Abstract
The transcription factor Nrf2 regulates expression of multiple cellular defence proteins through the antioxidant response element (ARE). Nrf2-deficient mice (Nrf2−/−) are highly susceptible to xenobiotic-mediated toxicity, but the precise molecular basis of enhanced toxicity is unknown. Oligonucleotide array studies suggest that a wide range of gene products is altered constitutively, however no equivalent proteomics analyses have been conducted. To define the range of Nrf2-regulated proteins at the constitutive level, protein expression profiling of livers from Nrf2−/− and wild type mice was conducted using both stable isotope labelling (iTRAQ) and gel electrophoresis methods. To establish a robust reproducible list of Nrf2-dependent proteins, three independent groups of mice were analysed. Correlative network analysis (MetaCore) identified two predominant groups of Nrf2-regulated proteins. As expected, one group comprised proteins involved in phase II drug metabolism, which were down-regulated in the absence of Nrf2. Surprisingly, the most profound changes were observed amongst proteins involved in the synthesis and metabolism of fatty acids and other lipids. Importantly, we show here for the first time, that the enzyme ATP-citrate lyase, responsible for acetyl-CoA production, is negatively regulated by Nrf2. This latter finding suggests that Nrf2 is a major regulator of cellular lipid disposition in the liver.
Keywords: Nrf2, Transgenic, Liver, Protein expression, iTRAQ, Lipid metabolism
Graphical abstract
1. Introduction
Exposure to electrophiles and reactive oxygen species (ROS) may result in intracellular damage to proteins, DNA and other macromolecules and can lead to the development of diseases, such as cancer, neurodegenerative disorders and cardiovascular disease [1–3]. To counteract the damage caused by electrophiles and ROS, higher animals have developed elaborate defence mechanisms [4,5], which include the coordinated induction of a battery of genes encoding phase II detoxifying enzymes and oxidative stress inducible proteins [6,7]. It is now well established that a principal regulator of the cellular defence response is the transcription factor termed nuclear factor erythroid-2 related factor 2 (Nrf2) [8–15]. Nrf2 been shown to play a key role in the transcriptional activation of multiple genes involved in cellular defence against ROS and electrophiles, such as NAD(P)H:quinone oxidoreductase (NQO1) [16], glutathione S-transferases (GSTs) [17,18], glutamate-cysteine ligase [19], haem oxygenase-1 (HO-1) [20], thioredoxin [12], and ferritin [21].
Nrf2 deficient transgenic mice have provided the most informative integrated model in which to examine the role of Nrf2 in regulating the defence response to chemical insults, particularly in the liver. Inactivation of the nrf2 gene results in no obvious phenotypic changes (except in aging female animals, where autoimmune diseases have been observed [22]), indicating that Nrf2 is not essential for normal growth and development [23]. Several studies focussing on individual proteins have shown that the presence of Nrf2 is essential for the enhanced expression of several antioxidant response proteins following administration of certain chemical inducers, however, constitutive expression of the same genes is often unaffected or only marginally reduced by deletion of the Nrf2 gene [9,11,14,23–29].
Acute exposure of Nrf2-deficient mice to a range of toxic chemical insults has been shown to result in enhanced toxicity compared to their wild type counterparts. Multiple studies show a reduced resistance to hepatotoxicity induced by a wide range of compounds, including paracetamol [25,30], carbon tetrachloride [31], pyrazole [32], ethanol [33] and pentachlorophenol [34].
Two possible explanations exist for the reduction in chemically-induced hepatotoxicity seen in each of these studies: first, the lack of Nrf2 may abrogate the animal's ability to up-regulate defence proteins in response to the chemical stimulus or, second, the enhanced toxicity may simply reflect a constitutive reduction in defence proteins due to the absence of Nrf2. Clearly, these two possible mechanisms are not mutually exclusive and each may contribute to a different degree, depending on the nature of the chemical insult. Nevertheless, it is important to understand the relative contribution of each mechanism for a given hepatotoxin in order to translate information gained in animal studies into improved clinical management of drug- or chemical-induced toxicity in man.
Oligonucleotide microarray analysis of Nrf2 null mice suggests that Nrf2 may regulate more than 200 genes, either constitutively or following exposure to a known inducer [29,35,36]. However, it is now well recognized that transcriptional up-regulation does not always equate to increased protein expression [37]. Until now, no equivalent proteomic analysis of Nrf2 null mice has been undertaken to substantiate the mRNA expression changes at the protein level. Here we report a global analysis of constitutive hepatic protein expression in Nrf2 null and wild type mice [10].
2. Materials and methods
2.1. Materials
Protein assay kits were from Bio-Rad (Hemel Hempstead, Herts, UK). Immobiline Dry Strips and associated buffers for 2-DE gels were obtained from GE Healthcare UK (Little Chalfont, Bucks, UK). 8-plex isobaric tags for relative and absolute quantification (iTRAQ) protein labelling kit/reagents were purchased from AB Sciex (Framingham, MA, USA). Sequencing grade trypsin was obtained from Promega UK (Southampton, Hants, UK). All other reagents were obtained from Sigma (Poole, Dorset, UK).
2.2. Animal studies
All experiments were undertaken in accordance with criteria outlined in a license granted under the Animals (Scientific Procedures) Act 1986, and approved by the Animal Ethics Committees of the University of Liverpool. Generation of the Nrf2 knockout mouse and genotyping of progeny have been described elsewhere [26,28]. Male mice of approximately 10 weeks of age were used throughout the study. Mice were housed at a temperature range of 19 °C–23 °C under 12-h light/dark cycles and given free access to food and water. Animals were killed by exposure to a rising concentration of CO2 followed by cervical dislocation. Livers were removed and snap-frozen immediately in liquid N2, before being stored at −80 °C.
Three groups of mice were used: for the first iTRAQ analysis (iTRAQ analysis 1), 8 mice (4 Nrf(+/+) and 4 Nrf(−/−)) were used, for the second iTRAQ analysis (iTRAQ analysis 2), 12 mice (6 Nrf(+/+) and 6 Nrf(−/−)) and for the 2DE gel analysis 8 mice were used (4 Nrf(+/+) and 4 Nrf(−/−)).
2.3. iTRAQ labelling of liver homogenates
Whole liver homogenates (75 μg protein) from Nrf2(+/+) and Nrf2(−/−) (n = 4), were prepared in TEAB/SDS. iTRAQ reagent labelling was then carried out according to the Applied Biosystems protocol for an 8plex procedure. Briefly, samples were denatured, reduced and capped with methylmethanethiosulfate (MMTS), before overnight digestion with trypsin and then labelled with isobaric tags. For the first three iTRAQ runs, Nrf2(+/+) samples were labelled with tags 113 to 116 while Nrf2(−/−) samples received the 117 to 121 tags. In the fourth experiment, the sample labelling was reversed such that the wild type animals had the heavier tags and the null mice the lighter tags, in order to control for labelling bias. iTRAQ-labelled peptides were then pooled and diluted to 4 mL with 10 mM potassium dihydrogen phosphate/25% acetonitrile (ACN; w/v). The pH of the samples was adjusted to < 3 using phosphoric acid prior to fractionation on a Polysulfoethyl A strong cation-exchange column (200 × 4.6 mm, 5 μm, 300 Å; Poly LC, Columbia, MD). A flow rate of 1 mL/min was applied and peptides eluted by increasing the concentration of KCl in the mobile phase to 0.5 M over 60 min. Fractions of 2 mL were collected and were dried by centrifugation under vacuum (SpeedVac, Eppendorf).
2.4. Mass spectrometric analysis of iTRAQ samples
For LC-MS/MS analysis of iTRAQ labelled samples, each cation exchange fraction was resuspended in 120 μL 5% ACN/0.05% trifluoroacetic acid (TFA) and 60 μL were loaded on column. Samples were analysed on a QSTAR® Pulsar i hybrid mass spectrometer (AB Sciex) and were delivered into the instrument by automated in-line liquid chromatography (integrated LCPackings System, 5 mm C18 nano-precolumn and 75 μm × 15 cm C18 PepMap column; Dionex, California, USA) via a nano-electrospray source head and 10 μm inner diameter PicoTip (New Objective, Massachusetts, USA). The precolumn was washed for 30 min at 30 μL/min with 5% ACN/0.05% TFA prior to initiation of the solvent gradient in order to reduce the level of salt in the sample. A gradient from 5% ACN/0.05% TFA (v/v) to 60% ACN/0.05% TFA (v/v) in 70 min was applied at a flow rate of 300 nL/min. The MS was operated in positive ion mode with survey scans of 1 s, and with an MS/MS accumulation time of 1 s for the three most intense ions. Collision energies were calculated on the fly based on the m/z of the target ion and the formula, collision energy = (slope × m/z) + intercept. The intercepts were increased by 3–5 V compared to standard data acquisition in order to improve the reporter ion intensities/quantitative reproducibility.
2.5. iTRAQ data analysis
Data analysis was performed using ProteinPilot software (Version 3, AB Sciex, Warrington, UK). The data were analysed with a fixed modification of MMTS-labelled cysteine, biological modifications allowed and with the confidence set to 10% to enable the False Discovery Rate to be calculated from screening the reversed SwissProt database. Ratios for each iTRAQ label were obtained, using a wild type mouse (WT mouse 1) sample as the denominator. The detected protein threshold (“unused protscore (conf)”) in the software was set to 1.3 to achieve 95% confidence.
2.6. Network analysis
The accession numbers of the 108 proteins identified as significantly different following Benjamini–Hochberg adjustment for multiple comparisons (p ≤ 0.2) were converted to Entrez gene IDs using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/conversion.jsp ) and analysed for evidence of network wide changes in cellular phenotype using MetaCore from GeneGo Inc., an integrated manually curated knowledge database for pathway analysis of gene lists (http://www.genego.com/metacore.php).
The gene list was analysed using the Pathway Maps tool, which maps the genes listed to defined signalling pathways that have been experimentally validated and are widely accepted. The proteins deemed Nrf2-regulated according to the criteria defined above were compared against a background file containing all of the identified proteins which had similarly been converted to a list of Entrez gene IDs using DAVID. The p values generated by the software were used to determine the statistical significance of the pathways identified. The p value represents the probability that a particular pathway will be represented by chance given the number of genes in the experiment and the number of genes in the pathway.
2.7. 2-DE of liver homogenates
Mouse livers were weighed and 0.3 g of tissue was homogenized in 1 mL lysis buffer [40 mM tris, 7 M urea, 2 M thiourea, 4% (w/v) 3-[(3-Cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS), 10 mM 1,4-dithiothreitol (DTT), 1 mM EDTA]. The homogenate was sonicated for 30 s and centrifuged at 150 000 g for 45 min. The supernatant was assayed for protein content [38] and stored at −80 °C. Aliquots of samples containing equal quantities of protein (0.5 mg) were diluted to 350 μL with rehydration buffer (9 M urea, 2% w/v CHAPS, bromophenol blue (trace), 2% v/v immobilized pH gradient (IPG) buffer, 0.28% w/v DTT) and incubated overnight with nonlinear Immobiline DryStrips (18 cm; pH 3–10 non-linear) in a re-swelling chamber. The samples were separated in the 1st dimension by isoelectric focussing (IEF) for 25 h at a constant temperature of 20 °C to achieve a total of 75 000 Vh (MultiPhor II, GE Healthcare UK, Little Chalfont, Bucks, UK). The IPG strips were then incubated with equilibration buffer (50 mM Tris, 6 M urea, 30% v/v glycerol, 2% w/v SDS, bromophenol blue (trace) containing 1% w/v DTT) for 15 min followed by incubation in the same buffer with the DTT replaced by 2.5% w/v iodoacetamide for a further 15 min. The strips were applied to the surface of 12% w/v SDS-PAGE gels and sealed with agarose [39]. The samples were subjected to electrophoresis at 20 W/gel and 25 °C for approximately 3 h (Ettan Dalt 12, GE Healthcare UK, Little Chalfont, Bucks, UK). The gels were then stained with colloidal Coomassie blue.
2.8. Image analysis of 2-DE gels
Colloidal Coomassie blue stained 2D gels were scanned using a GS710 calibrated imaging densitometer (BioRad, Hemel Hempstead, UK). TIFF images were generated and were analysed using ImageMasterTM 2D Elite software, version 4.01 (Amersham Pharmacia Biotech, Buckinghamshire, England). Altogether eight gels were analyzed (4 Nrf2(−/−) and 4 Nrf2(+/+)). An objective strategy for quantitative comparisons between wild type and null liver samples was adopted to exclude the possibility of bias, as follows. The gels were initially analysed using an automated procedure to identify spots. The authenticity and outline of each spot was validated by eye and edited where necessary. In each case approximately 500 validated spots were recorded from each gel. Spot matching was accomplished initially by automated fitting of the spots, followed by manual seeding of remaining spots that failed to match by automated fitting. A background value was subtracted for each gel and the spot volumes normalised against the total volume of all matched spots. For each spot, the ratio between its intensity and the sum of all spot intensities in the gel (normalized spot intensity) was determined and used for quantitative comparison. Visual and quantitative comparisons were only sought in spots that were matched in all four gels for a given treatment group.
2.9. Identification of proteins from 2DE gels
Protein spots of interest were excised from Colloidal Coomassie blue-stained 2DE gels by automated spot excision (Ettan Dalt Spot Picker, Amersham Biosciences) and were subjected to tryptic digestion [40]. Gel pieces were washed with 100 μl of 50% (v/v) ACN/50 mM ammonium bicarbonate (NH4CO3) (pH 7.8) for 15 min and were dried by centrifugation under vacuum (SpeedVac, Eppendorf). The dried gel pieces were rehydrated with 4–10 μl of digestion buffer (5 ng/μl of modified sequencing grade trypsin in 50 mM NH4CO3) and were incubated overnight at 37 °C. The resulting peptides were extracted by the addition of 30 μl of 60% ACN/1% TFA and incubation for 5 min in a sonicating water bath at 20 °C. The samples were briefly centrifuged and the supernatants were collected. A further 30 μl of 60% ACN/1% TFA was added to the gel plug and the sample was sonicated for 5 min. The supernatants were pooled and dried by centrifugation under vacuum. The peptides were resuspended in 10 μl of 5% ACN/0.05% TFA. 0.5 μl of the peptide mixture was spotted onto a 96-position stainless steel target and was mixed 1:1 with matrix [10 mg/ml α-cyano-4-hydroxycinnamic acid in 50% ACN/ 0.1%TFA]. Peptide mass finger prints were obtained on a Voyager DE Pro MALDI mass spectrometer (AB Sciex) and the resulting mass lists searched against the NCBInr database using Mascot software (Matrix Science). Scores of greater than 75 were regarded as sufficient for identification. Each significant identification was checked for consistency between its isoelectric point, molecular mass and mobility on the 2DE gel and, where possible, coincidence with a published mouse liver proteome 2DE database [41,42].
