Abstract
The flavin-dependent Quiescin-sulfhydryl oxidase (QSOX) inserts disulfide bridges into unfolded reduced proteins with the reduction of molecular oxygen to form hydrogen peroxide. This work investigates how QSOX and protein disulfide isomerase (PDI) cooperate in vitro to generate native pairings in two unfolded reduced proteins: ribonuclease A (RNase: 4 disulfide bonds and 105 disulfide isomers of the fully oxidized protein) and avian riboflavin binding protein (RfBP: 9 disulfide bonds and greater than 34 million corresponding disulfide pairings). Experiments combining avian or human QSOX with up to 200 μM of avian or human reduced PDI show that the isomerase is not a significant substrate of QSOX. Both reduced RNase and RfBP can be efficiently refolded in an aerobic solution containing micromolar concentrations of reduced PDI and nanomolar levels of QSOX without any added oxidized PDI or glutathione redox buffer. Refolding of RfBP is followed continuously using the complete quenching of the fluorescence of free riboflavin that occurs on binding to apo-RfBP. The rate of refolding is half-maximal at 30 μM reduced PDI when 1 μM of the reduced client protein is used in the presence of 30 nM QSOX. The use of high concentrations of PDI, in considerable excess over the folding protein client, reflect the concentration prevailing in the lumen of the endoplasmic reticulum and allow the redox poise of these in vitro experiments to be set with oxidized and reduced PDI. In the absence of either QSOX or redox buffer, the fastest refolding of RfBP is accomplished with excess reduced PDI and just enough oxidized PDI to generate 9- disulfides in the protein client. These in vitro experiments are discussed in terms of current models for oxidative folding in the endoplasmic reticulum.
Two distinct flavin-linked sulfhydryl oxidase families (Ero1 (1, 2) and Quiescin-sulfhydryl oxidase; QSOX1 (3, 4)), have been suggested to drive the generation of disulfide bonds during oxidative protein folding in higher eukaryotes (4–12). For Ero1, a series of pioneering investigations on yeast (13, 14), and complementary studies on mammalian cells (15, 16), have led to one model for oxidative folding shown in Figure 1A. Here, the immediate oxidant for an unfolded reduced protein client is oxidized protein disulfide isomerase (PDI). Reduced PDI is then reoxidized by the FAD-linked sulfhydryl oxidase Ero1 (13, 17, 18) with the eventual reduction of dioxygen to hydrogen peroxide (19). PDI intervenes again to catalyze the isomerization of incorrectly paired disulfide bonds (Figure 1A) either by redox neutral shuffling of disulfides or by net cycles of reduction and reoxidation ((20–25); see later).
FIGURE 1.
Oxidative folding models for Ero1 and QSOX. Panel A shows the PDI-first model of oxidative folding driven by Ero1p. QSOX oxidizes reduced unfolded proteins directly in panel B.
The flavin-linked QSOX enzymes, in particular their shorter isoforms, may also contribute to disulfide bond generation as proteins transit the secretory apparatus of higher eukaryotes (4, 9, 12, 26, 27). Unlike Ero1, QSOX enzymes can catalyze the direct oxidation of a wide range of unfolded proteins at rates between 500 and 2000 disulfides introduced per minute (9, 28–30). Unfolded reduced proteins input their reducing equivalents into a PDI-like CGHC motif in the N-terminal thioredoxin domain of QSOX, followed by a transfer of electrons to the Erv/ALR domain and eventual reduction of oxygen to hydrogen peroxide (30–32). The facility of these multidomain sulfhydryl oxidases at oxidizing unfolded proteins in vitro suggests that oxidative folding may also be driven by the direct oxidation pathway shown in Figure 1B.
In our initial attempts to recapitulate oxidative folding in vitro, Hoober et al. showed that avian QSOX1 can cooperate with protein disulfide isomerase to rapidly recover the native activity of reduced RNase (9). The present work explores the nature of this cooperation and shows that it does not involve a direct interaction between PDI and QSOX. While pancreatic RNase, with 4 disulfides and 105 fully oxidized disulfide isomers, has proved a particularly tractable and widely-used model protein for oxidative folding studies (33–36), we also wished to evaluate QSOX-mediated in vitro oxidative folding using a protein with a more complicated disulfide connectivity. Here we reintroduce (37, 38) riboflavin binding protein (a protein with 9 disulfides and hence >34 million pairings for the fully oxidized protein) as a stiffer test for an oxidative folding system. We show that the quenching of riboflavin fluorescence upon binding to the folded apoprotein (39) allows continuous monitoring of oxidative folding. This convenient and accessible in vitro system shows that glutathione redox buffers are unnecessary in refolding a protein with relatively complex disulfide connectivity, and suggests, again, that the redox poise of PDI will be a major determinant for efficient oxidative folding in vivo.
EXPERIMENTAL PROCEDURES
Materials
Bovine pancreatic RNase A, insulin, ampicillin, riboflavin, guanidine hydrochloride, GSH, GSSG, and lysozyme were obtained from Sigma-Aldrich. DTT was obtained from Acros Organics. IPTG was from Promega. EDTA was obtained from Fisher. Oligonucleotide primers were purchased from IDT Technologies.
Subcloning, Expression, and Purification of Human (gi 48735337) and Chicken (gi 30923135) PDI
A pET12a plasmid encoding human PDI insert between NdeI and BamHI restriction sites was a generous gift of Dr. Jakob Winther (Copenhagen). For ease of purification, PDI was sub-cloned into the pTrcHisA plasmid to incorporate an N-terminal hexahistidine tag using an N-terminal primer that replaced the NdeI site with an NheI site (see Supplemental Figure S1). The chicken PDI cDNA clone (pgm1cpk001j19 in the pCMV.SPORT6 plasmid) was obtained from the Chick EST library at the University of Delaware. During amplification, PCR primers were used that added an NheI N-terminal restriction site (excluding the signal sequence) and incorporated a BamHI site at the C-terminus. The modified chicken PDI insert was ligated into pre-cut pTrcHis A. All resulting sequences were confirmed by DNA sequencing. A comparison of the amino acid sequences of the human and avian PDI constructs is shown in supplementary Figure S1.
Both PDI constructs were transformed into BL 21 (DE3) cells (Invitrogen) for expression. Starter cultures were grown overnight at 37 °C in LB medium supplemented with 50 μg/mL ampicillin and used to inoculate four 2 L flasks containing 500 mL each of the same medium. Cells were grown at 37 °C to an absorbance of 0.4 at 600 nm and then induced with 0.5 mM IPTG for 6 h. Bacteria were harvested (2500 × g, 45 min, 4 °C) and re-suspended at 0.3 g wet weight per mL of lysis buffer (50 mM potassium phosphate buffer, pH 8.0, containing 300 mM NaCl, 0.1 mg/mL lysozyme, and an EDTA-free protease inhibitor cocktail (Roche). The suspension was passed through a French Pressure cell twice at 10,000 psi and then subject to five 30 s pulses of sonication separated by 1 min cooling intervals in an ice bath. The lysate was centrifuged at 4 °C for 45 min at 2500 × g and the supernatant (~ 60 mL) was removed and rocked for 2 h at 4 °C with 2 mL of Pro Bond Ni-NTA resin (Invitrogen). The resin was packed into a small chromatography column and then washed with 20 mL of 50 mM phosphate buffer, pH 8.0, containing 300 mM NaCl supplemented with 10 mM imidazole. The column was then developed with 10 mL of a 25 mM imidazole solution in water. Eluates were analyzed by UV absorbance and SDS-PAGE gels. Proteins were concentrated and washed into 50 mM potassium phosphate buffer, pH 7.5, containing 1 mM EDTA using a Centriprep YM-30 (Millipore) filtration device. Proteins were judged to be ~ 95% pure by SDS-PAGE stained with Coomassie Brilliant Blue. Human PDI was quantitated using a molar extinction coefficient of 56.4 mM−1 cm−1 at 280 nm for bovine PDI ((23), bovine and human PDI share 95% sequence). The concentration of avian PDI was determined using a molar extinction coefficient of 42.8 mM−1 cm−1 calculated from ProtParam (40). Chicken and human PDI were purified with typical yields of 40 and 50 mg/liter of culture respectively.
