Abstract
We have sequenced the genomes of 18 isolates of the closely related human pathogenic fungi Coccidioides immitis and Coccidioides posadasii to more clearly elucidate population genomic structure, bringing the total number of sequenced genomes for each species to 10. Our data confirm earlier microsatellite-based findings that these species are genetically differentiated, but our population genomics approach reveals that hybridization and genetic introgression have recently occurred between the two species. The directionality of introgression is primarily from C. posadasii to C. immitis, and we find more than 800 genes exhibiting strong evidence of introgression in one or more sequenced isolates. We performed PCR-based sequencing of one region exhibiting introgression in 40 C. immitis isolates to confirm and better define the extent of gene flow between the species. We find more coding sequence than expected by chance in the introgressed regions, suggesting that natural selection may play a role in the observed genetic exchange. We find notable heterogeneity in repetitive sequence composition among the sequenced genomes and present the first detailed genome-wide profile of a repeat-induced point mutation (RIP) process distinctly different from what has been observed in Neurospora. We identify promiscuous HLA-I and HLA-II epitopes in both proteomes and discuss the possible implications of introgression and population genomic data for public health and vaccine candidate prioritization. This study highlights the importance of population genomic data for detecting subtle but potentially important phenomena such as introgression.
Coccidioides spp. are dimorphic fungal pathogens, existing alternately as saprobes in the soil and as pathogens of living mammals. Coccidioidomycosis in humans typically begins as a pulmonary infection resulting from inhalation of airborne spores (arthroconidia) created by the asexual development of the soil-dwelling mycelial form of the fungus. It is estimated that 60% of Coccidioides infections are asymptomatic, but some patients exhibit influenza-like symptoms that may last many months (Arizona Department of Health Services 2007). Less than 1% of patients develop disseminated disease, which in some cases may require lifelong chemotherapeutic treatment with anti-fungal medication. The historic incidence of symptomatic Coccidioides infections in the United States has been approximately 30,000 cases per year, but recent increases in coccidioidomycosis in southern California and Arizona have been reported (Komatsu et al. 2003; Sunenshine et al. 2007; Kim et al. 2009; Vugla et al. 2009). Coccidioides was classified as a “Select Agent” of bioterrorism in response to the U.S. Antiterrorism and Effective Death Penalty Act of 1996 (Dixon 2001).
Coccidioides fungi are endemic to arid regions of the southwestern United States and northern Mexico, and patchily distributed through Central and South America. Once thought to be a monotypic genus, multilocus sequencing testing (MLST) and other genetic analyses have identified two genetically distinct cryptic species: Coccidioides immitis (San Joaquin Valley of California and Southern California) and Coccidioides posadasii (Arizona, Northern Mexico, Southern California, Texas, and parts of Central and South America) (Koufopanou et al. 1997; Fisher et al. 2001, 2002). This taxonomic splitting encountered resistance but has gained increasing acceptance in part due to concordant genetic evidence from MLST and evidence of differential thermotolerance between the species (BM Barker, C Wendel, JN Galgiani, and MJ Orbach, unpubl.).
A comparative genomics analysis of the first reference genome sequences for C. immitis and C. posadasii and related Ascomycota detected changes in gene family size, gene gain and loss, variation in rate of gene evolution, and nucleotide substitutions that alter protein sequence (Sharpton et al. 2009). These analyses assessed macroevolutionary events, including a shift from plant to animal hosts in the ancestral Coccidioides lineage, that could be detected among a group of closely related species using a single representative genome from each species. For assessment of microevolutionary events occurring on a more recent timescale, analysis of multiple genomes from related species may be more informative. For example, a population genomic analysis of more than 70 domestic and wild isolates of Saccharomyces cerevisiae and Saccharomyces paradoxus demonstrated the power of this approach for understanding functional and geographic variation not evident from the initial analyses of reference genomes for those species (Liti et al. 2009). The falling cost of genome sequencing is quickly making a population genomic analysis approach accessible for a broad array of organisms, including Drosophila (Sackton et al. 2009), Arabidopsis (Clark et al. 2007), and humans (Richard Durbin and David Altshuler, http://www.1000genomes.org/).
Here, by increasing the number of sequenced Coccidioides genomes to 20, we extend our evolutionary analysis within species to provide insights into the population biology, evolution, life cycle, and virulence of Coccidioides. We describe important new findings including the discovery of ample but unequal genetic diversity within both taxa, a novel form of transposon control, and a clear signal of recent introgression between C. immitis and C. posadasii, suggesting that these sister species are not reproductively isolated despite having diverged 5 million years ago (Sharpton et al. 2009). The genome-wide mapping of population-level diversity in both species will assist in the prioritization of vaccine candidates and improve our understanding of gene flow within the genus.
