Abstract
Nucleotide excision repair (NER) is the only mechanism in humans to repair UV-induced DNA lesions such as pyrimidine (6–4) pyrimidone photoproducts and cyclobutane pyrimidine dimers (CPD). In response to UV damage, the ataxia telangiectasia mutated and Rad3-related (ATR) kinase phosphorylates and activates several downstream effector proteins, such as p53 and XPA, to arrest cell cycle progression, stimulate DNA repair, or initiate apoptosis. However, following the completion of DNA repair, there must be active mechanisms that restore the cell to a prestressed homeostatic state. An important part of this recovery must include a process to reduce p53 and NER activity as well as to remove repair protein complexes from the DNA damage sites. Since activation of the damage response occurs in part through phosphorylation, phosphatases are obvious candidates as homeostatic regulators of the DNA damage and repair responses. Therefore, we investigated whether the serine/threonine wild-type p53-induced phosphatase 1 (WIP1/PPM1D) might regulate NER. WIP1 overexpression inhibits the kinetics of NER and CPD repair, whereas WIP1 depletion enhances NER kinetics and CPD repair. This NER suppression is dependent on WIP1 phosphatase activity, as phosphatase-dead WIP1 mutants failed to inhibit NER. Moreover, WIP1 suppresses the kinetics of UV-induced damage repair largely through effects on NER, as XPD-deficient cells are not further suppressed in repairing UV damage by overexpressed WIP1. Wip1 null mice quickly repair their CPD and undergo less UV-induced apoptosis than their wild-type counterparts. In vitro phosphatase assays identify XPA and XPC as two potential WIP1 targets in the NER pathway. Thus WIP1 may suppress NER kinetics by dephosphorylating and inactivating XPA and XPC and other NER proteins and regulators after UV-induced DNA damage is repaired.
Keywords: CPD, nucleotide excision repair, phosphatase, phosphorylation, PPM1D, WIP1
1. Introduction
Mammals have evolved sophisticated stress response and repair pathways to maintain the integrity of their DNA in response to constant endogenous and exogenous mutagenic insults. After exposure to genotoxic stress, the cell initiates an intricate DNA damage signaling cascade to activate numerous target proteins that arrest cell cycle progression, initiate DNA repair processes or instigate programmed cell death responses. One type of exogenous damage is ultraviolet (UV) light, which produces mainly two types of DNA lesions, cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6–4) pyrimidone photoproducts [1]. These DNA lesions hinder transcription and replication and may result in a decrease in RNA synthesis, cell cycle arrest, or apoptosis. If left unrepaired, the lesions may induce DNA mutations that may eventually lead to skin cancer [2,3]. In mammals, these UV-induced lesions are repaired by nucleotide excision repair (NER), which is carried out by a coordinated multi-protein complex consisting of over 30 proteins [3,4]. NER recognizes the DNA helix distorting lesion, remodels the chromatin surrounding the lesion, excises the oligonucleotide containing the lesion, and performs repair synthesis and ligation to complete the repair process [3,5]. NER is composed of two distinct subpathways that differ in their DNA damage recognition. Global genomic repair removes DNA lesions from the entire genome and transcription coupled repair (TCR) only removes DNA lesions from transcribed genes. As expected, defects in NER lead to several genetic disorders, notably xeroderma pigmentosum (XP), Cockayne’s syndrome (CS) and Trichothiodystrophy (TTD). XP patients exhibit hypersensitivity to UV-induced skin cancer and CS and TTD patients undergo premature ageing [3,5].
Data on how the upstream UV-induced DNA damage signaling cascade regulates NER is sparse. Recently the main regulator of the UV-induced DNA damage response, the ataxia telangiectasia mutated and Rad3-related (ATR) kinase [6,7], was shown to be required for global genomic NER during S-phase [8]. Furthermore, in response to UV, ATR becomes activated and phosphorylates a number of downstream substrates, such as p53 and XPA, to enhance NER [9,10] and cell survival after UV damage [11,12], respectively. Phosphorylated p53 becomes stabilized, which activates p53 to induce XPC [13] and DDB2 [14] gene expression, which are critical for DNA lesion recognition and recruitment of other repair proteins. Additionally, p53 may also have a direct role in NER by recruiting the XPB helicase to the sites of CPD damage [15]. However, following the completion of DNA repair, there must be active mechanisms that restore the cell to a prestressed homeostatic state. A fundamental part of this recovery must include a process to reduce p53 and NER activity as well as to remove repair protein complexes from the DNA damage sites. One plausible method involves phosphatases that dephosphorylate and inactivate p53 and/or NER proteins.
One phosphatase that acts on p53 and ATM/ATR signaling is the wild-type p53-induced phosphatase 1 (WIP1, also known as PPM1D). WIP1 is a member of the type 2C protein family of serine/threonine phosphatases (PP2C) that is transcriptionally upregulated in a p53-dependent manner in response to various types of DNA damage [16,17]. We previously showed that WIP1 can negatively regulate p53 by dephosphorylating p53 at serine 15, a site phosphorylated by the ATR kinase in response to UV damage [18]. Additionally, WIP1 can also negatively regulate p53 by dephosphorylating and inactivating positive regulators of p53 such as, ATM [19], CHK1 [18], CHK2 [20,21], and p38 MAPK [22] or by dephosphorylating and activating negative regulators of p53 such as, MDM2 [23] and MDMX [24]. Moreover, WIP1 can inhibit base excision repair [25] and DNA double strand break repair, through homologous recombination and non-homologous end joining [26]. Thus, we have proposed that the primary role of WIP1 is to dephosphorylate and deactivate DNA damage response-activated proteins [17,27].
The downregulation of p53 and the DNA damage and DNA repair responses by WIP1 has cancer implications, particularly since the DNA damage response has recently been shown to play a crucial role as an early anti-cancer barrier [28–30]. Not surprisingly, WIP1 has been implicated as an oncogene [17]. WIP1 gene amplification and/or protein overexpression have been reported in a variety of human tumors [17], including breast adenocarcinoma [31–33], neuroblastoma [34], and ovarian clear cell adenocarcinoma [35,36]. In vitro transformation assays demonstrate that Wip1 promotes transformation of primary rodent fibroblasts in cooperation with other oncogenes [31,37]. Taken together, among the mechanisms by which WIP1 contributes to tumorigenesis may be its ability to inactivate various stress response and repair pathways.
Here, we show that WIP1 can also regulate NER. WIP1 overexpression inhibits NER kinetics and CPD repair, whereas WIP1 depletion enhances NER kinetics and CPD repair. Additionally, inhibition of NER is dependent on the phosphatase activity of WIP1 but independent of p53. Wip1 null mice quickly repair their CPD and undergo less UV-induced apoptosis than their wild-type counterparts. In vitro phosphatase assays identify XPA and XPC as two potential WIP1 targets in the NER pathway. Thus WIP1 may suppress NER kinetics by dephosphorylating and inactivating XPA and XPC and other NER proteins and regulators after the DNA is repaired.
2. Materials and Methods
2.1 Wip1-deficient mice
Wip1 knockout mice were generated by ES cell targeting as previously described [38]. Chimeric offspring were mated to C57BL/6 mice resulting in mixed background founders who were analyzed for transmission of the mutant Wip1 allele. Once Wip1 germ line heterozygotes were obtained, these mixed 129/Sv X C57BL/6 mice were backcrossed at least three generations into C57BL/6. Wip1 heterozygotes were crossed to obtain Wip1+/+, Wip1+/−, and Wip1−/− offspring and genotyped by PCR as described below. All mice were bred and maintained in a specific pathogen free animal facility at Baylor College of Medicine, where cages, chow (Irradiated Harlan Teklad Diet 2920X), and water were changed weekly. All research with mice has been conducted in compliance with the Baylor Animal Protocol Committee (Baylor College of Medicine Animal Protocol AN336) and AAALAC recommendations as published in The Guide for the Care and Use of Laboratory Animals (NRC1996).