Insulin Reductase and Scrambled RNase Assays for PDI
The reductase activity of PDI (41) followed the increase in turbidity at 650 nm accompanying the reduction of 160 μM insulin by 1 mM DTT catalyzed by 1 μM PDI in 50 mM phosphate buffer, pH 7.5, 25 °C, containing 1mM EDTA (42). The activity of PDI as an isomerase was measured using 10 μM scrambled RNase in 20 mM Tris, pH 8.0, containing 100 mM KCl, 1 mM GSH/0.2 mM GSSG and 0.5 μM PDI. The return of RNase activity was assayed by following the hydrolysis of cyclic CMP at 296 nm (43). Scrambled RNase was prepared by incubating RNase with an equimolar amount of DTT (sufficient to reduce 1 of 4 disulfides) for 30 h under anaerobic conditions in 100 mM Tris, pH 8.0, containing 0.3 mM EDTA and 6 M guanidine hydrochloride. This solution was then opened to air and stirred in the dark at room temperature until less than 0.1 thiol per RNase molecule remained. Throughout this work the concentration of sulfhydryl groups was determined using Ellman's reagent (using ε412 14.15 mM−1 cm−1 for TNB (44)). The protein was exchanged into a solution of 50 mM NH4HCO3, containing 1 mM EDTA, pH 7.5, using a Centricon YM-10 device (Millipore). Samples were distributed in tubes and vacuum-centrifuged to dryness. Scrambled RNase was stored at −20 °C until use.
Preparation of reduced and oxidized proteins
Human PDI contains two CxxC redox-active disulfides (4 -SH equivalents upon reduction) and two additional cysteine residues that react very slowly with DTNB in the absence of denaturant (Figure S1; (45)). As purified from E. coli, human PDI rapidly released 2.7 TNB molecules showing that about 65% of the CxxC motifs were in their reduced state. Complete reduction of the human or avian PDI (yielding 4.1 ± 0.1 rapidly reacting -SH groups) was achieved by incubating PDI for 1 h at 25 °C in 50 mM phosphate buffer, pH 7.5, containing 1 mM EDTA and a 40-fold molar excess of DTT. Excess reductant was removed by gel-filtration on a PD-10 gel filtration column (1.45 × 5 cm; GE Healthcare) equilibrated with the same buffer in the absence of DTT. No greater than 0.5 mL of reduced protein was applied to these columns. Fractions (0.5 mL) were collected and small aliquots withdrawn and mixed with an equal volume of 10 mM DTNB to assess the separation between reduced PDI and DTT. Routinely reduced protein emerges in fractions 6–8 followed by two fraction containing insignificant thiol content. Excess free DTT begins to emerge in fractions 11 or 12. To make absolutely sure that no carryover of DTT had occurred, the resulting protein was concentrated to 0.5 mL and applied to a second freshly-equilibrated PD-10 as before. Reduced PDI fractions were pooled and standardized for protein concentration and thiol content. Final PDI solutions were concentrated and stored frozen at −20 C. Under these conditions PDI remained fully reduced for two weeks. The thiol titer of PDI was checked before each set of experiments to assure reduction. Fully oxidized PDI was prepared by treating the human protein, as isolated, with 10 mM GSSG for 16 h followed by gel filtration and characterization as before.
Reduced RNase was prepared by dissolving 20 mg protein and DTT (a 10-fold molar excess over RNase thiols) in 1 mL of degassed 100 mM Tris buffer, pH 8.0, containing 0.3 mM EDTA and 6M guanidine hydrochloride. After 1 h incubation at 37 °C, the protein was gel-filtered on a PD-10 column equilibrated with a degassed solution of 0.1% v/v acetic acid. Fractions were collected and characterized by absorbance at 278 nm (9.3 mM−1 cm−1 for reduced RNase (46)) and by thiol titer using DTNB as before.
Lyophilized egg white riboflavin binding protein was prepared as described earlier (47) and was a generous gift of Dr. Hal White (University of Delaware). This material contains 9 disulfide bonds (48, 49), between 7 and 8 phosphoserine residues located in a mobile loop close to the C-terminus (49, 50). Two sites of glycosylation are present (49, 51). The protein (8 mg) was dissolved in 0.5 mL of degassed solution containing a 10-fold excess of DTT over RfBP thiols in 100 mM potassium phosphate, pH 7.5, containing 1 mM EDTA and 6 M guanidine hydrochloride. The mixture was incubated for 2 h at 37 °C and desalted using a PD-10 column pre-equilibrated with the same buffer (containing guanidine hydrochloride and EDTA but without reductant) to remove released riboflavin and excess DTT. Fractions were analyzed as before using an extinction coefficient of 49.0 mM−1 cm−1 for apoRfBP (38).
Scrambled RfBP was prepared by incubating 141 μM reduced RfBP in 50 mM potassium phosphate buffer, pH 7.5, containing 1 mM EDTA and 3 M guanidine hydrochloride with 25 mM K3Fe(CN)6. The mixture was incubated for 2 h at 25 °C and the ferricyanide and guanidine hydrochloride removed from the oxidized protein by gel filtration using a PD-10 column equilibrated with 50 mM potassium phosphate buffer, pH 7.5, containing 1 mM EDTA.
Monitoring QSOX-mediated thiol oxidation
Disappearance of substrate thiols during the QSOX-mediated oxidation of reduced proteins was monitored discontinuously using DTNB as described previously (9, 32, 52).
Refolding of RNase
Reduced RNase (10 μM in 50 mM Tris, pH 7.5, containing 1 mM EDTA) was incubated at 25 °C in buffer alone, with 50 nM QSOX, with 50 nM QSOX and 5 μM reduced PDI, or with a glutathione redox buffer comprised of 1 mM GSH and 0.2 mM GSSG. Samples (50 μL) were removed every four minutes and added to 10 μL of a solution of NEM in water (to give final concentrations of 2 mM NEM when redox buffer was used, and 1 mM NEM in the absence of glutathione). The samples were stored capped at 25 °C and then diluted in a microcuvette with an equal volume of Tris buffer containing 2 mM cCMP.
Refolding of RfBP followed by fluorescence
In the standard refolding system, 1 μM reduced RfBP (18 μM free thiols) was mixed with 0.8 μM riboflavin, and various concentrations of QSOX and PDI in 50 or 100 mM potassium phosphate buffer, pH 7.5, 25 °C containing 1 mM EDTA. Disappearance of riboflavin fluorescence (39, 53) was monitored continuously using an Aminco Bowman Series 2 Luminescence Spectrophotometer (excitation 450 nm with a 2 nm slit width; emission 530 nm with a 16 nm slit width).
Characterization of reconstituted holo-RfBP
PDI was removed from reconstituted RfBP holoprotein by mixing with 1 mL of Ni-NTA resin and rocking for 3 h in a microcentrifuge tube at 4 °C. The resulting supernatant was concentrated by ultrafiltration and aliquots of the holoprotein were diluted with an equal volume of 6 M guanidine hydrochloride. Release of riboflavin was followed in the fluorimeter with settings as before and fit to first order kinetics (koff = 0.0125 ± .0001 s−1 compared to 0.0131 ± .0001 s−1 for the native holoprotein). The similarity between reconstituted and native protein was also evaluated by retention times on C18 reverse-phase HPLC (Hewlett Packard Series 1100 HPLC system). The gradient was 0–90% acetonitrile at 2%/min maintaining a concentration of 0.1% TFA. Refolded and native protein eluted at 18.31 ± 0.160 and 18.47 ± 0.06 min respectively.
