Abstract
The Snail transcription factor is a repressor and a master regulator of epithelial-mesenchymal transition events (EMT) in normal embryonic development and during tumor metastases. Snail directly regulates genes affecting cell adhesion, motility and polarity. Invasive tumor cells express high levels of Snail and it is a marker for aggressive disease and poor prognosis. Transcriptional repression and EMT induction by Snail requires binding to its obligate corepressor, the LIM protein Ajuba. It is unclear how this complex is assembled and maintained on Snail target genes. Here we define functional 14-3-3 binding motifs in Snail and Ajuba which selectively bind 14-3-3 protein isoforms. In Snail, a NH2-terminal motif in the repression domain cooperates with a COOH-terminal, high affinity motif for binding to 14-3-3 proteins. Coordinate mutation of both motifs abolishes 14-3-3 binding and inhibits Snail-mediated gene repression and EMT differentiation. Snail, 14-3-3 proteins, and Ajuba form a ternary complex which is readily detected via ChIP at the endogenous E-cadherin promoter. Collectively, these data show that 14-3-3 proteins are new components of the Snail transcriptional repression machinery and mediate its important biological functions.
Keywords: Snail, 14-3-3, Ajuba, epithelial-mesenchymal transition, transcriptional repression
Introduction
During tumor progression and metastases, EMT is characterized by down-regulation of E-cadherin and other cell adhesion molecules, which facilitates cell motility [1-2]. The Snail transcription factor is a master regulator of EMT during both normal embryonic development and tumor metastases [3-5]. Genetic deletion of the Snail gene in mice causes embryonic lethality due to defects in gastrulation and in the normal EMT process required for generation of the mesodermal layer [4]. Ectopic expression of Snail in epithelial cells induces the EMT transdifferentiation event accompanied by increased migration, motility and invasiveness [6-7]. Snail expression in immortalized human mammary epithelial cells results in the acquisition of mesenchymal traits and properties of stem cells [8]. In mice, Snail is spontaneously up-regulated during tumor recurrence in the mammary gland and high Snail expression strongly predicts decreased relapse-free survival in women with breast cancer [9]. Taken together, these studies demonstrate that Snail may play critical roles in both tumor metastases and recurrence.
The Snail protein contains four COOH-terminal tandem C2-H2 zinc finger motifs and a NH2-terminal SNAG (Snail/Slug, and Gfi-1) repression domain [10-11]. The DNA target sequence for zinc finger binding is the E-box, usually found in tandem in Snail target genes. The SNAG domain recruits the LIM domain protein Ajuba, which acts as an obligate corepressor for Snail mediated repression [12-13]. The Ajuba protein in turn recruits the protein arginine methyltranferase 5 (Prmt5) to form a ternary complex of Snail/Ajuba/Prmt5 which can be found at multiple Snail target genes [14]. The biochemical mechanisms which govern assembly and function of this complex are still largely unknown. In an effort to further define components of the Snail signaling system, we identified two putative 14-3-3 binding sites within the Snail protein.
The 14-3-3 proteins are a family of 28–33-kDa acidic polypeptides which are ubiquitously expressed in all eukaryotic organisms [15-17]. This family of proteins is highly conserved and consists of seven members in mammals denoted β, γ, ε, σ, ζ, τ and η, each encoded by a distinct gene. The 14-3-3 proteins form homo- or hetero-dimers, which appear to function as scaffolds for assembly of multi-protein complexes for a diverse array of proteins. These targets include transcription factors, enzymes, cytoskeletal proteins, signal transducers, apoptosis factors and histones [16-17]. Proteins which bind the 14-3-3 family contain the motifs RSXpS/TXP and/or RXXXpS/TXP (where pS/T represents phospho-serine or threonine, and X represents any amino acid residue except cysteine). Binding of 14-3-3 dimers to its partners can induce conformational changes in protein ligands that may alter their stability, catalytic activity, subcellular localization, protein-protein interactions and/or DNA binding [15-17]. Not surprisingly, the 14-3-3 proteins play crucial roles in diverse processes such as cell cycle regulation, DNA repair, apoptosis, cell differentiation, senescence and cell adhesion. Recently, 14-3-3 proteins have been shown to bind a phosphorylated form of histone H3, suggesting that 14-3-3 proteins also play a role in epigenetic chromatin remodeling [18-20]. Here, we report that the biological and transcriptional repression functions of Snail are dependent on 14-3-3 binding.
Materials and Methods
Plasmids, cell culture, transfections and luciferease assays
The pMEX-myc-Ajuba and pCMV-HA-Snail, pcDNA-Flag-14-3-3, and pGL2-E-cadherin luciferase reporter (E-cad-Luc) plasmids have been described [13-14, 21]. The pLU-Flag-Snail and mutants were constructed via PCR-based cloning [14]. The Sport6-CMV- Snail (murine) and pKLO1-shAjuba (human) plasmids were purchased from Open Biosystems.
