Abstract
DNA damage and DNA repair may mediate several cellular processes, like replication and transcription, mutagenesis and apoptosis and thus may be important factors in the development and pathology of an organism, including cancer. DNA is constantly damaged by reactive oxygen species (ROS) and reactive nitrogen species (RNS) directly and also by products of lipid peroxidation (LPO), which form exocyclic adducts to DNA bases. A wide variety of oxidatively-generated DNA lesions are present in living cells. 8-oxoguanine (8-oxoGua) is one of the best known DNA lesions due to its mutagenic properties. Among LPO-derived DNA base modifications the most intensively studied are ethenoadenine and ethenocytosine, highly miscoding DNA lesions considered as markers of oxidative stress and promutagenic DNA damage. Although at present it is impossible to directly answer the question concerning involvement of oxidatively damaged DNA in cancer etiology, it is likely that oxidatively modified DNA bases may serve as a source of mutations that initiate carcinogenesis and are involved in aging (i.e. they may be causal factors responsible for these processes). To counteract the deleterious effect of oxidatively damaged DNA, all organisms have developed several DNA repair mechanisms. The efficiency of oxidatively damaged DNA repair was frequently found to be decreased in cancer patients. The present work reviews the basis for the biological significance of DNA damage, particularly effects of 8-oxoGua and ethenoadduct occurrence in DNA in the aspect of cancer development, drawing attention to the multiplicity of proteins with repair activities.
Keywords: 8-oxoguanine; 1,N6-ethenoadenine; 3,N4-ethenocytosine; DNA repair; polymorphism; carcinogenesis
INTRODUCTION
Oxidative stress and free radicals
Oxidative stress is defined as an imbalance in the formation and decomposition of reactive oxygen species, in favor of ROS increase. Reactive oxygen species (ROS) are oxygen containing molecules which may be radical, for example, superoxide (O2•-) and hydroxyl radical (•OH) or non-radical, for example hydrogen peroxide (H2O2) and singlet oxygen (1O2). Free radicals are defined as any chemical moiety capable of existing with a single electron in an orbital i.e. an unpaired electron (denoted as •). It is this aspect which makes free radicals more reactive than non-radicals, since orbital pairing of electrons increases stability. Formation of reactive oxygen (ROS) and nitrogen (RNS) species is involved in many human pathologic conditions including certain types of human cancers, e.g., lung, breast and colon, as well as atherosclerosis, neurodegenerative diseases and aging [1, 2]. ROS are generated in organisms by γ, X and UV radiation, biotransformation of dietary chemicals, some diet components, e.g. transient metal ions, inflammatory reactions and during normal cellular metabolism. The resulting disturbance of the pro-oxidant/antioxidant balance leads to a condition of oxidative stress, with subsequent oxidation of cellular components, activation of cytoplasmic/nuclear signal transduction pathways, modulation of gene and protein expression and alteration of activities of DNA and RNA polymerases [3].
Normal cellular metabolism appears to be a primary source for endogenous ROS. The most reactive ROS are hydroxyl radicals. Their cellular formation may be mediated by transition metal ions [4]. Labile iron and copper pools mediate production of •OH [1] in the metal-catalysed Haber-Weiss and Fenton reactions:
 |
ROS attack all cellular components causing oxidation and fragmentation of nucleic acids, proteins and lipids.
OXIDATIVELY-DERIVED DNA BASE MODIFICATIONS
The attack of hydroxyl radicals on DNA results in single or double strand breaks, generation of abasic sites, base and sugar lesions. Hydroxyl radicals cause ionization of DNA bases as well as of other cellular components. Free radical attack upon DNA generates a whole series of DNA damage, among them a large number of pyrimidine- and purine-derived lesions in DNA [5]. Some of these modified DNA bases have considerable potential to affect the integrity of the genome [6, 7]. The principal products in oxidatively damaged DNA include 8-oxo-7,8-dihydroadenine (8-oxoAde), 8-oxo-7,8-dihydroguanine (8-oxoGua); and its deoxynu-cleoside equivalent, 8-oxodG, 5,6-dihydroxy-5,6-dihydrothymine (thymine glycol, Tg) and ring-opened lesions: 4,6-diamino-5-formamido-pyrimidine (FapyAde) and 2,6-diamino-4-hydroxy -5-formamidopyrimidine (FapyGua) [8, 9]. Examples of the most significant DNA lesions are presented in Figure 1.
Figure 1.

Major base lesions induced in DNA by oxidative stress and lipid peroxidation
8-oxo-7,8-dihydroguanine is one of the most widely studied lesions. The presence of 8-oxoGua residues in DNA leads to GC→TA transversions unless repaired prior to DNA replication [10]. Therefore, the presence of 8-oxoGua in cells may lead to point mutations. An elevated level of 8-oxoGua accompanies the development of several human diseases. High concentrations of 8-oxoGua have been found in DNA of blood leukocytes and in the urine of lung and colon cancer patients. 8-oxoGua was also increased in leukocytes of patients developing atherosclerosis and AIDS [11].
Oxidized DNA bases may be derived either from the direct attack of ROS on DNA or may be incorporated from nucleotide pool by DNA polymerases. Purines and pyrimidines are 100 to 1000-fold more susceptible to modification as mononucleosides and nucleotides or when they are present in ssDNA or RNA, than when they are protected in the ds helix structure. Nucleotide pool modification is an important source of nucleic acid damage. Both DNA and RNA polymerases can discriminate unchanged and damaged nucleotides, however, this discrimination is not complete and they incorporate damaged nucleotides into nucleic acids with a different frequency [12, 13].
Unsaturated fatty acids also play an important role, since lipid peroxidation (LPO) yields a plethora of stable derivatives, which add to nucleic acids forming exocyclic DNA adducts of high miscoding potential, as well as DNA-DNA and DNA-protein cross-links [14].
The major lipid peroxidation products are malondialdehyde, crotonaldehyde, trans-4-hydroxy-2- nonenal (HNE) and many other products [15]. The most extensively studied are 1,N6 -ethenoadenine (εA) and 3,N4-ethenocytosine (εC) (Figure 1). Although the precise pathway of ethenoadducts formation in the cell is unknown, it is assumed on the basis of in vitro experiments that they are the result of DNA bases interactions with LPO products. DNA-ethenoadducts are lesions of high miscoding potential in mammalian cells [16, 17], inducing also recombination and chromosomal aberrations [18], which are regarded as a good predictive biomarker of cancer risk [19]. Exocyclic DNA adducts were found to be increased in inflammatory bowel disease, Crohn disease, and pro-cancerous metal storage diseases such as Wilson disease and primary hemochromatosis [14, 20]. Recently, increasing attention has been focused on bulky DNA adducts formed by the long-chain LPO products, such as HNE. An increased level of HNE accumulation was observed in brain tissue and in the cerebrum of patients suffering from several neurodegenerative disorders like Alzheimer's (AD), Parkinson's, Pick's, amyotrophic lateral sclerosis and Huntington diseases [21]. Large amounts of HNE-modified proteins were found in the brains (globus pallidus) of Cockayne Syndrome patients [22]. The level of HNE-dG adducts in human brains (post mortem) was evaluated as 400 - 600 adducts per 109 normal nucleotides in the hippocampus [23]. These data unambiguously point to the link between oxidative DNA damage and human pathologies.
In view of the importance of DNA damage in carcinogenesis, it is conceivable that any agent capable of reacting with DNA and chemically modifying it could be carcinogenic. It is very likely that reactive oxygen species belong to this group. Moreover, many observations indicate a direct correlation between 8-oxoGua formation and carcinogenesis in vivo [6, 24] and that oxygen-derived radicals are known to induce mutations at hotspot codons of the human TP53 and Ha-ras genes [25, 26]. Therefore, oxidative damage to DNA may be critical to the development of cancer.
EFFECTS OF OXIDATIVELY DAMAGED DNA UPON THE CELL
The effects of a significant number of DNA base modifications upon replication and transcription have been described. Many oxidative base lesions are mutagenic, irrespective of whether they are formed in situ, or arise by misincorporation from the deoxynucleotide pool. Overall it seems that oxidatively-generated DNA lesions are best described as weakly mutagenic, for example, the frequency of mutations generated by 8-oxodG presence in mammalian cells DNA is 2.5 - 4.8%, although lesion formation, persistence and accumulation in vivo could increase this value. At least two cellular processes may diminish 8-oxoGua mutagenic potency: (i) DNA repair and (ii) translesion synthesis by Y family DNA polymerases. 8-oxoGua constitutes a moderate block for replicative DNA polymerases, which tend to incorporate A opposite 8-oxoGua. Accurate and efficient replication through the 8-oxoGua is ensured by DNA polymerase η [27], which can substitute replicative DNA polymerases upon replication inhibition. Mutations are not the only effect of oxidative DNA damage. Literature data suggest that events at the DNA level other than mutations are potentially involved in pathogenesis. Oxidants may affect gene expression either through ROS generation, or through interfering with transcription factor binding. Oxidants are known to modulate gene expression through alteration in cellular redox status. Redox status can alter transcription factor structure and binding of transcription factors to cognate DNA sequences through changes in cysteine reduction state and modification of zinc-finger domains [28]. Eucaryotic promoters usually contain GC-rich regions, either flanking the TATA-box or recognized by transcription factors [29]. 8-oxoGua formation within this region may affect promoter methylation since it was shown that transfer of the methyl group by methyltransferases is inhibited when the target sequence contains 8-oxoGua [30]. Thus may also change the mRNA synthesis level.
ROLE OF OXIDATIVELY DAMAGED DNA IN CARCINOGENESIS
It is clear that one of the consequences of oxidized base lesions persisting in DNA is mutation. DNA mutation is a crucial step in carcino-genesis and elevated levels of oxidatively-generated DNA lesions have been noted in many tumors, strongly implicating such damage in the etiology of cancer.
A potential role has been demonstrated for oxidative mechanisms in the initiation, promotion and malignant conversion (progression) stages of carcinogenesis. Given that cumulative cancer risk increases with age and is associated with an accumulation of DNA damage, oxidatively damaged DNA has been investigated in cancer.
Lesions such as 8-oxodG are established bio-markers of oxidative stress and coupled with their potential mutagenicity in mammalian cells. This has led to their proposed potential as intermediate markers of a disease endpoint, for example cancer. The findings that GC→TA transversions, potentially derived from 8-oxodG, have been observed in vivo, in the RAS oncogene and the TP53 tumour suppressor gene in lung and liver cancer [31, 32] support this proposal.
Ethenoadducts in mammalian cells cause a broad spectrum of transversions and transitions. εA induces AT->GC, AT->TA, AT->CG [33], and εC generates CG->AT, CG->TA, CG->GC base substitutions [34]. Such mutations are reported to be found in tumors associated with exposure to ethenoadduct forming chemicals in genes linked with cancer development e.g. TP53, Ha-ras, Ki-ras [35, 36].
Numerous studies have attempted to establish a relationship between levels of oxidatively damaged DNA and cancer. Elevated levels of damage are reported to arise as a consequence of an environment in the tumor, which is low in antioxidant enzymes and high in ROS generation [37].
FACTORS THAT SHAPE THE BACKGROUND LEVEL OF 8-OXOGUA IN CELLULAR DNA AND THEIR RELEVANCE TO CARCINOGENESIS
Antioxidant vitamins, uric acid versus the 8-oxoGua level in cellular DNA
Many epidemiological studies have reported an inverse association between vegetable and fruit consumption and occurrence of cancer and other degenerative diseases [38-40]. One of the possible mechanisms of this protective effect is by the antioxidative activities of such plant food constituents as vitamins A, C and E. These antioxidant vitamins are effective free radical scavengers. They should protect biomolecules such as proteins, lipids and nucleic acids from oxidative damage. Another effective scavenger of ROS is uric acid [41]. Uric acid at physiological concentration is regarded as the main antioxidant and not only does it efficiently scavenge free radicals but it has also been shown to stabilize ascorbic acid in human serum [42] and reduce consumption of α-tocopherol and β-carotene [41].
One of the possible mechanisms of the protective effect of antioxidant vitamins against cancer development may be by decreasing the amount of potentially mutagenic oxidatively-modified DNA bases. Duthie et al. [43] found that supplementation of healthy volunteers with vitamin C (100 mg/day), vitamin E (280 mg/ day) and β-carotene (25 mg/day) significantly reduced base damage in lymphocyte DNA. Collins at al. [44] demonstrated a significant negative correlation between basal concentration of serum carotenoids and oxidatively modified pyrimidines. Supplementation of patients with carotenoids did not influence the level of oxidatively damaged DNA. The authors did not find any correlation between the damage and concentration of vitamins E and C. Moreover, the majority of intervention studies have failed to show clearly decreased oxidatively damaged DNA or cancer risk [45]. In our recently published study, the relationship between the basal level of antioxidants (vitamins A, C and E and uric acid) and oxidatively damaged DNA represented by urinary excretion of 8-oxodG, 8-oxoGua as well as the level of oxidatively damaged DNA in leukocytes was analysed. Basal plasma levels of antioxidants may provide a better estimation of antioxidant status than supplementation data, taking into account not only the consumption, which may reflect a transient state, but also the absorption and utilization. Our results revealed a weak, statistically significant negative correlation between the analysed antioxidants and all the measured parameters of oxidatively damaged DNA. Therefore, the results suggest that the level of oxidative DNA lesions shows limited but significant response to antioxidants analysed in this study and is affected more by many other cellular functions like antioxidant enzymes or DNA repair enzymes as well as genetics [46].
In another study we have found that the endogenous levels of the analyzed antioxidant vitamins in the plasma of colon cancer patients were significantly lower than that in the control group [47]. Members of the studied groups were chosen randomly and in a way to match feeding habits and living conditions. Therefore, it is rather unlikely that the different concentrations of vitamins in their blood were the result of lifestyle. Severe oxidative stress, characteristic for colon cancer, resulting in the production of ROS is responsible for consumption of the anti-oxidant vitamins. The decreased amount of uric acid in blood plasma of colon cancer patients also supports this assumption. This prooxidative environment resulted in an increased amount of 8-oxodG in lymphocyte DNA of cancer patients, where the level of this lesion was significantly higher (p=0.0034) than in the DNA of the control group [48]. These findings suggest that oxidative stress may be characteristic not only for the diseased tissue but for some other tissues of the cancer patients.
Iron as possible cause of 8-oxoGua and ethenoadducts accumulation
Iron has the capacity to accept and donate electrons easily, changing between ferric (Fe2+) and ferrous (Fe3+) iron. Due to this feature it is a useful component of cytochromes and oxygen binding molecules like hemoglobin and myoglobin. However, inside the cell iron can exist in another form, as a “free” or “labile” iron (LIP, iron not bound to proteins). LIP- associated iron is in dynamic equilibrium with other sequestered iron forms in the cell and is bound to cytosolic low molecular weight ligands that have not yet been identified. This iron form is catalytically active and participates in the reaction involved in the production of harmful ROS (the Fenton reaction) and lipid peroxidation [49]. Iron ions circulate bound to plasma transferrin, whereas ferritin serves to accumulate them. We analyzed the broad spectrum of the components that affect iron metabolism and their possible association with the endogenous level of 8-oxodG [50,51]. No correlation has been found between the plasma concentration of ferritin or transferrin saturation and the amount of 8-oxodG in the DNA of lymphocytes. On the other hand, a positive correlation has been observed between LIP and 8-oxodG [51]. This suggests that under physiological conditions LIP is available for catalysing the Fenton type reaction in a close proximity to cellular DNA. Neither the exact chemical nature of the complex between iron and DNA is known, nor is it established how iron can get into the nucleus.
There are experimental data which demonstrate the existence of a free iron pool in patients with hemochromatosis [52]. Epidemiological data also show that elevation of the body iron level may increase the risk of liver cancer [53]. Iron overload may favor the persistence of harmful LIP, which can catalyse generation of the potentially carcinogenic 8-oxodG, as well as ethenoadducts in cellular DNA [54].
ACCUMULATION OF 8-OXOGUA AND ETHENOAD-DUCTS IN CANCER PATIENTS
Analyses of 8oxoGua in human samples
It is generally accepted that the products of 8-oxoGua repair in cellular DNA are excreted into the urine without further metabolism [55, 56]. The presence of the modified nucleoside (8-oxodG) in urine is commonly believed to represent either the primary repair product of the oxidative DNA damage in vivo, presumably via nucleotide excision repair (NER) [57-59] or is the effect of nucleotide pool sanitation by the MTH (Mut T Homolog) directed pathway (see also Nucleotide pool sanitation chapter). However, oxidatively damaged DNA bases probably appear in the urine as a consequence of the base excision repair (BER) activity [60, 61]. In our studies we have found that urinary excretion of 8-oxoGua and 8-oxodG does not depend on diet in the case of humans and may reflect involvement of different repair mechanisms (respectively BER and NER) [62].
Since the level of the modified nucleosides/ bases in urine may be a general marker of oxidative stress, we investigated whether the amount of 8-oxoGua and its nucleoside form (8-oxodG) excreted into urine was higher in cancer patients than in the control group. The amount of the modified base, but not the nucleoside, excreted into urine was found to be approximately 50% higher in cancer patients suffering from lung, breast or prostate cancer than in the control group [63].