Results
Genome sequencing and assembly
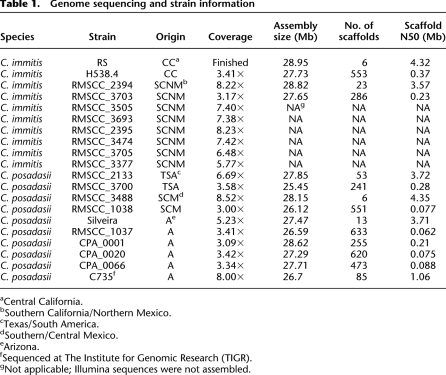
We performed targeted finish sequencing of the previously sequenced C. immitis RS reference genome to close assembly gaps and generate chromosome-scale supercontigs. We sequenced the genomes of nine additional isolates of C. immitis and nine additional isolates of C. posadasii (Table 1). The sequenced isolates represent lab strains and field isolates, and are derived from a broad geographic range. Sequencing of three C. immitis isolates and nine C. posadasii isolates was carried out using AB 3730 capillary sequencers. Sequencing depth for these isolates ranged from 3.4- to 8.52-fold coverage. Following observation of signs of introgression in C. immitis, six additional isolates of that species showing PCR-based sequencing evidence of introgression at the Mep4 locus (CIMG_00508) were sequenced with Illumina technology. One lane of unpaired 36-bp reads was generated for each isolate, yielding 5.8- to 8.2-fold average read coverage per isolate. Sequence assembly was performed for isolates sequenced via capillary sequencing, and final supercontig statistics are listed in Table 1. Genome assembly was not attempted for strains sequenced with Illumina technology; Illumina sequencing reads were used only for single nucleotide polymorphism (SNP) calling. Final assembly size was relatively consistent even for those genomes sequenced at lower depth of coverage, indicating that we have covered most of these genomes to a depth of at least one sequencing read.
Table 1.
Genome sequencing and strain information
aCentral California.
bSouthern California/Northern Mexico.
cTexas/South America.
dSouthern/Central Mexico.
eArizona.
fSequenced at The Institute for Genomic Research (TIGR).
gNot applicable; Illumina sequences were not assembled.
SNP calling
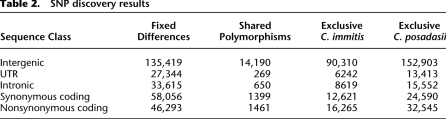
We identified 687,250 SNPs using the C. immitis RS finished assembly as a reference. Of these, 389,250 were intergenic, 58,054 were intronic, 46,791 were found in untranslated regions (UTRs), 96,505 were synonymous coding, and 95,235 were nonsynonymous coding. Table 2 illustrates further details about the distribution of these SNPs. The vast majority of polymorphisms are fixed differences or exclusive polymorphisms that segregate only within one of the two species, supporting earlier MLST results (Fisher et al. 2002) that C. immitis and C. posadasii are generally evolutionarily distinct lineages.
Table 2.
SNP discovery results
Population structure and divergence
We performed a principal components (PC) analysis of the SNP data using SMARTPCA (Patterson et al. 2006). Supplemental Figure 1A illustrates the distribution of each isolate according to eigenvectors 1, 2, and 3. Principal component 1 cleanly separates the 10 C. immitis genomes from the 10 C. posadasii genomes and indicates that the majority of genetic variation in this data set consists of inter-species fixed differences. More detailed PC analyses of population structure within each species are described in Supplemental Text 1 and Supplemental Figure 1, B and C.
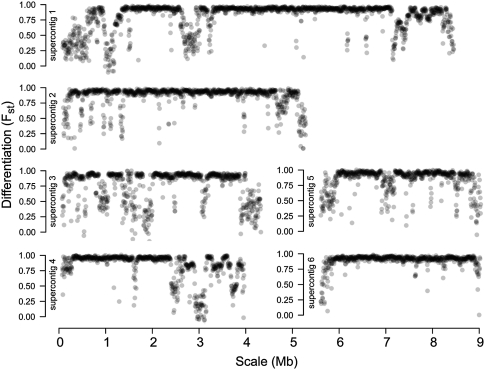
We analyzed interspecific divergence between the two species along each supercontig using Wright's Fst statistic and a sliding-window approach. Figure 1 illustrates substantial variation in divergence between C. immitis and C. posadasii along all six supercontigs of the reference assembly of C. immitis RS. Fst values close to 1 indicate reproductive isolation and a high degree of interpopulation divergence relative to intrapopulation diversity. Fst values close to 0 indicate a lack of interpopulation divergence. Inspection of genomic regions exhibiting low Fst values reveals a high incidence of SNPs that are segregating in both species, and in some cases, the sharing of long (>10 kb) haplotypes between species. The genome-wide distribution of Fst values is bimodal, with the highest peak at 0.95 and a second, smaller peak in Fst values at 0.40.
Figure 1.
Sliding-window analysis of species divergence (Fst). Each circle represents Fst calculated for a nonoverlapping 5-kb window. High Fst values indicate strong local divergence between C. immitis and C. posadasii.
Two scenarios may explain this regional heterogeneity in the Fst profile. First, as these two species are recently descended from a common ancestor, some genomic regions may exhibit incomplete lineage sorting. Alternatively, these two species diverged in the past, and recent genetic exchange through hybridization has occurred. To distinguish these two scenarios, we employed a coalescent approach (Wakeley and Hey 1997). Assuming a null hypothesis of no gene flow between populations, this approach analyzes batches of genetic loci to analytically determine the expected ratios of four classes of SNPs: shared polymorphisms, fixed differences, and polymorphisms exclusive to each of the two populations. A locus that has undergone exchange between populations more recently than the majority of the other loci will yield unexpected proportions of the four classes of mutations (namely, more shared polymorphisms and fewer fixed differences). Applying this analysis, we found that 1237 coding loci exhibit evidence of recent exchange between populations after q-value correction for multiple testing (Storey and Tibshirani 2003; q < 0.05). A full list of genes exhibiting evidence of recent introgression is included in Supplemental Table S1.