MCF7 cells and 293 cells were maintained in DMEM containing 10% FBS, 2 mM L-glutamine, and Penicillin (50 U/ml)/Streptomycin (50 μg/ml) at 37° C under 5% CO2 in a humidified chamber. MCF10A mammary epithelial cells were maintained in DMEM/F12 containing 5% horse serum, EGF (10ng/ml), Hydrocortisone (0.5μg/ml), Cholera toxin (100ng/ml), Insulin (10 μg/ml), and Penicillin (50 U/ml)/Streptomycin (50 μg/ml). Transfection and luciferase assays in 293 cells were performed as described [14].
The stable MCF10A-siAjuba and MCF10A-siLuc cells were established via a viral infection and puromycin selection. The MCF10A cells stably expressing Snail and its mutants were established via a viral infection. The Stable 293-Flag-Snail and 293-vector cells were previously described [14].
Immunoprecipitation, western blot, and antibodies, chromatin immunoprecipitation (ChIP)
Plasmids encoding Myc-Ajuba, HA-Snail and Flag-14-3-3 proteins were transiently transfected into 293 cells and 24 hours post-transfection, whole cell extracts were prepared as described [14]. These extracts were precleared with protein A/G beads and co-immunoprecipiation (co-IP) assays were performed with either α-Myc or α-HA antibodies. For endogenous co-IP assays, nuclear extracts were prepared from 293-Flag-Snail and 293-vector cells [14]. Mouse monoclonal α-Myc (Zymed), α-Flag (Sigma), α-HA (Santa Cruz), α-Vimentin (Sigma), rabbit polyclonal α-14-3-3 (Millipore) and α-Fibronectin (Santa Cruz) antibodies were purchased. The rabbit polyclonal α-Ajuba serum was previously described [14].
To prepare cells for ChIP, the MCF10A cells stably expressing Snail and its mutants were grown in 150 mm plates to 70-90% confluency and were processed as described [14]. The immunoprecipitated DNAs were detected and quantified with primer sets which amplify the DNA fragements flanking the known E-boxes in the E-cadherin promoter [22].
Transwell cell migration assays
The MCF10A cells expressing a GFP marker were sorted using FACS and serum-starved for 4 hours. The cells were harvested and resuspended in DMEM/F12 medium. Suspensions containing 1×105 cells were applied to 8-μm pore transwell filters (Corning). Then, DMEM/F12 medium containing 5% horse serum was added to the bottom chamber. After overnight incubation, the migrated cells at the bottom of the filter were fixed with 70% ethanol and cells in each chamber were quantified by counting three fields under 10× magnification using a fluorescent microscope. Each condition was performed in triplicate and the average number of cells per field is represented. Each Experiment was repeated twice.
Results
Snail contains two putative 14-3-3 protein binding motifs and selectively interacts with 14-3-3 protein isoforms
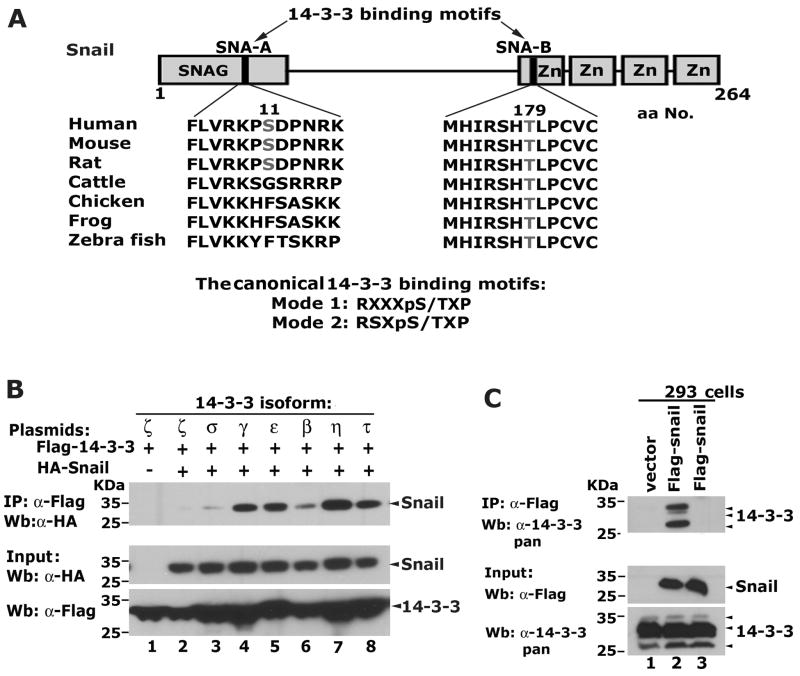
A comprehensive bioinformatic analysis of the Snail amino acid sequence revealed two putative 14-3-3 protein binding motifs within Snail designated SNA-A and SNA-B (Figure 1A). SNA-A is located in the SNAG repression domain, is found in human and rodent, but not in cattle, chicken, frog or zebrafish. In contrast, SNA-B is located in the first zinc-finger of Snail, is highly conserved and is found in all species compared.