2.2 Wip1 Allele Genotyping
High molecular weight genomic DNA was prepared from 1.5 cm mouse tail tips, 50 mg tissue, or 1 × 106 cells. Samples were digested overnight at 55–60°C in Tail Lysis Buffer (50 mM Tris pH 7.6, 50 mM EDTA pH 8.0, 100 mM NaCl, 1% SDS, 5 mM DTT, 0.1 mg Proteinase K). The DNA was phenol:chloroform extracted, precipitated with 100% ethanol, washed with 70% ethanol, and resuspended in 100 μl TE (10mM Tris-HCl pH 8.0, 1mM EDTA pH 8.0) buffer for three hours at 37°C or at room temperature overnight. All PCR primers were synthesized by Integrated DNA Technologies (Coralville, IA). Genotyping for the wild-type and mutant mouse Wip1 alleles was performed using primers in Intron 3 and Exon 4: Wip1 Intron 3: 5′-ACA AGC TTG CAG GGC TGT TTG TGG-3′ and PGK Promoter: 5′-CTT CCC AGC CTC TGA GCC CAG AAA GC-3′ and Wip1 Exon4 F: 5′-GTG GAG CTA TGA TTT CTT CAG TGG-3′ and Wip1 Exon4 R: 5′-GAT ACG ACA CAA GAC AAA CCT CC-3′. Using these primers, wild-type Wip1 allele produced a 300 bp fragment, whereas the mutant Wip1 allele produced a 500 bp fragment following PCR. PCR was performed under the following conditions combining 1 μl (or 500 ng) of genomic DNA, 5% DMSO, and 0.3125 μM primers in an AccuPower PCR PreMix tube (BioNeer, Inc., Alameda, CA): 95°C/5 min, [94°C/1 min, 50°C/1 min, 72°C/40 sec] x 35 cycles, 72°C/7 min. Samples were resolved on a 1% agarose gel.
2.3 Cell Culture
Saos-2 and U2-OS (two human osteosarcoma cell lines), HeLa, and XP17BE (XPD deficient) cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and cultured in DMEM, supplemented with 10% FBS and 1% Penicillin/Streptomycin. Wip1+/+, Wip1+/−, and Wip1−/− mouse embryonic fibroblasts (MEFs) were generated as previously described (Choi et al. 2002) and cultured in DMEM with 15% FBS and 1% Penicillin/Streptomycin. Wip1 MEFs were incubated in a modulator incubator chamber (Billups-Rothenberg, Inc, Del Mar, CA) containing a blood gas mixture of 5% carbon dioxide, 5.1% oxygen, and 90% balance nitrogen (Air Liquide American Corp., Houston, TX).
2.4 Plasmid Constructs
The mouse wild-type Wip1, phosphatase-dead Wip1 point mutants (mWip1-A95D, mWip1-D307A) and Wip1 truncation mutants (mWip1-ΔN(1-50), mWip1-ΔN(1-101), mWip1-ΔC(376-598)) were subcloned into pcDNA4/V5-His as previously described [25,37]. The human WIP1-D314A phosphatase-dead point mutant was generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) with either the WIP1-Flag-CMV-Neo-Bam or the WIP1-ΔExon6-His-pET-23a+ as a template. D314A point mutant primers were: 5′-TTATATTGGGGAGTGCTGGACTTTGGAATATGATTCCACC-3′ and 5′-GGTGGAATCATATTCCAAAGTCCAGCACTCCCCAATATAA-3′. All clones were sequenced to verify correct mutations.
The human WIP1-Flag-CMV-Neo-Bam plasmid was described previously [16] and obtained from Dr. E. Appella. The recombinant human WIP1 plasmid (WIP1-ΔExon6-His-pET-23a+) for bacterial expression was also described previously [39] and kindly provided by Dr. H. Yamaguchi. pRL-CMV was purchased from Promega and pGL3-CMV was obtained from Dr. A. Rice. The XPD (XPD-pCMV6-XL4) expression vector was purchased from Origene Technologies, Inc. (Rockville, MD).
2.5 Host Cell Reactivation Assay
Wip1−/−, Wip1+/−, and Wip1+/+ MEFs were seeded in 6-well plates and transfected with 500 ng of pGL3-CMV, which was damaged with 500 J/m2 UVC using a Stratalinker (Stratagene, La Jolla, CA), and 10 ng of pRL-CMV (Renilla luciferase marker plasmid for normalizing transfection efficiencies) using Lipofectamine Plus Reagent (Invitrogen). Twenty-four hours post transfection cells were rinsed once with phosphate-buffered saline (PBS) and lysed with reporter lysis buffer (Promega, Madison, WI) at room temperature. Luciferase activity from 20 μl of cell lysates was assayed using the Dual-Luciferase Reporter Assay System (Promega) in a Turner TD-20e Luminometer (Turner Designs, Sunnyvale, CA).
Host cell reactivation assays were also performed in U2-OS, SAOS-2, and XPD-deficient cells using the above protocol with some modifications as noted below. U2-OS cells were transfected with 500 ng of human WIP1, mouse (wild-type, point or truncation mutant) Wip1, or empty-CMV (pcDNA) expression constructs, 500 ng of 4000 J/m2 UVC-damaged pGL3-CMV and 10 ng of pRL-CMV using Lipofectamine Plus Reagent. SAOS-2 cells were transfected with 500 ng of human WIP1, WIP1-D314A or pcDNA expression constructs, 500 ng of 1000 J/m2 UVC-damaged pGL3-CMV and 10 ng of pRL-CMV using Lipofectamine Plus Reagent. XPD-deficient cells were transfected with 500 ng of human WIP1, WIP1-D314A, XPD, or pcDNA expression constructs, 500 ng of 500 J/m2 UVC-damaged pGL3-CMV and 10 ng of pRL-CMV using FuGENE 6 transfection reagent (Roche Applied Science, Indianapolis, IN). The total amount of DNA in each transfection was kept constant by complementing with empty-CMV (pcDNA) vector control DNA. WIP1 expression was verified by Western blotting.
2.6 Antibodies
Primary antibodies included antibodies against actin (I-19) (Santa Cruz Biotechnology, Santa Cruz, CA), cleaved caspase 3 (Asp175) (Cell Signaling Technology (CST), Danvers, MA), cleaved PARP (Asp214) (CST), DYKDDDDK (Flag) tag (BioLegend, San Diego, CA), thymine dimer and thymine dimer-HRP (Kamiya Biomedical Company, Seattle, WA), V5 (Invitrogen), WIP1/PPM1D (C-term) (Abgent, San Diego, CA). Secondary antibodies included donkey anti-goat IgG-HRP (sc-2020), goat anti-mouse IgG-HRP (sc-2302) and bovine anti-rabbit IgG-HRP (sc-2385) all from Santa Cruz, and highly cross-absorbed secondary Alexa Fluor 488 goat anti-mouse IgG (A11029) and Alexa Fluor 568 goat anti-rat IgG (A-11077) antibodies, both from Invitrogen.
2.7 Western blotting
Wip1+/+ and Wip1−/− MEFs were irradiated with 15 J/m2 of UVC (254 nm) using a Stratalinker (Stratagene). At indicated times after irradiation, cells were rinsed with PBS, scraped, and pelleted. Cell pellets were resuspended in lysis buffer (150mM NaCl, 50mM Tris pH 7.5, 1% NP-40, 0.1% SDS, 1% Na-deoxycholate, 10mM NaF, 1mM EDTA, Protease Inhibitor (Roche)) for 30 minutes on ice. Protein extracts were boiled at 100°C for 5 min and subjected to SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane using standard electroblotting procedures. The membrane was blocked with 5% milk in tris buffered saline containing 0.1% Tween-20 (TBST) for 1 hour and incubated with rabbit anti-cleaved caspase-3 and rabbit anti-cleaved PARP (both from CST), at 1:1000 dilution, and goat anti-actin (Santa Cruz) at 1:2000 dilution, primary antibodies at 4°C overnight. Protein was detected using SuperSignal Chemiluminescent substrate (Thermo Scientific).
HeLa, SAOS-2 and U2-OS cells transfected with WIP1 plasmids were washed once with PBS, scraped, and pelleted. Cell pellets were dissolved in Laemmli Sample Buffer (Bio-Rad Labs, Hercules, CA) supplemented with 5% 2-mercaptoethanol for 15 min at room temperature. Protein extracts were boiled at 100°C for 10 min and western blotted as above. The membrane was blocked with 5% milk-TBST and incubated with rabbit anti-WIP1 (Abgent), at 1:1000 dilution, and goat anti-actin (Santa Cruz) at 1:2000 dilution, primary antibodies at 4°C overnight. To detect mouse Wip1 expression, membranes were incubated with the mouse anti-V5 (Invitrogen) primary antibody and anti-mouse HRP (eBiosciences) secondary antibodies, both at 1:5000 dilution, for 1 hr each at room temperature.
