Abstract
Candida albicans is the most common cause of serious fungal disease in humans. Creation of isogenic null mutants of this diploid organism, which requires sequential gene targeting, allows dissection of virulence mechanisms. Published analyses of such mutants show a near-perfect correlation between C. albicans pathogenicity and the ability to undergo a yeast-to-hypha morphological switch in vitro. However, most studies used mutants constructed with a marker that is itself a virulence determinant and therefore complicates their interpretation. Using alternative markers, we created ~3000 homozygous deletion strains affecting 674 genes or roughly 11% of the C. albicans genome. Screening for infectivity in a mouse model and for morphological switching and cell proliferation in vitro, we identified 115 infectivity-attenuated mutants, of which nearly half demonstrated normal morphological switching and proliferation. Analysis of such mutants identified the glycolipid, glucosylceramide, as the first small molecule synthesized by this pathogen to be required specifically for virulence.
Introduction
Candida albicans is the major cause of serious fungal infections in the United States, and Candida species are the fourth most commonly cultured microbe from blood1. While some human fungal pathogens exist primarily as budding yeast cells (e.g. Cryptocococcus neoformans) or filamentous hyphal structures (e.g. Aspergillus spp.), C. albicans belongs to a group that freely alternates between these and other morphologies in response to specific environmental cues. Unlike other major fungal pathogens, C. albicans composes part of the normal human microbiome, inhabiting the skin and gastrointestinal tract as a commensal organism. However, it also produces serious disease in patients with immune deficits, who have undergone surgical instrumentation, or who have been treated with long courses of antibiotics. The attributable mortality from bloodstream C. albicans infections in adults is at least 15%2. Given the frequency and seriousness of these infections, there are surprisingly few tests and antifungal drugs to diagnose and treat them.
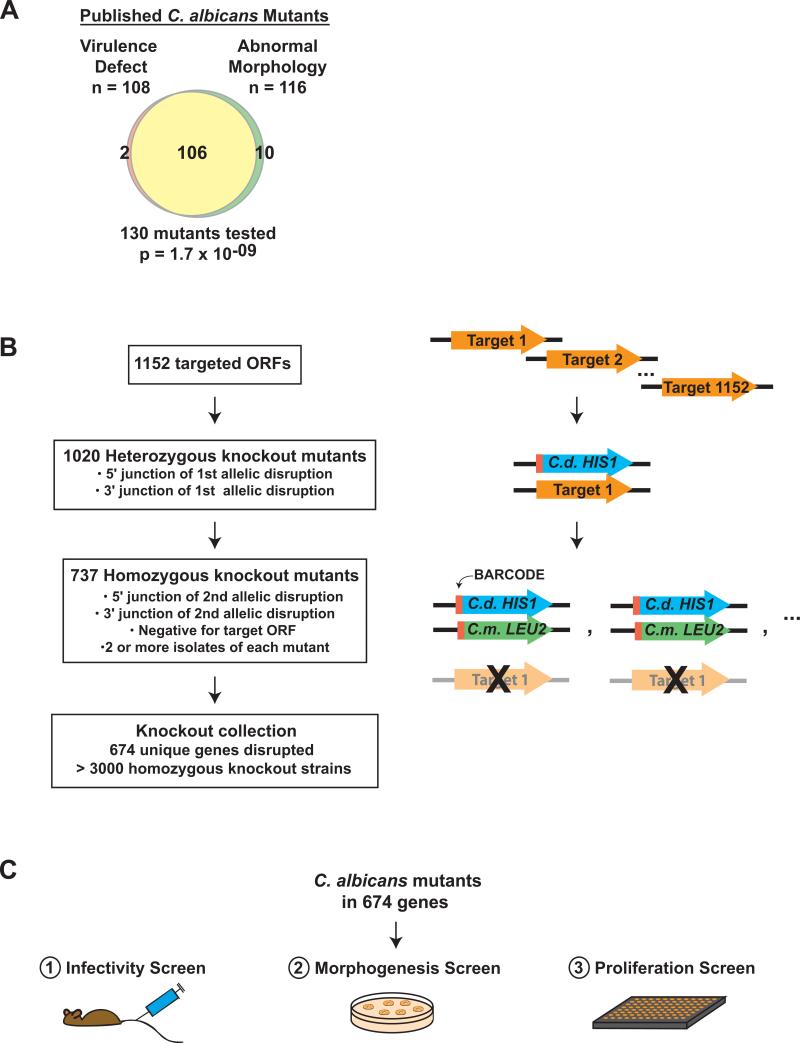
C. albicans has an obligate diploid genome and lacks a complete sexual cycle, two properties that have historically hindered genetic approaches to studying this organism. Nonetheless, numerous genes important for virulence have been identified through “reverse genetics,” in which both alleles of a selected gene are inactivated and the resulting mutant evaluated for ability to cause disease. These studies have found a strong concordance between virulence and the ability to alternate among three morphological forms—yeasts, hyphae, and pseudohyphae (Figure 1A and Table S13). Because the morphological switch is itself a complex transition involving changes in cellular shape, mechanical properties, interactions with other cells, and differential expression of myriad genes4-7, its close association with virulence has made it difficult to dissect out specific virulence mechanisms and effectors. Indeed, it remains uncertain whether the observed virulence defects result from defects in morphology per se or from genes whose expression is correlated with a particular morphological state8.
Figure 1. C. albicans mutants and screens.
A) Based on the published literature, Venn diagram depicting virtually complete overlap between C. albicans mutants with virulence defects and mutants with abnormal morphology (see Methods). Significance was calculated using the hypergeometric distribution. B) Construction of the homozygous gene disruption library by homologous recombination. One allele of each target gene was replaced by C. dubliniensis HIS1, and the second allele was replaced by C. maltosa LEU2. Red rectangles indicate the oligonucleotide barcodes. Successful knockout strains were defined as those with PCR-verified junctions at the 5′ and 3′ ends of the selectable markers, as well as absence of the target ORF. C) In vivo and in vitro screens of the C. albicans mutant library. As described in Methods, the 674 mutants were screened for infectivity in the mouse, for colony morphology on solid Spider medium at 30°C, and for proliferation in liquid SC at 37°C.
A second factor complicating our understanding of C. albicans virulence is the use of URA3 as a selectable marker in the majority of published mutants. Several studies have established that adequate expression of URA3 is essential for normal virulence as well as for efficient transitions between yeast and filamentous forms9-13. Because URA3 was used to replace the target gene of most published mutants and because its expression can vary significantly as a function of chromosomal position9-11, rigorous interpretation of published mutants has often proven difficult.
To investigate C. albicans virulence afresh, we created a large number of homozygous gene disruption mutants using auxotrophic markers that are neutral for virulence14 (Figure 1B). Because many C. albicans laboratory strains are aneuploid15,16, the parental strain SN152 was verified to have a diploid chromosome number by contour clamped homogeneous electric field (CHEF) gel analysis14 and comparative genome hybridization17 (CGH). Gene targets were selected to represent a wide variety of cellular processes, and the majority were previously unstudied. Each mutant was barcoded by embedding one of 48 unique oligonucleotide sequences in the gene disruption cassette. Rigorous quality control was implemented to verify deletion of both target alleles as well as the absence of a third copy of the open reading frame (ORF). To control for unwanted genetic rearrangements that sometimes arise during mutant construction in C. albicans17,18, two or more independent isolates were derived for each targeted gene. In total, we constructed ~3000 strains that correspond to deletions of 674 ORFs or ~11% of annotated coding genes in the C. albicans genome.
Three phenotypes that have been associated with virulence in animal models—infectivity in a mouse model of disseminated candidiasis, the yeast-to-hypha transition in vitro, and proliferation rates in vitro—were monitored across the entire deletion collection. Contrary to expectation, these traits were not strictly correlated with each other. In particular, many infectivity-defective mutants displayed normal morphological transitions and proliferation in vitro. We hypothesized that, unlike mutants with associated defects in morphogenesis and proliferation, those with isolated infectivity defects could be used to define discrete effectors of pathogenesis. We therefore pursued four such mutants with directed biochemical and virulence experiments, resulting in the identification of a biosynthetic pathway for glucosylceramide as a novel virulence factor that is important in the mouse, but dispensable for morphogenesis and proliferation in vitro. To our knowledge, glucosylceramide is the first small molecule synthesized by C. albicans to have these characteristics.