Figure 1.
Snail selectively interacts with 14-3-3 isoforms. (A) Snail contains conserved 14-3-3 binding motifs. The putative 14-3-3 binding motifs are designated SNA-A and SNA-B. The numbers correspond to the locations of the core serine and threonine residues within the 14-3-3 binding motifs of Snail. (B) Snail interacts with 14-3-3 isoforms when co-expressed in 293 cells. (C) Snail interacts with endogenous 14-3-3 proteins in 293 cells. Co-IP assays were carried out with the α-Flag antibody (lanes 1, 2), and a normal mouse IgG (Lane 3). Western blot was performed with a pan reactive antibody broadly recognizing endogenous 14-3-3 isoforms.
To determine if Snail interacts with 14-3-3 proteins, plasmids encoding HA- tagged Snail and Flag-tagged 14-3-3 proteins were transiently co-expressed in 293 cells followed by co-IP and western blot assays. Snail robustly interacted with 14-3-3γ, ε, τ, η isoforms, weakly associated with β and showed no interaction with 14-3-3 σ and ζ (Figure 1B). Snail also interacted with endogenous 14-3-3 proteins as shown via co-IP and western blot using a pan 14-3-3 reactive antibody (Figure 1C). These data demonstrate that Snail can strongly and selectively interact with 14-3-3 isoforms.
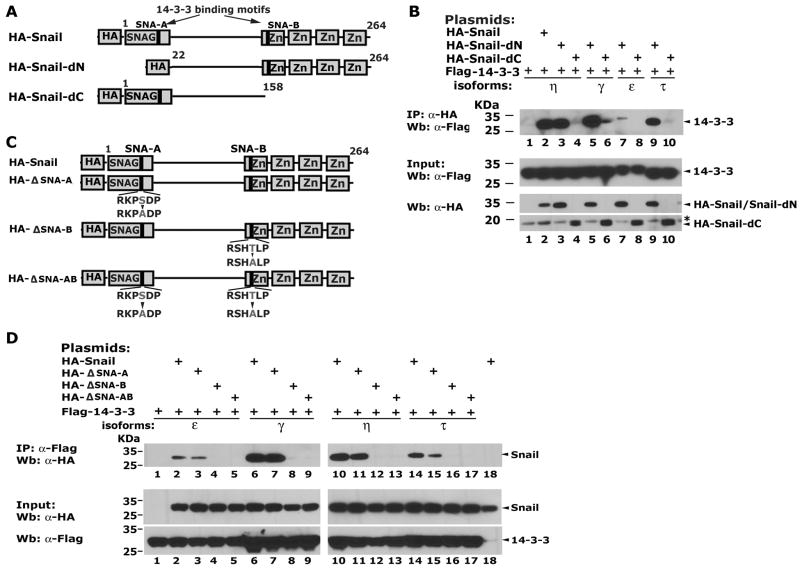
Both of the 14-3-3 binding motifs in Snail are required for maximal interaction with 14-3-3 proteins
The Snail-14-3-3 interaction was mapped using truncated forms of Snail: HA-Snail-dN (deletion of the SNAG domain) and HA-Snail-dC (deletion of the zinc finger region). Deletion of the SNAG domain (HA-Snail-dN) did not strongly affect 14-3-3 η binding (Figure 2B, lane 2 and 3), while HA-Snail-dC failed to bind all four 14-3-3 isoforms (Figure 2B) suggesting that the 14-3-3 binding motif located in the zinc finger region of Snail is most important for binding under these conditions. These motifs were mutated to alanines individually or in combination in the full-length protein (Figure 2C). The core serine in the NH2-terminal motif (ΔSNA-A) and the threonine in the COOH-terminal motif (ΔSNA-B) were targeted. Mutation of the NH2-terminal motif SNA-A slightly decreased its interaction with 14-3-3 isoforms (Figure 2D, lanes 3, 7, 11and 15). Strikingly, the single T179A substitution in the COOH motif SNA-B completely abolished the Snail-14-3-3 interaction (Figure 2D). Thus, the NH2-terminal motif shows weak binding activity alone. The COOH-terminal motif is the dominant site and is required for 14-3-3 protein binding. The full-length Snail with both motifs intact bound with the highest affinity.
Figure 2.
Both of the 14-3-3 binding motifs in Snail are required for optimal interaction with 14-3-3 proteins. (A) The truncated forms of Snail used. (B) Interaction of 14-3-3 proteins with truncated Snail variants. The “*” indicates a non-specific band. (C) Single amino acid substitutions in the 14-3-3 binding motifs of Snail. The core serine in the NH2-terminal motif and the threonine in the COOH-terminal motif were replaced with alanines, respectively. (D) The SNA-A and SNA-B sites are required for optimal 14-3-3 protein binding.