The higher level of 8-oxoGua in the urine of cancer patients may be explained by the reported oxidative stress in cancer tissue [37, 63-65]. However, the amount of the modified base/ nucleoside excreted into urine should represent the average rate of DNA damage in the whole body [56, 59]. It is doubtful that the elevated level of the base product in cancerous cells alone could account for the observed 50% increase of 8-oxoGua in urine. Our results suggest rather that oxidative stress, represented by the increased amount of the compound in urine, may be characteristic not only for the diseased tissue but also for some other tissues (or the whole organism) of cancer patients. Although the precise mechanism(s) of oxidative stress is still unknown, it has been recently documented that cancer patients showed signs of extensive granulocyte activation with a release of reactive oxygen species followed by a dramatic increase of 8-isoprostane, one of the biomarkers of oxidative stress [66]. Malignant cells have also been found to produce hydrogen peroxide at levels as high as those characteristic for stimulated polymorphonuclear leukocytes [67]. Therefore, one of the reasons for the observed oxidative stress in advanced stages of cancer may be the release of a large number of cancer cells into the blood stream [68] and their penetration into other tissues. Another reason for the observed phenomenon could be that some tumors may stimulate the defense systems of the body so that they react against the tumor to produce cytokines [69]. Some of the cytokines can be responsible for ROS production [70, 71]. On the other hand, it is also possible that a prooxidant environment is characteristic for advanced stages of cancer and that oxidative stress is rather a result of the disease development.
Cancer tissues
Elevated levels of typical free radical-induced DNA base modifications have been found to exist in human cancerous tissues when compared with the cancer-free surrounding tissue [63, 64, 72, 73]. The quantity of ethenoadducts in precancerous tissues was observed to increase in comparison to unaffected tissues. Polyps from FAP patients contain about twofold higher levels of εA and εC [74]. Significantly increased amounts of the abovementioned lesions have been reported in chronic pancreatitis, ulcerative colitis and Crohn's desease, which all are inflammatory disorders that present an elevated risk of cancer development [20].
It is not known whether these elevated levels of DNA lesions play a causative role in carcino-genesis or are merely the result of the disease. However, a treatment of laboratory animals with carcinogenic agents causes a similar pattern of oxidative base modification in their target organs before tumor formation occurs [75].
Our recent investigations of benign tumors showed that oxidative DNA damage might be a causative factor in cancer development. A higher endogenous level of 8-oxoGua in uterine myoma tissues was observed when compared to their respective tumor-free tissues [76]. One of the factors that may predispose to malignant transformation is the greater size of the tumor [77]. We have found a positive correlation between the size of the tumor and the amount of 8-oxoGua [76]. This suggests that the higher level of 8-oxoGua and possibly other base lesions may be a risk factor that can determine the transformation of benign tumors to malignant tumors. Conversely, the increased levels of modified DNA bases may contribute to the genetic instability and metastatic potential of tumor cells in fully developed cancer.
REPAIR OF OXIDATIVELY DAMAGED DNA
The major pathway to remove oxidized DNA bases and ethenoadducts is base excision repair. BER can be divided into five steps: (i) excision of the damaged base by the specific DNA glycosylase and formation of an apurinic/ apyrimidinic (AP) site; (ii) cleavage of the phos-phodiester bond at AP site by AP-endonuclease or AP-lyase; (iii) removal of chemical groups interfering with gap filling and ligation; (iv) gap filling; (v) ligation [78]. The first step of the BER pathway, recognition and excision of the damaged base by the specific DNA glycosylase, may be greatly influenced by the second BER pathway enzyme, AP endonuclease. The major human enzyme APE1 in vitro stimulates excision of 8-oxoGua and eC up to 400 fold by increasing enzyme turnover on damaged DNA [79, 80]. Other proteins that may affect the excision rate of oxidative DNA lesions are: XRCC1 (a platform protein, which is recruited to the site of damage by several DNA glycosylases and stays till ligation, regulating consecutive stages of the BER, PARP1 (polyADP ribose polymerase), which binds to free DNA ends and protects them against degradation, participates in chromatin relaxation and modulates binding of repair proteins to the site of damage by interaction with poly(ADP-ribose) chains [81-83]), PCNA (proliferating cell nuclear antigen, DNA polymerase processivity subunit in LP-BER), RFC (replication factor C, loading PCNA on DNA), WRN helicase (deficient in Werner syndrome, a premature aging disease [84]) or CSB (helicase/3’exonuclease, deficient in Cockayne syndrome, a neurodevelopmental and premature aging disease [85]).
System “GO” for 8-oxoGua excision from DNA
Repair of 8-oxoGua is a multistep process, which includes the activity of three proteins MTH, OGG1 and MYH acting in the so called “GO” system (Figure 2). MTH is pyrophospohy-drolase, which eliminates 8-oxodGTP from the cellular nucleotide pool and prevents its incorporation into DNA. When 8-oxoGua arises via DNA base oxidation, it is removed by the BER glycosylase OGG1 that excises 8-oxoGua paired with C or T. If repair is not completed before replication, replicative DNA polymerases frequently incorporate dA opposite 8-oxoGua, which results in GC→TA substitutions. Mispairs 8-oxoGua:A may be repaired via elimination of dA by MYH glycosylase. Subsequently DNA polymerase β fills the gap in the DNA strand with dCTP and generates an 8-oxoGua:C pair that can be processed by OGG1, leading to restoration of the initial G:C pair. If 8-oxoGua is incorporated into DNA from the nucleotide pool, and paired with A, it can be excised by two proteins: OGG2 and NEIL2. However, mechanisms of distinguishing between template and new synthesized DNA strand by repair glycosylases haven’t been proposed yet.
Figure 2.

“GO” system
Nucleotide pool sanitation
All cells from bacteria to humans are equipped with phosphohydrolases that hydrolyse triphosphates of damaged nucleotides to monophosphates. Nucleotide monophosphate kinases can discriminate between damaged and unchanged nucleotides, and damaged nucleotides are not recirculated to the cellular pool of nucleoside triphosphates, but instead they are dephos-phorylated by nucleotidases and extruded from the cell, which prevents their incorporation into DNA by DNA polymerases [86].
A role of MTH1 protein (8-oxodGTPase)
In E.coli the MutT protein, is a pyrophosphohy-drolase (i.e., 8-oxodGTPase) that hydrolyzes 8-oxodGTP to 8-oxodGMP and inorganic pyrophosphate, thus eliminating this damaged dGTP from the dNTP pool and preventing it from being incorporated into the DNA [87]. An Escherichia coli strain carrying a knockout mutation in the mutT gene coding for this enzyme demonstrates a very strong mutator phenotype [88] characterized by at least 1000-fold increase in the frequency of AT → CG point mutations [89, 90]. Maki and Sekiguchi found that MutT protein most effectively hydrolyzes 8-oxodGTP thereby preventing its incorporation into DNA during DNA replication [87]. Thus, an antimutagenic function of the MutT protein has been attributed to decomposing 8-oxodGTP which otherwise may cause AT → CG transversions. Mammalian homologues of the bacterial mutT gene have been cloned, characterized, and designated MTH1 genes (mutT homologue 1) [91]. Trans-fection of the human MTH1 gene into mutT- E. coli resulted in partial reversal of the mutator phenotype [92], and transfection of mouse and rat genes resulted in a complete reversal [93, 94] of the high AT → CG point mutation rate, typical for incorporational mutagenicity of 8-oxodGTP observed in these mutants. Therefore, the mammalian MTH1 proteins coded by these genes have been proposed to play the same role in sanitizing free nucleotide pools. Indeed, human [95], mouse [93, 96], rat [94, 97], and hamster [98, 99] MTH1 proteins are nucleoside 5′-triphosphate pyrophosphohyrolases that very effectively decompose 8-oxodGTP. This is why these mammalian enzymes are most frequently called 8-oxodGTPases. Although hMTH1 de-composes most effectively 2-hydroxy-2′-deoxyadenosine 5′-triphosphate (2-OH-dATP) [100], 2-hydroxyadenosine 5′-triphosphate (2-OH-ATP) [101], 8-oxodGTP [95], and 8-oxo-2′-deoxyadenosine 5′-triphosphate (8-oxodATP) [100], it is also capable of hydrolyzing less effectively 8-oxoguanosine 5′-triphosphate (8-oxoGTP) [102], 8-chloro-2′-deoxyguanosine 5′-triphosphate (8-Cl-dGTP) [103] and canonical deoxyribonucleoside and ribonucleoside 5’-triphosphates, such as dGTP [95]. Four isolated forms of hMTH1 protein (p18, p21, p22 and p26) demonstrate equal activity towards 8-oxodGTP [104]. Human 18-kDa 8-oxodGTPase was shown to be located mostly in cytosolic and mitochondrial soluble fractions [105], although rat tissues also revealed an apparent nuclear localization of the MTH1 protein [106].
MTH1 and carcinogenesis
Recent models of cancer development assume a formation of the mutator cell in the early stage of the cell transformation. A knockout of the MTH1 gene was anticipated to generate a mammalian mutator cell. However, such a knockout mutation in the MTH1 gene has not been discovered in mammalian cancer cells. Surprisingly, instead of that, a characteristic overex-pression of MTH1 has been noticed in different cancer cells and tissues as compared to their healthy counterparts [107-111]. Since this over-expression in the cancer cells was most frequently assigned to a state of persistent oxidative stress in these cells, it has also been proposed that MTH1 overexpression might be a marker of oxidative stress [107, 109]. Although an induction of MTH1 expression by high concentrations of hydrogen peroxide [111] and the free radical-generating crocidolite asbestos [112] has been recently demonstrated, different aspects of MTH1 gene expression regulation in normal and cancer cells still remain unclear.
A better insight into the role of the MTH1 protein has been acquired by the creation of MTH1 nullizygous cell lines and mice [113]. These transgenic mice, defective in MTH1 gene and devoid of 8-oxodGTP pyrophosphohydrolase activity, demonstrated higher incidence of lung, liver, and stomach cancers accompanied by a 2-fold increase in spontaneous mutation frequency in the Hprt gene, as compared to wild type mice [114]. Nevertheless, this slightly higher mutation rate has not been confirmed in a more recent study that showed the same level of mutation frequency in the rpsL reporter gene of E. coli, introduced into both MTH1+/+ and MTH1-/- backgrounds [115].
In our recent study we have found that the 8-oxoGua level in human DNA is determined not only by its excision rate, but also by the frequency of its incorporation from the oxidatively modified nucleotide pool into DNA by DNA polymerases, and the latter may be the most important contributor [116]. When studying the 8-oxoGua level in DNA, OGG1 repair activity and MTH1 activity in tumors and surrounding lung tissue, without histological changes (normal lung) of lung cancer patients, we found that the 8-oxoGua level was lower in tumor than in normal lung tissue, OGG1 activity was also lower in tumor, but MTH1 activity was higher in tumor than in normal lung. The activity of MTH1 was three orders of magnitude higher than that of OGG1. This great difference can be attributed mostly to differences in the turnover of these enzymes, since the expression of MTH1 and OGG1 mRNAs is similar [117]. The role of MTH1 protein is further highlighted by the observation that overexpression of MTH1 protein in mismatch repair deficient cell lines decreased the mutation rates to normal and reduced microsatellite instability which was accompanied by reduction of the 8-oxodG level in DNA [118]. Also the expression levels of MTH1 mRNA were inversely proportional to the levels of 8-oxodG in DNA in 11 human lung cancer cell lines and SV-transformed non-tumorigenic human bronchial epithelial cells [109]. The higher activity of 8-oxodGTPase also coincided with lower back-ground levels of 8-oxodG in DNA of fetal compared with maternal mouse organs [96].
BASE EXCISION REPAIR AS A RISK FACTOR IN CARCINOGENESIS
Functional studies
Epidemiological studies suggest that the etiology of some types of human cancers is closely related to chronic inflammations and infections. We have studied repair of oxidative DNA damage in lung cancer patients. Lung cancer is the most frequent cancer type all over the world [119, 120]. In the Caucasian population approximately 80 % of lung cancer cases are caused by tobacco smoking [119, 120]. To-bacco smoke contains over 4,000 compounds, and generates the formation of ROS as well as chronic lung inflammation [121]. ROS and RNS can cause lipid peroxidation, oxidation of DNA and protein thiols, as well as protein nitrosylation [15].
We have investigated repair of three oxidative DNA lesions, 8-oxoGua and two, which were induced by interaction of lipid peroxidation products with DNA, 1,N6-ethenoadenine (εA) and 3,N4-ethenocytosine (εC). These DNA lesions show high miscoding potential in mammalian cells [16, 17, 122] also inducing recombination and chromosomal aberrations [18].
In humans the major 8-oxoGua DNA glycosylase is OGG1 [123], while εA is eliminated from DNA by alkylpurine-DNA-N-glycosylase (ANPG) [124] and εC by mismatch specific thymine-DNA-glycosylase (TDG) [125]. Both latter enzymes are monofunctional DNA-glycosylases and require AP-endonuclease to incise DNA at the site of the removed base, while OGG1 is glycosy-lase/AP-lyase. OGG1 AP-lyase activity is, however 10-fold lower than that of N-glycosylase, so strand incision is mostly dependent on the availability of APE1 [79, 80].
TDG excises εC from the whole genome [125]. CpG sites are additionally processed by MBD4 glycosylase, which removes εC and T from G:T mismatches specifically from CpG sites [126]. It has also been suggested that ethenoadducts may also be repaired via oxidative dealkylation catalyzed by AlkB type protein(s) [127]. AlkB contribution to overall repair yield of this type of DNA damage has not been elucidated.
We have measured the level, and repair rates of 8-oxoGua, εA and εC in blood leukocytes of lung cancer patients, as well as in blood leukocytes of healthy individuals, matched with cancer patients for age, gender and lifestyle habits.
When comparing repair activities between cancer patients and controls, we have observed that repair capacity for 8-oxoGua and εA was significantly lower in blood leukocytes of lung cancer patients than in leukocytes of healthy volunteers [128, 129]. Studies of Livneh and coworkers also demonstrate decreased 8-oxoGua repair capacity in lung as well as in head and neck cancer patients [130, 131]. Consistently, the 8-oxoGua level in DNA from leukocytes of cancer patients was higher than that in healthy controls. Urinary excretion of 8-oxoGua was higher in smoking individuals, regardless of their health status, than in non-smokers. Since oxidatively-generated DNA insults represented by urinary excretion of oxidatively-derived DNA lesions was similar in both groups of subjects with similar smoking status, it appears likely that a higher rate of oxidative damage generation in cellular DNA of lung cancer patients is the result of the deficiency of repair mechanism(s) in this group.
Lung adenocarcinoma (AD) is a histological type of cancer whose etiology is linked to prolonged inflammation and healing of scars [132]. Repair activities for εC were lower than in healthy volunteers only in individuals with AD. Also the difference in εA repair rate between healthy volunteers and cancer patients was much greater for AD than for all cancer patients (Figure 3) No differences were observed in repair rates of 8-oxoguanine. This suggests that the etiology of lung adenocarcinoma may be related to inefficient repair of exocyclic DNA adducts, derived from lipid peroxidation (LPO). In only two other studies LPO has been linked to progression of lung cancer [133, 134]. However, this suggests that the development of different histological types of lung cancer occurs by different pathways, and that in the development of lung ade-nocarcinoma the deficiency of repair of lipid peroxidation derived DNA damage is one of the risk factors.
Figure 3.

Base excision activity toward ethenoadducts: (A,C) εA; (B,D) εC in lung cancer patients. SQ squamous cell carcinoma; AD – adenocarcinoma.
These functional studies show that decreased efficiency of BER to eliminate from DNA oxidatively-generated DNA lesions, 8-oxoGua, εA and εC may be a risk factor for the development of lung and other cancers. Molecular mechanisms responsible for this phenomenon are, however, only partially elucidated, and may include repair genes polymorphism, transcriptional activation/ down-regulation of specific repair genes by inflammatory processes and certain nutrients, post-translational modifications of repair enzymes and possibly other factors.
Polymorphism of Base Excision Repair genes
Several polymorphisms of DNA glycosylases responsible for excision of 8-oxoGua are known, and their presence in human genomes has been linked to the risk of developing specific types of cancers. It has been suggested that polymorphism in DNA repair genes may be associated with differences in the repair efficiency of DNA damage [135]. Some studies suggest involvement of the hOGG1 polymorphism in several human cancers [136].
Few hOGG1 polymorphisms have been described and the most common Ser326Cys. 326 Cys allele is found in approximately 40% of the Caucasian population. 326Cys OGG1 binds DNA lesions with significantly lower affinity and excises 8-oxoGua from duplex DNA and cleaves abasic sites at rates 2- to 6-fold lower than the wild-type enzyme, depending upon the base opposite the lesion. In contrast to the wild type enzyme the OGG1 326 Cys variant binds damaged DNA as a dimer, both in vitro and in human cells and is not significantly stimulated by the presence of AP-endonuclease [137], and it is not relocalized to nucleus during the S-phase of the cell cycle [138]. The altered substrate specificity, lack of stimulation by AP-endonuclease 1 (APE1) and anomalous DNA binding conformation of 326Cys OGG1 may be responsible to its linkage to cancer incidence [137]. A recent finding of Bravard and coworkers [139] indicates that the OGG1 Cys variant is more sensitive to inactivation by oxidizing agents than the Ser variant, and a reducing environment may restore excision activity of the Cys variant to that of Ser one. This might suggest that under oxidative stress individuals with the OGG1 326 Cys/Cys genotype may be under increased risk of cancer development.