2.8 Immuno Slot-Blot Analysis of CPD in Genomic DNA
Wip1+/+ and Wip1−/− MEFs were irradiated with 15 J/m2 of UVC (254 nm) using a Stratalinker (Stratagene). At indicated times after irradiation, cells were rinsed with PBS, scraped, and pelleted. Cell pellets were digested overnight at 55–60°C in Tail Lysis Buffer and genomic DNA was isolated using the method described above. The DNA concentration was determined by measurements of absorbance at 260 and 280 nm. For immuno slot-blot analysis, 2.5 μg of genomic DNA was denatured for 10 min at 100ºC, followed by the addition of one volume of 2M ammonium acetate to stabilize the denatured DNA. The denatured DNA was then loaded on a Nytran SuPerCharge nylon membrane (Schleicher & Schuell) using the Minifold II slot blot apparatus (Schleicher & Schuell). After fixing the DNA by baking the membrane at 80°C for 2 hr, the membrane was blocked in PBS containing 5% non-fat dried milk for 1 hr at room temperature. After three washes in PBS, the membrane was incubated with an anti-thymine dimer-HRP antibody (Kamiya) at 1:1000 dilution overnight at 4°C. After extensive washes in PBS, the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL) was added and the membrane was exposed to X-ray film. The membrane was briefly washed in deionized water (dH2O) and incubated in a DNA staining solution (0.1% methylene, 0.5M sodium acetate pH 5.2) for 15 min at room temperature. The DNA staining solution was removed and the membrane was destained extensively in dH2O. The CPD and DNA band intensities were scanned using a flat-bed Perfection 4180 Photo color scanner (Epson, Long Beach, CA) and the images were analyzed using Photoshop CS2 software (Adobe Systems Inc., San Jose, CA). The CPD content was normalized to the DNA content. The percent of remaining CPD was calculated by normalizing relative CPD amounts at various timepoints to the amount of CPD at 1hr after UV damage.
HeLa cells were seeded in 10 cm tissue culture plates and transfected with 6 μg of WIP1-Flag, WIP1-D314A-Flag or pcDNA empty vector using Lipofectamine Plus reagent (Invitrogen). Twenty-four hours post transfection cells were rinsed once with PBS and irradiated with 15 J/m2 of UVC. Genomic DNA isolation and slot blot analysis were performed as described above. WIP1 expression was verified by Western blotting.
2.9 Immunofluorescence
HeLa cells were grown on coverslips in 10 cm dishes 24 hr prior to transfection. 6 μg of WIP1-Flag expression vector or no DNA was transfected into the cells using Lipofectamine Plus reagent. Twenty-four hours post transfection cells were rinsed once with PBS and irradiated with 15 J/m2 of UVC. At indicated times after irradiation cells were rinsed twice in PBS then fixed with 3.8% formaldehyde in PBS for 10 min at room temperature. After fixation, cells were washed 3 times in PBS and permeabilized with 0.5% Triton X-100 in PBS for 15 min at room temperature. Cells were subsequently washed 3 times in PBS and incubated in 2M HCl for 5 min at 37°C to denature cellular DNA. After three washes in PBS cells were blocked in 3% BSA in PBS at 4°C overnight. Cells were costained with mouse anti-thymine dimer (Kamiya) and rat anti-DYKDDDDK (Flag) tag (BioLegend) primary antibodies, both at 1:1000 dilution, in 1% BSA-PBS at 37°C for 1 hr. Cells were then washed 3 times with PBS and stained with highly cross-absorbed secondary anti-mouse Alexa Fluor 488, at 1:500 dilution, and anti-rat Alexa Fluor 568, at 1:1000 dilution, (Invitrogen) antibodies along with the DNA counter stain 4′-6′-diamidino-2-phenylindole (DAPI), at 1:250 dilution, in 1% BSA-PBS at 37°C for 1 hr. Cells were washed 3 times in PBS and coverslips were mounted with Prolong Gold Antifade (Invitrogen).
2.10 Automated microscopy and analysis of microscopic images
Images were acquired with a Beckman-Coulter IC-100 microscope utilizing autofocus, a Nikon 20x 0.75 NA objective, a Hamamatsu Orca ER CCD camera, and appropriate filters for the three fluorophores, DAPI, Alexa Fluor 488, and Alexa Fluor 568. For each field, one image in each fluorescent channel was acquired. Image analysis was performed using Pipeline Pilot data pipelining software (Accelrys Inc., San Diego, CA). Prior to further analysis, each image was background-subtracted by subtracting the median pixel intensity of each image from each pixel within the image, and setting pixels that result in a negative value to zero. For each field, a binary mask was produced that marked the nuclear regions in the field, by performing a segmentation algorithm on the DAPI channel image of the field. Circularity, size, and DAPI channel sum of pixel intensities were calculated for each distinct nuclear region in the binary mask. Nuclear regions were eliminated from the binary mask if any of the following criteria were not met:
The region did not contact the edge of the field.
The region area was within the range of a typical interphase nucleus.
The region circularity was within the range of a typical interphase nucleus.
The region DNA content (represented by the DAPI sum of pixel intensities of the region) was within the range of 2C or 4C.
The DAPI channel 90th percentile pixel intensity of the region was within the range of a typical interphase nucleus.
These criteria largely excluded fluorescent debris, nuclear fragments, clustered nuclei, multi-nucleated cells, apoptotic cells, and mitotic cells from further analysis. For each remaining nuclear region, the underlying CPD channel sum of pixel intensities and WIP1 channel sum of pixel intensities were measured. Additionally, CPD channel and WIP1 channel mean pixel intensities were calculated for each region.
2.11 UVB irradiation of Wip1-deficient mice
The dorsal hair of 8 to 10 week old virgin female Wip1+/+ or Wip1−/− mice was first clipped using an electric clipper follow by treatment with Nair (Church & Dwight Co., Inc, Princeton, NJ) to completely remove all hair, at least 24 hr prior to UVB irradiation. Restrained mice were irradiated using an EL Series UV Lamp (UVP, LLC Upland, CA) that emits 100% UVB at 302 nm. The mice were treated with 2.5 kJ/m2 UVB given as a single exposure. Negative control mice were treated in a similar manner but only mock irradiated. Each group consisted of at least 3 animals. At indicated times after irradiation mice were sacrificed by cervical dislocation. One piece of dorsal skin from every mouse was fixed in 10% buffered formalin or digested in tail lysis buffer for the isolation of genomic DNA and CPD slot-blot analysis (as described above). Formalin-fixed tissues were processed, paraffin embedded, and sectioned to a thickness of 5 μm at the Baylor College of Medicine Comparative Pathology Laboratory. Mounted skin sections were stained with hematoxylin and eosin (H&E) for light microscopy. Images were obtained with a Nikon Eclipse 80i microscope, a Nikon 20x 0.75 NA objective, and a Nikon DS-5M color CCD camera (Nikon Inc, Melville, NY) using NIS Elements software.
2.12 TUNEL assay for detection of apoptotic cells
To assess the effects of Wip1 allele status on UVB-induced apoptosis, terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) staining was performed on the paraffin-embedded skin sections using the In Situ Cell Death Detection Kit, Fluorescein (Roche) following the manufacturer’s instructions. Sections were counterstained with DAPI (1 μg/ml) and mounted with Prolong Gold Antifade (Invitrogen). Sections were imaged with an epifluorescent Nikon Eclipse 80i microscope, a Nikon 20x 0.75 NA objective, and a Photometrics CoolSNAP cf monochrome CCD camera using NIS Elements software and appropriate filters for the two fluorophores, DAPI and Alexa Fluor 488. For each mouse, four random fields were imaged and the TUNEL and DAPI fluorescence intensity were quantitated. The TUNEL fluorescence intensity was normalized to the DAPI intensity and plotted.
2.13 Phosphopeptides and in vitro phosphatase assays
All phosphopeptides were custom synthesized by New England Peptide (Gardner, MA). The phosphopeptide sequences are as follows: substrates: XPA pS196 (Ac-LEVWGpSQEALE-Amide) and XPC pS892 (Ac-EEGTSpSQAEAA-Amide); negative control: UNG2 pT31 (Ac-AVQGpTGVAGV-Amide); and positive controls: p38-MAPK pT180 (Ac-TDDEMpTGpYVAT-Amide) and CHK1 pS345 (Ac-QGISFpSQPTCP-Amide). For in vitro phosphatase assays on phosphopeptides, recombinant human WIP1 ΔEx6 and phosphatase-dead WIP1-D314A ΔEx6 proteins were purified from bacteria as previously described [39]. 50 ng of purified protein was diluted in PP2C phosphatase buffer (50 mM Tris-HCl pH 7.5, 0.1 mM EGTA, and 0.02% 2-mercaptoethanol) and incubated with BSA (1 mg/ml) and 30 mM MgCl2 containing 100 μM phosphopeptide for 1 hr at 25ºC. Free phosphate was determined using a molybdate dye based Serine/Threonine Phosphatase Assay System (Promega) following protocols provided by the manufacturer. Absorbance was measured with a 630 nm filter in a Victor 2 1400 Multilabel 96-well Plate Reader (Perkin Elmer, Waltham, MA). The relative amount of phosphate released for the XPA and XPC substrates was normalized to the positive control of each phosphatase. Phosphatase assays were done in triplicate. In vitro phosphatase assays using PP1α, PP2A, and PP2Cα protein were also performed using the above protocol with some modifications as noted below. PP1α (cat# 539493) was purchased (Calbiochem, San Diego, CA) and diluted in PP1 phosphatase buffer (50 mM Tris-HCl, pH 7.0, 5 mM DTT, 200 μM MnCl2, 100 μM EDTA pH 7.0). The positive control for PP1α was the CHK1 pS345 phosphopeptide. PP2A (cat# 14-111) was also purchased (Millipore, Billerica, MA) and diluted in PP2A phosphatase buffer (50 mM Tris-HCl pH 7.0, 14 mM 2-mercaptoethanol, 100 μM EDTA pH 8.0). The RRA(pT)VA Ser/Thr phosphopeptide supplied with the Serine/Threonine Phosphatase Assay System served as the positive control for PP2A. PP2Cα (cat# 539569) was also purchased (Calbiochem) and diluted in the previously described PP2C phosphatase buffer. The p38-MAPK pT180 phosphopeptide also served as the PP2Cα positive control.