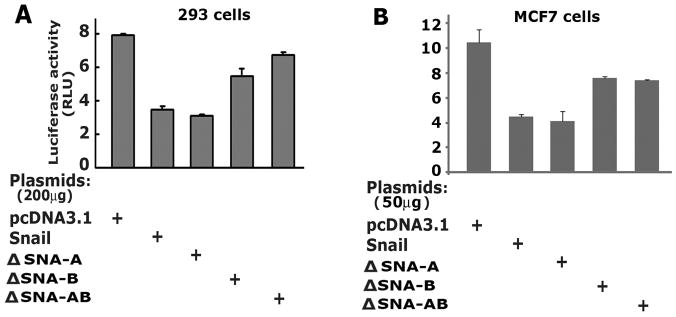
Snail-14-3-3 interaction is required for E-cadherin repression
E-cadherin is a direct target of Snail repressor and contains tandem Snail binding sites in the proximal promoter [23-24]. To test if 14-3-3 binding is crucial for Snail-mediated repression of E-cadherin, we performed luciferase reporter assays in 293 and MCF7 cells. An E-cadherin promoter reporter (E-cad-Luc) plasmid was transiently transfected with plasmids encoding Snail (wild-type or mutants) into 293 cells and luciferase asssays were performed. The wild-type Snail effectively repressed E-cad-Luc activity and the mutant, ΔSNA-A showed repression of E-cad-Luc comparable to the wild type Snail. The mutants ΔSNA-B and ΔSNA-AB were impaired in their repression activities compared to the wild type Snail in both 293 and MCF7 cells (Figure 3). Thus, the 14-3-3 binding motifs in Snail significantly affect Snail-mediated transcriptional repression.
Figure 3.
The COOH-terminal motif SNA-B is critical for Snail to repress E-cadherin. (A) The E-cadherin promoter reporter assays in 293 cells. The luciferase reporter construct E-cad-Luc, Snail and its mutants were co-transfected into 293 cells and normalized luciferase activity was determined. (B) The E-cadherin promoter reporter assays in MCF7 cells.
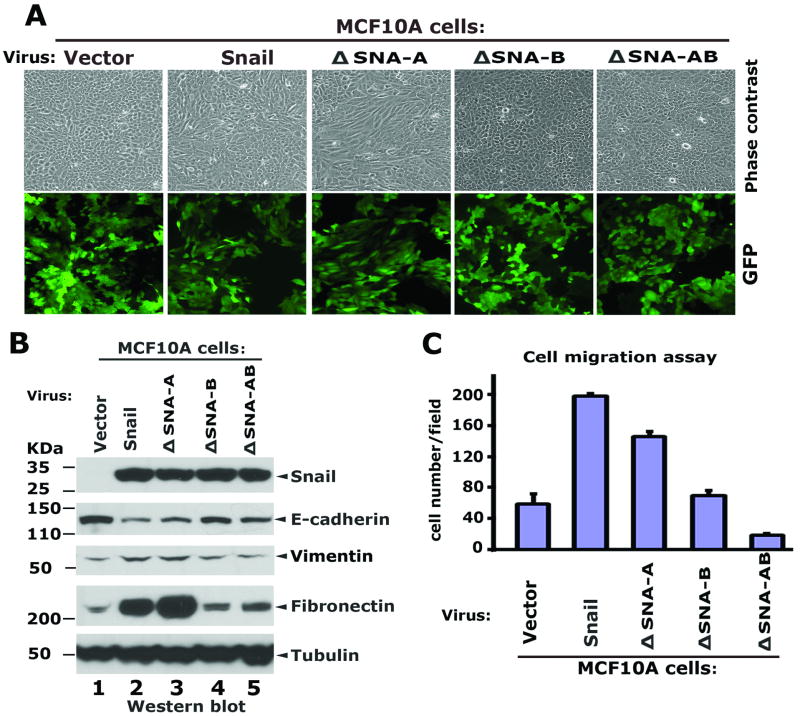
The COOH-terminal motif is required for Snail to induce EMT and cell migration
The process of Snail-induced EMT is characterized by dramatic phenotypic changes that include loss of the epithelial markers E-cadherin, γ-catenin/plakoglobin, α-catenin, and β-catenin, gain of the mesenchymal markers Fibronectin and Vimentin, and changes in cell shape and motility [1-2], all of which are readily observed in MCF10A mammary epithelial cells [7-8, 25]. To examine if these 14-3-3 binding sites in Snail are required for induction of EMT, MCF10A cells were infected with lentiviral vectors encoding wild type Snail, ΔSNA-A, ΔSNA-B or ΔSNA-AB proteins. Western blots were performed to examine the expression of the Snail variants, and the EMT markers E-cadherin, Fibronectin and Vimentin. The exogenous Snail proteins were expressed in MCF10A cells at similar levels (Figure 4A). The wild-type Snail and the ΔSNA-A mutant effectively repressed the endogenous E-cadherin, however, both the ΔSNA-B and ΔSNA-AB mutants were severely impaired in their abilities to repress E-cadherin (Figure 4A). Increased Fibronectin and Vimentin was observed in Snail and ΔSNA-A expressing cells, while expression of ΔSNA-B and ΔSNA-AB showed little effect on Fibronectin (Figure 4A). Concomitantly, expression of Snail and ΔSNA-A induced drastic morphological changes in MCF10A cells, indicative of EMT, whereas the mutants, ΔSNA-B and ΔSNA-AB which fail to interact with 14-3-3 proteins did not elicit EMT morphology (Figure 4B).