Results
Construction of a high-quality homozygous C. albicans gene deletion library
We designed our knockout library to include genes involved in a broad spectrum of cellular processes and to avoid a strong bias towards factors already known to affect C. albicans virulence. When this work was begun, the genome of clinical strain SC5314 had been sequenced and partially assembled by the Stanford Genome Technology Center19 We created a database that assembled the putative ORFs with results of pairwise BLAST comparisons to multiple sequenced genomes and predicted functional motifs (using the SMART algorithm). We next compiled the following nonexclusive sets of target genes: (1) Genes without clear homologs in two nonpathogenic yeasts, S. cerevisiae and S. pombe; (2) Genes apparently unique to C. albicans (i.e. not conserved in other sequenced genomes); (3) Genes with a functional motif potentially related to virulence (e.g. all genes with signatures of cell surface proteins such as signal sequences and GPI anchors); and (4) Additional ORFs of interest (e.g. genes whose transcripts are associated with the She3-dependent RNA transport system20). After excluding candidates that appeared unlikely to encode proteins, those in transposons, and ones that were homologous to essential genes in S. cerevisiae, we identified 1152 targets for disruption.
Mutants were constructed in C. albicans strain SN152, a derivative of SC5314 with auxotrophies for histidine, leucine, and arginine14 (Figure 1B). These nutritional markers are not required for virulence in the murine tail vein injection model that is most commonly used for virulence analysis14. Briefly, a fusion PCR technique was used to synthesize gene disruption cassettes containing either C. dubliniensis HIS1 or C. maltosa LEU2 flanked by ~350 nucleotides matching sequences upstream and downstream of the target gene. One of 48 20-nucleotide barcodes was included adjacent to the selectable markers. Heterozygous gene disruption strains were constructed by transformation of SN152 with a HIS1-marked gene disruption cassette; His+ transformants were screened by colony PCR for the presence of expected 5′ and 3′ junctions of the integrated DNA. Homozygous gene disruption strains were constructed by transforming the heterozygous knockout strain with a LEU2-marked gene disruption cassette; His+Leu+ transformants were screened for expected 5′ and 3′ junctions of the second disrupted allele and for absence of the original target ORF. At least two homozygous knockout isolates were obtained for each target gene.
As shown in Figure 1B, we were successful in obtaining homozygous knockouts of 64% of target genes, based on the original ORF designations. Some of the failures undoubtedly corresponded to essential genes. In the current C. albicans genome annotation, the disruption mutants—comprising more than 3000 strains—correspond to 674 unique genes. Below, we describe the characterization of these mutants by means of three genetic screens—an in vivo screen for infectivity, and in vitro screens for morphogenesis and rate of proliferation (Figure 1C).
Infectivity screen identifies 115 candidate virulence factors
Classical virulence analysis of C. albicans mutants typically involves infecting groups of mice with an individual mutant or a wild type strain and comparing the course of illness; virulence-defective mutants produce less disease or a delay in disease onset. To minimize the number of animals required to characterize our library of mutants, we adapted the Signature-Tagged Mutagenesis (STM) techique that has been highly successful in identifying novel virulence factors of bacterial pathogens21-23. The method involves simultaneous infection of an animal model with numerous mutants of interest, each marked by an oligonucleotide “signature tag” that allows for individual quantitation. Theoretically and in practice, mutants with virulence defects have reduced competitive fitness and are selectively depleted in the host.
In the mouse model of disseminated candidiasis, kidneys are the organs of greatest fungal proliferation24, and kidney tissue burden has been highly correlated with virulence among published mutants3 (see Table S2). We used pools of 48 barcoded C. albicans mutants and a matched wild type strain to infect BALB/c mice by tail vein injection (Figure 2A). After disease had progressed, the mice were euthanized, and the abundance of C. albicans strains recovered from host kidneys (R) and the infecting inoculum (I) was determined by real-time PCR (using primers to the signature tags). Each mutant was evaluated in at least 6 mice, and at least two independent isolates of each mutant were assessed.
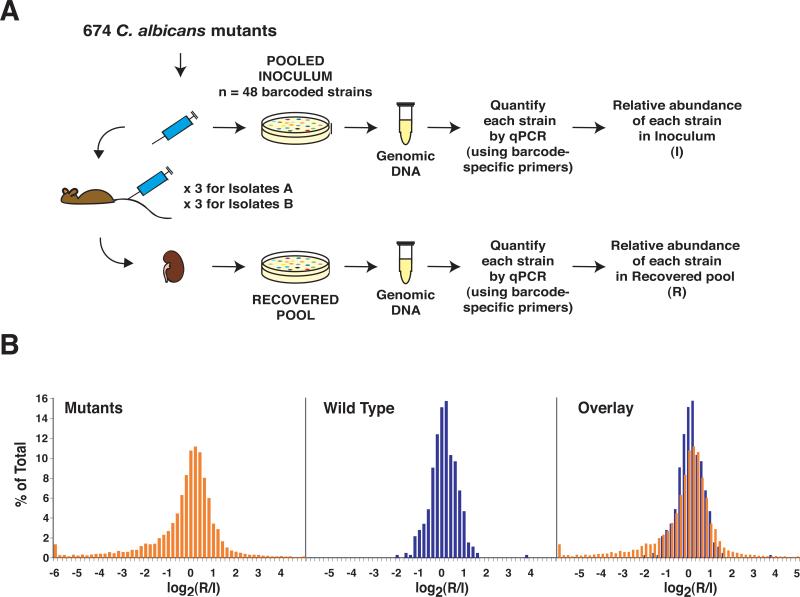
Figure 2. Infectivity screen.
A) Schematic of the Infectivity screen. Pools of up to 48 mutants and wild type were used to infect BALB/c mice. Samples of C. albicans from the infecting inoculum and recovered from mouse kidneys were plated on Sabouraud agar. Genomic DNA was recovered and the relative abundance of each strain in the inoculum (I) and the recovered pool (R) was determined by real-time PCR (qPCR), using primers specific to the DNA barcodes. B) Histograms depicting log2(R/I) values for C. albicans mutants (in orange), the wild type comparator (in blue), and the overlay between the two groups.
We screened a total of 118 pools, and the results—representing 6,917 real-time PCR measurements of C. albicans knockout mutants and 472 measurements of wild type—are presented in Figure 2B. The x-axis represents the log2 ratio of the abundance of each strain in the recovered pool (R) relative to the inoculum (I). A strain whose representation neither increased nor decreased in the infected mouse would have a ratio of 1, or a log2 ratio of 0. A strain that became underrepresented over the course of infection would be shifted to the left, and one that accumulated would be shifted to the right. Inspection of the histogram of wild type C. albicans, derived from data acquired from the majority of mice throughout the screen (blue values), revealed a bell-shaped distribution consistent with simple variation around a mean. In contrast, the distribution of the mutants (in orange) was asymmetric, being skewed to the left. We assumed that the prominence of the left-sided tail resulted from mutants with fitness defects in infecting or proliferating in mouse kidneys; we hypothesized that many of these mutants would also be defective in causing disease. Mutants shifted to the right may have had a fitness advantage resulting in hyperproliferation or a defect, for example, in exiting the kidney.
Although many mutants displayed obvious defects in infectivity (i.e. with log2(R/I) values shifted far to the left in Figure 2B), we used two systematic, statistics-based tests to also capture mutants with moderate defects. This analysis, along with estimates of false discovery rates and a discussion of our results with previously published virulence mutants, is described in detail in the Online Methods. Using these tests, we identified 115 mutants with significant defects in infectivity (Table S3). These disrupt genes involved in a broad range of cellular processes; as might be expected, they also affect a sizeable group of genes whose functions are completely unknown.
Systematic morphogenesis screen reveals determinants of the yeast-to-filament switch
As described in the Introduction, the yeast-to-hypha transition is closely linked to virulence in the C. albicans literature. Usually referred to as a “dimorphic” transition, this process actually involves interconversions among three types of cells: budding oval yeast cells that separate after cell division and two types of elongated cells that remain attached as filaments, called hyphae and pseudohyphae4,25 Interconversions among the three cell types are induced in vitro by modifying the pH, phosphate concentration, temperature, and/or serum concentration in the growth medium; these variables are thought to mimic environmental cues found within different niches of the host.
The macroscopic appearance of a C. albicans colony derives from the types of the cells that make it up, and changes in colony morphology have been used as a sensitive indicator of mutants with defects in the dimorphic transition26. On a variety of media, wild type C. albicans generates colonies with two distinct regions (Figure 3A): a central region (C) consisting of yeast, pseudohyphae, and true hyphae, and a peripheral region (P) consisting primarily of filamentous forms (hyphae and pseudohyphae). Mutants shifted towards filamentous forms produce colonies with increased central wrinkling and/or peripheral filamentation; mutants shifted towards the yeast morphology produce smooth colonies with decreased peripheral filamentation.
