CENP-C and CENP-N recognize distinct structural elements of CENP-A nucleosomes, providing a foundation for the assembly of other centromere and kinetochore components.
Abstract
Centromeres contain specialized nucleosomes in which histone H3 is replaced by the histone variant centromere protein A (CENP-A). CENP-A nucleosomes are thought to act as an epigenetic mark that specifies centromere identity. We previously identified CENP-N as a CENP-A nucleosome-specific binding protein. Here, we show that CENP-C also binds directly and specifically to CENP-A nucleosomes. Nucleosome binding by CENP-C required the extreme C terminus of CENP-A and did not compete with CENP-N binding, which suggests that CENP-C and CENP-N recognize distinct structural elements of CENP-A nucleosomes. A mutation that disrupted CENP-C binding to CENP-A nucleosomes in vitro caused defects in CENP-C targeting to centromeres. Moreover, depletion of CENP-C with siRNA resulted in the mislocalization of all other nonhistone CENPs examined, including CENP-K, CENP-H, CENP-I, and CENP-T, and led to a partial reduction in centromeric CENP-A. We propose that CENP-C binds directly to CENP-A chromatin and, together with CENP-N, provides the foundation upon which other centromere and kinetochore proteins are assembled.
Introduction
The accurate segregation of chromosomes during mitosis is essential for the reproduction and development of all organisms. Defects in chromosome segregation can cause cell death or chromosome aneuploidy, a condition that is associated with human diseases including cancer (Holland and Cleveland, 2009; Thompson et al., 2010). The fidelity of chromosome segregation depends on the assembly of a microtubule-binding site called the kinetochore on each sister chromatid of a replicated chromosome. Kinetochores attach to microtubules emanating from opposite poles of the mitotic spindle and are required for metaphase chromosome alignment and the anaphase chromosome movements that are necessary for accurate segregation (Cheeseman and Desai, 2008; Santaguida and Musacchio, 2009).
Centromeres are the specialized chromosomal regions present throughout the cell cycle upon which kinetochores are assembled during mitosis. In human cells, centromere DNA typically consists of megabase-length arrays of α-satellite repeats (Choo et al., 1991). However, in rare instances, functional centromeres have been observed at chromosomal locations that lack discernable α-satellite repeats (Voullaire et al., 1993; Marshall et al., 2008), which suggests that that DNA sequence is not required for centromere function in human cells. Fundamental to centromere function is the replacement of histone H3 with the histone H3 variant centromere protein A (CENP-A, also called CenH3) within centromeric nucleosomes (Palmer et al., 1987, 1991). CENP-A is localized to all active centromeres regardless of the underlying DNA sequence and is essential for kinetochore formation and chromosome segregation (Palmer et al., 1987; Stoler et al., 1995; Meluh et al., 1998; Buchwitz et al., 1999; Henikoff et al., 2000; Howman et al., 2000; Takahashi et al., 2000; Blower and Karpen, 2001; Oegema et al., 2001; Régnier et al., 2005). CENP-A nucleosomes are therefore thought to act as an epigenetic mark that specifies centromere identity.
In addition to CENP-A chromatin, human centromeres contain at least 16 nonhistone proteins (called CENP-C, CENP-H, CENP-I, CENP-K through CENP-U, CENP-W, and CENP-X) that are collectively referred to as the constitutive centromere-associated network (CCAN; Saitoh et al., 1992; Sugata et al., 2000; Goshima et al., 2003; Foltz et al., 2006; Izuta et al., 2006; Okada et al., 2006; Hori et al., 2008; Amano et al., 2009). CENP-C, CENP-H, CENP-N, CENP-M, and CENP-T copurify with CENP-A nucleosomes (Foltz et al., 2006), which suggests that CCAN proteins interact directly with centromeric chromatin during centromere and kinetochore assembly. Consistent with this possibility, electron microscopy and super-resolution light microscopy data have shown that CCAN proteins are the most chromatin-proximal elements of kinetochores in mitosis (Saitoh et al., 1992; Joglekar et al., 2009; Wan et al., 2009).
A systematic biochemical screen designed to identify proteins that are important for the recognition of centromeric chromatin showed that the CCAN protein CENP-N binds directly and specifically to CENP-A nucleosomes in a DNA sequence-independent manner (Carroll et al., 2009). Thus, CENP-N “reads” epigenetic information that is encoded within CENP-A nucleosomes. CENP-N is only required for targeting a subset of CCAN proteins to centromeres, however, which indicates that alternative mechanisms for centromeric chromatin recognition must exist.
CENP-C is a good candidate for having a direct role in the specific recognition of centromeric chromatin. CENP-A is required for CENP-C centromere localization, but other CCAN proteins, including CENP-N, CENP-K, and CENP-I, are not (Goshima et al., 2003; Okada et al., 2006; Cheeseman et al., 2008; Carroll et al., 2009). Furthermore, structure and function studies have identified overlapping regions of human CENP-C that bind directly to DNA and are sufficient for centromere localization in vivo (Sugimoto et al., 1994, 1997; Yang et al., 1996; Politi et al., 2002; Trazzi et al., 2002; Trazzi et al., 2009). CENP-C does not bind preferentially to centromere DNA (Yang et al., 1996), however, and it is unclear as to how the sequence-independent DNA-binding activity of CENP-C alone could specify its centromere-specific localization observed in human cells.
Here, we show that CENP-C preferentially binds to CENP-A nucleosomes over H3 nucleosomes or DNA alone. We identify the extreme C terminus of CENP-A as being necessary and sufficient for the interaction of nucleosomes with CENP-C, defining a novel structural element of centromeric chromatin likely to be important for centromere assembly and function. The recognition of CENP-A nucleosomes is critical for CENP-C centromere localization, which subsequently specifies the recruitment to centromeres of all other CCAN proteins tested. We propose a model for centromere assembly that integrates the dual recognition of CENP-A nucleosomes by CENP-N and CENP-C.
Results
CENP-C binds directly and specifically to CENP-A nucleosomes
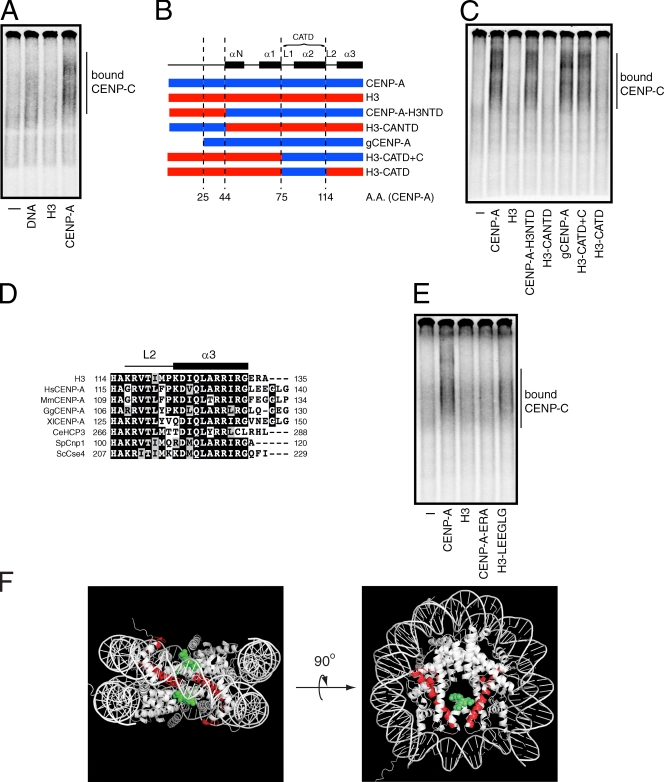
To understand the mechanisms by which CENP-C is recruited to centromeres, we reexamined the possibility that CENP-C interacts with CENP-A nucleosomes using our previously described gel-shift assay (Carroll et al., 2009). Full-length [35S]methionine-labeled CENP-C produced in reticulocyte extract was mixed with reconstituted nucleosomes that contained either CENP-A or histone H3 and a 186-bp fragment of α-satellite DNA, or with the α-satellite DNA alone (Fig. S1, A and B). Each mixture was resolved on a nondenaturing acrylamide gel, and the migration pattern of labeled CENP-C was compared with control reactions that contained no nucleosomes or DNA. A fast-migrating, albeit diffuse, CENP-C species was detected in the presence of α-satellite DNA (Fig. 1 A), which is consistent with previous studies showing that human CENP-C is a DNA-binding protein (Sugimoto et al., 1994, 1997; Yang et al., 1996). CENP-C did not bind to H3 nucleosomes, but did bind much more efficiently to CENP-A nucleosomes than it did to DNA alone (Fig. 1 A). About fivefold more CENP-A nucleosomes were typically required to detect CENP-C binding than were required to detect CENP-N binding. The comparatively weak binding of CENP-C to CENP-A nucleosomes and the poor resolution of the bound species likely contributed to the inability to identify CENP-C/CENP-A nucleosome binding in our previous study (Carroll et al., 2009). Nevertheless, CENP-C, like CENP-N, interacts directly and specifically with CENP-A nucleosomes.
