Abstract
The proper number of cells in developing tissues is achieved by coordinating cell division with apoptosis. In Drosophila, the adult wing is derived from wing imaginal discs, which undergo a period of growth and proliferation during larval stages without much programmed cell death. In this report, we demonstrate that the Drosophila casein kinase Iε/δ, known as Discs overgrown (Dco), is required for maintaining this low level of apoptosis. Expression of dco can suppress the apoptotic activity of Head involution defective (Hid) in the developing eye. Loss of dco in the wing disc results in a dramatic reduction in expression of the caspase inhibitor DIAP1 and a concomitant activation of caspases. The regulation of DIAP1 by Dco occurs by a post-transcriptional mechanism that is independent of hid. Mutant clones of dco are considerably smaller than controls even when apoptosis is inhibited, suggesting that Dco promotes cell division/growth in addition to its role in cell survival. The dco phenotype cannot be explained by defects Wingless (Wg) signaling. We propose that Dco coordinates tissue size by stimulating cell division/growth and blocking apoptosis via activation of DIAP1 expression.
Keywords: Drosophila, apoptosis, Dco, Casein Kinase I, DIAP1, Wing disc, Wnt, Wingless
Introduction
Apoptosis, or programmed cell death (PCD), is a tightly regulated mechanism of cell death that is essential to animal development and homeostasis of adult organs (Jacobson et al., 1997; Meier et al., 2000). In many developmental contexts, PCD is required for the removal of excess cells during morphogenesis. Apoptosis also serves as a quality control mechanism to eliminate abnormal or damaged cells from the organism. In addition, there is a link between cell growth/proliferation and PCD. Cells stimulated to grow and divide often also have an increased propensity for undergoing apoptosis (Abrams, 2002; Green and Evan, 2002). This relationship has been suggested to act as a “fail-safe” system, to prevent unwanted expansion of cell populations. Inhibition of this system is thought to occur during the multi-step process of oncogenesis (Evan and Vousden, 2001; Green and Evan, 2002; Hanahan and Weinberg, 2000).
PCD is executed by a family of cysteine proteases called caspases. Caspases are synthesized as inactive zymogens and are processed to their active form to cleave cellular substrates, thus promoting the apoptotic pathway. Caspases and many of their regulatory components, such as the Bcl2 family proteins, are evolutionarily conserved from nematodes to mammals (Kaufmann and Hengartner, 2001). However, additional strategies have evolved in insects and higher organisms to provide greater flexibility of regulation. One example is the Inhibitor of Apoptosis (IAP) family of proteins. IAPs can bind to caspases, inhibiting their processing, catalytic activity or the ability of active caspase to bind to substrates (Deveraux and Reed, 1999; Tenev et al. 2005). IAPs can also facilitate caspase degradation by targeting them for ubiquitination via their E3 ubiquitin ligase activity (Wilson et al., 2002; Yang et al., 2000).
Studies of a Drosophila member of the IAP family, DIAP1, encoded by the thread (th) locus, have provided critical insights into the function and regulation of IAPs. DIAP1 can be bound and inhibited by Head involution defective (Hid), Grim and Reaper (Rpr) (Hay, 2000). Overexpression of these pro-apoptotic factors can down regulate DIAP1 levels by promoting its degradation via the ubiquitin ligase activity of DIAP1 as well as inhibiting DIAP1 translation (Hays et al., 2002; Holley et al., 2002; Ryoo et al., 2002; Yoo et al., 2002). Once caspases are activated, they can cleave DIAP1, stimulating its ubiquitination and degradation (Ditzel et al., 2003). This multilevel regulation of DIAP1 reflects the critical role it plays in cell survival, as evidenced by the massive activation of caspases and apoptosis observed in DIAP1 mutants (Ryoo et al., 2004; Yoo et al., 2002).
The connection between cell growth/proliferation and cell death and pattern formation has been extensively studied in the Drosophila wing imaginal discs (Edgar, 1999; Serrano and O’Farrell, 1997; Tapon et al., 2001a). During the three larval stages, the wing discs undergo a rapid phase of cell growth and proliferation with very low levels of PCD (Milan et al., 1997). The Drosophila homologue of Casein Kinase Iε/δ (CKIε/δ), known as discs overgrown/double time (dco), was shown to be essential for imaginal tissue development (Jursnich et al., 1990; Zilian et al., 1999). Weak dco mutants exhibit a lag in disc growth, while stronger alleles have small or severely degenerated discs. Mitotic clones of dco null mutant cells in the wing disc did not grow or survive when induced during first or second larval stages, though small clones could be observed if induced at later stages (Zilian et al., 1999). Electron micrographs of dco mutant discs demonstrated the presence of basally extruded cells with nuclear condensation, suggesting that PCD was occurring. Based on these observations it was concluded that dco is required for cell proliferation and/or survival through an undetermined mechanism.
In this report we demonstrate that Dco is an essential cell survival factor in the wing imaginal disc. We initially identified Dco as a potent suppressor of Hid induced cell death in the eye. This Hid antagonism requires Dco kinase activity. Loss of dco in the wing results in massive PCD, which is a major cause of the small disc phenotype seen in dco mutants. When apoptosis is inhibited, dco mutant wings are normally patterned, suggesting that dco’s effect on cell survival is not an indirect consequence of abnormal cell specification. We demonstrate that dco mutant cells have a dramatic post-transcriptional reduction of DIAP1 protein levels, which is sufficient to explain the increase in PCD. This regulation of DIAP1 is independent of hid and not does appear to require the E3 ligase activity of DIAP1. We also provide results suggesting that Dco promotes cell growth/proliferation in the wing disc.
CKI family members, including CKIδ and CKIε, have been implicated in several developmentally important signaling pathways. In Drosophila, dco has been shown to act as a positive regulator of planar cell polarity in the larval eye and pupal wing (Strutt et al., 2006; Klein et al., 2006). In fly cell culture and the wing imaginal disc, dco has been shown to be required for Wnt/Wingless (Wg) signaling (Cong et al., 2004; Klein et al., 2006; Zhang et al., 2006). Wg signaling is known to promote cell survival in the wing imaginal discs (Giraldez and Cohen, 2003; Johnston and Sanders, 2003). However, we find that cells homozygous for a null dco allele have normal Wg signaling in the developing wing. Our data support a model where Dco promotes cell survival by activating DIAP1 protein levels independently of Wg signaling.
Materials and methods
Fly strains
The following fly strains were used. Oregon R was our wild type stock. The GMR-hid10, GMR-hid1M, GMR-rpr2, GMR-rpr5, GMR-grim2 and GMR-grim3 (Chen et al., 1996; Rodriguez et al., 1999) stocks were from J. Abrams. GMR-hidala5 (Bergmann et al., 1998) was from A. Bergmann. UAS-lacZ (Brand and Perrimon, 1993), UAS-p35 (Mergliano and Minden, 2003), enGal4 (Neufeld et al., 1998) and ptcGal4 (Johnson et al., 1995) and thj5c8 (Hay et al., 1995) were from the Bloomington Stock Center. UAS-dco (on the X chromosome; Price and Kalderon, 2002) was from M. Young. A wild-type UAS-dco stock and UAS-dcoD132N mutant (Klein et al., 2006) were obtained from M. Mlodzik and a UAS-dco and UAS-dcoK38R (Strutt et al., 2006) was provided by D. Strutt. GMRGal4 (Freeman, 1996) was from M. Freeman. The hid05014and hidX14 stocks (Grether et al., 1995) were from K. White, while the Df(3L)H99 and UAS-DIAP1 stocks (Hay et al., 1995) were from B. Hay. The dco mutant alleles dcoP915, dcoP1396 and dcole88 (Zilian et al., 1999) were from M. Noll. The th6B allele (Lisi et al., 2000) was from K. White. FRT82B P[w+, Ubi-GFPnls] and FRT82B P[w+, armlacZ] (Tapon et al., 2001b) were from I. Hariharan while all other chromosomes for clonal analysis, yw flp122, FRT82B (Xu et al., 1993) were from the Bloomington Stock Center. The UAS-dcoRNAi transgene was constructed by cloning an inverted repeat of the C-terminal non-kinase domain coding sequence using the PCR primers 5′CCAGTCTAGACAGCGGCCGCAGGACG GAGCGGAC3′ and 5′GCTATTTAGATCTAAACCGCAAGGGGTGCGGTGG3′ into the pWIZ vector (Lee and Carthew, 2003). Transgenic flies were generated following the standard P element methodology.