We further characterized the pattern of introgression using a sliding-window filter based on genetic distance and consensus haplotypes for each species (Methods). In total, we identified 70 discrete regions exhibiting evidence of introgression (Supplemental Table S2). The boundaries of these regions often vary among isolates, indicating that backcrossing may be occurring and altering the boundaries of the introgressed fragments over time. Of the 827 genes within these regions, 726 exhibited strong coalescent evidence of introgression (q < 0.05). Assuming that the most common haplotype in a particular region is the native one, the predominant direction of gene flow is from C. posadasii to C. immitis. However, there are five regions exhibiting bi-directional gene flow and two regions exhibiting possible gene flow from C. immitis to C. posadasii. All 10 C. immitis isolates exhibit evidence of introgression in multiple regions, with the H538.4 and RMSCC_3473 isolates exhibiting the least and greatest amounts of introgression, respectively. The reduced incidence of introgressed C. posadasii haplotypes in the H538.4 isolate of C. immitis may help to explain why that isolate is an outlier in the PC analysis of population structure (Supplemental Fig. 1B). Introgressed regions are found on all chromosomes, and 20 regions exhibit evidence of introgression in multiple isolates. A variety of functional processes and cellular components are represented in the 827 genes contained within these introgression regions. The two most-enriched Gene Ontology (GO) functional categories are DNA repair (GO 0006281; P = 0.009) and cell wall organization (GO 0007047; P = 0.03). A metalloproteinase (Mep4; CIMG_00508) similar to a gene known to contribute to host immune evasion (Hung et al. 2005) and a 1,3-β-glucanosyltransferase (CIMG_00181) known to be immunoreactive (Delgado et al. 2003) are among the genes found in the introgression regions, suggesting host immune selection as a possible reason for the recent genetic exchange between C. immitis and C. posadasii.
More generally, we find significantly more genes than expected by chance within the introgression regions using a simulation approach (P < 0.001). The genes located within the observed introgression regions cumulatively harbor 4228 nonsynonymous fixed differences between the nominal C. immitis and C. posadasii consensus haplotypes, suggesting the potential for substantial functional divergence in these regions and a possible substrate for natural selection to drive introgression.
Profile of introgression in a deeper population sample
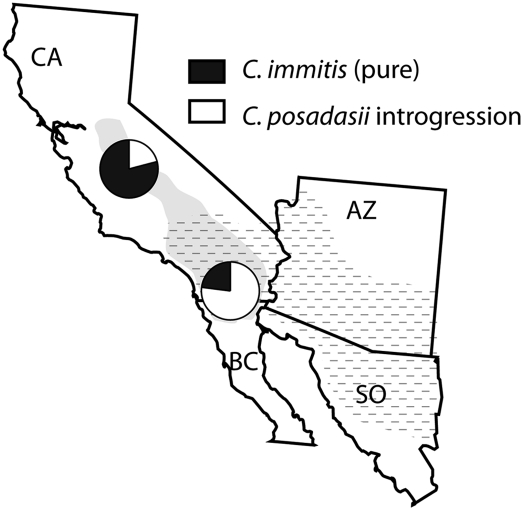
We pursued PCR-based sequence analysis of introgression at the Mep4 locus in a broader collection of C. immitis isolates to better understand the prevalence of introgression. Initial examination of the SNP profile in this region suggested that multiple sequenced C. immitis isolates exhibit signals of introgression, with variable boundaries indicative of backcrossing and recombination (Supplemental Fig. 2). PCR-based sequencing of this region in an additional 39 isolates confirms the pattern observed in the sequencing data (Supplemental Fig. 3). Phylogenetic and haplotypic analyses of multiple PCR amplicons in this region identify population-specific patterns of introgression, with more prevalent introgression into C. immitis occurring in populations more proximal to regions where C. posadasii occurs (Fig. 2; two-tailed P = 0.01, χ2 = 6.368). No C. posadasii strains tested have evidence of C. immitis haplotypes in this region, indicating unidirectional introgression at this locus.
Figure 2.
Geographic profile of introgression from a deeper population sample. The Southern California population of C. immitis, which is more proximal to the range of C. posadasii, exhibits a greater incidence of introgression near the MEP4 locus than the Central California population (χ2 test; P = 0.01). The approximated ranges of C. immitis and C. posadasii are indicated with gray shading and hashing, respectively.
Repetitive element diversity and mutational profile
Repetitive element content varies among the 14 assembled genomes, ranging from 8% to 21% of the genomes (Supplemental Fig. 4). The most predominant repetitive sequences are the gypsy retroelements, which comprise between 43% and 65% of all repetitive sequence and are responsible for most of the variation in repeat content. The distribution of the repetitive elements in each genome is also variable, with a varying mix of large clusters of nested elements and single element insertions into nonrepetitive sequence. We identified a total of 88 recent insertions in C. immitis and 321 in C. posadasii. Only 8%–10% of insertions were found in more than one strain, suggesting that the majority occurred after speciation. We also identified 41 genes interrupted by a recent repetitive element insertion (Supplemental Table S3). Further work will be needed to identify the biological consequence of these insertions.
Neurospora crassa is known to exhibit repeat induced mutation (RIP), which leads to the rapid degradation of repetitive sequences (Galagan and Selker 2004). Although the Coccidioides genomes contain more repetitive sequence than is typical for genomes with active RIP, there is evidence that Coccidioides repeats are subject to a distinct mutational profile, which may help to limit their proliferation. In all of the genome assemblies we find that CpG occurrence is negatively correlated with the copy number of the region (P < 2.2 × 10−16; Spearman's rank correlation), similar to the observation for the first Coccidioides genomes (Sharpton et al. 2009). CpG content is almost 30-fold lower in highly repetitive regions (mean of 1.8 CpGs per kilobase).