Calculation of redox state of a and a' domains of PDI in equilibrium with glutathione redox buffer
The rate constants of Darby and Creighton (54) for the interaction of a and a' domains of PDI with glutathione were used to simulate the approach to equilibrium with the following starting conditions: a ratio of GSH/GSSG of 5/1 (4.17 and 0.83 mM respectively) and 0.5 mM of the oxidized domains. Calculations were performed with Gepasi (55) or Chemical Kinetics Simulator (IBM). At equilibrium the reduced PDI/oxidized PDI ratio for a and a' domains were >11 and >4 respectively. The accumulation of protein-S-SG mixed disulfide was less than 2% of the total protein concentration.
RESULTS AND DISCUSSION
Human and avian PDI are poor substrates of avian QSOX1
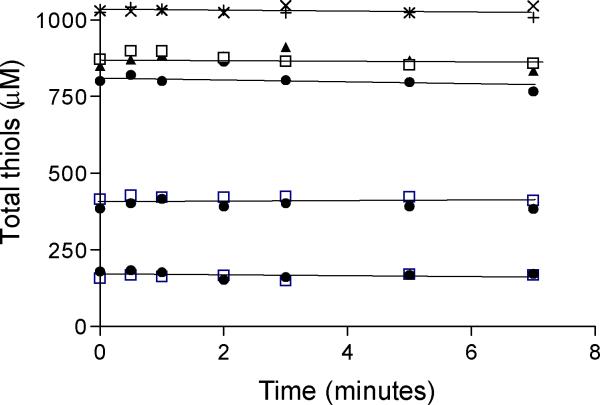
A key difference between the oxidative folding mechanisms in Figure 1 is that reduced PDI is believed to be the immediate substrate of Ero1p (13, 15–18, 56) whereas unfolded reduced proteins are proposed to be the direct substrates of QSOX enzymes (9, 11, 12, 26–30, 32). It was therefore important to test the predictions of Figure 1B directly. Figure 2 shows that reduced human PDI is a very poor substrate of avian QSOX1: with turnover numbers of less than about 6 thiols oxidized/min at PDI thiol concentrations up to 800 μM (corresponding to 200 μM of PDI; open squares). To ensure that this was not a consequence of non-cognate pairing, we sub-cloned and expressed avian PDI ((57) see Experimental Procedures). Figure 2 shows that reduced avian PDI was also a very sluggish substrate of avian QSOX (Figure 2 filled circles); the human pairings were similarly ineffective (filled triangles). Human and avian PDI ((gi 48735337 and gi 30923135 respectively) share ~85% sequence identity (Supplemental Figure S1) and showed comparable activities in the standard isomerase and reductase assays for PDI (Supplemental figure S2). To set the data of Figure 2 in perspective, a typical unfolded substrate of mammalian or avian QSOX1 enzymes shows kcat values of 500–2000 thiols oxidized per minute and a KM of 150 μM per thiol (9, 28–30). Since there appeared no substantive differences between cognate or non-cognate QSOX/PDI pairings, we have used the readily available avian QSOX1 and human PDI proteins for the remainder of this work.
FIGURE 2.
Reduced PDI as a substrate of QSOX. For each cognate and non-cognate PDI/QSOX pairing the oxidation of reduced PDI was evaluated by following the disappearance of CxxC thiols (using 50–200 μM PDI; 200–800 μM –SH groups; see Experimental Procedures) with 100 nM QSOX in 50 mM phosphate buffer, 1 mM EDTA, at pH 7.5 and 25 °C. Catalase (10 nM) was included in these incubations to discharge the hydrogen peroxide generated by QSOX. Aliquots were withdrawn at the indicated times for analysis of thiol content using DTNB (see Experimental Procedures). Open squares: avian QSOX and the indicated thiol titers of human PDI; filled circles: avian QSOX and avian PDI; filled triangles: human PDI and human QSOX. Crosses and checks indicate thiol consumption in an illustrative control reaction in the absence of QSOX using 250 μM of avian and human PDI respectively.
Previously we suggested, on the basis of indirect evidence, that reduced PDI was a substrate of QSOX (9). In this earlier experiment, a 16-fold excess of scrambled, fully oxidized, RNase was incubated with reduced PDI. The increasing inhibition of the refolding of scrambled RNase as QSOX was added to the mixture was taken to indicate a direct oxidation of reduced PDI by QSOX. Unfortunately, this conclusion is unwarranted because reduced PDI can efficiently transfer reducing equivalents to incorrect disulfide bonds in scrambled RNase (20) and QSOX, in turn, can rapidly and completely oxidize the RNase thiols so generated (9, 28, 32). Hence QSOX does not oxidize reduced PDI directly but indirectly via the mediation of an oxidized scrambled protein. To reiterate, the multiple direct experiments shown in Figure 2 show unequivocally that reduced PDI is a very poor substrate of QSOX.
Thioredoxin domains as potential substrates of QSOX
The essential non-reactivity of reduced PDI with QSOX raises some interesting questions. To what extent does the relatively positive redox poise of PDI (redox potentials quoted from −148 mV to −175 mV (measured over pH 7–8) for mammalian PDI (20, 54, 58)) play a role? PDI contains four thioredoxin domains, with the outer a and a' domains each containing a CxxC motif (25, 36, 59). To begin to address this question, we tested the single domain protein, E. coli thioredoxin. Even this potent and reactive cellular reductant (E'° −284 mV (60)) was a notably inferior substrate (in terms of KM values) compared to a typical unfolded reduced protein (kcat = 1170 thiols oxidized/min, KM = 1.1 mM, not shown; versus kcat = 1,220 thiols oxidized/min, KM = 115 μM for reduced RNase (30, 32)).
Protein substrates introduce reducing equivalents via a CxxC motif housed in a thioredoxin domain of QSOX (12, 30, 31). Why, then, is the communication between the thioredoxin domains of reduced PDI (or of thioredoxin itself) and the CxxC motif of the thioredoxin domain of QSOX ineffective? Beckwith and colleagues have noted the alternation of thioredoxin and non-thioredoxin folds in the five consecutive oxidoreductase modules that comprise the pathway for the reduction of periplasmic DsbD in E. coli (61, 62). Their suggestion, that structural factors might dictate an alternation in fold type, is consonant with the expected steric constraints for efficient thiol-disulfide exchange reactions (12, 63–65). Whatever the explanation for the observed non-reactivity of reduced PDI towards QSOX, it allows, in principle, a selective oxidation of unfolded reduced protein clients in the presence of a large excess of reduced PDI.