Generation of dco clones
To generate dco clones with p35 expressed in the posterior domain of the wing disc, the fly strain yw, flp122; enGal4; FRT82B P[armlacZ]/TM6 was crossed to w, UAS-p35; FRT82Bdcole88. For the DIAP-lacZ experiment, the following strain yw, flp122; enGal4; FRT82B P[w+, Ubi-GFPnls]/TM6 was crossed to UASp35; thj5c8, FRT82Bdcole88/TM6. The progenies were heat shocked during early 2nd instar larval stage for 1 hour at 37°C. The larvae were dissected and fixed with 4% formaldehyde 60 to 72 hours later.
Immunostainings, TUNEL and in situ hybridization
Immunostainings were performed as previously described (Parker et al., 2002). Mouse anti-DIAP1 (1:200) and rabbit anti-active Drice (1:2,000) (Yoo et al., 2002) were both provided by B. Hay. Guinea pig anti-Sen (1:5000) was made by immunization with GST-Sen as previously described (Nolo et al., 2000). Rabbit anti-Dll (1:200) was from J. Kohtz and rabbit anti-Spalt (1:100) (Kuhnlein et al., 1994) was from R. Schuh and B. Mollereau. Rabbit anti-β-galactosidase (Cappel, ICN, Costa Mesa, CA) and mouse anti-β-galactosidase (Sigma, St. Louis, MO) were both used at 1:500. Rabbit anti-phospho-Histone 3 (1:150) was from Upstate (Lake Placid, NY). Monoclonal anti-En (1:20) and anti-Wg (1:100) were from the Developmental Studies Hybridoma Bank (Iowa City, IA). Cy3- and Alexa Fluor 488-conjugated secondary antibodies were from Jackson Immunochemicals (West Grove, PA) and Molecular Probes (Eugene, OR), respectively. The TUNEL assay was performed using the Cell Death Detection kit (Roche Diagnostics, Indianapolis, IN) as previously described (Lin et al., 2004). In situ hybridization detection of hid, grim and rpr transcripts was performed as previously described (Lin et al., 2004). Pupal wings were stained as described (Axelrod, 2001) with Alexa568 phalloidin (Molecular Probes) at a concentration of 6.25U/ml.
Microscopy and image analysis
The adult eyes were photographed on a Leica M10 microscope as previously described (Parker et al., 2002). All fluorescent pictures were obtained with a Ziess Axiophot coupled to a Zeiss LSM510 confocal apparatus. The pixel values of the clone and twin areas in Fig. 8 were calculated using the histogram tool in Photoshop 6.
Fig. 8.
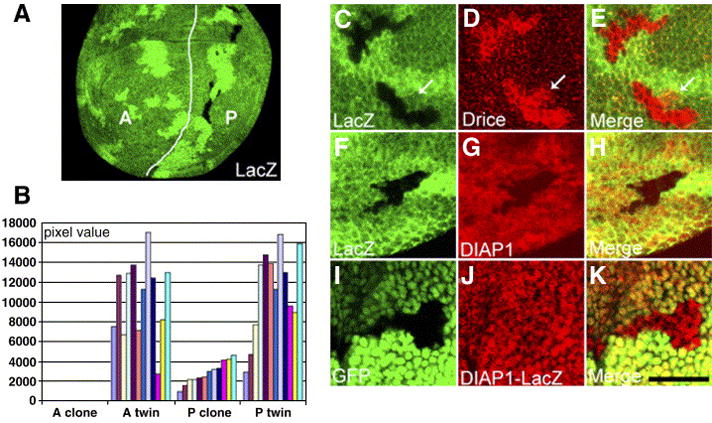
dco mutant clones are rescued by inhibition of caspases and exhibit increased Drice activation and decreased DIAP1 protein, but not DIAP-lacZ. dco null (dcole88) mutant clones were generated in an en-Gal4/UAS-p35 background and are marked by the absence of lacZ staining (green; A,C–H) or GFP (green; I–K) as indicated. (A) dco clones are found in the posterior (P) compartment where p35 is expressed by en-Gal4, but the anterior (A) clones are not observed. The white line marks the AP boundary. The presence of the twins at the anterior indicates that mitotic recombination had been induced. (B) Quantification of the dco clone/twin area. The pixel value of each bar represents the total area of clones or twins in A or P in one disc, and 12 discs containing a total of more than 30 individual clones were analyzed. The difference between the areas of A twins (average: 10451 pixels/disc) and P twins (average: 11138 pixels/disc) is not statistically significant (p>0.20 by the Mann-Whitney test). The average size of the P clones (2855 pixels/disc) is 25.6% of the size of the P twins. (C–E) Active Drice (red) is elevated in dco clones. Occasionally active Drice is observed adjacent to the clones (arrows). (F–H) DIAP1 protein (red) is decreased in dco clones. (I–K) dco clones marked by the absence of GFP were generated in a DIAP-lacZ background (thi5c8). DIAP-LacZ (red) is not altered in the clones. Scale bar: 20μm.
Results
Dco preferentially suppresses Hid induced cell death in the eye
dco was identified in an EP misexpression screen (Rorth et al., 1998) for genes that interact with the Wg signaling pathway. When placed under the control of the eye-specific GMR promoter, Wg causes a severe reduction in eye size (Parker et al., 2002). This is due in part to an increase in apoptosis in the late larval/early pupal eye (H. V. Lin, A. Rogulja and K. Cadigan, in preparation). We expected that some EP lines that could suppress the GMR/wg phenotype would be inhibitors of PCD. One such example was EP(3)3280, where the P-element is inserted into the first intron of dco (data not shown). Similar suppression of GMR/wg was obtained with a UAS-dco line, confirming that dco was responsible for the effect. Misexpression of dco in several contexts outside the eye revealed no effect on Wg signaling (data not shown). However, expression of dco in the eye strongly suppressed the activity of the pro-apoptotic factor Hid (Fig. 1B). dco had more subtle effects on the small eye phenotype caused by GMR-rpr (Figs. 1D, F) and moderately suppressed GMR-grim (Fig. 1H). Because hid, grim and rpr are expressed by the heterologous GMR promoter, the suppression is not due to transcriptional inhibition of these pro-apoptotic genes.
Fig. 1.

Ectopic expression of dco suppresses the small eye phenotype induced by overexpression of the proapoptotic genes hid, rpr and grim. Micrographs are of adult female eyes. All flies shown contain GMR-Gal4 and either UAS-lacZ (A, C, E, J) or UAS-dco (B, D, F, H). They contain two copies of GMR-hid1M (A, B), one copy of GMR-rpr2 (C, D), GMR-rpr5 (E, F) or a copy each of GMR-grim2 and GMR-grim3 (G, H). Overexpression of dco strongly suppresses GMR-hidIM and other GMR-hid strains (data not shown), while suppression of GMR-rpr and GMR-grim is weaker and variable.
