Na+,K+-ATPase polarity depends on the interaction between the β subunits of Na+,K+-ATPases located on neighboring cells. In the present work, we use energy transfer methods (FRET), in vivo to demonstrate that these β subunits interact directly at the intercellular space of epithelial cells.
Abstract
The very existence of higher metazoans depends on the vectorial transport of substances across epithelia. A crucial element of this transport is the membrane enzyme Na+,K+-ATPase. Not only is this enzyme distributed in a polarized manner in a restricted domain of the plasma membrane but also it creates the ionic gradients that drive the net movement of glucose, amino acids, and ions across the entire epithelium. In a previous work, we have shown that Na+,K+-ATPase polarity depends on interactions between the β subunits of Na+,K+-ATPases located on neighboring cells and that these interactions anchor the entire enzyme at the borders of the intercellular space. In the present study, we used fluorescence resonance energy transfer and coprecipitation methods to demonstrate that these β subunits have sufficient proximity and affinity to permit a direct interaction, without requiring any additional extracellular molecules to span the distance.
INTRODUCTION
Most epithelia, including cells cultured as monolayers that are used as model systems (Cereijido et al., 1978), express Na+,K+-ATPase in a polarized manner toward the basolateral side of the cell (Cereijido et al., 1981) and more specifically at cell borders facing the intercellular space (Contreras et al., 1995; Cereijido et al., 2001; Shoshani et al., 2005). This position is so resilient that when cells are harvested with trypsin/EDTA, although Na+,K+-ATPase is first randomized and redistributed over the whole plasma membrane, it recovers its polarity in a few hours upon replating at confluence (Cereijido et al., 1980). During this recovery, even pumps that were trapped on the apical (incorrect) surface because of the quick formation of the tight junction (TJ) are removed and relocated to the correct side (Contreras et al., 1989). The generation of cell surface polarity of most membrane proteins involves sorting signals encoded in their amino acid sequence (Matter and Mellman, 1994; Matter, 2000; Rodriguez-Boulan et al., 2005; Weisz and Rodriguez-Boulan, 2009), trafficking routes that include apical or basolateral recycling endosomes (Gonzalez and Rodríguez-Boulan, 2009), and interactions with epithelial-specific protein complexes such as AP-1B and clathrin, which may be regulated by small GTPases (Ellis et al., 2006; Gravotta et al., 2007; Tanos and Rodríguez-Boulan, 2008; Deborde et al., 2008; Mellman et al., 2008; González and Rodríguez-Boulan, 2009).
However, previous efforts to identify an amino acid sequence that functions as a basolateral polarity signal in Na+,K+-ATPase have failed (Muth et al., 1998; Dunbar and Caplan, 2000). Moreover, the basolateral localization of the pump is independent of AP-1B because the pump localizes to the basolateral surface in the μ1B-deficient cell line LLC-PK1 (Duffield et al., 2004) and in Madin-Darby canine kidney (MDCK) cells in which μ1B expression has been suppressed via RNA interference (Gravotta et al., 2007). In epithelial cells, newly synthesized Na+,K+-ATPase is sent directly to the basolateral membrane (Caplan et al., 1986; Contreras et al., 1989; Gottardi and Caplan, 1993a; Mays et al., 1995, Zurzolo and Rodríguez-Boulan, 1993; Gottardi and Caplan 1993b; Mays et al., 1995, Zurzolo and Rodríguez-Boulan, 1993). A recent study that used a SNAP tag system (New England Biolabs, Ipswich, MA) to determine the trafficking itinerary of the newly synthesized Na+ pump revealed that basolateral delivery of the Na+,K+-ATPase does not involve passage through recycling endosomes. Furthermore, because it is an AP-1B–independent cargo, it follows a different pathway en route to the basolateral domain of the epithelial plasma membrane, involving a distinct (and as yet unidentified) post-Golgi transport intermediate of the cellular sorting machinery (Farr et al., 2009). Once at the target domain, the asymmetric distribution of membrane proteins is reinforced by selective retention (Cereijido et al., 2003). Thus, most cell–cell adhesion proteins anchor to components of the cytoskeleton. Nevertheless, it has been shown that retention can also be achieved by cis (in the same plasma membrane) and/or trans (between adjacent cells) homotypic or heterotypic molecular interactions (Yoshida and Takeichi, 1982; Gallin et al., 1983; Nose et al., 1988; Contreras et al., 1995; Nagar et al., 1996; Nusrat et al., 2000, 2005; Arrate et al., 2001; Kostrewa et al., 2001; Niessen and Gumbiner 2002; Momose et al., 2002; Chen et al., 2005; Blasig et al., 2006; Patel et al., 2006). Na+,K+-ATPase has been shown to be retained at the basolateral membrane domain through binding to the ankyrin-fodrin cytoskeleton (Hammerton et al., 1991). Independent of the mechanisms that contribute to Na+,K+-ATPase targeting and based on the identification of the β2 isoform as an adhesion molecule on glia (AMOG; Gloor et al., 1990), we hypothesized that the β subunit is involved in Na+,K+-ATPase polarization (Contreras et al., 1995). Several lines of evidence suggest that the β1 subunit anchors the pump at the lateral borders of epithelial cells by a homotypic β–β interaction at the intercellular space: A given cell only expresses Na+,K+-ATPase at the cell border provided the neighboring cell expresses its own Na+,K+-ATPase at the contacting border (Contreras et al., 1995). MDCK cells (from dog kidney) cocultured with Chinese hamster ovary (CHO) cells only express Na+,K+-ATPase at contacting homotypic MDCK/MDCK borders, but not at heterotypic MDCK/CHO borders. However, if CHO cells are transfected beforehand with dog β subunits (CHO-β), then the dog Na+,K+-ATPase is expressed both at homo- and heterotypic contacts. This result is even more surprising considering that the Na+,K+-ATPases of these CHO-β cells contain the transfected dog β subunit and a hamster α subunit, which is dragged along to its correct position in the plasma membrane (Shoshani et al., 2005). However, it is presently unknown whether β subunits of neighboring cells are sufficiently close to allow for direct β–β interaction across the intercellular space, or if this is instead established through intermediate extracellular molecules. In the present work, we specifically addressed this question using in vitro and in vivo protein–protein interaction assays.
MATERIALS AND METHODS
Cell Culture
MDCK and normal rat kidney (NRK)-E52 cells were grown in DMEM containing 10% fetal calf serum (FCS), and CHO cells were cultured in a mixture of F-12/DMEM. Cells were harvested with trypsin-EDTA and plated on dishes with or without glass coverslips, or on glass-bottom dishes for live imaging (MatTek, Ashland, MA). Cells were transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol. After 5 d, cells were sorted by fluorescence-activated cell sorting by using a MoFlo Cell Sorter (Beckman Coulter, Fullerton, CA) for yellow and cyan fluorescence and were then selected with 0.8 mg/ml G418 to generate stable clones. Cell mixtures for immunofluorescence (IF) analysis were made as described previously (Contreras et al., 1995). In brief, NRK cells were prelabeled with 5-(and-6)-{[(4-chloromethyl)benzoyl]amino}-tetramethylrhodamine (Cell Tracker Orange CMTMR; Invitrogen) at a final concentration of 6.3 μM for 1 h at 36.5°C. The cells were then washed three times with phosphate-buffered saline (PBS) without calcium trypsinized and resuspended in CDMEM. Finally, the labeled cells were mixed with an equal number of MDCK cell, cocultured on glass coverslips for 48 h, and then processed for immunofluorescence assays.