2.10. Western immunoblotting for ATP-citrate lyase
Whole liver homogenate (25 µg of protein) was separated by denaturing electrophoresis on a 10% polyacrylamide gel (ProtoGel acrylamide solution and buffers, using Tris-Glycine-SDS running buffer) and transferred to a nitrocellulose membrane (GE healthcare). After transfer, a Ponceau Red stain was used to ensure equal loading and then the membrane was blocked using 10% milk in 1x TBS/0.1%Tween for 30 min at room temperature, before incubation with a rabbit monoclonal antibody to ATP citrate lyase (ab40793, Abcam plc, Cambridge, UK) at 1:2000 with 2% milk in 1xTBS/0.1%Tween at 4 °C overnight. The membrane was washed several times with TBS-Tween and then incubated with the secondary antibody (peroxidise-conjugated goat anti-rabbit immunoglobulin G, 1:10 000 in TBS-Tween containing 2% milk) for 1 h at room temperature. Enhanced Chemiluminescence Plus (GE Healthcare) was used to visualise the level of protein-antibody complex. Band volume was measured by densitometry using Biorad Quantity One 1D Analysis Software (BioRad).
2.11. Identification of antioxidant response elements in the promoter regions of Nrf2-regulated genes
ARE consensus sequences were sought in the 5′-flanking regions upstream of all genes identified in the initial iTRAQ analysis (iTRAQ analysis 1) as being Nrf2-regulated (p < 0.05, Student's t-test). Public domain software (Regulatory Sequence Analysis Tools, http://rsat.ulb.ac.be/rsat/) provided by the Service de Conformation des Macromolécules Biologiques et de Bioinformatique at the University Libre de Bruxelles [43] was used. 5′-flanking sequences (2000 bp upstream of the start codon) were retrieved directly from the ENSEMBL database from within the RSAT package. Promoter sequences were then interrogated for ARE or ARE-like sequences using both string-based and matrix-based protocols. String-based analysis was carried out using the programme ‘dna search’ available within the RSAT web resource. The search term used was RTGABNNNGCA (where R = G/C, B = G/C/T and N = any nucleotide) based on the consensus sequence derived by Nioi et al. [44]. In order to identify ARE-like sequences, a matrix-based pattern matching method was performed using the programme ‘patser’. A position-specific scoring matrix (PSSM) was created based on the core ARE (cARE) position-specific probability matrix published by Nerland [45]. In order to calculate the background base frequencies, A/T and C/G frequencies were determined within the upstream sequences of all the genes interrogated and a mean value for each base was defined (A/T 0.26, C/G 0.24). The derived PSSM was then used to scan each of the genes shown to be significantly different between Nrf2(+/+) and Nrf2(−/−) mice.
2.12. Statistical analysis
2.12.1. iTRAQ data
iTRAQ data for proteins within the 1% false discovery rate and for which full quantification data were obtained, were statistically analysed within the R computational environment [46]. R is an open source software environment for statistical computing and graphics (http://www.r-project.org/). Normality of data and equivalence of variance across the data sets was assessed by Shapiro–Wilk and F-tests, respectively, and also by inspection of histogram plots for all proteins identified. Data were then analysed by t-test using the module multtest, a package designed for re-sampling based multiple hypothesis testing. Benjamini–Hochberg corrections for multiple comparisons were performed on all raw p values generated [47]. Protein expression differences between wild type and Nrf2-null mice giving a p value of < 0.05 by t-test and a Benjamini–Hochberg value ≤ 0.2 were accepted for further correlative network analysis. The Benjamini–Hochberg cut-off was set at 0.2 to avoid the exclusion of correlated Nrf2-regulated proteins through application of too stringent a correction for multiple testing in accordance with multivariate modelling approaches to account for potential confounders [47].
2.12.2. 2DE gel data
Data are expressed as mean ± SEM for at least four separate experiments. All values were analysed for non-normality using the Shapiro–Wilk test. Normally distributed values were compared using Student's unpaired t-test whilst non-normal values were analysed using the Mann–Whitney test. These statistical analyses were performed using the SPSS statistical software package, version 12 (Chicago, IL, USA). Statistical significance was accepted at p values of < 0.05.
3. Results
3.1. iTRAQ analysis of Nrf2(+/+) and Nrf2(−/−) mouse liver proteins
Two independent sets of mice were analysed using iTRAQ stable isotope labelling. For the first analysis (iTRAQ analysis 1), samples from four Nrf2(+/+) and four Nrf2(−/−) mice were analysed using 8-plex iTRAQ reagents and the entire analysis was repeated on four separate occasions. For the second group of mice (iTRAQ analysis 2), six Nrf2(+/+) and six Nrf2(−/−) mice were compared on a single occasion using three sets of 4-plex iTRAQ reagents. The second set of mice was thus used as a validation cohort to challenge the reproducibility of the protein changes observed in the initial “training” set. In each case iTRAQ data were processed using Protein Pilot version 3, including FDR. Table 1 shows the numbers of proteins identified and quantified within the two independent iTRAQ analyses.
Table 1.
Total numbers of proteins identified and quantified with a false discovery rate (FDR) exclusion of 1% in iTRAQ analyses 1 and 2.
| iTRAQ analysis | LC-MS analysis | No. of proteins identified | No. of proteins identified above 1% global FDR | No. of proteins quantified |
|---|---|---|---|---|
| 1 | Run 1 | 486 | 265 | 162 |
| Run 2 | 1287 | 911 | 620 | |
| Run 3 | 1003 | 759 | 593 | |
| Run 4 | 726 | 563 | 426 | |
| Total | 1654 | 1109 | 769 | |
| 2 | Run 1 | 1068 | 825 | 654 |
| Run 2 | 1065 | 780 | 661 | |
| Run 3 | 1068 | 711 | 637 | |
| Total | 1717 | 1070 | 628 | |
Numbers are given for proteins identified with a confidence greater than 90% and for those characterized by at least 2 peptides. The number of proteins quantified relates to those proteins determined in all eight mouse liver samples.
3.1.1. iTRAQ analysis 1
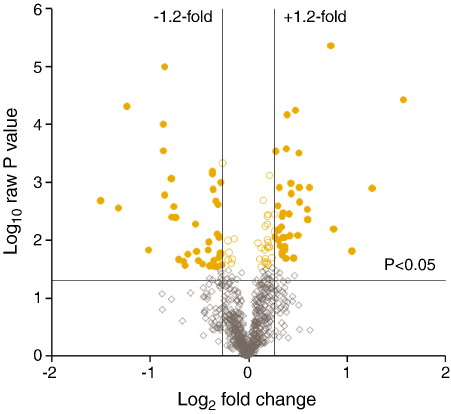
For iTRAQ analysis 1, each of the four runs represents a full repeat analysis of the same mouse liver sample. Thus, each protein expression value derived from iTRAQ analysis 1 represents the mean from four animals repeated on four occasions. In total, 1109 unique proteins were identified in at least one of the four runs within the FDR of 1% (Table 1); of these, 769 proteins had complete data sets in at least one run across all eight mice, and were consequently accepted for full quantitative analysis. Considerable variation was seen between the four runs, with the first run in particular giving relatively low proteome coverage. Nevertheless, all four runs were included for statistical analysis in order to maximise the number of proteins to include in the network analysis. Following statistical analysis, 108 proteins were found to be differentially expressed between Nrf2(+/+) and Nrf2(−/−) mouse livers using the criteria defined above and these are listed in Table 2. Fig. 1 shows a volcano plot of the entire data set highlighting proteins whose expression was significantly different (t-test p < 0.05) between wild type and Nrf2-null mice (open circles). Proteins that were significantly different by at least 20% are shown as filled circles. Approximately equivalent numbers of proteins were found to be up-regulated in Nrf2(−/−) mice as were down-regulated. Whilst those that were significantly less abundant in Nrf2 null animals corresponded mainly to proteins involved in phase II and phase III drug disposition, in line with previous oligonucleotide array and immunoblotting experiments, those that were up-regulated were mostly involved with lipid metabolism. Proteins whose function is identified within the Uniprot database (http://www.uniprot.org/) as lipid metabolism or lipid transport are summarized in Table 3.
Table 2.
Nrf2-regulated mouse hepatic proteins identified in iTRAQ analysis 1.
| Relative expression compared to WT 1 |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nrf2(+/+) |
Nrf2(−/−) |
Fold change | ||||||||||||||||||
| SwissProt acc. no. | Name | n | Average no. of peptides | Average coverage (%) | Mouse WT1 | MouseWT2 | Mouse WT3 | Mouse WT4 | Geometric mean | Lower 95% CI | Upper 95% CI | Mouse KO1 | Mouse KO2 | Mouse KO3 | Mouse KO4 | Geometric mean | Lower 95% CI | Upper 95% CI | Nrf2(−/−) Nrf2(+/+) | BH p |
| P02762 | Major urinary protein 6 | 4 | 19.8 | 54.9 | 1.00 | 1.35 | 1.29 | 1.54 | 1.28 | 1.07 | 1.53 | 0.47 | 0.28 | 0.48 | 0.64 | 0.45 | 0.32 | 0.63 | 0.35 | 0.057 |
| P17427 | AP-2 complex subunit alpha2 | 1 | 1.0 | 2.5 | 1.00 | 1.25 | 1.93 | 1.51 | 1.38 | 1.05 | 1.82 | 0.46 | 0.43 | 0.71 | 0.66 | 0.55 | 0.43 | 0.71 | 0.40 | 0.064 |
| P10649 | Glutathione S-transferase Mu 1 | 4 | 13.8 | 39.2 | 1.00 | 1.31 | 1.00 | 1.11 | 1.10 | 0.97 | 1.24 | 0.47 | 0.53 | 0.44 | 0.42 | 0.46 | 0.42 | 0.51 | 0.42 | 0.009 |
| Q61656 | Probable ATP-dependent RNA helicase DDX5 | 1 | 2.0 | 5.4 | 1.00 | 1.31 | 1.19 | 1.35 | 1.20 | 1.05 | 1.38 | 0.37 | 0.85 | 0.51 | 0.79 | 0.59 | 0.40 | 0.87 | 0.49 | 0.148 |
| Q91WG8 | Bifunctional UDP-N-acetylglucosamine 2-epimerase | 1 | 2.0 | 4.0 | 1.00 | 0.98 | 1.13 | 1.19 | 1.07 | 0.98 | 1.17 | 0.49 | 0.60 | 0.68 | 0.59 | 0.59 | 0.52 | 0.67 | 0.55 | 0.022 |
