Abstract
Ferlin proteins mediate membrane-fusion events in response to Ca2+. Myoferlin, a member of the ferlin family, is required for normal muscle development, during which it mediates myoblast fusion. We isolated both damaged and intact myofibers from a mouse model of muscular dystrophy using laser-capture microdissection and found that the levels of myoferlin mRNA and protein were increased in damaged myofibers. To better define the components of the muscle-injury response, we identified a discreet 1543-bp fragment of the myoferlin promoter, containing multiple NFAT-binding sites, and found that this was sufficient to drive high-level myoferlin expression in cells and in vivo. This promoter recapitulated normal myoferlin expression in that it was downregulated in healthy myofibers and was upregulated in response to myofiber damage. Transgenic mice expressing GFP under the control of the myoferlin promoter were generated and GFP expression in this model was used to track muscle damage in vivo after muscle injury and in muscle disease. Myoferlin modulates the response to muscle injury through its activity in both myoblasts and mature myofibers.
Keywords: NFAT, Myoferlin, Muscle regeneration, Myoblast, Myofiber
Introduction
Skeletal muscle is composed of multinucleated myofibers. Muscle growth and repair occur as a result of mononucleated myoblasts fusing with one another as well as myoblasts fusing to existing myotubes (Ontell et al., 1988). In addition, damaged muscle membranes also require fusion to reseal membrane disruption. Recently, two members of the ferlin family, dysferlin and myoferlin, have been implicated in muscle-membrane fusion. The gene encoding dysferlin is mutated in patients with Limb Girdle Muscular Dystrophy type 2B, a disorder of progressive muscle weakness (Bashir et al., 1998; Liu et al., 1998). Dysferlin encodes a 220-kDa membrane-associated protein with six C2 domains. The first C2 domain of dysferlin, C2A, binds negatively charged phospholipids, only in the presence of Ca2+ (Davis et al., 2002). In the absence of dysferlin, there is a marked delay of resealing of muscle-membrane tears (Bansal and Campbell, 2004; Lennon et al., 2003). This delay is associated with vesicle accumulation under the membrane and reflects defective vesicle-mediated resealing.
Myoferlin is highly related to dysferlin and, like dysferlin, contains six C2 domains. As with dysferlin, the C2A domain of myoferlin binds phospholipids in the presence of Ca2+ (Doherty et al., 2005). Myoferlin is broadly expressed and is highly expressed during muscle development, specifically in myoblasts undergoing fusion (Davis et al., 2002). In normal, mature skeletal muscle there is relatively low expression of myoferlin (Davis et al., 2000). However, in the adult mdx mouse – a mouse model of Duchenne muscular dystrophy, in which there is ongoing degeneration and regeneration – myoferlin mRNA is upregulated (Davis et al., 2000; Doherty et al., 2005). Myoferlin mRNA is also increased in human muscle affected by Duchenne muscular dystrophy (Haslett et al., 2003). The increase of myoferlin in muscular dystrophy supports a role for myoferlin not only in muscle regeneration, but also in muscle repair.
The defects in muscle growth and repair seen in myoferlin-null muscle are reminiscent of what was described in muscle lacking nuclear factor of activated T cells (NFAT)c2 (Horsley et al., 2001). NFATc2-null mice have defective myoblast fusion, resulting in fibers with a decreased cross-sectional area and delayed muscle repair (Horsley et al., 2001). Of the five NFAT isoforms, all are expressed in skeletal muscle and are required for proper muscle development and repair (Abbott et al., 1998; Calabria et al., 2009; Cho et al., 2007; Friday et al., 2000; Horsley et al., 2001; Kegley et al., 2001; O'Connor et al., 2007). NFATs 1-4 are responsive to calcineurin activation. Calcineurin, a serine/threonine protein phosphatase, responds to intracellular Ca2+ levels and regulates NFAT-dependent transcription. The activity of calcineurin A is elevated during myoblast differentiation and during muscle regeneration in mdx mice, a model of muscular dystrophy (Friday et al., 2000; Stupka et al., 2006b). Furthermore, regeneration of skeletal muscle fibers is enhanced in transgenic mice overexpressing calcineurin A (Stupka et al., 2007). Inhibition of calcineurin with the immunosuppressive drug cyclosporine A (CsA) causes inhibition of muscle repair in normal mice (Abbott et al., 1998; Sakuma et al., 2003) and mdx mice (Stupka et al., 2004).
We now used laser-guided microdissection to collect damaged and intact myofibers from a mouse model of muscular dystrophy. We examined differential gene-expression profiles in these two populations and found that myoferlin mRNA was upregulated sevenfold in damaged myofibers compared with neighboring intact myofibers. We characterized the sequences responsible for myoferlin gene expression and identified a 1543-bp promoter that drives expression 80-fold over baseline. We investigated the role of NFAT factors in driving myoferlin expression by identifying functional NFAT-binding consensus sequences in the 1543-bp myoferlin promoter. This myoferlin promoter sequence was sufficient to respond to calcineurin and NFAT activation, CsA inhibition, and to mediate the response to cardiotoxin-induced damage in multinucleate myotubes. Transgenic mice expressing GFP under the control of the myoferlin promoter express GFP in response to cardiotoxin exposure and in muscular dystrophy, providing a ‘damage sensor’.