Figure 1.
CENP-C binds directly to CENP-A nucleosomes. (A) [35S]methionine-labeled CENP-C was incubated alone (−) or in the presence of α-satellite DNA or nucleosomes (300 nM). The mixtures were resolved on a native gel, and [35S]methionine-labeled CENP-C was detected with a phosphorimager. The position of CENP-C bound to DNA or nucleosomes is indicated. (B) A schematic showing the junctions of the CENP-A (blue) and histone H3 (red) chimeras used in this study. The numbers below the diagram refer to amino acids positions (A.A.) within CENP-A. Helical elements (black boxes, labeled α) and loops (labeled L) found within the histone fold of CENP-A and the region encompassing the CATD are shown above for reference. (C) Wild-type nucleosomes or nucleosomes containing CENP-A/H3 chimeras (500 nM) were incubated with [35S]methionine-labeled CENP-C and resolved on a native gel. [35S]methionine-labeled CENP-C alone (−) was used as a negative control. The position of nucleosome-bound CENP-C is indicated. (D) Clustal W alignment of the C terminus of human histone H3 and CENP-A orthologues from humans (Hs), mice (Mm), chickens (Gg), X. laevis (Xl), C. elegans (CeHCP3), S. pombe (SpCnp1), and Saccharomyces cerevisiae (ScCse4). The identical residues within each protein are highlighted with a black background, whereas conservative amino acid changes are colored gray. The first and last amino acid positions of the residues contained within the alignment are shown on the left and right side of each sequence, respectively. The positions of loop 2 (L2) and helix 3 (α3) within CENP-A are shown above for reference. (E) [35S]methionine-labeled CENP-C was incubated alone (−) or in the presence of the indicated nucleosomes and analyzed as in A. (F) The crystal structure of histone H3 is shown. Amino acids positions analogous to the CENP-A CATD (red ribbon) and residues C-terminal to helix 3 (green dots) are highlighted. Only one of the C-terminal extensions is colored green on the right panel for clarity.
We reconstituted nucleosomes that contained histone H3/CENP-A chimeras to define the structural elements of CENP-A that are important for CENP-C binding (Figs. 1 B and S1, A and C). CENP-C bound efficiently to nucleosomes in which the N-terminal tail of H3 replaced the corresponding region of CENP-A (CENP-A–H3NTD), but did not bind to nucleosomes in which the CENP-A N-terminal tail was appended to the histone fold of H3 (H3-CANTD; Fig. 1 C). CENP-C also bound to CENP-A nucleosomes containing a deletion mutant that lacked the first 25 amino acids of the CENP-A N-terminal tail (gCENP-A; Fig. 1 C). These data indicate that the N-terminal tail of CENP-A is dispensable for CENP-C binding and that the histone fold of CENP-A is necessary and sufficient for the direct interaction of CENP-C with nucleosomes.
A region comprising loop 1 and helix 2 within the histone fold of CENP-A, called the CENP-A centromere-targeting domain (CATD; residues 75–114), is sufficient for CENP-N binding when substituted into the corresponding region of histone H3 (Black et al., 2004; Carroll et al., 2009). We asked whether the CATD region of CENP-A was also sufficient for CENP-C binding. Reconstituted nucleosomes containing the H3-CATD chimera did not interact with CENP-C (Fig. 1 C), which suggests that CENP-C and CENP-N recognize different structural elements within CENP-A nucleosomes. Nucleosomes that contained a histone chimera with the CATD and the remaining C-terminal amino acids from CENP-A (H3-CATD+C) did, however, bind to CENP-C as well as wild-type CENP-A nucleosomes (Fig. 1 C). Determinants within CENP-A that are C-terminal to the CATD are therefore required for CENP-C binding.
The region C-terminal to the CATD of CENP-A consists of loop 2 and helix 3 of the histone fold. Sequence alignments showed that, although this region is highly conserved between human CENP-A and histone H3, CENP-A contains a short nonconserved extension at its extreme C terminus (Fig. 1 D). The alignment of CENP-A orthologues from several species indicated that the addition of several amino acids and/or the divergence from H3 is common feature of CENP-A C-terminal residues (Fig. 1 D). To determine if the C-terminal extension of human CENP-A was important for CENP-C binding, we reconstituted nucleosomes with histone chimeras in which the residues after helix 3 of the histone fold were swapped between H3 and CENP-A (Fig. S1 C). Nucleosomes that contained CENP-A with the H3 C terminus (CENP-A-ERA) were unable to bind to CENP-C (Fig. 1 E). In contrast, nucleosomes that contained H3 with the C terminus of CENP-A (H3-LEEGLG) bound to CENP-C almost as well as wild-type CENP-A nucleosomes (Fig. 1 E). Thus, the LEEGLG motif at the extreme C terminus of CENP-A is both necessary and sufficient for the direct interaction of CENP-C with nucleosomes.
Together, these data show that the C-terminal extension of CENP-A is a novel functional element within centromeric nucleosomes that specifies binding to CENP-C. Based on the crystal structure of the H3 nucleosome (Luger et al., 1997), the CENP-A C-terminal extension is likely to present a solvent accessible surface extending above the face of the nucleosome (Fig. 1 F), which is consistent with the possibility that CENP-C interacts directly with CENP-A. Alternatively, the C terminus of CENP-A could impose unique structural properties upon nucleosomes that are recognized by CENP-C.
The minimal centromere localization domain of CENP-C is necessary and sufficient for CENP-A nucleosome binding
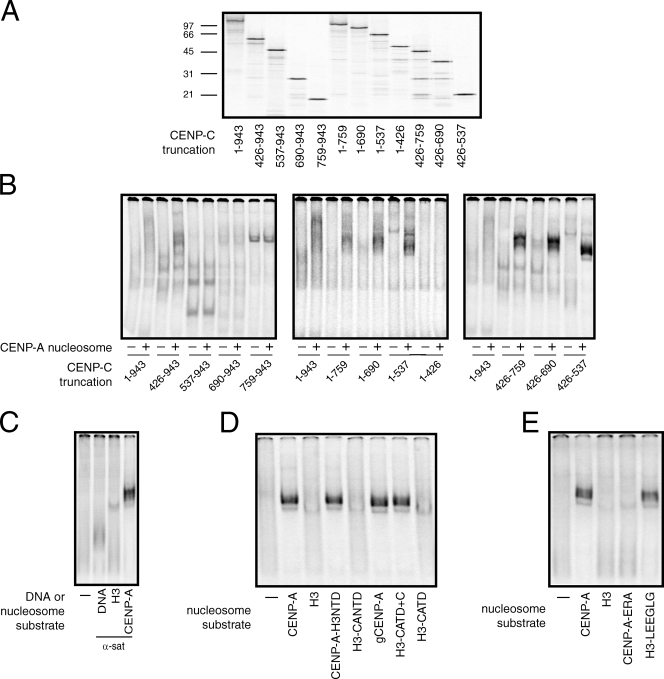
The analysis of CENP-C truncation mutants from numerous organisms has defined regions of CENP-C important for kinetochore assembly, self-association, sequence-independent DNA binding, and centromere localization (Fig. S2; Sugimoto et al., 1994, 1997; Lanini and McKeon, 1995; Yang et al., 1996; Politi et al., 2002; Trazzi et al., 2002, 2009; Cohen et al., 2008; Milks et al., 2009; Tanaka et al., 2009). To understand the relationship between the CENP-A nucleosome-binding activity of CENP-C and the previously defined functional regions of CENP-C, we generated a series of truncation mutants to identify a minimal region of CENP-C important for CENP-A nucleosome binding (Fig. 2 A). Because some CENP-C truncations migrate differently in native gels than full-length CENP-C, we assessed CENP-A nucleosome binding by comparing the migration pattern of each [35S]methionine-labeled CENP-C truncation protein alone to the pattern in the presence of reconstituted CENP-A nucleosomes (Fig. 2 B). In each case, the presence of novel CENP-C species in reactions that contained CENP-A nucleosomes indicated a direct interaction.