It was suggested that the presence of two hOGG1 326Cys alleles confers a 2-fold increased risk of lung cancer [140, 141], and also an elevated risk of prostate cancer and naso-pharyngeal carcinoma [142, 143], but not of colon cancer [144]. However, the results of other studies are contradictory. The 8-oxoGua excision rate probably depends on many other factors, since excision activities in human lymphocytes were reported not to be affected by the polymorphic status at codon 326 of the hOGG1 gene [145]. Since excision activity of OGG1 glycosylase may depend on several protein interactions among partners of the BER pathway, e.g. XRCC1 and APE1, further studies are needed to determine whether the polymorphism of Ser326Cys of hOGG1 is a significant risk factor for human cancers associated with oxidative DNA damage.
Less frequent OGG1 polymorphisms are Arg46Gln and Arg154His [146]. Both variants were found in human lung and gastric cancers, respectively, and the latter in blood leukocytes. Both have a reduced activity for excision of 8-oxoGua. In addition, the Arg 154 change to His relaxes the OGG1 requirement for a pyrimidine opposite 8-oxoGua. Since replicative DNA polymerases readily incorporate A opposite 8-oxoGua, such a change in substrate specificity confers a mutator character on this OGG1 variant. Due to the rarity of both polymorphisms their relation with human cancers was not established.
Bacterial MutY glycosylase and its human homolog hMYH excise adenine incorporated into DNA opposite 8-oxoGua by replicative DNA polymerases. In bacteria lack of MutY protein increases the spontaneous mutation rate 1000-fold, indicating the enzyme's role in correcting replicative errors [147]. In humans alternative splicing gives type 1 (535 amino acids) protein, which localizes in mitochondria and type 2 (521 amino acids) lacking the mitochondrial transport signal and localizing in nucleus. Seven different polymorphisms of the hMYH gene are known [146]. Polymorphisms Gln324His of type 1 protein and Gln310His of type 2 do not change the enzyme activity. Two other base sub-stitutions Gly328Asp and Tyr165Cys diminish the glycosylase activity towards 8-oxoGua:A and are risk factors for colorectal tumors. These hMYH polymorphisms in colorectal tumors were associated with GC→TA transversions in the APC gene [146]. Two nonsense mutations, Tyr90 to stop and Glu466 to stop are also associated with a possible risk of colorectal tumors. Two intronic polymorphisms were also found: G/ C in intron 1, which induces alternative splicing and reduces translation efficiency and A/G in intron 10, inducing production of truncated protein, which is not localized in the nucleus.
Surprisingly, very few polymorphisms were found in human genes coding for DNA glycosy-lases excising exocyclic DNA adducts εA (ANPG) and εC (TDG). The ones which were found are rare and do not change the efficiency of excision [148, 149].
Several sequence variants were identified in the APE1 gene, the major ones are Gln51His, Ile64Val and Asp148Glu. Asp148Glu was associated with hypersensitivity to ionizing radiation [150], and the presence of Ile64Val decreased lung cancer risk [151]. Association of the Asp148Glu polymorphism with lung and other cancers has not been demonstrated [152, 153].
More than 60 validated single nucleotide polymorphisms in the XRCC1 gene are listed in the Ensembl database. The most extensively studied are three genetic changes Arg194Trp, Ar-g280His, Arg399Gln [152]. The XRCC1 Arg399Gln genotype was linked with increased risk of tobacco-related cancers among light smokers, but decreased risk among heavy smokers. There are also controversial data on association of XRCC1 Arg280His and Arg399Gln polymorphisms with increased levels of benzo[a]pyrene and other DNA adducts as well as increased frequency of chromosomal aberrations [19, 152], a biomarker of cancer risk.
Extensive search for SNPs revealed that cancer risk may be increased in individuals bearing not one, but multiple polymorphisms in DNA repair genes, which if present separately have no effect on the frequency of cancer development. For example simultaneous presence of APE1 Asp148Glu and XRCC1 Arg194Trp polymorphisms increase the risk of pancreatic cancer, while each of these variants separately has no effect [153].
Transcriptional regulation of BER
Transcription of BER genes is regulated in the cell cycle and may also be affected by increased oxidative stress [154]. The mRNA level of al-kylpurine-DNA-N-glycosylase (ANPG), human thymine glycol-DNA-glycosylase (NTH), uracil-DNA-glycosylase (UDG), and human AP-endonuclease (APE) increase 2.5 – 3.5-fold during the G1 phase of the cell cycle, remain constant during the S phase, and decrease to the basal level after mitosis. In contrast, expression of the hOGG1 gene is not regulated during the cell cycle [155].
BER genes may be also activated by hydrogen peroxide and other ROS. Oxidative stress increases hMTH1 mRNA expression 2-3-fold and enzyme activity in cultured human fibroblasts [111], as well as hOGG1 mRNA and 8-oxoGua excising activity [112]. Transcription of the major human AP-endonuclease, APE1 is also augmented in response to ROS [156]. Since APE1 stimulates in vitro excision activity of OGG1 and TDG glycosylases several-fold [79, 80] stimulation of APE1 transcription will also stimulate 8-oxoGua and εC excision rate. Thus, inflammations and infections may exert a stimulatory effect on DNA repair by stimulating transcription of the BER system genes [157].
Cancer patients are usually characterized by increased oxidative stress, and one of the reasons is the depletion of antioxidant vitamins in these individuals [158]. Wilson and coworkers [159] showed that repair of 8-oxoGua was induced 5-6-fold by simultaneous treatment of cells with ascorbate and α-tocopherol. This increase was accompanied by the increase of the level of DNA polymerase β and this could result from induced de novo synthesis of the enzyme. Dietary vitamins intake and/or individual vitamin absorption limitations may then influence an individual's oxidative DNA damage repair capacity. Additional studies are necessary to elucidate the importance of antioxidant vitamin intake/absorption for repair of oxidative DNA damage in the whole organism.
Post-translational modifications of BER proteins
Several model studies demonstrate that repair activity of BER enzymes may be modulated by post-translational modifications, both non-enzymatic and enzymatic. The major enzymatic modifications include phosphorylation, acetylation and sumoylation.
OGG1 is phosphorylated by protein kinase C, Cdk4 and c-Abl kinases at several positions. Ser 326 phosphorylation triggers relocalisation of OGG1 from the cytoplasm to nucleoli during S-phase, but does not affect the enzyme's excision activity [138]. Phosphorylation by Cdk kinase increases 8-oxoGua excision rate over 2-fold [123]. Phosphorylation of hMYH increases excision rate of A from 8-oxoGua:A pair. Defective phosphorylation of wild type hMYH was observed in colon cancer cell lines [160]. Phos-phorylation of UNG2 glycosylase at Thr6 and Thr126 occurs 2 hrs after UV irradiation of mammalian cells and increases enzyme activity to remove from DNA uracil formed by UV-induced cytosine demination. Dephosphorylation is catalysed 8-10 hrs after UV treatment by TP53-induced magnesium dependent protein phosphatase 1D [161].
Acetylation by p300 protein is a common mechanism regulating the activity of several BER proteins, namely OGG1, TDG, NEIL2, polβ and APE1. OGG1 acetylation (Lys41 and Lys338) decreases OGG1 affinity for AP-sites and increases enzyme turnover, as well as increases its stimulation by APE1 [162]. In HeLa cells about 20% of OGG1 molecules are acetylated. The level of OGG1 acetylation is doubled upon oxidative stress. Acetylation of TDG occurs in an enzyme region responsible for interaction with APE1 and abolishes its stimulation by APE1, but not DNA binding [163]. TDG acetylation releases it from the complex CBP/p300 and may play the role of a molecular switch between two functions of TDG protein, DNA repair and the role of a transcription factor [163]. Acetylation of NEIL2 at Lys49 significantly decreases glycosylase and AP-lyase activity of the enzyme [164]. Acetylation of polβ at Lys47 decreases its dRP-lyase activity, but not DNA polymerase activity [165]. It is speculated that polβ acetylation may play a role in BER directing towards SP- or LP-BER.
Sumoylation is an important mechanism changing conformation and activity of TDG glycosy-lase. Unsumoylated TDG has high affinity for DNA containing G:T and G:U mismatches as well as εC and excises T, U and εC from DNA. The enzyme also has high affinity to AP-sites and does not leave the reaction product. Binding of SUMO1 and SUMO2/3 proteins at Lys 330 in C-terminal domain of the TDG glycosylase changes enzyme conformation in the N-terminal part. Change of conformation decreases enzyme affinity for DNA and facilitates its stimulation by APE1 [166].
The major known non-enzymatic modification of repair proteins is nitrosylation. Exogenous nitric oxide and peroxynitrite were shown to inhibit OGG1 [167], DNA ligase [168] formami-dopyrimidine-DNA-glycosylase [169] and O6-alkylguanine-DNA-alkyltransferase by direct ni-trosylation [170]. Inflammatory processes may then, on the one hand stimulate transcription, and on the other, directly inactivate some repair enzymes.
Although currently nothing is known whether the above mentioned modifications play a role in carcinogenesis induction or progression, such mechanisms cannot be excluded since the level of MutY protein phospohorylation can affect its activity within the cell. MutY is adenine glycosylase that removes adenines from A:8-oxoGua mispairs. Experiments using colon cancer cell lines which did not exhibit mutations in the MutY gene, showed that defective repair of A:8-oxoGua may be at least in part the consequence of alterations in endogenous phosphorylation of the MutY protein [160].
The influence of post-translational modifications of BER enzymes on repair efficiencies in the whole organism awaits further research.
DNA REPAIR IN CANCER TISSUES
Cancer cells are characterized by increased oxidative insult and great genomic instability, which results in changed metabolism, cell cycle frequency and loss of heterozygosity (LOH). We have analyzed the DNA damage level and the rate of εA, εC and 8-oxoGua repair in lung tumours and unaffected lung tissues from lung cancer patients. No difference in εA and εC level between tumor and unaffected lung was recorded, however, a significant increase in the excision rate of these two modified bases was observed in the tumor tissue, suggesting that oxidative stress is increased in cancer cells and that repair mechanisms may compensate it. Similarly, in colon benign adenomatous polyps the level of εA and εC was augmented, reflecting increased oxidative stress in disease development. However, disease progression to carcinoma is accompanied by a drastic decrease of the ethenoadduct level in DNA of tumors, even below that in unaffected colon [171]. Tumors have a higher content of cells in the S-phase. Expression of some DNA-glycosylases and AP-endonuclease genes was shown to be cell cycle dependent [155]. The mRNA levels of al-kylpurine-DNA-N-glycosylase (ANPG), human thymine glycol-DNA-glycosylase (NTH), uracil-DNA-glycosylase (UDG), and human AP-endonuclease (APE) increase 2.5 – 3.5-fold during the G1 phase of the cell cycle, remain constant during the S phase, and decrease to the basal level after mitosis. However, expression levels of the TDG gene is not regulated during the cell cycle [155]. It is thus possible that the increase of εC repair capacity in tumors was due to an increase in AP-endonuclease expression during the S phase of the cell cycle [155]. The enzyme transcript levels have been found to be elevated in a number of cancers [154, 172-174].
Surprisingly we observed decreased 8-oxoGua excision activity in tumor lung tissue in comparison with unaffected surrounding areas. The mechanism of this decrease is not known, but probably is not related to mutations in the OGG1 gene in the tumor, since they have been found in only 4% of human kidney cancers, and were also sporadic in lung cancers [175]. Some studies have shown frequent allelic loss in cancer tissue of chromosome fragments in the position, in which the OGG1 gene is located. Accordingly, a decrease of OGG1 expression was observed, e.g. in head and neck squamous cancer cases [176]. However, loss of heterozygosity in the OGG1 locus may vary between cancer types. No differences in OGG1 expression were observed between tumor and unaffected surroundings in human lung and kidney cancers [175]. In model systems OGG1 activity is stimulated by at least two proteins, APE and XRCC1 [79, 177]. Deregulation in tumor tissue of OGG1 cooperation with downstream partners of the BER pathway cannot be excluded, although APE expression was shown to increase in tumors.
The decrease of OGG1 activity may also be tumor-specific, driven by the loss of OGG1 activators in tumor tissue. One such gene may be a tumor suppressor protein – tuberin. In tuberin deficient Ekert rats, which spontaneously develop renal cancers, OGG1 expression and activity was reduced 3-fold [178]. Tumor-specific regulation of 8-oxoGua excision activity may also be due to mutations in the tumor suppressor gene TP53. It was recently shown that TP53 plays a central role in the cellular response to genotoxic stress and is associated with the DNA repair machinery which involves base excision repair (BER). In TP53 temperature-sensitive (ts) mutants of murine and human origin cell extracts overexpressing TP53 were found to exhibit an augmented BER activity measured in an in vitro assay. Depletion of TP53 from the nuclear extracts abolished this enhanced activity [179]. TP53 may also interact directly with the BER complex. For example it was found that recombinant TP53 protein stimulated an in vitro reconstituted BER assay, potentially by binding APE-1 and regulating DNA polymerase β (pol β) loading onto AP-sites [180]. On the other hand TP53 may also regulate genes involved in BER [181]. TP53 null cells treated with the base damaging alkylating agent, MMS exhibited slow BER, as measured in vivo using an alkaline comet assay. In this experimental system, pol β protein levels correlated with wild type TP3 status, though APE1 levels and activity were unaffected. In fact, previous studies have identified polβ as a DNA damage inducible gene [182], thus raising the possibility that it is tran-scriptionally regulated by TP53.
Another signaling protein that might regulate repair of oxidative DNA damage in cancer tissues is APC protein. APC tumor suppressor protein inhibits long patch BER (LP-BER, e.g. engaged in repair of εA and εC) via direct interaction with DNA polymerase β and FEN-1 endonuclease [183]. BER activity was inversely associated with APC expression in several breast cancer cell lines [184]. Since mutations in the APC gene are early events in colorectal carcinogenesis, and they are found in about 37 % of colon tumors [185], this mechanism may be important in regulating repair of ethenoadducts in colon tumors. Over-stimulating and unbalancing of BER may in turn favor genomic instability and in consequence cancer progression. It was shown that in tissues from non-cancerous colons of ulcerative colitis patients, methylpurine-DNA glycosylase (ANPG) and abasic site endonuclease (APE1) were significantly increased, and microsatellite instability (MSI) was positively correlated with their imbalanced repair enzymatic activities. These latter results were supported by mechanistic studies using yeast and human cell models in which overexpression of Mpg and/or Ape1 was associated with frameshift mutations and MSI [186].
Unbalanced expression of DNA repair enzymes may also affect cellular availability of signaling molecules. APE1 is a redox factor for several transcription factors including AP-1, HIF1-alpha, and TP53 [187]. Thymine-DNA glycosylase (TDG) also stimulates transcription of TP53 family proteins, TP53 and TP73, and stimulates growth repression, mediated by these proteins [188]. So overexpression of some BER enzymes, like APE1, on the one hand will stimulate genome instability, and on the other will repress cell growth to enable DNA repair.
OXIDATIVE PROCESSES IN AGING
Lipid peroxidation as an endogenous source of degeneration and aging
Lipid peroxidation is implicated in aging as well as in the pathogenesis of numerous human diseases, including atherosclerosis, cancer, diabetes and arthritis [189]. A significant increase in the LPO level was found in skeletal muscle of old individuals [190], as well as in hepatocytes isolated from old ovariectomized rats [191].
Peroxidation of cellular membrane lipids, or circulating lipoprotein molecules generates highly reactive aldehydes among which one of most important is 4-hydroxynonenal (HNE, Figure 4). The level of HNE is increased in brain tissue and cerebrospinal fluid of Alzheimer disease patients, and in the spinal cord of amyotrophic lateral sclerosis (ALS) patients. Increased levels of HNE in neurodegenerative disorders and immunohistochemical distribution of HNE in brain tissue indicate the pathophysiological role of oxidative stress in these diseases, and especially HNE in formation of abnormal filament deposits [21].
Figure 4.

Examples of HNE and its epoxide adducts to deoxyguanosine.
Physiological concentrations of HNE vary from 0.1-3 µM, and can increase up to 50 µM or even millimolar values under oxidative stress [192, 193]. Within cells, HNE binds primarily to thiols and proteins, depleting glutathione levels and forming protein-protein cross-links, which may accelerate formation of deposits [192]. Glutathione depletion by HNE is probably an important mechanism of aging. Lifespan and stress resistance of Caenorhabditis elegans are increased by expression of glutathione transferases capable of metabolizing HNE [194].
HNE also forms bulky adducts to DNA bases. These are exocylic propano- and etheno-type adducts, which bear six or seven carbon atom side chains (Figure 4) [195]. These adducts are relatively unstable, and may rearrange, forming DNA intra- and interstrand cross-links [196], as well as DNA-protein cross-links [197]. HNE-dG adducts were detected in DNA of unexposed humans and rodents, which indicates their endogenous origin [198, 199].