2.14 Statistical analysis
The results shown are means ± standard error. Statistical significance was assessed using Student’s t-test.
3. Results
3.1. Wip1-deficient MEFs exhibit enhanced nucleotide excision repair of UV-damaged DNA
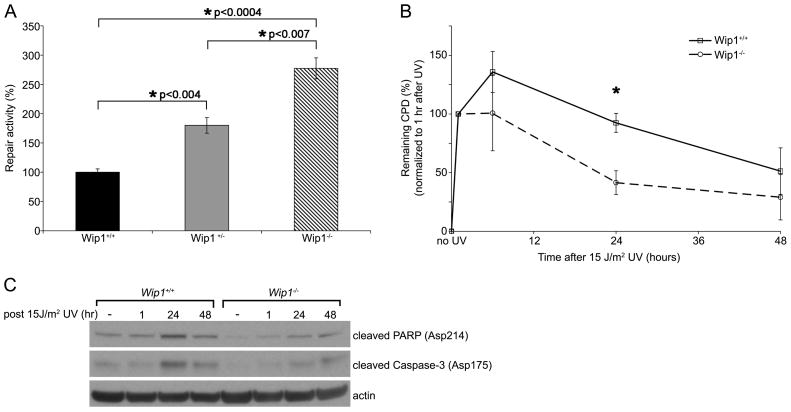
To assess whether WIP1 regulates NER, we utilized two established methods to analyze the repair of UV-damaged DNA. First we performed host cell reactivation (HCR) assays in mouse fibroblasts derived from Wip1+/+, Wip1+/−, and Wip1−/− embryos (Wip1 MEFs). UV-damaged firefly luciferase and undamaged Renilla luciferase plasmids were cotransfected into Wip1 MEFs and luciferase activity was measured in cell lysates 24 hours later. Given that UV-induced DNA lesions block transcription, expression of firefly luciferase provides an indicator of the ability of the transfected cells to repair the UV-damaged reporter plasmid, primarily through NER [40]. The Wip1−/− and Wip1+/− MEFs display a 2.8 and 1.8 fold increase, respectively, in NER activity as compared to the Wip1+/+ MEFs (Figure 1A).
Figure 1.
Wip1 depletion enhances nucleotide excision repair. (A) Wip1 null MEFs display increased NER activity as measured by host cell reactivation assays. A UV-damaged firefly luciferase reporter plasmid was cotransfected with an unirradiated Renilla luciferase plasmid into Wip1+/+, Wip1+/−, and Wip1−/− MEFs. Twenty-four hours after transfection, the cells were harvested and luciferase activity was measured. After correcting for transfection efficiency by dividing the firefly by Renilla luciferase activity, the Wip1+/+ MEFs cells were set to 100% and the normalized luciferase values for the other cells were reported as percent repair activity. Error bars indicate the standard error (n=6). Asterisks indicate statistical significance with P values indicated. (B) Wip1 null MEFs display enhanced kinetics of CPD repair. Kinetics of CPD repair in Wip1+/+ and Wip1−/− MEFs were determined by immuno slot-blot assay following 15 J/m2 UVC irradiation. The percent of remaining CPD was calculated by normalizing relative CPD amounts at various timepoints to the amount of CPD at 1 hour after UV damage. Each data point represents the mean of three experiments and the error bars represent the standard error. The asterisk indicates statistical significance (p<0.05). (C) Representative western blot showing reduced cleavage of apoptosis-facilitating proteins (caspase 3 and PARP) in Wip1−/− MEF compared to Wip1+/+ MEF after 15 J/m2 UV.
In a second assay we analyzed the repair of cyclobutane pyrimidine dimers (CPD), which are UV-induced lesions that occur when the double bonds between adjacent pyrimidines are saturated to create a four-membered ring [1]. Wip1 MEFs were irradiated with 15 J/m2 UV and genomic DNA was harvested from the cells at various times following UV irradiation. CPD repair was analyzed using an immuno slot-blot assay using antibodies specific for the UV-induced CPD. By 24 hours, Wip1−/− MEFs had repaired 59% of their CPD, in contrast to only 8% by the Wip1+/+ MEFs, as compared to the 1 hour timepoint (Figure 1B). A similar trend appears at 48 hours, although the difference in repair rates at this time point is not statistically significant. Taken together, our results show that loss of WIP1 enhances NER kinetics, thereby implicating WIP1 as an inhibitor of NER.
To determine whether the decreased CPD in the Wip1−/− MEF resulted in a concurrent increase in apoptosis, we next examined the levels of two pro-apoptotic proteins, cleaved caspase-3 and cleaved poly (ADP-ribose) polymerase (PARP). The Wip1−/− MEF had lower levels of cleaved caspase-3 and cleaved PARP l as compared to the Wip1+/+ MEF in response to UV (Figure 1C), indicating that the decreased CPD seen in the Wip1−/− MEF is not a result of increased apoptosis. On the contrary, these findings suggest that accumulation of CPD in the Wip1+/+ MEF may have triggered more UV-induced apoptosis.
3.2 WIP1 overexpression inhibits NER
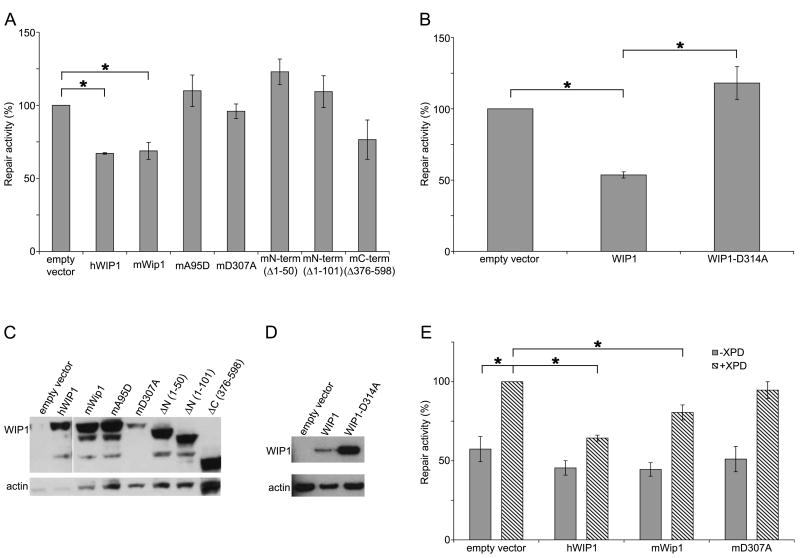
Given that reduction of Wip1 enhances NER activity, we wanted to investigate whether increased levels of WIP1 would diminish NER activity. We repeated the HCR assay in the U2-OS human osteosarcoma cells, which are p53-proficient, by cotransfecting the UV-damaged luciferase plasmid with plasmids expressing the WIP1 gene or an empty vector. U2-OS cells transfected with human or mouse Wip1 had a 33% and 31% reduction in NER, respectively, as compared to cells transfected with an empty vector (Figure 2A). To ascertain the domains of WIP1 that were responsible for regulating NER, we performed HCR assays using murine Wip1 mutants. Two N-terminal (phosphatase-domain) truncation mutants (Δ1-50 and Δ1-101) and two phosphatase-dead point mutants (A95D and D307A) of Wip1 had no effect on NER indicating that the phosphatase activity of Wip1 plays an important role in inhibiting NER. The C-terminal Wip1 truncation mutant (Δ376-598) had a minor inhibitory effect on NER, suggesting that the C-terminal domain outside of the phosphatase domain may also be involved in inhibiting NER. Western blot analysis of U2-OS cells show that the transfected WIP1 plasmids were expressed at robust levels (Figure 2C).
Figure 2.