It has been previously shown that ectopic expression of activated Ras preferentially suppresses Hid induced cell death in the eye (Bergmann et al., 1998; Kurada and White, 1998). However, the ability of activated Ras to suppress the GMR-hid small eye phenotype was blocked when five potential MAP kinase phosphorylation sites were mutated to alanine (GMR-hidala5) (Bergmann et al., 1998). Both EP(3)3280 and UAS-dco were able to suppress GMR-hidala5 to the same extent as wild type GMR-hid (Fig. 2B and data not shown), suggesting that dco overexpression is not working through a Ras/MAP kinase pathway.
Fig. 2.
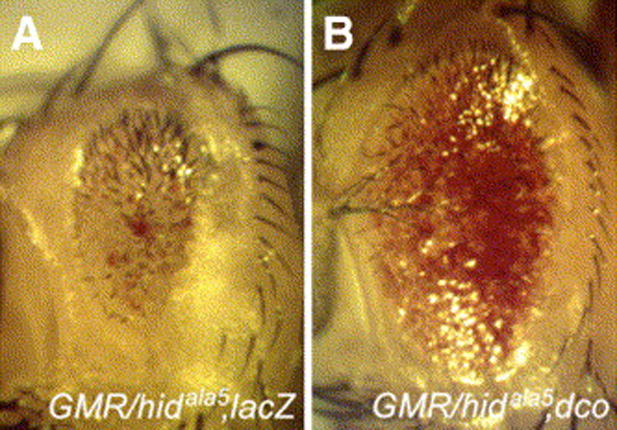
Ectopic expression of dco suppresses the small eye phenotype induced by a Ras/MAPK resistant form of Hid. Micrographs are of adult female eyes. Flies contain GMR-Gal4, GMR-hidala5 and either UAS-lacZ (A) or UAS-dco (B). Overexpression of dco strongly suppresses the GMR-hidala5 phenotype.
To determine whether endogenous dco regulates apoptosis in the eye, we reduced dco expression using RNAi via a dco hairpin construct (UAS-dcoRNAi). Expression of this hairpin with the GMR-Gal4 driver resulted in a roughened eye phenotype but did not reduce overall eye size (Fig. 3B). However, dcoRNAi enhanced the small eye phenotype of an intermediate GMR-hid1M construct (Figs. 3C, D). dcoRNAi had much smaller effects on GMR-rpr and GMR-grim constructs (data not shown). These data indicate that endogenous dco influences the level of apoptosis caused by misexpression of hid.
Fig. 3.
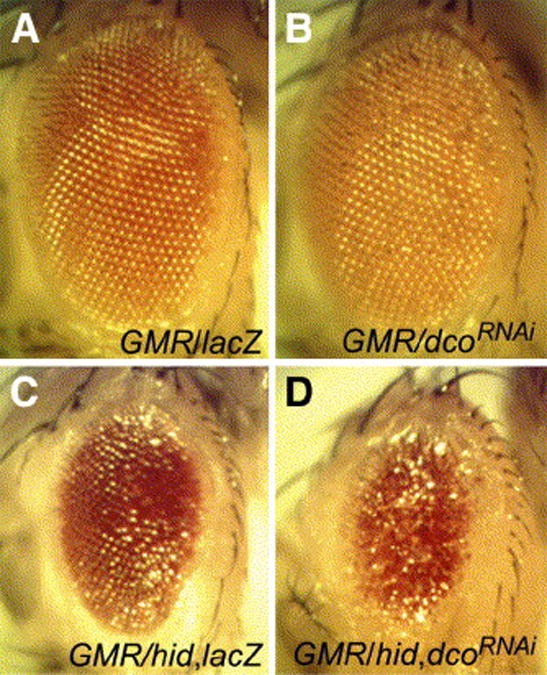
RNAi inhibition of dco enhances Hid-mediated apoptosis in the eye. Adult eyes from GMR-Gal4/UAS-lacZ (A, C), GMR-Gal4/UAS-dcoRNAi (B, D) without (A, B) or with one copy of GMR-hid1M (C, D). Depletion of dco roughens the eye but does not cause a reduction in size in an otherwise wild-type background (B), but does significantly enhance the GMR-hidIM phenotype (compare C and D).
To examine whether the kinase activity of Dco is required for Hid antagonism, two mutant forms of dco were co-expressed with hid in the eye. dcoD132N is predicted to compromise ATP binding (Peters et al., 1999; Zhu et al., 1998) while dcoK38R contains a mutation within the catalytic site (Peters et al., 1999; DeMaggio et al., 1992; Fish et al., 1995). Both mutants can act as dominant negatives for CKI function (Zhu et al., 1998; Peters et al., 1999; McKay et al., 2001; Klein et al., 2006; Strutt et al., 2006). However, DcoD132N was clearly able to suppress the GMR-hid small eye phenotype (Fig. 4B), while DcoK38R had the opposite effect. The K38R form of Dco enhanced both a moderate/strong GMR-hid (Fig. 4D) and a weaker GMR-hid transgene (Fig. 4F). When expressed by GMRGal4 in an otherwise wild-type eye, DcoK38R caused a roughening of the eye, but did not effect eye size (data not shown). Because the D132N mutation has been reported to reduce, but not eliminate CKI kinase activity (Zhu et al., 1998), the data suggest that a partially active Dco can still suppress Hid-mediated apoptosis but some Dco kinase activity is required for antagonizing Hid activity.
Fig. 4.

Kinase activity is required for dco to suppress Hid-mediated apoptosis in the eye. Adult eyes from GMR-Gal4, GMR-hidala5 and either UAS-GFP (A), UAS-dco (B), UAS-dcoD132N (C) or UAS-dcoK38R (D). While the ATP-binding mutant dcoD132N suppresses Hid activity as well as wild-type dco, the kinase dead mutant K38R enhances the small eye phenotype. (E, F) Enhancement was also observed with a weaker GMR-hid transgene. GMR-Gal4, GMR-hid1M and either UAS-GFP (E) or UAS-dcoK38R (F). Wild type dco and dcoD132N strongly suppressed GMR-hid1M (data not shown).
dco mutant imaginal discs have elevated apoptosis
Although dco overexpression and RNAi knockdown effect the ability of Hid to promote apoptosis, mitotic clones homozygous for a dco null allele (dcole88) are readily detectable in late larval eye discs and mid-pupal eyes (data not shown). dco mutant clones are not recovered in the adult eye, although some scarring near the twin spots was seen (data not shown). We have not been able to detect increased apoptosis in the larval or pupal clones. Apparantly, cells lacking dco are eliminated sometime in late pupal development.
To further explore whether dco is required for suppressing PCD, we examined several other imaginal discs. Using the TUNEL DNA fragmentation assay, we compared the level of apoptosis in imaginal discs from wild type and hypomorphic dco mutants (dcoP915/P1395). Throughout third instar larval development, TUNEL positive cells in the wing were detected at low levels in wild type, as shown in Figs. 5A & D. In contrast, we found a dramatic increase in TUNEL signal in the dco mutant wing discs (Figs. 5B, E). Note that the dco hypomorphs are developmentally delayed, and the discs were normalized for developmental stage by examining the pattern of Wg expression. Elevated TUNEL signal was also observed in the leg and haltere discs from dco mutants, but no increase was observed in eye imaginal discs (data not shown).
Fig. 5.