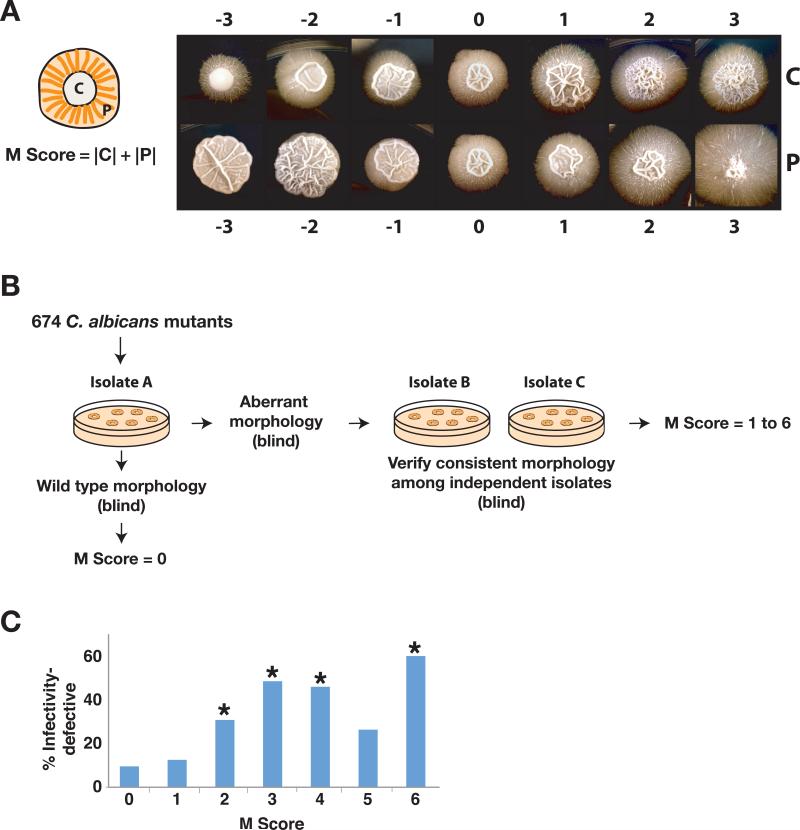
Figure 3. Colony morphology screen.
A) C. albicans colony morphology. As shown in the schematic on the left, C. albicans colonies are bipartite: a central region (C) comprises yeast, pseudohyphae, and true hyphae, and a peripheral region (P) comprises mainly pseudohyphae and hyphae. On the right are examples of C. albicans colonies that illustrate the spectrum of morphologies observed in the screen. Wild type morphology is designated as 0, and aberrant morphology is scored from 1 to 3 (increased relative to wild type) and −1 to −3 (decreased relative to wild type) for both C and P. The Morphology score (M score) of each mutant is calculated as the sum of the absolute values of C and P. B) Schematic of the colony morphology screen. One isolate of each of the 674 mutants and wild type was initially plated for single colonies on Spider medium. Mutants exhibiting morphology different from wild type underwent testing of one to three additional isolates (depending on availability) to confirm consistency of the phenotype. C) Histogram of Infectivity defects among mutants with different M scores. *Denotes significant association with infectivity defect (using the hypergeometric distribution); p=0.0077 for M score of 2, p=4.8x10−6 for M score of 3, p=4.8x10−6 for M score of 4, p=0.10 for M score of 5, p=0.027 for M score of 6.
We examined colony morphology across our mutant library. 5-10 cells of each mutant were plated on solid Spider medium and incubated at 30°C (Figure 3B). Colonies were photographed at 14 days, and central (C) and peripheral (P) regions were each scored on a scale of −3 (decreased morphology) to 3 (increased morphology), with 0 indicating wild type morphology. The overall Morphology score (M score) was the sum of the absolute values of C and P and ranged from 0 (wild type) to 6 (highly aberrant). Mutants with M scores of other than 0 underwent testing of additional isolates to confirm consistency of the phenotype. Ultimately, 504 mutants were indistinguishable from wild type (M score of 0), 16 had mildly altered morphology (M score of 1), and 133 had significant aberrations (M score of 2 or greater; Table 1). Four mutants had inconsistent defects (different abnormal morphologies), most likely resulting from unlinked mutations in at least one of the isolates, and 17 mutants grew slowly on Spider medium; these were excluded from subsequent analysis. Mutants with significant morphology defects (M of 2 or more) are presented in Table S4, and data for all of the mutants are in Table S5.
Table 1. Results of the morphogenesis and proliferation screens.
Morphogenesis results are presented as the number and percent of mutants with a given M score. Results were omitted for 4 mutants with inconsistent results and 17 mutants with poor growth on Spider medium. Proliferation results are presented as the number and percent of mutants with a given doubling time, also expressed as the number of standard deviations (s.d.) greater than that of wild type.
| Morphology |
Proliferation |
|||
|---|---|---|---|---|
| M Score | Number of Mutants (%) | Doubling Time (min) | # Standard Deviations > DTWT | Number of Mutants (%) |
| 0 | 504 (77) | <92.2 | 0 | 396 (59) |
| 1 | 16 (2) | 92.3-97.4 | 0.5 | 107 (16) |
| 2 | 39 (6) | 97.5-102.8 | 1 | 68 (10) |
| 3 | 33 (5) | 102.9-108.1 | 1.5 | 35 (5) |
| 4 | 37 (6) | 108.2-113.4 | 2 | 21 (3) |
| 5 | 19 (3) | 113.5-118.7 | 2.5 | 5 (<1) |
| 6 | 5 (<1) | >118.8 | 3 or more | 42 (6) |
We next examined the correlation between defects in infectivity and defects in the yeast-to-hypha transition by determining the frequency of infectivity defects among mutants with a given M score. If the ability to undergo the dimorphic transition were the major determinant of virulence, then the proportion of infectivity-defective mutants would be expected to increase as a function of the M score. This prediction was only partially borne out by our data (Figure 3C). Mutants with an M score >0 were more likely to have infectivity defects than those with wild type morphology (37% versus 10%, p-value of 6.5x10−14 calculated using the hypergeometric distribution), in keeping with the prevailing view that morphogenesis and infectivity are associated. However, among 24 mutants with the highest M scores of 5 and 6, two-thirds displayed normal infectivity. Likewise, among 103 infectivity-defective mutants that were scored for morphology, 48 or 47% had normal colony morphology on Spider medium. These results demonstrate that the relationship between infectivity and morphogenesis cannot be one of simple cause and effect.
Although colony morphology is a sensitive indicator of mutants with defects in the dimorphic switch, we also employed a specific test of the ability of yeast cells to form hyphae. We tested the group of 115 infectivity-defective mutants for a hyphal response to serum, which is the “germ tube test” used by clinical laboratories to distinguish C. albicans from other yeasts. Only 15 mutants failed to elaborate true hyphae (Table S6), all of which were also captured by the colony morphology assay (n=12) or grew too slowly on this medium to be scored (n=3).
Quantitative proliferation screen identifies mutants with slow growth phenotypes
It is a reasonable assumption, and one often made in the literature27, that a C. albicans mutant exhibiting slow proliferation in vitro will show a corresponding virulence defect in vivo. We addressed this assumption systematically by measuring proliferation rates across the mutant library and comparing them with the infectivity data. Wild type C. albicans and two isolates of each mutant were grown in chemically defined liquid medium at 37°C, and doubling times were measured (Table S5). The median doubling time of wild type under these conditions was 87 minutes.
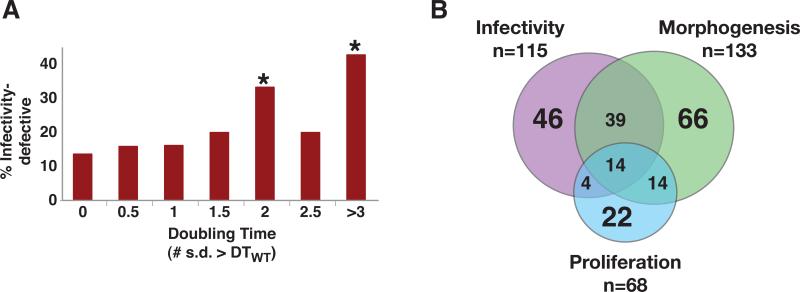
68 mutants displayed significant defects in proliferation, defined as a doubling time more than two standard deviations greater than that of wild type (Table 1). Contrary to our expectations, the majority (42 or 62%) of these proliferation-defective mutants were competent for infectivity (Figure 4A). Likewise, among the 115 infectivity-defective mutants, the vast majority (89 or 77%) exhibited a normal rate of proliferation in vitro (Table S5). The simplest interpretation of these results is that the requirements for proliferation in vitro are likely to differ substantially from those required for growth in the host. For example, nutrient limitation in the host plus direct attack by the immune system may result in very long doubling times in vivo, rendering irrelevant many of the differences that we observed under optimal conditions in vitro. At a minimum, these results invalidate the common assumption that poor growth in vitro necessarily correlates with decreased virulence.
Figure 4. Proliferation screen and venn diagram.
A) Histogram of infectivity defects among mutants with different doubling times. * Denotes significant association with infectivity defect (using the hypergeometric distribution); p=0.11 for 0.5 s.d.>DTWT, p=0.13 for 1 s.d.>DTWT, p=0.15 for 1.5 s.d.>DTWT, p=0.034 for 2 s.d.>DTWT, p=0.40 for 2.6 s.d.>DTWT,, and p= 3.3x10−5 for >3 s.d.>DTWT. B) Venn diagram illustrating the overlap of mutants from the three screens. Mutants with slow growth or inconsistent growth on Spider medium could not be scored for Morphogenesis and are not represented in the figure; these included 4 mutants with infectivity-specific defects, 8 with combined infectivity and proliferation defects, and 1 with a proliferation-specific defect.
