Abstract
The expression patterns of many protein-coding genes are orchestrated in response to exogenous stimuli, as well as cell-type-specific developmental programs. In recent years, researchers have shown that dynamic chromatin movements and interactions in the nucleus play a crucial role in gene regulation. In this review, we highlight our current understanding of the organization of chromatin in the interphase nucleus and the impact of chromatin dynamics on gene expression. We also discuss the current state of knowledge with regard to the localization of active and inactive genes within the three-dimensional nuclear space. Furthermore, we address recent findings that demonstrate the movements of chromosomal regions and genomic loci in association with changes in transcriptional activity. Finally, we discuss the role of intra-and interchromosomal interactions in the control of coregulated genes.
Keywords: nuclear organization, gene positioning, gene expression, chromosome territory
INTRODUCTION
Nuclear structures appear to be self-organizing and, unlike cytoplasmic organelles, are not enclosed by membranes (for review, see Reference 78). Therefore, they lack a physical compartmentalization of individual cellular functions and enzymatic activities. Even more surprising and enigmatic is the complex regulation of diverse functions such as cell-type-specific regulation of gene expression, DNA damage repair, DNA replication, and the maintenance of nuclear architecture upon successive cell divisions.
In interphase nuclei, each chromosome occupies a distinct territory (for review, see Reference 27). Increasing evidence indicates that the organization of chromatin in the nucleus is not static. Rather, dynamic rearrangements and repositioning relative to nuclear structures and other chromosomal loci on the levels of bulk chromatin, single chromosomes, and individual genes appear to be involved in the regulation of gene expression and differentiation. In this review, we highlight recent advances toward our understanding of the role of chromatin dynamics in these and other nuclear processes.
CHROMATIN DYNAMICS IN INTERPHASE NUCLEI
The dynamics of chromatin has long been of interest to geneticists and cell biologists. For example, the question of whether chromosome rearrangements observed during the pairing of meiotic homologs in maize and Neurospora require a special motile machinery, or whether they move by diffusion, was first raised by Barbara McClintock in 1945 (77). The development of fluorescent live cell imaging techniques in recent decades has allowed for chromatin dynamics to be studied in the live cells of a wide range of organisms from yeasts to insects and mammals.
When bulk chromatin and centromeres were studied on the scale of the whole nucleus, excluding apparent curvilinear chromosome movements that have been attributed to nuclear rotation, they appeared to be essentially stationary (31, 89, 105). Experiments in which bulk chromatin in Chinese hamster cells was irradiated with UV light, and the damaged region was tracked over time, also indicated that chromosomal regions exert no significant mobility (26). The mobility of bulk chromatin was also studied by fluorescence recovery after photobleaching (FRAP) experiments in Swiss 3T3 and HeLa cells (1). The photobleached area of the nucleus in these experiments was essentially immobile over distances greater than 0.4 µm and observation times greater than 1 h (1). However, chromatin movements might occur on a smaller scale and thus be inaccessible by such photobleaching experiments. Therefore, Sedat and colleagues applied submicrometer single-particle tracking of a fluorescently labeled yeast chromosomal region near the centromere of chromosome III (76). This study found that chromatin is free to undergo substantial Brownian diffusion but that a given chromatin segment is confined to a nuclear subregion. To rule out that nuclear rotation contributes to the apparent chromatin dynamics, the positions of two chromosomal loci relative to each other were measured in three dimensions. These measurements revealed a diffusion constant of yeast centromeric chromatin of D = 5 × 10−12 cm2 s−1 and a confinement radius of R = 0.3 µm, which is much smaller than the diploid yeast nucleus (∼2−3 µm diameter; 76). Interestingly, centromeric chromatin mobility was independent of active metabolism, with an almost identical diffusion constant upon a sodium-azide-induced block of cellular respiration (D = 3 × 10−12 cm2 s−1, R = 0.25 µm; 76). These findings indicate that for yeast centromeric chromatin, (a) mobility is independent of active processes, primarily caused by Brownian motion, and (b) the region is confined to a relatively small nuclear volume. Surprisingly, a 15-kb plasmid showed a mobility and confinement radius similar to centromeric chromatin (D = 3 × 10−12 cm2 s−1, R = 0.25 µm; 76). Because a smaller molecule should show a higher diffusion constant, the authors concluded that the mobility of the plasmid or bulk chromatin experiences similar constraints, presumably due to the attachment to an immobile nuclear structure. Interestingly, upon microtubule depolymerization by nocodazole treatment in yeast, chromatin was much less confined, which indicates that microtubules might play a role in the confinement of diffusion (76).
The diffusion constant of yeast centromeric chromatin is much lower than that of DNA in solution, which is on the order of 10−9 to 10−8 cm2 s−1 for bacteriophage DNA that is 4 to 300 kb in length (107). The diffusion of DNA in concentrated solutions (semidilute) has diffusion coefficients on the order of 10−9 to 10−8 cm2 s−1 (98, 119), which is approximately four orders of magnitude higher than that of chromatin. Similarly, the dynamics of DNA is lower by several orders of magnitude than the diffusion constant of proteins, with diffusion constants of 10−6 cm2 s−1 for proteins in aqueous solution and 10−8 to 10−7 cm2 s−1 in the cytoplasm (69). Marshall and colleagues compared their measured chromatin diffusion constant with the expected value from theoretical considerations of polymer dynamics and determined that chromatin is three orders of magnitude less mobile than expected. They concluded that crowding, entanglement, and/or association with immobile structures are, at least in part, responsible for the low mobility of chromatin.
Conversely, noncentromeric chromatin in yeast is highly mobile and microtubule independent, and its dynamics are cell cycle dependent (48). More specifically, early and late origins of DNA replication move rapidly at 0.5 µm s−1 in G1 phase. This dynamic appears to be energy dependent and is thus unlikely to result from simple Brownian diffusion (48). In addition to small-scale movements (≤0.2 µm), occasional larger displacements (≥0.5 µm) were detected during observation periods of 10 s (48). Interestingly, larger displacements ≥0.5 µm are five times less likely in early G1 phase daughter nuclei as compared to mother cell nuclei (48). Because daughter nuclei are 40% smaller than mother cell nuclei, this implies that nuclear volume and crowding effects might play a role in chromatin dynamics. The chromatin movements decrease in S-phase through a mechanism that involves DNA replication (48). Whether this is a direct effect of a change in chromatin attachment to nuclear structures during DNA replication remains to be determined. The confinement radius of 0.5 µm corresponds to approximately one-quarter of the diameter of a yeast nucleus and equals a distance of approximately 100 kb on the basis of a linear compaction ratio of ∼70-fold (43). Such relatively large motions are likely to involve a substantial intermingling of intra- and interchromosomal regions. In contrast to internal chromosomal regions, telomeric and centromeric chromatin are significantly more constrained in both G1 and S-phase in a replication-independent manner (48). This may be caused by yeast centromeres that appear to attach in a microtubule-dependent manner to the spindle pole body, whereas yeast telomeres associate with the nuclear envelope through the yeast yKu70/80 heterodimer or the silent regulatory 4 (Sir4) protein (46, 113).