Figure 4.
The COOH-terminal motif SNA-B is required for Snail to induce EMT and cell migration in MCF10 cells. (A) Mutation of SNA-B inhibits Snail-medidated repression on endogenous E-cadherin on induction of mesenchymal markers. MCF10A cells were infected with lentiviral vectors encoding the COOH-terminal Flag-tagged Snail wild type, its mutant variants and GFP. Whole cell extracts were prepared from exponentially growing MCF10A cells and western blots were performed using the indicated antibodies. (B) A functional SNA-B motif is essential for Snail to induce morphological changes in MCF10A cells. Images were taken under the phase-contrast microscope. GFP was used for the indicator of the infection efficiency. (C) Cell migration assays. Each condition was performed in triplicate: the average number of cells per field is represented.
To examine cell motility, MCF10A cells expressing Snail, ΔSNA-A, ΔSNA-B and ΔSNA-AB were sorted by FACS using GFP as the selection marker. The migration potential of these cells was assayed using standard transwell assays. Both Snail and the mutant ΔSNA-A promoted cell migration, while the mutants ΔSNA-B and ΔSNA-AB did not (Figure 4C). Taken together, these data indicate the 14-3-3 binding site located in the first zinc finger motif of Snail is required for Snail to induce motility in MCF10A cells.
The Snail corepressor Ajuba contains functional 14-3-3 binding motifs
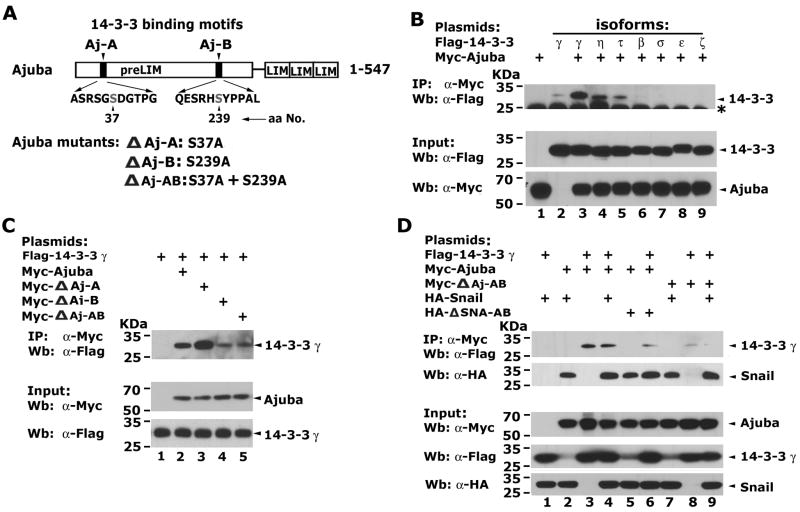
Since 14-3-3 proteins may function by mediating the assembly of multiprotein complexes, we asked if 14-3-3 proteins affect the potential of Snail to recruit its corepressor Ajuba. Two putative 14-3-3 binding motifs in the preLIM region of Ajuba were identified (Figure 5A). To verify the interaction between Ajuba and 14-3-3 isoforms, Myc-tagged Ajuba and Flag-tagged 14-3-3 isofoms were expressed in 293 cells followed by co-IP assays. Ajuba did not interact with 14-3-3 β, ε, σ, or ζ (Figure 5B). However, Ajuba strongly interacted with 14-3-3 γ, τ and η.
Figure 5.
Ajuba contains functional 14-3-3 binding motifs and selectively interacts with 14-3-3 isoforms. (A) Ajuba contains putative 14-3-3 binding motifs in the preLIM region: Aj-A and Aj-B. The numbers correspond to the locations of the core serine residues within 14-3-3 binding motifs of Ajuba and the alanine substitution mutations. (B) Ajuba interacts with 14-3-3 isoforms γ, η and τ when co-expressed in 293 cells. The “*” indicates a non-specific band. (C) Aj-B is the dominant site for 14-3-3 binding. Myc-Ajuba and its mutants, together with Flag-14-3-3γ were co-expressed in 293 cells and co-IP assays were performed. (D) Snail, Ajuba and 14-3-3 can form a ternary protein complex in living cells. Myc-Ajuba, Myc-ΔAj-AB, HA-Snail, HA-ΔSNA-AB and/or Flag-14-3-3γ were expressed in 293 cells, and co-IP assays were performed with α-Myc antibody and western blot with α-HA or α-Flag antibodies.