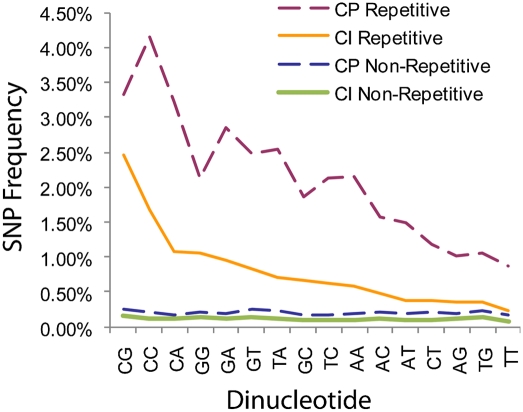
To determine whether repetitive elements are more mutable due to inherent compositional biases, or alternatively if they reflect the outcome of RIP-like mutational processes that alter their mutational profile, we compared mutation frequencies in repetitive and nonrepetitive regions for each dinucleotide. For both species, the normalized mutation rate is higher in repeats for every dinucleotide, although the effect is larger in C. posadasii (Fig. 3). CpG and CpC dinucleotides are the most biased, with a 16-fold higher mutation rate in repeats.
Figure 3.
Dinucleotide mutational profile of repetitive and nonrepetitive sequence, normalized by dinucleotide abundance. The vertical axis represents the fraction of SNP-callable locations that are polymorphic (averaged across all isolates for a species), relative to the appropriate reference genome (RS for C. immitis and RMSCC_3488 for C. posadasii). SNP rates for all dinucleotides are higher in repetitive sequence and also in C. posadasii isolates. In addition, CpG and CpC show the highest mutational rates.
In Neurospora and several other fungi, CpA dinucleotides are the preferred mutational target of RIP, but in Coccidioides we see a pronounced effect on CpG dinucleotides—a context that has also been reported in other fungi (Galagan and Selker 2004). We examined the frequencies of CpA and TpA dinucleotides in repetitive and nonrepetitive sequence. CpA frequencies are only slightly higher in repetitive regions relative to nonrepetitive regions (70.4/kb vs. 66.7/kb, respectively), whereas TpA frequencies are almost doubly higher in repetitive regions (86.2/kb vs. 45.6/kb). These two observations are consistent with an initial C:T transition converting a CpG to a CpA, followed by a second C:T transition converting the CpA to a TpA.
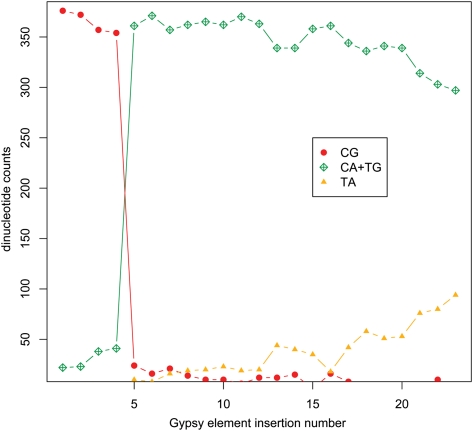
To further examine these mutational processes, we studied a family of gypsy element insertions present in the RS genome in 23 full-length copies. Four of the copies exhibit CpG levels similar to nonrepetitive sequence (49/kb) and appear unmodified. The remaining 19 elements show substantial modifications with 97.5% of all ancestral CpG sites modified. Interestingly, the detailed analysis of this single element family also shows evidence of a rapid initial modification of CpG sites to CpA or TpG and a more gradual modification of the resulting CpA sites to TpA sites (Fig. 4).
Figure 4.
Mutational profile across 23 gypsy element insertions. The C. immitis RS genome contains 23 full-length copies of a particular family of gypsy elements, 19 of which have substantial levels of modification at CpG residues. Analyzing only those locations that contain CpG in any of the 23 elements, this figure illustrates the two kinds of modifications seen. The green and red data points illustrate the almost complete modification of CpG to CpA (or TpG if on the other strand). There are no elements with intermediate levels of CpG content. The yellow data points illustrate the modification of CpA dinucleotides to a TpA occurring at a much more gradual pace, perhaps by a distinct process from that which makes the initial, high-efficiency CpG modifications.
Population genetics and natural selection
We compared the effective populations (Ne) of C. immitis and C. posadasii using Watterson's θ (Watterson 1975), which is based on the number of segregating sites in a population relative to the sequence length. When considering all 20 of the sequenced strains, we observe an Ne of 2.43 × 106 in C. immitis and 4.82 × 106 in C. posadasii, indicating that C. posadasii has a twofold larger effective population than C. immitis. We confirmed the larger effective population size in C. posadasii using Tajima's θ (π for the whole genome). Using this statistic, we observe a 2.06-fold larger effective population size in C. posadasii. We expected to observe a larger Ne in C. posadasii as it covers a larger geographical range and contains more subpopulations (Koufopanou et al. 1997; Fisher et al. 2001, 2002). Sampling from subpopulations confirms that Ne differences between species are not due to differences in the amount of population structure (Supplemental Text 1).
We have examined the profile of natural selection on the Coccidioides proteome using the McDonald-Kreitman test and the neutrality index (NI) (Supplemental Fig. 5, which compares synonymous and nonsynonymous polymorphism and divergence; Rand and Kann 1996). The median −log10 NI value out of the analyzed set of 8872 genes is less than zero (−0.1787), indicating that the majority of genes are subject to at least weak purifying selection and exhibit “excess” of amino acid variation within species relative to amino acid divergence between species. While 3043 genes exhibit a −log10 NI value > 0, indicating a “deficit” of amino acid variation within species relative to between species, we find no genes exhibiting a statistically significant McDonald-Kreitman signal for positive selection after correction for multiple testing. The NI value is only weakly positively associated with genes present in introgression regions (Mann-Whitney U = 62,562, P = 0.29). Genes exhibiting the strongest evidence of positive selection show no statistically significant functional enrichment, and 80% of these genes do not have homology with proteins or domains with known functions.