| P19157 | Glutathione S-transferase P 1 | 4 | 43.0 | 76.3 | 1.00 | 1.21 | 0.94 | 1.12 | 1.06 | 0.95 | 1.19 | 0.62 | 0.56 | 0.60 | 0.54 | 0.58 | 0.55 | 0.62 | 0.55 | 0.011 |
| P17717 | UDP-glucuronosyltransferase 2B5 | 4 | 5.8 | 15.5 | 1.00 | 1.16 | 0.99 | 1.08 | 1.05 | 0.98 | 1.13 | 0.59 | 0.57 | 0.56 | 0.61 | 0.58 | 0.56 | 0.61 | 0.55 | 0.004 |
| Q63836 | Selenium-binding protein 2 | 4 | 26.0 | 47.9 | 1.00 | 1.26 | 0.99 | 1.48 | 1.17 | 0.96 | 1.41 | 0.61 | 0.59 | 0.67 | 0.72 | 0.65 | 0.59 | 0.71 | 0.55 | 0.051 |
| Q8VCC2 | Liver carboxylesterase 1 | 3 | 2.3 | 4.6 | 1.00 | 1.34 | 1.06 | 0.94 | 1.08 | 0.93 | 1.25 | 0.62 | 0.60 | 0.58 | 0.70 | 0.62 | 0.58 | 0.68 | 0.58 | 0.042 |
| Q60991 | Cytochrome P450 7B1 | 1 | 2.0 | 7.1 | 1.00 | 1.43 | 1.66 | 1.65 | 1.40 | 1.11 | 1.77 | 0.85 | 0.82 | 0.82 | 0.79 | 0.82 | 0.80 | 0.84 | 0.58 | 0.073 |
| P46425 | Glutathione S-transferase P2 | 1 | 39.0 | 71.0 | 1.00 | 0.70 | 0.76 | 0.61 | 0.75 | 0.61 | 0.93 | 0.47 | 0.45 | 0.43 | 0.43 | 0.44 | 0.43 | 0.46 | 0.59 | 0.063 |
| P24472 | Glutathione S-transferase A4 | 2 | 2.5 | 17.6 | 1.00 | 1.01 | 0.99 | 0.92 | 0.98 | 0.94 | 1.02 | 0.49 | 0.62 | 0.76 | 0.50 | 0.58 | 0.48 | 0.72 | 0.60 | 0.073 |
| O35660 | Glutathione S-transferase M6 | 1 | 7.0 | 24.3 | 1.00 | 0.68 | 0.67 | 0.89 | 0.80 | 0.66 | 0.97 | 0.50 | 0.69 | 0.42 | 0.40 | 0.49 | 0.38 | 0.62 | 0.61 | 0.179 |
| P00186 | Cytochrome P450 1A2 | 3 | 3.0 | 10.9 | 1.00 | 1.14 | 1.26 | 1.21 | 1.15 | 1.04 | 1.27 | 0.59 | 0.61 | 0.91 | 0.86 | 0.73 | 0.58 | 0.91 | 0.63 | 0.186 |
| Q9EQU5 | Protein SET | 1 | 1.0 | 6.2 | 1.00 | 1.22 | 1.34 | 0.99 | 1.13 | 0.97 | 1.31 | 1.05 | 0.63 | 0.57 | 0.71 | 0.72 | 0.56 | 0.94 | 0.64 | 0.199 |
| Q91X77 | Cytochrome P450 2C50 | 3 | 6.0 | 16.5 | 1.00 | 1.30 | 1.29 | 1.33 | 1.22 | 1.07 | 1.40 | 0.67 | 0.67 | 1.03 | 0.87 | 0.80 | 0.65 | 0.98 | 0.65 | 0.162 |
| Q6XVG2 | Cytochrome P450 2C54 | 4 | 3.5 | 8.5 | 1.00 | 1.00 | 0.96 | 1.04 | 1.00 | 0.97 | 1.03 | 0.54 | 0.70 | 0.77 | 0.77 | 0.69 | 0.58 | 0.81 | 0.69 | 0.090 |
| Q91XE8 | Transmembrane protein 205 | 2 | 1.5 | 11.4 | 1.00 | 0.67 | 0.70 | 0.60 | 0.73 | 0.58 | 0.91 | 0.49 | 0.47 | 0.49 | 0.57 | 0.50 | 0.46 | 0.55 | 0.69 | 0.153 |
| P15105 | Glutamine synthetase | 4 | 9.8 | 25.1 | 1.00 | 1.16 | 1.06 | 1.29 | 1.12 | 1.01 | 1.25 | 0.70 | 0.67 | 0.99 | 0.83 | 0.79 | 0.66 | 0.93 | 0.70 | 0.182 |
| O55060 | Thiopurine S-methyltransferase | 2 | 1.0 | 5.4 | 1.00 | 0.85 | 0.99 | 0.75 | 0.89 | 0.78 | 1.02 | 0.49 | 0.71 | 0.71 | 0.70 | 0.65 | 0.54 | 0.77 | 0.72 | 0.194 |
| O35490 | Betaine-homocysteine S-methyltransferase 1 | 4 | 18.3 | 45.2 | 1.00 | 0.81 | 1.11 | 1.12 | 1.00 | 0.87 | 1.16 | 0.76 | 0.67 | 0.78 | 0.80 | 0.75 | 0.70 | 0.81 | 0.75 | 0.148 |
| P24549 | Retinal dehydrogenase 1 | 4 | 13.8 | 31.2 | 1.00 | 1.07 | 1.10 | 1.22 | 1.10 | 1.01 | 1.19 | 0.80 | 0.76 | 0.84 | 0.92 | 0.83 | 0.77 | 0.90 | 0.76 | 0.127 |
| P06801 | NADP-dependent malic enzyme | 3 | 8.0 | 20.5 | 1.00 | 1.32 | 1.16 | 1.22 | 1.17 | 1.04 | 1.31 | 0.75 | 0.93 | 1.06 | 0.84 | 0.89 | 0.77 | 1.02 | 0.76 | 0.201 |
| P62858 | 40 S ribosomal protein S28 | 4 | 1.0 | 17.4 | 1.00 | 1.03 | 1.08 | 1.11 | 1.05 | 1.01 | 1.10 | 0.87 | 0.76 | 0.81 | 0.82 | 0.82 | 0.77 | 0.86 | 0.77 | 0.038 |
| Q91VA0 | Acyl-coenzyme A synthetase ACSM1, mitochondrial | 3 | 6.3 | 20.4 | 1.00 | 0.95 | 1.03 | 0.90 | 0.97 | 0.91 | 1.03 | 0.80 | 0.71 | 0.75 | 0.75 | 0.75 | 0.72 | 0.79 | 0.78 | 0.039 |
| Q9JIF7 | Coatomer subunit beta | 2 | 3.0 | 3.9 | 1.00 | 0.86 | 0.92 | 0.94 | 0.93 | 0.87 | 0.99 | 0.77 | 0.74 | 0.65 | 0.74 | 0.72 | 0.67 | 0.77 | 0.78 | 0.044 |
| O55125 | Protein NipSnap homolog 1 | 3 | 1.0 | 4.0 | 1.00 | 0.76 | 0.80 | 0.93 | 0.87 | 0.77 | 0.98 | 0.62 | 0.68 | 0.68 | 0.73 | 0.68 | 0.64 | 0.72 | 0.78 | 0.201 |
| Q99JI4 | 26 S proteasome non-ATPase regulatory subunit 6 | 2 | 1.0 | 3.9 | 1.00 | 0.76 | 0.75 | 0.74 | 0.81 | 0.70 | 0.93 | 0.65 | 0.67 | 0.60 | 0.61 | 0.63 | 0.60 | 0.67 | 0.78 | 0.182 |
| Q99J99 | 3-mercaptopyruvate sulfurtransferase | 2 | 2.0 | 10.9 | 1.00 | 0.99 | 0.87 | 0.94 | 0.95 | 0.89 | 1.01 | 0.72 | 0.71 | 0.80 | 0.79 | 0.75 | 0.71 | 0.80 | 0.79 | 0.057 |
| Q76MZ3 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha | 2 | 1.0 | 3.4 | 1.00 | 0.76 | 0.94 | 0.83 | 0.88 | 0.78 | 0.99 | 0.65 | 0.73 | 0.60 | 0.82 | 0.70 | 0.61 | 0.80 | 0.80 | 0.204 |
| Q9Z0X1 | Apoptosis-inducing factor 1, mitochondrial | 2 | 1.0 | 2.1 | 1.00 | 0.99 | 0.89 | 0.86 | 0.93 | 0.87 | 1.00 | 0.67 | 0.78 | 0.76 | 0.80 | 0.75 | 0.69 | 0.81 | 0.80 | 0.114 |
| O70475 | UDP-glucose 6-dehydrogenase | 3 | 5.7 | 19.5 | 1.00 | 0.94 | 0.96 | 1.06 | 0.99 | 0.94 | 1.04 | 0.77 | 0.87 | 0.81 | 0.75 | 0.80 | 0.75 | 0.85 | 0.81 | 0.061 |
| Q8R1G2 | Carboxymethylene-butenolidase homolog | 2 | 2.5 | 12.9 | 1.00 | 0.85 | 0.96 | 1.06 | 0.96 | 0.88 | 1.06 | 0.71 | 0.75 | 0.82 | 0.86 | 0.78 | 0.72 | 0.85 | 0.81 | 0.178 |
| Q8VCU1 | Liver carboxylesterase 31-like | 3 | 10.0 | 20.4 | 1.00 | 0.95 | 0.90 | 0.83 | 0.92 | 0.85 | 0.99 | 0.74 | 0.69 | 0.79 | 0.78 | 0.75 | 0.70 | 0.79 | 0.81 | 0.117 |
| Q8VCA8 | Secernin-2 | 1 | 1.0 | 4.0 | 1.00 | 1.09 | 1.05 | 0.92 | 1.01 | 0.94 | 1.09 | 0.87 | 0.76 | 0.93 | 0.78 | 0.83 | 0.76 | 0.91 | 0.82 | 0.156 |
| Q91VS7 | Microsomal glutathione S-transferase 1 | 4 | 5.0 | 30.2 | 1.00 | 0.92 | 0.95 | 0.95 | 0.95 | 0.92 | 0.99 | 0.84 | 0.71 | 0.73 | 0.85 | 0.78 | 0.71 | 0.86 | 0.82 | 0.162 |
| Q9D6Y7 | Peptide methionine sulfoxide reductase | 3 | 3.0 | 16.7 | 1.00 | 1.08 | 1.04 | 1.06 | 1.04 | 1.01 | 1.08 | 0.83 | 0.82 | 0.93 | 0.85 | 0.86 | 0.81 | 0.91 | 0.82 | 0.044 |
| P70441 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | 3 | 1.7 | 5.7 | 1.00 | 0.82 | 0.84 | 0.73 | 0.84 | 0.74 | 0.96 | 0.75 | 0.68 | 0.69 | 0.68 | 0.70 | 0.67 | 0.73 | 0.83 | 0.198 |
| Q8VCW8 | Acyl-CoA synthetase family member 2, mitochondrial | 3 | 6.3 | 18.5 | 1.00 | 0.98 | 1.00 | 0.93 | 0.98 | 0.94 | 1.01 | 0.81 | 0.77 | 0.85 | 0.82 | 0.81 | 0.78 | 0.85 | 0.83 | 0.030 |
| P57776 | Elongation factor 1-delta | 3 | 3.7 | 23.5 | 1.00 | 0.90 | 0.87 | 0.83 | 0.90 | 0.83 | 0.97 | 0.84 | 0.76 | 0.78 | 0.74 | 0.78 | 0.74 | 0.82 | 0.87 | 0.180 |
| P07759 | Serine protease inhibitor A3K | 3 | 9.0 | 24.5 | 1.00 | 1.03 | 1.16 | 1.05 | 1.06 | 1.00 | 1.13 | 0.91 | 0.91 | 0.87 | 0.97 | 0.92 | 0.88 | 0.96 | 0.87 | 0.123 |
| Q91ZJ5 | UTP-glucose-1-phosphate uridylyltransferase | 3 | 3.0 | 8.0 | 1.00 | 0.99 | 1.08 | 1.06 | 1.03 | 0.99 | 1.08 | 0.86 | 0.89 | 0.96 | 0.88 | 0.90 | 0.85 | 0.94 | 0.87 | 0.156 |
| P11352 | Glutathione peroxidase 1 | 4 | 4.5 | 26.1 | 1.00 | 0.96 | 1.07 | 1.12 | 1.04 | 0.97 | 1.11 | 0.90 | 0.88 | 0.95 | 0.94 | 0.92 | 0.89 | 0.95 | 0.89 | 0.193 |
| P60867 | 40 S ribosomal protein S20 | 3 | 2.3 | 16.2 | 1.00 | 0.91 | 1.01 | 0.91 | 0.96 | 0.90 | 1.01 | 0.90 | 0.81 | 0.86 | 0.84 | 0.85 | 0.82 | 0.89 | 0.89 | 0.178 |
| Q9JII6 | Alcohol dehydrogenase [NADP+] | 4 | 5.5 | 25.0 | 1.00 | 0.95 | 0.97 | 0.89 | 0.95 | 0.91 | 1.00 | 0.85 | 0.89 | 0.86 | 0.84 | 0.86 | 0.84 | 0.88 | 0.90 | 0.121 |
| Q9DBJ1 | Phosphoglycerate mutase 1 | 2 | 8.0 | 44.3 | 1.00 | 0.98 | 1.02 | 1.04 | 1.01 | 0.98 | 1.03 | 1.09 | 1.06 | 1.04 | 1.10 | 1.07 | 1.05 | 1.10 | 1.07 | 0.128 |
| Q8BVI4 | Dihydropteridine reductase | 3 | 3.0 | 18.4 | 1.00 | 1.09 | 1.05 | 1.09 | 1.06 | 1.02 | 1.10 | 1.14 | 1.12 | 1.15 | 1.16 | 1.14 | 1.12 | 1.16 | 1.08 | 0.178 |
| Q8BH00 | Aldehyde dehydrogenase family 8 member A1 | 3 | 13.3 | 31.9 | 1.00 | 1.07 | 1.08 | 1.14 | 1.07 | 1.02 | 1.13 | 1.19 | 1.19 | 1.14 | 1.15 | 1.17 | 1.14 | 1.20 | 1.09 | 0.206 |
| Q8BFR5 | Elongation factor Tu, mitochondrial | 3 | 2.7 | 10.4 | 1.00 | 0.93 | 0.97 | 0.91 | 0.95 | 0.91 | 0.99 | 1.05 | 0.99 | 1.07 | 1.05 | 1.04 | 1.00 | 1.08 | 1.09 | 0.144 |
| Q3UQ44 | Ras GTPase-activating-like protein IQGAP2 | 3 | 3.3 | 3.4 | 1.00 | 1.04 | 0.99 | 0.96 | 1.00 | 0.97 | 1.03 | 1.11 | 1.15 | 1.09 | 1.08 | 1.10 | 1.08 | 1.13 | 1.11 | 0.057 |
| P21107 | Tropomyosin alpha-3 chain | 1 | 1.0 | 3.5 | 1.00 | 1.00 | 1.03 | 1.05 | 1.02 | 1.00 | 1.04 | 1.12 | 1.23 | 1.04 | 1.16 | 1.14 | 1.06 | 1.22 | 1.12 | 0.188 |
| Q64374 | Regucalcin | 4 | 13.8 | 42.6 | 1.00 | 1.07 | 1.02 | 1.08 | 1.04 | 1.01 | 1.08 | 1.12 | 1.22 | 1.24 | 1.09 | 1.17 | 1.10 | 1.24 | 1.12 | 0.148 |
| P45952 | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | 3 | 4.7 | 14.2 | 1.00 | 1.06 | 1.11 | 1.08 | 1.06 | 1.02 | 1.11 | 1.20 | 1.20 | 1.24 | 1.13 | 1.19 | 1.15 | 1.23 | 1.12 | 0.095 |
| P62991 | Ubiquitin | 4 | 4.8 | 50.3 | 1.00 | 1.08 | 1.03 | 1.15 | 1.06 | 1.00 | 1.13 | 1.17 | 1.27 | 1.18 | 1.15 | 1.19 | 1.14 | 1.24 | 1.12 | 0.178 |
| Q8CHT0 | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | 3 | 7.0 | 18.0 | 1.00 | 1.05 | 1.09 | 0.97 | 1.03 | 0.98 | 1.08 | 1.17 | 1.20 | 1.23 | 1.05 | 1.16 | 1.09 | 1.24 | 1.13 | 0.193 |
| Q99J08 | SEC14-like protein 2 | 3 | 6.3 | 25.5 | 1.00 | 1.08 | 1.12 | 1.21 | 1.10 | 1.02 | 1.19 | 1.27 | 1.18 | 1.32 | 1.23 | 1.25 | 1.19 | 1.31 | 1.14 | 0.186 |
| Q02053 | Ubiquitin-like modifier-activating enzyme 1 | 4 | 3.5 | 5.6 | 1.00 | 1.05 | 1.00 | 1.04 | 1.02 | 1.00 | 1.05 | 1.10 | 1.19 | 1.24 | 1.14 | 1.16 | 1.11 | 1.22 | 1.14 | 0.073 |
| O88569 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 4 | 4.5 | 12.5 | 1.00 | 0.98 | 1.03 | 1.08 | 1.02 | 0.98 | 1.07 | 1.22 | 1.17 | 1.10 | 1.16 | 1.16 | 1.12 | 1.21 | 1.14 | 0.090 |
| Q9QXD6 | Fructose-1,6-bisphosphatase | 4 | 16.0 | 46.5 | 1.00 | 0.93 | 0.98 | 0.92 | 0.96 | 0.92 | 0.99 | 1.16 | 1.14 | 1.05 | 1.01 | 1.09 | 1.02 | 1.16 | 1.14 | 0.144 |
| Q99JI6 | Ras-related protein Rap-1b | 2 | 1.0 | 6.5 | 1.00 | 1.02 | 0.99 | 1.01 | 1.00 | 1.00 | 1.01 | 1.21 | 1.20 | 1.08 | 1.10 | 1.14 | 1.08 | 1.21 | 1.14 | 0.121 |