Constructs and Recombinant Proteins
The full-length cDNA of the canine Na+,K+-ATPase β1 subunit was N-terminally tagged with a His6 epitope by polymerase chain reaction (PCR) amplification using forward primer 5′-GCGGCCGCAAAATGGCCCATCATCATCATCATCATCGCGGAAAAGCCAAGGAG-3′ and reverse primer 5′-CGCCGGCGTCAGCTCTTAACTTCAATTTTTACATC-3′. PCR products were cloned into a pCR 2.1 TOPO vector (Invitrogen) and sequenced with a capillary-based electrophoresis sequencer ABI Prism 310 (Genetic Analyzers, Applied Biosystems, Foster City, CA). Positive clones were digested with NotI, and the fragment was inserted into a similarly digested pIRESneo vector (Clontech, Palo Alto, CA) to generate the expression vector pIRESneo-βHis6. The chimera DPPβ (dog subunit) construct in a pCB6 vector was a kind gift of Dr. D. M. Fambrough and was generated as reported previously (Hamrick et al., 1993) for the chicken β1 subunit. To generate the fusion proteins (β1-cyan fluorescent protein [CFP] and β1-yellow fluorescent protein [YFP]), the full cDNA of the rat kidney Na+, K+-ATPase β1 subunit was amplified with forward primer 5′-TCGACTCGAGGAATGGCCCGCGGAAAAGCC-3′ and reverse primer 5′-GAATTCGGCTCTTAACTTCAATTTTTACATC-3′. The PCR products were cloned into a pBluescript II KS+/− vector (Stratagene, La Jolla, CA). The cDNA of the rat β1 subunit was digested with XhoI and KpnI and inserted into mammalian fluorescent expression vectors pAmCyan-N1 and pEYFP-N1 (Clontech) that had been previously digested with the same restriction enzymes. Several clones were subjected to plasmid purification using a GeneJET Plasmid Miniprep Kit (Fermentas, Burlington, ON, Canada), and the fusion protein constructs were confirmed by restriction digests and sequencing.
Immunofluorescence
The β1 subunit and its recombinant variants were assayed by immunofluorescence as described previously (Shoshani et al., 2005). After blocking, cells were incubated with primary antibodies against the β1 subunit, washed six times quickly with PBS-Tween (0.05%), and then incubated with the secondary antibodies (1:100) for 1 h at room temperature. The following secondary antibodies were used: Alexa 495-conjugated goat anti-mouse immunoglobulin G (Invitrogen) and cyanine 5-conjugated goat anti-mouse. Specimens were mounted with VECTASHIELD medium (Vector Laboratories, Burlingame, CA) and observed with a TCS SP2 confocal microscope (Leica, Hiena, Germany) or a FluoView FV1000 confocal microscope (Olympus, Center Valley, PA).
Immunoblotting
Western blot analysis of whole cell protein extract was performed as described previously (Shoshani et al., 2005). In brief, MDCK, NRK-E52, and CHO cells were washed with PBS and solubilized with radioimmunoprecipitation assay (RIPA) buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 7.4, 150 mM NaCl, 2 mM ethylenediamine-tetraacetic acid (EDTA), 1% Triton X-100, 0.5% sodium deoxicholate, and 10% glycerol] containing protease inhibitors (Complete Mini; Roche Diagnostics, Indianapolis, IN). The protein content of the cell lysate was measured (BCA protein assay reagent; Pierce Chemical, Rockford, IL) and prepared for SDS-polyacrylamide gel electrophoresis (PAGE) by boiling in sample buffer. The resolved proteins were electrotransferred to a polyvinylidene difluoride membrane (Hybond-P; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The proteins of interest were then detected with the specific polyclonal or monoclonal antibodies indicated in each case, followed by species-appropriate peroxidase-conjugated antibodies (Zymed Laboratories, South San Francisco, CA) and chemiluminescent detection (ECL PLUS; GE Healthcare).
Pull-Down Assay
CHO cells stably expressing βHis6 were cultured for 48 h and then lysed with RIPA buffer containing protease inhibitors and the protein content was determined. βHis6as bait was immobilized on nickel-nitrilotriacetic acid beads (His Trap FF column; GE Healthcare) previously equilibrated with 10 ml of RIPA containing protease inhibitors. In general, 4 mg of total protein extract was loaded and allowed to interact for at least 1 h at 4°C with gentle shaking. After 10 washes with 10 ml of 20 mM imidazole, the supernatant of SDβ-expressing CHO cells was loaded as prey, and interaction was allowed to occur overnight at 4°C. After washing with 10 and 20 mM imidazole, bound proteins were eluted with 500 mM imidazole. Eluates were loaded on a 10% SDS-PAGE gel and analyzed by immunoblotting using an anti-dog β1 antibody and a SuperSignal West HisProbe Kit (Thermo Fisher Scientific, Rockford, IL). For this assays, CHO SDβ cells were cultured in serum-free media for 4 d. The conditioned medium was then collected and further purified by fast-performance liquid chromatography chromatography.
Immunoprecipitation
MDCK cells stably expressing rat β1-YFP were cocultured with NRK-E52 cells. After 48–72 h, cells from the mixed monolayer were lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxicholate, and Complete Protease Inhibitor Cocktail 1× [Roche Diagnostics]). Immunoprecipitation was performed with a rabbit polyclonal antibody against the β1 subunit (Santa Cruz Biotechnology, Santa Cruz, CA) or with a chicken antibody against green fluorescent protein (GFP) (Aves Labs, Tigard, OR), followed by overnight incubation with AffiGel protein A agarose (Bio-Rad Laboratories, Hercules, CA). Immunoprecipitated proteins were eluted with freshly prepared buffer (10% glycerol, 50 mM Tris-HCl, pH 6.8, and 1 M NaCl), followed by removal of the agarose beads by centrifugation. The supernatant was then loaded on 8% SDS-PAGE gels. Western blot analysis was done using the following primary antibodies: mouse anti pan-species β1 antibody (1:1000 dilution; Santa Cruz); a mouse monoclonal antibody (mAb) against dog β1 subunit donated by Dr. M. Caplan (1:200 dilution; Yale University, New Haven, CT); and a mouse monoclonal anti-rat β1 subunit antibody (IEC 1/48 1:1000 dilution) donated by Dr. A. Quaroni (Cornell University, Ithaca, NY). Anti-mouse peroxidase-conjugated secondary antibodies (1:5000 dilution; Zymed Laboratories) were then applied.
Fluorescence Resonance Energy Transfer (FRET) Assay by Acceptor Photobleaching
For analyzing FRET efficiency, live MDCK cells stably expressing β1-CFP or β1-YFP were cocultured on glass-bottomed dishes (MatTek) for 48–72 h. The plates were mounted in a metal chamber at 37°C and 5% CO2 and analyzed with an FV-1000 confocal microscope (Olympus). The excitation ranges for the fluorophores were 390–425 nm for β1-CFP and 445–500 nm β1-YFP and the emission intensities were recorded at 475–525 nm for β1-CFP and 540–575 nm for β1-YFP. Photobleaching of enhanced YFP was performed on selected cell membrane areas. Assessment of the expected change in donor intensity corresponding to the nonbleached region of the acceptor provided a measure of background FRET or signal noise. Analysis of FRET data were based on the percentage increase of postbleach donor intensity compared with prebleach donor intensity. FRET efficiency (E) was calculated from the ratio of CFP fluorescence evaluated before (DPRE) and after (DPOST) photobleaching, using FV10-ASW version 1.7 (Olympus) and the following formula:
Cell cultures expressing only β1-CFP or β1-YFP were used for correction and determination of the laser intensity, as well as to determine the gain and offset settings. Emission intensities in several membrane regions were recorded at 475–525 nm for β1-CFP and at 540–575 nm for β1-YFP.
Statistical Analyses
Statistical analyses were performed with Prism 4 software (GraphPad Software, San Diego, CA). Results are expressed as means ± SE. The groups were compared by one-way analysis of variance (ANOVA) using the Bonferroni multiple-comparison test. Differences were considered significant if p < 0.05.