Epitope prediction and distribution
Given the limited therapeutic options for treatment of coccidioidomycosis, there is great interest in developing a preventative vaccine (Cutler et al. 2007). We used a bioinformatics approach to identify regions of the proteome that may have high affinity to HLA-I or HLA-II molecules (Methods). We identified 27,712 HLA-I epitopes and 30,188 HLA-II epitopes classified as “promiscuous,” or labile to a large fraction of HLA alleles. By ranking genes with extracellularly localized products according to their promiscuous epitope content and polymorphism level, we have generated a list of putatively immunoreactive extracellular proteins with low polymorphism that should be given high priority as novel vaccine candidates (Supplemental Table S4). We further identified two genes with extracellularly localized products (CIMG_11447 and CIMG_05736) containing monomorphic instances of promiscuous epitopes that are polymorphic in one or more other occurrences in the C. immitis proteome. Assuming these epitopes are polymorphic in some instances due to host immune pressure, but monomorphic in the aforementioned genes due to selective constraint, these loci may be excellent vaccine targets. CIMG_05736 has further been identified as a deuterolysin metalloprotease that a previous analysis has identified as rapidly evolving in the Coccidioides lineage (Sharpton et al. 2009).
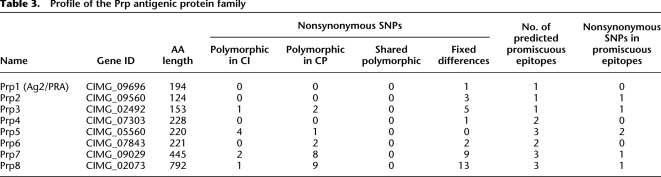
In addition to identifying novel antigenic loci in Coccidioides, the population genomic data also shed light on known antigens, including members of the Prp (proline rich proteins) family (Herr et al. 2007). One member of this family, Ag2/Pra (also known as Prp1), was originally a leading vaccine candidate, but vaccination of C57BL/6 mice with Ag2/Pra does not protect against Coccidioides infection over the long term (Cox and Magee 2004). Herr et al. (2007) identified an additional seven proteins (Prp2–Prp8) exhibiting homology with Ag2/Pra in the first Coccidioides genome to be sequenced (C735). Further analysis of Prp1–Prp8 has shown that Prp1 (Ag2/Pra), Prp2, and Prp7 have a conserved eight-cysteine-containing fungal extracellular (CFEM) domain, which has been identified in Ag2/Pra as a region that contains B-cell epitopes. It is evident that potential for ample functional diversity is exhibited among the members of this protein family. Ag2/Pra and Prp3–Prp6 are predicted to have a GPI anchor, while Prp2 does not. Prp2 does not protect over the short term. However, if Ag2/Pra and Prp2 are combined as a divalent vaccine, mice are well protected (Herr et al. 2007). Work is under way to compare expression levels of all eight of these proteins during different stages of the parasitic cycle. As Table 3 illustrates, the members of this family exhibit varying numbers of predicted promiscuous HLA-I and HLA-II epitopes, as well as varying degrees of nonsynonymous SNP diversity, which will inform future divalent or polyvalent vaccine combinations to test for protective efficacy.
Table 3.
Profile of the Prp antigenic protein family
Discussion
Both C. immitis and C. posadasii harbor substantial genetic variation in their populations. Assuming similar mutation rates in yeast and Coccidioides, our observations indicate a five- to 10-fold larger effective population size for Coccidioides than most populations of S. cerevisiae or S. paradoxus (Liti et al. 2009). Analysis of the first Coccidioides reference genomes illustrated the degree of interspecific divergence that had occurred over the previous 5 million yr (Sharpton et al. 2009); the additional 18 genomes presented here reveal important intraspecific functional variation in the form of SNPs and repetitive element insertions. These data give context to the interspecies divergence data and have allowed us to infer with high confidence that C. immitis and C. posadasii have recently hybridized and exchanged genes. The patchy distribution of genomic regions exhibiting introgression indicates that numerous backcrossing incidents have occurred since the generation of F1 hybrids. The mosaic pattern of introgression is similar to that observed in a recent global population genomic analysis of S. cerevisiae, in which genomic regions exhibiting closest ancestry with different geographic populations of yeast are juxtaposed within strains (Liti et al. 2009). Human activity is theorized to have played a role in the mixing of geographically distinct yeast populations, but it is unclear whether anthropogenic or natural causes are responsible for the recent genetic mingling observed between these two species of Coccidioides.
The predominant direction of gene flow is from C. posadasii into C. immitis, and at least 8% of the genes in the C. immitis population may be recently introgressed from C. posadasii in one or more isolates. The introgressed regions are significantly enriched for coding content, suggesting that natural selection may drive the introgression of some or many of the regions involved. That genes associated with immune evasion and with cell walls are enriched among introgressed genes may indicate that antigenic genes find a selective advantage when introduced into the genome of their sister species. The observation that several genomic regions exhibit C. posadasii haplotypes in multiple isolates of C. immitis is also suggestive of selection, as neutral diffusion of genes across the species boundary would be unlikely to lead to introgressed genes being found in more than one isolate in a sample of only 20 genomes, assuming they are not closely related by descent. PC analysis of the C. immitis samples (Supplemental Fig. 1B) fails to identify a correlation between co-occurrence of introgressed segments and overall relatedness of the isolates. For example, RMSCC_3377 and RMSCC_3505 are the most similar isolates in Supplemental Figure 1B; but out of the 38 genomic regions showing introgression in either isolate, only five regions show introgression in both.