In Drosophila embryos, a 359-bp heterochromatic repeat on the X-chromosome labeled with fluorescently tagged topoisomerase II exhibited low mobility, with a diffusion constant of D = 2.0 × 10−11 cm2 s−1 and a relatively large confinement radius of R = 0.9 µm (76). It has not yet been determined how much of the data obtained from Drosophila embryos can be directly compared to mammalian chromatin because Drosophila embryos have a unique, highly nonrandom radial and axial positioning of chromosomes in the nucleus. In this Rabl configuration, chromosomes display a polarized arrangement with telomeres at one pole of the nucleus and centromeres at the opposite pole. Interestingly, the Rabl configuration is lost at distinct stages of the cell cycle and during development (30, 33, 118). The structural basis for this organization is likely to be an association with the nuclear envelope of approximately 15 chromosomal regions per chromosome arm (75). These associations are likely to decrease chromatin dynamics in Drosophila embryos. In fact, in developing Drosophila spermatocytes that lack a Rabl configuration, chromatin has a high degree of local and long-range dynamics that is restricted to the size of a chromosome territory (124). In pre-meiotic mid-G2 spermatocyte nuclei, chromatin moves by constrained Brownian diffusion. Chromatin motion involved two components: (a) a short-range component over a 1–2 s timescale with an apparent diffusion constant of ≥1.3 × 10−2 µm2 s−1 and a confinement radius of R ≤ 0.3 µm and (b) a slower long-range movement over longer time intervals (10 min) with a 13 times smaller diffusion constant of 1 × 10−3 µm2 s−1 and a confinement radius of R = 0.6 µm (124). Such behavior indicates that chromatin mobility is severely constrained, and the entire confinement region is explored during the sampling interval. Interestingly, chromatin in mature spermatocytes in late G2 phase have an approximately 4 times lower short-range mobility (D = 3.4 × 10−3 µm2 s−1, R = 0.15 µm) and a 10 times lower long-range mobility (D = 9.4 × 10−5 µm2 s−1, R = 0.2 µm) compared with mid-G2 spermatocytes (124). These changes of chromatin dynamics during the cell cycle suggest the involvement of specific or nonspecific interactions of chromatin with other (immobile) components, such as the nuclear envelope or internal nuclear structures. How these differences in chromatin mobility reflect the spermatocyte’s progression towards meiosis remains to be determined. Chromatin movement does not appear to be directed or completely random. Rather, a large movement in one direction was often followed by a movement in the opposite direction, consistent with the random walk-on-a-chain model (48, 124).
In HeLa cells, centromeres are dispersed throughout the nucleus in interphase. This pattern is established by a nearly isometric expansion upon chromatin decondensation in telophase and early G1 (105). Therefore, the position of each centromere in early G1 is determined mainly by its position on the metaphase plate (105). Most interphase centromeres are immobile, although individuals or a small group of centromeres occasionally moved at slow rates of 7–10 µm h−1 (105). Using particle tracking by high-resolution two-photon microscopy with 20 nm and 30 ms spatial and temporal resolution, respectively, green fluorescent protein (GFP)-tagged Lac repressor protein that binds to a Lac operator repeat cassette was analyzed in interphase CHO cells (62). Interestingly, periods of rapid constrained diffusion of the labeled chromosomal locus were interrupted by relatively abrupt jumps of approximately 150 nm that lasted 0.3–2 s (62). The confinement radius of the rapid motion is ∼40 nm, which is on the order of the 30-nm fiber and could thus reflect local oscillations in chromatin folding (62). Two labeled chromosomal loci that are < 1–2 µm apart did not show correlated jumps, which indicates that these are local events that are not transmitted over longer distances through the chromatin fiber or the subnuclear region (62). Additionally, these movements were abolished by ATP depletion and lower temperature, which suggests an active mode of movement rather than passive Brownian diffusion (62). With a linker distance of ∼30 nm between nucleosomes in the beads-on-a-string structure (123), the reported jumps would account for the unfolding of one turn of the 30-nm chromatin fiber (6 nucleosomes). The authors argue that such spontaneous unfolding of chromatin is kinetically highly unlikely, which supports their proposed energy-driven mechanism (62). We currently know little about interphase chromatin packaging at or above the 30-nm fiber (7). The study above raises several intriguing questions:
What is the origin and function of these jumps? Are they related to transcription, and does transcriptional activity influence jump distance and/or frequency?
Do all chromosomal loci follow the same kinetics? One might predict that chromatin more closely associated with the nuclear envelope has less freedom for these motions, whereas genes at the surface of chromosome territories might jump farther and/or more frequently.
Are there specific (immobile) attachment sites that determine jump distance, and what are they?
In summary, bulk chromatin dynamics in the nucleus is several orders of magnitude slower than the diffusive mobility of DNA in solution. The diffusion of chromatin is likely to be influenced by various factors such as steric hindrance between large and possibly highly entangled chromatin regions, the degree of compaction of the chromatin fiber, the rigidity and density of the surrounding nucleoplasm, the attachment of chromatin to proteinaceous nuclear structures or possibly a yet ill-defined nonchromatin nuclear scaffold referred to as the nuclear matrix (90), the attachment of chromatin to subnuclear structures such as the nuclear envelope or the nucleolus, structural RNAs, or any combination of these. Variations in the density of such attachment sites might account for the observed differences in chromatin mobility during the cell cycle and cellular development (124). During interphase, there seems to be no major chromosomal rearrangement or translational mobility on the scale of individual chromosomes. However, chromatin dynamics appears to be fast enough to allow for intrachromosomal interactions, such as the association of an enhancer and a promoter in cis or in trans, over less than 1 µm distance within seconds (51, 68). Furthermore, long-range chromatin movements observed in Drosophila and mammalian cells suggest that individual chromosomal regions can, at least to some degree, invade neighboring chromosome territories (13, 30). Thus far, such long-range interactions have only rarely been observed in live mammalian cells. However, this may be caused by the limited number of loci examined by live cell imaging.