Figure 2.
Identification of a CENP-A recognition module within CENP-C. (A) Full-length (1–943) and various truncation mutants of [35S]methionine-labeled CENP-C were produced in rabbit reticulocyte extracts and resolved on a 12.5% SDS-PAGE gel. The amino acids within CENP-C that are included in each protein are indicated below the gel. The positions of molecular weight markers (kD) are indicated to the left of the gel. (B) Each CENP-C protein was incubated alone (−) or in the presence of 300 nM of CENP-A nucleosomes (+) and resolved on a native gel. (C) CENP-C426–537 was incubated alone (−) or in the presence of α-satellite DNA or the indicated nucleosome (10 nM) and resolved on a native gel. (D and E) CENP-C426–537 was incubated alone (−) or in the presence of the indicated nucleosomes (10 nM) and analyzed as above. The specific regions of CENP-A and H3 included in each chimera are schematized in Fig. 1 B.
A CENP-C fragment lacking the first 425 amino acids (426–943) efficiently bound to CENP-A nucleosomes (Fig. 2 B, left), which suggests that the region of CENP-C required for kinetochore assembly is not important for the binding of CENP-C to CENP-A nucleosomes (Milks et al., 2009; Tanaka et al., 2009). Deletion of the first 536 amino acids of CENP-C (537–943) or more (690–943, 759–943), however, resulted in a loss of CENP-A nucleosome binding. Systematic truncation from the C terminus indicated that a CENP-C mutant containing only amino acids 1–537 was able to bind to CENP-A nucleosomes (Fig. 2 B, center). Thus, the CENP-C dimerization domain (removed in the 1–759 truncation) and the Mif2 homology/CENP-C signature motif (removed in the 1–690 truncation) are not important for the recognition of CENP-A nucleosomes by CENP-C (Brown, 1995; Lanini and McKeon, 1995; Talbert et al., 2004; Cohen et al., 2008; Trazzi et al., 2009). Further deletion of the CENP-C C terminus to amino acid 426 (1–426) eliminated CENP-A nucleosome binding (Fig. 2 B, middle). We next generated a series of CENP-C truncation proteins with deletions in both N- and C-terminal residues. A CENP-C truncation mutant that contained only residues 426–537 (referred to CENP-C426–537 from here on) bound efficiently to CENP-A nucleosomes (Fig. 2 B, right), which indicates that this region represents a minimal CENP-A recognition module within CENP-C that is both necessary and sufficient for nucleosome binding.
Examination of the CENP-C truncation mutants that bound to CENP-A nucleosomes revealed that the number and distribution of bound species changed as different regions of CENP-C were removed (Fig. 2 B). Several distinct bound species were detectable for CENP-C proteins that contained the N or C terminus in addition to the minimal nucleosome-binding domain (1–537 and 426–943, respectively), whereas a single bound species is detected for CENP-C426–537. Previous work indicates that both the N- and C-terminal regions of CENP-C are capable of self-association (Fig. S2; Sugimoto et al., 1997; Cohen et al., 2008; Trazzi et al., 2009). The multiple bound species observed using larger CENP-C fragments in the presence of CENP-A nucleosomes most likely represent different oligomerization states of CENP-C bound to mononucleosomes.
The minimal CENP-A–binding module of CENP-C contained the same nucleosome specificity as full-length CENP-C. CENP-C426–537 bound weakly to α-satellite DNA when compared with CENP-A nucleosomes reconstituted with the same DNA fragment and did not bind at all to reconstituted H3 nucleosomes (Fig. 2 C). Moreover, analysis of the interaction of CENP-C426–537 with nucleosomes that contained the histone H3/CENP-A chimeras described in Fig. 1 showed that the CENP-A C-terminal LEEGLG motif was necessary and sufficient for both CENP-C426–537 and full-length CENP-C binding (Fig. 2, D and E).
The region of CENP-C identified here as being important for CENP-A nucleosome binding corresponds closely to regions of human CENP-C that bind directly to DNA and are sufficient for centromere localization (Fig. S2; Sugimoto et al., 1994, 1997; Yang et al., 1996; Politi et al., 2002; Trazzi et al., 2002). To date, it has been unclear as to how sequence-independent DNA binding could specify the centromere localization observed for CENP-C426–537, particularly because this region of CENP-C lacks domains important for interactions with other CENPs (Fig. S2; Milks et al., 2009; Tanaka et al., 2009). Our data suggest that the mechanism by which CENP-C426–537 is targeted to centromeres is not caused by sequence-independent DNA binding but rather by a direct and specific interaction with CENP-A nucleosomes.
CENP-A nucleosome binding and self-association independently contribute to vertebrate CENP-C centromere localization
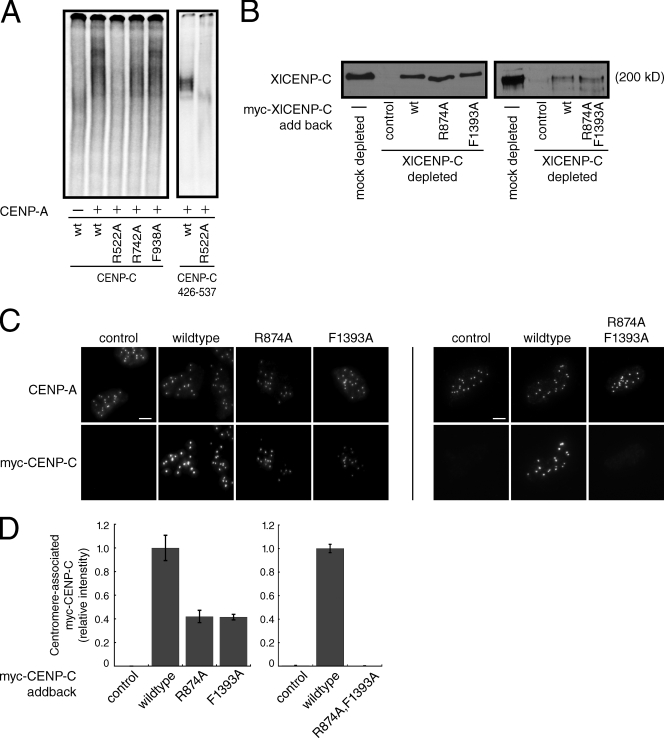
To further characterize the interaction between human CENP-C and CENP-A nucleosomes, we mutated three conserved residues—R522, R742, and F938—that are important for CENP-C function and determined the effects of these mutations on CENP-A nucleosome binding (Meluh and Koshland, 1995; Fukagawa et al., 2001b; Song et al., 2002; Heeger et al., 2005; Cohen et al., 2008; Milks et al., 2009). Mutation of R742 within the CENP-C motif to alanine had, at most, a small effect on nucleosome binding, whereas mutation of F938 to alanine, which is predicted to disrupt CENP-C dimerization (Cohen et al., 2008), had no effect on CENP-A nucleosome binding (Fig. 3 A, left). Thus, residues important for CENP-C function that lie outside the minimal CENP-A nucleosome recognition module are dispensable for nucleosome binding, which is consistent with our truncation analysis (Fig. 2 B). In contrast, mutation of R522A eliminated CENP-A nucleosome binding in the context of both full-length CENP-C and the CENP-C426–537 fragment (Fig. 3 A).