A Venn diagram summarizing the results of the three systematic screens is presented in Figure 4B. Overall, approximately 1/3 of the mutants in the library exhibited a defect in at least one screen. The majority had defects in a single phenotype.
Biochemical analysis of four mutants predicted to affect glycolipid biosynthesis
Notwithstanding the previous arguments, we focused on infectivity-defective mutants with normal colony morphology and normal proliferation in vitro (Table 2), hypothesizing that mutants with these characteristics would be most likely to reveal pathways used exclusively for virulence. Among the mutants meeting these criteria, two caught our attention because they were predicted to affect synthesis of sphingolipids—a class of molecules not previously associated with C. albicans virulence. Orf19.4831 was homologous to known sphingolipid methyltransferases/cyclopropane synthases, and Het1 was homologous to known sphingolipid transfer proteins. Inspection of mutants with less consistent infectivity defects revealed two additional mutants affecting ORFs that could be ordered with Orf19.4831 and Het1 into a hypothetical biosynthetic pathway for glucosylceramide (Figure 5A). Hsx11 had previously been demonstrated to possess sphingolipid glucosyltransferase activity28, and Orf19.260 was homologous to known sphingolipid desaturases.
Table 2. Mutants with defects in infectivity but not in morphogenesis or proliferation.
Gene names and proven or predicted protein functions are presented for mutants that exhibited defects in infectivity (as defined in text) but not in colony morphology (M scores were 1 or less) or proliferation (DTMUTANT < DTWT + 2 s.d.).
| Disrupted gene | Function of protein product |
|---|---|
| orf19.3384 | Cell wall component: protein |
| orf19.529 | Cell wall component: protein |
| PGA32/orf19.6784 | Cell wall component: protein |
| CHS4/orf19.7349 | Cell wall composition: carbohydrate |
| CHS7/orf19.2444 | Cell wall composition: carbohydrate |
| CHT2/orf19.3895 | Cell wall composition: carbohydrate |
| orf19.753 | Cell wall composition: carbohydrate |
| DUN1/orf19.4002 | DNA damage response |
| ESC4/orf19.1445 | DNA damage response |
| CRZ1/orf19.7359 | Gene expression: transcription factor |
| MTLA1/orf19.3201 | Gene expression: transcription factor |
| orf19.2315 | Gene expression: transcription factor |
| orf19.7397 | Gene expression: transcription factor |
| SEF1/orf19.3753 | Gene expression: transcription factor |
| orf19.2961 | Gene expression: transcription factor |
| CYB1/orf19.7049 | Lipid biosynthesis and regulation |
| HET1/orf19.6327 | Lipid biosynthesis and regulation |
| orf19.3283 | Lipid biosynthesis and regulation |
| orf19.4831 | Lipid biosynthesis and regulation |
| orf19.6411 | Lipid biosynthesis and regulation |
| MID1/orf19.3212 | Metal ion homeostasis: calcium |
| CFL2/orf19.1264 | Metal ion homeostasis: iron |
| HAK1/orf19.6249 | Metal ion homeostasis: potassium |
| orf19.3290 | Miscellaneous: aerobic respiration |
| CDC10/orf19.548 | Miscellaneous: cell cycle |
| NMD5/orf19.4188 | Miscellaneous: nuclear import of proteins |
| orf19.1392 | Miscellaneous: protein folding |
| CDC19/orf19.3575 | Nutrient acquisition and metabolism |
| HGT12/orf19.7094 | Nutrient acquisition and metabolism |
| HGT8/orf19.2021 | Nutrient acquisition and metabolism |
| orf19.449 | Nutrient acquisition and metabolism |
| orf19.4905 | Nutrient acquisition and metabolism |
| orf19.5499 | Nutrient acquisition and metabolism |
| orf19.7288 | Nutrient acquisition and metabolism |
| TRP2/orf19.10080 | Nutrient acquisition and metabolism |
| RGA2/orf19.4593 | Signaling: cell polarity |
| KAR2/orf19.2013 | Signaling: unfolded protein response |
| orf19.1276 | Unknown |
| orf19.2653 | Unknown |
| orf19.2726 | Unknown |
| orf19.3335 | Unknown |
| orf19.5509 | Unknown |
| orf19.557 | Unknown |
| orf19.5782 | Unknown |
| PDR6/orf19.2094 | Unknown |
| RBT4/orf19.6202 | Unknown |
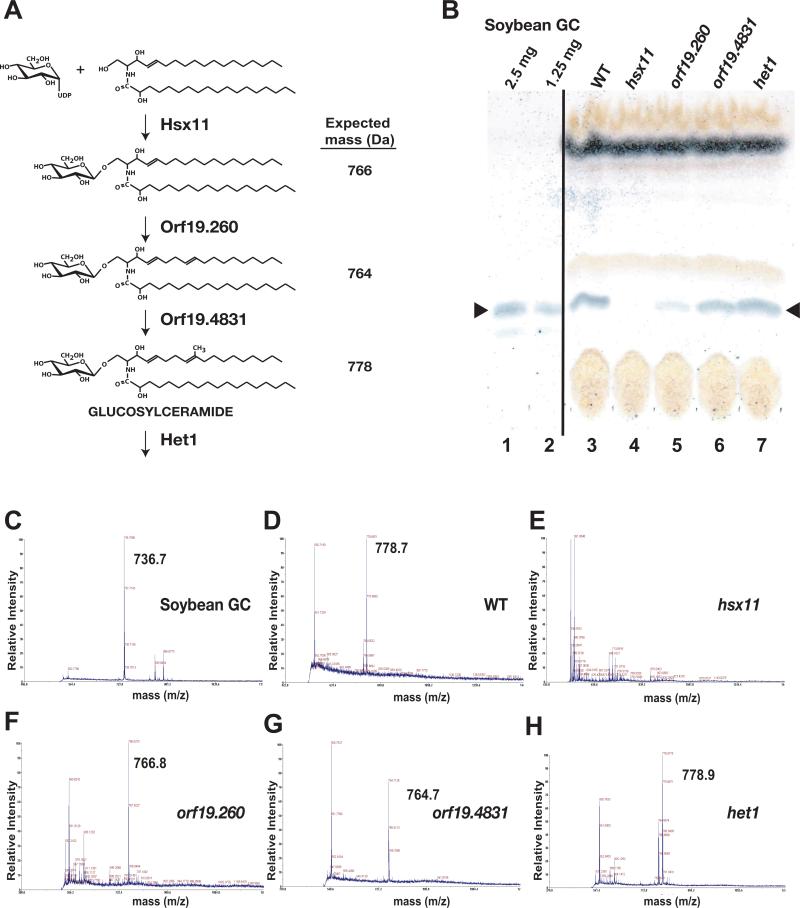
Figure 5. Characterization of mutants affecting glucosylceramide biosynthesis.
A) Schematic of the predicted glucosylceramide synthesis pathway and expected molecular masses. The position of the glucosyltransferase reaction mediated by Hsx1 has not been determined and may occur as depicted or elsewhere in the pathway. Note that the masses given are for the sodium salts of each molecule. B) TLC plate with glucosylceramide (GC) from soybean (lanes 1 and 2) and lipids extracted from equal masses of wild type (lane 3), hsx11 (lane 4), orf19.260 (lane 5), orf19.4831 (lane 6), and het1 (lane 7). The markers indicate the position of glucosylceramide, visualized with anthrone reagent. C,D,E,F,G, and H) MALDI-TOF spectra for a soybean glucosylceramide standard and lipid extracted from wild type, hsx11, orf19.260, orf19.4831, and het1, respectively.
To experimentally determine whether hsx11, orf19.260, orf19.4831, and het1 truly defined a glucosylceramide biosynthetic pathway in C. albicans, we compared membrane lipids from the four mutants and wild type. Crude lipid fractions prepared from equal masses of cells were resolved using thin layer chromatography (TLC), and glucosylceramide was visualized with anthrone reagent (Figure 5B); soybean glucosylceramide served as a mobility marker (lanes 1-2). The wild type strain synthesized a product that comigrated with bona fide glucosylceramide (lane 3). By comparison, hsx11 and orf19.260 produced significantly less comigrating product (lanes 4-5), orf19.4831 exhibited only a mild decrease (lane 6), and het1 produced as much comigrating product as wild type (lane 7).