To examine whether the two putative 14-3-3 binding motifs in Ajuba were required for 14-3-3 protein interaction, we mutated these two motifs individually or in combination (Figure 5A). Mutation of the NH2-terminal motif (Aj-A) enhanced the binding activity of Ajuba to 14-3-3 γ (Figure 5C). Mutation of the COOH-terminal motif (Aj-B) alone or mutation of Aj-A and Aj-B simultaneously significantly decreased, but did not completely abolish the interaction beween Ajuba and 14-3-3 γ (Figure 5C). These data together suggest that Aj-B is the dominant site for 14-3-3 binding, and that Ajuba may contain additional, weaker 14-3-3 binding sites at other locations.
To determine if Snail, 14-3-3 and Ajuba form a stable ternary complex, we co-expressed HA-Snail, Myc-Ajuba and Flag-14-3-3 γ in 293 cells. Ajuba readily co-immunoprecipitated Snail and 14-3-3 γ when these proteins were co-expressed (Figure 5D, lanes 2, 3). Mutation of the 14-3-3 binding motifs in Snail or in Ajuba did not significantly affect the Snail-Ajuba interaction. However, mutation of the 14-3-3 binding motifs in Ajuba drastically decreased its ability to bind 14-3-3 γ (Figure 5D, lanes 8, 9). These data suggest that a stable Snail-Ajuba-14-3-3 ternary complex can be assembled in cells
Ajuba is required for Snail to induce EMT in MCF10A cells
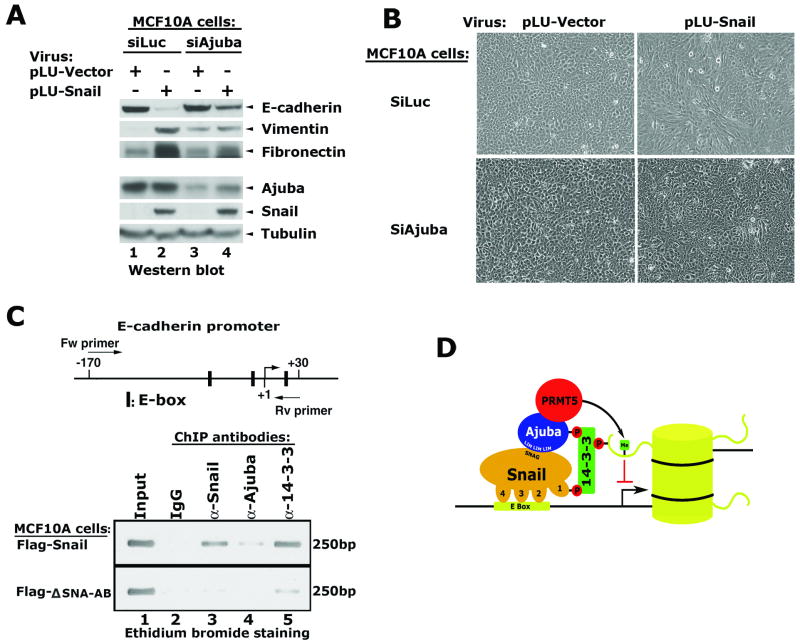
To examine the role of Ajuba in Snail-mediated repression and EMT, a viral vector containing a shRNA sequence targeting human ajuba was stably introduced into MCF10A cells to create MCF10A-siAjuba cells. A vector containing shRNA targeting luciferase was used as a control. Following puromycin selection, these populations were infected with viruses expressing Snail or empty vector. The expression of E-cadherin, Vimentin, Fibronectin, Ajuba and Snail were examined via western blot. Ajuba was efficiently knocked down but not abolished in MCF10-siAjuba cells (Figure 6A). Snail expression in MCF10A-siLuc cells repressed E-cadherin, and comcomitantly increased Vimentin and Fibronectin. Snail did not induce similar changes in MCF10A-siAjuba cells (Figure 6A). In further support of these observations, Snail failed to induce morphological changes in MCF10A-siAjuba cells (Figure 6B). Taken together, these data indicate that Ajuba is a critical co-factor for Snail-mediated EMT.
Figure 6.
14-3-3 proeins are recruited to the E-cadherin promoter via an interaction with Snail. (A) Depletion of Ajuba in MCF10A cells inhibits Snail-mediated repression on E-cadherin and induction of Fibronectin and Vimentin. (B) Snail does not induce morphological changes in MCF10A-siAjuba cells. (C) PCR analysis of the immunoprecipitated DNA fragments from MCF10A cells. ChIP assays were performed in the MCF10A-Snail and the MCF10A-ΔSNA-AB cells using the indicated antibodies. The PCR primers flanking the Snail binding sites in the proximal promoter produces a 200bp fragment detected by ethidium bromide staining. (D) The cartoon shows a model for how Snail, Ajuba and 14-3-3 may interact with target genes in chromatin.