We have shown that HNE-DNA adducts block replication, trigger recombination, base substitutions and frameshift mutations in a model system, ssM13 phage [195, 200]. Other studies showed that in mammalian cells HNE increases the frequency of micronuclei, chromosomal aberrations, sister-chromatid exchanges [201-203] and point mutations [204], already at low, physiological concentrations of 0.1-10 µM. HNE also exerts a clastogenic effect in human cells, possibly via inactivation of the functional SH groups in DNA polymerases [205].
Several degenerative diseases are related to malfunctioning of DNA repair. One of such diseases is Cockayne syndrome (CS), which is characterized by traits reminiscent of normal aging, such as neurological degeneration, cataracts and systemic growth failure. The majority of CS cases are caused by defects in the CS complementation group B (CSB) protein. The CSB gene encodes a 168 kDa protein belonging to the SWI2/SNF2 protein family. The CSB protein participates in both sub-pathways of nucleotide excision repair (NER), mainly in transcription-coupled repair (TCR), but also in global genome repair (GGR). CSB is also engaged in base excision repair (BER) of certain types of oxidative DNA damage, e.g. 8-oxoGua [206], in the poly(ADP-ribose) polymerase-1 (PARP-1) mediated response to oxidative DNA damage [207], in strand annealing and exchange, which might be engaged in mitotic recombination [208], as well as in general transcription.
We have found [209] that human CSB-deficient cells are hypersensitive to physiological concentrations (1-10 µM) of HNE, and in response to HNE they develop a higher level of sister chromatid exchanges in comparison to the wild type cells. We have also demonstrated that HNE -DNA adducts block in vitro transcription by T7 RNA polymerase, as well as by HeLa cell-free extracts. Transcription inhibition leads to stabilization of TP53 protein and, thus, triggers apoptosis [210]. This might at least partially explain accelerated aging and degeneration in CS patients. Treatment of wild type cells with low HNE concentrations, 1-20 µM, caused dephosphorylation of the CSB protein, which stimulates its ATPase activity necessary for TCR. However, high HNE concentrations (100-200 µM) inhibit in vitro CSB ATPase activity as well as the transcription machinery in HeLa cell-free extracts. These results suggest that HNE-DNA adducts are extremely toxic endogenous DNA lesions, and that their processing involves CSB. When these lesions are not removed from the transcribed DNA strand due to CSB gene mutation or CSB protein inactivation by high, pathological HNE concentrations, they may contribute to accelerated aging.
Inhibition of removal of UV dimers and benzo[a] pyrene adducts from DNA (performed mainly by the NER pathway) by the products of lipid peroxidation, HNE, malondialdehyde and acrolein was already reported [211-213]. What is the contribution of this mechanism in aging process on the level of the whole organism still is not clear.
Age-related changes in oxidative DNA damage in humans (see also Olinski R. et al. [214])
A number of research groups have reported the effects of aging on DNA oxidation in animal models [215, 216]. However, a summary of these kinds of studies shows no clear effect. It is difficult to explain why in some studies age-related increase in oxidative DNA damage was observed [217], whereas in others no effect was described [218]. It is possible that one reason for the discrepancies may be the reliability of the biomarkers used. Moreover, to date no comprehensive studies concerning age-related oxidative DNA damage in humans have been conducted.
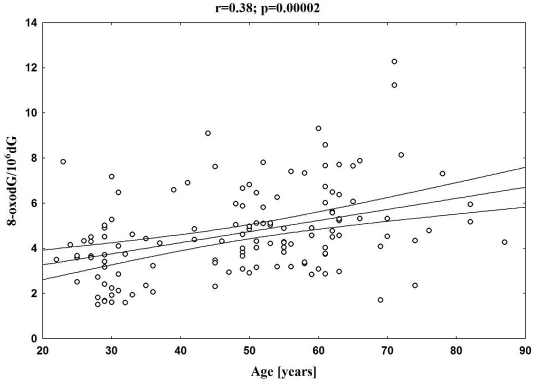
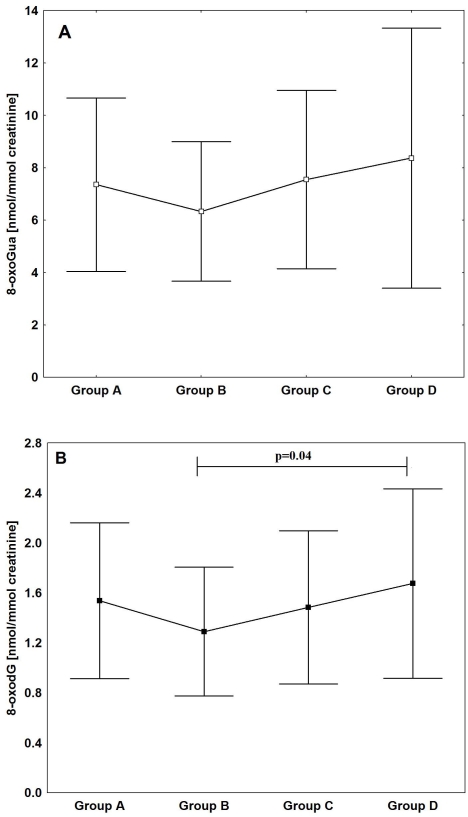
Therefore, the purpose of our recently published work [219] was to assess age-related changes in oxidative DNA damage in humans. For the first time, the broad spectrum of oxidative DNA damage biomarkers was analysed; urinary excretion of 8-oxodG and 8-oxoGua as well as the level of oxidative DNA damage in leukocytes. All parameters were determined in 255 healthy subjects divided into four age groups: group A - children (mean age 13), group B - adults (mean age 31), group C - middle age (mean age 50), and group D - elderly (mean age 67).
Antioxidant vitamins (A, C and E) and uric acid are effective free radical scavengers therefore they should protect biomolecules such as DNA. In addition to the aforementioned analyses the concentration of antioxidant vitamins A, C, E and uric acid was determined in blood serum.
There was a highly significant increase in the background level of 8-oxodG in leukocyte DNA in elderly and middle age groups in comparison with adults (Figure 5), and a statistically significant, positive correlation between age and 8-oxodG levels in leukocyte DNA.
Figure 5.
The mean level of 8-oxodG in leukocyte DNA in different age groups.
However, a steady increase of 8-oxodG levels in DNA isolated from leukocytes with age was seen only when the youngest group was excluded (Figure 6), since the level was significantly elevated in the group of youngest subjects (group A) when compared with the group representing “adults” (group B). It is likely that the unexpectedly high level of oxidative DNA damage in group A may reflect the higher metabolic rate of children. Children who are growing fast have a higher metabolic rate than adults. High metabolic rate, in turn, requires a high level of mito-chondrial respiration and subsequent elevated production of ROS, which are responsible for the formation of DNA modifications analyzed in our work. Indeed in our previous study highly significant, positive correlations between specific metabolic rates and urinary excretion rates for 8-oxodG and 8-oxoGua were found [220].
Figure 6.
Correlation between the level of 8-oxodG in leukocyte DNA and age with exclusion of group A.
The obvious question is why does oxidative DNA damage increase with age? The background level of 8-oxoGua in cellular DNA represents a dynamic equilibrium between the rate of oxidative DNA damage formation, and the rate of repair of the damage. Therefore, the observed age-related increase may be a result of deficiency in the ability of the cells from older subjects to remove the damage or it may mirror an intensification of processes responsible for the damage formation or both.
An age-related decrease in DNA repair capacity has been demonstrated mostly for nucleotide excision repair (NER) [221, 222]. However, base excision repair (BER) is primarily responsible for the removal of oxidatively-generated DNA base damage, and age-dependent reduction of hOGG1, the major enzyme involved in the removal of 8-oxoguanine, was also reported [223].
Urinary excretion rate, especially that measuring the level of 8-oxoGua is the most sensitive marker of the average oxidative stress to DNA of all body cells [224, 225]. Therefore besides analyses of the background level of 8-oxodG in leukocyte DNA also urinary excretion of the modified base and nucleoside was determined. Since both parameters showed a similar age-related pattern it is likely that their changes reflect, at least in part, age-dependent intensification of oxidative stress which resulted in DNA damage. However, since urinary excretion rates may also represent repair processes (see [226]), we cannot entirely exclude the possibility that the observed less distinct changes in age-dependent urinary excretion rates than of the background level of 8-oxodG in DNA (compare Figure 5 and 7A, 7B) may also reflect some deterioration of the repair mechanism(s) . Hence, age-related increase of oxidative stress appears to elevate oxidative DNA damage and the rate of repair represented by 8-oxoGua excretion although the activation of the repair process does not prevent accumulation of 8-oxodG in cellular DNA.
Figure 7.
The mean levels of 8-oxoGua (A) and 8-oxodG (B) in urine in the different age groups.
Impaired mitochondrial function is a factor which may be responsible for increased ROS production and therefore predispose to oxidative stress and DNA damage in the aged subjects. Indeed, several studies of the mitochondrial respiratory chain function in humans and animals have demonstrated an age-related decrease in respiration and increased production of ROS during aging [227, 228]. Further support for the age-related decline in mitochondrial function is provided by the demonstration that the amount of COX deficient muscle fibers increases in healthy aging humans [229].
Another source of age-related increase of oxidative stress may be the decline of antioxidant defense and age-dependent decline in the concentration of vitamin C in plasma was also observed (Figure 8). Vitamin C is a major aqueous-phase antioxidant. It should also be remembered that vitamin C acts in synergy with tocopherol by regenerating tocopheroxyl radical to tocopherol. One of the plausible explanations of the above-presented changes in vitamin concentrations is the sequential consumption of these antioxidants as a result of age-dependent intensification of oxidative stress. It was shown that during free radical mediated oxidation a decrease in vitamin E concentration in plasma can only be seen after the complete consumption of vitamin C. The sequential consumption of these antioxidants was also shown by the use of ESR spectroscopy [230].
Figure 8.
Relationship between the level of vitamin C and age in all studied subjects.
As can be seen in Figure 5 the “adult” group exhibits the lowest values of oxidative DNA damage in leukocytes. Evolution theory assumes that organisms are not programmed to age, instead evolution selects for survival and reproduction [231]. Therefore, it is possible that the lowest values of this harmful, potentially mutagenic, oxidatively-modified DNA in the aforementioned group may constitute proof of “specific concern” of evolution for humans of reproductive age. Individuals differ greatly in their rate of aging. There are also quite substantial inter-individual differences in the level of 8-oxodG in DNA (Fig 5 and 6). These differences can also be seen within the “adult” group with a subgroup where the values are around 2 modifications per 10 6 unmodified bases and a second subpopulation where the values are much higher than the mean level (Figure 5 and 6). It has been postulated that different factors which may affect the genome in adult life may influence the rate of subsequent functional decline of the organism [232]. Therefore, it is possible that one of these factors is oxidative DNA damage with genome destabilizing properties.
Why do different mammalian species age at different rates?
One of the intriguing issues concerning the aging process is the question why different mam-malian species age at different rates. One hypothesis that has attempted to explain these differences is once again the free radical theory of aging [233, 234]. All aerobic organisms utilize oxygen which is linked to the production of reactive oxygen species (ROS). The above mentioned differences may be explained, at least partially, by different metabolic rates that in turn are connected with oxygen consumption and ATP production during oxidative phosphorylation. The more ATP is required, the more oxygen must pass through the mitochondria and the more oxygen radicals are likely to be generated. The more oxygen radicals the greater will be the damage to cellular components including DNA. Oxidative DNA damage is removed via different repair pathways. Following excision from DNA, the oxidatively induced lesions are released into the blood stream and consequently into the urine, where their measurement has been acknowledged to be reflective of overall oxidative stress [235].
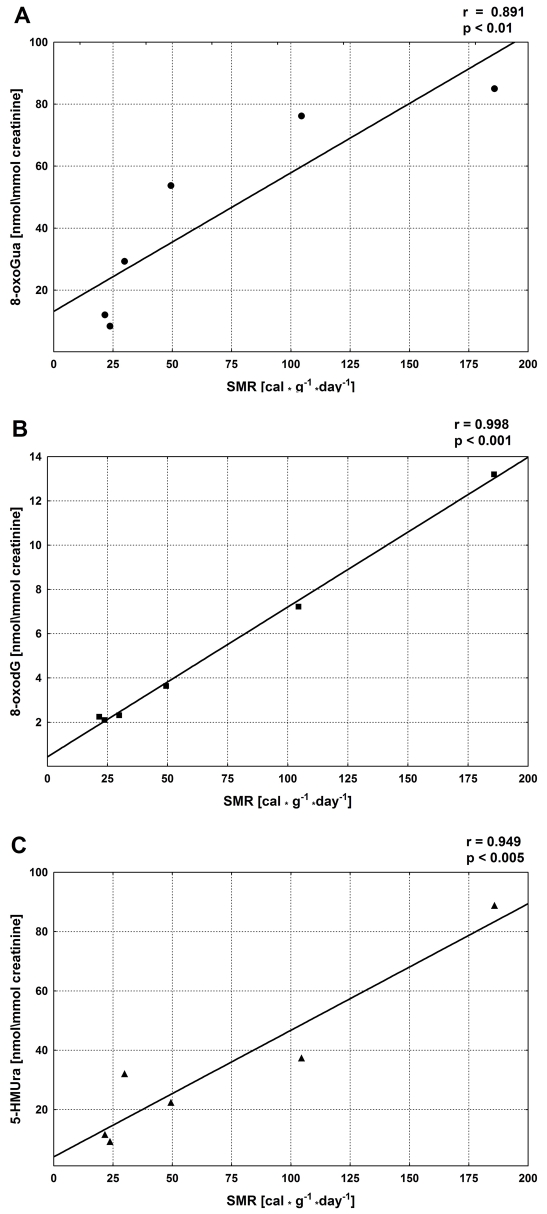
In our study we decided to analyze urinary excretion of possible repair products of oxidative DNA damage: 8-oxoGua, 8-oxodG and 5-(hydroxymethyl)uracil (5-HMUra), in mammalian species that differ substantially in metabolic rate and longevity, namely mice, rats, rabbits, dogs, pigs and humans [236].
The analyzed excretion rates should depend on oxygen consumption and metabolic rate. In turn, the metabolic rate may be described by specific metabolic rate (SMR) values [237, 238]. In agreement with these assumptions we have found good positive correlations between SMRs of different species and the excretion rates of all analyzed modifications (Figure 9).
Figure 9.
Relationship between the urinary excretion rates of the analyzed modifications and specific metabolic rates (SMR) of six different mammalian species.
Since metabolic rate may be associated with maximum life span (MLSP), we also determined whether there is some relationship between excretion rates of all analyzed modifications and the life span. Only 8-oxoGua excretion rate was found to significantly correlate with MLSP). 8-OxodG and 5-HMUra were also inversely correlated with MLSP. However, these relationships were not statistically significant (Figure 10). This in turn suggests that urinary excretion of 8-oxoGua reflects oxidative DNA damage better than the two other modifications. Likewise, in the case of cancer patients only urinary 8-oxoGua reflects oxidative stress associated with the disease [240, 241].
Figure 10.
Relationship between the urinary excretion rates of the analyzed modifications and natural logarithm of maximum life span (MLSP) of six different mammalian species.
The correlation of the excretion rate of 8-oxoGua with MLSP, found in our work, is in good agreement with previous studies, which demonstrated that oxidative damage to DNA is inversely related to MLSP of different mammals. However, in the aforementioned studies no humans were included and the assessment of DNA damage was restricted to certain organs [242-244]. In contrast, the analyses of the urinary base/nucleoside products presented in this work are reflective of oxidative DNA damage at the level of the whole organism. Our results demonstrated that ROS continually damage DNA and that this damage in vivo, in normal conditions is lower in long-lived species than in short-lived species. Incomplete repair of such damage would lead to its accumulation over time and eventually result in age-related deterioration.
Expression of the urinary excretion rates in nmol/kg/24h enables measurement of the number of the repaired lesions per day per cell [245]. Interestingly, urinary level of all measured modifications found in our study accounted for about 28,200 repaired events per average mouse cell per day and fits well with the estimation of Hamilton and co-workers who calculated that the DNA of the liver cell in mouse is exposed to about 47,000 8-oxoGua lesions in a 24 hour period [246] (taking into consideration that the liver is a high metabolic rate organ and that our values are an average for the whole organism). In contrast, the number of all lesions analyzed in our work, in humans accounts for about 2,800 repair events in the average cell per day. It is therefore possible that the high metabolic rate in mouse (or other short lived animals) may be responsible for severe everyday oxidative DNA insult that may be accumulated faster than in long-lived species. It is also noteworthy that the difference in urinary excretion of 8-oxoGua between mice and humans is very similar to the difference in reported oxygen consumption between these species /10 fold and 11 fold respectively [247].
To conclude, on the basis of the results presented above showing that urinary 8-oxoGua as well as the other modifications in different species is higher in rapidly aging mammalian species and the presented correlative association between oxidative DNA damage parameters and age in humans it seems reasonable to state that this damage may be one of the substantial factors in mammalian (including human) aging.