Figure 3.
A conserved arginine residue within CENP-C is required for nucleosome binding and centromere localization. (A) Wild-type (wt) human CENP-C or the indicated point mutant was incubated alone (−) or with CENP-A nucleosomes (+) and resolved on a native gel (left). Alternatively, the wild-type (wt) or R522A CENP-C426–537 fragments were incubated with CENP-A nucleosomes (10 nM). (B) Western blot using an anti–XlCENP-C antibody of mock-depleted or CENP-C–depleted Xenopus extracts containing reticulocyte-produced wild-type XlCENP-C (wt) or the indicated point mutants. Reticulocyte extract lacking myc-CENP-C (−) was used as a control. R874 and F1393 in Xenopus CENP-C are analogous to R522 and F938, respectively, in human CENP-C. The molecular weight of XlCENP-C is indicated. (C) Isolated sperm nuclei from CENP-C–depleted Xenopus extracts containing the indicated myc-CENP-C protein were stained with anti–XlCENP-A and anti-myc antibodies. Images are maximum-intensity projections of z stacks collected at 0.2-µM steps. Bars, 5 µM. (D) Quantification of centromere-associated myc-CENP-C from C. Error bars show the SEM from three independent experiments (>100 centromeres were counted for each condition in each experiment).
The current data are ambiguous with regards to the importance of R522 to CENP-C localization in human cells. R522 is required for the centromere localization of CENP-C fragments lacking the C terminus (Song et al., 2002); however, the ability of CENP-C fragments that include the C-terminal dimerization domain, but lack the CENP-A nucleosome recognition module, to localize to centromeres suggests that CENP-A nucleosome binding may be dispensable for centromere localization in the context of full-length CENP-C (Trazzi et al., 2002, 2009). The interpretation of these studies is further complicated by the presence of endogenous CENP-C in cells that may influence the localization efficiency of the transfected CENP-C constructs.
We have previously shown that Xenopus CENP-C (XlCENP-C) can be efficiently immunodepleted from egg extracts and replaced with exogenous protein (Milks et al., 2009), making this system ideal for the analysis of CENP-C mutants without complication from the endogenous protein. Furthermore, the amino acids that we mutated in human CENP-C are conserved in XlCENP-C, which suggests that the function of these residues is similar across species (Fig. S3 A). We therefore made mutations in R874 and F1393 within XlCENP-C, which correspond to R522 and F938 in human CENP-C, respectively, to determine the contributions of CENP-A nucleosome recognition and dimerization to CENP-C centromere localization (the effects of mutations in XlCENP-C R1210, which corresponds to HsCENP-C R742, have been described previously; Milks et al., 2009).
Western blotting and immunofluorescence showed that XlCENP-C levels were reduced >90% in CENP-C–depleted extracts when compared with mock-depleted extracts (Figs. 3 B and S3 B). Although the amount of myc-tagged XlCENP-C added back was lower than the amount of endogenous XlCENP-C normally present in these extracts (Fig. 3 B), the levels of myc-XlCENP-C protein in each case were identical, allowing a direct comparison of the localization efficiencies of the wild-type and mutant proteins. Quantification of the levels of centromere-associated XlCENP-C in isolated sperm nuclei, as judged by colocalization with CENP-A, indicated that the myc-tagged wild-type XlCENP-C was present on centromeres at ∼25–50% of the levels of endogenous XlCENP-C from mock-depleted extracts (Fig. S3, B and C). Most importantly, comparing the levels of the individual myc-tagged XlCENP-C proteins at centromeres revealed that the R874A and F1393A mutants were reduced ∼60% when compared with the wild-type myc-XlCENP-C (Fig. 3 C). Combining the mutations had an even greater effect; the myc-tagged XlCENP-C R874/F1393A double mutant was not detectable at centromeres above background (Figs. 3 C and S3, B and C). Thus, both CENP-A nucleosome binding and dimerization independently contribute to the localization of full-length XlCENP-C.
CENP-C forms a stoichiometric complex with CENP-A nucleosomes
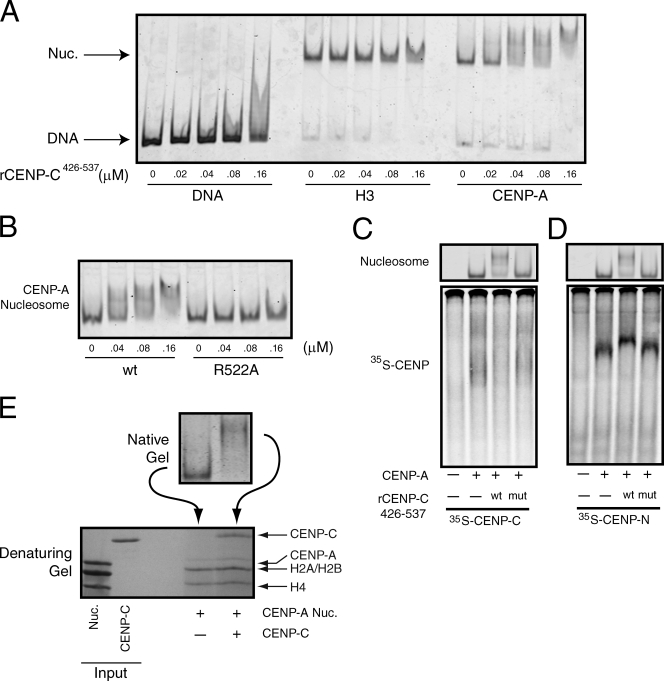
Our capacity to characterize the CENP-C–CENP-A nucleosome complex has thus far relied on using high concentrations of reconstituted CENP-A nucleosomes that are in vast excess of the comparatively low levels of CENP-C produced in reticulocyte extracts. We therefore expressed in bacteria and purified to near homogeneity recombinant wild-type CENP-C426–537 and CENP-C426–537 R522A (Fig. S4 A). Recombinant CENP-C426–537 (rCENP-C426–537) bound efficiently to CENP-A nucleosomes, but did not bind to the α-satellite DNA alone or to H3 nucleosomes (Fig. 4 A). Furthermore, mutation of R522A eliminated CENP-A nucleosome binding by rCENP-C426–537 (Fig. 4 B), which indicates that the rCENP-C426–537 recapitulated the behavior demonstrated for full-length CENP-C and the CENP-C426–537 fragment produced in reticulocyte extracts. Importantly, at the highest concentration of CENP-C used in this experiment (160 nM), the CENP-A nucleosomes were quantitatively shifted to a slower migrating species (Fig. 4, A and B). Further increasing the concentration of rCENP-C426–537 led to the progressive supershifting of the CENP-A nucleosomes (Fig. S4 B). However, at these concentrations of rCENP-C426–537 (400 nM and above), the specificity of rCENP-C426–537 for CENP-A over H3 nucleosomes was lost. Moreover, R522 was no longer required for CENP-A nucleosome binding by rCENP-C426–537 (Fig. S4 C). Collectively, these data suggest that the supershifting of CENP-A nucleosomes results from nonspecific association of rCENP-C426–537 with nucleosomes or DNA.
Figure 4.