Thin layer chromatography cannot resolve mature glucosylceramide from the preceding two intermediates (Figure 5A). Therefore, to determine the identities of the comigrating lipids, we used mass spectrometry. After a further purification step, we resolved lipids from wild type, hsx11, orf19.260, orf19.4831, and hsx11 strains by TLC, recovered species migrating at the position of mature glucosylceramide, and subjected the lipids to MALDI-TOF mass spectrometry. Soybean glucosylceramide was analyzed as a positive control and exhibited its expected molecular weight (736 for the sodium salt, Figure 5C). As shown Figure 5D, wild type C. albicans produced a product consistent with mature fungal glucosylceramide (778 sodium salt). The hsx11 mutant produced no product (Figure 5E), which was expected since an intermediate lacking the glucose moiety would not comigrate by TLC. orf19.260 and orf19.4831 accumulated products of molecular weights 766 and 764, respectively (Figures 5F and 5G); these are the weights of sodium salts of the final two intermediates in Figure 5A and indicate that, consistent with our model, Orf19.260 and Orf19.4831 mediate sphingolipid C8 desaturation and C9 methylation reactions, respectively. het1 accumulated a product consistent with mature glucosylceramide (Figure 5H); this mutant affects a predicted sphingolipid transfer protein expected to alter the localization but not synthesis of glucosylceramide. Taken together, these results confirm both the biosynthetic pathway of Figure 5A and our assignments of the genes responsible for each step.
Enzymes in the glucosylceramide biosynthesis pathway are required for virulence
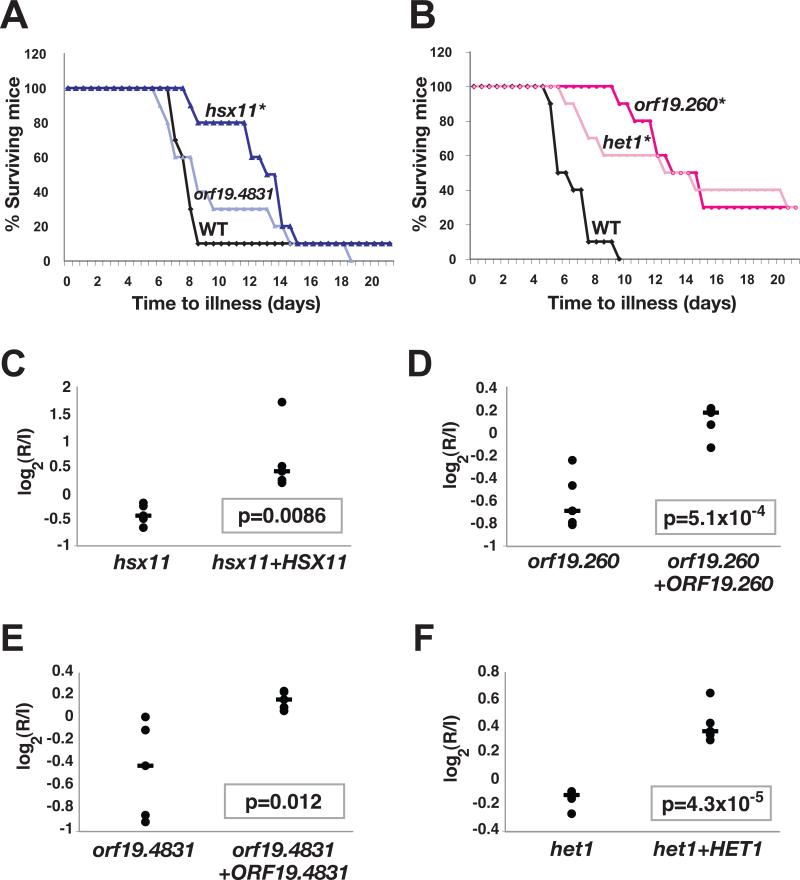
We next tested the virulence of mutants affecting the glucosylceramide pathway in single (as opposed to pooled) infections of BALB/c mice. 10 mice were infected with each mutant or wild type C. albicans, and time to illness was monitored. hsx11, orf19.260, and het1 demonstrated significant virulence defects (Figure 6A-6B), whereas orf19.4831 exhibited a trend towards reduced virulence that fell outside our significance threshold of 0.05 (Figure 6A). To establish whether deletion of the target genes was responsible for these phenotypes, we constructed “addback” strains into which one copy of the wild type gene was restored. When mice were coinfected with each mutant and its cognate gene addback strain, the addback strain exhibited superior fitness in each case (Figures 6C, 6D, 6E, and 6F). The observation that ORF19.4831 complemented the infectivity defect of the orf19.4831 mutant supports our previous surmise that this mutant is mildly attenuated for virulence. These results establish glucosylceramide biosynthesis as a novel and important determinant of C. albicans virulence.
Figure 6. Virulence analysis of mutants affecting glucosylceramide biosynthesis.
A and B) The glucosylceramide pathway is required for virulence. As described in the text, BALB/c mice were infected with wild type C. albicans or one of the glucosylceramide pathway mutants, and time to illness was monitored. *denotes a significant difference from wild type by the logrank test; p=0.0020 for hsx11, p<0.0001 for orf19.260, p>0.05 for orf19.4831, and p=0.032 for het1. C,D,E, and F) Wild type genes restore competitive fitness to the glucosylceramide pathway mutants. Two-strain infectivity assays were performed comparing each mutant to a gene addback strain in 5 BALB/c mice. Shown are the R/I values of each strain in individual mice (black dots), the median R/I value (black line), and the p-value of the observed differences.
The Cryptococcus neoformans homolog of hsx11 is also required for virulence of in a murine model, but this mutant has an in vitro growth defect at neutral or alkaline pH.29 We assessed the growth of the C. albicans glucosylceramide pathways mutants on solid laboratory medium buffered to pH 7, 7.5, 8, 9, and 10 and found no differences from wild type (Figure S1). Therefore, in contrast to Cryptococcus neoformans, C. albicans glucosylceramide likely mediates virulence through a mechanism other than facilitating growth at mammalian pH.
Discussion
Towards forward genetics in an ameiotic, diploid human fungal pathogen
Candida albicans is the most common fungal pathogen of humans, yet progress in understanding its virulence program has been difficult. Its diploid genome, chromosomal instability, and incomplete sexual cycle have made genetic approaches especially challenging. In this study, we describe the construction and analysis of homozygous gene deletion mutants that encompass 674 genes. The library contains at least two independent isolates affecting each gene of interest and was created without the use of URA3—an auxotrophic marker that has clouded the interpretation of most of the previously described mutants in this field. Accounting for ~11% of the C. albicans genome, our collection of ~3000 strains includes knockouts in genes predicted to affect a wide range of cellular processes, as well as ones whose functions are completely unknown. We detected no aneuploidy in a survey of 12 mutants using a quantitative PCR assay (6 with infectivity defects and 6 without; Table S7); however, because aneuploidy can occur during strain construction17, we recommend the use of complementation tests for mutants of special interest. Below, we summarize insights gained from systematic screens of the library for (1) host (kidney) colonization, (2) the yeast-to-hypha transition, and (3) rate of proliferation in vitro.
Decoupling C. albicans infectivity, morphogenesis, and proliferation
Prior to this work, fewer than 200 C. albicans mutants had been tested for virulence in the mouse model of disseminated disease3, and there was a near-perfect correlation between mutants with virulence defects and those affecting the yeast-tohypha transition (Figure 1A). This impression was based on myriad studies of individual gene disruption mutants that had been created in different laboratories, using different C. albicans strain backgrounds, and tested in different, nonstandardized assays for virulence and morphogenesis. An exception was an infectivity study of a large number of C. albicans mutants that had been created by disrupting one copy of each target gene and placing the other allele under control of a repressible promoter30. In this case, however, the identities of the disrupted genes were not revealed, the collection was not made freely available, and effects of the mutations on morphogenesis were not reported.
Using our library, we were able to directly compare the results of a standardized screen for infectivity to those for morphogenesis and proliferation in vitro, resulting in several general conclusions. First, across the mutant library, defects in the three properties were relatively common. Approximately 17% of mutants demonstrated reproducible defects in infectivity, 21% of mutants had abnormal morphology, and 10% of mutants were defective for proliferation (Table S5). The proportion of mutants with infectivity defects was somewhat higher than that reported in STM screens of bacterial pathogens21-23 and far lower than the >80% rate of virulence-defective mutants reported in the C. albicans literature3. Second, infectivity-defective mutants had a higher than average rate of morphology abnormalities (43% had M scores of 2 or more) and morphogenesis-defective mutants were similarly enriched for infectivity defects (40% by the Boolian test), supporting the well-described critical role of the yeast-to-hypha transition in virulence. Finally, our surprising discovery that a large group of mutants exhibited significant defects in infectivity without associated defects in morphogenesis or proliferation in vitro suggests that the process of virulence can be teased apart from the complex morphogenetic program, thereby providing a strategy to identify discrete effectors of virulence.