Results
Myoferlin is upregulated in dye-positive damaged myofibers and surrounding mononuclear cells
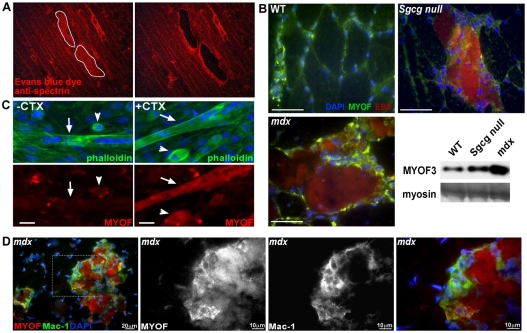
Evans Blue dye (EBD) is a reliable marker for identifying myofibers with disrupted sarcolemma (Matsuda et al., 1995; Straub et al., 1997). γ-sarcoglycan (Sgcg)-null mice, similar to other models of muscular dystrophy, develop focal regions of damage in muscle so that there is both degeneration and regeneration occurring within the same muscle (Hack et al., 1998). We isolated muscle from EBD-injected Sgcg-null mice and then rapidly stained it with an anti-spectrin antibody to identify those fibers that had taken up dye but retained an intact cytoskeleton. The same number of dye-positive and -negative myofibers was isolated by laser-capture microdissection (Fig. 1A), and total RNA was extracted and subjected to one round of linear amplification. Each sample of amplified RNA was hybridized to an Affymetrix Mouse 430A chip.
Fig. 1.
Myoferlin is focally upregulated in regions of degeneration and regeneration. (A) γ-sarcoglycan-null (Sgcg null) mice are a model of muscular dystrophy and develop focal areas of muscle damage (Hack et al., 1998). Sgcg-null mice were injected with EBD to mark compromised skeletal myofibers. The muscle was counterstained with an anti-spectrin antibody to mark myofibers that were both intact and positive for EBD, consistent with early-phase degeneration. Examples of dye-positive myofibers are shown before (left) and after (right) microdissection. (B) Myoferlin is focally upregulated in regions of degeneration and regeneration. Wild-type, Sgcg-null or mdx mice were injected with EBD, and muscles were studied. Increased myoferlin (green) is concentrated near or within EBD-positive fibers (red; co-staining appears yellow). DAPI staining of nuclei is shown in blue. (Bottom right) Immunoblot analysis demonstrates myoferlin protein is increased in Sgcg-null and mdx muscle compared with normal (WT) muscle. Myosin heavy chain is shown as a loading control. Scale bars: 50 μm. (C) Endogenous myoferlin expression is shown in a mixed cell culture containing myoblasts (arrowheads) and multinucleate myotubes (arrows; note linear arrangement of nuclei consistent with myotubes). In the absence of injury, myoferlin expression is readily detected in the mononuclear cells within the culture (left, −CTX) but is nearly absent from the neighboring multinucleate myotubes. With cardiotoxin exposure (+CTX), myoferlin expression was present in the elongated myotubes and appeared unchanged in the neighboring singly nucleated myoblasts. (D) Endogenous myoferlin (red) is also expressed in Mac-1-positive lymphocytes (green; coexpression appears yellow) that surround myoferlin-positive myofibers in mdx mouse quadriceps muscle. The right three panels are a magnified view of the white boxed area in the left panel.
We identified 43 differentially regulated genes (29 upregulated and 14 downregulated) in dye-positive compared with dye-negative myofibers (supplementary material Table S1). The genes include those expressed not only in myofibers, but also in invading inflammatory cells as well as neighboring fibroblasts. The genes with the greatest fold increases are predominately involved in modifying the cytoskeleton, in cell adhesion and in migration. The third highest differentially expressed gene in our data set, Fer13L, encoding myoferlin, was upregulated 6.8-fold in dye-positive damaged myofibers compared with dye-negative fibers. The upregulation of myoferlin mRNA in damaged myotubes is consistent with prior reports that surveyed gene expression in whole muscle from human or mouse models of muscular dystrophy (Davis et al., 2000; Haslett et al., 2002).
Immunolocalization of focally upregulated myoferlin in dystrophic muscle
Immunofluorescence microscopy with an anti-myoferlin antibody was performed on dye-injected muscle from wild-type mice and two different models of muscular dystrophy, Sgcg-null and mdx. In wild-type control skeletal muscle, myoferlin protein was found at low levels at the sarcolemma (Fig. 1B, top left). Myoferlin was upregulated similarly in both Sgcg-null and mdx dye-positive regenerating myofibers (Fig. 1B, top right and bottom left, respectively). Myoferlin staining was also increased in mononuclear cells surrounding dye-positive myotubes (Fig. 1B). Immunoblots of skeletal muscle lysates from Sgcg-null and mdx muscular-dystrophy models both showed an increase of myoferlin protein compared with wild-type muscle (Fig. 1B, bottom right). The C2C12 line is a myoblast-like cell line that can be induced to form multinucleated myotubes. C2C12 myotubes were treated with cardiotoxin to induce damage and to determine whether myoferlin was similarly upregulated in this cell-culture-based model. Cardiotoxin is a 60- to 65-amino-acid protein that induces microperforations along the plasma membrane leading to an increase in intracellular Ca2+ and cell injury in multiple cell types, including skeletal muscle. Endogenous myoferlin is expressed at low levels in healthy multinucleated myotubes (arrows in Fig. 1C, left) and is more abundantly detected in mononuclear myoblasts (arrowheads in Fig. 1C). After 1 μM cardiotoxin treatment, endogenous myoferlin was upregulated in multinucleated myotubes and myoblasts (Fig. 1C, right). These data demonstrate that myoferlin expression is increased in response to injury in both myoblasts and myotubes.