Identification of population structure and gene flow boundaries may have important implications for disease treatment and vaccine development. Although both species have similar disease etiologies, their coding sequences have accumulated thousands of differences, any one of which may cause important phenotypic variation relevant to future vaccine or therapeutic efficacy. For example, protein similarity between species or propensity for introgression between species might be important factors to consider when prioritizing vaccine candidates where protective efficacy against both species is desired.
The majority of genes in these Coccidioides genomes appear to be subject to purifying selection (Supplemental Fig. 5). We fail to find a prevalent signal of balancing selection in these genomes, as might be expected if a large portion of the proteome were under selection for immune evasion as in pathogens like Plasmodium. Rather than evading host immune detection, several studies have identified mechanisms by which Coccidioides fungi may engage in active immunomodulation of their hosts (Heitman et al. 2006). Work from the Cole laboratory and collaborators have shown that the spherule outer wall glycoprotein (SOWgp) is an immunodominant protein exposed at the surface of spherules prior to their endosporulation (Hung et al. 2007). The humoral and cellular immune responses to SOWgp exposure suggest that the host is stimulated to enhance a Th2 immune pathway, which has been shown to be ineffective in protection against Coccidioides infection.
Multiple genome sequences have illuminated the dynamics of repetitive element activity in the history of Coccidioides. Strain-specific insertions can be identified, indicating that element activity is ongoing, and some strains appear to have genes interrupted by their insertion. Therefore, it seems likely that repetitive elements are an ongoing agent for genome variation in spite of evidence for a mutation-based transposon control mechanism.
It has been postulated that mutational processes such as RIP play a role in defending the genome's integrity against the negative consequences of transposable element activity. In Coccidioides, the majority of repetitive regions of the genome have almost complete depletion of CpG content, suggesting that a process similar to RIP has been active in these genomes. However, evidence of plentiful recent transposable element activity in the strains sequenced and the relatively high amount of repetitive sequence in each of the strains suggests that this process may not be currently active, or is only activated rarely. This differs markedly from the canonical RIP profile observed in Neurospora, where repetitive sequences undergo rapid but not complete mutation at CpA dinucleotides, and where RIP is apparently more successful in controlling transposable element proliferation.
Whether or not the two processes are distinct variants is impossible to know without better mechanistic understanding, given that the sexual life cycle of Coccidioides is currently unknown. However, one piece of evidence that supports a distinct process at play is that the strong CpG mutational profile observed in Coccidioides appears to be masking a more modest CpA modification. Figure 4 shows that CpG dinucleotides can be modified to both CpA and TpA dinucleotides but at very different rates. Taken together, these observations suggest there may be two processes occurring at different time scales that rewrite the sequences of transposable elements. One process rapidly changes most of the CpG dinucleotides to CpA dinucleotides. Another process working at a reduced rate may gradually convert these CpAs to TpAs. The result is modification of virtually all CpG dinucleotides, but only a modest change in CpA levels.
The availability of multiple complete genome sequences for a eukaryotic pathogen is a resource that has only recently become attainable by the biological research community. We have begun to demonstrate the utility of such a resource for understanding the population biology and evolutionary history of a pair of pathogenic fungal species, and we believe there is great potential for this approach to prioritize vaccine candidates and inform control strategies in a broad array of pathogens.
Methods
Genome sequencing, assembly, alignment, and SNP calling
All Arachne scaffolds of the previously released C. immitis RS assembly were improved to meet the Bermuda standard using a combination of methods, including searching for assembler-unincorporated data, alternate chemistry resequencing, primer directed walks, transposition of spanning plasmids, and small insert (shatter) libraries constructed from PCR amplicons. Greater detail on these approaches is detailed by the International Human Genome Sequencing Consortium (2004).
Whole-genome shotgun sequencing of nine C. posadasii isolates and four C. immitis isolates was carried out using capillary-based ABI 3730 machines. Genome assembly was carried out using Arachne 2 software (Jaffe et al. 2003).
The genome assemblies of strains sequenced using capillary-based sequencing were aligned to the finished C. immitis RS reference assembly in several stages. We identified regions of pairwise alignment to the RS reference for each subject genome using Patternhunter II (Li et al. 2004). Subject genome sequences corresponding to regions of the RS reference were fetched from each genome assembly, and multiple alignment was performed using PECAN software (Paten et al. 2009).
We performed SNP calling from the multiple alignment using “neighborhood quality standard” (NQS) criteria (Altshuler et al. 2000) to control for base-calling errors and alignment artifacts. Briefly, to call a variant base an SNP, we required both alleles to exhibit a base quality score of at least Q20 (<1% chance of a base call error), and for no other variant bases to be present in the multiple alignment within 5 bp of the position in question. SNPs were called from strains sequenced using Illumina technology by aligning reads to C. immitis RS and calling SNPs using MAQ (Li et al. 2008). The C. posadasii “Silveira” strain was sequenced both by capillary-based and Illumina technologies. Concordance in SNP calls for genomic positions with calls from both data sets was 94%. Capillary-based SNP calls for C. posadasii “Silveira” were used in all analyses. Due to the evolutionary proximity of these isolates, orthology of coding regions for selection analyses was determined via unique alignment of assemblies or reads to C. immitis RS.
Population structure and introgression analysis
PCA analysis was performed with SMARTPCA (Patterson et al. 2006). For the combined species analysis only SNP loci with genotypes in at least nine C. immitis isolates and at least nine C. posadasii isolates (n = 319,452) were employed to avoid bias deriving from sequencing depth. For individual species analyses we also only included SNPs that were polymorphic within the species and that exhibited genotypes from at least nine out of 10 isolates (C. immitis, n = 55,431; C. posadasii, n = 121,139).