| P50580 | Proliferation-associated protein 2G4 | 2 | 2.0 | 6.6 | 1.00 | 1.01 | 1.07 | 0.98 | 1.01 | 0.97 | 1.05 | 1.10 | 1.19 | 1.25 | 1.10 | 1.16 | 1.09 | 1.23 | 1.14 | 0.127 |
| Q9R0Q7 | Prostaglandin E synthase 3 | 2 | 1.5 | 13.1 | 1.00 | 1.05 | 1.11 | 1.10 | 1.06 | 1.01 | 1.11 | 1.17 | 1.14 | 1.21 | 1.36 | 1.21 | 1.12 | 1.31 | 1.14 | 0.199 |
| Q9DCN2 | NADH-cytochrome b5 reductase 3 | 3 | 6.0 | 27.9 | 1.00 | 1.00 | 0.98 | 0.98 | 0.99 | 0.98 | 1.00 | 1.18 | 1.05 | 1.11 | 1.21 | 1.13 | 1.07 | 1.21 | 1.15 | 0.072 |
| Q99LP6 | GrpE protein homolog 1, mitochondrial | 2 | 1.0 | 6.5 | 1.00 | 1.18 | 1.10 | 1.09 | 1.09 | 1.02 | 1.17 | 1.31 | 1.22 | 1.20 | 1.31 | 1.26 | 1.20 | 1.32 | 1.15 | 0.142 |
| Q9JI75 | Ribosyldihydronicotinamide dehydrogenase [quinone] | 2 | 3.0 | 19.3 | 1.00 | 0.88 | 0.98 | 0.91 | 0.94 | 0.88 | 1.00 | 1.16 | 1.12 | 1.02 | 1.05 | 1.08 | 1.02 | 1.15 | 1.15 | 0.144 |
| P00329 | Alcohol dehydrogenase 1 | 4 | 13.0 | 32.8 | 1.00 | 1.04 | 1.10 | 1.05 | 1.05 | 1.01 | 1.09 | 1.26 | 1.20 | 1.18 | 1.20 | 1.21 | 1.18 | 1.24 | 1.16 | 0.039 |
| P06151 | L-lactate dehydrogenase A chain | 4 | 12.5 | 36.1 | 1.00 | 0.92 | 0.96 | 0.90 | 0.94 | 0.90 | 0.99 | 1.22 | 1.18 | 1.00 | 1.06 | 1.11 | 1.01 | 1.22 | 1.18 | 0.178 |
| Q8CHR6 | Dihydropyrimidine dehydrogenase [NADP+] | 2 | 2.0 | 2.6 | 1.00 | 0.96 | 1.00 | 1.01 | 0.99 | 0.97 | 1.01 | 1.33 | 1.13 | 1.16 | 1.14 | 1.18 | 1.10 | 1.28 | 1.20 | 0.072 |
| P00405 | Cytochrome c oxidase subunit 2 | 2 | 2.5 | 15.2 | 1.00 | 1.16 | 0.98 | 0.99 | 1.03 | 0.95 | 1.11 | 1.19 | 1.26 | 1.18 | 1.28 | 1.23 | 1.18 | 1.28 | 1.20 | 0.105 |
| Q9QXE0 | 2-hydroxyacyl-CoA lyase 1 | 3 | 2.7 | 7.5 | 1.00 | 1.02 | 0.89 | 0.83 | 0.93 | 0.84 | 1.02 | 1.10 | 1.17 | 1.13 | 1.07 | 1.12 | 1.08 | 1.16 | 1.20 | 0.117 |
| Q60932 | Voltage-dependent anion-selective channel protein 1 | 2 | 1.5 | 6.6 | 1.00 | 1.01 | 0.95 | 1.05 | 1.00 | 0.96 | 1.04 | 1.24 | 1.22 | 1.22 | 1.16 | 1.21 | 1.18 | 1.25 | 1.21 | 0.022 |
| Q61207 | Sulfated glycoprotein 1 | 3 | 2.0 | 2.8 | 1.00 | 1.08 | 1.17 | 1.24 | 1.12 | 1.02 | 1.23 | 1.35 | 1.50 | 1.32 | 1.30 | 1.37 | 1.28 | 1.46 | 1.22 | 0.121 |
| Q8VC12 | Probable urocanate hydratase | 4 | 8.0 | 14.9 | 1.00 | 1.20 | 1.12 | 1.09 | 1.10 | 1.02 | 1.18 | 1.36 | 1.39 | 1.30 | 1.33 | 1.35 | 1.31 | 1.39 | 1.23 | 0.063 |
| P80316 | T-complex protein 1 subunit epsilon | 1 | 4.0 | 14.4 | 1.00 | 1.22 | 1.02 | 1.03 | 1.06 | 0.97 | 1.16 | 1.37 | 1.30 | 1.23 | 1.34 | 1.31 | 1.25 | 1.37 | 1.23 | 0.103 |
| P50172 | Corticosteroid 11-beta-dehydrogenase isozyme 1 | 4 | 2.8 | 10.2 | 1.00 | 1.19 | 0.98 | 1.09 | 1.06 | 0.98 | 1.16 | 1.31 | 1.20 | 1.41 | 1.32 | 1.31 | 1.23 | 1.39 | 1.23 | 0.103 |
| Q8VCR7 | Abhydrolase domain-containing protein 14B | 3 | 3.7 | 22.1 | 1.00 | 1.17 | 1.01 | 1.24 | 1.10 | 0.99 | 1.22 | 1.36 | 1.36 | 1.31 | 1.41 | 1.36 | 1.32 | 1.40 | 1.24 | 0.123 |
| Q9DD20 | Methyltransferase-like protein 7B | 3 | 3.0 | 15.2 | 1.00 | 0.93 | 0.90 | 1.05 | 0.97 | 0.90 | 1.04 | 1.19 | 1.23 | 1.16 | 1.21 | 1.20 | 1.17 | 1.23 | 1.24 | 0.044 |
| Q61171 | Peroxiredoxin-2 | 3 | 1.7 | 10.4 | 1.00 | 1.11 | 0.94 | 1.21 | 1.06 | 0.95 | 1.18 | 1.26 | 1.29 | 1.38 | 1.35 | 1.32 | 1.27 | 1.37 | 1.25 | 0.142 |
| P24270 | Catalase | 4 | 12.3 | 25.9 | 1.00 | 1.25 | 1.02 | 1.14 | 1.10 | 0.99 | 1.21 | 1.39 | 1.33 | 1.41 | 1.40 | 1.38 | 1.35 | 1.42 | 1.26 | 0.096 |
| P16460 | Argininosuccinate synthase | 4 | 26.8 | 47.6 | 1.00 | 0.82 | 1.02 | 0.89 | 0.93 | 0.84 | 1.03 | 1.26 | 1.31 | 1.03 | 1.11 | 1.17 | 1.05 | 1.31 | 1.26 | 0.162 |
| P31786 | Acyl-CoA-binding protein | 4 | 4.8 | 39.1 | 1.00 | 0.85 | 0.92 | 0.83 | 0.90 | 0.83 | 0.97 | 1.22 | 1.20 | 1.05 | 1.09 | 1.14 | 1.06 | 1.22 | 1.26 | 0.073 |
| Q61425 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | 3 | 3.3 | 12.1 | 1.00 | 1.04 | 0.98 | 1.04 | 1.01 | 0.99 | 1.04 | 1.09 | 1.25 | 1.51 | 1.31 | 1.28 | 1.12 | 1.46 | 1.26 | 0.156 |
| A3KMP2 | Tetratricopeptide repeat protein 38 | 3 | 2.3 | 6.4 | 1.00 | 0.95 | 0.96 | 1.16 | 1.02 | 0.93 | 1.11 | 1.21 | 1.41 | 1.24 | 1.33 | 1.29 | 1.21 | 1.39 | 1.27 | 0.117 |
| Q99PG0 | Arylacetamide deacetylase | 3 | 3.3 | 12.8 | 1.00 | 1.13 | 1.13 | 1.12 | 1.09 | 1.03 | 1.16 | 1.44 | 1.25 | 1.36 | 1.54 | 1.39 | 1.28 | 1.52 | 1.27 | 0.072 |
| P12787 | Cytochrome c oxidase subunit 5A, mitochondrial | 2 | 3.0 | 36.6 | 1.00 | 1.12 | 0.85 | 1.12 | 1.02 | 0.89 | 1.16 | 1.18 | 1.27 | 1.45 | 1.33 | 1.31 | 1.20 | 1.42 | 1.29 | 0.148 |
| P32020 | Non-specific lipid-transfer protein | 4 | 11.8 | 25.1 | 1.00 | 1.34 | 1.09 | 1.22 | 1.15 | 1.02 | 1.31 | 1.53 | 1.41 | 1.45 | 1.54 | 1.48 | 1.42 | 1.55 | 1.29 | 0.117 |
| P55096 | ATP-binding cassette sub-family D member 3 | 3 | 2.3 | 5.8 | 1.00 | 1.29 | 1.08 | 1.29 | 1.16 | 1.02 | 1.32 | 1.46 | 1.37 | 1.53 | 1.64 | 1.50 | 1.39 | 1.61 | 1.29 | 0.142 |
| P05201 | Aspartate aminotransferase cytoplasmic | 3 | 5.7 | 19.4 | 1.00 | 0.83 | 1.02 | 0.90 | 0.94 | 0.85 | 1.03 | 1.34 | 1.39 | 1.04 | 1.12 | 1.22 | 1.06 | 1.39 | 1.30 | 0.178 |
| P19096 | Fatty acid synthase | 4 | 30.3 | 17.7 | 1.00 | 1.10 | 1.03 | 1.15 | 1.07 | 1.00 | 1.13 | 1.35 | 1.40 | 1.44 | 1.36 | 1.39 | 1.35 | 1.43 | 1.30 | 0.022 |
| Q9R0H0 | Peroxisomal acyl-coenzyme A oxidase 1 | 3 | 12.0 | 24.2 | 1.00 | 1.06 | 1.01 | 0.93 | 1.00 | 0.95 | 1.05 | 1.31 | 1.33 | 1.29 | 1.31 | 1.31 | 1.29 | 1.33 | 1.31 | 0.009 |
| P17665 | Cytochrome c oxidase subunit 7C, mitochondrial | 1 | 2.0 | 47.6 | 1.00 | 0.89 | 0.87 | 1.07 | 0.95 | 0.87 | 1.05 | 1.34 | 1.15 | 1.34 | 1.26 | 1.27 | 1.18 | 1.36 | 1.33 | 0.072 |
| Q9QXF8 | Glycine N-methyltransferase | 4 | 18.0 | 47.6 | 1.00 | 1.27 | 1.23 | 1.42 | 1.22 | 1.06 | 1.41 | 1.63 | 1.66 | 1.60 | 1.61 | 1.63 | 1.60 | 1.66 | 1.34 | 0.117 |
| P35492 | Histidine ammonia-lyase | 3 | 6.7 | 13.1 | 1.00 | 1.08 | 1.17 | 1.12 | 1.09 | 1.02 | 1.17 | 1.57 | 1.54 | 1.33 | 1.44 | 1.47 | 1.36 | 1.58 | 1.34 | 0.044 |
| P83940 | Transcription elongation factor B polypeptide 1 | 1 | 1.0 | 8.0 | 1.00 | 1.02 | 0.87 | 1.11 | 1.00 | 0.90 | 1.10 | 1.40 | 1.31 | 1.33 | 1.31 | 1.34 | 1.30 | 1.38 | 1.35 | 0.050 |
| P18242 | Cathepsin D | 2 | 5.0 | 17.7 | 1.00 | 1.20 | 1.00 | 1.42 | 1.14 | 0.97 | 1.35 | 1.49 | 1.86 | 1.47 | 1.48 | 1.57 | 1.40 | 1.75 | 1.37 | 0.178 |
| P25688 | Uricase | 4 | 7.5 | 27.4 | 1.00 | 1.11 | 0.98 | 1.04 | 1.03 | 0.98 | 1.09 | 1.39 | 1.42 | 1.43 | 1.50 | 1.43 | 1.39 | 1.48 | 1.39 | 0.009 |
| Q9QXD1 | Peroxisomal acyl-coenzyme A oxidase 2 | 1 | 2.0 | 3.8 | 1.00 | 1.38 | 1.28 | 1.32 | 1.23 | 1.07 | 1.42 | 1.68 | 1.73 | 1.57 | 2.01 | 1.74 | 1.57 | 1.93 | 1.41 | 0.117 |
| P62984 | 60 S ribosomal protein L40 | 1 | 1.0 | 19.2 | 1.00 | 0.96 | 0.93 | 1.07 | 0.99 | 0.93 | 1.05 | 1.42 | 1.52 | 1.41 | 1.27 | 1.40 | 1.31 | 1.51 | 1.42 | 0.022 |
| Q99P30 | Peroxisomal coenzyme A diphosphatase NUDT7 | 4 | 4.5 | 30.3 | 1.00 | 1.14 | 0.90 | 1.14 | 1.04 | 0.93 | 1.16 | 1.60 | 1.45 | 1.40 | 1.48 | 1.48 | 1.40 | 1.57 | 1.43 | 0.044 |
| Q9DBM2 | Peroxisomal bifunctional enzyme | 4 | 2.3 | 4.7 | 1.00 | 1.35 | 1.10 | 1.20 | 1.16 | 1.02 | 1.31 | 1.52 | 1.76 | 1.60 | 1.73 | 1.65 | 1.54 | 1.76 | 1.43 | 0.057 |
| O35423 | Serine-pyruvate aminotransferase, mitochondrial | 3 | 1.0 | 3.1 | 1.00 | 0.88 | 0.86 | 0.93 | 0.92 | 0.86 | 0.98 | 1.55 | 1.67 | 1.15 | 1.25 | 1.39 | 1.17 | 1.65 | 1.51 | 0.066 |
| Q8VBT2 | L-serine dehydratase | 3 | 4.3 | 22.3 | 1.00 | 0.72 | 0.97 | 0.90 | 0.89 | 0.77 | 1.03 | 1.47 | 1.58 | 1.18 | 1.20 | 1.35 | 1.17 | 1.55 | 1.51 | 0.078 |
| Q8JZR0 | Long-chain-fatty-acid-CoA ligase 5 | 2 | 3.5 | 7.7 | 1.00 | 0.97 | 0.99 | 0.88 | 0.96 | 0.91 | 1.02 | 1.58 | 1.56 | 1.20 | 1.59 | 1.47 | 1.29 | 1.69 | 1.53 | 0.044 |
| Q91V92 | ATP-citrate synthase | 3 | 11.3 | 14.4 | 1.00 | 1.13 | 1.05 | 1.09 | 1.07 | 1.01 | 1.12 | 1.97 | 2.02 | 1.84 | 1.79 | 1.90 | 1.80 | 2.01 | 1.78 | 0.003 |
| P62827 | GTP-binding nuclear protein Ran | 1 | 1.0 | 8.8 | 1.00 | 1.69 | 1.44 | 1.61 | 1.41 | 1.12 | 1.78 | 2.37 | 2.79 | 3.07 | 2.11 | 2.56 | 2.17 | 3.01 | 1.82 | 0.101 |
| P13516 | Acyl-CoA desaturase 1 | 1 | 2.0 | 9.0 | 1.00 | 1.43 | 1.09 | 1.19 | 1.17 | 1.00 | 1.36 | 4.04 | 2.12 | 1.53 | 2.58 | 2.41 | 1.62 | 3.59 | 2.07 | 0.153 |
| Q8VCH0 | 3-ketoacyl-CoA thiolase B, peroxisomal | 3 | 6.7 | 25.2 | 1.00 | 1.89 | 1.43 | 1.44 | 1.41 | 1.09 | 1.82 | 2.92 | 3.61 | 4.04 | 2.98 | 3.35 | 2.88 | 3.91 | 2.39 | 0.044 |
| Q05816 | Fatty acid-binding protein, epidermal | 4 | 1.8 | 17.0 | 1.00 | 1.24 | 1.01 | 0.85 | 1.02 | 0.87 | 1.18 | 3.64 | 3.17 | 2.74 | 2.62 | 3.02 | 2.61 | 3.50 | 2.97 | 0.009 |
Relative expression of hepatic proteins in livers of Nrf2 wild type (Nrf2(+/+)) and null (Nrf2(−/−)) mice determined in iTRAQ analysis 1. All values are expressed relative to a wild type control mouse (WT1). Proteins listed were significantly different in the null mice compared with wild type according to Student's t-test followed by Benjamini-Hochberg (BH) correction for multiple testing at a significance level of p ≤ 0.2. Four replicate iTRAQ analyses were conducted on each sample and the number of runs in which each protein appeared is designated by n in column 3. The values for each mouse thus represent the average of n replicates. The fold change was calculated from the geometric mean values obtained from the 4 individual mice. Variance of the geometric mean for the four animals in each group is expressed as upper and lower 95% confidence intervals (CI). Proteins are listed according to their expression in Nrf2(−/−) mice relative to wild type animals in ascending order of the fold-change value.