A glycolipid required for virulence but not for morphological switching
We tested the ability of our dataset to predict novel virulence effectors by focusing on four mutants with specific defects in infectivity but not in morphogenesis or proliferation. Using biochemical methods and mass spectrometry, we demonstrated that Hsx11, Orf19.260, and Orf19.4831 mediate sequential steps in the biosynthesis of fungal-specific glucosylceramide (Figure 5). Given its homology to lipid transfer proteins, Het1 likely localizes the final product. Hsx11 was previously known to possess glucosyltransferase activity28 and, recently, Orf19.260 (also known as Sld1) was independently shown to have sphingolipid desaturase activity31, consistent with our findings. In two-strain infectivity experiments, all four mutants exhibited defects in colonizing host kidneys that were complemented by restoration of the wild type genes; hsx11, orf19.260/sld1, and het1 were also significantly attenuated for virulence in classical monotypic infections (Figure 6). Thus, the glucosylceramide biosynthetic pathway is crucial for for causing disease as well as for proliferation in the kidney. Not all fungi retain this pathway; for example, S. cerevisiae and S. pombe lack all four genes involved in glucosylceramide synthesis and (proposed) localization. Although several small molecules have previously been associated with virulence of C. albicans10,32-42, glucosylceramide is the first such virulence effector to act independently of the yeast-tohypha transition.
Prospects for dissecting a unique host-pathogen relationship
C. albicans is unique in being both a normal human fungal commensal and the most common cause of serious fungal disease. As exemplified by the glucosylceramide pathway, our identification of 46 genes whose disruption leads to reproducible defects in infectivity in the host but to no observable changes in morphogenesis or proliferation in vitro (Table 2) offers opportunities for understanding the relationship between C. albicans and the mammalian host. The knockout collection should also facilitate investigations of other clinically relevant aspects of C. albicans biology. For example, little is known regarding how C. albicans (or any other organism) functions as a commensal or how the determinants of superficial epithelial infections differ from those required for invasive, disseminated infections. The development of experimental models of these broader aspects of Candida biology coupled with the genetic resource and strategies described here promise new insights into the biology of this important human pathogen.
Supplementary Material
Acknowledgements
We are extremely grateful to the Stanford Genome Technology Center for releasing the C. albicans genomic sequence prior to publication, and to the curators of the Candida Genome Database that has been an invaluable tool in our analysis of the published literature. We are also grateful to many colleagues at UCSF and elsewhere who were generous with advice, protocols, and reagents. Sean O'Brien constructed our C. albicans bioinformatics database. Bethann Hromatka assisted with construction of C. albicans mutants. Jing Zhe and Joe De Risi provided a database of putative unique C. albicans ORFs. Michael Fischbach suggested the link between our sphingolipid-associated mutants and the glucosylceramide pathway. Eric Chow and Jeff Cox provided expertise and equipment for lipid extraction. Pinar Erciyas Bailey and the Agard, Morgan, Shokat, and Walter labs provided training and access to their mass spectrometer. Bassam al-Sady and Geeta Narlikar shared their mass spectrometry plate. Hiten Madhani wrote software for data analysis. Sudarsi Desta prepared hundreds of liters of laboratory media. Hiten Madhani, Jeff Cox, Lynn Connolly, and Rich Locksley provided helpful comments on the manuscript. This work was supported by NIH RO1 A149187, NIH K08 AI062800, and the UCSF Program for Breakthrough Biomedical Research. S.M.N. was the recipient of a HHMI Postdoctoral Research Fellowship for Physicians and a Burroughs Wellcome Fund Career Award in the Biomedical Sciences during the course of these studies. Homozygous disruption mutants will be deposited with the Fungal Genetics Stock Center.
Online Methods
Analysis of published C. albicans mutants
Published C. albicans “features” and “phenotypes” were downloaded from the Candida Genome Database (CGD) website http://www.candidagenome.org/3 on 06/07/2009. Mutants affecting virulence, morphogenesis, and organ colonization were identified through searches for: “virulence;” “filamentation” and “hyphal;” and “kidney,” “tissue,” and “organ,” respectively. Phenotypes were associated with homozygous knockout or transcriptional depletion mutants. Contradictory reports were omitted unless one report addressed a deficiency in the other. Because phenotypes can differ between model systems, virulence was limited to time to illness in the mouse tail vein model of disseminated disease. Morphology phenotypes describe colony morphology on Spider medium; reports of defective germ tube formation were also included since this phenotype reliably predicts defects on Spider medium (e.g.Table S6).
C. albicans annotation database and target selection
Predicted ORFs in C. albicans Assembly 19 (Stanford Genome Technology Center)19 were compared by BLAST to the genomes of Saccharomyces cerevisiae, Schizosaccharomyces pombe, Aspergillus nidulans, Magnaporthe grisea, Neurospora crassa, and the Nonredundant Protein Database (NR). They were also analyzed with the SMART motif finding algorithm (EMBL).
Targets were identified by multiple, partially overlapping criteria: (1) ORFs without homologs in S. cerevisiae and S. pombe, n=352. These were similar to an ORF in NR with a chance probability <10−6 and similar to ORFs in S. cerevisiae or S. pombe with p>10−2. (2) ORFs without known homologs, n=1374. The C. albicans genome was compared by BLAST to genomes of numerous bacteria as well as Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Rattus norvegicus, Plasmodium falciparum, Homo sapiens, Saccharomyces bayanus, Saccharomyces cerevisiae, Saccharomyces paradoxus, Saccharomyces mikatae, Saccharomyces castellii, Schizosaccharomyces pombe, Saccharomyces kluyveri, and Saccharomyces mikatae. C. albicans ORFs that appeared unique after cluster analysis were selected. (3) ORFs with motifs potentially related to virulence, n=1215. These included ORFs with SMART motifs associated with immunity (e.g. “LRR”), pathogenesis (e.g. “pathogen,” “toxin”), transcription (e.g. “ZnF”) signaling (e.g. “G alpha”), and cell surface and secreted proteins (e.g. “signal sequence”). (4) ORFs encoding She3-associated mRNAs, n=5120. (The She3-dependent RNA transport system localizes mRNAs to the tips of hyphal cells, and we were interested to know whether any of these is required for infectivity.) ORFs were eliminated if they appeared to be false ORFs (e.g. short ORF within a larger predicted ORF), were associated with transposons, or were recognized homologs of essential genes in S. cerevisiae.
Construction of the mutant library
Primers used for mutant construction are listed in Table S8. Fusion PCR was used to produce gene disruption fragments as described.14 Two alleles of each target gene were sequentially disrupted in parental strain SN152 (Table S9) with disruption fragments marked by C. dubliniensis HIS1 and C. maltosa LEU2, respectively. His+Leu+ transformants were screened by colony PCR for the four expected 5′ and 3′ gene disruption junctions and absence of the target ORF.
Infectivity Screen
Procedures involving animals were approved by the UCSF Institutional Animal Care and Use Committee (IACUC). Overnight cultures of 48 C. albicans mutants and the isogenic wild type reference strain QMY2343 (Table S9) were diluted 1:20 into fresh YEPD and incubated for ~4 hours at 30°C. The log phase cultures were pooled, washed twice with sterile normal saline, and ~1.1x106 CFUs was injected by tail vein into18-20 gm female BALB/c mice. The inoculum was also plated onto Sabouraud medium (with ampicillin 50 ug/ml, gentamicin 15 ug/ml) and incubated at 30°C. Mice were monitored closely and euthanized when morbid (defined as weight loss >15%, hunched posture, reduced activity). C. albicans was recovered by plating homogenates of mouse kidneys onto Sabouraud/Amp/Gent (antibiotics were included to suppress contaminating bacteria) and incubation at 30°C. Genomic DNA was prepared from yeast scraped off the plates44.
Abundance of individual mutants in each pool was determined using real-time PCR and primers specific to each signature tag; the second, common primer hybridized to C. dubliniensis HIS1. Reactions included 200 ng of genomic DNA, 2 μM each primer, 2 mM MgCl2, 50 mM KCl, 10 mM Tris 8.3, 0.075× SYBR Green (Sigma), and 2 units Taq polymerase. Wild type C. albicans was quantified using the C. dubliniensis HIS1 primer and primer SNO183 (that hybridizes upstream of LEU2, where C.d.HIS1 is integrated in this strain). Cycling was performed in an Opticon (Biorad) using the following parameters: 92°C for 6 min followed by 40 cycles of (92°C for 30 sec, 52°C for 45 sec, 72°C for 1 min), and finally 72°C for 10 min.
C(T) values were converted to a linear scale by the equation, linear value=2−C(T). Experiments comparing 48 strains resulted in 48 values for the inoculum (I) and another 48 for the recovered pool (RRAW). RRAW values were multiplied by median(I)/median(RRAW) to generate normalized R values. Finally, R/I was calculated for each mutant and wild type; these are presented in Tables S10 and S11. Please see “Statistical Analysis” below for an explanation of statistical methods. Plots and calculations were based on median centered data.