RESULTS
The β–β Interaction Is Species Specific
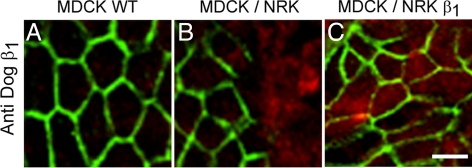
Interactions between adjacent cells from different animal species are not unique or universal but depend on the type of cell–cell junction involved. TJs are promiscuous, because those established between cells belonging to different animal species have a degree of sealing that can be predicted from the transepithelial electrical resistance of each cell type and their proportions in the mixed monolayer (González-Mariscal et al., 1989). In contrast, adherens junctions are homotypic and can be seldom, if ever, established between epithelial cells from different animal species (Contreras et al., 2002; Halbleib and Nelson, 2006). We have found previously that the homotypic requirement for cell–cell junctions extends to the distribution of Na+,K+-ATPase (Contreras et al., 1995). In keeping with our previous observations (Shoshani et al., 2005), Figure 1A shows that MDCK cells present a typical “chicken fence” pattern of Na+,K+-ATPase distribution in monolayers of pure MDCK cells; however, when the MDCK cells are cocultured with NRK-E52 epithelial cells from NRK (Figure 1B), Na+,K+-ATPase is only detected in homotypic interactions. In contrast, when the NRK cells are transfected with dog β1 (NRKβ), MDCK cells express Na+,K+-ATPase in all cell–cell borders (Figure 1C).
Figure 1.
MDCK cells express β1 subunits at homotypic but not at heterotypic contacts. Localization of the dog β1 subunit (A–C, green) of Na+,K+-ATPase was studied by immunofluorescence assay in MDCK cells derived from dog kidney. (A) Monolayer of pure MDCK cells showing that the β1 subunit is only expressed at the plasma membranes in the lateral domain where cells contact each other. (B) MDCK cells cocultured with NRK cells (derived from normal rat kidney) that were labeled beforehand with CMTMR (red). In the mixed monolayer, the β1 subunit is only expressed at homotypic borders (MDCK/MDCK) but not at heterotypic ones (MDCK/NRK). (C) Monolayer of mixed MDCK/NRK cells transfected with dog β1 subunit shows that the β subunit (green) is concentrated at both homotypic MDCK/MDCK contacts and heterotypic MDCK/NRK contacts. Bars, 20 μm.
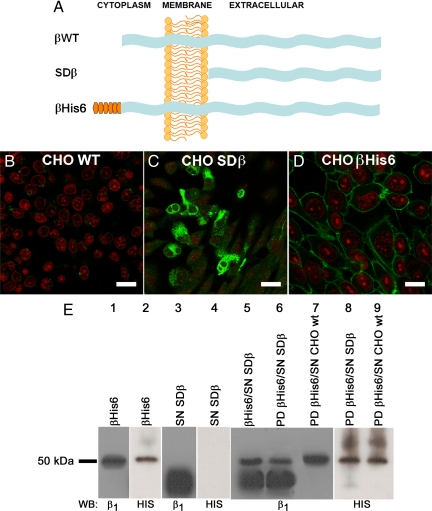
Dog β1 Subunits Interact In Vitro
The β–β interaction in vitro was studied with pull-down assay using the wild-type dog β1 subunit (βWT), its extracellular domain (SDβ) and the complete dog β1 tagged with a hexahistidine repeat epitope tag (βHis6) (Figure 2A). Wild-type CHO cells express a Na+,K+-ATPase that is barely recognized by a specific anti-dog β1 subunit antibody (Figure 2B). These cells were transfected with the cDNA for dog SDβ and βHis6 to generate stable clones. Figure 2, C and D, shows CHO cells expressing cytoplasmic SDβ in transit to be secreted and βHis6 in the plasma membrane (green), respectively. Western blots of lysates of βHis6-expressing CHO cells present a band of ∼50 kDa corresponding to the fully glycosylated form of the dog β1 subunit detected with an antibody against this species (Figure 2E, lane 1). As shown in Figure 2E (lane 2), this band corresponds to βHis6, because it is also detected using a His6 detection kit (His probe). We then immobilized purified βHis6 on a nickel matrix column, and after extensive washing, loaded it with SDβ supernatant (SN SDβ). Figure 2E (lane 3) shows a broad band of soluble β1 ectodomain that, as expected, is lighter because it lacks the cytoplasmic and transmembrane domains. Notably, a His probe does not recognize any protein when applied to SN SDβ (lane 4). After an incubation period, bound proteins were eluted with imidazole buffer, separated on SDS-PAGE, and analyzed by Western blotting using an antibody against dog β1 subunit or His probe. The pattern of bands observed in Figure 2E (lane 5) corresponds to the in vitro mixture of the soluble extracellular domain of the dog β1 subunit and βHis6 and is similar to the pattern observed in lane 6, which corresponds to the eluted (pulled down) proteins, indicating that the soluble extracellular domain of dog β1 subunit and βHis6 were interacting. For comparison, lane 7 shows a negative pull-down assay in which wild-type CHO cell supernatant was used. Lanes 8 and 9 are analogous to lanes 6 and 7, except that this time the membrane was blotted with His probe. As expected, bands corresponding to the β1 subunit are observed only where βHis6 is present. Together, these results suggest that β–β interaction can be reconstituted in vitro. This interaction probably involves the extracellular domains, although this experiment does not show that the interaction is direct.
Figure 2.
β Subunits interact in vitro. (A) Scheme of the wild-type and recombinant β subunits used in this study. Shown are the wild-type β subunit (βWT, top), the soluble β subunit consisting only of the extracellular domain (SDβ, middle), and the β subunit tagged with a hexahistidine repeat (βHis6, bottom). (B–D) Immunofluorescence images of the dog β1 subunit expressed in CHO cells. CHO WT cells (B), CHO cells expressing SDβ (C), and CHO cells expressing βHis6 in their plasma membranes (D) are depicted. Nuclei were stained with propidium iodide (red). Bars, 40, 30, and 20 μm, respectively. (E) Pull-down assay analysis. Western blots using an antibody against the dog β1 subunit and a His probe reveal, in both cases, the recombinant βHis6 protein (lane 1 and 2), whereas only the dog β1 antibody recognizes the soluble extracellular domain of dog β1 (SDβ, lanes 3 and 4). A mixture of purified SDβ with βHis6 produces the mixed pattern shown in lane 5. The proteins eluted from a pull-down (PD) assay by using βHis6 as bait and soluble SDβ as prey (lane 6) show the same pattern of bands. Only the band corresponding to the bait protein (βHis6) is observed when the pull-down is performed with CHO WT cell supernatant (lane 7). This is confirmed when both pull-downs are blotted to detect the His6 repeat (lanes 8 and 9).
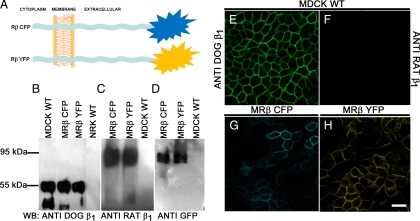
Molecular Tools to Study β–β Interactions In Vivo
As mentioned in the Introduction, the aim of this work was to study the interaction of the β subunit of a given Na+,K+-ATPase with that of another Na+,K+-ATPase in the plasma membrane of a cell across the intercellular space. Our first approach, using an in vitro protein–protein interaction assay (Figure 2), suggested that such an interaction is possible, although it did not confirm a direct interaction. To test this possibility in vivo, we constructed recombinant rat β1 subunits fused to the fluorescent proteins CFP or YFP, which we will refer to as Rβ CFP and Rβ YFP, respectively (Figure 3A). These constructs were expressed in MDCK cells. Using a specific antibody against the dog β1 subunit in a Western blot assay, the only band detected is, of course, the band representing the endogenous β1 subunit of MDCK cells (Figure 3B, lanes 1–3). Whereas this antibody detects no band at all in NRK cells (Figure 3B, lane 4), a specific antibody against the rat β1 subunit shows a single band of 95 kDa in cell extracts of MDCK cells transfected with either Rβ CFP or Rβ YFP (MR βYFP and MR βCFP, Figure 3C, lanes 1 and 2). No band is observed in wild-type MDCK cells (Figure 3C, lane 3). An antibody against GFP that recognizes all GFP-derived fluorescent proteins confirms that the 95-kDa band corresponds to the rat β1YFP (Figure 3D, lane 1) and rat β1CFP (Figure 3D, lane 2). As expected, this high-molecular-weight band is not detected in nontransfected MDCK cells (Figure 3D, lane 3). To make sure that the fluorescent β1 subunit is expressed in a membrane position that would allow us to study β-β interactions in cells, we used immunofluorescence and confocal microscopy to visualize the localization of Rβ CFP and Rβ YFP. Figure 3E shows the typical chicken fence pattern of the β1 subunit as seen in nontransfected MDCK cells using an antibody against the dog β1 subunit. This pattern is not observed if the anti-rat β1 subunit antibody is used (Figure 3F). As shown in Figure 3, G and H, both fluorescent proteins are expressed at the lateral plasma membrane of transfected MDCK cells. This fluorescent signal colocalizes with the rat β1, as seen by anti rat β1 antibody staining (Supplemental Figure S1, A and B) and staining of the endogenous dog β1 subunit (Supplemental Figure S1, C and D). Moreover, nontransfected MDCK cells are excited in the same conditions as the YFP-transfected ones (Supplemental Figure S2, A and C), and when observed with confocal microscopy, they do not exhibit a fluorescent signal. Nonetheless, cadherin expression, as assayed by an anti-pan-cadherin antibody, is well conserved in MDCK cells expressing Rβ YFP (Supplemental Figure S2, B and D). Therefore, the molecular tools we have developed to study the interaction between β subunits (Rβ CFP and Rβ YFP) are localized to the plasma membrane and do not perturb the distribution of other membrane molecules.