CENP-C and CENP-N bind to different sites on the same CENP-A nucleosomes. (A) α-Satellite DNA or reconstituted nucleosomes (10 nM) were incubated with the indicated concentration of rCENP-C426–537 and resolved on native gels. DNA or nucleosomes were visualized after staining with SYBR gold. (B) CENP-A nucleosomes (10 nM) were incubated with the indicated concentration of wild-type (wt) or mutant (R522A) rCENP-C426–537 and resolved on a native gel. (C) [35S]methionine-labeled CENP-C was incubated alone (−) or in the presence of 150 nM of CENP-A nucleosomes (+). In addition, binding reactions contained wild-type (wt) or the R522A mutant (mut) rCENP-C426–537 (300 nM), or buffer alone (−). The gel was stained with ethidium bromide to visualize nucleosomes (top) and was subsequently dried and scanned on a phosphorimager to visualize the labeled CENP-C (bottom). (D) An identical experiment to C except that [35S]methionine-labeled CENP-N was used. (E) Isolated nuclei were digested with micrococcal nuclease, and wild-type or R11A mutant GFP–CENP-N was immunoprecipitated using anti-GFP antibodies. Each immunoprecipitate was probed with the indicated antibodies. Control cells don’t express GFP–CENP-N.
We determined whether the rCENP-C426–537 would engage nucleosomes in the same manner as full-length CENP-C using competition assays. [35S]methionine-labeled CENP-C was incubated with CENP-A nucleosomes either alone or in the presence of wild-type or R522A rCENP-C426–537, and each mixture was resolved on a native gel. The gel was first stained with ethidium bromide to visualize the nucleosomes, then dried and exposed to a phosphorimager screen to visualize the full-length CENP-C. The CENP-A nucleosomes behaved as expected in that addition of wild-type rCENP-C426–537 shifted the nucleosomes to a slow migrating species when compared with reactions containing no rCENP-C426–537 or the rCENP-C426–537 R522A mutant (Fig. 4 C, top). Binding of the labeled CENP-C to CENP-A nucleosomes was substantially reduced in reactions that contained wild-type rCENP-C426–537 but not in control reactions (Fig. 4 C, bottom), which indicates that the rCENP-C426–537 competed for binding with the labeled full-length CENP-C. Thus, rCENP-C426–537 does engage CENP-A nucleosomes by a mechanism that is similar to full-length CENP-C. Increasing the concentration of wild-type or R522A rCENP-C426–537 in an otherwise identical experiment resulted in supershifted CENP-A nucleosomes, as described (Fig. S4 C). However, the [35S]methionine-labeled full-length CENP-C still bound to the supershifted CENP-A nucleosomes in the presence of the rCENP-C426–537 R522A mutant, which confirms that the supershifting of CENP-A nucleosomes at high concentrations of CENP-C426–537 results from nonspecific binding.
We performed a similar competition experiment except that labeled CENP-N was used instead of labeled CENP-C to directly test whether CENP-N and CENP-C bind to different sites on CENP-A nucleosomes. The labeled CENP-N still bound efficiently to the slow migrating CENP-A nucleosomes in the presence of wild-type rCENP-C426–537 (Fig. 4 D), which demonstrates that rCENP-C426–537 does not compete with CENP-N for binding to CENP-A nucleosomes. CENP-N and the rCENP-C426–537 therefore bind to different sites on CENP-A nucleosomes, which is consistent with our previous data showing that the determinants that specify CENP-N and CENP-C binding to nucleosomes are distinct (Fig. 1, C and E; Carroll et al., 2009).
We next determined the stoichiometry of the rCENP-C426–537–CENP-A nucleosome complex under conditions in which the CENP-A nucleosomes were quantitatively bound by rCENP-C426–537. CENP-A nucleosomes alone or in the presence of rCENP-C426–537 were first resolved on a native gel (Fig. 4 E). We then isolated from the native gel the unbound and rCENP-C426–537–bound CENP-A nucleosomes, and subsequently resolved each in a denaturing gel. The recovery of CENP-A was poor after isolation from the native gel regardless of whether rCENP-C426–537 was included in the reaction, which precluded a direct measurement of the ratio of CENP-A to CENP-C in the bound species. However, quantification of the ratio of CENP-C to H4 and CENP-C to H2A/H2B in the bound species revealed a mean relative stoichiometry of 1.2:1 and 0.6:1 (n = 2), respectively, which suggests that two molecules of rCENP-C426–537 bind to each CENP-A nucleosome.
CENP-C is required for centromere assembly
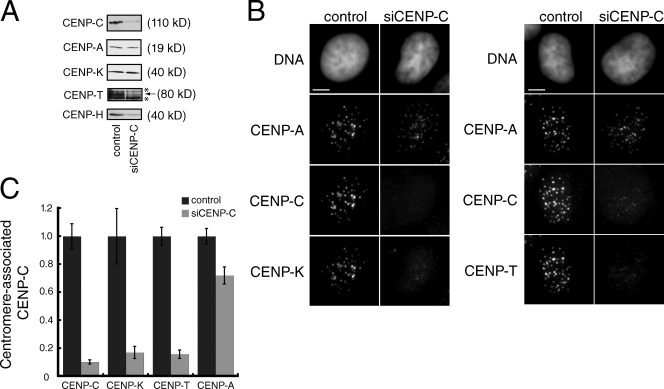
Current models for centromere assembly in human cells suggest that CENP-A specifies the recruitment of CENP-C to centromeric chromatin independently of other CCAN proteins (Goshima et al., 2003; Liu et al., 2006). Indeed, CENP-C is still present at centromeres, albeit at reduced levels, after depletion of CENP-N, CENP-I, or CENP-K (Goshima et al., 2003; Liu et al., 2006; Cheeseman et al., 2008; Carroll et al., 2009). The localization dependency of other CCAN proteins on CENP-C, however, has not been comprehensively determined. We depleted CENP-C with siRNA and examined the localization of several other CCAN proteins. CENP-C depletion was efficient as indicated by a ∼90% reduction in both total CENP-C protein levels and in centromere-localized CENP-C when compared with mock-depleted control cells (Fig. 5, A and B). All of the other nonhistone CENPs tested here, including CENP-H, CENP-I, CENP-K, and CENP-T, depended on CENP-C for their localization in interphase and mitosis (Fig. 5 B; and Fig. S5, A and B). Quantification of the centromere-associated levels of CENP-K and CENP-T showed that both proteins were reduced to the same extent as CENP-C itself in CENP-C–depleted cells when compared with mock-depleted cells (Fig. 5 C). Furthermore, Western blots showed that, although the abundance of total CENP-K did not change after CENP-C depletion, both CENP-H and CENP-T protein levels were dramatically reduced in cells lacking CENP-C (Fig. 5 A). Although we could not examine CENP-N directly in this experiment, CENP-N localization to the centromere has previously been shown to require CENP-H (McClelland et al., 2007), which suggests that CENP-C is also required for the localization of CENP-N to centromeres. In addition, we observed a modest reduction both in the overall levels of CENP-A protein by Western blotting and in the levels of CENP-A associated with centromeric chromatin (Fig. 5, A and B). An identical phenotype was observed for cells depleted of CENP-N, which results from a partial defect in targeting newly synthesized CENP-A to existing centromeres (Carroll et al., 2009). Together, our data suggest that CENP-C is required for the localization and stability of CCAN proteins, and that CENP-C contributes to the maintenance of centromere identity by ensuring normal levels of CENP-A within centromeric chromatin.
Figure 5.
CENP-C is required for centromere assembly. (A) Western blotting of whole-cell extracts prepared from mock-depleted (control) or CENP-C–depleted HeLa cells. The antibody used for each Western blot is indicated on the left. Background bands (*) were present above and below the CENP-T band, which is indicated with an arrow. The molecular weight of each protein is indicated. (B) Representative images from control or mock-depleted cells stained with Hoechst (DNA) or the indicated antibody. Images are maximum-intensity projections of z stacks collected at 0.2-µM steps. Bars, 5 µM. (C) Quantification of the relative levels at centromeres of the indicated protein in mock- or CENP-C–depleted cells. For each antigen, >20 cells and >200 centromeres were quantified. Error bars indicate the SEM from three independent experiments.