Oxidative refolding of pancreatic RNase with QSOX, PDI and redox buffers
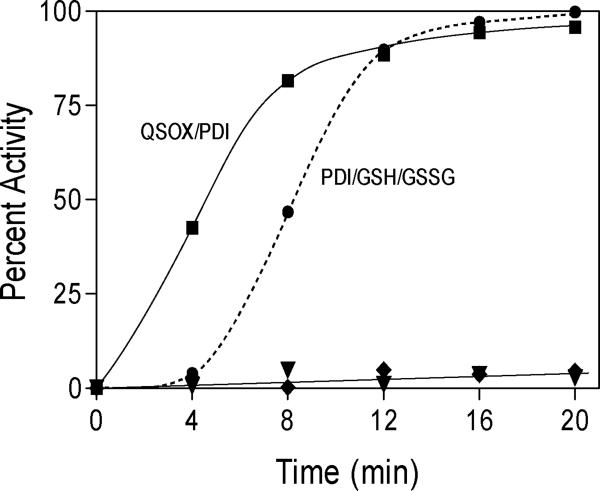
We first conducted a limited series of experiments with reduced ribonuclease A (RNase) because it has been so widely used in in vitro studies of oxidative protein folding. In preliminary work we, with H.F. Gilbert, showed efficient recovery of RNase activity using 0.5μM PDI in the presence of 1 mM GSH after the addition of 10 nM QSOX (9). GSH served to maintain the PDI in an isomerase-competent reduced state. A potential complexity of this experimental design is that GSH might have additional functions (e.g. in the resolution of mixed disulfides between client protein and PDI or QSOX, or via a non-enzymatic reduction of mis-paired disulfides in RNase). We thus investigated QSOX-driven refolding of RNase in the absence of glutathione in Figure 3. Here, we sampled the recovery of RNase activity discontinuously, after quenching aliquots of refolding cocktail with excess NEM (see Experimental Procedures). In our hands, this procedure proved easier to implement than following the regain of RNase activity continuously in a cuvette containing components of both refolding and assay cocktails (66). Figure 3 shows that while reduced RNase is an excellent substrate of QSOX (9, 32), very low levels of activity are recovered when reduced RNase is mixed with QSOX in the absence of PDI. The inclusion of 5 μM reduced PDI (20 μM CxxC thiols, with undetectable levels of oxidized PDI) leads to a rapid return of the native disulfide pairings in RNase. No oxidized or reduced glutathione is present in this experiment. Figure 3 also compares these data with results from two additional oxidative protein folding protocols. The glutathione redox buffer, (1.0 mM GSH and 0.2 mM GSSG) leads to a very slow regain of activity (amounting to 19% over 80 min in the absence of PDI, not shown). The inclusion of 5 μM PDI with this redox buffer strongly accelerates folding after a significant lag phase (Figure 3). These data show that glutathione is not needed for the efficient refolding of RNase using mixtures of QSOX and reduced PDI. Several other reports describe PDI-mediated oxidative folding in the absence of glutathione. Gilbert and coworkers have shown that oxidized PDI drives the refolding of reduced RNase in the absence of glutathione (67). Weissman and colleagues have demonstrated folding of reduced RNase in the absence of glutathione using PDI and Ero1 (18). Finally, glutathione is not required for oxidative folding of carboxypeptidase Y in the yeast endoplasmic reticulum (2).
FIGURE 3.
Oxidative refolding of reduced RNase. Reduced RNase 10 μM (80 μM –SH) in 50 mM Tris buffer, pH 7.5, 25 °C was incubated with 5 μM reduced PDI and 50 nM QSOX (squares); 50 nM QSOX alone (triangles); 5 μM reduced PDI and 1 mM GSH and 0.2 mM GSSG (circles); or 1 mM GSH and 0.2 mM GSSG alone (diamonds). At the times indicated samples were quenched with NEM and assayed for RNase using cCMP (see Experimental Procedures).
Oxidative refolding of reduced riboflavin binding protein (RfBP)
Pancreatic RNase, with 4-disulfides, has a much simpler disulfide complexity than many proteins of the metazoan secretome. Hence we wished to find a stiffer test for the QSOX/PDI folding system. Avian riboflavin binding protein (RfBP, Figure 4) provides a readily obtainable and useful model system (37, 38, 47, 68). It contains 9 disulfides and therefore has >34 million disulfide isomers of the fully oxidized protein (48, 49). Apo-riboflavin binding protein binds riboflavin rapidly and tightly (Kd = 1 nM) with a pronounced red shift in the absorbance envelope and a complete quenching of the flavin fluorescence exciting at 450 nm (39, 53, 68). Hence this system permits refolding to be continuously monitored spectroscopically over a wide range of RfBP concentrations in mixtures with high background protein absorbance (such as those that would be encountered in the lumen of the ER). RfBP has been used to study oxidative protein folding in two prior investigations. Using discontinuous sampling, Pattanaik et al. (37) observed that riboflavin binding, and the creation of an epitope associated with the native protein, only occurred after almost complete disulfide bond formation. In the second study, McClelland et al. (38) first explored the effects of PDI on the oxidative refolding of RfBP using a redox buffer (see later).
FIGURE 4.
Stereoview of the crystal structure of riboflavin binding protein. The isoalloxazine ring of the bound riboflavin (gold) is sandwiched between Tyrosine 75 and Tryptophan 156 (grey). None of the 9 disulfide bonds come closer than 9 Å to the isoalloxazine ring. The N-terminus is at the bottom left, and the C-terminus is at the end of the helix at the bottom right of the structure.
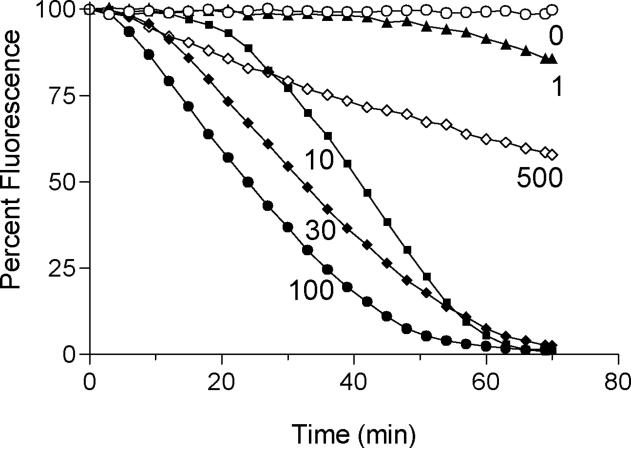
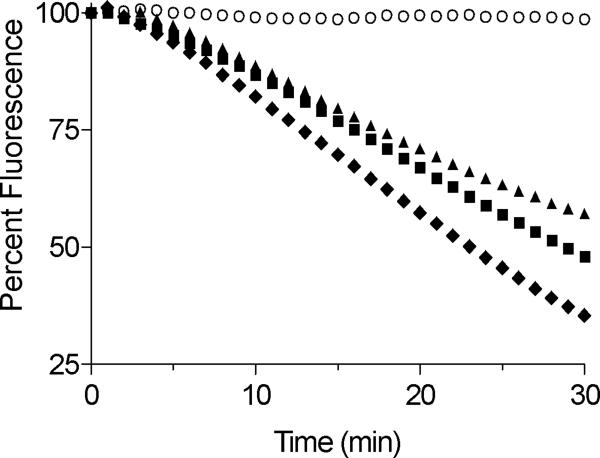
Reduced RfBP was prepared and rigorously freed of excess reductant by gel-filtration in 6 M guanidine hydrochloride (see Experimental Procedures). The unfolded reduced protein (18 -SH groups) was added to a solution of aerobic phosphate buffer (pH 7.5, 25 °C) to give final concentration of 1–5 μM. Flavin binding was first followed by the reappearance of the characteristic resolved absorbance spectrum of holo-RfBP (see Supplementary Figure S3). However the complete fluorescence quenching of riboflavin provides a more sensitive approach for assessing the return of the functional binding protein (39, 53, 68). In the experiments that follow, we used a slightly sub-stoichiometric level of riboflavin (0.8 μM flavin and 1.0 μM binding protein) to ensure that complete binding of the vitamin could occur. Thus under these conditions the yield of functional re-folded protein is at least 80%. Figure 5 shows that increasing concentrations of QSOX (from 1–100 nM) both increase the maximal rate of riboflavin binding and shorten the lag time of the fluorescence decrease. Very high concentrations of QSOX (500 nM) leads to a pronounced slowing of riboflavin-binding ability probably because the rapid insertion of disulfide bonds overwhelms the ability of reduced PDI to isomerize them. This slowing may have parallels in RNase because scrambled RNase (fully oxidized incorrectly paired protein) is a poor substrate of PDI when compared to partially oxidized intermediates (36, 66, 69). Logically optimal rates for the recovery of activity would represent a balance between the oxidative and isomerization components of oxidative folding.
FIGURE 5.