We calculated Wright's fixation index (Fst) using the method of Hudson et al. (1992). Sliding-window Fst analyses were conducted using all SNPs found within 5-kb nonoverlapping windows. Coalescent analyses of introgression using the method of Wakeley and Hey (1997) were conducted using the WH software available from the Hey laboratory (Jody Hey, http://genfaculty.rutgers.edu/hey/software). As the software handles a maximum of 20 loci at one time, we conducted 11 replicate analyses for each gene using a random selection of 19 other loci. We computed χ2 values for each replicate based on the observed versus expected numbers of fixed differences relative to other polymorphisms for each locus, and took the median χ2 value from the set of replicates as a measure of the degree to which each locus conformed to the degree of divergence observed in the rest of the genome. We performed false discovery rate correction using the q-value approach (Storey and Tibshirani 2003). We also identified regions of recent introgression used a sliding-window filter based on genetic distances between the sequences of each isolate and the consensus haplotypes for each species (Supplemental Text 1).
Fine-scale introgression analysis in the Mep4 region
DNA was prepped from liquid cultures grown overnight or by scraping mycelium from a young colony and extracted as described previously (Kellner et al. 2005). Primers were developed in the primer design module of MacVector, and primer sequences are listed in Supplemental Text S1. PCR was performed using 2× Promega Hotmaster mix following standard protocol. Alignments of sequenced PCR products were visually inspected using MEGA 4.0 (Tamura et al. 2007). Species affiliation of haplotypes was inferred using the neighbor-joining method (Saitou and Nei 1987).
Transposable element analysis
LTRharvest (Ellinghaus et al. 2008) and RepeatScout (Price et al. 2005) were both used for initial transposable element identification and the results were compared. Based on these results, LTRharvest was used with a similarity setting of 70 to produce a repeat element library from each Coccidioides genome. Individual libraries were then combined to form a master repeat database that was characterized by comparing to the Repbase fungal repeat database (Jurka et al. 2005), and was used with RepeatMasker (http://www.repeatmasker.org/) to delineate repeats in each genome. Repeat density and clustering were analyzed using custom Perl scripts measuring the number of repetitive 1-kb slices in each 100-kb sliding window across each genome. Recent transposable element insertions were identified and characterized with custom Perl scripts using within-species pairwise alignments of each genome against either RS or RMSCC_3488 made using MUMmer (Delcher et al. 2002).
Mutational bias analysis
Initial characterization of CpG content was measured by counting CpG content in 1-kb windows. Repetitive sequence was defined as a 1-kb window that has matches longer than 500 bp to multiple locations in the genome. Mutational bias analysis used custom Perl scripts with the combined SNP data set described above to compare repetitive and nonrepetitive locations.
Genetic diversity, natural selection analyses
Effective population size (Ne) was determined from the parameter θ = 2Neμ, based on two separate estimators of θ, Watterson's and Tajima's θ and an estimated mutation rate (μ) of 1 × 10−9. All coding SNPs were divided into four categories by comparing the alleles between the C. immitis and C. posadasii strains: nonsynonymous fixed (FN), nonsynonymous polymorphic (PN), synonymous fixed (FS), and synonymous polymorphic (PS). We used these counts to conduct the McDonald-Kreitman test of selection for all genes with at least five coding region SNPs. P-values for each gene were calculated using Fisher's exact test and corrected using q-values (Storey and Tibshirani 2003). The neutrality index (NI) was calculated as (PN/FN)/(PS/FS). One pseudocount was added to each mutation class for each gene to eliminate counts of 0 and enable NI calculation for all genes, making the test more conservative by reducing the power to reject neutrality. Gene Ontology (GO) functional enrichment analysis was performed on the tails of the NI distribution. The GO term associations were determined through orthology with S. cerevisiae, Schizosaccharomyces pombe, and N. crassa; and overrepresented terms in the gene sets were determined using the hypergeometric distribution.
Epitope analysis
We predicted epitopes using position-specific scoring matrices (PSSM) available on the BIMAS website (http://www-bimas.cit.nih.gov/). Using a sliding-window approach, we scored each 9-mer peptide in the C. immitis RS proteome for every HLA allele with an available PSSM (HLA-I, n = 27; HLA-II, n = 50). We considered a 9-mer peptide to be a promiscuous HLA-I epitope if its PSSM score was in at least the 80th percentile of all scores across the proteome for 80% of the HLA-I alleles. Similarly, we considered a 9-mer peptide to be a promiscuous HLA-II epitope if its PSSM score was in at least the 95th percentile of all scores across the proteome for 80% of the tested HLA-II alleles. Thresholds levels for qualification as promiscuous were chosen to achieve a roughly comparable count of promiscuous epitopes for HLA-I and HLA-II and to register ∼5% of the amino acid positions in the genome as promiscuous. For each protein, we calculated the promiscuous epitope density for HLA-I and HLA-II as the quotient of the number of amino acid residues predicted to be contained in promiscuous epitopes and the total number of amino acids in the protein.
Acknowledgments
Genome sequencing at the Broad Institute was supported by NIAID through the Broad Institute's Microbial Sequencing Center (contract no. HHSN266200400001C). Illumina sequencing was funded by support from the BIO5 Institute to S.D.R. Analysis was supported in part by NIH/NIAID R01AI70891 (J.W.T. and G.T.C.) and U54-AI65359 (J.W.T). B.M.B. was supported by an NSF grant to the University of Arizona for the BioME program. J.E.S. was supported by a postdoctoral fellowship from the Miller Institute for Basic Research in Science at the University of California, Berkeley. T.J.S. was supported by the Chang-lin Tien graduate fellowship at the University of California, Berkeley.