CONCLUSIONS
It is becoming increasingly apparent that oxidative damage plays a role in numerous pathological conditions [49]. However, greater knowledge of whether oxidative DNA damage initiates the disease process or is merely a byproduct of disease development is of critical importance. On the basis of the presented data and literature reports it seems reasonable to postulate that oxidative DNA damage/oxidative stress is probably a contributing factor to aging. however, mechanisms that underlie aging are highly complex and may depend on different factors like genetic background, dietary behavior, life style, to name a few. Thus, oxidative stress may contribute to a limited extent to the aging of some individuals and could be a major factor in others. It should be also remembered that association between oxidative stress and aging is complicated by the considerations that there is no general agreement as to what aging is, when aging begins and what triggers its onset and that oxidative stress occurs by multiple mechanisms.
While many details regarding the role of ROS induced DNA damage, in the etiology of complex multifactoral diseases like cancer are yet to be discovered, it is evident that oxidants act at several stages in malignant transformation since they can induce permanent DNA sequence changes [248]. In the light of the presented data it is likely that severe oxidative stress is a consequence of development of many types of cancer. However, at present it is impossible to directly answer the question concerning involvement of oxidative stress in the origin of cancer since full development of the disease in response to carcinogen exposure takes 20-40 years. Therefore, it is very difficult to prove directly that the DNA lesion responsible for carcinogenic process is the lesion present in tumors many generations later. Nevertheless, it should be remembered that DNA damage, altered gene expression and mutations are required participants in the process of carcinogenesis. Although these events may be driven by different mechanisms a commonality is the involvement of oxidants in all these phenomena.
Glossary
ABBREVIATIONS
- 8-oxoGua
8-oxo-7,8-dihydroguanosine
- 8-oxodG
8-oxo-7,8-dihydrodeoxyguanosine
- εA
1,N6-ethenoadenine
- εC
3,N4-ethenocytosine
REFERENCES
- 1.von Sontag C. London: Taylor and Francis; 1987. The Chemical Basis of Radiation Biology. [Google Scholar]
- 2.Migliore L, Coppede F. Genetic and environmental factors in cancer and neurodegenerative diseases. Mutat Res. 2002;512(2-3):135–153. doi: 10.1016/s1383-5742(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 3.Wiseman H, Kaur H, Halliwell B. DNA damage and cancer: measurement and mechanism. Cancer Lett. 1995;93(1):113–120. doi: 10.1016/0304-3835(95)03792-U. [DOI] [PubMed] [Google Scholar]
- 4.Halliwell B, Cross CE. Oxygen-derived species: their relation to human disease and environmental stress. Environ Health Perspect. 1994;102(Suppl 10):5–12. doi: 10.1289/ehp.94102s105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dizdaroglu M. Oxidative damage to DNA in mammalian chromatin. Mutat Res. 1992;275(3-6):331–342. doi: 10.1016/0921-8734(92)90036-o. [DOI] [PubMed] [Google Scholar]
- 6.Floyd RA. The role of 8-hydroxyguanine in carcinogenesis. Carcinogenesis. 1990;11(9):1447–1450. doi: 10.1093/carcin/11.9.1447. [DOI] [PubMed] [Google Scholar]
- 7.Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477(1-2):7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- 8.Dizdaroglu M. Quantitative determination of oxidative base damage in DNA by stable isotope-dilution mass spectrometry. FEBS Lett. 1993;315(1):1–6. doi: 10.1016/0014-5793(93)81120-o. [DOI] [PubMed] [Google Scholar]
- 9.Dizdaroglu M. Free-radical-induced formation of an 8,5'-cyclo-2'-deoxyguanosine moiety in deoxyribonucleic acid. Biochem J. 1986;238(1):247–254. doi: 10.1042/bj2380247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A →C substitutions. J Biol Chem. 1992;267(1):166–172. [PubMed] [Google Scholar]
- 11.Olinski R, Gackowski D, Foksinski M, Rozalski R, Roszkowski K, Jaruga P. Oxidative DNA damage: assessment of the role in carcinogenesis, atherosclerosis, and acquired immunodeficiency syndrome. Free Radic Biol Med. 2002;33(2):192–200. doi: 10.1016/s0891-5849(02)00878-x. [DOI] [PubMed] [Google Scholar]
- 12.Revich GG, Beattie KL. Utilization of 1,N6-etheno-2'-deoxyadenosine 5'-triphosphate during DNA synthesis on natural templates, catalyzed by DNA polymerase I of Escherichia coli. Carcinogenesis. 1986;7(9):1569–1576. doi: 10.1093/carcin/7.9.1569. [DOI] [PubMed] [Google Scholar]
- 13.Kamiya H, Kasai H. 2-Hydroxy-dATP is incorporated opposite G by Escherichia coli DNA poly-merase III resulting in high mutagenicity. Nucleic Acids Res. 2000;28(7):1640–1646. doi: 10.1093/nar/28.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartsch H, Nair J. Oxidative stress and lipid peroxidation-derived DNA-lesions in inflammation driven carcinogenesis. Cancer Detect Prev. 2004;28(6):385–391. doi: 10.1016/j.cdp.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Bartsch H. Exocyclic adducts as new risk markers for DNA damage in man. In: Singer B, Bartsch H, editors. Exocyclic DNA Adducts in Mutagenesis and Carcinogenesis. Lyon: IARC Scientific Publication; 1999. pp. 1–16. [PubMed] [Google Scholar]
- 16.Levine RL, Yang IY, Hossain M, Pandya GA, Grollman AP, Moriya M. Mutagenesis induced by a single 1,N6-ethenodeoxyadenosine adduct in human cells. Cancer Res. 2000;60(15):4098–4104. [PubMed] [Google Scholar]
- 17.Moriya M, Pandya GA, Johnson F, Grollman AP. Cellular response to exocyclic DNA adducts. IARC Sci Publ. 1999;150:263–270. [PubMed] [Google Scholar]
- 18.Bartsch H, Barbin A, Marion MJ, Nair J, Guichard Y. Formation, detection, and role in car-cinogenesis of ethenobases in DNA. Drug Metab Rev. 1994;26(1-2):349–371. doi: 10.3109/03602539409029802. [DOI] [PubMed] [Google Scholar]
- 19.Tuimala J, Szekely G, Wikman H, Järventaus H, Hirvonen A, Gundy S, Norppa H. Genetic poly-morphisms of DNA repair and xenobiotic-metabolizing enzymes: effects on levels of sister chromatid exchanges and chromosomal aberrations. Mutat Res. 2004;554(1-2):319–333. doi: 10.1016/j.mrfmmm.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Nair J, Gansauge F, Beger H, Dolara P, Winde G, Bartsch H. Increased etheno-DNA adducts in affected tissues of patients suffering from Crohn's disease, ulcerative colitis, and chronic pancreatitis. Antioxid Redox Signal. 2006;8(5-6):1003–1010. doi: 10.1089/ars.2006.8.1003. [DOI] [PubMed] [Google Scholar]
- 21.Zarkovic K. 4-hydroxynonenal and neurode-generative diseases. Mol Aspects Med. 2003;24(4-5):293–303. doi: 10.1016/s0098-2997(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi M, Itoh M, Araki S, Kumada S, Shioda K, Tamagawa K, Mizutani T, Morimatsu Y, Minagawa M, Oda M. Oxidative stress and disturbed glutamate transport in hereditary nucleotide repair disorders. J Neuropathol Exp Neurol. 2001;60(4):350–356. doi: 10.1093/jnen/60.4.350. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Lovell MA, Lynn BC. Detection and quantification of endogenous cyclic DNA adducts derived from trans-4-hydroxy-2-nonenal in human brain tissue by isotope dilution capillary liquid chromatography nanoelectrospray tandem mass spectrometry. Chem Res Toxicol. 2006;19(5):710–718. doi: 10.1021/tx0502903. [DOI] [PubMed] [Google Scholar]
- 24.Feig DI, Reid TM, Loeb LA. Reactive oxygen species in tumorigenesis. Cancer Res. 1994;54(7 Suppl):1890s–1894s. [PubMed] [Google Scholar]
- 25.Yu D, Berlin JA, Penning TM, Field J. Reactive oxygen species generated by PAH o-quinones cause change-in-function mutations in p53. Chem Res Toxicol. 2002;15(6):832–842. doi: 10.1021/tx010177m. [DOI] [PubMed] [Google Scholar]
- 26.Du MQ, Carmichael PL, Phillips DH. Induction of activating mutations in the human c-Ha-ras-1 proto-oncogene by oxygen free radicals. Mol Carcinog. 1994;11(3):170–175. doi: 10.1002/mc.2940110308. [DOI] [PubMed] [Google Scholar]
- 27.Haracska L, Washington MT, Prakash S, Prakash L. Inefficient bypass of an abasic site by DNA polymerase eta. J Biol Chem. 2001;276(9):6861–6866. doi: 10.1074/jbc.M008021200. [DOI] [PubMed] [Google Scholar]
- 28.Webster KA, Prentice H, Bishopric NH. Oxidation of zinc finger transcription factors: physiological consequences. Antioxid Redox Signal. 2001;3(4):535–548. doi: 10.1089/15230860152542916. [DOI] [PubMed] [Google Scholar]
- 29.Evans MD, Cooke MS. Factors contributing to the outcome of oxidative damage to nucleic acids. Bioessays. 2004;26(5):533–542. doi: 10.1002/bies.20027. [DOI] [PubMed] [Google Scholar]
- 30.Weitzman SA, Turk PW, Milkowski DH, Kozlowski K. Free radical adducts induce alterations in DNA cytosine methylation. Proc Natl Acad Sci U S A. 1994;91(4):1261–1264. doi: 10.1073/pnas.91.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teufel A, Staib F, Kanzler S, Weinmann A, Schulze-Bergkamen H, Galle PR. Genetics of hepatocellular carcinoma. World J Gastroenterol. 2007;13(16):2271–2282. doi: 10.3748/wjg.v13.i16.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robles AI, Linke SP, Harris CC. The p53 network in lung carcinogenesis. Oncogene. 2002;21(45):6898–6907. doi: 10.1038/sj.onc.1205563. [DOI] [PubMed] [Google Scholar]
- 33.Pandya GA, Moriya M. 1,N6-ethenodeoxyadenosine, a DNA adduct highly mutagenic in mammalian cells. Biochemistry. 1996;35(35):11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 34.Moriya M, Zhang W, Johnson F, Grollman AP. Mutagenic potency of exocyclic DNA adducts: marked differences between Escherichia coli and simian kidney cells. Proc Natl Acad Sci U S A. 1994;91(25):11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanley LA. Molecular aspects of chemical carcinogenesis: the roles of oncogenes and tumour suppressor genes. Toxicology. 1995;96(3):173–194. doi: 10.1016/0300-483x(94)02991-3. [DOI] [PubMed] [Google Scholar]
- 36.Hollstein M, Marion MJ, Lehman T, Welsh J, Harris CC, Martel-Planche G, Kusters I, Montesano R. p53 mutations at A:T base pairs in angiosarcomas of vinyl chloride-exposed factory workers. Carcinogenesis. 1994;15(1):1–3. doi: 10.1093/carcin/15.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Toyokuni S, Okamoto K, Yodoi J, Hiai H. Persistent oxidative stress in cancer. FEBS Lett. 1995;358(1):1–3. doi: 10.1016/0014-5793(94)01368-b. [DOI] [PubMed] [Google Scholar]
- 38.Block G, Patterson B, Subar A. Fruit, vegetables, and cancer prevention: a review of the epidemiological evidence. Nutr Cancer. 1992;18(1):1–29. doi: 10.1080/01635589209514201. [DOI] [PubMed] [Google Scholar]
- 39.Ames BN, Gold LS, Willett WC. The causes and prevention of cancer. Proc Natl Acad Sci U S A. 1995;92(12):5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willett WC. Diet and health: what should we eat? Science. 1994;264(5158):532–537. doi: 10.1126/science.8160011. [DOI] [PubMed] [Google Scholar]
- 41.Schlotte V, Sevanian A, Hochstein P, Weithmann KU. Effect of uric acid and chemical analogues on oxidation of human low density lipoprotein in vitro. Free Radic Biol Med. 1998;25(7):839–847. doi: 10.1016/s0891-5849(98)00160-9. [DOI] [PubMed] [Google Scholar]
- 42.Sevanian A, Davies KJ, Hochstein P. Conservation of vitamin C by uric acid in blood. J Free Radic Biol Med. 1985;1(2):117–124. doi: 10.1016/0748-5514(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 43.Duthie SJ, Ma A, Ross MA, Collins AR. Antioxidant supplementation decreases oxidative DNA damage in human lymphocytes. Cancer Res. 1996;56(6):1291–1295. [PubMed] [Google Scholar]
- 44.Collins AR, Olmedilla B, Southon S, Granado F, Duthie SJ. Serum carotenoids and oxidative DNA damage in human lymphocytes. Carcino-genesis. 1998;19(12):2159–2162. doi: 10.1093/carcin/19.12.2159. [DOI] [PubMed] [Google Scholar]
- 45.Moller P, Loft S. Oxidative DNA damage in human white blood cells in dietary antioxidant intervention studies. Am J Clin Nutr. 2002;76(2):303–310. doi: 10.1093/ajcn/76.2.303. [DOI] [PubMed] [Google Scholar]
- 46.Foksinski M, Gackowski D, Rozalski R, Siomek A, Guz J, Szpila A, Dziaman T, Olinski R. Effects of basal level of antioxidants on oxidative DNA damage in humans. Eur J Nutr. 2007;46(3):174–180. doi: 10.1007/s00394-006-0642-7. [DOI] [PubMed] [Google Scholar]
- 47.Gackowski D, Kruszewsk M, Banaszkiewicz Z, Jawien A, Olinski R. Lymphocyte labile iron pool, plasma iron, transferrin saturation and ferritin levels in colon cancer patients. Acta Biochim Pol. 2002;49(1):269–272. [PubMed] [Google Scholar]
- 48.Jaruga P, Jaruga B, Gackowski D, Olczak A, Halota W, Pawlowska M, Olinski R. Supple-mentation with antioxidant vitamins prevents oxidative modification of DNA in lymphocytes of HIV-infected patients. Free Radic Biol Med. 2002;32(5):414–420. doi: 10.1016/s0891-5849(01)00821-8. [DOI] [PubMed] [Google Scholar]
- 49.Halliwell B, Gutteridge JM. 3nd ed. Oxford ed. New York, NY: Oxford University Press; 1999. Free radicals in biology and medicine. [Google Scholar]
- 50.Gackowski D, Kruszewski M, Jawien A, Ciecierski M, Olinski R. Further evidence that oxidative stress may be a risk factor responsible for the development of atherosclerosis. Free Radic Biol Med. 2001;31(4):542–547. doi: 10.1016/s0891-5849(01)00614-1. [DOI] [PubMed] [Google Scholar]
- 51.Gackowski D, Kruszewski M, Bartlomiejczyk T, Jawien A, Ciecierski M, Olinski R. The level of 8-oxo-7,8-dihydro-2'-deoxyguanosine is positively correlated with the size of the labile iron pool in human lymphocytes. J Biol Inorg Chem. 2002;7(4-5):548–550. doi: 10.1007/s00775-001-0335-x. [DOI] [PubMed] [Google Scholar]
- 52.de Valk B, Addicks MA, Gosriwatana I, Lu S, Hider RC, Marx JJ. Non-transferrin-bound iron is present in serum of hereditary haemochromatosis heterozygotes. Eur J Clin Invest. 2000;30(3):248–251. doi: 10.1046/j.1365-2362.2000.00628.x. [DOI] [PubMed] [Google Scholar]
- 53.Nelson RL, Davis FG, Sutter E, Sobin LH, Kikendall JW, Bowen P. Body iron stores and risk of colonic neoplasia. J Natl Cancer Inst. 1994;86(6):455–460. doi: 10.1093/jnci/86.6.455. [DOI] [PubMed] [Google Scholar]
- 54.Stevens RG, Graubard BI, Micozzi MS, Neriishi K, Blumberg BS. Moderate elevation of body iron level and increased risk of cancer occurrence and death. Int J Cancer. 1994;56(3):364–369. doi: 10.1002/ijc.2910560312. [DOI] [PubMed] [Google Scholar]
- 55.Shigenaga MK, Gimeno CJ, Ames BN. Urinary 8-hydroxy-2'-deoxyguanosine as a biological marker of in vivo oxidative DNA damage. Proc Natl Acad Sci U S A. 1989;86(24):9697–9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki J, Inoue Y, Suzuki S. Changes in the urinary excretion level of 8-hydroxyguanine by exposure to reactive oxygen-generating sub-stances. Free Radic Biol Med. 1995;18(3):431–436. doi: 10.1016/0891-5849(94)00152-a. [DOI] [PubMed] [Google Scholar]
- 57.Cooke MS, Evans MD, Herbert KE, Lunec J. Urinary 8-oxo-2'-deoxyguanosine–source, significance and supplements. Free Radic Res. 2000;32(5):381–397. doi: 10.1080/10715760000300391. [DOI] [PubMed] [Google Scholar]
- 58.Loft S, Fischer-Nielsen A, Jeding IB, Vistisen K, Poulsen HE. 8-Hydroxydeoxyguanosine as a urinary biomarker of oxidative DNA damage. J Toxicol Environ Health. 1993;40(2-3):391–404. doi: 10.1080/15287399309531806. [DOI] [PubMed] [Google Scholar]