WIP1 inhibits NER. (A) WIP1, but not mutant murine Wip1, overexpression inhibits NER in U2-OS cells. Human WIP1, mouse (wild-type, point or truncation mutant) Wip1, or empty-CMV expression constructs were cotransfected with a UV-damaged firefly luciferase reporter plasmid and an unirradiated Renilla luciferase plasmid into U2-OS (p53 proficient cells). Twenty-four hours after transfection, the cells were harvested and luciferase activity was measured. After correcting for transfection efficiency by normalizing to firefly Renilla luciferase activity, the empty vector transfected cells were set to 100% and the normalized luciferase values for the other cells were reported as percent repair activity. Error bars indicate the standard error (n=3). Asterisks indicate statistical significance (p<0.05). (B) p53-independent inhibition of NER in Saos-2 cells by WIP1. Human wild-type or phosphatase-dead WIP1 or empty-CMV expression constructs were cotransfected with luciferase plasmids in p53 null Saos-2 cells as described above. Error bars indicate the standard error (n=4) and asterisks indicate statistical significance (p<0.05). (C) Western blot analysis of WIP1 expression in U2-OS cells transfected with human or mouse Wip1 as analyzed in (A). A monoclonal Flag or V5 antibody was used to detect human or mouse Wip1, respectively, which is the top band in each lane. β-actin was used as loading control. (D) Western blot analysis of WIP1 expression in Saos-2 cells transfected with human WIP1, as analyzed in (B). (E) WIP1 suppression of UV-damage repair is largely dependent on a functioning NER pathway. WIP1 overexpression inhibits NER in XP17BE (XPD deficient) cells complemented with an XPD plasmid. Human wild-type or phosphatase-dead WIP1 or empty-CMV expression constructs were cotransfected with luciferase plasmids in the presence or absence of exogenous XPD in XP17BE cells as described above. After correcting for transfection efficiency by dividing the firefly by Renilla luciferase activity, the empty vector + exogenous XPD transfected cells were set to 100% and the normalized luciferase values for the other cells were reported as percent repair activity. Error bars indicate the standard error (n=3). Asterisks indicate statistical significance (p<0.05).
Since p53 enhances NER [9,10,13–15] and WIP1 negatively regulates p53 (as part of a negative feedback regulatory loop) [18,23], it might be that the increased NER activity in Wip1−/− MEFs and the decreased NER activity in WIP1 overexpressed U2-OS cells was simply due to increased and decreased p53 activity, respectively, and not due to the direct effects of WIP1 on NER functions. To determine whether WIP1 regulation of NER is p53 independent, we examined the effects of overexpressed WIP1 on NER in Saos-2 cells, which are p53-null human osteosarcoma cells. Saos-2 cells transfected with human WIP1 had a 46% reduction in NER activity as compared to the empty vector transfected control cells (Figure 2B). As expected, the phosphatase-dead human WIP1 mutant (D314A) had no effect on NER. Western blot analysis showed that the transfected WIP1 plasmids were expressed (Figure 2D). Altogether, these results demonstrate that inhibition of NER by WIP1 is p53 independent.
As a negative control, HCR assays were also performed in XP17BE cells that are XPD-deficient. Overexpression of either human or mouse Wip1 in the XP17BE cells had no effect on NER as compared to the empty vector control as these cells are unable to efficiently repair the damaged luciferase plasmid (Figure 2E). In contrast, when the XP17BE cells were complemented with an XPD expression vector, overexpression of human or mouse Wip1 inhibited NER by 36% and 19%, respectively, as compared to empty vector controls. Overexpression of the phosphatase-dead Wip1 again had no effect on NER. In summary, our results show that WIP1 overexpression inhibits repair of UV-damaged DNA templates, and that the inhibition is dependent on a functioning NER pathway. Moreover, WIP1 phosphatase activity is required for the inhibition, and that inhibition is p53 independent.
3.3 WIP1 overexpression inhibits the repair of CPD
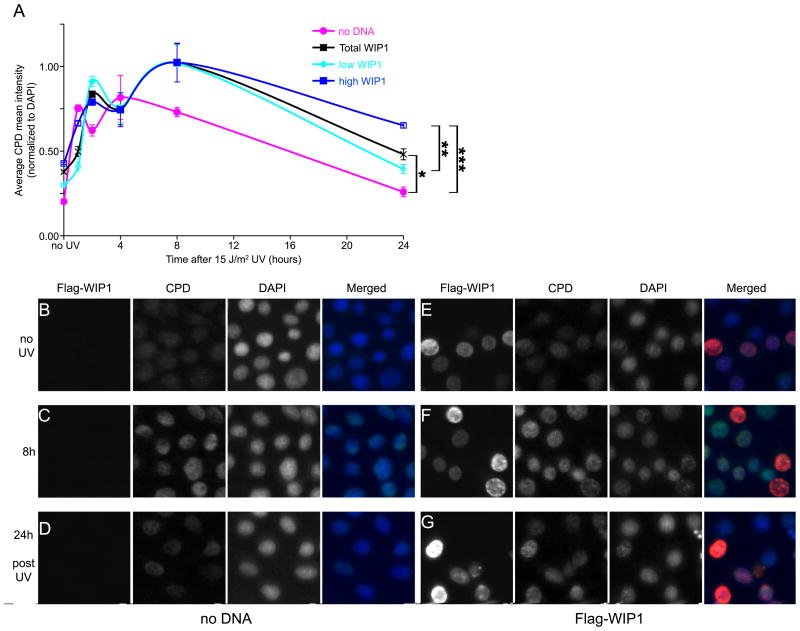
To confirm the HCR assay results, we also examined the effects of WIP1 overexpression on the repair of CPD. HeLa cells were transfected with wild-type WIP1, phosphatase-dead WIP1, or empty vector 24 hours prior to irradiation with 15 J/m2 UV. At various times following UV irradiation, genomic DNA was harvested from the transfected cells and CPD repair was analyzed using an immuno slot-blot assay. Surprisingly, the difference in the repair of CPD between empty vector or phosphatase-dead WIP1 and WIP1 transfected cells was not statistically significant (data not shown). Considering that the transfection efficiency of this experiment was less than 50% (data not shown), we reasoned that a significant effect by the transfected cells might be masked in this assay by the presence of untransfected cells.
To remedy this problem and distinguish between cells that express low or high levels of WIP1 we immunostained for CPD and WIP1. HeLa cells transfected with no DNA or Flag-WIP1 were uniformly irradiated with 15 J/m2 UV and immunostained for CPD and Flag (to detect WIP1 expression) at various time points after irradiation. The coverslips were imaged using a high through-put microscope with appropriate filters for DAPI, CPD, and WIP1 (Figure 3B–G). The sum and mean nuclear fluorescence of each channel was measured. Nuclei clusters, mitotic cells, and apoptotic cells were filtered from the total cell population. Analysis of the total HeLa cells transfected with either no DNA or WIP1 showed that these cells were able to repair 90% and 83% of their CPD, respectively, within 24 hours after irradiation (Figure 3A). However, when the total WIP1 transfected HeLa cell population was divided into two subpopulations, low and high WIP1, the difference in CPD repair rates were clearly visible. Cells with low WIP1 levels repaired 87% of their CPD by 24 hours as compared to cells with high WIP1 levels, which only repaired 61% of their CPD. Thus, quantitative analysis of CPD and WIP1 fluorescence by high throughput microscopy revealed a highly significant reduction in CPD repair in cells with high WIP1 levels as compared to cells with low WIP1 (26% difference between low and high WIP1) or no DNA (29% difference between no DNA and high WIP1), indicating that higher WIP1 levels inhibit CPD repair. Examination of individual transfected UV-treated Hela cells (Figure 3B–G) shows that cells with high levels of WIP1 at 24 hours post UV exhibit higher levels of CPD fluorescence compared to their non-WIP1 expressing counterparts (Figure 3G).
Figure 3.
WIP1 inhibits repair of CPD. (A) Quantitative analysis of CPD repair in UV irradiated HeLa cells with or without overexpressed WIP1. The CPD fluorescence intensity (normalized to DAPI) from the mock-transfected and WIP1-transfected cells were measured by high-throughput microscopic imaging and plotted. Each data point represents the average of two (open boxes or circles) or three (filled boxes or circles) coverslips, each containing at least 3,500 independent cells. Error bars represent the standard error and the asterisk indicates statistical significance: (* = p<0.0078; ** = p<0.0072; *** = p<0.0011). (B–G) WIP1 overexpression inhibits the repair of CPD. HeLa cells transfected with no DNA (B–D) or wild-type WIP1 (E–G) were irradiated with 15 J/m2 UVC 24 hours after transfection. Cells were fixed at various timepoints after irradiation and immunostained with anti-Flag (red fluorescence) and anti-CPD antibodies (green fluorescence) to assess CPD repair. DAPI staining (blue fluorescence) was used to identify nuclei. (B, E) no UV, (C, F) 8 hours post UV, (D, G) 24 hours post UV irradiation.