Fig. 1.
Volcano plot of the entire set of proteins quantified during iTRAQ analysis 1. Each point represents the difference in expression (fold-change) between Nrf2(+/+) and Nrf2(−/−) mice plotted against the level of statistical significance. Solid lines represent differential expression differences of ± 20% and a significance level of p < 0.05 (Student's t-test). Proteins represented by diamonds were not differentially expressed. Circles represent proteins that gave a raw p value of < 0.05 and Benjamini–Hochberg value of ≤ 0.2.
Table 3.
Differentially up-regulated proteins listed in the UniProt database as involved in lipid synthesis or metabolism in ITRAQ analysis 1.
| SwissProt acc. no. | Name | Subcellular location | Relative expression compared to Nrf2(+/+) mouse 1 |
|||||
|---|---|---|---|---|---|---|---|---|
| Nrf2(+/+) |
Nrf2(−/−) |
Fold change Nrf2(−/−) Nrf2(+/+) | p | |||||
| Geometric mean | 95% CI | Geometric mean | 95% CI | |||||
| Q05816 | Fatty acid-binding protein, epidermal | C | 1.02 | (0.87–1.18) | 3.02 | (2.61–3.50) | 2.97 | 0.009 |
| Q8VCH0 | 3-Ketoacyl-CoA thiolase B, peroxisomal | P | 1.41 | (1.09–1.82) | 3.35 | (2.88–3.91) | 2.39 | 0.044 |
| P13516 | Acyl-CoA desaturase 1 | ER | 1.17 | (1.00–1.36) | 2.41 | (1.62–3.59) | 2.07 | 0.153 |
| Q91V92 | ATP-citrate synthase | C | 1.07 | (1.01–1.12) | 1.90 | (1.80–2.01) | 1.78 | 0.003 |
| Q8JZR0 | Long-chain-fatty-acid-CoA ligase 5 | ER, Mi | 0.96 | (0.91–1.02) | 1.47 | (1.29–1.69) | 1.53 | 0.044 |
| Q9DBM2 | Peroxisomal bifunctional enzyme | P | 1.16 | (1.02–1.31) | 1.65 | (1.54–1.76) | 1.43 | 0.057 |
| Q99P30 | Peroxisomal coenzyme A diphosphatase NUDT7 | P | 1.04 | (0.93–1.16) | 1.48 | (1.40–1.57) | 1.43 | 0.044 |
| Q9QXD1 | Peroxisomal acyl-coenzyme A oxidase 2 | P | 1.23 | (1.07–1.42) | 1.74 | (1.57–1.93) | 1.41 | 0.117 |
| Q9R0H0 | Peroxisomal acyl-coenzyme A oxidase 1 | P | 1.00 | (0.95–1.05) | 1.31 | (1.29–1.33) | 1.31 | 0.009 |
| P19096 | Fatty acid synthase | C | 1.07 | (1.00–1.13) | 1.39 | (1.35–1.43) | 1.30 | 0.022 |
| P32020 | Non-specific lipid-transfer protein | C | 1.15 | (1.02–1.31) | 1.48 | (1.42–1.55) | 1.29 | 0.117 |
| Q61425 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | Mi | 1.01 | (0.99–1.04) | 1.28 | (1.12–1.46) | 1.26 | 0.156 |
| P31786 | Acyl-CoA-binding protein | Mi | 0.90 | (0.83–0.97) | 1.14 | (1.06–1.22) | 1.26 | 0.073 |
| P50172 | Corticosteroid 11-beta-dehydrogenase isozyme 1 | ER | 1.06 | (0.98–1.16) | 1.31 | (1.23–1.39) | 1.23 | 0.103 |
| Q9QXE0 | 2-Hydroxyacyl-CoA lyase 1 | P | 0.93 | (0.84–1.02) | 1.12 | (1.08–1.16) | 1.20 | 0.117 |
3.1.2. iTRAQ analysis 2
For iTRAQ analysis 2, each run represents a single analysis of a different set of wild-type and Nrf2-null liver samples. In this case, 1070 proteins were initially identified with a FDR below 1%, of which 628 were associated with full quantitative datasets (Table 1). There was little variation between the three runs with respect to protein numbers identified, however the final number of unique proteins quantified was slightly lower than in iTRAQ analysis 1. Following Student's t-test analysis with adjustment for multiple testing by Benjamini–Hochberg analysis, thirty eight proteins were found to be differentially regulated between Nrf2(+/+) and Nrf2(−/−) liver samples, as shown in Table 4.
Table 4.
Nrf2-regulated mouse hepatic proteins determined in iTRAQ analysis 2 (test set).
| SwissProt acc. no. | Name | Relative expression compared to Nrf2(+/+) mouse 1 |
Fold change Nrf2(−/−) /Nrf2(+/+) | p | |||||
|---|---|---|---|---|---|---|---|---|---|
| Nrf2(+/+) |
Nrf2(−/−) |
||||||||
| Geometric mean | Lower 95% CI | Upper 95% CI | Geometric mean | Lower 95% CI | Upper 95% CI | ||||
| P10649 | Glutathione S-transferase Mu 1 | 1.00 | 0.91 | 1.10 | 0.44 | 0.40 | 0.47 | 0.44 | 0.001 |
| P17717 | UDP-glucuronosyltransferase 2B5 | 0.99 | 0.93 | 1.06 | 0.55 | 0.52 | 0.57 | 0.55 | 0.001 |
| Q8VCC2 | Liver carboxylesterase 1 | 1.06 | 0.96 | 1.17 | 0.59 | 0.54 | 0.64 | 0.56 | 0.001 |
| Q91X77 | Cytochrome P450 2C50 | 0.97 | 0.87 | 1.08 | 0.56 | 0.46 | 0.69 | 0.58 | 0.001 |
| P19157 | Glutathione S-transferase P 1 | 0.95 | 0.89 | 1.01 | 0.58 | 0.54 | 0.63 | 0.62 | 0.001 |
| Q9D379 | Epoxide hydrolase 1 | 0.97 | 0.90 | 1.05 | 0.63 | 0.59 | 0.68 | 0.65 | 0.001 |
| Q64458 | Cytochrome P450 2C29 | 1.08 | 0.90 | 1.29 | 0.75 | 0.62 | 0.90 | 0.69 | 0.001 |
| P30115 | Glutathione S-transferase A3 | 1.03 | 0.98 | 1.08 | 0.72 | 0.66 | 0.78 | 0.70 | 0.001 |
| P24549 | Retinal dehydrogenase 1 | 0.94 | 0.86 | 1.04 | 0.68 | 0.58 | 0.80 | 0.72 | 0.021 |
| O70475 | UDP-glucose 6-dehydrogenase | 1.09 | 0.97 | 1.23 | 0.79 | 0.63 | 0.99 | 0.73 | 0.183 |
| Q62452 | UDP-glucuronosyltransferase 1-9 | 0.99 | 0.93 | 1.06 | 0.73 | 0.58 | 0.92 | 0.74 | 0.183 |
| Q91VA0 | Acyl-coenzyme A synthetase ACSM1, mitochondrial | 0.97 | 0.91 | 1.04 | 0.79 | 0.74 | 0.84 | 0.81 | 0.001 |
| Q64442 | Sorbitol dehydrogenase | 1.02 | 0.92 | 1.13 | 0.84 | 0.78 | 0.91 | 0.83 | 0.081 |
| P97494 | Glutamate-cysteine ligase catalytic subunit | 1.15 | 1.06 | 1.25 | 0.95 | 0.89 | 1.02 | 0.83 | 0.021 |
| Q8CG76 | Aflatoxin B1 aldehyde reductase member 2 | 1.06 | 1.01 | 1.11 | 0.88 | 0.81 | 0.96 | 0.83 | 0.013 |
| Q9CQX2 | Cytochrome b5 type B | 1.01 | 0.91 | 1.13 | 0.85 | 0.76 | 0.94 | 0.83 | 0.197 |
| Q9JII6 | Alcohol dehydrogenase [NADP+] | 1.01 | 0.98 | 1.04 | 0.86 | 0.80 | 0.92 | 0.85 | 0.003 |
| Q8VCW8 | Acyl-CoA synthetase family member 2, mitochondrial | 1.00 | 0.93 | 1.08 | 0.86 | 0.79 | 0.93 | 0.86 | 0.132 |
| O55022 | Membrane-associated progesterone receptor component 1 | 1.03 | 0.96 | 1.11 | 0.89 | 0.81 | 0.98 | 0.86 | 0.207 |
| P47738 | Aldehyde dehydrogenase, mitochondrial | 1.02 | 0.99 | 1.06 | 0.88 | 0.85 | 0.92 | 0.86 | 0.000 |
| Q8QZS1 | 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial | 1.07 | 1.00 | 1.13 | 0.94 | 0.89 | 1.00 | 0.89 | 0.084 |
| Q9ET01 | Glycogen phosphorylase, liver form | 0.98 | 0.96 | 1.00 | 0.87 | 0.80 | 0.94 | 0.89 | 0.081 |
| O35945 | Aldehyde dehydrogenase, cytosolic 1 | 0.97 | 0.93 | 1.01 | 0.86 | 0.83 | 0.89 | 0.89 | 0.024 |
| Q8VDJ3 | Vigilin | 1.10 | 1.05 | 1.15 | 0.99 | 0.94 | 1.04 | 0.90 | 0.069 |
| Q9EQ20 | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial | 1.00 | 0.98 | 1.03 | 0.92 | 0.87 | 0.98 | 0.92 | 0.140 |
| Q9Z2I8 | Succinyl-CoA ligase [GDP-forming] subunit beta, mitochondrial | 1.02 | 0.99 | 1.06 | 0.96 | 0.92 | 0.99 | 0.93 | 0.121 |
| Q99P30 | Peroxisomal coenzyme A diphosphatase NUDT7 | 0.98 | 0.93 | 1.03 | 1.10 | 1.05 | 1.17 | 1.13 | 0.039 |
| Q9CW42 | MOSC domain-containing protein 1, mitochondrial | 0.95 | 0.90 | 1.00 | 1.09 | 1.04 | 1.15 | 1.15 | 0.095 |
| Q9QXD6 | Fructose-1,6-bisphosphatase 1 | 1.02 | 0.96 | 1.09 | 1.21 | 1.10 | 1.33 | 1.18 | 0.117 |
| P31786 | Acyl-CoA-binding protein | 1.01 | 0.94 | 1.07 | 1.19 | 1.06 | 1.34 | 1.18 | 0.207 |
| P24369 | Peptidyl-prolyl cis-trans isomerase B | 0.95 | 0.89 | 1.01 | 1.12 | 1.05 | 1.20 | 1.18 | 0.017 |
| Q8VDM4 | 26 S proteasome non-ATPase regulatory subunit 2 | 0.90 | 0.80 | 1.02 | 1.08 | 1.00 | 1.17 | 1.20 | 0.183 |
| P06151 | L-lactate dehydrogenase A chain | 0.97 | 0.89 | 1.05 | 1.16 | 1.06 | 1.28 | 1.20 | 0.086 |
| Q61207 | Sulfated glycoprotein 1 | 0.94 | 0.84 | 1.05 | 1.14 | 1.07 | 1.21 | 1.21 | 0.057 |
| P16460 | Argininosuccinate synthase | 1.02 | 0.93 | 1.13 | 1.27 | 1.16 | 1.40 | 1.25 | 0.038 |
| Q3THE2 | Myosin regulatory light chain MRLC2 | 1.13 | 1.03 | 1.24 | 1.46 | 1.25 | 1.70 | 1.29 | 0.117 |
| Q8VBT2 | L-serine dehydratase | 1.02 | 0.91 | 1.15 | 1.37 | 1.13 | 1.67 | 1.34 | 0.183 |
| Q05816 | Fatty acid-binding protein, epidermal | 1.17 | 0.96 | 1.43 | 2.10 | 1.69 | 2.60 | 1.79 | 0.005 |
All values are expressed relative to a wild type control mouse (WT1). Proteins listed were significantly different in the null mice compared with wild type controls (Benjamini–Hochberg; p ≤ 0.2).
3.1.3. Cellular defence and lipid metabolism are the primary biochemical functions regulated by Nrf2
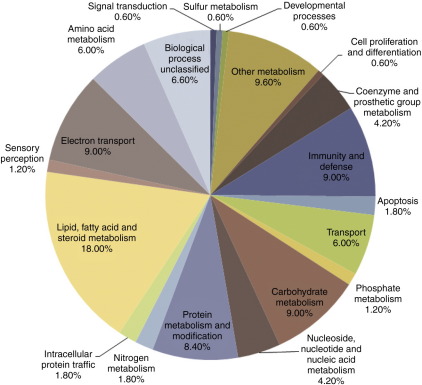
The proteins identified as Nrf2-regulated by the two iTRAQ analyses were independently subjected to correlative network analysis. The 108 proteins obtained from the initial iTRAQ analysis were submitted to the PANTHER database for alignment to specific cellular pathways. Fig. 2 is a pie chart indicating the pathways identified along with the percentage of the proteins corresponding to each fraction. The most prominent class of proteins were those involved in lipid, fatty acid and steroid metabolism (18%). Other functional groupings included metabolism (protein, carbohydrate and amino acid), electron transport, immunity and defence, and transport. In order to mine further into the specific pathways influenced by Nrf2, the 108 proteins identified as Nrf2-regulated from iTRAQ analysis 1 were subjected to pathway analysis using the software MetaCore. Of the 108 proteins, 104 were recognised by MetaCore and 68 had been mapped to pathways. The significant proteins were analysed against a background file containing all proteins quantified across the four replicate runs. Of the 769 proteins in the background file, 752 were recognised by the software and 504 had been mapped to pathways. Ten pathways were identified as significantly different (p < 0.05) between wild type and Nrf2 null mice, as shown in Table 5: seven of these are involved in fatty acid metabolism or other lipid-related processes. In particular, several of the pathways are associated with peroxisomes, suggesting that non-mitochondrial fatty acid metabolism may be a specific target for Nrf2-associated protein expression. Amongst the ten significant pathways identified only one, the glutathione metabolism pathway, is directly involved in cellular defence against reactive oxygen species or electrophiles. Nevertheless, this pathway was populated by five differentially regulated proteins (Table 5).
Fig. 2.
Panther functional classification of proteins shown to be differentially regulated in the Nrf2(−/−)mouse model.
Table 5.