CHROMOSOME TERRITORIES
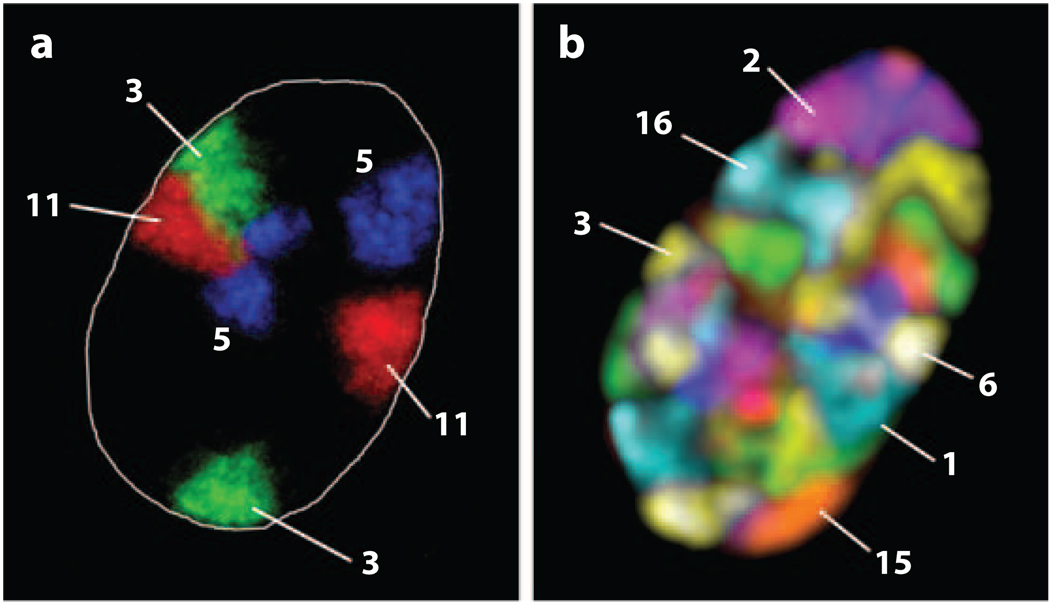
The idea that chromosomes, visible during mitosis, occupy distinct regions in the inter-phase nucleus when they are not as condensed was first suggested by Carl Rabl in 1885 (93) and later refined in 1909 by Theodor Boveri, who coined the term chromosome territory (11). Theoretical considerations and indirect evidence during the following eight decades (for a detailed review, see References 24 and 25) led to the first indirect visualization of chromosome territories, by the clustering on metaphase chromosomes, of laser UV-microirradiated interphase chromatin (26, 136). Advancements in in situ hybridization techniques allowed the visualization of distinct, nonoverlapping chromosome territories in interspecies somatic hybrid cells (74, 99). The territorial organization of chromosomes in nonhybrid cells was demonstrated by whole chromosome fluorescence in situ hybridization (chromosome painting) of mammalian cells (28, 36, 64, 65, 92) and in plants (9, 61, 71) (Figure 1). In live cells, chromosome territories were visualized by incorporating labeled nucleotide analogs during S-phase and following the segregation of labeled and nonlabeled sister chromatids during several rounds of mitosis (10, 73, 134). Live cell analysis of such labeled chromosome regions in neuroblastoma or HeLa cells revealed only small Brownian diffusion-like dynamics with diffusion constants on the order of D = 10−11 cm2 s−1 during an observation period of several hours (10, 35). However, chromatin dynamics might depend on the cell type, the observation period, and the differentiation state of the cell. For instance, fluorescently labeled chromatin domains in neuronal interphase cells exhibited significant motion by nuclear rotation (31). Similarly, gross chromosomal movements were observed in Drosophila larval CNS cells, with an increased separation of distal positions of a long chromosome arm at the onset of S-phase (30). In Drosophila imaginal discs, chromatin movement is more constrained in differentiated cells than in undifferentiated cells (diffusion constant D = 2.1 × 10−12 cm2 s−1 and 3.3 × 10−12 cm2 s−1, respectively), which suggests a link between a more restricted gene expression profile and the confinement of chromatin to a smaller nuclear space (117).
Figure 1.
Projections of mid-optical sections of human fibroblast nuclei that highlight chromosome territories. Three (a) and all 23 (b) pairs of chromosomes were detected using 3D-FISH with chromosome paint probes obtained by flow-sorting. Individual chromosomes are indicated. Image in panel a courtesy of Irina Solovei. Image in panel b courtesy of Andreas Bolzer and Irina Solovei, University of Munich, Germany.
Whether the spatial order of interphase chromosomes is propagated through mitosis has been quite controversial. Initial FRAP analysis indicated that global chromosome positions are transmitted from mother to daughter cells (45). However, further analysis of smaller chromosomal regions indicated that the exact positions of chromatin were not transmitted (120, 127). In fact, increased chromatin dynamics was observed in early G1 cells (127). Chromosomal positions in adult human fibroblasts with diploid or triploid karyotype or trisomy 21 may be established early in development and retained in prometaphase rosettes and through mitosis (84). Conversely, chromosome positions in the prometaphase rosette in normal human diploid embryonic fibroblasts were relatively random (4). However, the position of a chromosome on the metaphase plate carried through anaphase into telophase (4). Chromosomal positions were described to be cell type specific, similar to the positions of human chromosomes 3, 7, 8, 13, 17, 21, X, and Y, which were found to be different in Sertoli cells versus blood lymphocytes by whole chromosome painting (21). Human chromosome territory positions have also been reported to be size dependent, with smaller chromosomes generally located toward the interior of the nucleus and larger chromosomes at the periphery (112).
The positions of human chromosomes 18 and 19, which are similar in DNA content with 85 Mb and 67 Mb, respectively, have been examined in the nuclei of several cell lines (29). The gene-poor chromosome 18, with low CpG island density, late replicating DNA, and little hyperacetylated histone H4, localized to more peripheral sites; whereas the gene-rich chromosome 19, with a high density of CpG islands, early replicating DNA, and abundant hyperacetylated H4, localized preferentially to the nuclear interior (29). The nuclear positions of these chromosomes appeared to be established early in the cell cycle and were maintained throughout interphase (29). The chromosomal arrangements of chromosomes 18 and 19 have been conserved between humans and New World monkey species (116), as well as Old World monkeys (115). Similarly, in chicken cell nuclei, chromosomes occupy highly consistent radial chromatin arrangements with gene-rich, early-replicating microchromo-somes in the nuclear interior, and gene-poor, late-replicating macrochromosomes at the periphery (114). Interestingly, the chicken macrochromosomes 2 and Z contain genes homologous to human chromosome 18, whereas the chicken microchromosomes correspond to human chromosome 19. Therefore, researchers have proposed that despite profound changes in the karyotype, the radial positioning of chromosomes has been conserved over 300 million years of evolution between chickens and humans (114). The correlation between gene density and the nuclear position of chromosomes has been confirmed with other chromosomes in diploid human lymphoblasts and primary fibroblasts (12). Intriguingly, the rod cells of the retinas of nocturnal, but not diurnal, mammals have recently been shown to possess an inverted chromatin organization, with euchromatin as well as nascent transcripts and the splicing machinery lining the nuclear periphery while heterochromatin is localized to an internal nuclear region (108).