QSOX cooperates with PDI to refold RfBP. Avian QSOX (0–500 nM) in 50 mM phosphate buffer (pH 7.5, 1 mM EDTA, 25 °C), was mixed with 0.8 μM riboflavin and 30 μM reduced human PDI (120 μM CxxC thiols; see Experimental Procedures). The reaction was initiated by the addition of reduced RfBP (1 μM; 18 μM cysteine thiols with a carry-over of ~ 50 mM guanidine hydrochloride). Binding of riboflavin was assessed by the decrease in riboflavin fluorescence emission.
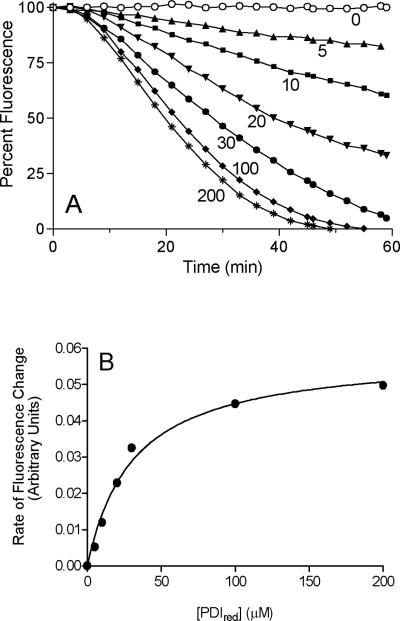
Figure 5 illustrates that nanomolar concentrations of QSOX, combined with 30 μM reduced PDI (in the absence of either glutathione or oxidized PDI), are effective in oxidative refolding of a client protein with a reasonably complicated disulfide network. The dependence of refolding on the concentration of reduced PDI is followed in Figure 6A keeping QSOX at 30 nM. Without the isomerase the recovery of riboflavin binding is undetectable, but increasing concentrations of reduced PDI accelerate folding. The maximal rate of fluorescence quenching obtained for each concentration of reduced PDI gives a hyperbolic dependence, with a half-saturation at a concentration of about 30 μM (Figure 6B). No evidence of slowing of the rate of oxidative folding is observed up to 200 μM reduced PDI. The effectiveness of oxidative folding can also be qualitatively represented by the time taken to reach 50% fluorescence quenching. Plotting these times as a function of PDI concentration leads to a limiting half-time of 19 min (not shown). While a determination of the molecular factors that limit the rate of riboflavin binding in this folding system at saturating PDI concentrations has not yet been investigated, they might include a rate-limiting protein conformational change. One of 8 proline residues in RfBP participates in a cis-peptide bond (49).
FIGURE 6.
The effect of increasing concentrations of reduced PDI on the refolding of RfBP driven by QSOX. Panel A: the conditions of Figure 5 were used except that a concentration of 30 nM QSOX was used and the reduced PDI concentration was varied from 0 to 200 μM. Panel B: the maximal rates of riboflavin rebinding attained for each curve in panel A show a hyperbolic dependence with half-saturation at 30 μM reduced PDI.
Previous work by McClelland et al. reported the oxidative refolding of reduced RfBP catalyzed by PDI in a glutathione redox buffer in the absence of riboflavin (38). Here oxidative folding was assessed discontinuously by withdrawing aliquots and evaluating the fluorescence quenching observed upon the addition of riboflavin. These data showed slow refolding with 43% recovery of riboflavin binding in 100 h. We were thus interested in evaluating whether vitamin binding influenced the course of oxidative refolding of RfBP in the QSOX/PDI system. Figure S4 shows that refolding proceeds slightly slower (by about 50%) in the absence of riboflavin. RfBP can be folded effectively in the absence of riboflavin in vivo because it is found to be typically less than half-saturated in chicken eggs (68).
Refolding of reduced RfBP in the absence of QSOX
Figure 7 compares the QSOX/PDI system with conventional oxidative folding systems. Although glutathione redox buffers (typically with a GSH/GSSG ratio of 5/1) are widely used in refolding studies, they are ineffective with reduced RfBP over 30 min (Figure 7 top) in agreement with earlier studies (37, 38). Conceivably the complexity of a 9-disulfide client protein, with millions of potential disulfide isomers, may limit the effectiveness of simple redox buffers when compared to a 4-disulfide containing protein (with a maximum of 105 isomers). As expected, supplementing the redox buffer with 30 μM reduced PDI led to a marked acceleration of riboflavin binding (Figure 7). The further inclusion of QSOX to this redox buffer/PDI mixture provides a further modest increase in rate (Figure 7, squares). None of these combinations are faster than the simple mixture of QSOX and reduced PDI (Figure 7, diamonds).
FIGURE 7.
Comparison of the effectiveness of different oxidative refolding systems for reduced RfBP. Reduced RfBP was incubated with riboflavin (1 μM and 0.8 μM, respectively) in 50 mM phosphate buffer, pH 7.5, 25 °C in the presence of: a redox buffer of 5 mM GSH/1 mM GSSG (open circles); the redox buffer and 30 μM reduced PDI (triangles); redox buffer, reduced PDI and 30 nM QSOX (squares); or QSOX and reduced PDI alone (diamonds).
Oxidative refolding of reduced RfBP using PDI alone
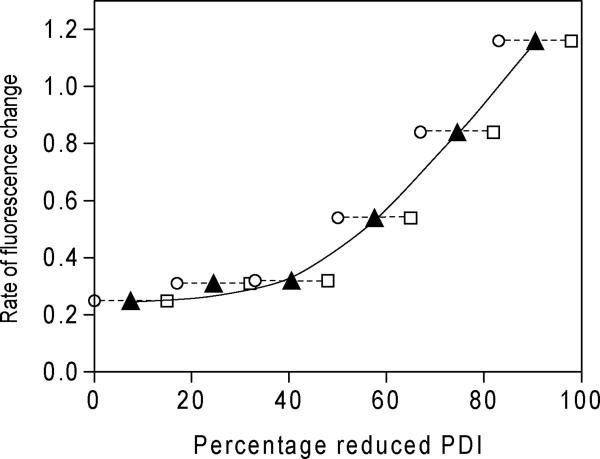
We next wanted to examine the rate of refolding reduced RfBP as a function of the redox state of PDI in the absence of QSOX and glutathione redox buffer. As mentioned earlier, glutathione redox buffers not only poise the ratio of oxidized to reduced PDI but might also participate directly in oxidative folding e.g. by forming and resolving mixed disulfide intermediates. Mixtures of fully oxidized and reduced PDI were combined to give a total concentration of 30 μM PDI (see Experimental Procedures) and mixed with 1 μM reduced RfBP and 0.8 μM riboflavin. Since oxidized PDI is now the sole oxidant in this experiment, the initial ratio of reduced to oxidized PDI will increase as disulfide bonds accumulate in riboflavin binding protein. For example, the complete oxidative refolding of 1 μM RfBP (18 μM – SH) would require 4.5 μM oxidized PDI (2 CxxC each). Circles and squares in Figure 8 thus represent the nominal and final ratios of reduced/oxidized PDI. Figure 8 shows that, over the concentration range used, the fastest refolding of reduced RfBP occurs when just enough oxidized PDI is added to provide for stoichiometric oxidation of reduced RfBP. In our studies the redox poise of PDI is substantially buffered by using an aggregate concentration of oxidized and reduced PDI that is 50-fold higher than those of reduced RfBP. Using a different experimental design, in which oxidized PDI is added as the sole oxidant of reduced RNase without additional reduced PDI, Lyles and Gilbert also found that the fastest folding rate occurred under conditions where all the added PDI would be eventually reduced by RNase (67). Adding more oxidized PDI than required for stoichiometric oxidation slowed the folding of RNase appreciably (67).
FIGURE 8.
The influence of PDI redox poise on the refolding rate of reduced RfBP. Reduced RfBP (1 μM) was added to 0.8 μM riboflavin and 30 μM PDI at six redox ratios in the absence of QSOX or glutathione (see Experimental Procedures): the initial percentage of reduced PDI in the mixtures is depicted by the open circles. Reduced RfBP will transfer reducing equivalents to oxidized PDI as each experiment progresses. Open squares would represent stoichiometric transfer; see the Text. The solid triangles are the average between these two percentages.