Metacore network analysis of data from iTRAQ analysis 1.
| Pathway name | Negative log p value | Number of pathway objects | |
|---|---|---|---|
| 1 | n-6 Polyunsaturated fatty acid biosynthesis | 2.52 | 5 |
| 2 | n-3 Polyunsaturated fatty acid biosynthesis | 2.52 | 5 |
| 3 | Regulation of lipid metabolism_Regulation of lipid metabolism via LXR, NF-Y and SREBP | 2.44 | 3 |
| 4 | Vitamin E (alfa-tocopherol) metabolism | 1.98 | 5 |
| 5 | Regulation of metabolism_Bile acids regulation of glucose and lipid metabolism via FXR | 1.89 | 4 |
| 6 | Fatty Acid Omega Oxidation | 1.64 | 4 |
| 7 | Peroxysomal straight-chain fatty acid beta-oxidation | 1.64 | 4 |
| 8 | CFTR-dependent regulation of ion channels in Airway Epithelium (norm and CF) | 1.62 | 2 |
| 9 | Cell cycle_Role of SCF complex in cell cycle regulation | 1.62 | 2 |
| 10 | Glutathione metabolism/Rodent version | 1.3 | 5 |
Proteins identified in iTRAQ analysis 1 as being differentially expressed (Benjamini-Hochberg p ≤ 0.2) were interrogated for pathway perturbation using the pathway analysis software Metacore. The total list of all quantified proteins was applied as a background for the analysis.
3.1.4. ATP-citrate lyase is negatively regulated by Nrf2 in mice
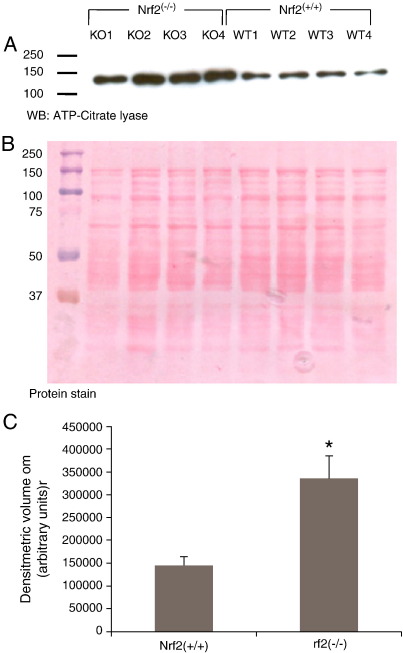
Since a role for Nrf2 as a negative regulator of proteins involved in lipid metabolism has only recently been suggested [48,49], an attempt was made to verify some of the changes observed by immunoblotting. One of the most significant differences observed in the experiments in iTRAQ analysis 1 involved ATP-citrate lyase. This showed a mean 1.75-fold increase but in the “test” cohort a value of 1.2-fold was seen, which failed to reach significance (data not shown). Consequently, a comparison between the wild type and knockout animals was conducted by Western immunoblotting in order to validate the original iTRAQ observation. Due to the difficulty in identifying a suitable ‘housekeeping’ protein (we have found both actin and GAPDH to be unreliable when comparing whole liver homogenates from Nrf2(−/−) and Nrf2(+/+) mice) the total protein (Ponceau) stain was used as a loading control. The immunoblot (Fig. 3) confirmed that ATP-citrate lyase was indeed considerably over-expressed in Nrf2-null mouse liver with densitometric analysis indicating a 2.6-fold enhancement. As far as we are aware, this association between ATP-citrate lyase and Nrf2 has not been shown previously.
Fig. 3.
Western immunoblot of ATP-citrate lyase in liver homogenate from Nrf2(−/−) and Nrf2(+/+) mice. (A) Immunoblot for ATP-citrate lyase in liver homogenate from four Nrf2(−/−) mice (KO1–KO4) and four Nrf2(+/+) mice (WT1–WT4). The molecular mass of ATP-citrate lyase is approximately 120 kDa. (B) Ponceau protein stain of the transfer membrane shown in A) indicating approximately equal loading across the gel. Lane KO1 shows slightly decreased loading which is consistent with the lower level of ATP-citrate lyase in the blot above. (C) Densitometric analysis of immunoblot showing a statistically significant (p < 0.05; Student's t-test) elevation of expression in the Nrf2(−/−) mice compared with the wild type controls.
3.2. Identification of Nrf2-dependent liver proteins: 2-DE studies
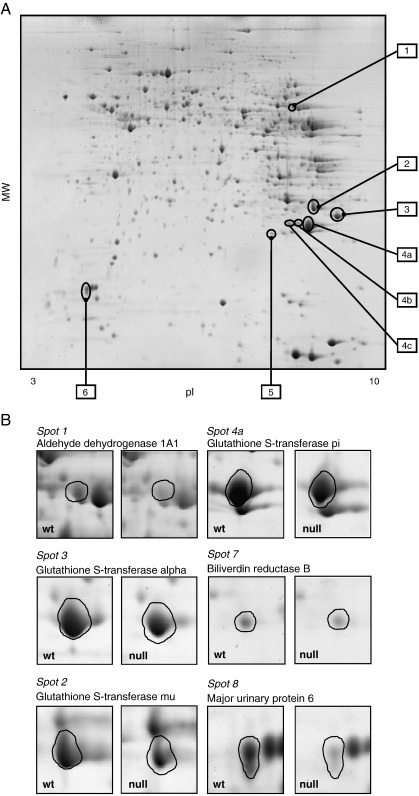
Mouse liver protein extracts were separated by 2-DE and the protein spots were visualized following staining with colloidal Coomassie blue. A total of 8 gels were produced (4 Nrf2(+/+) , 4 Nrf2(−/−)). Approximately 500 spots/gel were detected by the automated spot detection algorithm across the 8 gels. Using the criteria defined above (statistically significant difference within one or more comparisons and spot detected in all four gels for any treatment group) 8 spots were differentially expressed indicating a role for Nrf2 in their regulation. These spots are labelled in the representative gel image (from a wild type control mouse liver) shown in Fig. 4a. Montage images of the differentially expressed spots across the four treatment groups are shown in Fig. 4b. Table 6 lists all the gel spots whose intensity varied in one or more of the treatment groups. Proteins associated with each gel spot were identified by MALDI mass spectrometric analysis. One protein (glutathione S-transferase pi) was identified in three of the differentially regulated spots.
Fig. 4.
2D gel electropherograms of Nrf2 null and wild type mouse liver proteins. (A) Representative 2DE gel of liver homogenate prepared from a wild type mouse annotated with the spot reference numbers of proteins that were found to be regulated by Nrf2. (B) Expanded montages of differentially expressed protein spots from Nrf2(+/+) and Nrf2(−/−) mouse liver homogenates.
Table 6.
Proteins regulated by Nrf2 identified by 2DE analysis.
| Protein spot | SwissProt acc. No. | Protein | Mr/pI | Normalized spot Intensity (% total spot intensity) |
Fold change |
p | |
|---|---|---|---|---|---|---|---|
| Nrf2(+/+) | Nrf2(−/−) | (Nrf2(−/−)/ Nrf2(+/+)) | |||||
| 1 | P24549 | Aldehyde dehydrogenase family 1, subfamily A1 | 54468/7.91 | 0.19 ± 0.01 | 0.15 ± 0.01 | 0.78 | 0.0025 |
| 2 | P10649 | Glutathione S-transferase, mu 1 | 25970/7.7 | 0.88 ± 0.20 | 0.53 ± 0.07 | 0.60 | 0.0158 |
| 3 | P30115 | Glutathione S-transferase, alpha 3 | 25361/8.76 | 0.85 ± 0.09 | 0.67 ± 0.02 | 0.80 | 0.0098 |
| 4a | P19157 | Glutathione S-transferase, pi 1 | 23609/7.69 | 2.06 ± 0.17 | 1.20 ± .50 | 0.58 | 0.0170 |
| 4b | P19157 | Glutathione S-transferase, pi 1 | 23609/7.69 | 0.29 ± 0.03 | 0.017 ± 0.05 | 0.57 | 0.0040 |
| 4c | P19157 | Glutathione S-transferase, pi 1 | 23609/7.69 | 0.21 ± 0.01 | 0.11 ± 0.05 | 0.50 | 0.0073 |
| 5 | Q923D2 | Biliverdin reductase B | 22197/6.49 | 0.13 ± 0.01 | 0.010 ± 0.01 | 0.83 | 0.0155 |
| 6 | P11588 | Major urinary protein 6 | 20648/5.0 | 0.46 ± 0.06 | 0.17 ± 0.05 | 0.36 | 0.0002 |
Proteins from the livers of individual mice were separated by 2-DE as described in the Materials and methods. The protein spots were quantified from colloidal Coomassie blue-stained gels using ImageMasterTM 2D Elite software. Spot intensities were normalized to the total spot intensity for each gel and expressed as the mean percentage value ± SD (n = 4 for each group). Proteins that were significantly different (Student's t-test; p < 0.05) between the wild type and Nrf2 null mice are shown.
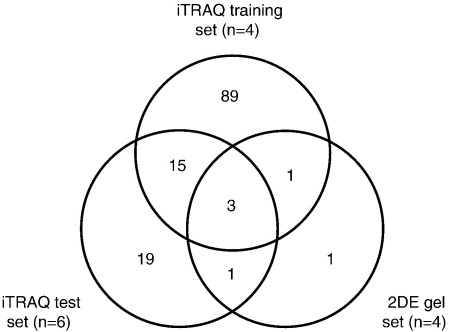
Attempts to relate the iTRAQ data with the 2DE gel protein expression changes were hampered by the small number of proteins identified by the gel-based approach. This may reflect the fact that Nrf2-regulated proteins have properties that are not amenable to 2DE gel analysis, e.g. membrane bound, low abundance or incompatible pKa values. Only two of the identified proteins, glutathione S-transferases Mu1 and Pi1, were shown to be Nrf2-regulated in both the 2DE analysis and the two iTRAQ analyses (Table 7). A summary of the overlap between the three different analyses is provided by Venn diagram in Fig. 5.
Table 7.
Proteins identified as Nrf2 dependent in two or more analyses.
| SwissProt acc. no. | Protein name | iTRAQ Analysis 1 |
iTRAQ Analysis 2 |
2DE gel analysis |
|||
|---|---|---|---|---|---|---|---|
| Fold-change | p | Fold-change | p | Fold-change | p | ||
| Q8VCW8 | Acyl-CoA synthetase family member 2, mitochondrial | 0.83 | 0.030 | 0.86 | 0.132 | ||
| P31786 | Acyl-CoA-binding protein | 1.26 | 0.073 | 1.18 | 0.207 | ||
| Q91VA0 | Acyl-coenzyme A synthetase ACSM1, mitochondrial | 0.78 | 0.039 | 0.81 | 0.001 | ||
| Q9JII6 | Alcohol dehydrogenase [NADP+] | 0.90 | 0.121 | 0.85 | 0.003 | ||
| P24549 | Aldehyde dehydrogenase family 1, subfamily A1 | 0.76 | 0.127 | 0.72 | 0.021 | 0.78 | 0.003 |
| P16460 | Argininosuccinate synthase | 1.26 | 0.162 | 1.25 | 0.038 | ||
| Q91X77 | Cytochrome P450 2C50 | 0.65 | 0.162 | 0.58 | 0.001 | ||
| Q05816 | Fatty acid-binding protein, epidermal | 2.97 | 0.009 | 1.79 | 0.005 | ||
| Q9QXD6 | Fructose-1,6-bisphosphatase 1 | 1.14 | 0.144 | 1.18 | 0.117 | ||
| P30115 | Glutathione S-transferase, alpha 3 | 0.70 | 0.001 | 0.80 | 0.010 | ||
| P10649 | Glutathione S-transferase, mu 1 | 0.42 | 0.009 | 0.44 | 0.001 | 0.60 | 0.016 |
| P19157 | Glutathione S-transferase, pi 1 | 0.55 | 0.011 | 0.62 | 0.001 | 0.55 | 0.009 |
| Q8VCC2 | Liver carboxylesterase 1 | 0.58 | 0.042 | 0.56 | 0.001 | ||
| P06151 | L-lactate dehydrogenase A chain | 1.18 | 0.178 | 1.20 | 0.086 | ||
| Q8VBT2 | L-serine dehydratase | 1.51 | 0.078 | 1.34 | 0.183 | ||
| P02762 | Major urinary protein 6 | 0.35 | 0.057 | 0.36 | 0.001 | ||
| Q99P30 | Peroxisomal coenzyme A diphosphatase NUDT7 | 1.43 | 0.044 | 1.13 | 0.039 | ||
| Q61207 | Sulfated glycoprotein 1 | 1.22 | 0.121 | 1.21 | 0.057 | ||
| O70475 | UDP-glucose 6-dehydrogenase | 0.81 | 0.061 | 0.73 | 0.183 | ||
| P17717 | UDP-glucuronosyltransferase 2B5 | 0.55 | 0.004 | 0.55 | 0.001 | ||
Each protein was significantly (p < 0.05, Student t-test) over- or underexpressed in Nrf2(−/−) mice compared with the wild type controls in at least two out of the three independent. Fold changes are the ratios of the mean expression changes from 4 to 6 mice.
Fig. 5.
Venn diagram indicating the overlap between the proteins identified as Nrf2-regulated across the three different analyses.
3.3. Identification of putative antioxidant response elements (ARE) and ARE-related motifs in the promoters of the Nrf2-regulated genes
Each of the genes encoding proteins identified as Nrf2-regulated was interrogated for ARE or ARE-like enhancer elements in their promoter regions. Using a string-based searching algorithm with the input term RTGABNNNTCA (representing the consensus sequence derived by Nioi et al. [44]), several ARE sequences were identified across the panel of genes shown to be differentially expressed between Nrf2(+/+) and Nrf2(−/−) mice. Table 8 shows the number of consensus sequences identified in the 2000 bp promoter regions of the genes encoding nine representative proteins whose expression showed the greatest differential expression (> 0.4-fold difference) between the two mouse strains. It is apparent that there is little correlation between the number of perfect AREs identified and the fold-change in expression. Indeed, the average number of consensus ARE sequences identified by string-based searching across the entire panel of proteins identified was 1.21 compared with a value of 1.25 for those shown to be Nrf2 regulated. The complete data set for promoter analysis of all proteins significantly altered before correction for multiple testing is given in Table 1 of the on-line supplementary data.
Table 8.