3.4 Wip1 null mice exhibit increased CPD repair and decreased apoptosis in response to UV
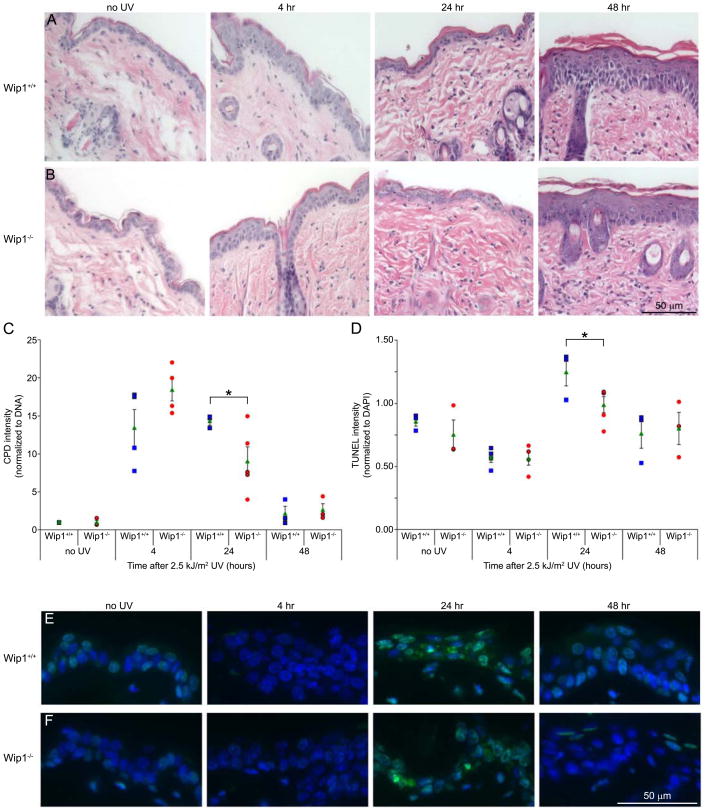
To investigate whether the principal target of UV light in animals, the skin, also benefits from Wip1 depletion, we analyzed the repair of CPD in the epidermis of Wip1 null mice in response to UVB irradiation. The depilated dorsal skin of female Wip1+/+ and Wip1−/− mice was exposed to 2.5 kJ/m2 UVB and harvested at 4, 24, and 48 hours post irradiation. Hematoxylin and eosin (H & E) staining of skin sections revealed no overt differences between the two genotypes (Figure 4A & B). Neutrophils infiltrated the subcutaneous fat and dermis of the Wip1+/+ skin after UV irradiation, whereas both neutrophils and monocytes infiltrated the Wip1−/−skin. Epidermal hyperplasia, or thickening, was visible in both genotypes by 48 hours post irradiation. Analysis of the epidermal thickness revealed no statistical difference between the two genotypes (data not shown). Immuno slot-blot analysis of genomic DNA isolated from irradiated skin sections showed CPD accumulation by 4 hours and almost all CPD clearance by 48 hours post irradiation in both genotypes (Figure 4C). Interestingly, Wip1−/− mice displayed a 50% decrease in their CPD between the 4 and 24 hour timepoints, whereas the Wip1+/+ mice maintained the same amount of CPD at both timepoints. Moreover, Wip1−/− mice have 63% less CPD than their Wip1+/+ counterparts by 24 hours post irradiation.
Figure 4.
Effects of UVB exposure on Wip1 deficient mice. Skin sections from Wip1+/+ and Wip1−/− mice exposed to 2.5 kJ/m2 UVB were collected at the times indicated. (A, B) Epidermal morphology in Wip1+/+ (A) and Wip1−/− (B) mice. Skin sections were stained with hematoxylin and eosin. (C) Wip1 null mice displayed increased CPD repair activity. Genomic DNA was isolated from irradiated mouse skin and CPD were detected by immuno slot-blot assay. Each data point (blue squares = Wip1+/+, red circles = Wip1−/−) represents an individual mouse. The green triangle represents the average of the genotype and the error bars represent the standard error. The asterisk indicates statistical significance (p<0.05). (D) Wip1 null mice are less sensitive to UV-induced apoptosis. A TUNEL apoptosis assay was performed on skin sections. TUNEL fluorescence intensity was normalized to DAPI staining and plotted. Each data point (blue squares = Wip1+/+, red circles = Wip1−/−) represents an individual mouse. The green triangle represents the average of the genotype and the error bars represent the standard error. The asterisk indicates statistical significance (p<0.05). (E, F) TUNEL staining of the epidermis of Wip1+/+ (E) and Wip1−/− (F) mice.
A TUNEL assay was also performed on the skin sections to examine whether apoptosis was induced (Figure 4D–F). While apoptotic cells were visible in the epidermis of both genotypes by 24 hours post UV irradiation, Wip1+/+ mice had significantly more TUNEL staining than the Wip1−/− mice. These results suggest that the decreased CPD seen in Wip1−/− mice was not a result of apoptosis, but rather accumulation of CPD in the Wip1+/+ mice may have triggered more UV-induced apoptosis. In summary, Wip1 null mice can more rapidly remove their CPD, which may reduce their sensitivity to UV-induced apoptosis.
3.5 XPA S196 and XPC S892 are potential WIP1 targets
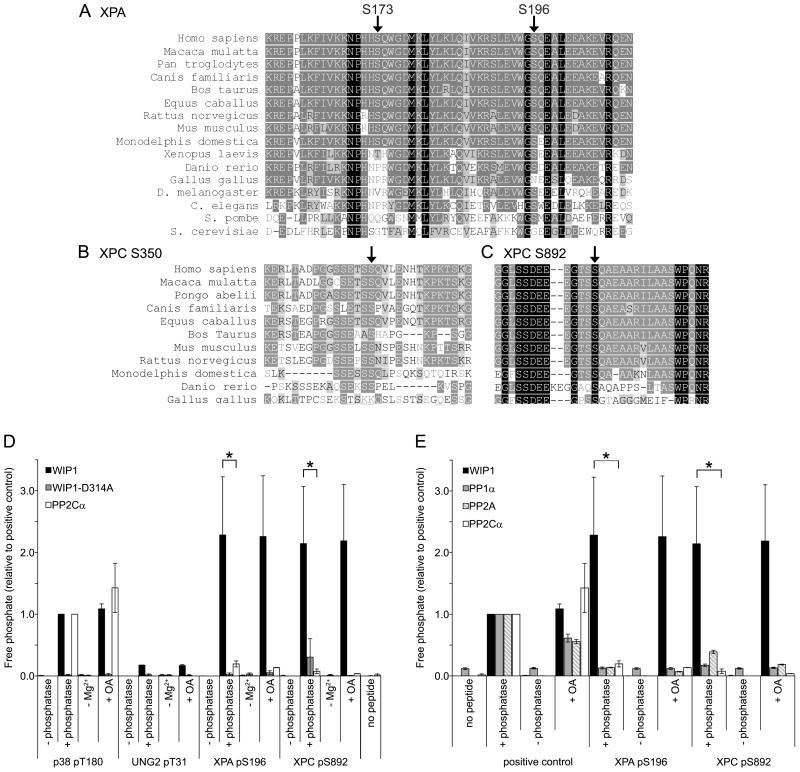
To determine a mechanism by which WIP1 might inhibit NER, potential WIP1 targets in the NER pathway were identified. Recently, the ATM/ATR kinases were reported to phosphorylate two NER proteins, XPA and XPC (xeroderma pigmentosum complementation group A and C, respectively) in response to DNA damage [12,41]. A sequence alignment containing the identified XPA sites, serines (Ser) 173 and 196, and XPC sites, serines 350 and 892, showed that XPA Ser196 and XPC Ser892 are highly conserved from zebrafish to humans (Figure 5A–C). Overall the XPA Ser196 site, which has an 87.5% identity and a 100% similarity, is more highly conserved from yeast to humans than the Ser173 site, which has a 56.3% identity and a 68.8% similarity. XPC Ser892, with a 100% identity and similarity, is slightly more conserved from chickens to humans than Ser350, with a 90.9% identity and similarity.
Figure 5.