Footnotes
[Supplemental material is available online at http://www.genome.org. Genome assemblies from this study have been submitted to NCBI (http://www.ncbi.nlm.nih.gov/) under Genome Project IDs 10735 and 12821. SNPs have been submitted to dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) under submitter handle BROAD_GENOMEBIO and submission batch CI_0001 (ss numbers are available in the Supplemental material). Sequencing data have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession nos. SRX012717–SRX012723.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.103911.109.
References
- Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, Lander ES 2000. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407: 513–516 [DOI] [PubMed] [Google Scholar]
- Arizona Department of Health Services 2007. Valley Fever Annual Report Arizona Department of Health Services, Phoenix [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342 [DOI] [PubMed] [Google Scholar]
- Cox RA, Magee DM 2004. Coccidioidomycosis: Host response and vaccine development. Clin Microbiol Rev 17: 804–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler JE, Deepe GS Jr, Klein BS 2007. Advances in combating fungal diseases: Vaccines on the threshold. Nat Rev Microbiol 5: 13–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Phillippy A, Carlton J, Salzberg SL 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 30: 2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado N, Xue J, Yu JJ, Hung CY, Cole GT 2003. A recombinant β-1,3-glucanosyltransferase homolog of Coccidioides posadasii protects mice against coccidioidomycosis. Infect Immun 71: 3010–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DM 2001. Coccidioides immitis as a select agent of bioterrorism. J Appl Microbiol 91: 602–605 [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9: 18 doi: 10.1186/1471-2105-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Koenig GL, White TJ, San-Blas G, Negroni R, Alvarez IG, Wanke B, Taylor JW 2001. Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proc Natl Acad Sci 98: 4558–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Rannala B, Chaturvedi V, Taylor JW 2002. Disease surveillance in recombining pathogens: Multilocus genotypes identify sources of human Coccidioides infections. Proc Natl Acad Sci 99: 9067–9071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Selker EU 2004. RIP: The evolutionary cost of genome defense. Trends Genet 20: 417–423 [DOI] [PubMed] [Google Scholar]
- Heitman J, Edward JE, Filler SG, Mitchell AP 2006. Molecular principles of fungal pathogenesis ASM Press, Washington, DC [Google Scholar]
- Herr RA, Hung CY, Cole GT 2007. Evaluation of two homologous proline-rich proteins of Coccidioides posadasii as candidate vaccines against coccidioidomycosis. Infect Immun 75: 5777–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Seshan KR, Yu JJ, Schaller R, Xue J, Basrur V, Gardner MJ, Cole GT 2005. A metalloproteinase of Coccidioides posadasii contributes to evasion of host detection. Infect Immun 73: 6689–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CY, Xue J, Cole GT 2007. Virulence mechanisms of coccidioides. Ann NY Acad Sci 1111: 225–235 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- Jaffe DB, Butler J, Gnerre S, Mauceli E, Lindblad-Toh K, Mesirov JP, Zody MC, Lander ES 2003. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res 13: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467 [DOI] [PubMed] [Google Scholar]
- Kellner EM, Orsborn KI, Siegel EM, Mandel MA, Orbach MJ, Galgiani JN 2005. Coccidioides posadasii contains a single 1,3-β-glucan synthase gene that appears to be essential for growth. Eukaryot Cell 4: 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MM, Blair JE, Carey EJ, Wu Q, Smilack JD 2009. Coccidioidal pneumonia, Phoenix, Arizona, USA, 2000-2004. Emerg Infect Dis 15: 397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Vaz V, McRill C, Colman T, Comrie A, Sigel K, Clark T, Phelan M, Hajjeh R, Park B 2003. Increase in coccidioidomycosis—Arizona, 1998-2001 (reprinted from MMWR 52: 109–112). JAMA 289: 1500–150212672753 [Google Scholar]
- Koufopanou V, Burt A, Taylor JW 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci 94: 5478–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Ma B, Kisman D, Tromp J 2004. Patternhunter II: Highly sensitive and fast homology search. J Bioinform Comput Biol 2: 417–439 [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B, Herrero J, Beal K, Birney E 2009. Sequence progressive alignment, a framework for practical large-scale probabilistic consistency alignment. Bioinformatics 25: 295–301 [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D 2006. Population structure and eigenanalysis. PLoS Genet 2: e190 doi: 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA 2005. De novo identification of repeat families in large genomes. Bioinformatics 21: i351–i358 [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM 1996. Excess amino acid polymorphism in mitochondrial DNA: Contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol 13: 735–748 [DOI] [PubMed] [Google Scholar]
- Sackton TB, Kulathinal RJ, Bergman CM, Quinlan AR, Dopman EB, Carneiro M, Marth GT, Hartl DL, Clark AG 2009. Population genomic inferences from sparse high-throughput sequencing of two populations of Drosophila melanogaster. Genome Biol Evol 2009: 449–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, Maiti R, Kodira CD, Neafsey DE, Zeng Q, et al. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res 19: 1722–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunenshine RH, Anderson S, Erhart L, Vossbrink A, Kelly PC, Engelthaler D, Komatsu K 2007. Public health surveillance for Coccidioidomycosis in Arizona. Ann NY Acad Sci 1111: 96–102 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Vugla DJ, Wheeler C, Cummings KC, Karon A 2009. Increase in Coccidioidomycosis—California, 2000-2007 (reprinted from MMWR 58: 105–109). JAMA 301: 1760–1762 [Google Scholar]
- Wakeley J, Hey J 1997. Estimating ancestral population parameters. Genetics 145: 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA 1975. On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7: 256–276 [DOI] [PubMed] [Google Scholar]