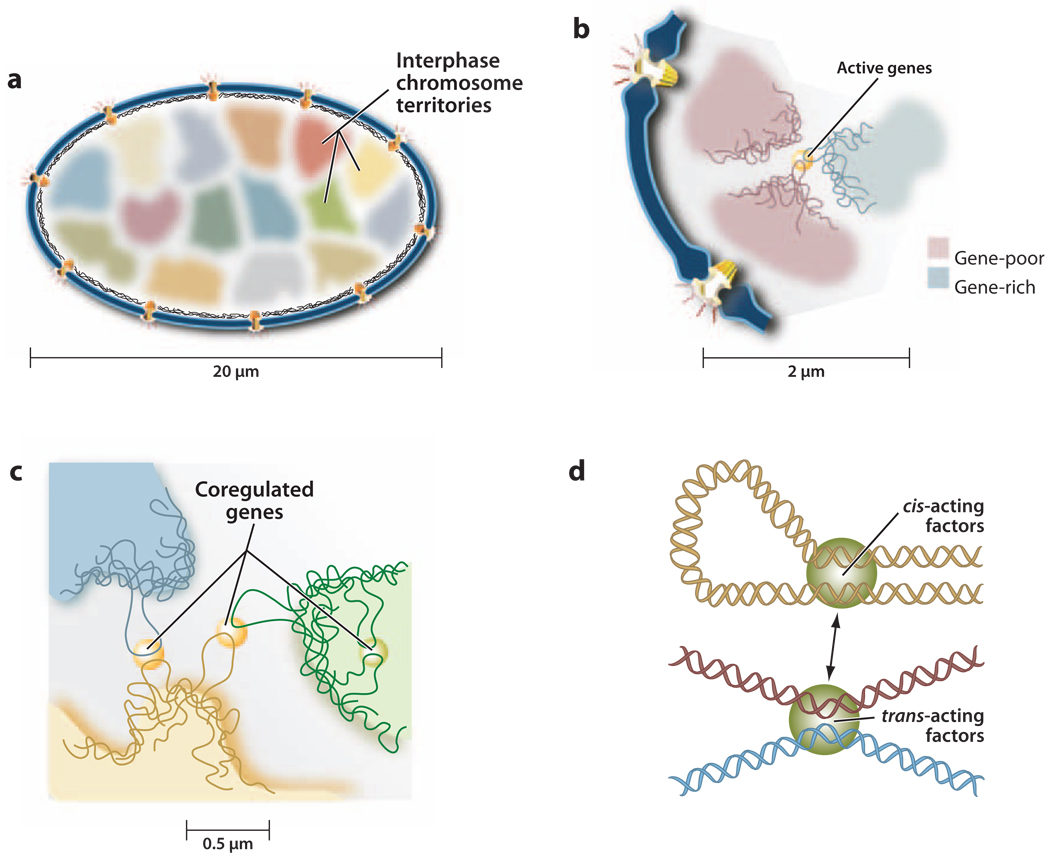
On the basis of the nuclear distribution of DNA hypersensitive sites at the nuclear periphery, active genes transcribed by RNA polymerase II (RNA Pol II) were originally suggested to have a preferentially peripheral localization (52, 56, 88). However, the direct visualization of incorporated 3H-uridine or Bromo-UTP (for review, see Reference 109), or of proteins involved in transcription and RNA processing, has not revealed a predominant localization at the nuclear periphery (17, 53, 128). Rather, RNA transcripts are concentrated at the surface of chromosome territories, and snRNPs involved in pre-mRNA splicing locate predominantly outside of chromosome territories (135). Interestingly, protein coding genes, irrespective of their transcriptional activity, localize predominantly at the periphery of chromosome territories, whereas noncoding sequences assume more interior positions or are randomly positioned in the territory (59). It was therefore proposed that genes preferentially localize to the surface of chromosome territories where they are accessible to transcription factors and splicing components that reside in the interchromatin domain (ICD) between territories (23, 125, 135) (Figure 2a,b). However, invaginations within chromosome territories likely also make internal regions of the chromosome accessible to the transcription machinery. Evidence for a role of transcription in the frequency of intermingling of chromosome territories has come from high-resolution studies in human lymphoblasts (13). Importantly, the degree of intermingling correlated with the frequency of chromosome translocations, which implies that DNA double strand breaks formed within an intermingling region are more likely to cause interchromosomal rearrangements (13).
Figure 2.
(a) Chromosomes are organized into territories in the interphase nucleus. (b) Gene-rich chromosomes assume more interior positions in the nucleus, whereas gene-poor chromosomes are more peripheral. (c) Actively transcribed genes tend to locate at the surface of chromosome territories. Coregulated genes can form intra- and interchromosomal contacts and colocalize with foci rich in proteins involved in transcription and splicing. (d) Intra- and interchromosomal associations play a role in the regulation of gene expression by bringing enhancers in contact with protein coding genes.
The discovery of chromosome territories leads to several intriguing questions:
What is the basis for chromosome territory formation and (self-) organization?
Is the nuclear position, and the position relative to each other, an inherent property of chromosomes, cell types and tissues?
Is chromosome position a cause or consequence of their gene expression state?
The development of new high-resolution imaging techniques and high-throughput chromatin-interaction networks will help us to address these questions in the near future.
DYNAMICS WITHIN CHROMOSOME TERRITORIES
It was widely accepted that the nuclear periphery is a generally repressive compartment and harbors mainly gene-poor chromosome regions in the yeast Saccharomyces cerevisiae (for review, see Reference 3). Surprisingly, a genome-wide analysis in S. cerevisiae revealed that several highly transcribed genes associate with nuclear pore proteins and that GAL genes relocate to the nuclear periphery upon transcriptional activation (19). Similarly, the INO1 gene relocates to the nuclear periphery when activated (14). The nuclear pore basket structure may play a role in the gene activation process in yeast because it binds to the promoter region of genes early during transcriptional upregulation (100). The gene gating hypothesis proposed by Blobel in 1985 might help explain the localization of some activated genes to the nuclear periphery (8). According to this hypothesis, a subset of transcribed genes associates with nuclear pores, which facilitates the export of RNPs from the nucleus into the cytoplasm.
Thus far, it has not been conclusively determined whether the transcriptional activity of a gene is the cause or the consequence of, or is independent of, its location within a chromosome territory or within the three-dimensional nuclear space. Interesting insight comes from a study of the nuclear localization of the X-chromosome-linked adenine nucleotide translocase genes ANT2 and ANT3 in human female amniotic fluid cells (34). ANT3 escapes X-chromosome inactivation, and both transcriptionally active alleles locate at the exterior of the two homologous X-chromosome territories. In contrast, the transcriptionally silent ANT2 allele on the inactive X-chromosome (Xi) locates to the interior of the chromosome territory, whereas the expressed ANT2 allele on the active X-chromosome (Xa) locates to the periphery of the chromosome territory (34). On human chromosome 6, large chromatin loops that contain several megabases of DNA protruded outwards from the surface of the chromosome territory (126). The frequency with which a genomic region was observed at the periphery of the territory was cell type specific and appeared to be related to the number of active genes in that region (126). Importantly, the upregulation by interferon-gamma of the major histocompatibility class II complex (MHC II) on chromosome 6 led to an increase in the frequency with which the gene cluster was found on a protruding chromatin loop (13, 126). Similarly, retinoic acid–induced differentiation of mouse embryonic stem cells resulted in the decondensation and relocalization of activated Hoxb genes away from their chromosome 11 territory (20). Conversely, in the case of Hoxd genes, decondensed alleles can be found in the interior of a chromosome territory, whereas looped-out gene loci can still be condensed, indicating that looping and chromatin decondensation might not be causally linked (80).
During B and T lymphocyte development in mouse, large-scale spatial reorganization of the nucleus and genome may play a role in maintaining differentiation-induced heritable gene silencing (15, 16). In particular, several lymphoid genes move close to centromeric heterochromatin clusters upon transcriptional shutdown (15). Because this relocation appears temporally delayed due to transcriptional shutdown, it is likely to be the consequence rather than the cause of gene silencing (15). Conversely, the transcriptional activation of the human β-globin locus involves movement of the locus control region (LCR) away from the centromeric heterochromatin (102). Therefore, this relocation may be required to achieve histone H3-H4 hyperacetylation and an open chromatin structure of the locus but is not sufficient for high-level transcription (102). Similarly, during murine B lymphocyte development, Igh and Igκ loci preferentially locate at the nuclear periphery in hematopoietic progenitors and pro T-cells, but they assume more central positions in pro-B nuclei (55). On the basis of these studies, large-scale chromatin rearrangement and repositioning during lymphocyte differentiation may play a role in transcriptional regulation, replication timing, and the recombination of Igh and Igκ loci (55, 132). Similarly, the T-helper-cell cytokines IFN-γ on chromosome 10 and the regulatory regions of the TH2 locus on chromosome 11 associate in trans in naïve T-cells but less frequently in differentiated TH1 or TH2 cells (111). Furthermore, the beta-globin locus undergoes a relocalization during mouse erythroid differentiation, in which the locus progressively moves away from the nuclear periphery (94). The LCR-dependent association of the beta-globin locus with transcriptionally engaged RNA Pol II foci may be the driving force for locus movement (94). Interestingly, transcriptional activation at the periphery precedes locus movement (94). This indicates that the nuclear periphery is not a totally repressive compartment in mammalian cells (3, 104). In fact, the nuclear periphery may be divided into subdomains associated with active or silent gene regions (for review, see Reference 58).