In addition to myoblast activation, muscle injury is associated with an infiltration of inflammatory cells. Given the increase in myoferlin seen in damaged muscle and the known broad expression pattern of myoferlin, we queried whether the mononuclear cells seen surrounding the damaged fibers were at least partly immunogenic in nature. Quadriceps muscle from the mdx mouse model of muscular dystrophy was analyzed by immunofluorescence microscopy. Myoferlin-positive fibers were surrounded by macrophage-1 antigen (Mac-1; also known as integrin αMβ2 and Cd11b)-positive mononuclear cells that also express myoferlin (Fig. 1D). Mac-1 marks macrophages, natural killer cells, monocytes and granulocytes (Ault and Springer, 1981). We conclude that myoferlin is expressed in myoblasts, injured myotubes and the surrounding inflammatory cells seen in injured muscle.
The myoferlin promoter contains conserved, active NFAT-binding sites
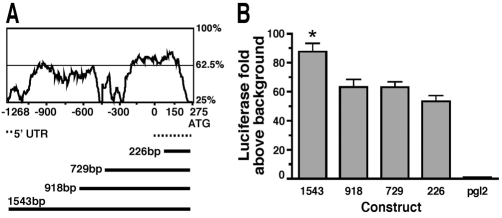
To characterize the molecular signature of muscle injury and repair, we dissected the myoferlin promoter. Sequences directly upstream of the myoferlin translational start site were compared between the mouse and human genomes up to 20 kb (Fig. 2A). The first 1543 bp of the myoferlin promoter showed high conservation across species and was inserted into the pGL2-basic vector to test the ability of the promoter to drive expression in C2C12 cells. In C2C12 myoblasts, the 1543-bp–pGL2 construct induced luciferase activity 80-fold above background. Smaller constructs induced high-level promoter activity ranging from 50- to 60-fold above background, but each of these smaller constructs was consistently less effective than the full-length 1543-bp–pGL2 construct (Fig. 2B).
Fig. 2.
The 1543-bp myoferlin promoter drives high-level expression. (A) Sequence conservation of the first 1500 bp of the myoferlin promoter from humans and mice. ‘0’ refers to the transcriptional start site and ‘275’ marks the initiator methionine. Regions as high as 75% similarity were identified. Below is a schematic of four different promoter constructs generated and tested. (B) Activity of the myoferlin promoter fragments shown in A in C2C12 myoblasts. The 1543-bp–pGL2 reporter activates luciferase expression 82-fold above background.
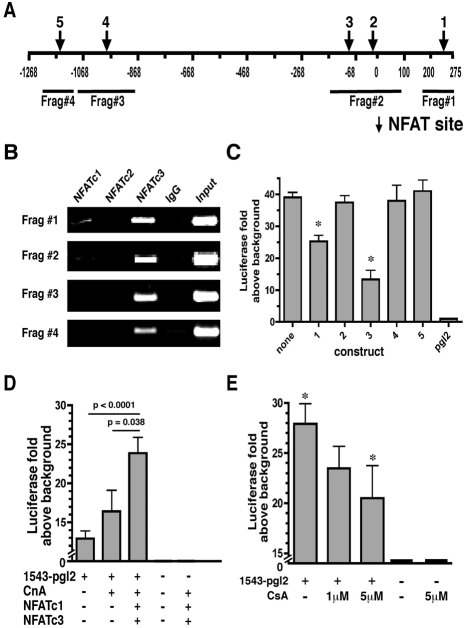
On the basis of the known role of NFAT proteins in muscle development, the 1543-bp sequence upstream of the myoferlin translational start site was analyzed for NFAT-binding consensus sequences (WGGAAANH) (Rao et al., 1997) (Fig. 3A). Multiple additional conserved transcription-factor-binding sequences were also identified, including for Myc, MEF2, CEBP, Sp1 and AP1 (data not shown). Chromatin immunoprecipitation (ChIP) assays were performed using cross-linked chromatin from confluent C2C12 cells in growth media to confirm that these sequences bound endogenous NFATc1 and NFATc3. IgG and NFATc2 were used as controls because NFATc2 translocates to the nucleus in nascent myotubes and not myoblasts (Abbott et al., 1998). As expected, interactions with endogenous NFATc2 or IgG were not detected. NFATc4 was not analyzed because the majority of NFATc4 translocates to the nucleus 48 hours after differentiation, not in growth media conditions (Cho et al., 2007). NFATc5 was not studied because NFATc5 is constitutively nuclear in a variety of cell types and is calcineurin insensitive (Lopez-Rodriguez et al., 1999). We found that NFATc3 bound to all four promoter fragments, whereas only fragments 1 and 2 were found to interact with NFATc1 (Fig. 3B).