- 59.Loft S, Poulsen HE. Estimation of oxidative DNA damage in man from urinary excretion of repair products. Acta Biochim Pol. 1998;45(1):133–144. [PubMed] [Google Scholar]
- 60.Girard PM, Boiteux S. Repair of oxidized DNA bases in the yeast Saccharomyces cerevisiae. Biochimie. 1997;79(9-10):559–566. doi: 10.1016/s0300-9084(97)82004-4. [DOI] [PubMed] [Google Scholar]
- 61.Dianov G, Bischoff C, Piotrowski J, Bohr VA. Repair pathways for processing of 8-oxoguanine in DNA by mammalian cell extracts. J Biol Chem. 1998;273(50):33811–33816. doi: 10.1074/jbc.273.50.33811. [DOI] [PubMed] [Google Scholar]
- 62.Gackowski D, Rozalski R, Roszkowski K, Jawien A, Foksinski M, Olinski R. 8-Oxo-7,8-dihydroguanine and 8-oxo-7,8-dihydro-2'-deoxyguanosine levels in human urine do not depend on diet. Free Radic Res. 2001;35(6):825–832. doi: 10.1080/10715760100301321. [DOI] [PubMed] [Google Scholar]
- 63.Olinski R, Zastawny T, Budzbon J, Skokowski J, Zegarski W, Dizdaroglu M. DNA base modifications in chromatin of human cancerous tissues. FEBS Lett. 1992;309(2):193–198. doi: 10.1016/0014-5793(92)81093-2. [DOI] [PubMed] [Google Scholar]
- 64.Jaruga P, Zastawny TH, Skokowski J, Dizdaroglu M, Olinski R. Oxidative DNA base damage and antioxidant enzyme activities in human lung cancer. FEBS Lett. 1994;341(1):59–64. doi: 10.1016/0014-5793(94)80240-8. [DOI] [PubMed] [Google Scholar]
- 65.Malins DC, Haimanot R. Major alterations in the nucleotide structure of DNA in cancer of the female breast. Cancer Res. 1991;51(19):5430–5432. [PubMed] [Google Scholar]
- 66.Schmielau J, Finn OJ. Activated granulocytes and granulocyte-derived hydrogen peroxide are the underlying mechanism of suppression of t-cell function in advanced cancer patients. Cancer Res. 2001;61(12):4756–4760. [PubMed] [Google Scholar]
- 67.Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51(3):794–798. [PubMed] [Google Scholar]
- 68.De Vita JVT, Hellman S, Rosenberg SA. Principles & practice of oncology. Philadelphia: >Lippincott Williams & Wilkins; 2001. Cancer. [Google Scholar]
- 69.Franks LM, Teich NM. New York, Tokyo: Oxford University Press; 1997. Introduction to the cellular and molecular biology of cancer. [Google Scholar]
- 70.Ohba M, Shibanuma M, Kuroki T, Nose K. Production of hydrogen peroxide by transforming growth factor-beta 1 and its involvement in induction of egr-1 in mouse osteoblastic cells. J Cell Biol. 1994;126(4):1079–1088. doi: 10.1083/jcb.126.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kayanoki Y, Fujii J, Suzuki K, Kawata S, Matsuzawa Y, Taniguchi N. Suppression of antioxidative enzyme expression by transforming growth factor-beta 1 in rat hepatocytes. J Biol Chem. 1994;269(22):15488–15492. [PubMed] [Google Scholar]
- 72.Malins DC, Holmes EH, Polissar NL, Gunselman SJ. The etiology of breast cancer. Characteristic alteration in hydroxyl radical-induced DNA base lesions during oncogenesis with potential for evaluating incidence risk. Cancer. 1993;71(10):3036–3043. doi: 10.1002/1097-0142(19930515)71:10<3036::aid-cncr2820711025>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 73.Okamoto K, Toyokuni S, Uchida K, Ogawa O, Takenewa J, Kakehi Y, Kinoshita H, Hattori-Nakakuki Y, Hiai H, Yoshida O. Formation of 8-hydroxy-2'-deoxyguanosine and 4-hydroxy-2-nonenal-modified proteins in human renal-cell carcinoma. Int J Cancer. 1994;58(6):825–829. doi: 10.1002/ijc.2910580613. [DOI] [PubMed] [Google Scholar]
- 74.Schmid K, Nair J, Winde G, Velic I, Bartsch H. Increased levels of promutagenic etheno-DNA adducts in colonic polyps of FAP patients. Int J Cancer. 2000;87(1):1–4. doi: 10.1002/1097-0215(20000701)87:1<1::aid-ijc1>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 75.Kasprzak KS, Jaruga P, Zastawny TH, North SL, Riggs CW, Olinski R, Dizdaroglu M. Oxidative DNA base damage and its repair in kidneys and livers of nickel(II)-treated male F344 rats. Carcinogenesis. 1997;18(2):271–277. doi: 10.1093/carcin/18.2.271. [DOI] [PubMed] [Google Scholar]
- 76.Foksinski M, Kotzbach R, Szymanski W, Olinski R. The level of typical biomarker of oxidative stress 8-hydroxy-2'-deoxyguanosine is higher in uterine myomas than in control tissues and correlates with the size of the tumor. Free Radic Biol Med. 2000;29(7):597–601. doi: 10.1016/s0891-5849(00)00358-0. [DOI] [PubMed] [Google Scholar]
- 77.Schwartz LB, Diamond MP, Schwartz PE. Leio-myosarcomas: clinical presentation. Am J Obstet Gynecol. 1993;168(1 Pt 1):180–183. doi: 10.1016/s0002-9378(12)90910-2. [DOI] [PubMed] [Google Scholar]
- 78.Fortini P, Pascucci B, Parlanti E, D'Errico M, Simonelli V, Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85(11):1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 79.Hill JW, Hazra TK, Izumi T, Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP -endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29(2):430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Privezentzev CV, Saparbaev M, Laval J. The HAP1 protein stimulates the turnover of human mismatch-specific thymine-DNA-glycosylase to process 3,N4-ethenocytosine residues. Mutat Res. 2001;480-481:277–284. doi: 10.1016/s0027-5107(01)00186-5. [DOI] [PubMed] [Google Scholar]
- 81.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Ménissierde Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2(PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277(25):23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 82.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24(22):4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leppard JB, Dong Z, Mackey ZB, Tomkinson AE. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol Cell Biol. 2003;23(16):5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harrigan JA, Wilson DM, 3rd, III, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase beta. Nucleic Acids Res. 2006;34(2):745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tuo J, Chen C, Zeng X, Christiansen M, Bohr VA. Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA Repair (Amst) 2002;1(11):913–927. doi: 10.1016/s1568-7864(02)00116-7. [DOI] [PubMed] [Google Scholar]
- 86.Colussi C, Parlanti E, Degan P, Aquilina G, Barnes D, Macpherson P, Karran P, Crescenzi M, Dogliotti E, Bignami M. The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr Biol. 2002;12(11):912–918. doi: 10.1016/s0960-9822(02)00863-1. [DOI] [PubMed] [Google Scholar]
- 87.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355(6357):273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 88.Treffers HP, Spinelli V, Belser NO. A Factor (or Mutator Gene) Influencing Mutation Rates in Escherichia Coli. Proc Natl Acad Sci U S A. 1954;40(11):1064–1071. doi: 10.1073/pnas.40.11.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yanofsky C, Cox EC, Horn V. The unusual mutagenic specificity of an E. Coli mutator gene. Proc Natl Acad Sci U S A. 1966;55(2):274–281. doi: 10.1073/pnas.55.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schaaper RM, Dunn RL. Escherichia coli mutT mutator effect during in vitro DNA synthesis. Enhanced A.G replicational errors. J Biol Chem. 1987;262(34):16267–16270. [PubMed] [Google Scholar]
- 91.Nakabeppu Y. Molecular genetics and structural biology of human MutT homolog, MTH1. Mutat Res. 2001;477(1-2):59–70. doi: 10.1016/s0027-5107(01)00096-3. [DOI] [PubMed] [Google Scholar]
- 92.Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic sub-strate for DNA synthesis. J Biol Chem. 1993;268(31):23524–23530. [PubMed] [Google Scholar]
- 93.Kakuma T, Nishida J, Tsuzuki T, Sekiguchi M. Mouse MTH1 protein with 8-oxo-7,8-dihydro-2'-deoxyguanosine 5'-triphosphatase activity that prevents transversion mutation. cDNA cloning and tissue distribution. J Biol Chem. 1995;270(43):25942–25948. doi: 10.1074/jbc.270.43.25942. [DOI] [PubMed] [Google Scholar]
- 94.Cai JP, Kakuma T, Tsuzuki T, Sekiguchi M. cDNA and genomic sequences for rat 8-oxo-dGTPase that prevents occurrence of spontaneous mutations due to oxidation of guanine nucleotides. Carcinogenesis. 1995;16(10):2343–2350. doi: 10.1093/carcin/16.10.2343. [DOI] [PubMed] [Google Scholar]
- 95.Mo JY, Maki H, Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protesanitization of nucleotide pool. Proc Natl Acad Sci U S A. 1992;89(22):11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bialkowski K, Bialkowska A, Anderson LM, Kasprzak KS. Higher activity of 8-oxo-2'-deoxyguanosine 5'-triphosphate pyrophospho-hydrolase (8-oxo-dGTPase) coincides with lower background levels of 8-oxo-2'-deoxyguanosine in DNA of fetal compared with maternal mouse organs. Free Radic Biol Med. 1999;27(1-2):90–94. doi: 10.1016/s0891-5849(99)00040-4. [DOI] [PubMed] [Google Scholar]
- 97.Bialkowski K, Bialkowska A, Kasprzak KS. Cadmium(II), unlike nickel(II), inhibits 8-oxo-dGTPase activity and increases 8-oxo-dG level in DNA of the rat testis, a target organ for cad-mium(II) carcinogenesis. Carcinogenesis. 1999;20(8):1621–1624. doi: 10.1093/carcin/20.8.1621. [DOI] [PubMed] [Google Scholar]
- 98.Bialkowski K, Kasprzak KS. A novel assay of 8-oxo-2'-deoxyguanosine 5'-triphosphate pyro-phosphohydrolase (8-oxo-dGTPase) activity in cultured cells and its use for evaluation of cadmium(II) inhibition of this activity. Nucleic Acids Res. 1998;26(13):3194–3201. doi: 10.1093/nar/26.13.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bialkowski K, Kasprzak KS. Activity of the antimutagenic enzyme 8-oxo-2'-deoxyguanosine 5'-triphosphate pyrophosphohydrolase (8-oxo-dGTPase) in cultured chinese hamster ovary cells: effects of cell cycle, proliferation rate, and population density. Free Radic Biol Med. 2000;28(3):337–344. doi: 10.1016/s0891-5849(99)00246-4. [DOI] [PubMed] [Google Scholar]
- 100.Fujikawa K, Kamiya H, Yakushiji H, Fujii Y, Nakabeppu Y, Kasai H. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J Biol Chem. 1999;274(26):18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- 101.Fujikawa K, Kamiya H, Yakushiji H, Nakabeppu Y, Kasai H. Human MTH1 protein hydrolyzes the oxidized ribonucleotide, 2-hydroxy-ATP. Nucleic Acids Res. 2001;29(2):449–454. doi: 10.1093/nar/29.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hayakawa H, Hofer A, Thelander L, Kitajima S, Cai Y, Oshiro S, Yakushiji H, Nakabeppu Y, Kuwano M, Sekiguchi M. Metabolic fate of oxidized guanine ribonucleotides in mammalian cells. Biochemistry. 1999;38(12):3610–3614. doi: 10.1021/bi982361l. [DOI] [PubMed] [Google Scholar]
- 103.Fujikawa K, Yakushiji H, Nakabeppu Y, Suzuki T, Masuda M, Ohshima H, Kasai H. 8-Chloro-dGTP, a hypochlorous acid-modified nucleotide, is hydrolyzed by hMTH1, the human MutT homolog. FEBS Lett. 2002;512(1-3):149–151. doi: 10.1016/s0014-5793(02)02240-8. [DOI] [PubMed] [Google Scholar]
- 104.Oda H, Taketomi A, Maruyama R, Itoh R, Nishioka K, Yakushiji H, Suzuki T, Sekiguchi M, Nakabeppu Y. Multi-forms of human MTH1 polypeptides produced by alternative translation initiation and single nucleotide polymorphism. Nucleic Acids Res. 1999;27(22):4335–4343. doi: 10.1093/nar/27.22.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kang D, Nishida J, Iyama A, Nakabeppu Y, Furuichi M, Fujiwara T, Sekiguchi M, Takeshige K. Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J Biol Chem. 1995;270(24):14659–14665. doi: 10.1074/jbc.270.24.14659. [DOI] [PubMed] [Google Scholar]
- 106.Kasprzak KS, Nakabeppu Y, Kakuma T, Sakai Y, Tsuruya K, Sekiguchi M, Ward JM, Diwan BA, Nagashima K, Kasprzak BH. Intracellular distribution of the antimutagenic enzyme MTH1 in the liver, kidney and testis of F344 rats and its modulation by cadmium. Exp Toxicol Pathol. 2001;53(5):325–335. doi: 10.1078/0940-2993-00201. [DOI] [PubMed] [Google Scholar]
- 107.Okamoto K, Toyokuni S, Kim WJ, Ogawa O, Kakehi Y, Arao S, Hiai H, Yoshida O. Overex-pression of human mutT homologue gene messenger RNA in renal-cell carcinoma: evidence of persistent oxidative stress in cancer. Int J Cancer. 1996;65(4):437–441. doi: 10.1002/(SICI)1097-0215(19960208)65:4<437::AID-IJC7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 108.Okochi E, Ichimura S, Sugimura T, Ushijima T. The absence of Mth1 inactivation and DNA polymerase kappa overexpression in rat mammary carcinomas with frequent A:T to C:G transversions. Jpn J Cancer Res. 2002;93(5):501–506. doi: 10.1111/j.1349-7006.2002.tb01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kennedy CH, Cueto R, Belinsky SA, Lechner JF, Pryor WA. Overexpression of hMTH1 mRNA: a molecular marker of oxidative stress in lung cancer cells. FEBS Lett. 1998;429(1):17–20. doi: 10.1016/s0014-5793(98)00505-5. [DOI] [PubMed] [Google Scholar]
- 110.Wani G, Milo GE, D'Ambrosio SM. Enhanced expression of the 8-oxo-7,8-dihydrodeoxyguanosine triphosphatase gene in human breast tumor cells. Cancer Lett. 1998;125(1-2):123–130. doi: 10.1016/s0304-3835(97)00507-7. [DOI] [PubMed] [Google Scholar]
- 111.Meyer F, Fiala E, Westendorf J. Induction of 8-oxo-dGTPase activity in human lymphoid cells and normal fibroblasts by oxidative stress. Toxicology. 2000;146(2-3):83–92. doi: 10.1016/s0300-483x(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 112.Kim HN, Morimoto Y, Tsuda T, Ootsuyama Y, Hirohashi M, Hirano T, Tanaka I, Lim Y, Yun IG, Kasai H. Changes in DNA 8-hydroxyguanine levels, 8-hydroxyguanine repair activity, and hOGG1 and hMTH1 mRNA expression in human lung alveolar epithelial cells induced by crocidolite asbestos. Carcinogenesis. 2001;22(2):265–269. doi: 10.1093/carcin/22.2.265. [DOI] [PubMed] [Google Scholar]
- 113.Tsuzuki T, Egashira A, Kura S. Analysis of MTH1 gene function in mice with targeted mutagenesis. Mutat Res. 2001;477(1-2):71–78. doi: 10.1016/s0027-5107(01)00108-7. [DOI] [PubMed] [Google Scholar]
- 114.Tsuzuki T, Egashira A, Igarashi H, Iwakuma T, Nakatsuru Y, Tominaga Y, Kawate H, Nakao K, Nakamura K, Ide F, Kura S, Nakabeppu Y, Katsuki M, Ishikawa T, Sekiguchi M. Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc Natl Acad Sci U S A. 2001;98(20):11456–11461. doi: 10.1073/pnas.191086798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Egashira A, Yamauchi K, Yoshiyama K, Kawate H, Katsuki M, Sekiguchi M, Sugimachi K, Maki H, Tsuzuki T. Mutational specificity of mice defective in the MTH1 and/or the MSH2 genes. DNA Repair (Amst) 2002;1(11):881–893. doi: 10.1016/s1568-7864(02)00113-1. [DOI] [PubMed] [Google Scholar]
- 116.Speina E, Arczewska KD, Gackowski D, Zielińska M, Siomek A, Kowalewski J, Oliński R, Tudek B, Kuśmierek JT. Contribution of hMTH1 to the maintenance of 8-oxoguanine levels in lung DNA of non-small-cell lung cancer patients. J Natl Cancer Inst. 2005;97(5):384–395. doi: 10.1093/jnci/dji058. [DOI] [PubMed] [Google Scholar]
- 117.Kennedy CH, Pass HI, Mitchell JB. Expression of human MutT homologue (hMTH1) protein in primary non-small-cell lung carcinomas and histologically normal surrounding tissue. Free Radic Biol Med. 2003;34(11):1447–1457. doi: 10.1016/s0891-5849(03)00176-x. [DOI] [PubMed] [Google Scholar]
- 118.Russo MT, Blasi MF, Chiera F, Fortini P, Degan P, Macpherson P, Furuichi M, Nakabeppu Y, Karran P, Aquilina G, Bignami M. The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells. Mol Cell Biol. 2004;24(1):465–474. doi: 10.1128/MCB.24.1.465-474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000;50(1):7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- 120.Didkowska J, Wojciechowska W, Tarnowski W, Zatonski WA. Cancer In Poland. Warsaw, Poland: Centre of Oncol. Scien. Publ. 2002 [Google Scholar]