CONCLUSIONS
An unresolved issue in the mechanism of action of PDI is the extent to which the isomerase activity occurs via redox-neutral shuffling or net cycles of reoxidation and reduction, in which oxidized PDI is a discrete intermediate (20–22, 24, 25, 70). The relative importance of shuffling and redox-cycling modes might be protein dependent and might change as folding of a particular client protein progresses. These models for PDI function have been largely developed using proteins of comparative simplicity and so the more complex disulfide connectivity of RfBP provides an additional perspective. With RfBP, the fastest oxidative refolding occurs when PDI is largely in its reduced state (e.g. Figure 8) in agreement with an earlier study with RNase (67). At first sight, these data would seem to favor a redox-neutral shuffling model for PDI action. However they are just as consistent with an oxidoreductase mode providing that small levels of oxidized PDI can match the reductase activity of reduced PDI as disulfide bonds are exchanged iteratively. Whatever the relative importance of these two modes of action, PDI functions most efficiently in these two systems when it is substantially reduced.
Consistent with this finding, the redox state of PDI in the mammalian ER is largely reduced (16, 71). This observation aligns with recently re-determined values for the GSH/GSSG ratio in the ER (72). By suppressing post-isolation oxidation of the ER lumenal contents, Hagen and colleagues found the ratio of GSH/GSSG to be (5.1 ± 0.4)/1 compared to earlier widely-quoted estimates of 1.5/1 to 3.3/1 (73). Using this more reducing ratio, an overall glutathione concentration of 5 mM, and the kinetic parameters of Darby and Creighton for the reaction of glutathione and PDI (54):
suggest that equilibration is attained rapidly (t1/2 < 1 s) with a ratio of reduced/oxidized PDI of greater than 11 and 4 for a and a' domains of PDI respectively (see Experimental Procedures). The redox poise for ER-lumen components in equilibrium with glutathione and PDI may be more reducing than generally believed.
The QSOX/PDI system reported here proves an efficient in vitro system for the generation of native pairing in two dissimilar proteins. In vivo, a sulfhydryl oxidase that communicates poorly with both reduced PDI and GSH (9, 29, 30, 32) might allow for selective oxidation of client proteins without directly impacting the PDI redox pools; and without undue collateral oxidation of cellular glutathione via the equilibrium shown above (11). However it should be emphasized that the contribution of QSOX enzymes to the global disulfide output of the mammalian ER remains unclear. A key unresolved issue is whether the short form of QSOX, a relatively abundant protein found in a variety of extracellular locales, is active during transit through the ER (11, 12).
In an interesting study, Bulleid and coworkers have shown that a V5-tagged long-form of QSOX (with the C-terminal single membrane-spanning region intact) accumulates in the Golgi of mammalian cells (10). They suggested that QSOX long-form may participate in late-stage disulfide bond formation and in the assembly of disulfide bridged multimers (10). While we have no in vitro information on the catalytic potential of these longer forms of QSOX, the view expressed by these authors that the facile and error-prone oxidation of unfolded proteins by QSOX would make them potentially unsuitable residents of the ER (10) can be addressed by our present data. Low nanomolar concentrations of QSOX, combined with micromolar levels of reduced PDI, can rapidly and almost quantitatively refold two widely different client proteins. Indeed a mixture of QSOX and reduced PDI generates native pairings faster than either mixtures of oxidized and reduced PDI alone or of PDI whose redox poise is maintained by a glutathione redox buffer. In terms of disqualifying a candidate oxidant because it is error-prone, it should be noted that no known oxidant of reduced unfolded proteins can infallibly generate the correct disulfide pairings de novo. This is perhaps most clearly seen for PDI itself. PDI does not “know” which disulfides are the correct ones: it reaches the ensemble of native pairings by iteration (21). Oxidized PDI, at levels beyond those needed for the stoichiometric generation of disulfides, impedes the attainment of the native state in both RNase (67) and RfBP (Figure 8).
The present work emphasizes the apparent plasticity of oxidative folding systems. The observation that glutathione redox buffers are unimportant for efficient oxidative folding in RNase (18, 67) and now RfBP (this work) is consistent with the important finding that glutathione is not necessary for the secretion of disulfide containing proteins in yeast (2). By using amounts of PDI more in line with those found in the ER, the requirement for a redox buffer is circumvented allowing us to demonstrate that all that is needed for oxidative refolding of a protein with a huge number of disulfide isomers is a stoichiometric amount of oxidant and a liberal supply of reduced PDI. Whether oxidizing equivalents are generated by QSOX, Ero1, or other cellular oxidants, the universal additional requirement for efficient oxidative folding is reduced PDI.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joan Burnside and Deborah Fass for gifts of plasmid containing avian PDI and E. coli thioredoxin respectively, Erin Heckler for recombinant human QSOX, and Hal White for riboflavin binding protein. We acknowledge Danny Ramadan for useful discussions and Hugo Monaco for the coordinates for RfBP. We also thank Jakob Winther for a gift of a plasmid containing human PDI and for very useful discussions.
This work was supported in part by National Institutes of Health Grant (GM26643) and USPHS Training Grant 1-T32-GM08550 (P.C.R.). The content of this work is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Abbreviations: cCMP, cytidine 2',3' cyclic monophosphate; DTNB, 5,5′-dithiobis(2-nitrobenzoate); DTT, dithiothreitol; ER, endoplasmic reticulum; GSH, reduced glutathione; GSSG, oxidized glutathione; IPTG, isopropyl-D-thiogalactopyranoside; NEM, N-ethylmaleimide; PDI, protein disulfide isomerase; QSOX, Quiescin-sulfhydryl oxidase; RfBP, riboflavin binding protein; RNase, ribonuclease A.