Fig. 3.
NFAT binds the myoferlin promoter and regulates its activity. (A) The position of the five predicted NFAT-binding sites are designated by arrows. The four different regions amplified in the ChIP assay are indicated below the line. (B) ChIP assays using anti-NFAT antibodies for endogenous NFAT proteins from confluent C2C12 cells. NFATc1, NFATc2 and NFATc3 were tested, and NFATc3 was detected occupying all NFAT sites tested. Low-level occupancy of NFATc1 was also detected. NFATc2 was not detected at the myoferlin promoter. (C) pGL2 constructs were generated containing mutations in five predicted NFAT-binding sites in the 1543-bp myoferlin promoter labeled NFAT site 1 through 5. Constructs were transfected into C2C12 myoblasts and luciferase expression analyzed. Mutagenesis of NFAT sites 1 and 3 reduced luciferase expression by approximately 30% and 50%, respectively, compared with the 1543-bp–pGL2 construct or mutants 2, 4 or 5 (*P<0.004). (D) The 1543-bp–pGL2 reporter construct or pGL2-basic control vectors were transfected into C2C12 myoblasts in combination with activated calcineurin A (CnA), NFATc1 and/or NFATc3 expression plasmids. Luciferase expression was increased by twofold with the addition of CnA, NFATc1 and NFATc3 compared with the 1543-bp–pGL2 vector alone (P<0.0001). (E) The 1543-bp–pGL2 construct was transfected into C2C12 myoblasts and, 18 hours post-transfection, 1 μM or 5 μM CsA was added to the cultures. After 24 hours, luciferase expression was analyzed. The addition of 5 μM CsA reduced the 1543-bp myoferlin-promoter activity by 30% compared with 1543-bp–pGL2 (*P<0.05).
To examine further the role of NFATs in modulating myoferlin expression, we mutated five NFAT-binding consensus sites that were identified by the TESS and VISTA algorithms (Fig. 3C). Mutation of NFAT site 1 or NFAT site 3 significantly, but incompletely, reduced luciferase induction to 25-fold or 12-fold above background, respectively (P<0.004). An electrophoretic mobility shift assay (EMSA) was performed to verify further that mutating NFAT sites 1 and 3 causes a reduction in binding to the 1543-bp myoferlin promoter (supplementary material Fig. S1). Mutating NFAT sites 2, 4 or 5 in the 1543-bp–pGL2 vector had no effect on luciferase induction as compared to the 1543-bp–pGL2 construct (Fig. 3C). Together these results demonstrate that NFATc3 and NFATc1 interact with the myoferlin promoter and that NFAT sites 1 and 3 of the myoferlin promoter are crucial for full promoter function.
The myoferlin promoter responds to activated calcineurin A and is inhibited by CsA
We next investigated the calcineurin responsiveness of the myoferlin promoter. In C2C12 cells, transfection with 1543-bp–pGL2 resulted in a 12-fold induction of luciferase expression above background (Fig. 3D). Because of the need for co-transfection, these experiments used fivefold less of the 1543-bp–pGL2 plasmid than previous experiments and therefore have a lower level of induction (Fig. 2B). Co-transfection of 1543-bp–pGL2 along with constitutively active calcineurin plus NFATc1 and NFATc3 further promoted luciferase expression 25-fold above background. Similarly, co-transfection with 1543-bp–pGL2 and either NFATc1 alone or NFATc3 alone resulted in expression that was less than the combined transfection (not shown). The induction caused by the addition of constitutively active calcineurin, NFATc1 and NFATc3 was statistically significant compared with 1543-bp–pGL2 (P<0.0001) and 1543-bp–pGL2 plus constitutively active calcineurin (P=0.038).
CsA blocks the Ca2+-calcineurin-dependent activation of NFAT (Schreiber and Crabtree, 1992). C2C12 cells were transfected with 1543-bp–pGL2 in the presence of 1 μM or 5 μM CsA. The addition of CsA caused a dose-dependent decrease in promoter activity as seen by the reduction of luciferase expression from 28-fold above background in the 1543-bp–pGL2-transfected cultures to 23-fold and 20-fold above background in the 1 μM or 5 μM CsA treated cultures, respectively (Fig. 3E; P=0.05). These results provide additional support that the calcineurin-NFAT pathway modulates the activity of the 1543-bp myoferlin promoter. The inability of CsA to fully suppress myoferlin-reporter expression suggests that other factors regulate basal myoferlin expression.