14-3-3 proeins are recruited to the E-cadherin promoter via an interaction with Snail
To examine how mutation of the 14-3-3 binding motifs affects the ability of Snail and its associated proteins to localize to the promoters of endogenous Snail target genes, we applied ChIP analysis to the Snail target gene E-cadherin. Antibodies specific to Snail, Ajuba and 14-3-3 proteins were used for the ChIP assays and the immunoprecipitated DNA fragments were quantified by PCR amplification using a primer set flanking the three Snail-binding sites located in the proximal promoter region of the E-cadherin gene (Figure 6C). The proximal promoter of the E-cadherin gene was highly enriched by antibodies to Snail and 14-3-3 (Figure 6C, lanes 3 & 5), and modestly enriched by antibody to Ajuba in MCF10A-Flag-Snail cells (Figure 6C, lane 4). However, binding of Snail, Ajuba and 14-3-3 proteins was significantly diminished in the MCF10A cells expressing the ΔSNA-AB mutant (Figure 6C). Taken together, these data suggest that the association of Snail, Ajuba and 14-3-3 with the endogenous E-cadherin promoter is strongly influenced by the 14-3-3 binding motif.
Discussion
Expression of Snail is directly associated with acquisition of the metastatic and invasive phenotype in human cancers [5-6, 8]. Thus, definition of the machinery which mediates Snail functions is of great interest. In this study, we show that 14-3-3 proteins are new components of this machinery. The 14-3-3 proteins bind Snail directly, and mutation of the 14-3-3 binding motif in the zinc finger domain of Snail significantly impairs Snail-mediated repression and EMT. Snail, 14-3-3 proteins, and Ajuba are readily detected at the endogenous E-cadherin promoter in a Snail-dependent manner. Collectively, these data suggest that 14-3-3 proteins may function as co-factors for Snail and directly modulate Snail functions.
The 14-3-3 proteins are often implicated in cytoplasmic signaling cascades. However, there is less evidence that they regulate gene transcription via interactions with transcription factors [16-17]. In these few reported cases, binding of 14-3-3 proteins to these transcription factors can either sequester them in the cytoplasm or affect their DNA binding activities. For example, the forkhead transcription factor Foxo1 has two functional 14-3-3 binding motifs and upon phosphorylation can bind 14-3-3 proteins [26]. 14-3-3 proteins tether the Foxo1 protein to other cytoplasmic factors and may also inhibit the Foxo1-DNA interaction. 14-3-3 proteins interact with p53 via a 14-3-3 binding motif located in the DNA binding domain of p53. However, this interaction can increase p53's DNA binding activity and activates gene expression [27]. Here, we demonstrate that a functional 14-3-3 binding motif in the DNA binding domain of Snail is essential for its repression functions and mutation of this motif results in dissociation of Snail-associated protein complexes from target genes in chromatin. Thus, binding of 14-3-3 proteins to Snail may either stablize the Snail repression complex and/or direct the Snail complexes to chromatin.
Recently, 14-3-3 proteins have been described as “interpreters” of the histone code by binding to phosphorylated serine 10 in histone H3, a modification that is associated with mitotic chromatin compaction [18-20]. However, interphase phosphorylation on Serine 10 in histone H3 has been linked to transcriptional activation of target genes. The ability of 14-3-3 proteins to recognize multiple post-translational modifications, and their ability to homo/hetero-dimerize suggests that they may have broad capabilities to recognize combinatorial marks in chromatin and thus control diverse nuclear processes. A working model for the role of 14-3-3 proteins in Snail-mediated repression is shown in Figure 6D. Snail recognizes the E-box DNA sequence in target genes and serves as the targeting component of this complex. 14-3-3 homo/heterodimeric proteins may bridge and stabilize a bipartite Snail-Ajuba complex via recognition of the pS/pT motifs in each component. The 14-3-3 components then may utilize a different surface to bind modifications in histone tails as that has been shown in the case of histone H3 scenario, thereby anchoring the complex to chromatin. This scenario would also facilitate access of the arginine mehtyltransferase Prmt5 for chromatin, allowing the placement of additional recognition marks, which are readily observed in nucleosomes repressed by Snail [14]. It is also possible that the 14-3-3 proteins directly recruit chromatin modulators like HDACs to Snail target genes [28]. Regardless of the exact mechanims, our data clearly show that the 14-3-3 binding motif in Snail is required to repress target genes during the EMT transdifferentiation process. These observations are consistent with other reports of 14-3-3 protein functions and cancer [16, 29-30]. Thus, biological or pharmacological strategies to target the Snail-14-3-3 repression machinery may prove useful for cancer therapeutics and metastasis prevention.