dco mutant wing discs have elevated cell death and growth delay that is largely suppressed by removing one copy of hid, rpr and grim. Confocal images of wing discs stained for TUNEL (green) to label apoptotic cells and Wg (red, apical optical section) to identify the developmental age of each disc. The discs in panels A–C are early third instar and those in panels D-F are late third instar. (A, D) Wild type discs 3 and 5 days after egg laying. (B, E) dco mutant discs (dcoP915/dcoP1396), 6 and 8 days after egg laying. (C, F) Df(3L)H99/+, dco (dcoP915/dcoP1396) discs 4 and 6 days after egg laying. H99 is a deficiency that removes a chromosomal region including hid, rpr and grim. The green signals at the edge of the wing discs in D and F are non-specific staining of nuclei from the peripodal membrane. (G, H) Wild type (G) and Df(3L)H99/+, dco (H) pupal wings stained with phalloidin. The wings are 33 hr and 38 hr after the white prepupal stage, respectively. The white arrows denote the position of the longitudinal veins. The overall pattern of the H99/+; dco wings is identical to wild-type. Scale bar: 100μm.
To determine whether the elevated PCD we observe is related to hid, grim and/or rpr function, we lowered the dose of these genes in a dco mutant background. Removal of one copy of hid, grim and rpr (using the Df(3L)H99 deficiency) caused a significant reduction of TUNEL signal in dco mutants (Figs. 5E, F). Removal of one copy of hid alone did not significantly reduce the level of apoptosis, but a dramatic reduction was achieved by removing both copies of hid (data not shown). These data suggest that in dco mutant discs endogenous hid, grim and/or rpr are required for the elevated cell death.
As reported previously, dcoP915/P1395 mutant animals have a prolonged larval phase with slow imaginal disc growth (Zilian et al., 1999). These animals die in late larval or early pupal stages. We found that the decrease in cell death in H99, dcoP915/+, dcoP1395 discs was correlated with marked acceleration in disc growth and larval development (from 8–10 days to 5–6 days). These H99/+, dcoP915/P1395 animals have increased survival into late pupal stages, and the veination pattern of the majority of these animals are similar to wildtype (Figs. 5G, H). Occasional escapers survive to adulthood displaying normal legs and wrinkled or blistered wings (data not shown). These results suggests that the developmental delay in disc growth and larval development seen in dco mutants is largely caused by elevated apoptosis. In addition, the data in Fig. 5H indicate that patterning of the wing is normal in dco mutants when PCD is reduced.
Dco activates DIAP1 expression in the wing imaginal disc
Both overexpression and loss of function studies with dco suggest that it encodes an anti-apoptotic factor in Drosophila imaginal tissues. Our initial observation that overexpression of dco differentially suppresses GMR-hid and GMR-rpr is reminiscent of DIAP1 alleles containing mutations in its RING domain (Lisi et al., 2000; Wilson et al., 2002). It has been shown that these DIAP1 mutants lack ubiquitin ligase activity and may be present at higher levels due to stabilization of DIAP1. These findings prompted us to look at DIAP1 protein levels when dco expression is elevated (with UAS-dco) or reduced (UAS-dcoRNAi). Both UAS transgenes were driven by patched (ptc)-Gal4, which is active in a stripe on the anterior side of the anterior/posterior boundary of the wing imaginal disc (Johnson et al., 1995).
Overexpression of dco led to a moderate increase of DIAP1 protein levels in the ptc expression domain (Fig. 6B). Conversely, DIAP1 expression was decreased upon RNAi-mediated knockdown of dco (Fig. 6C). Using an antibody specific for the activated form of the effector caspase Drice (Yoo et al., 2002), we found that dco knockdown also caused a marked increased in Drice activation (Fig. 6E). Z-axis projection of ptc-Gal4/UAS-dcoRNAi wing discs reveals that the Drice-positive cells are found basally in the epithelial sheet (Fig. 6G) and that DIAP1 levels are low both apically and basally (Fig. 6H). Loss of DIAP1 is known to cause apoptosis (Ryoo et al. 2004; Wang et al. 1999; Yoo et al. 2002), and the data suggest that Dco promotes cell survival by maintaining DIAP1 expression.
Fig. 6.

Dco regulates DIAP1 protein levels in the wing imaginal disc. Confocal images of late third instar larval wing discs stained for DIAP1 in red (A–D,H,I) and activated Drice in green (E–G,I). ptcGal4 is expressed at an anterior strip next to the anterior-posterior boundary (indicated by arrows in B–D). All discs are oriented anterior to the left. (A) Wild type. (B) ptc-Gal4/UAS-dco. (C, E) ptc-Gal4/UAS-dcoRNAi. (D, F) ptc-Gal4/UAS-dcoRNAi; hid05014/hidX14. (G–I) Z axis projection of ptc-Gal4/UAS-dcoRNAi disc showing basal localization of Drice positive cells. Arrows indicate the reduction in DIAP1 staining in the ptc expression domain.
To determine whether hid was required for Dco regulation of DIAP1, the ptc-Gal4/UAS-dcoRNAi experiments were repeated in a hid mutant background. An obvious decrease in DIAP1 levels was still observed (Fig. 6D) indicating that Dco can activate DIAP1 independently of hid. The reduction in DIAP1 was also seen in a H99/+ background (data not shown). In contrast, the activation of Drice by dcoRNAi was greatly suppressed in hid mutants (Fig. 6F) or H99 heterozygotes (data not shown). This is consistent with the results from dco hypomorphic mutants (Figs. 5C, F).
To rule out the possibility that Dco regulates the transcript levels of hid, grim or rpr, in situ hybridization was performed on ptc-Gal4/UAS-dcoRNAi wing imaginal discs. hid expression was not detectably effected (Fig. 7A), but a small reduction in grim expression was sometimes observed within the Ptc domain (Fig. 7B) while a small increase in rpr transcripts was also consistently seen (Fig. 7C). These minor differences were not observed in wild-type discs (data not shown). We consider it highly unlikely that these minor and contradictory differences can explain the dramatic effect observed on DIAP1 protein levels.
Fig. 7.
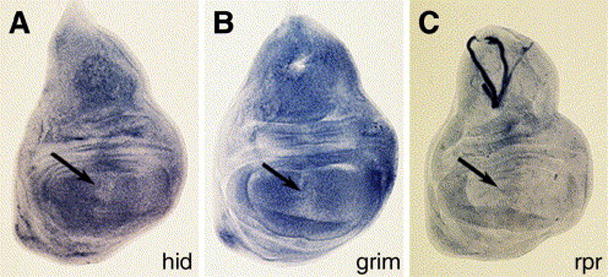
Inhibition of dco has little effect on hid, grim or rpr transcript levels. ptc-Gal4/UAS-dcoRNAidiscs stained for hid (A), grim (B) and rpr (C) transcripts. The location of the ptc expression domain is indicated by the arrows. There is no consistent change in hid expression by dcoRNAi, but a small decrease in grim was sometimes observed, as was a small increase in rpr expression.
We also examined the phenotype of dco null (dcole88) mutant cells induced by mitotic recombination. As reported previously (Zilian et al., 1999), clones of this allele were not obtained when recombination was induced before third larval instar (data not shown). This experiment was repeated in discs where the baculovirus caspase inhibitor p35 was constantly expressed in the posterior compartment via engrailed (en)-Gal4. As shown in Fig.8A, dco mutant clones were not detected in the anterior compartment (lacking p35), but were found in the posterior compartment expressing p35. Twin spots were visible in both compartments, indicating that mitotic recombination had occurred. These data indicate that caspase-dependent PCD contributes significantly to the loss of cells in dco mutant clones.