The myoferlin promoter recapitulates endogenous myoferlin expression during development and repair in C2C12 cells
To determine whether expression from the myoferlin promoter fragment mirrored endogenous myoferlin expression, the 1543-bp region was ligated to a promoterless GFP expression plasmid. The 1543-bp–GFP plasmid was transfected into C2C12 cells and GFP expression analyzed. At 24 hours after transfection, strong GFP expression was seen in the singly-nucleated myoblasts transfected with 1543-bp–GFP (Fig. 4B). The level of induction produced from the myoferlin promoter was qualitatively equivalent to that seen from the strong CMV viral promoter (Fig. 4A). Transfected C2C12 cells were allowed to differentiate into multinucleated myotubes for 3 days, then the cultures were analyzed for GFP expression. Myotubes transfected with 1543-bp–GFP displayed reduced levels of GFP expression compared with myoblasts, whereas singly-nucleated cells within these same cultures continued to express high levels of GFP (Fig. 4D). By contrast, CMV-GFP-transfected myotubes retained a high level of GFP expression in both multinucleate and singly nucleated cells (Fig. 4C). These results were corroborated by immunoblotting for GFP (supplementary material Fig. S2). This pattern of GFP expression, in which high-level expression is seen in myoblasts followed by reduction after fusion to multinucleated myotubes, parallels what is seen for myoferlin protein expression in development (Doherty et al., 2005).
Fig. 4.
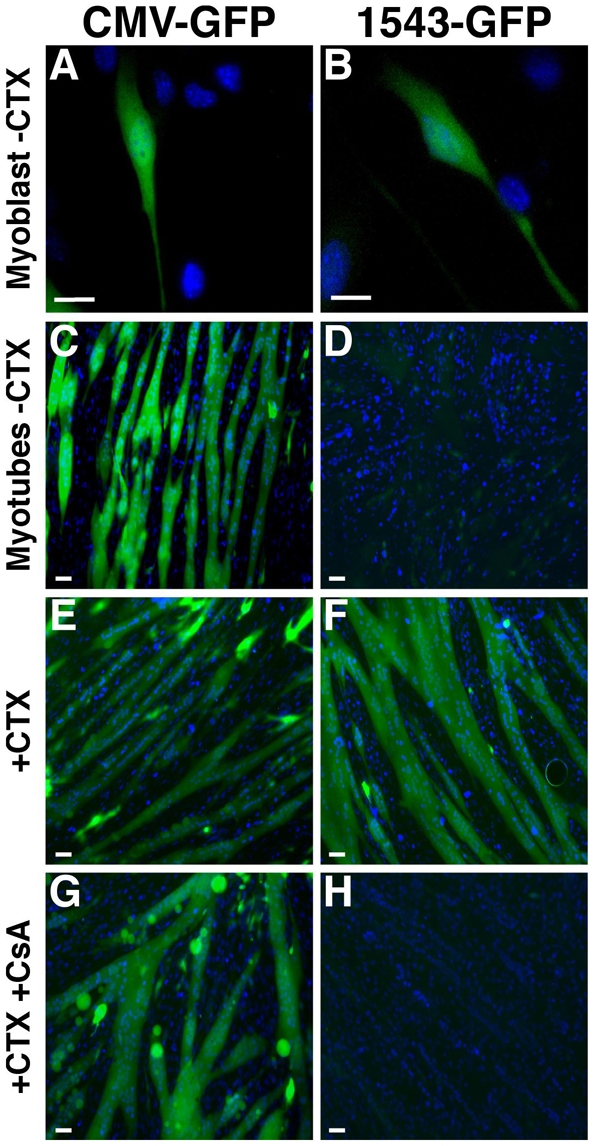
The 1543-bp myoferlin promoter drives GFP expression in C2C12 myotubes in response to cardiotoxin-induced damage. (A-H) The CMV-GFP or 1543-bp–GFP constructs were transfected into C2C12 cells and cells were imaged for GFP expression by live-cell microscopy. (A,B) Mononuclear myoblasts transfected with CMV-GFP or 1543-bp–GFP expressed high levels of GFP, similar to endogenous myoferlin expression. (C,D) After differentiation and fusion into myotubes, the CMV promoter, but not the myoferlin 1543-bp promoter, drives expression of GFP in multinucleate myotubes. (E,F) Myotubes were treated with CTX in the absence of CsA (E,F) or in the presence of CsA (G,H). The 1543-bp myoferlin promoter expresses GFP in cardiotoxin-treated myotubes (F), whereas GFP expression is blocked by the presence of CsA (H). Cardiotoxin or cardiotoxin and CsA did not alter CMV-GFP expression (C,E,G). Scale bars: 10 μm.
We treated transfected myotubes with cardiotoxin to induce damage. After cardiotoxin exposure, the 1543-bp–GFP-transfected myotubes demonstrated high-level GFP expression (Fig. 4F) as compared with the untreated 1543-bp–GFP-transfected myotubes (Fig. 4D). Notably, CsA blocked the cardiotoxin-induced activity of the myoferlin promoter (Fig. 4H). As a control, cardiotoxin or cardiotoxin plus CsA did not affect the level of GFP expression in the CMV-GFP-containing myotubes, because GFP levels remained high in all conditions (Fig. 4C,E,G). GFP expression levels were verified by immunoblot using an anti-GFP antibody (data not shown). Thus, these results suggest that the 1543-bp promoter contains the region responsible for upregulating myoferlin in response to cardiotoxin-induced damage.