Acknowledgments
We thank Dr. Gregory Longmore for sharing the Ajuba and Snail plasmids. F.J.R. is supported, in part, by NIH grants (CA095561 and CA12988), and the Samuel Waxman Cancer Research Foundation. Z.H. is supported by the NIH training grant (CA09171). We acknowledge the National Cancer Institute Supported Wistar Institute Cancer Center Shared Facilities: Genomics, Protein Expression, Flow Cytometry, Proteomics and Hybridoma, and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
Abbreviations
- Prmt5
protein arginine methyltransferase 5
- EMT
epithelial-mesenchymal transition
- ChIP
Chromatin immunoprecipitation
References
- 1.Nieto MA. Epithelial-Mesenchymal Transitions in development and disease: old views and new perspectives. Int J Dev Biol. 2009;53:1541–7. doi: 10.1387/ijdb.072410mn. [DOI] [PubMed] [Google Scholar]
- 2.Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008;27:6958–69. doi: 10.1038/onc.2008.346. [DOI] [PubMed] [Google Scholar]
- 3.Alberga A, Boulay JL, Kempe E, Dennefeld C, Haenlin M. The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development. 1991;111:983–92. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- 4.Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–61. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 6.De Craene B, Gilbert B, Stove C, Bruyneel E, Van Roy F, Berx G. The transcription factor Snail induces tumor cell invasion through modulation of the epithelial cell differentiation program. Cancer Res. 2005;65:6237–44. doi: 10.1158/0008-5472.CAN-04-3545. [DOI] [PubMed] [Google Scholar]
- 7.Medici D, Hay ED, Olsen BR. Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol Biol Cell. 2008;19:4875–87. doi: 10.1091/mbc.E08-05-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moody SE, Perez D, Pan TC, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol. 1996;16:6263–72. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zweidler-Mckay PA, Grimes HL, Flubacher MM, Tsichlis PN. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol. 1996;16:4024–34. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayyanathan K, Peng H, Hou Z, et al. The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic Shuttling. Cancer Res. 2007;67:9097–106. doi: 10.1158/0008-5472.CAN-07-2987. [DOI] [PubMed] [Google Scholar]
- 13.Langer EM, Feng Y, Zhaoyuan H, Rauscher FJ, 3rd, Kroll KL, Longmore GD. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14:424–36. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou Z, Peng H, Ayyanathan K, et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 2009;8:1009–17. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obsilová V, Silhan J, Boura E, Teisinger J, Obsil T. 14-3-3 proteins: a family of versatile molecular regulators. Physiol Res. 2008;57 3:S11–21. doi: 10.33549/physiolres.931598. [DOI] [PubMed] [Google Scholar]
- 18.Macdonald N, Welburn JP, Noble ME, et al. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 19.Walter W, Clynes D, Tang Y, Marmorstein R, Mellor J, Berger SL. 14-3-3 interaction with histone H3 involves a dual modification pattern of phosphoacetylation. Mol Cell Biol. 2008;28:2840–9. doi: 10.1128/MCB.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winter S, Simboeck E, Fischle W, et al. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 2008;27:88–99. doi: 10.1038/sj.emboj.7601954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet. 1998;19:175–8. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 22.Cardamone MD, Bardella C, Gutierrez A, et al. ERalpha as ligand-independent activator of CDH-1 regulates determination and maintenance of epithelial morphology in breast cancer cells. Proc Natl Acad Sci U S A. 2009;106:7420–5. doi: 10.1073/pnas.0903033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cano A, Perez-Moreno MA, Rodrigo I, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 24.Batlle E, Sancho E, Franci C, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–9. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 25.Kim HJ, Litzenburger BC, Cui X, et al. Constitutively active type insulin-like growth factor receptor causes transformation and xenograft growth of immortalized mammary epithelial cells and is accompanied by an epithelial-to-mesenchymal transition mediated by NF-kappa B and snail. Mol Cell Biol. 2007;27:3165–75. doi: 10.1128/MCB.01315-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X, Gan L, Pan H, et al. Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: characterization of phosphorylation- and 14-3-3-dependent and -independent mechanisms. Biochem J. 2004;378(Pt 3):839–49. doi: 10.1042/BJ20031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR. 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res. 2008;36:5983–91. doi: 10.1093/nar/gkn598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–40. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tzivion G, Gupta VS, Kaplun L, Balan V. 14-3-3 proteins as potential oncogenes. Semin Cancer Biol. 2006;16:203–13. doi: 10.1016/j.semcancer.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Hermeking H, Benzinger A. 14-3-3 proteins in cell cycle regulation. Semin Cancer Biol. 2006;16:183–92. doi: 10.1016/j.semcancer.2006.03.002. [DOI] [PubMed] [Google Scholar]