10.1074/jbc.M700423200. [DOI] [PubMed] [Google Scholar]
- 28.Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Smit-McBride Z, Pan X, Rheinhardt J, Hershey JW. An oncogenic role for the phosphorylated h-subunit of human translation initiation factor eIF3. J Biol Chem. 2008;283:24047–24060. doi: 10.1074/jbc.M800956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin YM, Chen YR, Lin JR, Wang WJ, Inoko A, et al. eIF3k regulates apoptosis in epithelial cells by releasing caspase 3 from keratin-containing inclusions. J Cell Sci. 2008;121:2382–2393. doi: 10.1242/jcs.021394. [DOI] [PubMed] [Google Scholar]
- 31.Fish JE, Yan MS, Matouk CC, St Bernard R, Ho JJ, Jr, et al. Hypoxic repression of endothelial nitric-oxide synthase transcription is coupled with eviction of promoter histones. J Biol Chem. 2010;285:810–826. doi: 10.1074/jbc.M109.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, et al. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. Embo J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallant P. Control of transcription by Pontin and Reptin. Trends Cell Biol. 2007;17:187–192. doi: 10.1016/j.tcb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, et al. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 37.Ameri K, Lewis CE, Raida M, Sowter H, Hai T, et al. Anoxic induction of ATF-4 through HIF-1-independent pathways of protein stabilization in human cancer cells. Blood. 2004;103:1876–1882. doi: 10.1182/blood-2003-06-1859. [DOI] [PubMed] [Google Scholar]
- 38.Koumenis C. ER stress, hypoxia tolerance and tumor progression. Curr Mol Med. 2006;6:55–69. doi: 10.2174/156652406775574604. [DOI] [PubMed] [Google Scholar]
- 39.Sontheimer EJ, Carthew RW. Molecular biology. Argonaute journeys into the heart of RISC. Science. 2004;305:1409–1410. doi: 10.1126/science.1103076. [DOI] [PubMed] [Google Scholar]
- 40.Irisarri M, Lavista-Llanos S, Romero NM, Centanin L, Dekanty A, et al. Central role of the oxygen-dependent degradation domain of Drosophila HIFalpha/Sima in oxygen-dependent nuclear export. Mol Biol Cell. 2009;20:3878–3887. doi: 10.1091/mbc.E09-01-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 42.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belaiba RS, Bonello S, Zahringer C, Schmidt S, Hess J, et al. Hypoxia up-regulates hypoxia-inducible factor-1alpha transcription by involving phosphatidylinositol 3-kinase and nuclear factor kappaB in pulmonary artery smooth muscle cells. Mol Biol Cell. 2007;18:4691–4697. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, et al. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 45.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. Rna. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl Acad Sci U S A 2000 Jun 6;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HRE-luciferase reporter induction in cells exposed to hypoxia or DFO. (A) Schematic representation of the HIF-responsive firefly luciferase reporter element used in this study (HRE-Luc). A dimerized regulatory sequence derived from the murine lactate dehydrogenase enhancer was cloned upstream of a firefly luciferase gene in a pGL3 plasmid bearing a fly hsp70 minimal promoter. Each 51 bp sequence contains two HIF responsive elements (HREs) and one cyclic AMP responsive element (CRE). (B) S2-HRE-luc cells were seeded in 96-wells tissue culture plates (1×104 cells per well), grown for 3 days, and stimulated with DFO (100 µM), or exposed to hypoxia (1% O2) for 20 hours. Strong induction of luciferase activity was observed in cells stimulated with DFO or hypoxia. Results are expressed as fold induction of luciferase activity respect to normoxic untreated cells. (C) Scatter plot of the duplicate results (Z-scores; see Materials and Methods) of the primary screen, showing the overall reproducibility of the data.
(0.03 MB PDF)
miRNAs and the response to hypoxia. (A) Upper panel, schematic representation of the miRNA reporter CG10011-luc; the miR-12 miRNA binds to the 3′ UTR of the luciferase mRNA, thereby inhibiting translation. Over-expression of miR-12 is therefore expected to provoke strong inhibition of translation. Lower panel, S2 cells were co-transfected with the CG10011-luc reporter and the pAC-miR-12 over-expression plasmid, or with an empty vector (pAC) as a control, and exposed to ago1 or gfp dsRNA treatments during 4 days. miR-12 over-expression inhibits 80% of luciferase expression in the control cells treated with gfp dsRNA, whereas in cells depleted from ago1 (ago1.1 or ago1.2 dsRNAs) miR-12 over-expression failed to inhibit luciferase expression to a large extent. (B) S2-HRE-luc cells were treated with dsRNA against gfp (control), sima, ago1, ago2, piwi, or dicer-2, grown during 4-8 days, and stimulated with DFO (100 µM). Cells depleted from ago1 or sima showed strong reduction of reporter activity, whereas cells depleted from ago2, dicer-2, or piwi exhibited normal induction of the reporter upon DFO exposure.
(0.01 MB PDF)
Regulation of Sima subcellular localization is not affected in Ago1 homozygous mutant embryos. We have analyzed Sima subcellular localization in en-Gal4/UAS-sima transgenic embryos carrying a homozygous mutation in the Ago1 locus (ago1k0208), and compared with Sima localization in en-Gal4/UAS-sima wild type individuals. The analysis was carried out as we reported previously (Dekanty et al., 2005) [15]. Three categories of Sima subcellular localization were defined for quantitative purposes: “Nuclear” (black color), “Ubiquitous” (grey) and “Cytoplasmic” (white). The Ago1 mutation does not impinge on Sima subcellular localization neither in normoxia nor in hypoxia.
(0.01 MB PDF)
Summary of the three phases of the overall screen for genes required for HIF activity.
(0.04 MB PDF)
Results of the primary screen carried out in cells exposed to DFO are shown. The screen was performed in duplicate; genes in which at least one of the two Z scores values was below −2.5 are depicted in the table. Under this criterion, 603 genes scored as positives in this initial phase of the screen.
(0.43 MB PDF)
The data obtained at the primary screen (Table S2) were filtered against the results of a cell viability screen previously carried out at the DRSC (Boutros et al. 2004) [23]. Sequences from the “Sanger collection” were also eliminated from the study; the 225 genes that remained as positive hits of the primary screen are depicted.
(0.27 MB PDF)
The secondary screen was also carried out in cells exposed to DFO. A second-generation library (DRSC 2.0 library) was used, in which most genes are represented by more than one dsRNA. Normalized luciferase activity (firefly/renilla luciferase activity ratio) for each well was calculated and expressed as a percentage of the inhibition respect to control cells treated with dsRNA against GFP that were exposed to DFO. The screen was carried out in duplicate and the mean percentage of inhibition is depicted.
(0.26 MB PDF)