Promoter analysis for the mouse genes encoding Nrf2-regulated proteins.
| String search (dna-pattern) |
Matrix analysis (patser) |
Highest scoring ARE |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SwissProt acc. no. | Protein name | Fold-change | Number of consensus sequences (RTGABNNNGCA) | Number of matching sequences | Highest score | Mean score | SD | Location from to | Sequence | |
| P02762 | Major urinary protein 6 | 0.35 | 0 | 14 | 4.89 | 2.03 | 1.07 | −1935 | −1923 | ttccCTGTCACTAAGCAtgtt |
| P10649 | Glutathione S-transferase Mu 1 | 0.41 | 4 | 15 | 4.40 | 2.42 | 1.09 | −56 | −44 | gtggGCAGGACAAAACAgcgg |
| P19157 | Glutathione S-transferase P 1 | 0.54 | 0 | 13 | 4.02 | 2.11 | 0.98 | −68 | −56 | aacgTGTTGAGTCAGCAtccg |
| Q91WG8 | Bifunctional UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase | 0.55 | 0 | 12 | 5.95 | 2.50 | 1.70 | −387 | −375 | gcagGGGTGGCAAAGCTtaaa |
| P17717 | UDP-glucuronosyltransferase 2B5 | 0.55 | 1 | 13 | 5.59 | 2.40 | 1.23 | −398 | −386 | cagtCCATGACTGAGTTtgaa |
| Q99P30 | Peroxisomal coenzyme A diphosphatase NUDT7 | 1.41 | 1 | 8 | 4.68 | 2.49 | 1.14 | −848 | −836 | caagGCATTACACAGCCcagg |
| Q8JZR0 | Long-chain-fatty-acid-CoA ligase 5 | 1.57 | 1 | 10 | 7.66 | 2.56 | 1.90 | −1234 | −1222 | cttaGAATGACCCAGCCcttg |
| Q91V92 | ATP-citrate synthase | 1.75 | 1 | 9 | 10.02 | 3.26 | 2.58 | −1899 | −1887 | agaaAAATGACTAAGCAggta |
| Q8VCH0 | 3-ketoacyl-CoA thiolase B, peroxisomal | 2.21 | 2 | 15 | 5.84 | 2.55 | 1.44 | −137 | −125 | tgggGGAAGACTCAGGAagag |
| Q05816 | Fatty acid-binding protein, epidermal | 2.81 | 0 | 15 | 4.37 | 2.59 | 0.86 | −1728 | −1716 | agtgGGATGTCGCAGCTcagg |
| Mean values for all Nrf2-regulated proteins | 1.26 | 1.25 | 13.69 | 5.62 | 2.50 | 1.33 | ||||
| Mean values for all down-regulated Nrf2-dependent proteins | 0.57 | 1.00 | 15.40 | 5.20 | 2.54 | 1.23 | ||||
| Mean values for all up-regulated Nrf2-dependent proteins | 1.57 | 1.36 | 12.91 | 5.81 | 2.49 | 1.37 | ||||
| Mean values for all proteins identified | 1.21 | 13.20 | 6.48 | 2.03 | 1.62 | |||||
Sequences of the genes of Nrf2-regulated proteins were obtained from the ENSMBL mouse genome database and interrogated for ARE and ARE/like consensus sequences using the RSAT analysis software (http://rsat.ulb.ac.be/rsat/). Both string-based (dna-pattern) and matrix-based (patser) pattern searching strategies were adopted (see text for details). For the dna-pattern analysis, returned sequences were rated against the ‘perfect’ consensus sequence RTGABNNNGCA. For the patser analysis, the number of sequences matching the position specific scoring matrix with a score > 1 are given, along with the highest score attained. For comparison, equivalent data from the entire set of identified proteins is included at the foot of the table.
For the matrix analysis, the patser algorithm assigns a score for each region within the promoter that matches the position-specific probability matrix. The score is based on the degree of similarity to the most frequently observed sequence within a series of known Nrf2 target genes [45]. To define a reference score, promoters for all the 769 quantified proteins were searched for putative ARE sequences. The mean patser score across the entire panel of genes was 2.50 whilst that for the Nrf2-regulated genes was 2.03.
4. Discussion
Nrf2 deficient mice are highly susceptible to liver damage evoked by a range of chemical hepatotoxins [25,30–34]. The cause of this predisposition may be either reduced constitutive expression of Nrf2-regulated genes or the loss of ability to respond to the noxious stimulus by up-regulation of cellular defence proteins. It is likely that both of these potential mechanisms plays a role in counteracting the damage caused by exposure to chemical toxins, however, as yet the relative importance of each pathway has not been established for individual hepatotoxins. Therapeutically, it is important to understand how each mechanism contributes to the cellular defence process since the Nrf2/Keap1 system provides a potential focus for the development of therapeutic strategies for management of drug or chemical-induced liver pathologies.
In most of the studies conducted with hepatotoxins in Nrf2 null mice, the chemical was administered either acutely or over a short dosing period. For example, loss of protection against liver damage from paracetamol can be observed in Nrf2 null mice following a single hepatotoxic dose. Although we have previously shown that paracetamol can activate hepatic Nrf2, even at doses that do not give rise to overt toxicity [50], it seems unlikely that such transcriptional activation, and the consequent up-regulation of cellular defence proteins, could occur sufficiently rapidly to afford protection against a massive acute chemical insult. Overt liver damage can be seen within 5 h following a toxic dose of paracetamol in mice: a timescale inconsistent with the up-regulation of proteins involved in the defence response [51]. Whilst the induction of Nrf2 may play a role in damage limitation and repair, it seems likely that constitutive differences between the wild type and knockout animals represent the major factor in protecting against the initial hepatotoxic response. Although several transcriptomic studies incorporate a comparison between Nrf2 wild type and null mice, none has specifically addressed the differences at the constitutive level. Consequently, this study represents the first comprehensive global analysis of the role of Nrf2 in the basal regulation of proteins in the liver.
The main aim of the study was to identify protein networks that are perturbed at the constitutive level in Nrf2 null mice compared with wild type controls. In addition, we attempted to produce a list of differentially expressed proteins that could be employed as definitive indices of Nrf2 activity and, thus, provide a pool of potential biomarkers for application in preclinical drug safety assessment. Such potential biomarkers might ultimately provide a translational bridge for assessment of Nrf2 activity in man. By using an experimental approach incorporating three independent sample cohorts we identified twenty proteins that were Nrf2-regulated in at least two of the three independent analyses. Of these, twelve proteins were down-regulated in Nrf2 null mice, seven of which are involved in drug metabolism and, predominantly, phase II metabolism. The reproducible and substantial reduction of proteins such as glutathione S-transferases mu and pi, and the UDP-glucuronosyl transferase 2B5 in Nrf2(−/−) mice clearly indicates that protection against chemical toxins that undergo bioactivation to chemically reactive species, such as electrophiles, may be severely compromised due to lack of Nrf2 under basal conditions. The constitutive deficiency in such protective proteins will almost certainly play a part in the enhanced susceptibility to chemical toxins seen upon acute administration.
Somewhat surprisingly, many proteins that were significantly different between the null and wild-type animals were up-regulated in the absence of Nrf2, suggesting a negative regulation of their expression. Inspection of these proteins indicated that the majority were primarily involved in lipid metabolism. Consequently, an attempt was made to categorise the differentially expressed proteins with respect to biochemical function by pathway analysis. MetaCore. These analyses identified lipid metabolic pathways as being highly overrepresented within the lists of significantly altered proteins when compared against the entire list of proteins identified. Indeed, seven of the ten pathways shown to be significantly perturbed in the Nrf2(−/−) mice related to the regulation of lipid biochemistry — in particular with respect to lipogenesis.
A potential role for Nrf2 in the regulation of lipid biochemistry, and more specifically in the disposition of fatty acids, has only recently been recognised [48,49,52]. This probably reflects the fact that earlier studies with transgenic animals predominantly concentrated on proteins that are directly correlated with Nrf2 activity, which comprises principally proteins involved in cellular antioxidant defence. Three recent studies have, however, noted the reciprocal relationship between Nrf2 function and the expression of multiple lipid-related gene products. Studies by Tanaka et al. [53], involving Nrf2(−/−) mice fed a high fat diet, and Yates et al. [54], which compared Keap1 knockout mice with mice exposed to a potent Nrf2 inducer, utilized transcriptomic approaches to define gene expression profiles. Both studies noted that a high proportion of the up-regulated mRNAs coded for proteins involved in lipid homeostasis. In the former study, 4 weeks on a high fat diet resulted in a marked increase in the mRNAs for several cholesterol synthetic and up-take genes, including LDL receptor, HMGcoA reductase, HMGCoA synthase and SR-B1. Interestingly, the mRNA for Nrf2 itself was substantially reduced following this diet, suggesting that the Keap1/Nrf2 pathway may be directly regulated by certain dietary lipids. This must be balanced against the fact that some terpenoids, which are also lipids and share synthetic pathways with cholesterol, are among the most potent activators of Nrf2 in mouse models [55,56]. For example, the synthetic triterpenoid CDDO-Im has been shown to reduce hepatic lipid accumulation in mice on a high fat diet through activation of Keap1/Nrf2 signalling [49]. Clearly, the role of Nrf2 in lipid homeostasis is complex and requires further clarification. One of the most recent demonstrations that Nrf2 is negatively linked to the expression of lipid-related genes results from a study by Chowdary et al. [48] investigating the influence of Nrf2 on non-alcoholic hepatosteatosis (NASH). This disease is characterised by macro- and/or micro-vesicular vacuolization of hepatocytes and can be induced by administering a methionine/choline deficient diet. The histopathological symptoms of the condition were exacerbated in Nrf2 deficient mice and was accompanied by up-regulation of proteins involved in lipid metabolism including Adrp, a fatty acid- and cholesterol-binding protein that promotes accumulation of triacylglycerols and stimulates the uptake of fatty acids [57].
One of the proteins involved in lipid metabolism shown to be strongly enhanced in Nrf2(−/−) mice in the current study was ATP citrate synthase (also known as ATP-citrate lyase). The almost two-fold increase in this enzyme indicated by the initial iTRAQ analysis (Table 2) was confirmed by Western immunoblotting (Fig. 3). As far as we are aware, regulation of this protein by Nrf2 has not been previously demonstrated. ATP-citrate lyase plays a critical role in acetyl CoA production within the cytoplasm of most cells, and is especially active in liver [58]. In the presence of ATP and Coenzyme A, ATP-citrate lyase is able to cleave citrate to form acetyl-CoA and oxaloacetic acid. The latter is a precursor for pyruvate which sits at the crossroads of multiple biochemical pathways, such as amino acid synthesis, glycogenolysis and lipogenesis. Furthermore, it has recently been shown that ATP-citrate lyase is a key enzyme in the acetylation of histones, and may therefore play a major role in gene transcription [59,60]. The demonstration that loss of Nrf2 results in such a large up-regulation of this already abundant protein may therefore have significant implication for multiple cellular functions, and this is the subject of further investigation in our laboratories.
Of all the statistically significant changes in protein expression between wild type and Nrf2 null mice, numerically the largest fold decrease was seen with major urinary protein 6 (MUP6; 0.35 relative to wild type) whilst the biggest increase was seen with epidermal fatty acid binding protein (FABP5; 2.97). MUP6 is a member of a species and sex specific class of secreted proteins synthesised in the liver but used by male mice for a variety of behavioural purposes, including territorial marking and mate attraction [61]. Major urinary proteins (MUPs) are lipid binding molecules that are specifically tailored to the transport of pheromones: following urinary excretion the pheromones are released only slowly from the MUP to prolong their signalling properties. Curiously, fatty acid binding proteins belong to the same class of proteins as the MUPs — both are lipocalins — and they fulfil a similar function, both being involved in lipid transport. Consequently, the two proteins whose expression was most disparately affected by loss of Nrf2 were of a similar type, again emphasising the complexity of Nrf2 regulation of lipid metabolism. It is possible that up regulation of FABP5 occurred at the expense of MUP6 synthesis, which then showed a marked fall in expression; further work is required to understand the interaction between these proteins and the precise role of Nrf2. Perturbation of the expression of FABPs and MUPs has been shown in other mouse models involving antioxidant proteins. CuZn superoxide dismutase (Sod1) deficient mice showed a marked decrease in MUP11 and MUP8 expression, but in this case FABP1 was also down-regulated [62].
A requisite property of all genes identified thus far to be Nrf2-responsive is that they contain an ARE sequence. Consequently, it was important to seek the presence of such AREs within the promoter regions of the genes encoding the identified proteins, particularly those not previously reported to be Nrf2-dependent. This was accomplished using software available in the public domain that allows both multiple string searching and pattern recognition analysis of 5′-flanking regions. String searching based on the ‘perfect’ ARE sequence of RTGABNNNGA as defined by Rushmore [63–66] indicated that a wide variety of potential AREs was present across the 16 genes significantly altered in Nrf2 null animals. However, only GSTM1 among the most highly Nrf2 regulated genes had more than two perfect ARE sequences within their promoter regions. Overall, there was little difference in the average number of AREs found in the Nrf2-regulated genes (mean = 1.25) compared with the number found right across the pool of identified proteins (1.21). Analysis based on a pattern recognition algorithm (patser) similarly showed little difference between up-or down-regulated genes when compared against the total protein pool. These results suggest that prediction of Nrf2-regulated genes based on regulatory sequence analysis may be an unreliable approach and a molecular-based promoter analysis is required to define the precise site of Nrf2 activation.
In summary, this study has identified a panel of Nrf2-dependent hepatic proteins that is statistically robust and demonstrated both decreased and enhanced expression in the absence of the Nrf2 gene. Twenty proteins were identified as Nrf2-regulated in at least two of the three analyses providing further confidence that these proteins might provide useful candidate biomarkers for future translational studies. The number of proteins that can be interrogated by currently available proteomic technology is far fewer than the number of genes interrogated in the oligoarray studies carried out by various groups [37–39]. This reflects, at least in part, the much lower analyte coverage of proteomic approaches and the fact that enhanced mRNA levels are often not mirrored by an equivalent up-regulation of the corresponding protein [41]. From a global perspective, there was little concordance between the proteins identified here and in all the various microarray studies presented previously. However, there was some overlap with well-established Nrf2-regulated mouse genes, such as glutathione transferases and glucuronyl transferases. Overall, the number of phase II proteins shown in the study to be Nrf2-regulated was surprisingly small and only one pathway (glutathione metabolism) involved in cellular defence was identified as Nrf2 regulated from the MetaCore analysis. iTRAQ suffers from the same limitation as all global proteomic methodologies in that, although it can sample around 1000 proteins simultaneously, high abundance species still dominate. Furthermore, the inherent variability associated with any technique that relies on the quantification of a protein based on the analysis of multiple peptide mass spectra (where the number of peptides identified may vary between analyses) means that fold changes less than 20% are unlikely to yield statistically robust quantitative data. In biological terms, however, a 10–20% decrease in protein expression could have a marked bearing on the toxicological consequence of chemical exposure. However, the strength of such global approaches is that they can reveal hitherto unrecognised roles for key cellular regulators, as evidenced here by the demonstration that lipid homeostasis is a key function of Nrf2. In conclusion, this study has identified a reliable panel of proteins that are reproducibly Nrf2-regulated at the constitutive level. This paves the way for the future translational applications in the pursuit of a human biomarker that would enable the assessment of the role of Nrf2 in man. The identification of two principal sets of seemingly unrelated proteins — one involved in cellular defence, the other in lipid homeostasis — that are reciprocally regulated by Nrf2 was an unexpected finding of this study, and may provide a powerful reservoir of diagnostic biomarkers for development in both animal and human studies of Nrf2 activity.
Acknowledgements
This work was supported by The Wellcome Trust. Azman Abdullah would like to thank the Ministry of Science, Technology and Innovation of the Government of Malaysia as well as the National University of Malaysia for sponsoring his doctoral studies at the University of Liverpool. JW and RS are BBSRC-funded PhD students with further support from AstraZeneca Pharmaceuticals. We are grateful to Brian Lane (Department of Medical Microbiology, School of Infection and Host Defence, University of Liverpool) and to Tracey Farragher (Centre for Medical Statistics and Health Evaluation, University of Liverpool) for the statistical advice and support.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jprot.2010.03.018.