SUPPORTING INFORMATION AVAILABLE Figures S1–S4 show sequence comparisons of human and avian PDI; a comparison of the reductase and isomerase activities of PDI; the refolding of RfBP monitored by absorbance; and the effect of riboflavin on RfBP refolding. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplamic reticulum. Mol. Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- 2.Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- 3.Hoober KL, Glynn NM, Burnside J, Coppock DL, Thorpe C. Homology between egg white sulfhydryl oxidase and quiescin Q6 defines a new class of flavin-linked sulfhydryl oxidases. J. Biol. Chem. 1999;274:31759–31762. doi: 10.1074/jbc.274.45.31759. [DOI] [PubMed] [Google Scholar]
- 4.Benayoun B, Esnard-Fève A, Castella S, Courty Y, Esnard F. Rat seminal vesicle FAD-dependent sulfhydryl oxidase:biochemical characterization and molecular cloning of a member of the new sulfhydryl oxidase/quiescin Q6 gene family. J. Biol. Chem. 2001;276:13830–13837. doi: 10.1074/jbc.M010933200. [DOI] [PubMed] [Google Scholar]
- 5.Dias-Gunasekara S, Gubbens J, van Lith M, Dunne C, Williams JA, Kataky R, Scoones D, Lapthorn A, Bulleid NJ, Benham AM. Tissue-specific expression and dimerization of the endoplasmic reticulum oxidoreductase Ero1beta. J. Biol. Chem. 2005;280:33066–33075. doi: 10.1074/jbc.M505023200. [DOI] [PubMed] [Google Scholar]
- 6.Cabibbo A, Pagani M, Fabbri M, Rocchi M, Farmery MR, Bulleid NJ, Sitia R. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J. Biol. Chem. 2000;275:4827–4833. doi: 10.1074/jbc.275.7.4827. [DOI] [PubMed] [Google Scholar]
- 7.Pagani M, Fabbri M, Benedetti C, Fassio A, Pilati S, Bulleid NJ, Cabibbo A, Sitia R. Endoplasmic reticulum oxidoreductin 1-lbeta (ERO1-Lbeta), a human gene induced in the course of the unfolded protein response. J. Biol. Chem. 2000;275:23685–23692. doi: 10.1074/jbc.M003061200. [DOI] [PubMed] [Google Scholar]
- 8.Papp E, Nardai G, Mandl J, Banhegyi G, Csermely P. FAD oxidizes the ERO1-PDI electron transfer chain: The role of membrane integrity. Biochem. Biophys. Res. Commun. 2005;338:938–945. doi: 10.1016/j.bbrc.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 9.Hoober KL, Sheasley SS, Gilbert HF, Thorpe C. Sulfhydryl oxidase from egg white: a facile catalyst for disulfide bond formation in proteins and peptides. J. Biol. Chem. 1999;274:22147–22150. doi: 10.1074/jbc.274.32.22147. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarthi S, Jessop CE, Willer M, Stirling CJ, Bulleid NJ. Intracellular catalysis of disulphide bond formation by the human sulphydryl oxidase, QSOX1. Biochem. J. 2007;404:403–411. doi: 10.1042/BJ20061510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorpe C, Coppock DL. Generating disulfides in multicellular organisms: Emerging roles for a new flavoprotein family. J. Biol. Chem. 2007;282:13929–13933. doi: 10.1074/jbc.R600037200. [DOI] [PubMed] [Google Scholar]
- 12.Heckler EJ, Rancy PC, Kodali VK, Thorpe C. Generating disulfides with the Quiescin-sulfhydryl oxidases. Biochim. Biophys. Acta. 2008;1783:567–577. doi: 10.1016/j.bbamcr.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frand AR, Kaiser CA. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell. 1999;4:469–477. doi: 10.1016/s1097-2765(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 14.Kulp MS, Frickel EM, Ellgaard L, Weissman JS. Domain architecture of protein disulfide isomerase facilitates its dual role as an oxidase and an isomerase in ERO1P-mediated disulfide formation. J. Biol. Chem. 2006;281:876–884. doi: 10.1074/jbc.M511764200. [DOI] [PubMed] [Google Scholar]
- 15.Benham AM, Cabibbo A, Fassio A, Bulleid N, Sitia R, Braakman I. The CXXCXXC motif determines the folding, structure and stability of human Ero1-Lalpha. EMBO J. 2000;19:4493–4502. doi: 10.1093/emboj/19.17.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001;20:6288–6296. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frand AR, Kaiser CA. Two pairs of conserved cysteines are required for the oxidative activity of Ero1p in protein disulfide bond formation in the endoplasmic reticulum. Mol. Biol. Cell. 2000;11:2833–2843. doi: 10.1091/mbc.11.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–1574. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- 19.Gross E, Sevier CS, Heldman N, Vitu E, Bentzur M, Kaiser CA, Thorpe C, Fass D. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. U S A. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwaller M, Wilkinson B, Gilbert HF. Reduction-reoxidation cycles contribute to catalysis of disulfide isomerization by protein-disulfide isomerase. J. Biol. Chem. 2003;278:7154–7159. doi: 10.1074/jbc.M211036200. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert HF. Protein disulfide isomerase and assisted protein folding. J. Biol. Chem. 1997;272:29399–29402. doi: 10.1074/jbc.272.47.29399. [DOI] [PubMed] [Google Scholar]
- 22.Laboissiere MC, Sturley SL, Raines RT. The essential function of protein-disulfide isomerase is to unscramble non-native disulfide bonds. J. Biol. Chem. 1995;270:28006–28009. doi: 10.1074/jbc.270.47.28006. [DOI] [PubMed] [Google Scholar]
- 23.Darby NJ, Creighton TE. Functional properties of the individual thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:11725–11735. doi: 10.1021/bi00037a009. [DOI] [PubMed] [Google Scholar]
- 24.Darby NJ, Freedman RB, Creighton TE. Dissecting the mechanism of protein disulfide isomerase: catalysis of disulfide bond formation in a model peptide. Biochemistry. 1994;33:7937–7947. doi: 10.1021/bi00191a022. [DOI] [PubMed] [Google Scholar]
- 25.Freedman RB, Klappa P, Ruddock LW. Protein disulfide isomerases exploit synergy between catalytic and specific binding domains. EMBO Rep. 2002;3:136–140. doi: 10.1093/embo-reports/kvf035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coppock DL, Thorpe C. Multidomain flavin-dependent sulfhydryl oxidases. Antioxid. Redox Signal. 2006;8:300–311. doi: 10.1089/ars.2006.8.300. [DOI] [PubMed] [Google Scholar]
- 27.Thorpe C, Hoober K, Raje S, Glynn N, Burnside J, Turi G, Coppock D. Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch. Biochem. Biophys. 2002;405:1–12. doi: 10.1016/s0003-9861(02)00337-5. [DOI] [PubMed] [Google Scholar]
- 28.Hoober KL, Thorpe C. Egg white sulfhydryl oxidase: Kinetic mechanism of the catalysis of disulfide bond formation. Biochemistry. 1999;38:3211–3217. doi: 10.1021/bi9820816. [DOI] [PubMed] [Google Scholar]
- 29.Jaje J, Wolcott HN, Fadugba O, Cripps D, Yang AJ, Mather IH, Thorpe C. A flavin-dependent sulfhydryl oxidase in bovine milk. Biochemistry. 2007;46:13031–13040. doi: 10.1021/bi7016975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heckler EJ, Alon A, Fass D, Thorpe C. Human quiescin-sulfhydryl oxidase, QSOX1: probing internal redox steps by mutagenesis. Biochemistry. 2008;47:4955–4963. doi: 10.1021/bi702522q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raje S, Thorpe C. Inter-domain redox communication in flavoenzymes of the quiescin/sulfhydryl oxidase family: role of a thioredoxin domain in disulfide bond formation. Biochemistry. 2003;42:4560–4568. doi: 10.1021/bi030003z. [DOI] [PubMed] [Google Scholar]
- 32.Hoober KL, Joneja B, White HB, III, Thorpe C. A Sulfhydryl Oxidase from Chicken Egg White. J. Biol. Chem. 1996;271:30510–30516. doi: 10.1074/jbc.271.48.30510. [DOI] [PubMed] [Google Scholar]
- 33.Ahmed AK, Schaffer SW, Wetlaufer DB. Nonenzymic reactivation of reduced bovine pancreatic ribonuclease by air oxidation and by glutathione oxidoreduction buffers. J. Biol. Chem. 1975;250:8477–8482. [PubMed] [Google Scholar]
- 34.Narayan M, Welker E, Wedemeyer WJ, Scheraga HA. Oxidative folding of proteins. Acc. Chem. Res. 2000;33:805–812. doi: 10.1021/ar000063m. [DOI] [PubMed] [Google Scholar]