Figure 3.
Cellular expression of molecular tools. (A) Scheme of the β subunits fused to donor or acceptor fluorophores. Shown are the rat β1 subunit fused to CFP in the extracellular domain (Rβ CFP, top) rat β1 subunit fused to YFP in the extracellular domain (Rβ YFP). (B–D) Western blot assays to detect the expression of both constructs in transfected MDCK cells (MRβ CFP or MRβ YFP). (B) Detection of β1 subunits with a specific antibody against the dog subunit. No band is detected in rat NRK-E52 cells. (C) Detection of the rat β1 subunit with a specific antibody for this species. No band corresponding to the rat β1 subunit is observed in WT MDCK cells. (D) Immunodetection using an anti-GFP antibody. A 95-kDa band corresponding to the fluorescent constructs (MRβ CFP or MRβ YFP) is visible. This band coincides with the band recognized by the rat-specific antibody and does not appear in MDCK WT cells. (E–H) Confocal images of WT MDCK cells and MDCK cells transfected with Rβ CFP or YFP. (E) IF of the endogenous dog β1 and rat β1 (F) subunits in WT MDCK cells. (G) Fluorescence imaging of Rβ CFP obtained by exciting at 390–425 nm and recording its recovery at 475–525 nm. (H) Fluorescence imaging of the Rβ YFP obtained by exciting at 475–515 nm and recording its emission at 540–575 nm. Bar, 20 μm.
β–β Interactions Are Strong and Specific
To analyze whether the β1 subunits of Na+,K+-ATPases of neighboring cells interact with each other (transinteraction), we applied an immunoprecipitation (IP) assay. For this strategy, MDCK cells transfected with Rβ YFP and wild-type NRK cells were cocultured. Cell lysates of nontransfected MDCK cells and of wild-type NRK cells were separated on SDS-PAGE gels and immunoblotted with a mAb that recognizes the β1 subunit of various animal species. A clear band corresponding to the endogenous β1 subunit was detected in MDCK cell extract (Figure 4A, lane 1) and in NRK cell extract (Figure 4A, lane 2). To analyze the possible β–β interaction between the rat β1 subunits of neighboring cells, immunoblotting of the cell lysate obtained from cocultures (Figure 4A, lane 3) and immunoprecipitation with an anti-β1 subunit antibody (Figure 4A, lane 4) were performed. Two bands were observed: a band of high molecular weight (95 kDa), corresponding to the exogenous rat β1 subunit (Rβ YFP) and a lower weight band (55 kDa), corresponding to the endogenous dog and rat β1 subunits (which cannot be distinguished by molecular weight). Conversely, immunoprecipitation of the transfected Rβ YFP by using an antibody against GFP produced a 55-kDa coimmunoprecipitated protein (Figure 4A, lane 5), indicating that these two proteins strongly interact. Nevertheless, the possibility exists that the immunoprecipitated recombinant rat β1 subunit could coprecipitate the dog β1 subunit of a neighboring Na+,K+-ATPase present in the same cell membrane (cis interaction). To rule out this possibility, we analyzed the anti-GFP immunoprecipitates by immunoblotting, using specific antibodies against either the dog β1 subunit or the rat β1 subunit. As shown in Figure 4B, the dog-specific antibody detected neither the low-molecular-weight (WT) nor the high-molecular-weight (recombinant) rat β1 subunits (lane 1) when cocultures were IPed with an anti-GFP antibody, whereas the rat-specific antibody recognized both proteins (lane 2). On the other hand, the antibody against the dog subunit barely recognized the endogenous dog β1 in immunoprecipitates obtained with an antibody against β1 from several species (lane 3), whereas the anti rat-β1 antibody recognized both the endogenous and YFP-tagged proteins (lane 4). Immunoprecipitates of pure MRβ YFP cells immunoblotted with antibodies against both the rat β1 and dog β1 subunits show a band corresponding to the dog β1 subunit (lane 5) and a band corresponding to the rat recombinant protein (lane 6), suggesting that β–β interaction between dog subunits may occur naturally in MDCK cells. As negative controls for immunoprecipitation, we used cocultures of WT MDCK and NRK and blotted them with an anti-β1 antibody (Figure 4C). As expected, dog and rat β1 are detected in the immunoprecipitates for the β1 subunit (Figure 4C, lanes 1 and 2) but are not detected in GFP immunoprecipitates (lane 3). The overall conclusion from this group of experiments is that rat β1 subunits of a given cell interact with the homotypic β1 subunits of neighboring cells.
Figure 4.
Interaction between β subunits of neighboring cells. (A) Immunodetection of β1 subunits with the pan-species β1 subunit antibody. Characteristic bands of MDCK WT (lane 1), NRK WT (lane 2), and cocultures of NRK WT and MRβ YFP cells (lane 3) blotted with the pan-species β1 subunit antibody, dog (Dβ1) and rat subunits (Rβ1). The low-molecular-weight endogenous β1 and the high-molecular-weight β1-YFP are both recognized by the same antibody (lane 4). Immunoprecipitated cocultures of NRK WT and MRβ YFP cells by using an antibody against GFP reveal a band corresponding to the recombinant Rβ YFP (95-kDa band; lane 5) and a coimmunoprecipitated lighter band corresponding to the native β1 subunit (55-kDa band in lane 5). (B) Immunoprecipitation with an anti-GFP antibody shows no bands when blotted with an antibody against Dβ1 (lane 1). If the blot is instead performed with the pan-species β1 subunit antibody, both the endogenous and recombinant proteins are revealed (lane 2). The same pattern is seen if the immunoprecipitations are performed with the pan-species β1 antibody (lanes 3 and 4). Immunoprecipitation with the β1 antibody of MDCK cells expressing recombinant rat β1 (MRβ YFP) shows the endogenous β1 (lane 5) when blotted with the Dβ1 antibody and the transfected one when detected with the Rβ1 antibody (lane 6). (C) As negative controls, we immunoprecipitated cocultures of MDCK WT with NRK cells and probed them with Dβ1, Rβ1, and GFP antibodies. As expected, the endogenous dog (lane 1) and rat (lane 2) subunits were detected, whereas YFP-β1 was not (lane 3).