Consistent with a role of Dco in promoting cell survival by maintaining DIAP1 levels, we found elevated active Drice and decreased DIAP1 in dco clones (Figs. 8C–H). The activation of Drice is largely cell autonomous, though we occasionally see some additional signal adjacent to the clones (arrows in Figs. 8C–E). We speculate that this might represent dying cells engulfed by surrounding cells or macrophages. It has been reported in fly embryos that phagocytosis of cells containing activated caspases is not blocked by p35 expression (Mergliano and Minden, 2003). The dco null clones have decreased DIAP1 levels regardless of the area in the wing discs (hinge, pouch, notum) where the clones are found, indicating that Dco regulates DIAP1 throughout the wing discs.
When dco clones were generated in a Minute/+ background, a strong decrease in DIAP1 levels were observed, but only a small increase in activated Drice was seen, most prominently in the basal area of the clone (Supplementary Fig. 1). This suggests that cell competition is involved in promoting PCD in dco mutant cells, though not in the primary effect on DIAP1 protein expression.
To examine whether this activation of DIAP1 by dco is at the transcriptional level, we looked at the expression of a lacZ reporter under the control of the DIAP1 control region (DIAP1-lacZ) in dco clones and found that is was not altered (Figs. 8I–K). This suggests that Dco regulates DIAP1 by a post-transcriptional mechanism.
dco mutant clones are significantly smaller than their twins
A role for Dco in cell proliferation was suggested by the size of dco clones in the presence of the caspase inhibitor p35, which were consistently smaller than the twin spots (e.g., Fig. 8A). When this was quantitated, we found that the dco clones were on average 4 times smaller than the twin spots in the posterior compartment (Fig. 8B). Because the twins and clones originate from the two daughter cells of a single cell that underwent mitotic recombination, they grow for the same period of time in a similar location. Therefore the approximate 1:4 ratio of clone/twin size in dco mutants suggests a defect in the rate of growth/proliferation. To examine this further, we performed bromo deoxyuridine incorporation and immunostaining for phospho-Histone 3, which label cells that are undergoing DNA replication or mitosis, respectively. Neither analysis revealed any obvious differences between the clones and surrounding tissues (data not shown). These techniques are probably not sensitive enough to reveal subtle differences in cell proliferation that could contribute to the reduced clone size after several generations of cell division. The cells and nuclei in the clones also appear to be of similar size and shape as those in the surrounding tissue, suggesting that reduced cell size cannot account for the small size of dco clones.
The E3 ubiquitin ligase activity of DIAP1 is not required for its regulation by Dco
The C-terminus of DIAP1 contains a RING domain that has intrinsic E3 ubiquitin ligase activity which can promote its own degradation via auto-ubiquitination. Mutations that abolish this activity suppress Hid induced cell death but do not affect or enhance Rpr killing (Lisi et al., 2000; Wilson et al., 2002; Yokokura et al., 2004). This is consistent with the differential suppression of Hid and Rpr by ectopic expression of Dco (Figs. 1A–F). It is conceivable that Dco regulates DIAP1 level through modulating the auto-ubiquitination process.
To explore the relationship between Dco and DIAP1 E3 ligase activity, we examined whether a point mutation in one of the essential cysteine residues in the RING domain (th6B) disrupted the decrease in DIAP1 protein levels upon RNAi depletion of dco. One complication of monitoring DIAP1 levels in the RING domain mutant is the possibility that the mutant DIAP1 proteins are unusually stable compared to wild-type, due to lack of degradation by Hid or perhaps Rpr/Grim, since there are reports that these factors can also effect DIAP1 stability (Hays et al. 2002; Ryoo et al. 2002). However, mitotic clones of Df(3L)H99 did not contain higher levels of DIAP1 immunostaining in the wing imaginal disc (Figs. 9A–C). This was true of clones located throughout the wing pouch and notum (data not shown). While much of the tissue surrounding these clones are heterozygous for the hid, grim and rpr deficiency, the +/+ twins clones also had similar levels of DIAP1 staining compared to the H99 clones (data not shown). These data indicate that under normal conditions, endogenous Hid, Grim and Rpr do not significantly regulate the expression of DIAP1 in the wing.
Fig. 9.

Hid, Grim and Rpr do not regulate DIAP1 expression in the wing disc and DIAP1 E3 ligase activity is not required for Dco regulation of DIAP1. (A–C) Confocal images of late third wing imaginal discs containing clones of Df(3L)H99 (A–C), stained for DIAP1 (red). Clones were marked by GFP (green) and the location of the D/V boundary is indicated by the white arrow. The expression pattern of DIAP1 is unaffected in the clones. (D–F) Confocal images of wing imaginal discs stained for DIAP1 (red) and activated Drice (green) from ptc-Gal4/UAS-dcoRNAi; th6B/+ animals. As in ptc-Gal4/UAS-dcoRNAi controls (see Fig. 4), a strong decrease in DIAP1 levels and increase in activated Drice is observed in the ptc stripe. The scale bars are 20 μM in C and 100μM in F.
As previously described, expression of dcoRNAi in the ptc domain of the wing disc results in a significant decrease in DIAP1 expression (Fig. 6C) and an increase in Drice activation (Fig. 6E). In a th6B/+ background, similar results were obtained (Figs. 9D–E), suggesting that the effect of Dco on DIAP1 protein levels does not require DIAP1’s E3 ligase activity.
dco mutant clones have normal Wg and Dpp signaling
CKIε has been proposed to act both as a positive and negative regulator of Wnt signaling (Price, 2006). A positive role for Dco in Wg signaling has recently been demonstrated in cultured Drosophila S2 cells (Cong et al., 2004) and wing imaginal discs (Klein et al., 2006; Zhang et al., 2006). Wg signaling regulates cell proliferation and promotes cell survival in the wing imaginal discs (Giraldez and Cohen, 2003; Johnston and Sanders, 2003). Therefore, the phenotypes we have observed in dco mutant clones could be explained by a reduction in Wg signaling. However, we find that two targets of Wg signaling in wing imaginal discs, Senseless (Sen) and Distal-less (Dll) (Parker et al., 2002) are unaffected in cells (expressing p35) that are homozygous for the dco null mutation (Figs. 10A–F). We conclude that the cell survival function of Dco in wing imaginal discs is not linked to the Wg pathway and that Dco is dispensable for Wg signaling under the conditions used in our study.
Fig. 10.

The dco mutant cells have unaffected Wg and Dpp signaling. Confocal images of late third instar wing imaginal discs containing dcole88 null clones, generated as described in Fig. 5. Clones are marked by the lack of lacZ staining (green) and the Wg targets Sens and Dll and Dpp target Spalt are stained in red. Sens (A–C) and Dll (D–F) and Spalt (G–I) expression are normal in dco clones.
Like Wg, Dpp signaling has also been shown to promote cell proliferation and cell survival in the wing imaginal disc (Martin-Castellanos and Edgar, 2002; Moreno et al., 2002). CKIε has been shown to influence TGF-β signaling in mammalian cell culture, both negatively and positively, depending on the assay (Waddell et al. 2004). However, Dpp signaling appears to be normal in cells lacking dco, as judged by expression of the Dpp target Spalt (Figs. 10G–I).
Discussion
Dco promotes cell survival by activating DIAP1 expression
Dco is required for suppression of PCD in several developing tissues. Hypomorphic dco mutants have elevated apoptosis in the wing, leg and haltere discs (Fig. 5 and data not shown). Reduction of dco with either RNAi or with mitotic clones of a null allele also caused activation of the effector caspase Drice in wing discs (Figs. 6E and 8D). In the eye disc and early pupal eye, clones of the null dco allele did not result in obvious activation of Drice or PCD (data not shown). However, reduction of endogenous dco with RNAi did enhance the ability of Hid to promote cell death in the eye (Figs. 3C, D), indicating that Dco promotes cell survival in this tissue as well. It is possible that more subtle phenotype in the eye is caused by other CKI family members acting redundantly with Dco in this tissue.