- 35.Raines RT. Ribonuclease A. Chem. Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim. Biophys. Acta-Proteins and Proteomics. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 37.Pattanaik P, Sooryanarayana, Adiga PR, Visweswariah SS. Refolding of native and recombinant chicken riboflavin carrier (or binding) protein: evidence for the formation of non-native intermediates during the generation of active protein. Eur. J. Biochem. 1998;258:411–418. doi: 10.1046/j.1432-1327.1998.2580411.x. [DOI] [PubMed] [Google Scholar]
- 38.McClelland DA, McLaughlin SH, Freedman RB, Price NC. The refolding of hen egg white riboflavin-binding protein: effect of protein disulphide isomerase on the reoxidation of the reduced protein. Biochem. J. 1995;311(Pt 1):133–137. doi: 10.1042/bj3110133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becvar J, Palmer G. The binding of flavin derivatives to the riboflavin-binding protein of egg white. A kinetic and thermodynamic study. J. Biol. Chem. 1982;257:5607–5617. [PubMed] [Google Scholar]
- 40.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundstrom J, Holmgren A. Protein disulfide-isomerase is a substrate for thioredoxin reductase and has thioredoxin-like activity. J. Biol. Chem. 1990;265:9114–9120. [PubMed] [Google Scholar]
- 42.Smith AM, Chan J, Oksenberg D, Urfer R, Wexler DS, Ow A, Gao L, McAlorum A, Huang SG. A high-throughput turbidometric assay for screening inhibitors of protein disulfide isomerase activity. J. Biomol. Screen. 2004;9:614–620. doi: 10.1177/1087057104265292. [DOI] [PubMed] [Google Scholar]
- 43.Crook EM, Mathias AP, Rabin BR. Spectrophotometric assay of bovine pancreatic ribonuclease by the use of cytidine 2':3'-phosphate. Biochem. J. 1960;74:234–238. doi: 10.1042/bj0740234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eyer P, Worek F, Kiderlen D, Sinko G, Stuglin A, Simeon-Rudolf V, Reiner E. Molar absorption coefficients for the reduced Ellman reagent: reassessment. Anal. Biochem. 2003;312:224–227. doi: 10.1016/s0003-2697(02)00506-7. [DOI] [PubMed] [Google Scholar]
- 45.Walker KW, Lyles MM, Gilbert HF. Catalysis of oxidative protein folding by mutants of protein disulfide isomerase with a single active -site cysteine. Biochemistry. 1996;35:1972–1980. doi: 10.1021/bi952157n. [DOI] [PubMed] [Google Scholar]
- 46.Schaffer SW, Ahmed AK, Wetlaufer DB. Salt effects in the glutathione-facilitated reactivation of reduced bovine pancreatic ribonuclease. J. Biol. Chem. 1975;250:8483–8486. [PubMed] [Google Scholar]
- 47.Miller MS, White HB., 3rd Isolation of avian riboflavin-binding protein. Methods Enzymol. 1986;122:227–234. doi: 10.1016/0076-6879(86)22174-6. [DOI] [PubMed] [Google Scholar]
- 48.Hamazume Y, Mega T, Ikenaka T. Positions of disulfide bonds in riboflavin-binding protein of hen egg white. J. Biochem. 1987;101:217–223. doi: 10.1093/oxfordjournals.jbchem.a121894. [DOI] [PubMed] [Google Scholar]
- 49.Monaco HL. Crystal structure of chicken riboflavin-binding protein. EMBO J. 1997;16:1475–1483. doi: 10.1093/emboj/16.7.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller MS, Mas MT, White HB., 3rd Highly phosphorylated region of chicken riboflavin-binding protein: chemical characterization and 31P NMR studies. Biochemistry. 1984;23:569–576. doi: 10.1021/bi00298a027. [DOI] [PubMed] [Google Scholar]
- 51.Mega T, Ikenaka T. Methanolysis products of asparagine-linked N-acetylglucosamine and a new method for determination of N- and O-glycosidic N-acetylglucosamine in glycoproteins that contain asparagine-linked carbohydrates. Anal. Biochem. 1982;119:17–24. doi: 10.1016/0003-2697(82)90659-5. [DOI] [PubMed] [Google Scholar]
- 52.Hoober KL, Thorpe C. Flavin-dependent sulfhydryl oxidases in protein disulfide bond formation. Methods Enzymol. 2002;348:30–34. doi: 10.1016/s0076-6879(02)48622-3. [DOI] [PubMed] [Google Scholar]
- 53.Rhodes MB, Bennett N, Feeney RE. The flavoprotein-apoprotein system of egg white. J. Biol. Chem. 1959;234:2054–2060. [PubMed] [Google Scholar]
- 54.Darby NJ, Creighton TE. Characterization of the active site cysteine residues of the thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:16770–16780. doi: 10.1021/bi00051a027. [DOI] [PubMed] [Google Scholar]
- 55.Mendes P. Biochemistry by numbers: simulation of biochemical pathways with Gepasi 3. Trends Biochem. Sci. 1997;22:361–363. doi: 10.1016/s0968-0004(97)01103-1. [DOI] [PubMed] [Google Scholar]
- 56.Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J. Cell. Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parkkonen T, Kivirikko KI, Pihlajaniemi T. Molecular cloning of a multifunctional chicken protein acting as the prolyl 4-hydroxylase beta-subunit, protein disulphide-isomerase and a cellular thyroid-hormone-binding protein. Comparison of cDNA-deduced amino acid sequences with those in other species. Biochem. J. 1988;256:1005–1011. doi: 10.1042/bj2561005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lundstrom J, Holmgren A. Determination of the reduction-oxidation potential of the thioredoxin-like domains of protein disulfide-isomerase from the equilibrium with glutathione and thioredoxin. Biochemistry. 1993;32:6649–6655. doi: 10.1021/bi00077a018. [DOI] [PubMed] [Google Scholar]
- 59.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng Z, Arscott LD, Ballou DP, Williams CH., Jr. The relationship of the redox potentials of thioredoxin and thioredoxin reductase from Drosophila melanogaster to the enzymatic mechanism: reduced thioredoxin is the reductant of glutathione in Drosophila. Biochemistry. 2007;46:7875–7885. doi: 10.1021/bi700442r. [DOI] [PubMed] [Google Scholar]
- 61.Katzen F, Beckwith J. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell. 2000;103:769–779. doi: 10.1016/s0092-8674(00)00180-x. [DOI] [PubMed] [Google Scholar]
- 62.Ritz D, Beckwith J. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 2001;55:21–48. doi: 10.1146/annurev.micro.55.1.21. [DOI] [PubMed] [Google Scholar]
- 63.Fernandes PA, Ramos MJ. Theoretical insights into the mechanism for thiol/disulfide exchange. Chemistry. 2004;10:257–266. doi: 10.1002/chem.200305343. [DOI] [PubMed] [Google Scholar]
- 64.Houk J, Whitesides GM. Structure Reactivity Relations for Thiol Disulfide Interchange. J. Amer. Chem. Soc. 1987;109:6825–6836. [Google Scholar]
- 65.Bach RD, Dmitrenko O, Thorpe C. The Mechanism of Thiolate-disulfide Interchange Reactions in Biochemistry. J. Org. Chem. 2008;73:12–21. doi: 10.1021/jo702051f. [DOI] [PubMed] [Google Scholar]
- 66.Lyles MM, Gilbert HF. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry. 1991;30:613–619. doi: 10.1021/bi00217a004. [DOI] [PubMed] [Google Scholar]
- 67.Lyles MM, Gilbert HF. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: pre-steady-state kinetics and the utilization of the oxidizing equivalents of the isomerase. Biochemistry. 1991;30:619–625. doi: 10.1021/bi00217a005. [DOI] [PubMed] [Google Scholar]
- 68.White HB, 3rd, Merrill AH., Jr. Riboflavin-binding proteins. Annu. Rev. Nutr. 1988;8:279–299. doi: 10.1146/annurev.nu.08.070188.001431. [DOI] [PubMed] [Google Scholar]
- 69.Freedman RB. Protein Folding in the Cell. In: Creighton TE, editor. Protein Folding. WH Freeman; New York: 1992. pp. 455–539. [Google Scholar]
- 70.Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem. Sci. 2006;31:455–464. doi: 10.1016/j.tibs.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 71.Appenzeller-Herzog C, Ellgaard L. In vivo reduction-oxidation state of protein disulfide isomerase: the two active sites independently occur in the reduced and oxidized forms. Antioxid. Redox Signal. 2008;10:55–64. doi: 10.1089/ars.2007.1837. [DOI] [PubMed] [Google Scholar]
- 72.Dixon BM, Heath SH, Kim R, Suh JH, Hagen TM. Assessment of Endoplasmic Reticulum Glutathione Redox Status Is Confounded by Extensive Ex Vivo Oxidation. Antioxid. Redox Signal. 2008;10:963–972. doi: 10.1089/ars.2007.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.