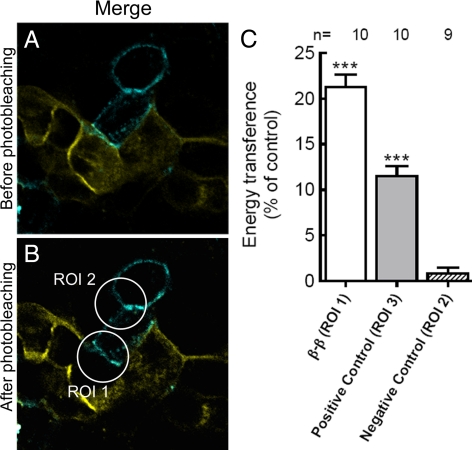
Rat β1 Subunits Located in Neighboring Cells Interact In Vivo
To further demonstrate that rat β1 subunits of neighboring cells interact in situ, two stable clones of MDCK cells, one expressing Rβ CFP and the other Rβ YFP, were generated (Figure 3). Cocultures of MDCK cells expressing CFP or YFP fusion proteins were used to perform FRET by acceptor photobleaching assays. FRET quenches donor fluorescence; therefore, the rebound of donor fluorescence after photodestruction of acceptors provides a straightforward means to measure the apparent FRET efficiency, Eapp. The raw data consist of two images: donor fluorescence taken before (DPRE) and after (DPOST) acceptor photobleaching. A representative confocal image of an experiment in which photobleaching of YFP was performed on a selected domain of cell membrane is shown in Figure 5. Merged images of CFP and YFP fluorescence are shown in Figure 5, A and B. A strong laser line of 515 nm fades YFP but not CFP (Figure 5B). Photobleaching of YFP increased the fluorescence intensity of CFP in the region of interest (ROI) 1 (Figure 5B) by 26.6% relative to its intensity before photobleaching and the calculated energy transference efficiency (%E) of 10 experiments was 21.3%. This %E suggests that the β1 subunits tagged with CFP and YFP must be located at <10 nm from each other (Kenworthy, 2005). As an internal control, fluorescence intensity was measured in ROI 2 (Figure 5B), where CFP does not have YFP in its proximity; therefore, its intensity does not change (Supplemental Table 1). As a positive control, a fused CFP-YFP molecule was expressed in the cytoplasm of MDCK cells. In this construct, the two proteins are located close enough to transfer resonance energy (Supplemental Figure S3, A–C). Photobleaching of YFP increased the fluorescence intensity of CFP (ROI 3 in Supplemental Figure S3C) by 13.4% relative to its intensity before photobleaching, representing a %E equal to 11.5% (Supplemental Table 1). Figure 5C summarizes these results. The first bar represents the energy transfer between two β1 subunits placed in the plasma membrane of neighboring (living) MDCK cells. The shaded bar corresponds to the positive control (fused CFP-YFP). The third bar shows the endogenous negative control (ROI 2). The %E values of bar 1 and 2 are significantly higher (p < 0.001) than that of bar 3. To validate the system, a region was chosen in which CFP-expressing cell was separated from YFP-expressing cell by an MDCK cell that did not express either protein (ROI 4 in Supplemental Figure S3F). Photobleaching of this area showed no energy transfer effect (Supplemental Table 1). As an additional negative control, MDCK cells expressing the CFP plasmid alone (without the β1 subunit) in contact with cells expressing Rβ YFP were analyzed. When region ROI 5 was photobleached (Supplemental Figure S3I), there was no energy transfer effect, as expected (Supplemental Table 1). Together, these results suggest that the β1 subunits of Na+,K+-ATPases in neighboring MDCK cells are close enough to directly interact. This was a basic assumption of the model proposed by Shoshani et al. (2005) to account for the maintenance of the polarized distribution of the Na+,K+-pump in epithelial cells.
Figure 5.
Interaction between β1 subunits observed by FRET after the in vivo acceptor photobleaching assay. (A) Fluorescence image captured before acceptor photobleaching in cocultures of MRβ CFP (cyan) and MRβ YFP (yellow) living cells. The optical merge shows the colocalization of both fluorescent proteins in cell-cell contact areas (green). (B) Fluorescence of the same optical section after photobleaching of the YFP in a membrane section that possessed both CFP and YFP expression. ROI 1 was the region used for quantification of %E. Bleaching of YFP in this region increased the fluorescence of CFP. ROI 2 was the region used as an internal negative control, in which the change of CFP fluorescence was also measured. Bar, 20 μm. (C) Quantification of %E. The percentage of energy transference obtained in 10 experiments was averaged. Bars represent SEM and asterisks indicate p < 0.001 with respect to negative control. ROI 3 is from MDCK cells expressing a CFP-YFP tandem construct (positive control) and ROI 1 and 2 are the same as in B. Data are summarized in Supplemental Table 1.
DISCUSSION
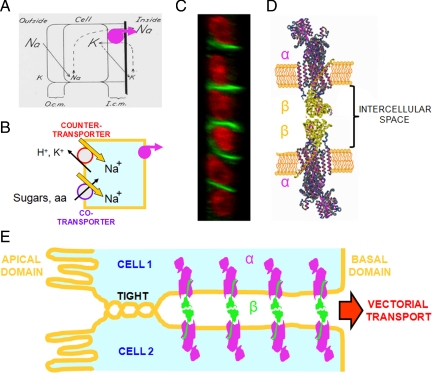
Half a century ago, Koefoed-Johnsen and Ussing (1958) put forward a plausible explanation for the transepithelial transport of Na+ (Figure 6A) that was subsequently used as a model for the transport of substances across all epithelia (Figure 6B). Ironically, although a central element of the KJ-U model is the asymmetric distribution of Na+,K+-ATPase and this asymmetry was later supported by the fact that ouabain inhibits Na+ transport when added to the inner bathing solution but not to the outer one, the intrinsic mechanism for producing this polarity remained unknown (Cereijido et al., 2001). Even today, although it is known that the polarized expression of proteins in epithelial cell membranes is influenced by sorting and retention signals in the protein sequence and by the series of events preceding their insertion in the membrane, the polarized expression of Na+,K+-ATPase has not been directly attributed to these factors (Matter, 2000; Mellman and Nelson, 2008; Farr et al., 2009). Because the transport and ATP-hydrolysis capabilities of Na+,K+-ATPase are primarily due to its α subunit, it was assumed that this subunit would also be responsible for the asymmetric distribution of the enzyme. However, we have shown previously that the polarity is also due to interactions between β subunits of Na+,K+-ATPases in neighboring cells. By binding to each other across the intercellular space, these subunits anchor the entire enzyme at the cell's lateral membrane (Shoshani et al., 2005; Figure 6, C–E). Subsequently, the question arose as to whether the distance between these enzymes is close enough to permit β–β binding without the participation of an intermediate molecule. The present work addressed this question, with the following results: 1) two β1 subunits can interact directly in vitro; 2) this β–β interaction is also established in vivo between β1 subunits located in the plasma membranes of neighboring cells; 3) the β–β interaction is species-specific because it is observed between rat/rat and dog/dog β1 subunits, but not between rat/dog subunits; and 4) in cultured monolayers of MDCK cells, the extracellular domains of β1 subunits on neighboring cells are close enough to allow energy transference in FRET assays. Together, our results indicate that β–β interactions have all the necessary properties to anchor the Na+,K+-ATPase at the lateral membrane of MDCK cells. However, this does not exclude the possibility that other molecular species might participate.
Figure 6.
Polarized expression of epithelial Na+,K+-ATPase. (A) Seminal model of Koefoed-Johnsen and Ussing (1958), in which the Na+,K+-ATPase (reinforced in magenta) was assumed to occupy the basal side of the cell, which constitutes the inner-facing barrier. (B) From this position, the pump transports Na+ toward the interstitial side of the cell, producing a net decrease in the cytoplasmic concentration of this ion and setting up an electrochemical force that drives counter- and cotransporters of sugars, amino acids, and ions and possible the existence of metazoans. (C) Confocal transverse section of a monolayer of MDCK cells. The nuclei are stained with propidium iodide (red) and the β subunits of Na+,K+-ATPase are stained with a specific antibody (green), showing that this subunit is localized to the lateral surfaces of cells, but not to the apical (left) or basal sides. (D) Na+,K+-ATPase molecules of two neighboring epithelial cells with interacting β subunits (green), as shown previously (Shoshani et al., 2005) and in the present work. (E) Na+,K+-ATPase molecules anchored to the lateral membranes. Because of the tight junction, Na+ ions pumped into the intercellular space can only diffuse inwards, generating vectorial transport across the epithelium.