We used two methods to determine which mutants displayed significant infectivity defects. First, the t-test was used to compare the log2(R/I) values of mutants to those of the wild type dataset, using a significance threshold of p<7.4x10−5 (i.e. p<0.05, corrected for 674 comparisons) to identify mutants that were significantly different from wild type. The t-test captured all of the infectivity mutants that were obviously depleted in Figure S2A (e.g. mutants 7, 9, 23, 29, 39, and 47) and had a very low false discovery rate, estimated at <0.04%. However, close inspection of the data revealed that this test—based on the average log2(R/I) of each mutant—missed a class of mutants that displayed a mix of aberrant phenotypes. Certain mutants (e.g. mutants 6,17, 30, 31, and 41 in Figure S2A) were alternately increased or decreased in kidneys of different hosts, such that the mean log2(R/I) ratios were normal even though individual log2(R/I) ratios were well outside the mean. This phenomenon is reflected by mutants with high standard deviations around the mean in Figure S2B and may reflect the presence of multiple fitness deficits that are manifested stochastically in different infected mice. To include candidates with this phenotype, we devised a Boolean test for aberrant infectivity, defined as having a preponderence (>70%) of R/I measurements that are abnormally high and/or low (<0.7, >2). When applied to 106 random sets of 6 values from the wild type dataset, the false discovery rate of this test was 1.3%.
We compared the ability of the t-test versus the Boolean test to discriminate among 57 previously published C. albicans mutants—all previously tested in a mouse tail vein model of disseminated disease—that were included in our library. We note that the published mutants are not a bona fide gold standard for assessing virulence because most were constructed using URA3, and ~30% of reported virulence defects are thought to result from deficient URA3 expression rather than deletion of the target genes9. Despite these caveats, the Boolean test exhibited superior sensitivity and specificity, properly identifying 21 of 47 mutants previously reported as virulence-defective (Figure S2A and Table S12) and only one of 10 mutants previously reported as competent for virulence (in GPA245;Table S12). The Boolean method also captured all four virulence-defective mutants that had been constructed without the use of URA346-49. Results of both statistical tests are presented in Table S5.
Morphogenesis Screen
Mutants and wild type reference strain SN250 (Table S9) were plated onto Spider agar at a density of 5-10 CFUs/plate. Colonies were photographed after 7 and 14 days of incubation at 30°C. Morphology scores were based on colony appearance at 14 days, with consideration of appearance at 7 days in questionable cases.
Initial assignments were based on the first isolate of every mutant in the library. The center (C) and periphery (P) were scored separately on a scale from −3 (fewer cntral wrinkles or fewer peripheral filaments) to 3 (increased central wrinkles or peripheral filaments); wild type morphology was defined as 0. Scoring was blind in that the mutants were numerically coded. One to three additional isolates (depending on availability) of mutants with abnormal phenotypes was analyzed by a different investigator. If an additional isolate had wild type morphology, the mutant was scored as wild type (with the presumption that the isolate with abnormal morphology had acquired an additional mutation). Three mutants exhibited inconsistent abnormal morphologies and were scored as “IC”; 18 mutants had severe growth defects on Spider (“S”) and were not scored.
Proliferation Screen
Two isolates of every mutant and wild type reference strain QMY23 (Table S9) were diluted 1:30 into Synthetic Complete liquid medium in 96-well optically clear plates (Nunc) and incubated with shaking at 37°C. A600 was monitored every 60-90 minutes for ~10 hours with a plate reader (Molecular Devices). Doubling times were calculated based on the peak rate of growth between any two time points, and the average of the doubling times of the two isolates was reported.
Virulence Analysis
Cultures of SN285 (orf19.459=hsx11), SN255 (orf19.260), SN290 (orf19.4831), SN297 (orf19.632=het1), and the isogenic reference strain SN250 (wild type) were prepared in liquid SC and grown for 2 days at room temperature. Mid to late log phase cells were washed two times with sterile normal saline and counted using a hemacytometer. ~4.5 × 105 CFUs of each strain used to infect 10 female 18-20 gm BALB/c mice by tail vein injection. Mice were monitored twice daily and euthanized when morbid by criteria described above.
Complementation Analysis
Plasmids for complementation were constructed using PCR and homologous recombination in S. cerevisiae50 The plasmids (pSN103=HSX11, pSN104=orf19.260, pSN101=orf19.4831, and pSN102=HET1) were engineered to sandwich a wild type copy of each test gene and C. dubliniensis ARG4 between sequences upstream and downstream of C. albicans LEU2 gene. Inserts were sequenced to verify the absence of mutations. Disruption fragments were liberated by digestion with PmeI and introduced into the appropriate mutants (SN285 [orf19.4592=hsx11], SN255 [orf19.260], SN290 [orf19.4831], and SN297 [orf19.6327=het1], Table S9). An additional plasmid (pSN105) was created with just the C. d. ARG4 gene and C.a.LEU2 flanking sequences; this was used to create auxotrophic marker-matched versions of the homozygous deletion mutants. Strain designations appear in Table S9.
Two-strain infectivity experiments were performed by infecting female 18-20 gm BALB/c mice by tail vein with ~8×105 CFUs of a 1:1 pool of each mutant and its gene addback comparitor. Five mice were infected with each pool, and the abundance of strains in the inoculum vs. infected kidneys was determined by real-time PCR. Standard curves were created using dilutions of genomic DNA from individual strains.
Lipid Extraction and Mass Spectrometry
100 ml cultures (in YEPD) of SN425 (wild type), SN442 (hsx11), SN444 (orf19.260), SN446 (orf19.4831), and SN448 (het1) were incubated with shaking at 30°C overnight. Cell pellets were frozen in liquid nitrogen and lyophilized overnight. 10 mg of each lyophilized cell pellet was extracted for 48 hours with chloroform/methanol (2:1, v/v). The extract was mixed with 0.2 volume 0.34% MgCl2. The aqueous layer was discarded and the organic phase mixed with 5 ml of 2 N KCl/methanol (4:1, v/v). The aqueous layer was discarded, and the organic phase was washed six times with 5 ml artificial upper phase (chloroform/methanol/water, 3:48:47, v/v/v). The organic phase was dried under N2 vapor and then dissolved in 500 μl CHCl3/methanol (2:1, v/v). 30 μl of each preparation was then applied to a silica gel 60 HPTLC plate (Whatman); soybean glucosylceramide (Polar Lipids) was included as a mobility marker and chloroform/methanol (90:25, v/v) was the mobile phase. Glucosylceramide was visualized with anthrone reagent (in concentrated H2SO4) and baking.
For mass spectrometry, 450 μl of each lipid preparation was resolved on silica gel 60 plates with divided lanes (J.T. Baker). For each test sample, the region of TLC resin that alignined with soybean glucosylceramide was scraped into 5 ml CHCl3/methanol (2:1). Samples were dried under N2 vapor, solubilized in 5 ml of 0.1 N NaOH/methanol, then incubated at 55°C for 1 hour. Mixtures were acidified with 1 ml of 1 N HCl/methanol, mixed with 5 drops of water and 5 ml hexane, and centrifuged. The lower phase was removed and evaporated under N2 vapor, and the lipid residue dissolved in 2 ml theoretical lower phase (chloroform/methanol/water 86:14:1). 8 ml of theoretical upper phase (chloroform/methanol/water 3:48:47) was added to create a Folch partition. After centrifugation, the lower phase was recovered, evaporated to dryness, and solubilized in 200 μl chloroform/methanol (2:1, v/v). Samples were mixed 1:1 with 10 mg/ml 2,5-DHB (in chloroform/methanol 2:1, v/v) and assessed with a Voyager-DE MALDI-TOF mass spectrometer (Applied Biosystems) in reflector mode.
Statistical Analysis
The hypergeometric distribution was used to evaluate the significance of associations of virulence with morphogenesis among published mutants, virulence with kidney burden among published mutants, previously published virulence defects and infectivity defects in our reconstructed mutants, and all combinations of defects in infectivity, morphogenesis, and proliferation in our screens. A p value of <0.05 was considered significant.
The t-test (2 tailed, comparison of unpaired samples) was used to evaluate the significance of differences between: a) log2(R/I) values of mutants in the infectivity screen versus the wild type reference strain; a p value of <0.05/674 was considered significant (Bonferroni correction for multiple comparisons); and b) log2(R/I) values of the glucosylceramide mutants and their cognate gene addback strains in the paired infectivity experiments; a p value of <0.05 was considered significant.
The logrank test was compare the time-to-illness curves of mice infected with wild type C.albicans vs. mutants in the glucosylceramide pathway; a p value of <0.05 was considered significant.
False discovery rates for the t-test and Boolean methods of identifying infectivity mutants were determined by applying these tests to 1000 random sets of 6 values from the wild type dataset 1000 times and calculating the median % positive.
Footnotes
Competing Interests
The authors state that they have no competing financial interests.