In the wing disc, the loss of dco resulted in a dramatic reduction in DIAP1 expression (Figs. 6, 8, 9). The loss of DIAP1 in either fly embryos or the wing imaginal discs leads to massive activation of caspases and cell death (Ryoo et al., 2004; Wang et al., 1999; Yoo et al., 2002). Therefore, the reduction in DIAP1 levels observed in dco mutants is sufficient to explain the elevated caspase activation and apoptosis in these cells. The reduction in DIAP1 expression is not a secondary consequence of activating the apoptotic machinery, since a strong reduction in DIAP1 levels was observed in the presence of a caspase inhibitor (Fig. 8G), or in mutant backgrounds where active caspases are barely detectable (Figs. 6D, F). These results argue that reduction in dco leads to a loss of DIAP1 expression, which results in caspase activation and PCD.
Though our data indicate that Dco activation of DIAP1 is important to suppress apoptosis, the possibility existed that this regulation was due to non-specific effects of mispatterning in dco mutants. However, the phenotype of H99, dcoP915/+, dcoP1395 mutants argues strongly against this scenario. Heterozygosity of H99 suppressed the elevated PCD in the wing (Figs. 5C, F), shortened the developmental lag observed in dco hypomorphic mutants and increased the lethal phase of these mutants from early pupation to late pupal stages, with some escapers eclosing. The flies that emerged had normally patterned legs, but the pattern of the wings was difficult to analyze due to lack of unfolding and blistering. However, the veination pattern of the pupal wings (prior to folding) appeared normal (Fig. 5H). This data, combined with the fact that Dpp and Wg signaling appear normal in dco null clones (Fig. 10) strongly suggests that Dco does not have a significant role in patterning, but rather is a direct repressor of apoptosis.
The role of the IAP antagonists Hid, Grim and Rpr
dco interacts genetically with hid, grim and rpr in the eye (Figs. 1–4) and the wing (Fig. 5 and 6), suggesting a link between Dco and these pro-apoptotic factors. Dco appears to regulate DIAP1 at a post-transcriptional level (Fig. 8J) and it has been reported that DIAP1 is inhibited by Hid, Grim and Rpr at both the translational level and by increased protein turnover (Hays et al., 2002; Holley et al., 2002; Ryoo et al., 2002; Yoo et al., 2002). This raises the possibility that Dco could activate DIAP1 expression through repression of one or more of these IAP inhibitors. Alternatively, Dco could activate DIAP1 though a pathway that acts in parallel to Hid, Grim and Rpr.
If Dco regulates DIAP expression by repressing IAP inhibitor activity, it cannot work solely through Hid. The elevated caspase activation and PCD observed upon reduction of dco gene activity is greatly suppressed in a hid mutant background (Fig. 6F and data not shown). However, DIAP1 levels are still reduced with dco RNAi in hid mutants (Fig. 6D). The apoptotic phenotype in dco mutant wing discs is suppressed by removal of one copy of hid, grim and rpr (Figs. 5C,F), but not by removal of one copy of hid (data not shown). Therefore Dco would have to act by inhibiting Hid and Grim and/or Rpr. While Dco overexpression most effectively suppresses Hid activity (Fig. 1B), it also moderately suppresses Grim (Fig. 1H) and has a small effect on Rpr-induced PCD (Fig. 1F).
If Dco regulates DIAP1 in parallel to the IAP inhibitors, then how to explain the suppression of caspase activation and PCD in dco mutants when hid is mutated (Fig. 6F) or in Df(3L)H99 heterozygotes (Figs. 5C, F)? These data can be explained by a balance between negative regulators of apoptosis (Dco) and positive inputs (Hid/Grim/Rpr). Reduction of dco lowers DIAP1 levels, promoting caspase activation, however, the IAP inhibitors are known to bind to DIAP1 and sequester it from its normal function of caspase inhibition (Hay, 2000). So reduction of these DIAP1 inhibitors allows the lower levels of DIAP1 in the dco mutants to bind to caspases and prevent their activation.
In addition to binding to DIAP1 and inhibiting its ability to repress caspases, Hid, Grim and Rpr have been shown to down regulate DIAP1 expression (Hays et al., 2002; Holley et al., 2002; Ryoo et al., 2002; Yoo et al., 2002). Interestingly, we found that clones lacking hid, grim and rpr had no effect on DIAP1 protein levels in the wing disc (Figs. 9A–C). These results suggest that under normal conditions, these IAP inhibitors are not major regulators of DIAP1 expression in the wing imaginal disc. A more informative experiment would be to examine DIAP1 expression in clones lacking hid, grim, rpr and dco, to determine whether the decrease in DIAP1 requires the three pro-apoptotic factors. Attempts to perform this experiment have thus far not been possible, due to technical difficulties.
Hid, Grim and Rpr have been suggested to downregulate DIAP1 though its intrinsic E3 ubiquitin ligase activity (Hays et al., 2002; Ryoo et al., 2002; Yoo et al., 2002). However, a mutation that disrupts enzymatic activity (th6B) is still downregulated in dcoRNAi depleted cells (Figs. 9D–F). This argues against DIAP1 auto-ubiquitination playing an important role in Dco regulation of DIAP1. This experiment could only be performed in a th6B heterozygous background, because th6B clones do not survive/grow.
Dco may promote cell growth/proliferation
In addition to its role in promoting cell survival, our clonal analysis indicates that dco may promote cell growth/proliferation. In the presence of p35 caspase inhibitor, dco mutant clones are about four times smaller than their twins (Fig. 8B). One explanation for this is that p35 does not completely suppress the elevated apoptosis in the dco clone. However, the UAS-p35 transgene used is able to fully suppress a strong GMR-hid phenotype in the eye, and no TUNEL positive cells are observed in our dco clones in the presence of p35 (data not shown).
The conclusion that dco mutant cells grow/divide slower than controls is tempered by several caveats. Recent findings show that apoptotic cells in the wing disc can stimulate their neighbors to proliferate (de la Cova et al., 2004; Huh et al., 2004; Moreno and Basler, 2004; Ryoo et al., 2004). This phenomenon has been observed in cells expressing p35, suggesting that caspase activation but not PCD itself generates the proliferative signal (Huh et al., 2004; Ryoo et al., 2004; Perez-Garijo et al. 2005). Induced expression of Wg in the dying cells has been proposed to be part of the mitogenic signal for this compensatory proliferation (Ryoo et al., 2004). While we observed substantial ectopic Wg staining in dco mutant wing discs (Supplementary information Fig. 2), ectopic Wg was not observed in the p35 expressing dco mutant clones (n=30, data not shown). Despite this, our current data cannot rule out that the low clone/twin ratio we observe for dco clones in Fig. 8B could at least be partially due to increased proliferation of the twins.
Relation of Dco to Wg and other pathways affecting PCD/proliferation
In our study, no defect in Wg signaling was observed in mitotic clones for a dco null allele in wing imaginal discs (Fig. 10A–F). A similar result has been recently reported, though a hypomorphic dco allele was used (Strutt et al., 2006). In contrast, another recent study generated large clones with a strong dco allele (dcodbt-P) in a Minute/+ background, and reported no activation of caspases and a loss of Sens and adult wing margin, two readouts of Wg signaling (Klein et al., 2006). We have made dcole88 clones with the Minute technique, and find that they are infrequent and similar in size to the ones reported in Fig. 8. The few at the dorsal/ventral boundary expressed Sens normally (data not shown), while clones throughout the wing pouch had a dramatic reduction in DIAP1 expression and only a small increase in activated Drice (Supplementary information Fig. 1). Another report generating very large dcole88 clones using MS1096-Gal4/UAS-FLP, Minute and p35 expression (Zhang et al. 2006) observed a strong decrease in Sens expression. This report also found evidence that other CKI family members could promote Wg signaling (Zhang et al., 2006). Clone size and redundancy/compensation may explain the differences between our results and those of Klein et al. (2006) and Zhang et al. (2006). However, the primary phenotype observed when dco is removed under the conditions used in this study is a decrease in DIAP1 levels.