Until recently, the three-dimensional (3D) structure of the Na+,K+-ATPase β subunit and the length of its extracellular moiety was unknown. Nevertheless, there was evidence suggesting that isoforms of the Na+,K+-ATPase β subunit had properties of adhesion proteins. For example, the β2 isoform was originally identified in the rat brain as an AMOG (Gloor et al., 1990). It was also shown that the β1 subunit has an intrinsic glycan-binding capacity and might be involved in neural cell interactions (Kitamura et al., 2005). Fortunately, the atomic structure of Na+,K+-ATPase, including its three subunits (α, β, and γ), was recently resolved to 3.5 and then 2.4 Å (Morth et al., 2007; Shinoda et al., 2009). Therefore, we now have a more precise idea about the 3D organization of the extracellular domain of the β subunit and its adhesive properties. As suggested previously by biochemical approaches, part of the extracellular domain of β subunit interacts with the α subunit (Noguchi et al., 1987) and is essential for its catalytic function (Geering et al., 1996). Accordingly, Bab-Dinitz et al. (2009) have shown that the C-terminal lobe has an independent structure that is analogous to those of other cell–cell adhesion molecules. Much of the surface of the ectodomain, including the majority of the immunoglobulin-like structure, is not in direct contact with the α subunit and thus could interact with other proteins.
It is possible that the ligation of β subunits on neighboring cells activates cytoplasmic interactions and signaling pathways that modulate cell adhesion and cytoskeleton organization. This possibility is in keeping with experiments showing that Na+,K+-ATPase participates in signaling pathways that modulate different cell functions (Efendiev et al., 2006; Liang et al., 2006; Tian et al., 2006) and demonstrating its involvement in cell–cell adhesion (Gloor et al., 1990; Belusa et al., 1997; Contreras et al., 1999, 2004, 2006; Vagin et al., 2006, 2007; Bab-Dinitz et al., 2009).
Three isoforms of the β-subunit (β1, β2, and β3) have been identified. The Na+,K+-ATPase β2 subunit has multiple N-glycosylation sites (up to nine sites), whereas the β1 or β3 isoforms have only two or three N-glycosylation sites. The high degree of glycosylation of the β2 subunit might imply a role in the apical sorting of the corresponding α/β complexes. Moreover, it has been demonstrated that the polarized distribution of Na+,K+-ATPase is correlated with the β subunit isoform expressed by a given cell. Hence, β1 is directed to the lateral membrane in epithelial MDCK cells (Shoshani et al., 2005; Vagin et al., 2005) and β2 is delivered to the apical membrane in ADPKD (Wilson et al., 2000) and HGT-1 cells (Vagin et al., 2005). In agreement with the idea that the multiple N-glycosylation sites in the β2 isoform function as an apical localization determinant, Vagin et al. (2005) have shown that the introduction of extra N-glycosylation sites into the β1 isoform results in apical delivery of the mutated β1 in HGT-1 cells. Accordingly, Wilson et al. (2000) transfected MDCK cells with the β2 isoform and demonstrated that the α1/β2 complex is delivered to the apical domain. In contrast, Liang et al. (2006) have shown that deglycosylation treatments in well-polarized hepatic cells by deglycosylation drugs, or by site-directed mutagenesis of the N-linked-glycosylation residues, cause the β1 subunit to traffic from the native basolateral domain to the apical/canalicular domain.
The polarized delivery of Na+,K+-ATPases to the membrane has been shown to be strictly dependent on the assembly of αβ dimers in the endoplasmic reticulum (Geering et al., 1996, Tokhtaeva et al., 2009). Nevertheless, it is not clear whether these dimers can disassemble and their subunits persist as functional independent proteins once they are inserted in the plasma membrane. In this respect, Moreno et al. (2002) have shown that the removal of a postsynaptic density 95/disc-large/zona occludens group from the cytoplasmic C-terminal end of the Shaker K+-channel does not prevent polarization but dramatically reduces the protein's residence time in the cell membrane. Likewise, binding between β subunits might anchor the two Na+,K+-ATPases to which they belong to the cell membrane.
Sodium transport across epithelia is important, not only because of the physiological role of this ion, but also because the Na+ pumped out of the cytoplasm by Na+,K+-ATPase creates an asymmetry between cytoplasmic Na+ and extracellular Na+. This asymmetry establishes an electrochemical gradient across the plasma membrane that provides the driving force and increases the substrate affinity for a wide variety of Na+/glucose, Na+/amino acid, Na+/K+, and Na+/Ca2+ cotransporters (usually, carriers only acquire an affinity for the cotransported substance once they combine with Na+) (see Cereijido and Rotunno, 1971). The net movement of substances such as glucose, amino acids and ions across epithelia is, in turn, a key requirement for the existence of metazoan life. Not surprisingly, Na+ transport is closely modulated in response to a multitude of physiological, pharmacological, and pathological conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Douglas Fambrough (Johns Hopkins University) for providing the cDNA for the soluble dog Na+,K+-ATPase β1 subunit and Dr. Ingolf Blasig (FMP Berlin) for the CFP-YFP tandem construct. We also thank Rosalia Aguirre, Raul Bonilla, Gabriel Orozco, Pablo Reyes, Victor Garcia, and Minerva Maldonado for technical assistance. This work was supported by research grants and research fellowships from Consejo Nacional de Ciencia y Tecnologia (National Research Council of México).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0081) on May 5, 2010.
REFERENCES
- Arrate M. P., Rodriguez J. M., Tran T. M., Brock T. A., Cunningham S. A. Cloning of human junctional adhesion molecule 3 (JAM3) and its identification as the JAM2 counter-receptor. J. Biol. Chem. 2001;276:45826–45832. doi: 10.1074/jbc.M105972200. [DOI] [PubMed] [Google Scholar]
- Bab-Dinitz E., Albeck S., Peleg Y., Brumfeld J., Gottschalk K. E., Karlish S. A C-terminal lobe of the β subunit of Na, K-ATPase and H, K-ATPase resembles cell adhesion molecules. Biochemistry. 2009;48:8684–8691. doi: 10.1021/bi900868e. [DOI] [PubMed] [Google Scholar]
- Belusa R., Wang Z., Matsubara T., Sahingren B., Dulubova I., Nair A. C., Ruoslathi E., Greengard P., Aperia A. “Mutation of the protein kinase C phosphorylation site on rat α1 Na+, K+-ATPase alters regulation of intracellular Na+ and pH and influences Cell shape and adhesiveness”. J. Biol. Chem. 1997;272:20179–20184. doi: 10.1074/jbc.272.32.20179. [DOI] [PubMed] [Google Scholar]
- Blasig I. E., Winkler L., Lassowski B., Mueller S. L., Zuleger N., Krause E., Krause G., Gast K., Kolbe M., Piontek J. On the self-association potential of transmembrane tight junction proteins. Cell. Mol. Life. Sci. 2006;63:505–514. doi: 10.1007/s00018-005-5472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan M. J., Anderson H. C., Palade G. E., Jamieson J. D. Intracellular sorting and polarized cell surface delivery of (Na+, K+)ATPase, an endogenous component of MDCK cell basolateral plasma membranes. Cell. 1986;46:623–631. doi: 10.1016/0092-8674(86)90888-3. [DOI] [PubMed] [Google Scholar]