- 121.Saetta M, Baraldo S, Corbino L, Turato G, Braccioni F, Rea F, Cavallesco G, Tropeano G, Mapp CE, Maestrelli P, Ciaccia A, Fabbri LM. CD8+ve cells in the lungs of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1999;160(2):711–717. doi: 10.1164/ajrccm.160.2.9812020. [DOI] [PubMed] [Google Scholar]
- 122.Parlanti E, Fortini P, Macpherson P, Laval J, Dogliotti E. Base excision repair of adenine/8-oxoguanine mispairs by an aphidicolin-sensitive DNA polymerase in human cell extracts. Oncogene. 2002;21(34):5204–5212. doi: 10.1038/sj.onc.1205561. [DOI] [PubMed] [Google Scholar]
- 123.Hu J, Imam SZ, Hashiguchi K, Souza-Pinto NC, Bohr VA. Phosphorylation of human oxoguanine DNA glycosylase (alpha-OGG1) modulates its function. Nucleic Acids Res. 2005;33(10):3271–3282. doi: 10.1093/nar/gki636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Saparbaev M, Kleibl K, Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23(18):3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Saparbaev M, Laval J. 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary sub-strate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc Natl Acad Sci U S A. 1998;95(15):8508–8513. doi: 10.1073/pnas.95.15.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bader SA, Walker M, Harrison DJ. A human cancer-associated truncation of MBD4 causes dominant negative impairment of DNA repair in colon cancer cells. Br J Cancer. 2007;96(4):660–666. doi: 10.1038/sj.bjc.6603592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mishina Y, Yang CG, He C. Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J Am Chem Soc. 2005;127(42):14594–14595. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Speina E, Zielinska M, Barbin A, Gackowski D, Kowalewski J, Graziewicz MA, Siedlecki JA, Oliński R, Tudek B. Decreased repair activities of 1,N6-ethenoadenine and 3,N4-ethenocytosine in lung adenocarcinoma patients. Cancer Res. 2003;63(15):4351–4357. [PubMed] [Google Scholar]
- 129.Gackowski D, Speina E, Zielinska M, Kowalewski J, Rozalski R, Siomek A, Paciorek T, Tudek B, Olinski R. Products of oxidative DNA damage and repair as possible bio-markers of susceptibility to lung cancer. cancer Res. 2003;63(16):4899–4902. [PubMed] [Google Scholar]
- 130.Paz-Elizur T, Krupsky M, Blumenstein S, Elinger D, Schechtman E, Livneh Z. DNA repair activity for oxidative damage and risk of lung cancer. J Natl Cancer Inst. 2003;95(17):1312–1319. doi: 10.1093/jnci/djg033. [DOI] [PubMed] [Google Scholar]
- 131.Paz-Elizur T, Ben Yosef R, Elinger D, Vexler A, Krupsky M, Berrebi A, Shani A, Schechtman E, Freedman L, Livneh Z. Reduced repair of the oxidative 8-oxoguanine DNA damage and risk of head and neck cancer. Cancer Res. 2006;66(24):11683–11689. doi: 10.1158/0008-5472.CAN-06-2294. [DOI] [PubMed] [Google Scholar]
- 132.Madri JA, Carter D. Scar cancers of the lung: origin and significance. Hum Pathol. 1984;15(7):625–631. doi: 10.1016/s0046-8177(84)80286-5. [DOI] [PubMed] [Google Scholar]
- 133.Ambs S, Bennett WP, Merriam WG, Ogunfusika MO, Oser SM, Khan MA, Jones RT, Harris CC. Vascular endothelial growth factor and nitric oxide synthase expression in human lung cancer and the relation to p53. Br J cancer. 1998;78(2):233–239. doi: 10.1038/bjc.1998.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zieba M, Nowak D, Suwalski M, Piasecka G, Grzelewska-Rzymowska I, Tymińska K, Kroczyńska-Bednarek J, Kwiatkowska S. Enhanced lipid peroxidation in cancer tissue homogenates in non-small cell lung cancer. Monaldi Arch Chest Dis. 2001;56(2):110–114. [PubMed] [Google Scholar]
- 135.Butkiewicz D, Rusin M, Enewold L, Shields PG, Chorazy M, Harris CC. Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis. 2001;22(4):593–597. doi: 10.1093/carcin/22.4.593. [DOI] [PubMed] [Google Scholar]
- 136.Le Marchand L, Donlon T, Lum-Jones A, Seifried A, Wilkens LR. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11(4):409–412. [PubMed] [Google Scholar]
- 137.Hill JW, Evans MK. Dimerization and opposite base-dependent catalytic impairment of poly-morphic S326C OGG1 glycosylase. Nucleic Acids Res. 2006;34(5):1620–1632. doi: 10.1093/nar/gkl060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Luna L, Rolseth V, Hildrestrand GA, Otterlei M, Dantzer F, Bjørås M, Seeberg E. Dynamic relo-calization of hOGG1 during the cell cycle is disrupted in cells harbouring the hOGG1-Cys326 polymorphic variant. Nucleic Acids Res. 2005;33(6):1813–1824. doi: 10.1093/nar/gki325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bravard A, Vacher M, Moritz E, Vaslin L, Hall J, Epe B, Radicella JP. Oxidation status of human OGG1-S326C polymorphic variant determines cellular DNA repair capacity. Cancer Res. 2009;69(8):3642–3649. doi: 10.1158/0008-5472.CAN-08-3943. [DOI] [PubMed] [Google Scholar]
- 140.Sugimura H, Kohno T, Wakai K, Nagura K, Genka K, Igarashi H, Morris BJ, Baba S, Ohno Y, Gao C, Li Z, Wang J, Takezaki T, Tajima K, Varga T, Sawaguchi T, Lum JK, Martinson JJ, Tsugane S, Iwamasa T, Shinmura K, Yokota J. hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol Bio-markers Prev. 1999;8(8):669–674. [PubMed] [Google Scholar]
- 141.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11(12):1513–1530. [PubMed] [Google Scholar]
- 142.Chen L, Elahi A, Pow-Sang J, Lazarus P, Park J. Association between polymorphism of human oxoguanine glycosylase 1 and risk of prostate cancer. J Urol. 2003;170((6 Pt 1)):2471–2474. doi: 10.1097/01.ju.0000087498.23008.bb. [DOI] [PubMed] [Google Scholar]
- 143.Cho EY, Hildesheim A, Chen CJ, Hsu MM, Chen IH, Mittl BF, Levine PH, Liu MY, Chen JY, Brinton LA, Cheng YJ, Yang CS. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer Epidemiol Biomarkers Prev. 2003;12(10):1100–1104. [PubMed] [Google Scholar]
- 144.Hansen R, Saebo M, Skjelbred CF, Nexø BA, Hagen PC, Bock G, Bowitz Lothe IM, Johnson E, Aase S, Hansteen IL, Vogel U, Kure EH. GPX Pro198Leu and OGG1 Ser326Cys polymor-phisms and risk of development of colorectal adenomas and colorectal cancer. Cancer Lett. 2005;229(1):85–91. doi: 10.1016/j.canlet.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 145.Janssen K, Schlink K, Gotte W, Hippler B, Kaina B, Oesch F. DNA repair activity of 8-oxoguanine DNA glycosylase 1 (OGG1) in human lymphocytes is not dependent on genetic polymorphism Ser326/Cys326. Mutat Res. 2001;486(3):207–216. doi: 10.1016/s0921-8777(01)00096-9. [DOI] [PubMed] [Google Scholar]
- 146.Nohmi T, Kim SR, Yamada M. Modulation of oxidative mutagenesis and carcinogenesis by polymorphic forms of human DNA repair enzymes. Mutat Res. 2005;591(1-2):60–73. doi: 10.1016/j.mrfmmm.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 147.Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci U S A. 1992;89(15):7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rusin M, Samojedny A, Harris CC, Chorazy M. Novel genetic polymorphisms in DNA repair genes: O(6)-methylguanine-DNA methyltrans-ferase (MGMT) and N-methylpurine-DNA glyco-sylase (MPG) in lung cancer patients from Poland. Hum Mutat. 1999;14(3):269–270. doi: 10.1002/(SICI)1098-1004(1999)14:3<269::AID-HUMU13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 149.Krzesniak M, Butkiewicz D, Samojedny A, Chorazy M, Rusin M. Polymorphisms in TDG and MGMT genes - epidemiological and functional study in lung cancer patients from Poland. Ann Hum Genet. 2004;68((Pt 4)):300–312. doi: 10.1046/j.1529-8817.2004.00079.x. [DOI] [PubMed] [Google Scholar]
- 150.Hu JJ, Smith TR, Miller MS, Mohrenweiser HW, Golden A, Case LD. Amino acid substitution variants of APE1 and XRCC1 genes associated with ionizing radiation sensitivity. Carcinogenesis. 2001;22(6):917–922. doi: 10.1093/carcin/22.6.917. [DOI] [PubMed] [Google Scholar]
- 151.Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, Phillips DH, Canzian F, Haugen A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27(3):560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 152.Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162(10):925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 153.Jiao L, Bondy ML, Hassan MM, Wolff RA, Evans DB, Abbruzzese JL, Li D. Selected poly-morphisms of DNA repair genes and risk of pancreatic cancer. Cancer Detect Prev. 2006;30(3):284–291. doi: 10.1016/j.cdp.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dizdaroglu M, Olinski R, Doroshow JH, Akman SA. Modification of DNA bases in chromatin of intact target human cells by activated human polymorphonuclear leukocytes. Cancer Res. 1993;53(6):1269–1272. [PubMed] [Google Scholar]
- 155.Bouziane M, Miao F, Bates SE, Somsouk L, Sang BC, Denissenko M, O'Connor TR. Promoter structure and cell cycle dependent expression of the human methylpurine-DNA gly-cosylase gene. Mutat Res. 2000;461(1):15–29. doi: 10.1016/s0921-8777(00)00036-7. [DOI] [PubMed] [Google Scholar]
- 156.Pines A, Perrone L, Bivi N, Romanello M, Damante G, Gulisano M, Kelley MR, Quadrifoglio F, Tell G. Activation of APE1/Ref-1 is dependent on reactive oxygen species generated after purinergic receptor stimulation by ATP. Nucleic Acids Res. 2005;33(14):4379–4394. doi: 10.1093/nar/gki751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Touati E, Michel V, Thiberge JM, Avé P, Huerre M, Bourgade F, Klungland A, Labigne A. Deficiency in OGG1 protects against inflammation and mutagenic effects associated with H. pylori infection in mouse. Helicobacter. 2006;11(5):494–505. doi: 10.1111/j.1523-5378.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- 158.Gackowski D, Banaszkiewicz Z, Rozalski R, Jawien A, Olinski R. Persistent oxidative stress in colorectal carcinoma patients. Int J Cancer. 2002;101(4):395–397. doi: 10.1002/ijc.10610. [DOI] [PubMed] [Google Scholar]
- 159.Chen KH, Srivastava DK, Singhal RK, Jacob S, Ahmed AE, Wilson SH. Modulation of base excision repair by low density lipoprotein, oxidized low density lipoprotein and antioxidants in mouse monocytes. Carcinogenesis. 2000;21(5):1017–1022. doi: 10.1093/carcin/21.5.1017. [DOI] [PubMed] [Google Scholar]
- 160.Parker AR, O'Meally RN, Sahin F, Su GH, Racke FK, Nelson WG, DeWeese TL, Eshleman JR. Defective human MutY phosphorylation exists in colorectal cancer cell lines with wildtype MutY alleles. J Biol Chem. 2003;278(48):47937–47945. doi: 10.1074/jbc.M306598200. [DOI] [PubMed] [Google Scholar]
- 161.Lu X, Bocangel D, Nannenga B, Yamaguchi H, Appella E, Donehower LA. The p53-induced oncogenic phosphatase PPM1D interacts with uracil DNA glycosylase and suppresses base excision repair. Mol Cell. 2004;15(4):621–634. doi: 10.1016/j.molcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 162.Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol Cell Biol. 2006;26(5):1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9(2):265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 164.Bhakat KK, Hazra TK, Mitra S. Acetylation of the human DNA glycosylase NEIL2 and inhibition of its activity. Nucleic Acids Res. 2004;32(10):3033–3039. doi: 10.1093/nar/gkh632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hasan S, El Andaloussi N, Hardeland U, Hassa PO, Bürki C, Imhof R, Schär P, Hottiger MO. Acetylation regulates the DNA end-trimming activity of DNA polymerase beta. Mol Cell. 2002;10(5):1213–1222. doi: 10.1016/s1097-2765(02)00745-1. [DOI] [PubMed] [Google Scholar]
- 166.Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr Biol. 2005;15(7):616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 167.Jaiswal M, LaRusso NF, Nishioka N, Nakabeppu Y, Gores GJ. Human Ogg1, a protein involved in the repair of 8-oxoguanine, is inhibited by nitric oxide. Cancer Res. 2001;61(17):6388–6393. [PubMed] [Google Scholar]
- 168.Graziewicz M, Wink DA, Laval F. Nitric oxide inhibits DNA ligase activity: potential mechanisms for NO-mediated DNA damage. Carcinogenesis. 1996;17(11):2501–2505. doi: 10.1093/carcin/17.11.2501. [DOI] [PubMed] [Google Scholar]
- 169.Wink DA, Laval J. The Fpg protein, a DNA repair enzyme, is inhibited by the biomediator nitric oxide in vitro and in vivo. Carcinogenesis. 1994;15(10):2125–2129. doi: 10.1093/carcin/15.10.2125. [DOI] [PubMed] [Google Scholar]
- 170.Laval F, Wink DA. Inhibition by nitric oxide of the repair protein, O6-methylguanine-DNA-methyltransferase. Carcinogenesis. 1994;15(3):443–447. doi: 10.1093/carcin/15.3.443. [DOI] [PubMed] [Google Scholar]
- 171.Bartsch H, Nair J, Owen RW. Exocyclic DNA adducts as oxidative stress markers in colon carcinogenesis: potential role of lipid peroxidation, dietary fat and antioxidants. Biol Chem. 2002;383(6):915–921. doi: 10.1515/BC.2002.098. [DOI] [PubMed] [Google Scholar]
- 172.Kelley MR, Cheng L, Foster R, Tritt R, Jiang J, Broshears J, Koch M. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin Cancer Res. 2001;7(4):824–830. [PubMed] [Google Scholar]
- 173.Robertson KA, Bullock HA, Xu Y, Tritt R, Zimmerman E, Ulbright TM, Foster RS, Einhorn LH, Kelley MR. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 2001;61(5):2220–2225. [PubMed] [Google Scholar]
- 174.Langie SA, Knaapen AM, Brauers KJ, van Berlo D, van Schooten FJ, Godschalk RW. Development and validation of a modified comet assay to phenotypically assess nucleotide excision repair. Mutagenesis. 2006;21(2):153–158. doi: 10.1093/mutage/gel013. [DOI] [PubMed] [Google Scholar]
- 175.Chevillard S, Radicella JP, Levalois C, Lebeau J, Poupon MF, Oudard S, Dutrillaux B, Boiteux S. Mutations in OGG1, a gene involved in the repair of oxidative DNA damage, are found in human lung and kidney tumours. Oncogene. 1998;16(23):3083–3086. doi: 10.1038/sj.onc.1202096. [DOI] [PubMed] [Google Scholar]
- 176.Fan CY, Liu KL, Huang HY, Barnes EL, Swalsky PA, Bakker A, Woods J, Finkelstein SD. Frequent allelic imbalance and loss of protein expression of the DNA repair gene hOGG1 in head and neck squamous cell carcinoma. Lab Invest. 2001;81(10):1429–1438. doi: 10.1038/labinvest.3780356. [DOI] [PubMed] [Google Scholar]
- 177.Marsin S, Vidal AE, Sossou M, Ménissierde Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J Biol Chem. 2003;278(45):44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 178.Habib SL, Phan MN, Patel SK, Li D, Monks TJ, Lau SS. Reduced constitutive 8-oxoguanine-DNA glycosylase expression and impaired induction following oxidative DNA damage in the tuberin deficient Eker rat. Carcinogenesis. 2003;24(3):573–582. doi: 10.1093/carcin/24.3.573. [DOI] [PubMed] [Google Scholar]
- 179.Offer H, Wolkowicz R, Matas D, Blumenstein S, Livneh Z, Rotter V. Direct involvement of p53 in the base excision repair pathway of the DNA repair machinery. FEBS Lett. 1999;450(3):197–204. doi: 10.1016/s0014-5793(99)00505-0. [DOI] [PubMed] [Google Scholar]
- 180.Zhou J, Ahn J, Wilson SH, Prives C. A role for p53 in base excision repair. EMBO J. 2001;20(4):914–923. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Seo YR, Fishel ML, Amundson S, Kelley MR, Smith ML. Implication of p53 in base excision DNA repair: in vivo evidence. Oncogene. 2002;21(5):731–737. doi: 10.1038/sj.onc.1205129. [DOI] [PubMed] [Google Scholar]