XPA and XPC phosphopeptides are dephosphorylated by WIP1 in vitro. (A) Protein sequence alignment of various orthologues of XPA, containing serines 173 and 196, indicated by arrows. Identical amino acids are highlighted with a black background while conservative amino acid substitutions are indicated with a gray background. (B, C) Protein sequence alignment of various orthologues of XPC, containing serines 350 (B) and 892 (C), indicated by arrows. (D) XPA S196 and XPC S892 phosphopeptides are dephosphorylated by WIP1 in vitro. XPA pS196 and XPC pS892 phosphopeptides were incubated with human recombinant WIP1, phosphatase-dead WIP1-D314A, or PP2Cα protein. p38 pT180 and UNG2 pT31 phosphopeptides were used as a positive and negative control, respectively. −Mg2+ indicates incubation in the absence of magnesium and +OA indicates incubation with the PP1α and PP2A phosphatase inhibitor, okadaic acid. Free phosphate released from the phosphopeptide was measured by malachite green phosphate assay to determine relative phosphatase activities on each phosphopeptide. Error bars represent the standard error (n=3) and the asterisk indicates statistical significance (p<0.05). (E) WIP1, but not other serine/threonine phosphatases, dephosphorylates XPA S196 and XPC S892 phosphopeptides in vitro. XPA pS196 and XPC pS892 phosphopeptides were incubated with human recombinant WIP1, PP1α, PP2A, or PP2Cα protein. Positive controls were as follows: p38 pT180 for WIP1 and PP2Cα, CHK1 pS345 for PP1α, and the generic serine/threonine phosphopeptide, RRA(pT)VA, for PP2A. Phosphatase activity was measured as described above. Error bars represent the standard error (n=3) and the asterisk indicates statistical significance (p<0.05).
We next investigated whether WIP1 can dephosphorylate the phosphoserines on XPA and XPC in vitro. XPA Ser196 and XPC Ser892 phosphopeptides were synthesized and incubated with bacterially purified WIP1 protein and Mg2+ in an in vitro phosphatase assay. Release of free phosphate from the phosphopeptides was measured and normalized against the positive control. The phosphatase assay showed that WIP1 can robustly dephosphorylate both XPA Ser196 and XPC Ser892 phosphopeptides in vitro as compared to the p38 MAPK positive control (Figure 5D). Like other PP2C members, WIP1 activity was Mg2+ dependent and okadaic acid insensitive [16,17]. In addition, incubation of WIP1 with a negative control phosphopeptide, UNG2 Thr31, displayed minimal phosphatase activity. Finally, incubation with the WIP1-D314A phosphatase-dead mutant exhibited no activity against the XPA Ser196 and XPC Ser892 phosphopeptides.
To demonstrate the specificity of WIP1 dephosphorylation, we incubated the XPA Ser196 and XPC Ser892 phosphopeptides with four different purified serine/threonine phosphatases (Figure 5E). Only the WIP1 phosphatase dephosphorylated these phosphopeptides, whereas PP1α, PP2A, and the closely related PP2Cα displayed only minimal phosphatase activity. In summary, these results suggest that XPA and XPC are possible NER targets of WIP1 for further investigation.
4. Discussion
Post-translational modifications such as phosphorylation and dephosphorylation may play a role in regulating DNA repair protein activities since mounting evidence suggests that repair processes are modulated in this manner [12,25,42–45]. For example, PP2A has been shown to activate NER [46], possibly by dephosphorylating XPB at S751 to stimulate 5′ incision activity [47]. Here we propose that phosphatases may also play a role in deactivating DNA repair pathways.
We show that WIP1 decreases the rate of NER kinetics by reducing the repair of UV-damaged DNA as well as CPD. In accord with the previous results, Wip1 null MEFs have enhanced NER activity and CPD repair relative to their wild-type counterparts. In addition, Wip1 heterozygous MEFs also have increased NER activity as compared to the wild-type MEFs, suggesting that depletion of just one allele of Wip1 can confer an enhancement in NER activity. In vivo, Wip1 null mice displayed decreased CPD accumulation and apoptosis relative to Wip1 wild-type mice in response to UVB irradiation. CPDs have been proposed to be the major contributor to UV-induced apoptosis and hyperplasia in the epidermis of murine skin [48,49]. Accordingly, these results suggest that CPD repair is increased in Wip1 null mice, thus reducing UV-induced apoptosis in the skin of these mice.
Two possible NER protein candidates that WIP1 may inactivate include XPA and XPC. WIP1 may dephosphorylate XPA S196 and XPC S892, which are well conserved from zebrafish to humans and have been shown to be substrates of the ATM/ATR kinases in response to DNA damage [12,41]. Following UV damage, ATR interacts with and phosphorylates XPA on S196, which moderately enhances cell survival [11,12,50]. In contrast, the physiological relevance of XPC phosphorylation on S892 in response to damage remains to be elucidated. However, phosphorylation of XPA and XPC may increase the binding affinity of these proteins for UV-damaged DNA and/or their respective binding partners.
Based on the data presented here, we propose a model in which UV damage activates ATR, which phosphorylates and activates XPC, XPA, and other proteins to facilitate NER. In addition, ATR activates p53 (through phosphorylation of p53 ser15). Activated p53 will transcriptionally activate a number of target genes, including WIP1. Once UV-damaged DNA has been repaired, accumulated WIP1 will then dephosphorylate and inactivate XPC and XPA (and perhaps other NER proteins), thus inactivating NER and returning the cell to a normal prestress state. In addition, WIP1 dephosphorylates and downregulates p53 and a number of targets in the ATM/ATR DNA damage response pathway, indicating that WIP1 plays a global role in downregulating the DNA damage response, as well as multiple DNA repair pathways that include base excision repair [25] as well as DNA double strand break repair [26]. In normal cells, WIP1 thus plays a critical homeostatic role in the recovery from DNA damage-induced stress.
Acknowledgments
We are grateful to Tajhal Dayaram, Cathy Gatza, Alan Herron, Yingjun Jiang, Ou Ma, Neha Parikh, Thanh Quach, and Thuan Vo for technical assistance, and to Ivan Uray at the Baylor College of Medicine Integrated Microscopy Core for help with the high-throughput microscopy imaging. We thank Lei Li, Susan Marriott, Jun Qin, Zhou Songyang, and Tse-Hue Tan for helpful discussions. We also thank Hiroshi Yamaguchi and Ettore Appella for providing the WIP1 bacterial expression plasmid. This study was supported by grants to T.-A.N. from the Department of Defense Breast Cancer Research Program’s Predoctoral Traineeship Award (BC050781), S.D.S from the Huffington Foundation, X.L from the National Cancer Institute (CA136549), and to L.A.D. from the National Cancer Institute (CA100420).
Abbreviations
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia mutated and Rad3-related
- CHK1
checkpoint kinase 1
- CHK2
checkpoint kinase 2
- CPD
cyclobutane pyrimidine dimers
- CS
Cockayne’s syndrome
- DAPI
4′-6′-diamidino-2-phenylindole
- DDB2
damaged DNA binding gene 2
- MAPK
mitogen-activated protein kinase
- MDM2
murine double minute 2
- MDMX
murine double minute X
- MEF
mouse embryonic fibroblast
- Mg2+
magnesium
- NER
nucleotide excision repair
- OA
okadaic acid
- PARP
poly (ADP-ribose) polymerase
- PP1
protein phosphatase type 1
- PP2A
protein phosphatase type 2A
- PP2C
protein phosphatase type 2C
- PPM1D
protein phosphatase 1D magnesium-dependent, delta isoform
- Ser
serine
- TTD
Trichothiodystrophy
- TUNEL
terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling
- UNG2
uracil DNA glycosylase 2
- UV
ultraviolet
- WIP1
wild-type p53-induced phosphatase 1
- XP
xeroderma pigmentosum
- XPA
xeroderma pigmentosum complementation group A
- XPC
xeroderma pigmentosum complementation group C
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Thuy-Ai Nguyen, Email: thuyaitn@yahoo.com.
Scott D. Slattery, Email: scott.slattery@bcm.edu.
Sung-Hwan Moon, Email: sm148045@bcm.edu.
Yolanda F. Darlington, Email: darlingt@bcm.edu.
Xiongbin Lu, Email: xlu@biol.sc.edu.
Lawrence A. Donehower, Email: larryd@bcm.edu.