- Cereijido M., Rotunno C. A. The effect of antidiuretic hormone on Na+ movement across frog skin. J. Physiol. 1971;213:119–133. doi: 10.1113/jphysiol.1971.sp009372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereijido M., Robbins E. S., Dolan W. J., Rotunno C. A., Sabatini D. D. Polarized monolayers formed by epithelial cells on a permeable and translucent support. J. Cell Biol. 1978;77, 3:853–880. doi: 10.1083/jcb.77.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereijido M., Stefani E., Palomo A. M. Occluding junctions in a cultured transporting epithelium: structural and functional heterogeneity. J. Membr. Biol. 1980;53:19–32. doi: 10.1007/BF01871169. [DOI] [PubMed] [Google Scholar]
- Cereijido M., Meza I., Martínez-Palomo A. Occluding junctions in cultured epithelial monolayers. Am. J. Physiol. 1981;240:C96–C102. doi: 10.1152/ajpcell.1981.240.3.C96. [DOI] [PubMed] [Google Scholar]
- Cereijido M., Contreras R. G., Shoshani L., García-Villegas M. R. “Membrane targeting.” Prog. Biophys. Mol. Biol. 2003;81:81–115. doi: 10.1016/s0079-6107(02)00047-0. [DOI] [PubMed] [Google Scholar]
- Cereijido M., Shoshani L., Contreras R. G. The polarized distribution of Na+, K+-ATPase and active transport across epithelia. J. Membr. Biol. 2001;184:299–304. doi: 10.1007/s00232-001-0097-y. [DOI] [PubMed] [Google Scholar]
- Chen C. P., Posy S., Ben-Shaul A., Shapiro L., Honig B. H. Specificity of cell–cell adhesion by classical cadherins: critical role for low-affinity dimerization through β-strand swapping. Proc. Natl. Acad. Sci. USA. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras R. G., Avila G., Gutiérrez C., Bolívar J. J., González-Mariscal L., Darzon A., Beaty G., Rodríguez-Boulan E., Cereijido M. Repolarization of Na+-K+ pumps during establishment of epithelial monolayers. Am. J. Physiol. 1989;257:C896–C905. doi: 10.1152/ajpcell.1989.257.5.C896. [DOI] [PubMed] [Google Scholar]
- Contreras R. G., Lázaro A., Bolívar J. J., Flores-Maldonado C., Sánchez S. H., González-Mariscal L., García-Villegas M. R., Valdés J., Cereijido M. A novel type of cell–cell cooperation between epithelial cells. J. Membr. Biol. 1995;145:305–310. doi: 10.1007/BF00232722. [DOI] [PubMed] [Google Scholar]
- Contreras R. G., Shoshani L., Flores-Maldonado C., Lázaro A., Cereijido M. Relationship between Na(+),K(+)-ATPase and cell attachment. J. Cell Sci. 1999;112:4223–4232. doi: 10.1242/jcs.112.23.4223. [DOI] [PubMed] [Google Scholar]
- Contreras R. G., Shoshani L., Flores-Maldonado C., Lázaro A., Monroy A. O., Roldán M., Fiorentino R., Cereijido M. E-Cadherin and tight junctions between epithelial cells of different animal species. Pflugers Arch. 2002;444:467–475. doi: 10.1007/s00424-002-0827-8. [DOI] [PubMed] [Google Scholar]
- Contreras R. G., Flores-Maldonado C., Lázaro A., Shoshani L., Flores-Benitez D., Larré I., Cereijido M. Ouabain binding to Na+,K+-ATPase relaxes cell attachment and sends a specific signal (NACos) to the nucleus. J. Membr. Biol. 2004;198:147–158. doi: 10.1007/s00232-004-0670-2. [DOI] [PubMed] [Google Scholar]
- Contreras R. G., Flores-Benítez D., Flores-Maldonado. C., Larre I., Shoshani L., Cereijido M. Na+,K+-ATPase and hormone ouabain: new roles for an old enzyme and an old inhibitor. Cell. Mol. Biol. 2006;52:31–40. [PubMed] [Google Scholar]
- Deborde S., Perret E., Gravotta D., Deora A., Salvarezza S., Schreiner R., Rodríguez-Boulan E. Clathrin is a key regulator of basolateral polarity. Nature. 2008;452:719–723. doi: 10.1038/nature06828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield A., Fölsch H., Mellman I., Caplan M. J. Sorting of H, K-ATPase β-subunit in MDCK and LLC-PK cells is independent of mu 1B adaptin expression. Traffic. 2004;5:449–461. doi: 10.1111/j.1398-9219.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- Dunbar L. A., Caplan M. J. The cell biology of ion pumps: sorting and regulation. Eur. J. Cell Biol. 2000;79:557–563. doi: 10.1078/0171-9335-00079. [DOI] [PubMed] [Google Scholar]
- Efendiev R., Cinelli A. R., Leibiger I. B., Bertorello A. M., Pedemonte C. H. “FRET analysis reveals a critical conformational change within the Na, K-ATPase α1 subunit N-terminus during GPCR-dependent endocytosis”. FEBS Lett. 2006;580:5067–5070. doi: 10.1016/j.febslet.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Ellis M. A., Potter B. A., Cresawn K. O., Weisz O. A. Polarized biosynthetic traffic in renal epithelial cells: sorting, sorting, everywhere. Am. J. Physiol. Renal Physiol. 2006;291:F707–F713. doi: 10.1152/ajprenal.00161.2006. [DOI] [PubMed] [Google Scholar]
- Farr G. A., Hull M., Mellman I., Caplan M. J. Membrane proteins follow multiple pathways to the basolateral cell surface in polarized epithelial cells. J. Cell Biol. 2009;186:269–282. doi: 10.1083/jcb.200901021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallin W. J., Edelman G. M., Cunningham B. A. Characterization of L-CAM, a major cell adhesion molecule from embryonic liver cells. Proc. Natl. Acad. Sci. USA. 1983;80:1038–1042. doi: 10.1073/pnas.80.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geering K., Beggah P., Good S., Girardet S., Roy D., Schaer D., Jaunin P. Oligomerization and maturation of Na,K-ATPase: functional interaction of the cytoplasmic NH2 terminus of the beta subunit with the alpha subunit. J. Cell Biol. 1996;133:1193–1204. doi: 10.1083/jcb.133.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor S., Antonieck H., Sweadner K. J., Pagliusi S., Frank R., Moos M., Schachner M. The adhesion molecule on glia (AMOG) is a homologue of the b-subunit of the Na, K-ATPase. J. Cell Biol. 1990;110:165–174. doi: 10.1083/jcb.110.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., Rodríguez-Boulan E. Clathrin and AP1B: key roles in basolateral trafficking through trans-endosomal routes. FEBS Lett. 2009;583:3784–3795. doi: 10.1016/j.febslet.2009.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Mariscal L., Chávez de Ramírez B., Lázaro A., Cereijido M. Establishment of tight junctions between cells from different animal species and different sealing capacities. J. Membr. Biol. 1989;107:43–56. doi: 10.1007/BF01871082. [DOI] [PubMed] [Google Scholar]
- Gottardi C. J., Caplan M. J. Delivery of Na+,K(+)-ATPase in polarized epithelial cells. Science. 1993a;260:552–554. doi: 10.1126/science.8386395. [DOI] [PubMed] [Google Scholar]
- Gottardi C. J., Caplan M. J. Molecular requirements for the cell-surface expression of multisubunit ion-transporting ATPases. Identification of protein domains that participate in Na, K-ATPase and H, K-ATPase subunit assembly. J. Cell Biol. 1993b;268:14342–14427. [PubMed] [Google Scholar]
- Gravotta D., Deora A., Perret E., Oyanadel C., Soza A., Schreiner R., González A., Rodríguez-Boulan E. AP1B sorts basolateral proteins in recycling and biosynthetic routes of MDCK cells. Proc. Natl. Acad. Sci. USA. 2007;104:1564–1569. doi: 10.1073/pnas.0610700104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbleib J. M., Nelson W. J. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;23:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- Hammerton R. W., Krzemisnski K., Mays R. W., Ryan T. A., Wollner D. A., Nelson J. Mechanism for regulating cell surface distribution of Na+, K+-ATPase in polarized epithelial cells. Science. 1991;254:847–850. doi: 10.1126/science.1658934. [DOI] [PubMed] [Google Scholar]
- Hamrick M., Renaud K. J., Fambrough D. M. Assembly of the extracellular domain of the Na, K-ATPase beta subunit with the alpha subunit. Analysis of beta subunit chimeras and carboxyl-terminal deletions. J. Biol. Chem. 1993;268:24367–24373. [PubMed] [Google Scholar]