- 182.Fornace AJ, Jr, Zmudzka B, Hollander MC, Wilson SH. Induction of beta-polymerase mRNA by DNA-damaging agents in Chinese hamster ovary cells. Mol Cell Biol. 1989;9(2):851–853. doi: 10.1128/mcb.9.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jaiswal AS, Balusu R, Armas ML, Kundu CN, Narayan S. Mechanism of adenomatous poly-posis coli (APC)-mediated blockage of long-patch base excision repair. Biochemistry. 2006;45(51):15903–15914. doi: 10.1021/bi0607958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Narayan S, Jaiswal AS, Balusu R. Tumor suppressor APC blocks DNA polymerase beta-dependent strand displacement synthesis during long patch but not short patch base excision repair and increases sensitivity to methylmethane sulfonate. J Biol Chem. 2005;280(8):6942–6949. doi: 10.1074/jbc.M409200200. [DOI] [PubMed] [Google Scholar]
- 185.Luchtenborg M, Weijenberg MP, Wark PA, Saritas AM, Roemen GM, van Muijen GN, de Bruïne AP, van den Brandt PA, de Goeij AF. Mutations in APC, CTNNB1 and K-ras genes and expression of hMLH1 in sporadic colorectal carcinomas from the Netherlands Cohort Study. BMC Cancer. 2005;5:160. doi: 10.1186/1471-2407-5-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hofseth LJ. The adaptive imbalance to genotoxic stress: genome guardians rear their ugly heads. Carcinogenesis. 2004;25(10):1787–1793. doi: 10.1093/carcin/bgh196. [DOI] [PubMed] [Google Scholar]
- 187.Bhakat KK, Mantha AK, Mitra S. Transcriptional Regulatory Functions of Mammalian AP-endonuclease (APE1/Ref-1), an Essential Multifunctional Protein. Antioxid Redox Signal. 2008 doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kim EJ, Um SJ. Thymine-DNA glycosylase interacts with and functions as a coactivator of p53 family proteins. Biochem Biophys Res Commun. 2008;377(3):838–842. doi: 10.1016/j.bbrc.2008.10.058. [DOI] [PubMed] [Google Scholar]
- 189.Halliwell B. Free radicals, reactive oxygen species and human disease: a critical evaluation with special reference to atherosclerosis. Br J Exp Pathol. 1989;70(6):737–757. [PMC free article] [PubMed] [Google Scholar]
- 190.Pansarasa O, Bertorelli L, Vecchiet J, Felzani G, Marzatico F. Age-dependent changes of antioxidant activities and markers of free radical damage in human skeletal muscle. Free Radic Biol Med. 1999;27(5-6):617–622. doi: 10.1016/s0891-5849(99)00108-2. [DOI] [PubMed] [Google Scholar]
- 191.Castillo C, Salazar V, Ariznavarreta C, Vara E, Tresguerres JA. Effect of isoflavone admini-stration on age-related hepatocyte changes in old ovariectomized femal Wistar rats. Phytomedicine. 2006;13(7):468–476. doi: 10.1016/j.phymed.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 192.Schaur RJ. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol Aspects Med. 2003;24(4-5):149–159. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 193.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42(4):318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 194.Ayyadevara S, Engle MR, Singh SP, Dandapat A, Lichti CF, Benes H, Shmookler Reis RJ, Liebau E, Zimniak P. Lifespan and stress resistance of Caenorhabditis elegans are increased by expression of glutathione transferases capable of metabolizing the lipid peroxidation product 4-hydroxynonenal. Aging Cell. 2005;4(5):257–271. doi: 10.1111/j.1474-9726.2005.00168.x. [DOI] [PubMed] [Google Scholar]
- 195.Kowalczyk P, Ciesla JM, Komisarski M, Kusmierek JT, Tudek B. Long-chain adducts of trans-4-hydroxy-2-nonenal to DNA bases cause recombination, base substitutions and frameshift mutations in M13 phage. Mutat Res. 2004;550(1-2):33–48. doi: 10.1016/j.mrfmmm.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 196.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA inter-chain cross-links formed by acrolein and crotonaldehyde. J Am Chem Soc. 2003;125(1):50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 197.Kurtz AJ, Lloyd RS. 1,N2-deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4 -hydroxynonenal cross-link to peptides via Schiff base linkage. J Biol Chem. 2003;278(8):5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 198.Chung FL, Zhang L, Ocando JE, Nath RG. Role of 1,N2-propanodeoxyguanosine adducts as endogenous DNA lesions in rodents and humans. IARC Sci Publ. 1999;(150):45–54. [PubMed] [Google Scholar]
- 199.Wacker M, Schuler D, Wanek P, Eder E. Development of a 32P-postlabeling method for the detection of 1,N2-propanodeoxyguanosine adducts of trans-4-hydroxy-2-nonenal in vivo. Chem Res Toxicol. 2000;13(11):1165–1173. doi: 10.1021/tx000058r. [DOI] [PubMed] [Google Scholar]
- 200.Janowska B, Komisarski M, Prorok P, Sokołowska B, Kuśmierek J, Janion C, Tudek B. Nucleotide excision repair and recombination are engaged in repair of trans-4-hydroxy-2-nonenal adducts to DNA bases in Escherichia coli. Int J Biol Sci. 2009;5(6):611–620. doi: 10.7150/ijbs.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Esterbauer H, Eckl P, Ortner A. Possible mutagens derived from lipids and lipid precursors. Mutat Res. 1990;238(3):223–233. doi: 10.1016/0165-1110(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 202.Eckl PM, Ortner A, Esterbauer H. Genotoxic properties of 4-hydroxyalkenals and analogous aldehydes. Mutat Res. 1993;290(2):183–192. doi: 10.1016/0027-5107(93)90158-c. [DOI] [PubMed] [Google Scholar]
- 203.Karlhuber GM, Bauer HC, Eckl PM. Cytotoxic and genotoxic effects of 4-hydroxynonenal in cerebral endothelial cells. Mutat Res. 1997;381(2):209–216. doi: 10.1016/s0027-5107(97)00170-x. [DOI] [PubMed] [Google Scholar]
- 204.Cajelli E, Ferraris A, Brambilla G. Mutagenicity of 4-hydroxynonenal in V79 Chinese hamster cells. Mutat Res. 1987;190(2):169–171. doi: 10.1016/0165-7992(87)90050-9. [DOI] [PubMed] [Google Scholar]
- 205.Emerit I, Khan SH, Esterbauer H. Hydroxynonenal, a component of clastogenic factors? Free Radic Biol Med. 1991;10(6):371–377. doi: 10.1016/0891-5849(91)90045-5. [DOI] [PubMed] [Google Scholar]
- 206.Licht CL, Stevnsner T, Bohr VA. Cockayne syndrome group B cellular and biochemical functions. Am J Hum Genet. 2003;73(6):1217–1239. doi: 10.1086/380399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Thorslund T, von Kobbe C, Harrigan JA, Indig FE, Christiansen M, Stevnsner T, Bohr VA. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol Cell Biol. 2005;25(17):7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Muftuoglu M, Sharma S, Thorslund T, Stevnsner T, Soerensen MM, Brosh RM, Jr, Bohr VA. Cockayne syndrome group B protein has novel strand annealing and exchange activities. Nucleic Acids Res. 2006;34(1):295–304. doi: 10.1093/nar/gkj410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Maddukuri L, Speina E, Christiansen M, Dudzińska D, Zaim J, Obtułowicz T, Kabaczyk S, Komisarski M, Bukowy Z, Szczegielniak J, Wójcik A, Kuśmierek JT, Stevnsner T, Bohr VA, Tudek B. Cockayne syndrome group B protein is engaged in processing of DNA adducts of lipid peroxidation product trans-4-hydroxy-2-nonenal. Mutat Res. 2009;666(1-2):23–31. doi: 10.1016/j.mrfmmm.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.D'Errico M, Lemma T, Calcagnile A, Proietti De SL, Dogliotti E. Cell type and DNA damage specific response of human skin cells to environmental agents. Mutat Res. 2007;614(1-2):37–47. doi: 10.1016/j.mrfmmm.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 211.Feng Z, Hu W, Tang MS. Trans-4-hydroxy-2-nonenal inhibits nucleotide excision repair in human cells: a possible mechanism for lipid peroxidation-induced carcinogenesis. Proc Natl Acad Sci U S A. 2004;101(23):8598–8602. doi: 10.1073/pnas.0402794101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Feng Z, Hu W, Marnett LJ, Tang MS. Malondialdehyde, a major endogenous lipid peroxidation product, sensitizes human cells to UV-and BPDE-induced killing and mutagenesis through inhibition of nucleotide excision repair. Mutat Res. 2006;601(1-2):125–136. doi: 10.1016/j.mrfmmm.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 213.Feng Z, Hu W, Hu Y, Tang MS. Acrolein is a major cigarette-related lung cancer agent: Preferential binding at p53 mutational hot-spots and inhibition of DNA repair. Proc Natl Acad Sci U S A. 2006;103(42):15404–15409. doi: 10.1073/pnas.0607031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Olinski R, Siomek A, Rozalski R, Gackowski D, Foksinski M, Guz J, Dziaman T, Szpila A, Tudek B. Oxidative damage to DNA and antioxidant status in aging and age-related diseases. Acta Biochim Pol. 2007;54(1):11–26. [PubMed] [Google Scholar]
- 215.Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, Walter CA, Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001;98(18):10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2'-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci U S A. 1990;87(12):4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic Res. 1998;29(6):573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- 218.Anson RM, Senturker S, Dizdaroglu M, Bohr VA. Measurement of oxidatively induced base lesions in liver from Wistar rats of different ages. Free Radic Biol Med. 1999;27(3-4):456–462. doi: 10.1016/s0891-5849(99)00091-x. [DOI] [PubMed] [Google Scholar]
- 219.Siomek A, Gackowski D, Rozalski R, Dziaman T, Szpila A, Guz J, Olinski R. Higher leukocyte 8 -oxo-7,8-dihydro-2’ -deoxyguanosine and lower plasma ascorbate in aging humans? Antioxid Redox Signal. 2007;9(1):143–150. doi: 10.1089/ars.2007.9.143. [DOI] [PubMed] [Google Scholar]
- 220.Foksinski M, Rozalski R, Guz J, Ruszkowska B, Sztukowska P, Piwowarski M, Klungland A, Olinski R. Urinary excretion of DNA repair products correlates with metabolic rates as well as with maximum life spans of different mammalian species. Free Radic Biol Med. 2004;37(9):1449–1454. doi: 10.1016/j.freeradbiomed.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 221.Hart RW, Setlow RB. Correlation between de-oxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc Natl Acad Sci U S A. 1974;71(6):2169–2173. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Goukassian D, Gad F, Yaar M, Eller MS, Nehal US, Gilchrest BA. Mechanisms and implications of the age-associated decrease in DNA repair capacity. FASEB J. 2000;14(10):1325–1334. doi: 10.1096/fj.14.10.1325. [DOI] [PubMed] [Google Scholar]
- 223.Chen SK, Hsieh WA, Tsai MH, Chen CC, Hong AI, Wei YH, Chang WP. Age-associated decrease of oxidative repair enzymes, human 8-oxoguanine DNA glycosylases (hOgg1), in human aging. J Radiat Res (Tokyo) 2003;44(1):31–35. doi: 10.1269/jrr.44.31. [DOI] [PubMed] [Google Scholar]
- 224.Cooke MS, Evans MD, Herbert KE, Lunec J. Urinary 8-oxo-2'-deoxyguanosine–source, significance and supplements. Free Radic Res. 2000;32(5):381–397. doi: 10.1080/10715760000300391. [DOI] [PubMed] [Google Scholar]
- 225.Shigenaga MK, Gimeno CJ, Ames BN. Urinary 8-hydroxy-2'-deoxyguanosine as a biological marker of in vivo oxidative DNA damage. Proc Natl Acad Sci U S A. 1989;86(24):9697–9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Cooke MS, Evans MD, Dove R, Rozalski R, Gackowski D, Siomek A, Lunec J, Olinski R. DNA repair is responsible for the presence of oxidatively damaged DNA lesions in urine. Mutat Res. 2005;574(1-2):58–66. doi: 10.1016/j.mrfmmm.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 227.Martin GM, Loeb LA. Ageing: mice and mitochondria. Nature. 2004;429(6990):357–359. doi: 10.1038/429357a. [DOI] [PubMed] [Google Scholar]
- 228.Takasawa M, Hayakawa M, Sugiyama S, Hattori K, Ito T, Ozawa T. Age-associated damage in mitochondrial function in rat hearts. Exp Gerontol. 1993;28(3):269–280. doi: 10.1016/0531-5565(93)90034-b. [DOI] [PubMed] [Google Scholar]
- 229.Kopsidas G, Kovalenko SA, Kelso JM, Linnane AW. An age-associated correlation between cellular bioenergy decline and mtDNA rear-rangements in human skeletal muscle. Mutat Res. 1998;421(1):27–36. doi: 10.1016/s0027-5107(98)00150-x. [DOI] [PubMed] [Google Scholar]
- 230.Sharma MK, Buettner GR. Interaction of vitamin C and vitamin E during free radical stress in plasma: an ESR study. Free Radic Biol Med. 1993;14(6):649–653. doi: 10.1016/0891-5849(93)90146-l. [DOI] [PubMed] [Google Scholar]
- 231.Kirkwood TB, Austad SN. Why do we age? Nature. 2000;408(6809):233–238. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- 232.McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36(2):197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 233.Sohal RS, Mockett RJ, Orr WC. Mechanisms of aging: an appraisal of the oxidative stress hypothesis. Free Radic Biol Med. 2002;33(5):575–586. doi: 10.1016/s0891-5849(02)00886-9. [DOI] [PubMed] [Google Scholar]
- 234.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11(3):298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 235.Loft S, Poulsen HE. Estimation of oxidative DNA damage in man from urinary excretion of repair products. Acta Biochim Pol. 1998;45(1):133–144. [PubMed] [Google Scholar]
- 236.Foksinski M, Rozalski R, Guz J, Ruszkowska B, Sztukowska P, Piwowarski M, Klungland A, Olinski R. Urinary excretion of DNA repair products correlates with metabolic rates as well as with maximum life spans of different mammalian species. Free Radic Biol Med. 2004;37(9):1449–1454. doi: 10.1016/j.freeradbiomed.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 237.Kleiber M. New York: John Wiley; 1961. The fire of life. Ref Type: Serial (Book,Monograph) [Google Scholar]
- 238.Sohal RS, Mockett RJ, Orr WC. Mechanisms of aging: an appraisal of the oxidative stress hypothesis. Free Radic Biol Med. 2002;33(5):575–586. doi: 10.1016/s0891-5849(02)00886-9. [DOI] [PubMed] [Google Scholar]
- 239.Sohal RS, Mockett RJ, Orr WC. Mechanisms of aging: an appraisal of the oxidative stress hypothesis. Free Radic Biol Med. 2002;33(5):575–586. doi: 10.1016/s0891-5849(02)00886-9. [DOI] [PubMed] [Google Scholar]
- 240.Rozalski R, Gackowski D, Roszkowski K, Foksinski M, Olinski R. The level of 8-hydroxyguanine, a possible repair product of oxidative DNA damage, is higher in urine of cancer patients than in control subjects. cancer Epidemiol Biomarkers Prev. 2002;11(10):1072–1075. [PubMed] [Google Scholar]
- 241.Gackowski D, Speina E, Zielinska M, Kowalewski J, Rozalski R, Siomek A, Paciorek T, Tudek B, Olinski R. Products of oxidative DNA damage and repair as possible bio-markers of susceptibility to lung cancer. cancer Res. 2003;63(16):4899–4902. [PubMed] [Google Scholar]
- 242.Barja G, Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J. 2000;14(2):312–318. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- 243.Cutler RG. Antioxidants and aging. Am J Clin Nutr. 1991;53(1 Suppl):373S–379S. doi: 10.1093/ajcn/53.1.373S. [DOI] [PubMed] [Google Scholar]
- 244.Cutler RG. Human longevity and aging: possible role of reactive oxygen species. Ann N Y Acad Sci. 1991;621:1–28. doi: 10.1111/j.1749-6632.1991.tb16965.x. [DOI] [PubMed] [Google Scholar]
- 245.Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN. DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc Natl Acad Sci U S A. 1998;95(1):288–293. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hamilton ML, Guo Z, Fuller CD, Van Remmen H, Ward WF, Austad SN, Troyer DA, Thompson I, Richardson A. A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA. Nucleic Acids Res. 2001;29(10):2117–2126. doi: 10.1093/nar/29.10.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Shigenaga MK, Gimeno CJ, Ames BN. Urinary 8-hydroxy-2'-deoxyguanosineas a biological marker of in vivo oxidative DNA damage. Proc Natl Acad Sci U S A. 1989;86(24):9697–9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477(1-2):7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]