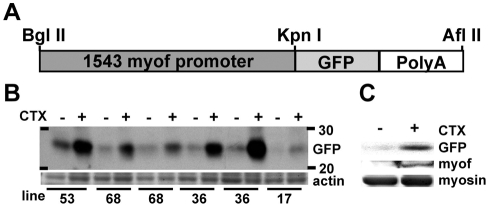
The myoferlin promoter is responsive to cardiotoxin-induced damage in vivo
Multiple independent lines of transgenic mice were generated carrying the 1543-bp myoferlin promoter driving GFP (Fig. 5A). Four lines were tested for transgene expression before and after muscle damage (lines 17, 36, 53 and 68). Consistent with the known pattern of endogenous myoferlin expression, relatively low GFP expression was present in uninjured adult muscle (Fig. 5B). However, after intramuscular cardiotoxin injection to induce injury, each of these lines demonstrated an increase in GFP protein (Fig. 5B). To verify that the GFP response mirrored myoferlin expression, equal amounts of protein from uninjured and cardiotoxin-injured skeletal muscle from a transgene-positive mouse were immunoblotted for both GFP and myoferlin. As expected, the levels of both GFP and myoferlin increased in response to cardiotoxin-induced muscle damage (Fig. 5C). Thus, the 1543-bp myoferlin promoter contains the region responsible for upregulating myoferlin in response to cardiotoxin-induced damage in vivo.
Fig. 5.
The 1543-bp myoferlin promoter drives GFP expression in damaged muscle in vivo. (A) Schematic of the 1543-bp–GFP transgene construct used to generate multiple lines of transgenic mice. (B) Immunoblot analysis of four transgenic lines showing induction of GFP in response to muscle damage induced by cardiotoxin (CTX) injection into the muscle. (C) Immunoblot analysis of transgene-positive muscle lysates with and without cardiotoxin-induced muscle damage, confirming that GFP expression mirrors endogenous myoferlin expression in response to muscle injury.
The 1543-bp myoferlin promoter drives GFP expression in myoblasts and damaged myofibers in vivo
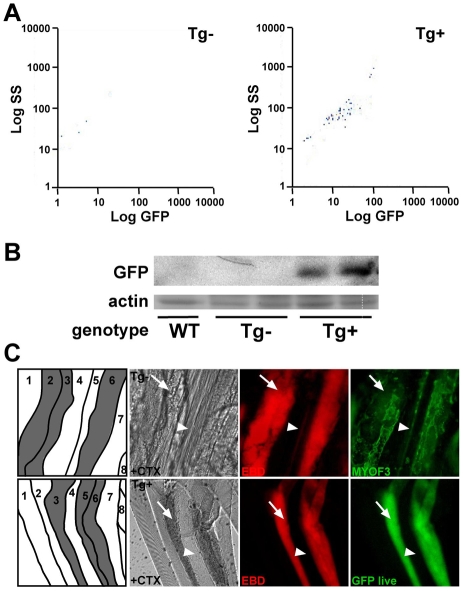
To identify the cell type expressing GFP in the transgenic 1543-bp–GFP mice, we enzymatically digested muscle from newborn uninjured mice and isolated only the mononuclear cells for FACS analysis. FACS analysis detected GFP fluorescence in the 1543-bp–GFP suspension, indicating that GFP positivity derives from mononuclear muscle-derived cells (Fig. 6A). This culture protocol from newborn uninjured muscle typically yields cultures that are more than 90% myoblasts and less than 10% fibroblasts (Rando and Blau, 1994; Doherty et al., 2008). Myoblasts in culture are distinguished from fibroblasts on the basis of their refractive properties and elongated shape (Klamut et al., 1986). GFP expression in transgene-positive mononuclear muscle-derived myoblasts was confirmed by immunoblotting with an anti-GFP antibody. GFP expression was not detected in transgene-negative wild-type littermates or a wild-type control line, confirming myoferlin-promoter activity in myoblasts (Fig. 6B). We injected skeletal muscle of transgenic and littermate control mice with cardiotoxin to induce local muscle damage and with EBD to mark damaged muscle fibers. At 3 days post-injection, tissue was harvested. Schematics of isolated muscle are included in Fig. 6C identifying healthy fibers (white) and damaged fibers (gray). Dissected fibers were fixed on glass slides and stained with an anti-myoferlin antibody. Myoferlin was upregulated in dye-positive damaged fibers (numbered 2, 3 and 6 in Fig. 6C, upper panel), whereas little to no myoferlin expression was seen in healthy dye-negative fibers (numbered 1, 4, 5, 7 and 8 in Fig. 6C, upper panel). Similarly, live imaging of myofibers revealed GFP expression in dye-positive damaged myofibers (numbered 3, 5 and 6 in Fig. 6C, bottom panel), whereas little or no expression of GFP was seen in neighboring undamaged, EBD-negative myofibers (numbered 1, 2, 4, 7 and 8 in Fig. 6C, bottom panel). Therefore, GFP expression mirrors endogenous myoferlin expression; both are upregulated in dye-positive damaged muscle fibers.