In our misexpression studies in the eye, Dco was able to suppress the ability of a Ras/MAPK resistant version of Hid (Bergmann et al., 1998) to induce apoptosis (Fig. 2). At least in the eye, Dco appears to control cell survival independently from Ras/MAPK signaling.
The phenotype of dco is similar to that of yorkie, a nuclear protein that is required for DIAP1 expression and proliferation (Huang et al., 2005). Yorkie is inhibited by a complex containing the Lats and Hippo kinases, and the adaptor protein Salvador (Sav). Clones mutant for lats, sav or hippo display an opposite phenotype to dco, i.e., dramatic increases in proliferation and a block in PCD (see Edgar, 2006 for a review). Double mutant clones for sav and dco do not survive/grow (data not shown), suggesting that Dco may act downstream of the Wts/Sav/Hippo complex. However, the expression of DIAP1-lacZ is upregulated in hippo clones (Wu et al., 2003) and decreased in yorkie clones (Huang et al., 2005) but we observe no effect on this transgene in dco clones (Figs. 8I–K). In addition, Wts/Sav/Hippo represses Cyclin E expression, but Dco does not (data not shown). Therefore, we consider it unlikely that Dco acts with Wts/Sav/Hippo in a simple linear pathway. Hippo has also been reported to phosphorylate DIAP1, leading to instability (Harvey et al., 2003; Pantalacci et al., 2003), so the possibility remains that Dco may interact with Hippo in this regard.
CKIδ/ε in other systems
Our data supports a model where Dco promotes cell survival in fly imaginal discs by maintaining expression of DIAP1 through a novel pathway. Do casein kinase Iδ/ε family members have similar functions in other organisms? The yeast CKI gene Hrr25 is involved in DNA repair, cell growth and viability (Hoekstra et al., 1991). In mammalian cell lines, CKIε plays a protective role against apoptosis induced by extrinsic death signals (Desagher et al., 2001; Izeradjene et al. 2004). This cell survival activity was correlated with CKIε phosphorylation of the pro-apoptotic factor Bid (Desagher et al., 2001). Inhibition of CKIδ induces apoptosis in trophoblast cells (Stoter et al. 2005). Further studies will be required to determine whether there are commonalities between these different systems. The ability of Dco to inhibit apoptosis and possibly promote cell proliferation makes it an attractive candidate for functioning as an oncogene in tumorigenesis.
Supplementary Material
Supplement Fig. 1. Mitotic clones of dco in a Minute/+ background result in loss of DIAP1, but only modest Drice activation. A apical (A–D) and basal (E–H) optical slice through a clone of dcole88 stained for DIAP1 (B,F) and active Drice (C,G). Clonal boundaries were marked with GFP (A,D). A strong reduction in DIAP1 is observed in the clones, while only a modest elevation of active Drice was observed in the basal portion of the clones. Note that there is appreciable Drice activation outside of the clones in the basal portion of the disc.
Supplement Fig. 2. The dying cells in dco mutant wing discs are basally extruded and express Wg. TUNEL (green) labels dying cells and Wg is stained in red. All four panels are confocal images of the same wing disc from a dcoP915/dcoP1396 larva. (A, C) Basal focal plane. (B, D) Apical focal plane. Wg retains a relatively normal pattern in the apical view but is ectopically expressed in the dying cells at the basal epithelium. Scale bar: 100μm.
Acknowledgments
We would like to thank all the researchers who provided the reagents described in the Methods section, especially M. Noll for the dco stocks, J. Abrams for the GMR-hid, GMR-rpr and GMR-grim stocks, K. White for the hid alleles, H. Steller, K. White and B. Hay for th alleles. Special thanks to D. Strutt and M. Mlodzik for providing UAS-dco mutant stocks shortly after publication. Thanks also to J. Kennell for generating one of the UAS-dcoRNAi stocks used in this study. We also would like to thank B. Hay for DIAP1 and active Drice antibodies, P. Meier for DIAP1 antibody, L. Saez and M. Young for the Dco antibody and the UAS-dco stock, and H. Richardson for the CycE antibodies. This work was supported by NIH grants RO1 GM59846 and RO1 CA95869 to K.M.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams JM. Competition and compensation: Coupled to death in development and cancer. Cell. 2002;110:403–406. doi: 10.1016/s0092-8674(02)00904-2. [DOI] [PubMed] [Google Scholar]
- Axelrod JD. Unipolar membrane association of Dishevelled mediates Frizzled planar cell polarity signaling. Genes Dev. 2001;15:1182–1187. doi: 10.1101/gad.890501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1994;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Cong F, Schweizer L, Varmus H. Casein kinase Iepsilon modulates the signaling specificities of dishevelled. Mole Cell Biol. 2004;24:2000–2011. doi: 10.1128/MCB.24.5.2000-2011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila Myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- DeMaggio AJ, Lindberg RA, Hunter T, Hoekstra MF. The budding yeast HRR25 gene product is a casein kinase I isoform. Proc Natl Acad Sci USA. 1992;89:7008–7012. doi: 10.1073/pnas.89.15.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desagher S, Osen-Sand A, Montessuit S, Magnenat E, Vilbois F, Hochmann A, Journot L, Antonsson B, Martinou JC. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol Cell. 2001;8:601–611. doi: 10.1016/s1097-2765(01)00335-5. [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Reed JC. IAP family proteins--suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, Meier P. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- Edgar BA. From small flies come big discoveries about size control. Nat Cell Biol. 1999;1:E191–E193. doi: 10.1038/70217. [DOI] [PubMed] [Google Scholar]
- Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- Fish KJ, Cegielska A, Getman ME, Landes GM, Virshup DM. Isolation and Characterization of Human Casein Kinase Iε (CKI), a Novel Member of the CKI Gene Family. J Biol Chem. 1995;270:14875–14883. doi: 10.1074/jbc.270.25.14875. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Gerether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cohen SM. Wingless and Notch signaling provide cell survival cues and control cell proliferation during wing development. Development. 2003;130:6533–6543. doi: 10.1242/dev.00904. [DOI] [PubMed] [Google Scholar]
- Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Haung J, Wu S, Barrera J, Matthews K, Pan D. The Hippo Signaling Pathway Coordinately Regulates Cell Proliferation and Apoptosis by Inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor and apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Hay BA. Understanding IAP function and regulation: a view from Drosophila. Cell Death Differ. 2000;7:1045–1056. doi: 10.1038/sj.cdd.4400765. [DOI] [PubMed] [Google Scholar]
- Hays R, Wickline L, Cagan R. Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nat Cell Biol. 2002;4:425–431. doi: 10.1038/ncb794. [DOI] [PubMed] [Google Scholar]
- Hoekstra MF, Liskay RM, Ou AC, Demaggio AJ, Burbee DG, Heffron F. Hrr25, a putative protein-kinase from budding yeast - association with repair of damaged DNA. Science. 1991;253:1031–1034. doi: 10.1126/science.1887218. [DOI] [PubMed] [Google Scholar]
- Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol. 2002;4:439–444. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol. 2004;14:1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Izeradjene K, Douglas L, Delaney AB, Houghton JA. Casein Kinase I attenuates Tumor Necrosis Factor-related apoptosis-inducing ligand-induced apoptosis by regulating the recruitment of Fas-associated death domain and procaspase-8 to the death-inducing signal complex. Cancer Res. 2004;64:8036–8044. doi: 10.1158/0008-5472.CAN-04-0762. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Grenier JK, Scott MP. patched overexpression alters wing disc size and pattern: transcriptional and post-transcriptional effects on hedgehog targets. Development. 1995;121:4161–4170. doi: 10.1242/dev.121.12.4161. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Sanders AL. Wingless promotes cell survival but constrains growth during Drosophila wing development. Nat Cell Biol. 2003;5:827–833. doi: 10.1038/ncb1041. [DOI] [PubMed] [Google Scholar]
- Jursnich VA, Fraser DE, Held LIJ, Ryerse J, Bryant PJ. Defective gap-junctional communication associated with imaginal disc overgrowth and degeneration caused by mutations of the dco gene in Drosophila. Dev Biol. 1990;140:413–429. doi: 10.1016/0012-1606(90)90090-6. [DOI] [PubMed] [Google Scholar]
- Kaufmann SH, Hengartner MO. Programmed cell death: alive and well in the new millennium. Trends Cell Biol. 2001;11:526–534. doi: 10.1016/s0962-8924(01)02173-0. [DOI] [PubMed] [Google Scholar]
- Klein TJ, Jenny A, Djiane A, Mlodzik M. CKIε/discs overgrown promotes both Wnt/b-catenin and Fz/PCP signaling in Drosophila. Curr Biol. 2006;16:1337–1343. doi: 10.1016/j.cub.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Kuhnlein RP, Frommer G, Friedrich M, Gonzalez-Gaitan M, Weber A, Wagner-Bernholz JF, Gehring WJ, Jackle H, Schuh R. splat encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 1994;13:168–179. doi: 10.1002/j.1460-2075.1994.tb06246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. Making a better RANi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Lin HV, Rogulja A, Cadigan KM. Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development. 2004;131:2409–2418. doi: 10.1242/dev.01104. [DOI] [PubMed] [Google Scholar]
- Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos C, Edgar BA. A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development. 2002;129:1003–1013. doi: 10.1242/dev.129.4.1003. [DOI] [PubMed] [Google Scholar]
- McKay RM, Peters JM, Graff J. The Casein Kinase I Family: Roles in Morphogenesis. Dev Biol. 2001;235:378–387. doi: 10.1006/dbio.2001.0307. [DOI] [PubMed] [Google Scholar]
- Meier P, Finch A, Evan G. Apoptosis in development. Nature. 2000;407:796–801. doi: 10.1038/35037734. [DOI] [PubMed] [Google Scholar]
- Mergliano J, Minden JS. Caspase-independent cell engulfment mirrors cell death pattern in Drosophila embryos. Development. 2003;130:5779–5789. doi: 10.1242/dev.00824. [DOI] [PubMed] [Google Scholar]
- Milan M, Campuzano S, Garcia-Bellido A. Developmental parameters of cell death in the wing disc of Drosophila. Proc Natl Acad Sci USA. 1997;94:5691–5696. doi: 10.1073/pnas.94.11.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E, Basler K, Morata G. Cells compete for Decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature. 2002;416:755–759. doi: 10.1038/416755a. [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Parker DS, Jemison J, Cadigan KM. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development. 2002;129:2565–2576. doi: 10.1242/dev.129.11.2565. [DOI] [PubMed] [Google Scholar]
- Perez-Garijo A, Martin FA, Struhl G, Morata G. Dpp signaling and the induction of neoplastic tumors by caspase-inhibited apoptotic cells in Drosophila. Proc Natl Acad Sci USA. 2005;102:17664–17669. doi: 10.1073/pnas.0508966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, McKay RM, McKay JP, Graff JM. Casein kinase I transduces Wnt signals. Nature. 1999;401:345–350. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase I. Cell. 2002;108:823–835. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- Price MA. CKI, there’s more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 2006;20:399–410. doi: 10.1101/gad.1394306. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Oliver H, Zou H, Chen P, Abrams J. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- Rorth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milan M, Benes V, Ansorge W, Cohen SM. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–57. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Serrano N, O’Farrell PH. Limb morphogenesis: connections between patterning and growth. Curr Biol. 1997;7:R186–R195. doi: 10.1016/s0960-9822(97)70085-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoter M, Bamberger AM, Aslan B, Kurth M, Speidel D, Loning T, Frank HG, Kaufmann P, Lohler J, Henne-Bruns D, Deppert W, Knippschild U. Inhibition of casein kinase I delta alters mitotic spindle formation and induces apoptosis in trophoblast cells. Oncogene. 2005;24:7964–7975. doi: 10.1038/sj.onc.1208941. [DOI] [PubMed] [Google Scholar]
- Strutt H, Price MA, Strutt D. Planar Polarity is Positively Regulated by Casein Kinase Iε in Drosophila. Curr Biol. 2006;16:1329–1336. doi: 10.1016/j.cub.2006.04.041. [DOI] [PubMed] [Google Scholar]
- Tapon N, Moberg KH, Hariharan IK. The coupling of cell growth to the cell cycle. Curr Opin Cell Biol. 2001a;13:731–737. doi: 10.1016/s0955-0674(00)00284-2. [DOI] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001b;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol. 2005;7:70–77. doi: 10.1038/ncb1204. [DOI] [PubMed] [Google Scholar]
- Waddell DS, Liberati NT, Guo X, Frederick JP, Wang XF. Casein Kinase Iε plays a functional role in the Transforming Growth Factor-β signaling pathway. J Biol Chem. 2004;279:29236–29246. doi: 10.1074/jbc.M400880200. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HAJ, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang JB, Dong JX, Pan DJ. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- Yokokura T, Dresnek D, Huseinovic N, Lisi S, Abdelwahid E, Bangs P, White K. Dissection of DIAP1 functional domains via a mutant replacement strategy. J Biol Chem. 2004;279:52603–52612. doi: 10.1074/jbc.M409691200. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang LJ, Wang SL, Feldman RMR, Clem RJ, Muller HAJ, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Zhang L, Jia J, Wang B, Amanai K, Wharton KA, Jiang J. Regulation of wingless signaling by the CKI family in Drosophila limb development. Dev Biol. 2006;299:221–237. doi: 10.1016/j.ydbio.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Shibasaki F, Price R, Guillernot JC, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Intramolecular masking of nuclear import signal on NF-AT4 by Casein Kinase I and MEKK1. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]
- Zilian O, Frei E, Burke R, Brentrup D, Gutjahr T, Bryant PJ, Noll M. double-time is identical to discs overgrown, which is required for cell survival, proliferation and growth arrest in Drosophila imaginal discs. Development. 1999;126:5409–5420. doi: 10.1242/dev.126.23.5409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Fig. 1. Mitotic clones of dco in a Minute/+ background result in loss of DIAP1, but only modest Drice activation. A apical (A–D) and basal (E–H) optical slice through a clone of dcole88 stained for DIAP1 (B,F) and active Drice (C,G). Clonal boundaries were marked with GFP (A,D). A strong reduction in DIAP1 is observed in the clones, while only a modest elevation of active Drice was observed in the basal portion of the clones. Note that there is appreciable Drice activation outside of the clones in the basal portion of the disc.
Supplement Fig. 2. The dying cells in dco mutant wing discs are basally extruded and express Wg. TUNEL (green) labels dying cells and Wg is stained in red. All four panels are confocal images of the same wing disc from a dcoP915/dcoP1396 larva. (A, C) Basal focal plane. (B, D) Apical focal plane. Wg retains a relatively normal pattern in the apical view but is ectopically expressed in the dying cells at the basal epithelium. Scale bar: 100μm.