When functionally unrelated neighboring genes on chromosome 7 in several human cell types were studied, active genes preferentially located with euchromatin in the nuclear interior, whereas inactive genes located mainly to perinuclear heterochromatin at the nuclear periphery (133). Upon activation, the cystic fibrosis transmembrane conductance regulator (CFTR) gene on chromosome 7 moved from a peripheral position toward the nuclear interior (133). The Mash1 locus, which is repressed and located at the nuclear periphery in mouse embryonic stem cells, was shown to move toward the nuclear interior when it becomes activated during neuronal differentiation (129). Interestingly, the same group found that the nuclear periphery is not a universally repressive compartment because the IFNγ gene remains at the periphery upon transcriptional induction during T-helper cell differentiation (49).
The effect of transcriptional induction on the nuclear position of single genetic loci has been studied in live cells by using transgenes whose integration sites can be visualized with fluorescent proteins. In Chinese hamster ovary (CHO) cells, a transgene that contains a Lac operator (LacO) repeat cassette that preferentially localizes at the nuclear periphery relocated predominantly to the nuclear interior when constitutively activated by the VP16 acidic activation domain (122). In another study, a transcriptionally activated locus exerted intermittent directional movement toward the nuclear interior at velocities of 0.1–0.9 µm min−1 over distances of 1–5 µm in an actin- and/or myosin I-dependent fashion (22). On the other hand, two other inducible loci in baby hamster kidney (BHK) cells, or on human chromosome 1p36 in U2OS cells, did not significantly change their nuclear position upon transcriptional upregulation (54, 121). Interestingly, this human locus is integrated near a telomere that may serve an anchoring function. Upon induction, a significant chromatin decondensation, as well as the recruitment of RNA Pol II, splicing factors, and histone H3.3, were visualized (54). This indicates that transcriptional induction and chromatin decondensation can be independent of intranuclear localization and are likely locus and/or chromatin context specific. Further evidence that localization to the nuclear periphery does not preclude transcription came from a study in which an inducible reporter gene was tethered to the nuclear periphery via a Lac repressor-lamin B fusion. The locus targeted to the nuclear lamina was inducible with kinetics similar to the untargeted locus (57). Interestingly, the targeting of the locus to the lamina required passage through mitosis (57), which indicates that a gross chromatin reorganization might require chromosome condensation and nuclear breakdown and reformation. In a similar approach, an emerin fusion protein was used to target an active gene to the nuclear lamina in a process that also involved mitosis (95). Interestingly, this study showed a silencing of the reporter gene, as well as neighboring genes (95). Additionally, Lac repressor-mediated tethering of portions of human chromosomes 4 and 11 to the nuclear membrane protein Lap2β led to the silencing of some genes located near the tethering site or even further away, whereas many other genes were not affected (37). The differences between these results might be caused by the different targeting approaches to potentially different microdomains at the nuclear periphery. Furthermore, the nuclear periphery might play different roles in the transcriptional activation of a silent gene or the maintenance of ongoing transcription.
In some cases, DNA motifs have been implicated in transcription-dependent gene positioning. For instance, a functional enhancer antagonized the transcriptional silencing of a transgene by preventing its localization close to centromeric heterochromatin (39). In contrast, the Gypsi chromatin insulator element caused sequences from separate chromosomal loci to colocalize in insulator bodies at the nuclear periphery and thereby regulate gene expression (44). In S. cerevisiae, gene recruitment sequences (GRS) in the promoter region of the INO1 gene are sufficient to target an internal locus to the nuclear periphery and confer physical contact with the nuclear pore complex, which is required for full induction of the INO1 gene (2).
In addition to a role in transcription, the nuclear periphery has recently been shown to play a role in DNA repair in budding yeast. The Msp3 protein that spans the inner nuclear membrane sequestered persistent double strand DNA breaks (DSBs), as well as telomeres, to the nuclear periphery (87, 101). Additionally, irreparable DSBs were recruited to the nuclear periphery through association with the nucleo-porin Nup 84 and the nuclear-pore-associated SUMO ligase Slx5/Slx8 (82).
INTRA- AND INTERCHROMOSOMAL GENE ASSOCIATIONS
Several scenarios have been suggested with regard to how the transcription machinery and target genes meet up in the nucleoplasm. On the one hand, transcription factors have been shown to move throughout the nucleoplasm by diffusion, thereby gaining accessibility to target loci (91). On the other hand, it has been suggested that, rather than recruiting and assembling transcription complexes, genes can migrate to preassembled foci termed transcription factories (18, 85). These foci exist in the absence of transcription, but transcription initiation is necessary to tether distal genes to shared foci (79). Genes on different chromosomes such as the Myc proto-oncogene on chromosome 15 and the Igh locus on chromosome 12 in mouse B lymphocytes colocalize in the same transcription factory, which suggests a high mobility of individual chromosomal regions (86) (Figure 2c). However, the direct movement of genes to transcription factories has not been demonstrated in living cells.
Intra- and interchromosomal interactions have also been studied by chromosome confirmation capture (3C) (32) and fluorescent in situ hybridization (FISH) techniques. Upon transcriptional activation by the female steroid hormone estradiol, the estrogen receptor alpha (ERα) target genes TFF1 (pS2) and GREB1 colocalize in foci rich in transcription factors and splicing components (51). This interchromosomal interaction depends on the nuclear actin/myosin-I machinery and dynein light chain 1 (51). Interestingly, the histone H3 lysine 4 demethylase LSD1 is essential for estradiol-dependent gene activation (42) and is involved in the association of the target genes with interchromatin granules but not their interchromosomal interaction (51). It remains to be elucidated if the TFF1 and GREB1 genes show a directed movement toward a pre-formed transcription factory where coregulated genes associate or if transcription complexes are recruited to the site where these coregulated genes become associated. Similarly, the Hoxb1 locus shows increased intra- and interchromosomal interactions upon transcriptional activation and looping-out of its chromosome territory during mouse embryonic stem cell differentiation (130). Recently, the 3D architecture of the human genome has been investigated using a variation of 3C, referred to as Hi-C. Using this methodology, Lieberman-Aiden et al. (66) reported that chromatin conformation at the megabase scale is consistent with a fractal globule where interactions are more likely within a compartment than between compartments.