- Kenworthy A. K. Photobleaching FRET microscopy. In: Periassamy A., editor. Molecular Imaging. New York: Oxford University Press; 2005. pp. 146–164. [Google Scholar]
- Kitamura N., Ikekita M., Sato T., Akimoto Y., Hatanaka Y., Kawakami H., Inomata M., Furukawa K. Mouse Na+/K+-ATPase beta1-subunit has a K+-dependent cell adhesion activity for beta-GlcNAc-terminating glycans. Proc. Natl. Acad. Sci. USA. 2005;102:2796–2801. doi: 10.1073/pnas.0409344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koefoed-Johnsen V., Ussing H. The nature of frog skin potential. Acta Physiol. Scand. 1958;42:298–308. doi: 10.1111/j.1748-1716.1958.tb01563.x. [DOI] [PubMed] [Google Scholar]
- Kostrewa D., Brockhaus M., D'Arcy A., Dale G. E., Nelboeck P., Schmid G., Mueller F., Bazzoni G., Dejana E., Bartfai T., Winkler F. K., Hennig M. X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. EMBO J. 2001;20:4391–4398. doi: 10.1093/emboj/20.16.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Cai T., Tian J., Qu W., Xie Z. J. Functional characterization of Src-interacting Na/K-ATPase using RNA interference assay. J. Biol. Chem. 2006;281:19709–19719. doi: 10.1074/jbc.M512240200. [DOI] [PubMed] [Google Scholar]
- Matter K., Mellman I. “Mechanisms of cell polarity: sorting and transport in epithelial cells.” Curr. Opin. Cell Biol. 1994;6:545–554. doi: 10.1016/0955-0674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- Matter K. Epithelial polarity: sorting out the sorters. Curr. Biol. 2000;10:R39–R42. doi: 10.1016/s0960-9822(99)00256-0. [DOI] [PubMed] [Google Scholar]
- Mays R. W., Siemers K. A., Fritz B. A., Lowe A. W., van Meer G., Nelson W. J. Hierarchy of mechanisms involved in generating Na/K-ATPase polarity in MDCK epithelial cells. J. Cell. Biol. 1995;130:1105–1115. doi: 10.1083/jcb.130.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I., Nelson W. J. Coordinated protein sorting, targeting and distribution in polarized cells. Nat. Rev. Mol. Cell Biol. 2008;9:833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose Y., Honda T., Inagaki M., Shimizu K., Irie K., Nakanishi H., Takai Y. Role of the second immunoglobulin-like loop of nectin in cell-cell adhesion. Biochem. Biophys. Res. Commun. 2002;293:45–49. doi: 10.1016/S0006-291X(02)00183-3. [DOI] [PubMed] [Google Scholar]
- Moreno J., Cruz-Vera L. R., García-Villegas M. R., Cereijido M. Polarized expression of Shaker channels in epithelial cells. J. Membr. Biol. 2002;190:175–187. doi: 10.1007/s00232-002-1034-4. [DOI] [PubMed] [Google Scholar]
- Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- Muth T. R., Gottardi C. J., Roush D. L., Caplan M. J. A basolateral sorting signal is encoded in the alpha-subunit of Na-K-ATPase. Am. J. Physiol. 1998;274:C688–C696. doi: 10.1152/ajpcell.1998.274.3.C688. [DOI] [PubMed] [Google Scholar]
- Nagar B., Overduin M., Ikura M., Rini J. M. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- Niessen C. M., Gumbiner B. M. Cadherin-mediated cell sorting not determined by binding or adhesion specificity. J. Cell Biol. 2002;156:389–399. doi: 10.1083/jcb.200108040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi S., Mishina M., Kawamura M., Numa S. “Expression of functional (Na+, K+)-ATPase from cloned cDNAs. FEBS Lett. 1987;225:27–32. doi: 10.1016/0014-5793(87)81125-0. [DOI] [PubMed] [Google Scholar]
- Nose A., Nagafuchi A., Takeichi M. Expressed recombinant cadherins mediate cell sorting in model systems. Cell. 1988;54:993–1001. doi: 10.1016/0092-8674(88)90114-6. [DOI] [PubMed] [Google Scholar]
- Nusrat A., Chen J. A., Foley C. S., Liang T. W., Tom J., Cromwell M., Quan C., Mrsny R. J. The coiled-coil domain of occludin can act to organize structural and functional elements of the epithelial tight junction. J. Biol. Chem. 2000;275:29816–29822. doi: 10.1074/jbc.M002450200. [DOI] [PubMed] [Google Scholar]
- Nusrat A., Brown G. T., Tom J., Drake A., Bui T. T., Quan C., Mrsny R. J. Multiple protein interactions involving proposed extracellular loop domains of the tight junction protein occludin. Mol. Biol. Cell. 2005;16:1725–1734. doi: 10.1091/mbc.E04-06-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. D., Ciatto C., Chen; C. P, Bahna F., Rajebhosale M., Arkus N., Schieren I., Jessell T. M., Honig B., Price S. R. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Kreitzer G., Musch A. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 2005;6:233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- Shinoda T., Ogawa H., Cornelius F., Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- Shoshani L., Contreras R. G., Roldán M. L., Moreno J., Lázaro A., Balda M. S., Matter K., Cereijido M. The polarized expression of Na+,K+-ATPase in epithelia depends on the association between beta-subunits located in neighboring cells. Mol. Biol. Cell. 2005;16:1071–1081. doi: 10.1091/mbc.E04-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos B., Rodríguez-Boulan E. The epithelial polarity program: machineries involved and their hijacking by cancer. Oncogene. 2008;27:6939–6957. doi: 10.1038/onc.2008.345. [DOI] [PubMed] [Google Scholar]
- Tian J., Cai T., Yuan Z., Wang H., Liu L., Haas M., Maksimova E., Huang X. Y., Xie Z. J. Binding of Src to Na+/K+-ATPase forms a functional signaling complex. Mol. Biol. Cell. 2006;17:317–326. doi: 10.1091/mbc.E05-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokhtaeva E., Sachs G., Vagin O. Assembly with the Na,K-ATPase alpha(1) subunit is required for export of beta(1) and beta(2) subunits from the endoplasmic reticulum. Biochemistry. 2009;48:11421–11431. doi: 10.1021/bi901438z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin O., Turdikulova S., Sachs G. Recombinant addition of N-glycosylation sites to the basolateral Na, K-ATPase beta1 subunit results in its clustering in caveolae and apical sorting in HGT-1 cells. J. Biol. Chem. 2005;280:43159–43167. doi: 10.1074/jbc.M508262200. [DOI] [PubMed] [Google Scholar]
- Vagin O., Tokhtaeva E., Sachs G. The role of β subunit of the Na+, K+ ATPase and its glycosilation in cell-cell adhesion. J. Biol. Chem. 2006;281:39573–39587. doi: 10.1074/jbc.M606507200. [DOI] [PubMed] [Google Scholar]
- Vagin O., Turdikulova S., Tokhtaeva E. “Polarized membrane distribution of potassium-dependent ion pumps in epithelial cells: different roles of the N-glycans of their β subunits.”. Cell Biochem. Biophys. 2007;47:376–391. doi: 10.1007/s12013-007-0033-6. [DOI] [PubMed] [Google Scholar]
- Weisz O. A., Rodriguez-Boulan E. Apical trafficking in epithelial cells: signals, clusters and motors. J. Cell Sci. 2009;122:4253–4266. doi: 10.1242/jcs.032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson P. D., Devuyst O., Li X., Gatti L., Falkenstein D., Robinson S., Fambrough D., Burrow C. R. Apical plasma membrane mispolarization of Na, K-ATPase in polycystic kidney disease epithelia is associated with aberrant expression of the beta2 isoform. Am. J. Pathol. 2000;156:253–268. doi: 10.1016/s0002-9440(10)64726-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C., Takeichi M. Teratocarcinoma cell adhesion: identification of a cell-surface protein involved in calcium-dependent cell aggregation. Cell. 1982;28:217–224. doi: 10.1016/0092-8674(82)90339-7. [DOI] [PubMed] [Google Scholar]
- Zurzolo C., Rodríguez-Boulan E. Delivery of Na+,K+-ATPase in polarized epithelial cells. Science 260. 1993;5107:550–552. doi: 10.1126/science.8386394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.