Fig. 6.
In response to in vivo muscle damage, the 1543-bp myoferlin promoter drives GFP expression in myoblasts. (A) Muscle from newborn mice was digested to release mononuclear cells and then subjected to FACS analysis gated to detect GFP. Mononuclear cells from transgene (Tg)-positive newborn muscle express GFP, reflecting expression in myoblasts. (B) Immunoblot with an anti-GFP antibody, demonstrating GFP expression in myoblasts cultured from transgene-positive neonatal mice. (C) Transgene-negative and transgene-positive mice were injected with cardiotoxin (CTX) into skeletal muscle to induce focal damage. The animals were also injected with EBD to mark damaged myofibers. A schematic of individual fibers (white, healthy; gray, damaged) is shown, indicating that the myofibers run vertical in the images. At 3 days post-injection, transgene-negative muscle was harvested, and single live fibers were isolated and stained with an anti-myoferlin antibody (upper panels). The image shows aligned fibers running longitudinally from top to bottom. Myoferlin is upregulated in EBD-positive fibers numbered 2, 3 and 6, and not in the adjacent, intact EBD-negative myofibers numbered 1, 4, 5, 7 and 8 (upper panels). In transgene-positive muscle, GFP expression parallels myoferlin expression. Arrows indicate damaged fibers; arrowheads indicate healthy fibers.
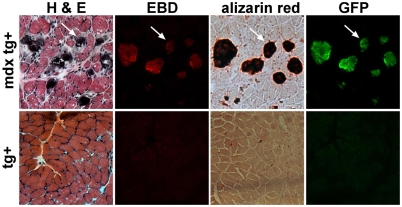
To assess the expression of myoferlin in damaged myotubes in a more physiological form of muscle damage, we bred mice containing the 1543-bp–GFP transgene to the mdx mouse model of Duchenne muscular dystrophy. mdx mice have continual muscle degeneration that exceeds the capacity for muscle regeneration, resulting in muscle wasting similar to what is observed in the Sgcg-null mice. The 1543-bp–GFP:mdx muscle displays signs of muscle degeneration and regeneration, namely centrally nucleated fibers (Fig. 7). Furthermore, basophilic degenerating and/or regenerating fibers are also evident (bluish fibers in Fig. 7, top left). These basophilic fibers are the same fibers that stain positive for EBD, indicating membrane damage (Fig. 7, top row). With membrane disruption, there is an increase in intracellular Ca2+, visualized by Alizarin Red. 1543-bp–GFP:mdx mice accumulated EBD-positive, Alizarin-Red-positive fibers within the diaphragm muscle, and GFP expression was present in the same myofibers (Fig. 7, top row). No basophilic, EBD-positive, Alizarin-Red-positive or GFP-positive myofibers were seen in 1543-bp–GFP mice in the absence of the mdx allele (Fig. 7, bottom row). These data show that the 1543-bp promoter is activated in vivo in a mouse model of muscular dystrophy in damaged, Ca2+-loaded myofibers, which is consistent with the role of myoferlin in injury response.
Fig. 7.
GFP expression from the myoferlin promoter drives expression in the mdx mouse model of muscular dystrophy. Serial sections of 1543-bp–GFP:mdx transgene (tg)-positive (top row) and wild-type transgene-positive (bottom row) diaphragm were stained with hematoxylin and eosin (H&E) to show gross muscle pathology. EBD-labeled fibers are shown in red. Alizarin Red staining identifies increased Ca2+ within damaged myofibers and this correlates with those fibers expressing GFP from the myoferlin promoter. No positive fibers were ever seen in the wild-type transgene-positive tissue. Arrows indicate the identical fiber.
Discussion
Muscle development is a multi-step process. During the first phase of muscle development, termed myoinitiation, mononucleated myoblasts fuse to each other to generate myotubes. Following this, myoaugmentation occurs, in which myoblasts fuse with existing myotubes, adding to the multinucleated syncytium and causing muscle growth. After injury, muscle regeneration is also a multi-step process. As in muscle development, activated myoblasts can fuse with one another, generating new myofibers, or they can fuse with existing myotubes, promoting muscle growth and repair (Antonio and Gonyea, 1993; Kelley, 1996; Pena et al., 1995). Cycles of degeneration and regeneration of skeletal myofibers are a hallmark of the muscular dystrophy phenotype. In the muscular dystrophies, as well as in other types of muscle damage, degeneration and regeneration are tightly linked. Muscle repair entails extensive cytoskeletal remodeling, cellular fusion, and differentiation. We determined the molecular signature of damaged versus intact myofibers in dystrophic muscle. In this setting, differentially expressed genes are independent of the primary genetic defect and are directly related to myofiber function and/or repair. We detected a large number of upregulated genes that mediate cytoskeletal remodeling and cell-cell signaling, suggesting that these myofibers are attempting repair along with regeneration.