Long-distance chromosomal interactions in cis and in trans (Figure 2d) have been described for the transcriptional regulation of olfactory receptor genes, which are the largest gene family in mammals. The regulatory 2.1-kb H-element on chromosome 14 associates with only one of approximately 1300 odorant receptor (OR) genes on different chromosomes. This leads to the monoallelic and mutually exclusive expression of one OR gene per olfactory neuron (68, 103). However, the H-element is probably not the only element that confers gene activation to OR genes because the deletion of the H-element in mice has a graded effect on gene expression with distance from the H-element in cis and had no effect on OR gene expression in trans (40).
Intrachromosomal interactions also play a role in gene imprinting in mouse. The LCR of the imprinted insulin-like growth factor Igf/H19 locus on chromosome 7 interacts with the upstream differentially methylated region 1 (DMR1) of the Igf2 gene, as well as an intergenic region flanked by the Wsb1 and Nf1 genes of chromosome 11 and a chromosome 6 locus (67, 81). The association requires CCCTC-binding factor (CTCF), which recruits Polycomb repressive complex 2 through its interaction with Suz12 (63). Interestingly, the LCR association is specific for the maternal Igf2/H19 locus and the paternal Wsb1/Nf1 locus, despite biallelic expression of Wsb1 and Nf1 genes (67). The association of the LCR with the Wsb1/Nf1 region occurred in approximately 40% of the cells studied (67). It remains to be elucidated why this interaction is not detected in all cells and if the interaction is cell cycle dependent. Furthermore, it is unknown if the loci interact directly or if they only colocalize with the same nuclear substructure.
The pairing of homologous chromosomes in somatic cells has been observed in plants, as well as in Drosophila and other dipterans (for review, see References 47 and 50). In contrast, chromosome pairing occurs infrequently and is presumably a random event in human cells. A few exceptions exist: In human T-lymphocytes, a spatial and temporal association of the maternal and paternal chromosome 15q11-q13 region has been observed during S-phase (60). Interestingly, this region contains an imprinted region that contains the Prader-Willi syndrome (PWS) and the Angelman syndrome (AS) loci. Cells from PWS and AS patients lack the association of homologous loci, and therefore the mutual recognition and pairing of chromosome 15 may be involved in the imprinting process (60).
Female mammalian cells randomly inactivate one of the X-chromosomes in an inheritable manner early in development to equalize the dosage of X-linked genes. At the onset of X-chromosome inactivation, two homologous X-chromosome inactivation centers (XICs) transiently colocalize (5, 6). The non-coding Xist RNA is upregulated on one of the X-chromosomes, coats the entire chromosome, and acts as a transcriptional repressor in cis (6). It is not yet known if and how Xist RNA is involved in the association of XICs. Interestingly, the inactive X-chromosome contacts the per-inucleolar region in 80%–90% of mid-to-late S-phase cells in an Xist RNA-dependent manner (131). Therefore, the inactive X-chromosome may need to visit the perinucleolar region repeatedly to maintain gene silencing (131). However, Xist RNA is not the only noncoding RNA involved in gene silencing. The 91.5-kb Kcnq1ot1 RNA is involved in the imprinting of the Kcnq1 locus (38, 72), and the antisense Igf2r RNA (Airn) plays a role in the imprinting of the Igf2r locus (4, 70, 83). It will be interesting to find out if these noncoding RNAs play a role similar to Xist in the pairing of homologous loci to induce monoallelic gene silencing.
Intra- and interchromosomal interactions are not restricted to genes that are transcriptionally coregulated. For example, during V(D)J recombination in developing B- and T-cells, the distant variable (V) and (diversity)-joining constant (D)JC gene segments colocalize in a stochastic manner to facilitate chromosomal rearrangements, which lead to a diverse antigen receptor repertoire (for review, see References 41, 55, 96, 97, and 110). Similarly, the T-cell receptor β (Tcrb) and T-cell receptor αδ (Tcra-Tcrd) loci undergo long-range interactions by locus contraction and looping of the 655-kb Trcb locus (106). Interestingly, once recombination is complete, the loci separate, and the interaction is no longer observed in subsequent developmental stages, which probably prevents further Vβ−DJβ recombination (96, 106).
CONCLUSIONS
It has been a little over a century since chromosomes have been postulated to localize in distinct territories in interphase nuclei. Over the past three decades, we have gained significant insight, with increasing spatial and temporal resolution into the dynamics of bulk chromatin, chromosomal loci, and individual genes. Nevertheless, some essential questions have not been conclusively answered yet: Is there a subnuclear structure that serves as a scaffold for the organization of the genetic material, or are chromosomes self-organizing? If so, what is the basis of this self-organization, and how are complex processes such as transcription, DNA replication, DNA repair, and cell division regulated in regard to nuclear position? Do genes move to specialized compartments for efficient transcription, and do coregulated genes colocalize in these structures? Are changes in nuclear localization a cause or a consequence of gene activation? Currently, the development and utilization of high-resolution microscopes and high-throughput methods to detect gene interactions are vastly expanding our knowledge of the role of gene position and dynamics in the regulation of gene expression. Research over the next decade is certain to provide many unexpected surprises in regard to these and other outstanding questions about how the genome functions within the context of the cell nucleus.
SUMMARY POINTS
Chromosomes occupy distinct territories in the interphase nucleus.
Chromatin movements are generally on the scale of 0.3–0.5 µm.
Chromatin position is not precisely transmitted through mitosis.
Gene-poor chromosomes tend to be on the nuclear periphery, and gene-rich chromosomes localize to more internal nuclear regions.
Both silent and active genes can be found at the nuclear periphery.
Examples of chromatin movement have been documented in regard to both transcriptional induction and gene silencing.
Intra- and interchromosomal gene associations have been shown to influence gene expression.
FUTURE ISSUES
What is the nuclear organization of chromatin, and what role does it play in transcriptional regulation?
What are the dynamic processes of chromatin at higher spatial and temporal resolution?
Is the position of chromosome territories and chromosomal loci a cause or a consequence of their transcriptional activity?
Are individual chromosome territories organized by a scaffold-type structure?
Is the colocalization of coregulated genes a common process, and how is their motion accomplished?
ACKNOWLEDGMENTS
We thank Irina Solovei, Andreas Bolzer, and Thomas Cremer (University of Munich, Germany) for the images in Figure 1, and James Duffy (CSHL) for drawing Figure 2. Research in the Spector laboratory is supported by NIGMS 42694, NIH/EY 18244, and 5PO1CA013106–38. M.R.H. received fellowships from the European Molecular Biology Organization (EMBO; ALTF 160-2005) and the German Academic Exchange Service (DAAD).
Glossary
- FRAP
fluorescence recovery after photobleaching
- Chromosome territory
interphase equivalent of a chromosome
- GFP
green fluorescent protein
- RNA Pol II
RNA polymerase II
- LCR
locus control region
- Transcription factory
nuclear foci rich in Ser5-phosphorylated RNA Pol II
- FISH
fluorescent in situ hybridization
- XIC
X-chromosome inactivation center
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Michael R. Hübner, Email: huebner@cshl.edu.
David L. Spector, Email: spector@cshl.edu.
LITERATURE CITED
- 1.Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J. Cell Biol. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, et al. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 4.Allison DC, Nestor AL. Evidence for a relatively random array of human chromosomes on the mitotic ring. J. Cell Biol. 1999;145:1–14. doi: 10.1083/jcb.145.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Augui S, Filion GJ, Huart S, Nora E, Guggiari M, et al. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- 6. Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365.Demonstrates the transient Xist noncoding RNA-dependent colocalization of X-chromosomes at the onset of X inactivation.