Discussion
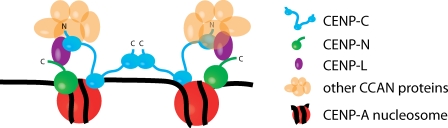
We propose a model for human centromere assembly based on the data presented here and in the existing literature (Fig. 6). A critical first step is the direct recognition of CENP-A chromatin by CENP-C. Because dimerization is important for CENP-C centromere targeting, but not for mononucleosome binding, we speculate that each CENP-C dimer binds to two different, possibly adjacent, CENP-A nucleosomes within centromeric chromatin. A CENP-C dimer in conjunction with centromeric chromatin then provides the foundation upon which the rest of the CCAN is assembled. CENP-N also binds directly to CENP-A nucleosomes, thus providing additional CENP-A nucleosome contacts that reinforce the specificity of the centromere assembly process (Carroll et al., 2009). We discuss aspects of this model and their implications for centromere assembly and structure below.
Figure 6.
A model for centromere assembly in human cells. CENP-A (red), CENP-C (cyan), CENP-N (green), and CENP-L (magenta) nucleosomes are shown. Other CCAN proteins are colored orange. The N and C termini of CENP-C are indicated, as is the C terminus of CENP-N.
Is the interaction of CENP-C with CENP-A nucleosomes conserved in vertebrates?
CENP-C has long been thought to make direct contacts with centromeric chromatin in vivo, but the centromere-specific elements recognized by CENP-C have not previously been defined. We show here that CENP-C binds preferentially to CENP-A nucleosomes over H3 nucleosomes or DNA alone. The region of CENP-C that is necessary and sufficient for CENP-A nucleosome binding corresponds closely to a region of CENP-C previously demonstrated to be sufficient for centromere localization (Yang et al., 1996; Trazzi et al., 2002). Moreover, mutation of R522 within human CENP-C, which reduces the efficiency of centromere localization for CENP-C fragments lacking the C terminus (Song et al., 2002), disrupts CENP-A nucleosome binding in vitro. A mutation in the analogous residue of XlCENP-C (R874) also reduced its targeting to centromeres in Xenopus egg extracts. Together, these data indicate that CENP-A nucleosomes are the chromatin substrate important for CENP-C centromere localization in vertebrates.
A different conclusion was recently reached in chicken DT40 cells (Hori et al., 2008). In that study, immunopurified CENP-C was associated with H3 nucleosomes, but not CENP-A nucleosomes, after the complete digestion of chromatin into mononucleosomes. This result may reflect the evolution of species-specific differences in the chromatin-binding specificity of CENP-C orthologues. An alternative explanation is that the arrays of CENP-A nucleosomes in vertebrate centromeres provide the opportunity for multivalent interactions that could stabilize the specific association of CENP-C oligomers with centromeric chromatin (Zinkowski et al., 1991; Blower et al., 2002; Chueh et al., 2005). It has previously been shown that digestion of chromatin to mononucleosomes disrupts the interactions between CENP-A nucleosomes and CENP-C (Ando et al., 2002). Removing multivalent interactions between CENP-C oligomers and CENP-A nucleosome arrays may allow CENP-C to equilibrate between H3 and CENP-A mononucleosomes present in the extract. The combination of a vast excess of H3 nucleosomes relative to CENP-A nucleosomes and a weak affinity of CENP-C for DNA may explain the apparent association of CENP-C to H3 nucleosomes in this case. A definitive test of the chromatin specificity of chicken CENP-C in vivo would be to isolate the nucleosomal DNAs associated with immunopurified CENP-C and determine their chromosomal origins.
Identification of a novel structural element within human CENP-A nucleosomes that is likely to be important for centromere assembly
Our data show that the C-terminal LEEGLG motif of CENP-A is both necessary and sufficient to specify CENP-C binding to nucleosomes. Given that CENP-C is essential for centromere assembly and function, our data predict that the C-terminal LEEGLG motif of CENP-A will be an essential structural element of centromeric chromatin. Consistent with this possibility, residues C terminal to helix 3 of Cse4 (the budding yeast CENP-A orthologue) are essential for accurate chromosome segregation (Keith et al., 1999). Furthermore, Cse4 can be replaced with a histone chimera whose histone fold domain contains only the regions in Cse4 that correspond to the CATD and C terminus (Black et al., 2007). Interestingly, in human cells, a histone chimera that contains only the CATD region of CENP-A is sufficient to support kinetochore assembly and chromosome segregation after depletion of >90% of the endogenous CENP-A (Black et al., 2007). The levels of CENP-C at centromeres are normal in these cells, which suggests that mechanisms in addition to direct binding of CENP-A nucleosomes may contribute to the localization of CENP-C to centromeres. Alternatively the remaining wild-type CENP-A present within centromeric chromatin in these cells may be sufficient for CENP-C localization, as it has previously been shown that 10% of endogenous CENP-A can support efficient CENP-I localization (Liu et al., 2006). The genetic replacement of CENP-A with histone H3/CENP-A chimeras in vertebrate cell lines will ultimately be required to determine if the C-terminal LEEGLG motif of CENP-A is essential for vertebrate centromere assembly and function (Régnier et al., 2005).
How are distinct CENP-A recognition modules integrated in vivo?
We show here that depletion of human CENP-C with siRNA caused the mislocalization of CENP-H, CENP-I, CENP-K, and CENP-T (Figs. 5 and S5), which presumably also results in the loss of CENP-N from centromeres (McClelland et al., 2007). Furthermore, although other CCAN proteins are not required for CENP-C localization, the levels of CENP-C at centromeres are partially reduced in cells depleted of CENP-N, CENP-I, or CENP-K (Goshima et al., 2003; Cheeseman et al., 2008; Carroll et al., 2009), Thus, the CENP-A nucleosome recognition activities of CENP-C and CENP-N are functionally linked in vivo.
The molecular mechanisms by which CENP-C and CENP-N mutually stimulate each other’s localization to centromeres are unclear. Our in vitro analysis indicates that CENP-N and CENP-C426–537 can bind to the same CENP-A nucleosome in vitro (Fig. 4 D); however, we did not observe any stimulation of CENP-N binding to CENP-A nucleosomes in the presence of saturating amounts of CENP-C426–537. Thus, additional regions of CENP-C and/or other CCAN proteins may be required to couple the binding of CENP-C and CENP-N to CENP-A mononucleosomes. Consistent with this possibility, CENP-L, which binds directly to the C terminus of CENP-N (Carroll et al., 2009), was recently shown to bind to CENP-C in Schizosaccharomyces pombe (Tanaka et al., 2009). Alternatively, the binding to different CENP-A nucleosomes within centromeric chromatin may be required in vivo for CENP-C and CENP-N to enhance one another’s centromere localization. The continued effort to elucidate the architecture of the CCAN and the distribution of CCAN proteins within centromeric chromatin in vivo, coupled with the reconstitution of assemblies with more complex chromatin substrates, will contribute greatly to our understanding of the structure of centromeres.
That all the CCAN proteins tested here required CENP-C for their centromere localization suggests that CENP-C binding to CENP-A nucleosomes is perhaps the dominant mode of centromeric chromatin recognition in human cells. However, data from other model systems indicates that there is plasticity with regard to the role of CENP-C in centromere assembly. In chicken cells, CENP-H, CENP-I, and CENP-K are required for the localization of CENP-C to centromeres in interphase, but the reverse is not true, a situation that is opposite of that found in human cells (Fukagawa et al., 2001a; Nishihashi et al., 2002; Kwon et al., 2007). Similarly, mutations in S. pombe genes encoding Mis15 (CENP-N), Mis17 (CENP-M), Mis6 (CENP-I), and Sim4 (CENP-K) all cause identical phenotypes that suggest they function upstream of the nonessential Cnp3 (CENP-C) in centromere assembly (Takahashi et al., 2000; Pidoux et al., 2003; Hayashi et al., 2004; Tanaka et al., 2009). A common theme between the centromere assembly pathways in these organisms is that the dependency relationships among CCAN proteins are remarkably similar. CENP-N, CENP-H, CENP-I, and CENP-K (and others) characteristically cluster in one epistasis group, whereas CENP-C typically forms its own epistasis group. We suggest that the fundamental difference in centromere assembly in eukaryotic organisms is not the physical connectivity of the individual proteins but rather the relative importance of the CENP-N– and CENP-C–dependent chromatin recognition modules. Interestingly, both Drosophila and Caenorhabditis elegans have clear CENP-C orthologues that are required for centromere function but do not appear to have orthologues of many other CCAN proteins, including CENP-N (Oegema et al., 2001; Heeger et al., 2005). These examples may represent extremes in which one centromere recognition module has become so dominant that the other was ultimately lost.