Reference List
- 1.Gillet LCJ, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chemical Reviews. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 3.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 4.Lazzaro F, Giannattasio M, Puddu F, Granata M, Pellicioli A, Plevani P, Muzi-Falconi M. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair (Amst) 2009;8:1055–1067. doi: 10.1016/j.dnarep.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 5.Nouspikel T. DNA repair in mammalian cells: Nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 8.Auclair Y, Rouget R, Affar eB, Drobetsky EA. ATR kinase is required for global genomic nucleotide excision repair exclusively during S phase in human cells. Proc Natl Acad Sci U S A. 2008;105:17896–17901. doi: 10.1073/pnas.0801585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helton ES, Chen X. p53 modulation of the DNA damage response. J Cell Biochem. 2007;100:883–896. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- 10.Smith ML, Seo YR. p53 regulation of DNA excision repair pathways. Mutagenesis. 2002;17:149–156. doi: 10.1093/mutage/17.2.149. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Shell SM, Liu Y, Zou Y. ATR-dependent checkpoint modulates XPA nuclear import in response to UV irradiation. Oncogene. 2007;26:757–764. doi: 10.1038/sj.onc.1209828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu XM, Shell SM, Yang ZG, Zou Y. Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group a by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation. Cancer Research. 2006;66:2997–3005. doi: 10.1158/0008-5472.CAN-05-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc Natl Acad Sci U S A. 2002;99:12985–12990. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci U S A. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang YC, Jan KY, Cheng CA, Liao CB, Liu YC. Direct involvement of the tumor suppressor p53 in nucleotide excision repair. DNA Repair (Amst) 2008;7:751–761. doi: 10.1016/j.dnarep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Fiscella M, Zhang HL, Fan SJ, Sakaguchi K, Shen SF, Mercer WE, VandeWoude GF, Connor PMO, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008;27:123–135. doi: 10.1007/s10555-008-9127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu XB, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes & Development. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, Minami Y, Appella E, Bulavin DV. Wip1 phosphatase modulates ATM-dependent signaling pathways. Molecular Cell. 2006;23:757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Fujimoto H, Onishi N, Kato N, Takekawa M, Xu X, Kosugi A, Kondo T, Imamura M, Oishi I, Yoda A, Minami Y. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death and Differentiation. 2006;13:1170–1180. doi: 10.1038/sj.cdd.4401801. [DOI] [PubMed] [Google Scholar]
- 21.Oliva-Trastoy M, Berthonaud V, Chevalier A, Ducrot C, Marsolier-Kergoat MC, Mann C, Leteurtre F. The Wip1 phosphatase (PPM1D) antagonizes activation of the Chk2 tumour suppressor kinase. Oncogene. 2007;26:1449–1458. doi: 10.1038/sj.onc.1209927. [DOI] [PubMed] [Google Scholar]
- 22.Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, Taya Y, Imai K. p53-inducible Wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. Embo Journal. 2000;19:6517–6526. doi: 10.1093/emboj/19.23.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–354. doi: 10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Lin L, Guo H, Yang J, Jones SN, Jochemsen A, Lu X. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res. 2009;69:7960–7968. doi: 10.1158/0008-5472.CAN-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu XB, Bocangel D, Nannenga B, Yamaguchi H, Appella E, Donehower LA. The p53-induced oncogenic phosphatase PPM1D interacts with uracil DNA glycosylase and suppresses base excision repair. Molecular Cell. 2004;15:621–634. doi: 10.1016/j.molcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Moon SH, Lin L, Zhang X, Nguyen TA, Darlington Y, Waldman AS, Lu X, Donehower LA. Wildtype p53-induced phosphatase 1 dephosphorylates histone variant {gamma}-H2AX and suppresses DNA double strand break repair. J Biol Chem. 2010 doi: 10.1074/jbc.M109.071696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu XB, Nguyen TA, Donehower LA. Reversal of the ATM/ATR-mediated DNA damage response by the oncogenic phosphatase PPM1D. Cell Cycle. 2005;4:1060–1064. [PubMed] [Google Scholar]
- 28.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 29.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, DiTullio RA, Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 30.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 31.Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, Ambrosino C, Sauter G, Nebreda AR, Anderson CW, Kallioniemi A, Fornace AJ, Appella E. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nature Genetics. 2002;31:210–215. doi: 10.1038/ng894. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven G, Nguyen KCQ, Gabriele T, McCurrach ME, Marks JR, Hoey T, Lowe SW, Powers S. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nature Genetics. 2002;31:133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 33.Rauta J, Alarmo EL, Kauraniemi P, Karhu R, Kuukasjarvi T, Kallioniemi A. The serine-threonine protein phosphatase PPM1D is frequently activated through amplification in aggressive primary breast tumours. Breast Cancer Research and Treatment. 2006;95:257–263. doi: 10.1007/s10549-005-9017-7. [DOI] [PubMed] [Google Scholar]
- 34.Saito-Ohara F, Imoto I, Inoue J, Hosoi H, Nakagawara A, Sugimoto T, Inazawa J. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Research. 2003;63:1876–1883. [PubMed] [Google Scholar]
- 35.Hirasawa A, Saito-Ohara F, Inoue J, Aoki D, Susumu N, Yokoyama T, Nozawa S, Inazawa J, Imoto I. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clinical Cancer Research. 2003;9:1995–2004. [PubMed] [Google Scholar]
- 36.Tan DS, Lambros MB, Rayter S, Natrajan R, Vatcheva R, Gao Q, Marchio C, Geyer FC, Savage K, Parry S, Fenwick K, Tamber N, Mackay A, Dexter T, Jameson C, McCluggage WG, Williams A, Graham A, Faratian D, El Bahrawy M, Paige AJ, Gabra H, Gore ME, Zvelebil M, Lord CJ, Kaye SB, Ashworth A, Reis-Filho JS. PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin Cancer Res. 2009;15:2269–2280. doi: 10.1158/1078-0432.CCR-08-2403. [DOI] [PubMed] [Google Scholar]
- 37.Nannenga B, Lu XB, Dumble M, Van Maanen M, Nguyen TA, Sutton R, Kumar TR, Donehower LA. Augmented cancer resistance and DNA damage response phenotypes in PPM1D null mice. Molecular Carcinogenesis. 2006;45:594–604. doi: 10.1002/mc.20195. [DOI] [PubMed] [Google Scholar]
- 38.Choi J, Nannenga B, Demidov ON, Bulavin DV, Cooney A, Brayton C, Zhang YX, Mbawuike IN, Bradley A, Appella E, Donehower LA. Mice deficient for the wild-type p53-induced phosphatase gene (Wip1) exhibit defects in reproductive organs, immune function, and cell cycle control. Molecular and Cellular Biology. 2002;22:1094–1105. doi: 10.1128/MCB.22.4.1094-1105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi H, Minopoli G, Demidov ON, Chatterjee DK, Anderson CW, Durell SR, Appella E. Substrate specificity of the human protein phosphatase 2C delta Wip1. Biochemistry. 2005;44:5285–5294. doi: 10.1021/bi0476634. [DOI] [PubMed] [Google Scholar]
- 40.Johnson JM, Latimer JJ. Analysis of DNA repair using transfection-based host cell reactivation. Methods Mol Biol. 2005;291:321–335. doi: 10.1385/1-59259-840-4:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao ZM, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 42.Christmann M, Tomicic MT, Kaina B. Phosphorylation of mismatch repair proteins MSH2 and MSH6 affecting MutSalpha mismatch-binding activity. Nucleic Acids Res. 2002;30:1959–1966. doi: 10.1093/nar/30.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang HC, Chou WC, Shieh SY, Shen CY. Ataxia telangiectasia mutated and checkpoint kinase 2 regulate BRCA1 to promote the fidelity of DNA end-joining. Cancer Res. 2006;66:1391–1400. doi: 10.1158/0008-5472.CAN-05-3270. [DOI] [PubMed] [Google Scholar]
- 44.Zhao S, Weng YC, Yuan SS, Lin YT, Hsu HC, Lin SC, Gerbino E, Song MH, Zdzienicka MZ, Gatti RA, Shay JW, Ziv Y, Shiloh Y, Lee EY. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- 45.Zhuang J, Zhang J, Willers H, Wang H, Chung JH, van Gent DC, Hallahan DE, Powell SN, Xia F. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer Res. 2006;66:1401–1408. doi: 10.1158/0008-5472.CAN-05-3278. [DOI] [PubMed] [Google Scholar]
- 46.Ariza RR, Keyse SM, Moggs JG, Wood RD. Reversible protein phosphorylation modulates nucleotide excision repair of damaged DNA by human cell extracts. Nucleic Acids Res. 1996;24:433–440. doi: 10.1093/nar/24.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coin F, Auriol J, Tapias A, Clivio P, Vermeulen W, Egly JM. Phosphorylation of XPB helicase regulates TFIIH nucleotide excision repair activity. EMBO J. 2004;23:4835–4846. doi: 10.1038/sj.emboj.7600480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jans J, Schul W, Sert YG, Rijksen Y, Rebel H, Eker AP, Nakajima S, van Steeg H, de Gruijl FR, Yasui A, Hoeijmakers JH, van der Horst GT. Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol. 2005;15:105–115. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 49.Schul W, Jans J, Rijksen YM, Klemann KH, Eker AP, de Wit J, Nikaido O, Nakajima S, Yasui A, Hoeijmakers JH, van der Horst GT. Enhanced repair of cyclobutane pyrimidine dimers and improved UV resistance in photolyase transgenic mice. EMBO J. 2002;21:4719–4729. doi: 10.1093/emboj/cdf456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shell SM, Li Z, Shkriabai N, Kvaratskhelia M, Brosey C, Serrano MA, Chazin WJ, Musich PR, Zou Y. Checkpoint kinase ATR promotes nucleotide excision repair of UV-induced DNA damage via physical interaction with xeroderma pigmentosum group A. J Biol Chem. 2009;284:24213–24222. doi: 10.1074/jbc.M109.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]