- 7.Belmont AS, Dietzel S, Nye AC, Strukov YG, Tumbar T. Large-scale chromatin structure and function. Curr. Opin. Cell Biol. 1999;11:307–311. doi: 10.1016/S0955-0674(99)80041-6. [DOI] [PubMed] [Google Scholar]
- 8.Blobel G. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157.Presents a quantitative 3D map of all human chromosome territories.
- 10.Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C. Quantitative motion analysis of sub-chromosomal foci in living cells using four-dimensional microscopy. Biophys. J. 1999;77:2871–2886. doi: 10.1016/S0006-3495(99)77119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boveri T. Die Blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenin-dividualität. Arch. Zellforsch. 1909;3:181–268. [Google Scholar]
- 12.Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 13.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 16.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 17.Carmo-Fonseca M, Tollervey D, Pepperkok R, Barabino SM, Merdes A, et al. Mammalian nuclei contain foci which are highly enriched in components of the premRNA splicing machinery. EMBO J. 1991;10:195–206. doi: 10.1002/j.1460-2075.1991.tb07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter DRF, Eskiw C, Cook PR. Transcription factories. Biochem. Soc. Trans. 2008;36:585–589. doi: 10.1042/BST0360585. [DOI] [PubMed] [Google Scholar]
- 19. Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9.Provides evidence for the association of actively transcribed genes with the nuclear pore in yeast.
- 20.Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- 21.Chandley AC, Speed RM, Leitch AR. Different distributions of homologous chromosomes in adult human Sertoli cells and in lymphocytes signify nuclear differentiation. J. Cell Sci. 1996;109:773–776. doi: 10.1242/jcs.109.4.773. [DOI] [PubMed] [Google Scholar]
- 22.Chuang C-H, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr. Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 23.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 24.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. Eur. J. Histochem. 2006;50:161–176. [PubMed] [Google Scholar]
- 25.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur. J. Histochem. 2006;50:223–272. [PubMed] [Google Scholar]
- 26.Cremer T, Cremer C, Baumann H, Luedtke EK, Sperling K, et al. Rabl’s model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Hum. Genet. 1982;60:46–56. doi: 10.1007/BF00281263. [DOI] [PubMed] [Google Scholar]
- 27.Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories—a functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Cremer T, Lichter P, Borden J, Ward DC, Manuelidis L. Detection of chromosome aberrations in metaphase and interphase tumor cells by in situ hybridization using chromosome-specific library probes. Hum. Genet. 1988;80:235–246. doi: 10.1007/BF01790091. [DOI] [PubMed] [Google Scholar]
- 29. Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119.Describes the relationship between gene density of a chromosome and its nuclear position.
- 30.Csink AK, Henikoff S. Large-scale chromosomal movements during interphase progression in Drosophila. J. Cell Biol. 1998;143:13–22. doi: 10.1083/jcb.143.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Boni U, Mintz AH. Curvilinear, three-dimensional motion of chromatin domains and nucleoli in neuronal interphase nuclei. Science. 1986;234:863–866. doi: 10.1126/science.3775367. [DOI] [PubMed] [Google Scholar]
- 32.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 33.Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, et al. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 34.Dietzel S, Schiebel K, Little G, Edelmann P, Rappold GA, et al. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity. Exp. Cell Res. 1999;252:363–375. doi: 10.1006/excr.1999.4635. [DOI] [PubMed] [Google Scholar]
- 35.Edelmann P, Bornfleth H, Zink D, Cremer T, Cremer C. Morphology and dynamics of chromosome territories in living cells. Biochim. Biophys. Acta. 2001;1551:M29–M39. doi: 10.1016/s0304-419x(01)00023-3. [DOI] [PubMed] [Google Scholar]
- 36.Ferguson M, Ward DC. Cell cycle dependent chromosomal movement in premitotic human T-lymphocyte nuclei. Chromosoma. 1992;101:557–665. doi: 10.1007/BF00660315. [DOI] [PubMed] [Google Scholar]
- 37.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 39.Francastel C, Walters MC, Groudine M, Martin DIK. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centromeric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- 40.Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Bassets I, Kwon Y-S, Telese F, Prefontaine GG, Hutt KR, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gasser SM. Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- 44.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 45.Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 46.Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- 47.Henikoff S. Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr. Opin. Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- 48. Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366.Provides a quantitative analysis of chromatin dynamics during the cell cycle in yeast.
- 49.Hewitt SL, High FA, Reiner SL, Fisher AG, Merkenschlager M. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur. J. Immunol. 2004;34:3604–3613. doi: 10.1002/eji.200425469. [DOI] [PubMed] [Google Scholar]
- 50.Hilliker AJ, Appels R. The arrangement of interphase chromosomes: structural and functional aspects. Exp. Cell Res. 1989;185:267–318. doi: 10.1016/0014-4827(89)90301-7. [DOI] [PubMed] [Google Scholar]
- 51.Hu Q, Kwon Y-S, Nunez E, Cardamone MD, Hutt KR, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc. Natl. Acad. Sci. USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hutchison N, Weintraub H. Localization of DNAase I-sensitive sequences to specific regions of interphase nuclei. Cell. 1985;43:471–482. doi: 10.1016/0092-8674(85)90177-1. [DOI] [PubMed] [Google Scholar]
- 53.Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 56.Krystosek A, Puck TT. The spatial distribution of exposed nuclear DNA in normal, cancer, and reverse-transformed cells. Proc. Natl. Acad. Sci. USA. 1990;87:6560–6564. doi: 10.1073/pnas.87.17.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060.Demonstrates that mitosis is involved in the repositioning of genomic loci.
- 58.Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132:929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurz A, Lampel S, Nickolenko JE, Bradl J, Benner A, et al. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- 61.Leitch AR, Mosgoller W, Schwarzacher T, Bennett MD, Heslop-Harrison JS. Genomic in situ hybridization to sectioned nuclei shows chromosome domains in grass hybrids. J. Cell Sci. 1990;95:335–341. doi: 10.1242/jcs.95.3.335. [DOI] [PubMed] [Google Scholar]
- 62. Levi V, Ruan Q, Plutz M, Belmont AS, Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 2005;89:4275–4285. doi: 10.1529/biophysj.105.066670.Describes chromatin dynamics at high spatial and temporal resolution and describes periods of constrained diffusion interrupted by abrupt jumps.