Materials and methods
Histone expression and nucleosome reconstitution
Expression and purification of soluble CENP-A/H4, CENP-A–H3NTD/H4, H3-CANTD/H4, gCENP-A/H4, H3-CATD/H4, CENP-A–CATD+C, and CENP-A–ERA/H4 tetramers was performed as described previously (Carroll et al., 2009). Histone H3, H4, H3-LEEGLG, H2A, and H2B were expressed, purified, and refolded as described except that soluble H3/H4 and H3-LEEGLG/H4 tetramers and H2A/H2B dimers were refolded independently and frozen on liquid nitrogen for later use (Luger et al., 1999). Nucleosome reconstitution and purification was performed as described using a 186-bp fragment of α-satellite DNA generated by PCR (Luger et al., 1999; Carroll et al., 2009). All the purified nucleosomes used in this study contain similar histone-to-DNA ratios (Fig. S1, B and C). α-Satellite DNA was initially cloned from human genomic DNA by PCR using oligos 5′-CTTGCTAGCAATCTGCAAGTGG-3′ and 5′-CTTGTCGACTACAAAAAGAGTG-3′.
Gel shift nucleosome binding assays
CENP-C or CENP-N were expressed in rabbit reticulocyte extracts in the presence of 10 mCi/ml [35S]methionine (PerkinElmer) using the TnT Quick-Coupled Transcription/Translation system (Promega) according the manufacturer’s instructions. Typical binding reactions (10 µl total volume) contained 1 µl of transcription/translation mix and 300–500 nM of reconstituted nucleosomes in 10 mM Tris-HCl, and 20% glycerol, pH 7.4, unless indicated otherwise. After 30 min at room temperature, each reaction was loaded directly onto a 5% acrylamide gel in 0.5× Tris/Borate/EDTA and run for 80 min at 10 mA. Gels were stained with Coomassie blue, dried, and imaged using a phosphorimager (GE Healthcare) to visualize labeled CENP-N or CENP-C. Alternatively, gels were stained with ethidium bromide (1 µg/ml) or SYBR gold (Invitrogen) to visualize nucleosomes. The CENP-C truncation mutants used for binding assays were constructed by PCR. CENP-C point mutants were made with the QuikChange site-directed mutagenesis kit (Agilent Technologies).
For the gel extraction experiments in Fig. 4 F, nucleosomes were resolved on native gels as and stained with ethidium bromide. The unbound or rCENP-C426–537–bound nucleosomes were then excised and minced with a clean scalpel, and resuspended in 500 µl of isolation buffer (10 mM Tris-HCl, 1 mM EDTA, 0.1% SDS, pH 7.4). After agitating overnight at 4°C, the gel pieces were removed by centrifugation through a low-retention Durapore membrane (Millipore), and the extracted proteins were precipitated with 10% trichloroacetic acid. Precipitates were resuspended, run on a 17.5% polyacrylamide gel, and stained with Coomassie blue or SYPRO ruby (Invitrogen). The intensity of each band was quantified using imageJ software.
Expression and purification of CENP-C426–537
The region of CENP-C encoding amino acids 426–537 was cloned into the Asc1 and Pac1 sites of a modified version of pGex6P-1 (GE Healthcare) for expression of GST–CENP-C426–537 in bacteria. 2L of BL21 cells harboring the GST–CENP-C426–537 expression plasmid were grown in 2× YT at 37°C to an OD600 of ∼0.2, then switched to 23°C until reaching an OD600 of ∼0.6. GST–CENP-C426–537 expression was induced by the addition of IPTG to 0.3 mM for 6 h, after which the cells were harvested and frozen directly on liquid nitrogen. Cell pellets were ground to a fine powder with a mortar and pestle on liquid nitrogen and stored at −80°C. Cell powder was resuspended in 25 ml of lysis buffer (1× PBS, 0.5 M NaCl, 0.2% Triton X-100, 1 mM EDTA, 2mM DTT, 1 mM PMSF, and 25 µg/ml lysozyme), incubated on ice for 30 min, and sonicated. Crude extract was clarified by centrifugation at 100,000 g, and the supernatant was loaded onto a 2-ml glutathione agarose column (Sigma-Aldrich) equilibrated in lysis buffer. The column was then washed with 50 ml of wash buffer (1× PBS, 0.25 M NaCl, 0.1% Triton X-100, and 1 mM DTT) followed by 10 ml of PreScission Protease elution buffer (20 mM Tris-HCl, 200 mM NaCl, 0.5 mM EDTA, and 1 mM DTT, pH 7.4). After washing, the glutathione agarose was resuspended in 2 ml of PreScission Protease elution buffer containing 50 µg of purified GST–PreScission Protease and rocked at 4°C for 12 h. CENP-C426–537 was eluted from the glutathione column, concentrated, and run over a gel filtration column (Sephadex 75; GE Healthcare) in gel filtration buffer (20 mM Hepes and 200 mM NaCl, pH 7.4) to remove any remaining contaminating proteins.
siRNA and immunofluorescence
HeLa cells were grown in DME containing 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen). CENP-C (5′-GCGAAUAGAUUAUCAAGGAUU-3′, 5′-GAACAGAAUCCAUCACAAAUU-3′, 5′-CGAAGUUGAUAGAGGAUGAUU-3′, and 5′-UCAGGAGGAUUCGUGAUUAUU-3′) siRNA pools (Thermo Fisher Scientific) were used according to the manufacturer’s instructions. Mock transfections contained no siRNA. Western blotting and immunofluorescence of HeLa cells was performed as described previously (Carroll et al., 2009). Antibodies to CENP-H and CENP-I were provided by S.-T. Liu (University of Toledo, Toledo, OH). Preparation of Xenopus extracts, immunodepletion, and immunoblotting were performed as described previously (Milks et al., 2009). Images were collected on a microscope (Eclipse 80i; Nikon) with a 60×, 1.4 NA oil immersion lens using MetaMorph software (MDS Analytical Technologies) and a charge-coupled device camera (CoolSnapHQ; Photonics).
Online supplemental material
Fig. S1 shows a characterization of reconstituted nucleosomes used in this study. Fig. S2 shows the domain architecture of CENP-C. Fig. S3 shows that XlCENP-C is efficiently depleted from Xenopus extracts. Fig. S4 shows that recombinant CENP-C426–537 binds nonspecifically to nucleosomes at high concentrations. Fig. S5 shows that CENP-C is required for centromere assembly. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001013/DC1.
Acknowledgments
The authors would like to thank the Straight Laboratory for many thoughtful discussions and comments on the manuscript. We are grateful to Song-Tao Liu for CENP-H and CENP-I antibodies, and DNA 2.0 for gene synthesis.
C.W. Carroll was generously supported by postdoctoral fellowships from the Helen Hay Whitney Foundation and the American Heart Association. K.J. Milk was supported by graduate fellowships from the Department of Defense and the National Science Foundation. A.F. Straight is the Gordon Family Scholar of the Damon Runyon Cancer Research Foundation. This work was supported by National Institutes of Health grant R01GM074728 to A.F. Straight.