Appendix A. Supplementary data
Promoter analysis for the mouse genes encoding Nrf2-regulated proteins. Sequences of the genes of Nrf2-regulated proteins were obtained from the ENSMBL mouse genome database and interrogated for ARE and ARE/like consensus sequences using the RSAT analysis software (http://rsat.ulb.ac.be/rsat/). Both matrix-based (patser) and string-based (dna-pattern) pattern searching strategies were adopted (see text for details). For the patser analysis, the number of sequences matching the position specific scoring matrix with a score > 1 are given, along with the highest score attained. For the dna-pattern analysis, returned sequences were rated against the ‘perfect’ consensus sequence RTGABNNNGCA. For comparison, equivalent data from the entire set of identified proteins is included at the foot of the table.
References
- 1.Ames B.N. Dietary carcinogens and anticarcinogens. Oxygen radicals and degenerative diseases. Science. 1983;221:1256–1264. doi: 10.1126/science.6351251. [DOI] [PubMed] [Google Scholar]
- 2.Miller J.A. Carcinogenesis by chemicals: an overview–G. H. A. Clowes memorial lecture. Cancer Res. 1970;30:559–576. [PubMed] [Google Scholar]
- 3.Sims P., Grover P.L., Swaisland A., Pal K., Hewer A. Metabolic activation of benzo(a)pyrene proceeds by a diol-epoxide. Nature. 1974;252:326–328. doi: 10.1038/252326a0. [DOI] [PubMed] [Google Scholar]
- 4.Prestera T., Zhang Y., Spencer S.R., Wilczak C.A., Talalay P. The electrophile counterattack response: protection against neoplasia and toxicity. Adv Enzyme Regul. 1993;33:281–296. doi: 10.1016/0065-2571(93)90024-8. [DOI] [PubMed] [Google Scholar]
- 5.Primiano T., Sutter T.R., Kensler T.W. Antioxidant-inducible genes. Adv Pharmacol. 1997;38:293–328. doi: 10.1016/s1054-3589(08)60989-8. [DOI] [PubMed] [Google Scholar]
- 6.Buetler T.M., Gallagher E.P., Wang C., Stahl D.L., Hayes J.D., Eaton D.L. Induction of phase I and phase II drug-metabolizing enzyme mRNA, protein, and activity by BHA, ethoxyquin, and oltipraz. Toxicol Appl Pharmacol. 1995;135:45–57. doi: 10.1006/taap.1995.1207. [DOI] [PubMed] [Google Scholar]
- 7.Hayes J.D., Pulford D.J. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 8.Alam J., Stewart D., Touchard C., Boinapally S., Choi A.M., Cook J.L. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 9.Chanas S.A., Jiang Q., McMahon M., McWalter G.K., McLellan L.I., Elcombe C.R. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem J. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes J.D., Chanas S.A., Henderson C.J., McMahon M., Sun C., Moffat G.J. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem Soc Trans. 2000;28:33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- 11.Ishii T., Itoh K., Takahashi S., Sato H., Yanagawa T., Katoh Y. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 12.Kim YC, Masutani H., Yamaguchi Y., Itoh K., Yamamoto M., Yodoi J. Hemin-induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. J Biol Chem. 2001;276:18399–18406. doi: 10.1074/jbc.M100103200. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.M., Moehlenkamp J.D., Hanson J.M., Johnson J.A. Nrf2-dependent activation of the antioxidant responsive element by tert-butylhydroquinone is independent of oxidative stress in IMR-32 human neuroblastoma cells. Biochem Biophys Res Comm. 2001;280:286–292. doi: 10.1006/bbrc.2000.4106. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen T., Huang H.C., Pickett C.B. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J Biol Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 15.Wild A.C., Moinova H.R., Mulcahy R.T. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J Biol Chem. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 16.Favreau L.V., Pickett C.B. The rat quinone reductase antioxidant response element. Identification of the nucleotide sequence required for basal and inducible activity and detection of antioxidant response element-binding proteins in hepatoma and non-hepatoma cell lines. J Biol Chem. 1995;270:24468–24474. doi: 10.1074/jbc.270.41.24468. [DOI] [PubMed] [Google Scholar]
- 17.Rushmore T.H., King R.G., Paulson K.E., Pickett C.B. Regulation of glutathione S-transferase Ya subunit gene expression: identification of a unique xenobiotic-responsive element controlling inducible expression by planar aromatic compounds. Proc Natl Acad Sci U S A. 1990;87:3826–3830. doi: 10.1073/pnas.87.10.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rushmore T.H., Pickett C.B. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. Characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J Biol Chem. 1990;265:14648–14653. [PubMed] [Google Scholar]
- 19.Mulcahy R.T., Wartman M.A., Bailey H.H., Gipp J.J. Constitutive and beta-naphthoflavone-induced expression of the human gamma-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J Biol Chem. 1997;272:7445–7454. doi: 10.1074/jbc.272.11.7445. [DOI] [PubMed] [Google Scholar]
- 20.Alam J., Wicks C., Stewart D., Gong P., Touchard C., Otterbein S. Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells. Role of p38 kinase and Nrf2 transcription factor. J Biol Chem. 2000;275:27694–27702. doi: 10.1074/jbc.M004729200. [DOI] [PubMed] [Google Scholar]
- 21.Orino K., Lehman L., Tsuji Y., Ayaki H., Torti S.V., Torti F.M. Ferritin and the response to oxidative stress. Biochem J. 2001;357:241–247. doi: 10.1042/0264-6021:3570241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoh K., Itoh K., Enomoto A., Hirayama A., Yamaguchi N., Kobayashi M. Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int. 2001;60:1343–1353. doi: 10.1046/j.1523-1755.2001.00939.x. [DOI] [PubMed] [Google Scholar]
- 23.Chan K., Lu R., Chang J.C., Kan Y.W. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan K., Han X.D., Kan Y.W. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci U S A. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enomoto A., Itoh K., Nagayoshi E., Haruta J., Kimura T., O'Connor T. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 26.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 27.Martin F., van Deursen J.M., Shivdasani R.A., Jackson C.W., Troutman A.G., Ney P.A. Erythroid maturation and globin gene expression in mice with combined deficiency of NF-E2 and nrf-2. Blood. 1998;91:3459–3466. [PubMed] [Google Scholar]
- 28.McMahon M., Itoh K., Yamamoto M., Chanas S.A., Henderson C.J., McLellan L.I. The cap 'n' collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 29.Thimmulappa R.K., Mai K.H., Srisuma S., Kensler T.W., Yamamato M., Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Research. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 30.Chan K.M., Han X.D., Kan Y.W. An important function of Nrf2 in combating oxidative stress: Detoxification of acetaminophen. Proc Natl Acad Sci U S A. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu W.H., Hellerbrand C., Kohler U.A., Bugnon P., Kan Y.W., Werner S. The Nrf2 transcription factor protects from toxin-induced liver injury and fibrosis. Lab Invest. 2008;88:1068–1078. doi: 10.1038/labinvest.2008.75. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y., Gong P., Cederbaum A.I. Pyrazole induced oxidative liver injury independent of CYP2E1/2A5 induction due to Nrf2 deficiency. Toxicology. 2008;252:9–16. doi: 10.1016/j.tox.2008.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamle J., Marhenke S., Borlak J., Von Wasielewski R., Eriksson C.J.P., Geffers R. Nuclear factor-eythroid 2-related factor 2 prevents alcohol-induced fulminant liver injury. Gastroenterology. 2008;134:1159–1168. doi: 10.1053/j.gastro.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Umemura T., Kuroiwa Y., Kitamura Y., Ishii Y., Kanki K., Kodama Y. A crucial role of Nrf2 in in vivo defense against oxidative damage by an environmental pollutant, pentachlorophenol. Toxicol Sci. 2006;90:111–119. doi: 10.1093/toxsci/kfj076. [DOI] [PubMed] [Google Scholar]
- 35.Kwak M.K., Wakabayashi N., Itoh K., Motohashi H., Yamamoto M., Kensler T.W. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 36.Lee J.M., Calkins M.J., Chan K., Kan Y.W., Johnson J.A. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 37.Abbott A. A post-genomic challenge: learning to read patterns of protein synthesis. Nature. 1999;402:715–720. doi: 10.1038/45350. [DOI] [PubMed] [Google Scholar]
- 38.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 39.Gorg A., Obermaier C., Boguth G., Harder A., Scheibe B., Wildgruber R. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000;21:1037–1053. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 40.Courchesne P.L., Patterson S.D. Identification of proteins by matrix-assisted laser desorption/ionization mass spectrometry using peptide and fragment ion masses. Methods Mol Biol. 1999;112:487–511. doi: 10.1385/1-59259-584-7:487. [DOI] [PubMed] [Google Scholar]
- 41.Fountoulakis M., Berndt P., Boelsterli U.A., Crameri F., Winter M., Albertini S. Two-dimensional database of mouse liver proteins: changes in hepatic protein levels following treatment with acetaminophen or its nontoxic regioisomer 3-acetamidophenol. Electrophoresis. 2000;21:2148–2161. doi: 10.1002/1522-2683(20000601)21:11<2148::AID-ELPS2148>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 42.Fountoulakis M., Juranville J.F., Berndt P., Langen H., Suter L. Two-dimensional database of mouse liver proteins. An update. Electrophoresis. 2001;22:1747–1763. doi: 10.1002/1522-2683(200105)22:9<1747::AID-ELPS1747>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 43.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J.D. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H: quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nerland D.E. The antioxidant/electrophile response element motif. Drug Metab Rev. 2007;39:235–248. doi: 10.1080/03602530601125000. [DOI] [PubMed] [Google Scholar]
- 46.R_Development_Core_Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2009. [Google Scholar]
- 47.Katz M.H. Multivariable analysis: a primer for readers of medical research. Ann Intern Med. 2003;138:644–650. doi: 10.7326/0003-4819-138-8-200304150-00012. [DOI] [PubMed] [Google Scholar]
- 48.Chowdhry S., Nazmy MH, Meakin P.J., Dinkova-Kostova A.T., Walsh SV, Tsujita T. Loss of Nrf2 markedly exacerbates non-alcoholic steatohepatitis. Free Radic Biol Med. 2009 doi: 10.1016/j.freeradbiomed.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Shin S., Wakabayashi J., Yates M.S., Wakabayashi N., Dolan P.M., Aja S. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-Imidazolide. Eur J Pharmacol. 2009;620:138–144. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldring C.E., Kitteringham N.R., Elsby R., Randle L.E., Clement Y.N., Williams D.P. Activation of hepatic Nrf2 in vivo by acetaminophen in CD-1 mice. Hepatology. 2004;39:1267–1276. doi: 10.1002/hep.20183. [DOI] [PubMed] [Google Scholar]
- 51.Kitteringham N.R., Powell H., Clement Y.N., Dodd C.C., Tettey J.N.A., Pirmohamed M. Hepatocellular response to chemical stress in CD-1 mice: Induction of early genes and gamma-glutamylcysteine synthetase. Hepatology. 2000;32:321–333. doi: 10.1053/jhep.2000.9602. [DOI] [PubMed] [Google Scholar]
- 52.Sugimoto H., Okada K., Shoda J., Warabi E., Ishige K., Ueda T. Deletion of nuclear factor-E2-related factor-2 leads to rapid onset and progression of nutritional steatohepatitis in mice. Am J Physiol Gastrointest Liver Physiol. 2010;298:G283–G294. doi: 10.1152/ajpgi.00296.2009. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka Y., Aleksunes L.M., Yeager R.L., Gyamfi M.A., Esterly N., Guo G.L. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J Pharmacol Exp Ther. 2008;325:655–664. doi: 10.1124/jpet.107.135822. [DOI] [PubMed] [Google Scholar]
- 54.Yates M.S., Tran Q.T., Dolan P.M., Osburn W.O., Shin S., McCulloch C.C. Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis. 2009;30:1024–1031. doi: 10.1093/carcin/bgp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X.P., Goldring C.E.P., Wang H.Y., Copple I.M., Kitteringham N.R., Park B.K. Extract of Ginkgo biloba induces glutathione-S-transferase subunit-P1 in vitro. Phytomedicine. 2009;16:451–455. doi: 10.1016/j.phymed.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 56.Thimmulappa R.K., Fuchs R.J., Malhotra D., Scollick C., Traore K., Bream J.H. Preclinical evaluation of targeting the Nrf2 pathway by triterpenoids (CDDO-Im and CDDO-Me) for protection from LPS-induced inflammatory response and reactive oxygen species in human peripheral blood mononuclear cells and neutrophils. Antioxid Redox Signal. 2007;9:1963–1970. doi: 10.1089/ars.2007.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin S., Parton R.G. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 58.Elshourbagy N.A., Near J.C., Kmetz P.J., Wells T.N., Groot P.H., Saxty B.A. Cloning and expression of a human ATP-citrate lyase cDNA. Eur J Biochem. 1992;204:491–499. doi: 10.1111/j.1432-1033.1992.tb16659.x. [DOI] [PubMed] [Google Scholar]
- 59.Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., Thompson C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cousins RJ, Aydemir T.B., Lichten L.A. Plenary Lecture 2 Transcription factors, regulatory elements and nutrient-gene communication. Proc Nutr Soc. 2009:1–4. doi: 10.1017/S0029665109991790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beynon R.J., Hurst J.L. Urinary proteins and the modulation of chemical scents in mice and rats. Peptides. 2004;25:1553–1563. doi: 10.1016/j.peptides.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 62.Elchuri S., Naeemuddin M., Sharpe O., Robinson W.H., Huang T.T. Identification of biomarkers associated with the development of hepatocellular carcinoma in CuZn superoxide dismutase deficient mice. Proteomics. 2007;7:2121–2129. doi: 10.1002/pmic.200601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellis E.M., Slattery C.M., Hayes J.D. Characterization of the rat aflatoxin B-1 aldehyde reductase gene, AKR7A1. Structure and chromosomal localization of AKR7A1 as well as identification of antioxidant response elements in the gene promoter. Carcinogenesis. 2003;24:727–737. doi: 10.1093/carcin/bgg016. [DOI] [PubMed] [Google Scholar]
- 64.Motohashi H., O'Connor T., Katsuoka F., Engel J.D., Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 65.Rushmore T.H., Morton M.R., Pickett C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 66.Wasserman W.W., Fahl W.E. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Promoter analysis for the mouse genes encoding Nrf2-regulated proteins. Sequences of the genes of Nrf2-regulated proteins were obtained from the ENSMBL mouse genome database and interrogated for ARE and ARE/like consensus sequences using the RSAT analysis software (http://rsat.ulb.ac.be/rsat/). Both matrix-based (patser) and string-based (dna-pattern) pattern searching strategies were adopted (see text for details). For the patser analysis, the number of sequences matching the position specific scoring matrix with a score > 1 are given, along with the highest score attained. For the dna-pattern analysis, returned sequences were rated against the ‘perfect’ consensus sequence RTGABNNNGCA. For comparison, equivalent data from the entire set of identified proteins is included at the foot of the table.