- 63.Li T, Hu JF, Qiu X, Ling J, Chen H, et al. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- 65.Lichter P, Cremer T, Tang CJ, Watkins PC, Manuelidis L, Ward DC. Rapid detection of human chromosome 21 aberrations by in situ hybridization. Proc. Natl. Acad. Sci. USA. 1988;85:9664–9668. doi: 10.1073/pnas.85.24.9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 68.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 69.Luby-Phelps K, Lanni F, Taylor DL. Behavior of a fluorescent analogue of calmodulin in living 3T3 cells. J. Cell Biol. 1985;101:1245–1256. doi: 10.1083/jcb.101.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, et al. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- 71.Lysak MA, Fransz PF, Ali HB, Schubert I. Chromosome painting in Arabidopsis thaliana. Plant J. 2001;28:689–697. doi: 10.1046/j.1365-313x.2001.01194.x. [DOI] [PubMed] [Google Scholar]
- 72.Mancini-DiNardo D, Steele SJS, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manders EM, Kimura H, Cook PR. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J. Cell Biol. 1999;144:813–821. doi: 10.1083/jcb.144.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manuelidis L. Individual interphase chromosome domains revealed by in situ hybridization. Hum. Genet. 1985;71:288–293. doi: 10.1007/BF00388453. [DOI] [PubMed] [Google Scholar]
- 75.Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol. Biol. Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- 77.McClintock B. Cytogenetic studies of maize and Neurospora. Annu. Rep. Carnegie Inst. Wash. 1945;44:108–112. [Google Scholar]
- 78.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 79.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- 81.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 82.Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 84.Nagele RG, Freeman T, Fazekas J, Lee KM, Thomson Z, Lee HY. Chromosome spatial order in human cells: evidence for early origin and faithful propagation. Chromosoma. 1998;107:330–338. doi: 10.1007/s004120050315. [DOI] [PubMed] [Google Scholar]
- 85.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 86.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park PC, De Boni U. Transposition of DNase hypersensitive chromatin to the nuclear periphery coincides temporally with nerve growth factor-induced up-regulation of gene expression in PC12 cells. Proc. Natl. Acad. Sci. USA. 1996;93:11646–11651. doi: 10.1073/pnas.93.21.11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parvinen M, Soderstrom KO. Chromosome rotation and formation of synapsis. Nature. 1976;260:534–535. doi: 10.1038/260534a0. [DOI] [PubMed] [Google Scholar]
- 90.Pederson T. Half a century of “the nuclear matrix”. Mol. Biol. Cell. 2000;11:799–805. doi: 10.1091/mbc.11.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, et al. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, et al. Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc. Natl. Acad. Sci. USA. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rabl C. Über die Zelltheilung. Morphol. Jahrb. 1885;10:214–330. [Google Scholar]
- 94. Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506.Shows that the LCR of the beta-globin locus changes its nuclear position upon transcriptional activation.
- 95. Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727.Shows that targeting to the nuclear periphery can result in gene silencing.
- 96.Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, et al. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sayegh CE, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scalettar BA, Hearst JE, Klein MP. FRAP and FCS studies of self-diffusion and mutual diffusion in entangled DNA solutions. Macromolecules. 1989;22:4550–4559. [Google Scholar]
- 99.Schardin M, Cremer T, Hager HD, Lang M. Specific staining of human chromosomes in Chinese hamster × man hybrid cell lines demonstrates interphase chromosome territories. Hum. Genet. 1985;71:281–287. doi: 10.1007/BF00388452. [DOI] [PubMed] [Google Scholar]
- 100.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 101.Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schubeler D, Francastel C, Cimbora DM, Reik A, Martin DI, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 103.Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, et al. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 104.Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat. Struct. Mol. Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 105.Shelby RD, Hahn KM, Sullivan KF. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat. Immunol. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 107.Smith DE, Perkins TT, Chu S. Dynamical scaling of DNA diffusion coefficients. Macromolecules. 1996;29:1372–1373. [Google Scholar]
- 108.Solovei I, Kreysing M, Lanctôt C, Kösem S, Peichl L, et al. Nuclear architecture of rod photore-ceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 109.Spector DL. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- 110.Spicuglia S, Franchini DM, Ferrier P. Regulation of V(D)J recombination. Curr. Opin. Immunol. 2006;18:158–163. doi: 10.1016/j.coi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 111.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 112.Sun HB, Shen J, Yokota H. Size-dependent positioning of human chromosomes in interphase nuclei. Biophys. J. 2000;79:184–190. doi: 10.1016/S0006-3495(00)76282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 2004;23:1301–1312. doi: 10.1038/sj.emboj.7600144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tanabe H, Habermann FA, Solovei I, Cremer M, Cremer T. Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat. Res. 2002;504:37–45. doi: 10.1016/s0027-5107(02)00077-5. [DOI] [PubMed] [Google Scholar]
- 115.Tanabe H, Kupper K, Ishida T, Neusser M, Mizusawa H. Inter- and intraspecific gene-density-correlated radial chromosome territory arrangements are conserved in Old World monkeys. Cytogenet. Genome Res. 2005;108:255–261. doi: 10.1159/000080824. [DOI] [PubMed] [Google Scholar]
- 116.Tanabe H, Müller S, Neusser M, von Hase J, Calcagno E, et al. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. USA. 2002;99:4424–4429. doi: 10.1073/pnas.072618599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thakar R, Csink AK. Changing chromatin dynamics and nuclear organization during differentiation in Drosophila larval tissue. J. Cell Sci. 2005;118:951–960. doi: 10.1242/jcs.01684. [DOI] [PubMed] [Google Scholar]
- 118.Thakar R, Gordon G, Csink AK. Dynamics and anchoring of heterochromatic loci during development. J. Cell Sci. 2006;119:4165–4175. doi: 10.1242/jcs.03183. [DOI] [PubMed] [Google Scholar]
- 119.Thomas JC, Allison SA, Schurr JM, Holder RD. Dynamic light scattering studies of internal motions in DNA. II. Clean viral DNAs. Biopolymers. 1980;19:1451–1474. doi: 10.1002/bip.1980.360190804. [DOI] [PubMed] [Google Scholar]
- 120.Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr. Biol. 2004;14:166–172. doi: 10.1016/j.cub.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 121.Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat. Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- 122.Tumbar T, Belmont AS. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat. Cell Biol. 2001;3:134–139. doi: 10.1038/35055033. [DOI] [PubMed] [Google Scholar]
- 123.van Holde K, Zlatanova J. What determines the folding of the chromatin fiber? Proc. Natl. Acad. Sci. USA. 1996;93:10548–10555. doi: 10.1073/pnas.93.20.10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Vazquez J, Belmont AS, Sedat JW. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 2001;11:1227–1239. doi: 10.1016/s0960-9822(01)00390-6.Describes cell-cycle-dependent changes in chromatin dynamics in Drosophila.
- 125.Verschure PJ, Van Der Kraan I, Manders EMM, van Driel R. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 1999;147:13–24. doi: 10.1083/jcb.147.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000;113:1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- 127.Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 2003;160:685–697. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wansink DG, Schul W, Van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Williams RRE, Azuara V, Perry P, Sauer S, Dvorkina M, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 130.Wurtele H, Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res. 2006;14:477–495. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- 131.Zhang L-F, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 132.Zhou J, Ermakova OV, Riblet R, Birshtein BK, Schildkraut CL. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol. Cell. Biol. 2002;22:4876–4889. doi: 10.1128/MCB.22.13.4876-4889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zink D, Cremer T, Saffrich R, Fischer R, Trendelenburg MF, et al. Structure and dynamics of human interphase chromosome territories in vivo. Hum. Genet. 1998;102:241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]
- 135.Zirbel RM, Mathieu UR, Kurz A, Cremer T, Lichter P. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1993;1:93–106. doi: 10.1007/BF00710032. [DOI] [PubMed] [Google Scholar]
- 136.Zorn C, Cremer T, Cremer C, Zimmer J. Laser UV microirradiation of interphase nuclei and post-treatment with caffeine. A new approach to establish the arrangement of interphase chromosomes. Hum. Genet. 1976;35:83–89. doi: 10.1007/BF00295622. [DOI] [PubMed] [Google Scholar]