Footnotes
Abbreviations used in this paper:
- CATD
- CENP-A centromere-targeting domain
- CCAN
- constitutive centromere-associated network
- CENP
- centromere protein
References
- Amano M., Suzuki A., Hori T., Backer C., Okawa K., Cheeseman I.M., Fukagawa T. 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 186:173–182 10.1083/jcb.200903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando S., Yang H., Nozaki N., Okazaki T., Yoda K. 2002. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 22:2229–2241 10.1128/MCB.22.7.2229-2241.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L., Jr., Cleveland D.W. 2004. Structural determinants for generating centromeric chromatin. Nature. 430:578–582 10.1038/nature02766 [DOI] [PubMed] [Google Scholar]
- Black B.E., Jansen L.E., Maddox P.S., Foltz D.R., Desai A.B., Shah J.V., Cleveland D.W. 2007. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 25:309–322 10.1016/j.molcel.2006.12.018 [DOI] [PubMed] [Google Scholar]
- Blower M.D., Karpen G.H. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3:730–739 10.1038/35087045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D., Sullivan B.A., Karpen G.H. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2:319–330 10.1016/S1534-5807(02)00135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.T. 1995. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 160:111–116 10.1016/0378-1119(95)00163-Z [DOI] [PubMed] [Google Scholar]
- Buchwitz B.J., Ahmad K., Moore L.L., Roth M.B., Henikoff S. 1999. A histone-H3-like protein in C. elegans. Nature. 401:547–548 10.1038/44062 [DOI] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C., Godek K.M., Jansen L.E., Straight A.F. 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11:896–902 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo K.H., Vissel B., Nagy A., Earle E., Kalitsis P. 1991. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 19:1179–1182 10.1093/nar/19.6.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh A.C., Wong L.H., Wong N., Choo K.H. 2005. Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum. Mol. Genet. 14:85–93 10.1093/hmg/ddi008 [DOI] [PubMed] [Google Scholar]
- Cohen R.L., Espelin C.W., De Wulf P., Sorger P.K., Harrison S.C., Simons K.T. 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 19:4480–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., III, Cleveland D.W. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469 [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., Ikemura T. 2001a. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 20:4603–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Regnier V., Ikemura T. 2001b. Creation and characterization of temperature-sensitive CENP-C mutants in vertebrate cells. Nucleic Acids Res. 29:3796–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu T., Yoda K., Yanagida M. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 118:715–729 [DOI] [PubMed] [Google Scholar]
- Heeger S., Leismann O., Schittenhelm R., Schraidt O., Heidmann S., Lehner C.F. 2005. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 19:2041–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Platero J.S., van Steensel B. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA. 97:716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Cleveland D.W. 2009. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10:478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 [DOI] [PubMed] [Google Scholar]
- Howman E.V., Fowler K.J., Newson A.J., Redward S., MacDonald A.C., Kalitsis P., Choo K.H. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA. 97:1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K. 2006. Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 11:673–684 [DOI] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K., Salmon E.D. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19:694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith K.C., Baker R.E., Chen Y., Harris K., Stoler S., Fitzgerald-Hayes M. 1999. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell. Biol. 19:6130–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., Fukagawa T. 2007. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 18:2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanini L., McKeon F. 1995. Domains required for CENP-C assembly at the kinetochore. Mol. Biol. Cell. 6:1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T., Rattner J.B., Jablonski S.A., Yen T.J. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53 10.1083/jcb.200606020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 389:251–260 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Luger K., Rechsteiner T.J., Richmond T.J. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304:3–19 10.1016/S0076-6879(99)04003-3 [DOI] [PubMed] [Google Scholar]
- Marshall O.J., Chueh A.C., Wong L.H., Choo K.H. 2008. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82:261–282 10.1016/j.ajhg.2007.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S.E., Borusu S., Amaro A.C., Winter J.R., Belwal M., McAinsh A.D., Meraldi P. 2007. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 26:5033–5047 10.1038/sj.emboj.7601927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Koshland D. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 6:793–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Yang P., Glowczewski L., Koshland D., Smith M.M. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 94:607–613 10.1016/S0092-8674(00)81602-5 [DOI] [PubMed] [Google Scholar]
- Milks K.J., Moree B., Straight A.F. 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell. 20:4246–4255 10.1091/mbc.E09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihashi A., Haraguchi T., Hiraoka Y., Ikemura T., Regnier V., Dodson H., Earnshaw W.C., Fukagawa T. 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2:463–476 10.1016/S1534-5807(02)00144-2 [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A.A. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., III, Desai A., Fukagawa T. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Palmer D.K., O’Day K., Wener M.H., Andrews B.S., Margolis R.L. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805–815 10.1083/jcb.104.4.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D.K., O’Day K., Trong H.L., Charbonneau H., Margolis R.L. 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA. 88:3734–3738 10.1073/pnas.88.9.3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L., Richardson W., Allshire R.C. 2003. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 161:295–307 10.1083/jcb.200212110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi V., Perini G., Trazzi S., Pliss A., Raska I., Earnshaw W.C., Della Valle G. 2002. CENP-C binds the alpha-satellite DNA in vivo at specific centromere domains. J. Cell Sci. 115:2317–2327 [DOI] [PubMed] [Google Scholar]
- Régnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., Earnshaw W., Brown W. 2005. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25:3967–3981 10.1128/MCB.25.10.3967-3981.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C.A., Ratrie H., III, Maurer M., Rothfield N.F., Earnshaw W.C. 1992. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 70:115–125 10.1016/0092-8674(92)90538-N [DOI] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Gronemeyer B., Lu W., Eugster E., Tomkiel J.E. 2002. Mutational analysis of the central centromere targeting domain of human centromere protein C, (CENP-C). Exp. Cell Res. 275:81–91 10.1006/excr.2002.5495 [DOI] [PubMed] [Google Scholar]
- Stoler S., Keith K.C., Curnick K.E., Fitzgerald-Hayes M. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573–586 10.1101/gad.9.5.573 [DOI] [PubMed] [Google Scholar]
- Sugata N., Li S., Earnshaw W.C., Yen T.J., Yoda K., Masumoto H., Munekata E., Warburton P.E., Todokoro K. 2000. Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere—kinetochore complexes. Hum. Mol. Genet. 9:2919–2926 10.1093/hmg/9.19.2919 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Yata H., Muro Y., Himeno M. 1994. Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J. Biochem. 116:877–881 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Kuriyama K., Shibata A., Himeno M. 1997. Characterization of internal DNA-binding and C-terminal dimerization domains of human centromere/kinetochore autoantigen CENP-C in vitro: role of DNA-binding and self-associating activities in kinetochore organization. Chromosome Res. 5:132–141 10.1023/A:1018422325569 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Chen E.S., Yanagida M. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 288:2215–2219 10.1126/science.288.5474.2215 [DOI] [PubMed] [Google Scholar]
- Talbert P.B., Bryson T.D., Henikoff S. 2004. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 3:18 10.1186/jbiol11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Chang H.L., Kagami A., Watanabe Y. 2009. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell. 17:334–343 10.1016/j.devcel.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Thompson S.L., Bakhoum S.F., Compton D.A. 2010. Mechanisms of chromosomal instability. Curr. Biol. 20:R285–R295 10.1016/j.cub.2010.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S., Bernardoni R., Diolaiti D., Politi V., Earnshaw W.C., Perini G., Della Valle G. 2002. In vivo functional dissection of human inner kinetochore protein CENP-C. J. Struct. Biol. 140:39–48 10.1016/S1047-8477(02)00506-3 [DOI] [PubMed] [Google Scholar]
- Trazzi S., Perini G., Bernardoni R., Zoli M., Reese J.C., Musacchio A., Della Valle G. 2009. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS One. 4:e5832 10.1371/journal.pone.0005832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voullaire L.E., Slater H.R., Petrovic V., Choo K.H. 1993. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am. J. Hum. Genet. 52:1153–1163 [PMC free article] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.H., Tomkiel J., Saitoh H., Johnson D.H., Earnshaw W.C. 1996. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 16:3576–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski R.P., Meyne J., Brinkley B.R. 1991. The centromere-kinetochore complex: a repeat subunit model. J. Cell Biol. 113:1091–1110 10.1083/jcb.113.5.1091 [DOI] [PMC free article] [PubMed] [Google Scholar]