References
- 1.Edmond MB, et al. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29:239–44. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- 2.Zaoutis TE, et al. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis. 2005;41:1232–9. doi: 10.1086/496922. [DOI] [PubMed] [Google Scholar]
- 3.Skrzypek MS, et al. New tools at the Candida Genome Database: biochemical pathways and full-text literature search. Nucleic Acids Res. 2010;38:D428–32. doi: 10.1093/nar/gkp836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–24. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Enjalbert B, Whiteway M. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryot Cell. 2005;4:1203–10. doi: 10.1128/EC.4.7.1203-1210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell. 2005;16:2903–12. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–53. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saville SP, et al. Inhibition of filamentation can be used to treat disseminated candidiasis. Antimicrob Agents Chemother. 2006;50:3312–6. doi: 10.1128/AAC.00628-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brand A, MacCallum DM, Brown AJ, Gow NA, Odds FC. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell. 2004;3:900–9. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng S, et al. Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect Immun. 2003;71:6101–3. doi: 10.1128/IAI.71.10.6101-6103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lay J, et al. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect Immun. 1998;66:5301–6. doi: 10.1128/iai.66.11.5301-5306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharkey LL, Liao WL, Ghosh AK, Fonzi WA. Flanking direct repeats of hisG alter URA3 marker expression at the HWP1 locus of Candida albicans. Microbiology. 2005;151:1061–71. doi: 10.1099/mic.0.27487-0. [DOI] [PubMed] [Google Scholar]
- 13.Sundstrom P, Cutler JE, Staab JF. Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect Immun. 2002;70:3281–3. doi: 10.1128/IAI.70.6.3281-3283.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Magee BB, Dawson D, Magee PT, Kumamoto CA. Chromosome 1 trisomy compromises the virulence of Candida albicans. Mol Microbiol. 2004;51:551–65. doi: 10.1046/j.1365-2958.2003.03852.x. [DOI] [PubMed] [Google Scholar]
- 16.Selmecki A, Bergmann S, Berman J. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol Microbiol. 2005;55:1553–65. doi: 10.1111/j.1365-2958.2005.04492.x. [DOI] [PubMed] [Google Scholar]
- 17.Arbour M, et al. Widespread occurrence of chromosomal aneuploidy following the routine production of Candida albicans mutants. FEMS Yeast Research. 2009;9:1070–7. doi: 10.1111/j.1567-1364.2009.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouchonville K, Forche A, Tang KE, Selmecki A, Berman J. Aneuploid chromosomes are highly unstable during DNA transformation of Candida albicans. Eukaryot Cell. 2009 doi: 10.1128/EC.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones T, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A. 2004;101:7329–34. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elson SL, Noble SM, Solis N, Filler SG, Johnson AD. An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS Genet. 2009;5:e1000664. doi: 10.1371/journal.pgen.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mecsas J. Use of signature-tagged mutagenesis in pathogenesis studies. Curr Opin Microbiol. 2002;5:33–7. doi: 10.1016/s1369-5274(02)00282-5. [DOI] [PubMed] [Google Scholar]
- 22.Saenz HL, Dehio C. Signature-tagged mutagenesis: technical advances in a negative selection method for virulence gene identification. Curr Opin Microbiol. 2005;8:612–9. doi: 10.1016/j.mib.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Shea JE, Santangelo JD, Feldman RG. Signature-tagged mutagenesis in the identification of virulence genes in pathogens. Curr Opin Microbiol. 2000;3:451–8. doi: 10.1016/s1369-5274(00)00120-x. [DOI] [PubMed] [Google Scholar]
- 24.MacCallum DM, Odds FC. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses. 2005;48:151–61. doi: 10.1111/j.1439-0507.2005.01121.x. [DOI] [PubMed] [Google Scholar]
- 25.Bastidas RJ, Heitman J. Trimorphic stepping stones pave the way to fungal virulence. Proc Natl Acad Sci U S A. 2009;106:351–2. doi: 10.1073/pnas.0811994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhl MA, Biery M, Craig N, Johnson AD. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C.albicans. Embo J. 2003;22:2668–78. doi: 10.1093/emboj/cdg256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rieg G, et al. Unanticipated heterogeneity in growth rate and virulence among Candida albicans AAF1 null mutants. Infect Immun. 1999;67:3193–8. doi: 10.1128/iai.67.7.3193-3198.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leipelt M, et al. Glucosylceramide synthases, a gene family responsible for the biosynthesis of glucosphingolipids in animals, plants, and fungi. J Biol Chem. 2001;276:33621–9. doi: 10.1074/jbc.M104952200. [DOI] [PubMed] [Google Scholar]
- 29.Rittershaus PC, et al. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest. 2006;116:1651–9. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roemer T, et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol. 2003;50:167–81. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- 31.Oura T, Kajiwara S. Disruption of the sphingolipid Delta8-desaturase gene causes a delay in morphological changes in Candida albicans. Microbiology. 2008;154:3795–803. doi: 10.1099/mic.0.2008/018788-0. [DOI] [PubMed] [Google Scholar]
- 32.Arguelles JC, Rodriguez T, Alvarez-Peral FJ. Trehalose hydrolysis is not required for human serum-induced dimorphic transition in Candida albicans: evidence from a tps1/tps1 mutant deficient in trehalose synthesis. Res Microbiol. 1999;150:521–9. doi: 10.1016/s0923-2508(99)00105-9. [DOI] [PubMed] [Google Scholar]
- 33.Cao F, et al. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol Biol Cell. 2006;17:295–307. doi: 10.1091/mbc.E05-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donovan M, et al. Virulence of a phosphoribosylaminoimidazole carboxylase-deficient Candida albicans strain in an immunosuppressed murine model of systemic candidiasis. Infect Immun. 2001;69:2542–8. doi: 10.1128/IAI.69.4.2542-2548.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hornby JM, et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol. 2001;67:2982–92. doi: 10.1128/AEM.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huh WK, Kim ST, Kim H, Jeong G, Kang SO. Deficiency of D-erythroascorbic acid attenuates hyphal growth and virulence of Candida albicans. Infect Immun. 2001;69:3939–46. doi: 10.1128/IAI.69.6.3939-3946.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia N, et al. Candida albicans sterol C-14 reductase, encoded by the ERG24 gene, as a potential antifungal target site. Antimicrob Agents Chemother. 2002;46:947–57. doi: 10.1128/AAC.46.4.947-957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirsch DR, Whitney RR. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect Immun. 1991;59:3297–300. doi: 10.1128/iai.59.9.3297-3300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mio T, Kokado M, Arisawa M, Yamada-Okabe H. Reduced virulence of Candida albicans mutants lacking the GNA1 gene encoding glucosamine-6-phosphate acetyltransferase. Microbiology. 2000;146(Pt 7):1753–8. doi: 10.1099/00221287-146-7-1753. [DOI] [PubMed] [Google Scholar]
- 40.Pedreno Y, et al. Disruption of the Candida albicans ATC1 gene encoding a cell-linked acid trehalase decreases hypha formation and infectivity without affecting resistance to oxidative stress. Microbiology. 2007;153:1372–81. doi: 10.1099/mic.0.2006/003921-0. [DOI] [PubMed] [Google Scholar]
- 41.Rocha CR, et al. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell. 2001;12:3631–43. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaragoza O, Blazquez MA, Gancedo C. Disruption of the Candida albicans TPS1 gene encoding trehalose-6-phosphate synthase impairs formation of hyphae and decreases infectivity. J Bacteriol. 1998;180:3809–15. doi: 10.1128/jb.180.15.3809-3815.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitrovich QM, Tuch BB, Guthrie C, Johnson AD. Computational and experimental approaches double the number of known introns in the pathogenic yeast Candida albicans. Genome Res. 2007;17:492–502. doi: 10.1101/gr.6111907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ausubel FM, et al. Current protocols. John Wiley & Sons; New York, N.Y.: 1990. [Google Scholar]
- 45.Miwa T, et al. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot Cell. 2004;3:919–31. doi: 10.1128/EC.3.4.919-931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Badrane H, et al. The Candida albicans phosphatase Inp51p interacts with the EH domain protein Irs4p, regulates phosphatidylinositol-4,5-bisphosphate levels and influences hyphal formation, the cell integrity pathway and virulence. Microbiology. 2008;154:3296–308. doi: 10.1099/mic.0.2008/018002-0. [DOI] [PubMed] [Google Scholar]
- 47.Kelly MT, et al. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 2004;53:969–83. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- 48.Warenda AJ, Kauffman S, Sherrill TP, Becker JM, Konopka JB. Candida albicans septin mutants are defective for invasive growth and virulence. Infect Immun. 2003;71:4045–51. doi: 10.1128/IAI.71.7.4045-4051.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu W, Lockhart SR, Pujol C, Srikantha T, Soll DR. Heterozygosity of genes on the sex chromosome regulates Candida albicans virulence. Mol Microbiol. 2007;64:1587–604. doi: 10.1111/j.1365-2958.2007.05759.x. [DOI] [PubMed] [Google Scholar]
- 50.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–2. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.