The identification of myoferlin as a focally upregulated gene and protein product in damaged myofibers is consistent with muscle regeneration because myoferlin is upregulated in pre-fusion myoblasts and downregulated after fusion is complete (Davis et al., 2002; Doherty et al., 2005). Immunolocalization studies confirmed that myoferlin is expressed in the repairing multinucleate myofibers and surrounding mononuclear cells. Nearly all of the mononuclear cells in dye-positive regions, including Mac-1-positive cells, showed upregulation of myoferlin, as did myofibers themselves. These findings are important because dysferlin, the gene responsible for limb girdle muscular dystrophy 2B, has been shown to mediate resealing of damaged myofibers after laser injury. Given the upregulation of myoferlin in response to muscle damage, it is also possible that myoferlin contributes to membrane resealing in the damaged fibers. Both myoferlin and dysferlin can bind negatively charged phospholipids (Davis et al., 2002). Therefore, it is likely that both of these proteins contribute to repair of the multinucleate myofiber, although the molecular mechanisms might not be completely overlapping. Recently, a role for myoferlin in endocytic recycling was uncovered (Doherty et al., 2008); whether dysferlin also participates in this process is unknown.
Owing to its role in repair and its complex expression profile, modulation of myoferlin is tightly regulated. NFATc1 and NFATc3 regulate the myoferlin promoter. These data are consistent with the expression of NFATc1 and NFATc3 in myoblasts, in which they have the ability to translocate from the cytoplasm to the nucleus (Delling et al., 2000). Consistent with this pathway, the myoferlin promoter is sensitive to calcineurin activation. At the onset of myogenic differentiation, calcineurin phosphatase activity is increased (Delling et al., 2000). This increase in activation coincides temporally with the onset of myoferlin expression (Doherty et al., 2005), as well as NFATc3 translocation to the nucleus (Delling et al., 2000). The Ca2+-sensitive calcineurin-NFAT pathway is crucial not only in muscle development but also in muscle repair. Both in vivo and in vitro data have shown that blocking calcineurin and/or NFAT function causes defects in muscle differentiation and repair (Abbott et al., 1998; Friday et al., 2000; Horsley et al., 2001; O'Connor et al., 2007). The addition of CsA, a calcineurin inhibitor, to myoblast cultures causes a decrease in differentiation in a dose-dependent fashion, with a maximum inhibition of 75-90% (Abbott et al., 1998). This phenomenon is recapitulated in muscle of CsA-treated mice. Specifically, CsA-treated muscle displays a lack of centrally located nuclei, a distinct marker of muscle regeneration (Abbott et al., 1998). We found that the addition of CsA decreased myoferlin promoter activity. We expect that at least some of the negative effect of CsA on muscle regeneration can be explained by the downregulation of myoferlin.
The five NFAT isoforms are expressed in muscle; however, each isoform is activated at different stages throughout muscle growth and differentiation (Abbott et al., 1998; Cho et al., 2007; O'Connor et al., 2007). The loss of individual NFAT isoforms results in unique muscle defects, suggesting specific functions for each NFAT isoform. Interestingly, the phenotype of the myoferlin-null mouse is similar to the NFATc2-null mouse (Doherty et al., 2005; Horsley et al., 2001). Both the myoferlin-null and NFATc2-null mice have decreased fiber cross-sectional area, fusion defects and delayed muscle repair. Our data suggests that NFATc3 binds to the myoferlin promoter in the early myoblast growth stage, allowing for high expression of myoferlin, preparing the myoblasts for the upcoming fusion events. Under our experimental growth conditions, NFATc3 can translocate to the nucleus, whereas NFATc2 and NFATc4 are not active until 48 hours after differentiation media is added (Abbott et al., 1998; Cho et al., 2007). Later in differentiation, when large multinucleate myotubes are present, NFATc2 and NFATc4 are no longer active (Abbott et al., 1998; Cho et al., 2007). This decrease in NFAT activity correlates with decreased myoferlin expression in myotubes, suggesting that NFATs might differentially bind the myoferlin promoter throughout muscle development.
Recently, myoferlin has been implicated in regulating growth-factor signaling that affects myoblast fusion and muscle growth (Demonbreun et al., 2009). NFATc2 activates growth-factor promoters such as that expressing IL-4, a protein that is secreted by myotubes to recruit myoblasts and is crucial for myoaugmentation (Horsley et al., 2003). We hypothesize that the phenotypes of the NFATc2-null and myoferlin-null mice are similar owing to the loss of these fusion signals. Both loss of NFATc2, resulting in decreased production of growth factors, or the loss of myoferlin, resulting in a decreased response to growth factors, would result in similar muscle defects in growth and repair. The role of NFATc1 has yet to be determined because NFATc1-null mice die in utero from cardiac defects (de la Pompa et al., 1998; Ranger et al., 1998). However, NFATc1 expression is upregulated in the mdx mouse model of muscular dystrophy and after locally induced injury in rat, suggesting a role for NFATc1 in muscle repair similar to the endogenous myoferlin expression profile (Sakuma et al., 2003; Stupka et al., 2006a). It has also been reported that the NFATc3-null mouse has a decreased number of myofibers (Kegley et al., 2001). Therefore, a combinatorial effect of multiple NFATc factors probably regulates myoferlin expression in vivo, consistent with our in-vitro-based studies.
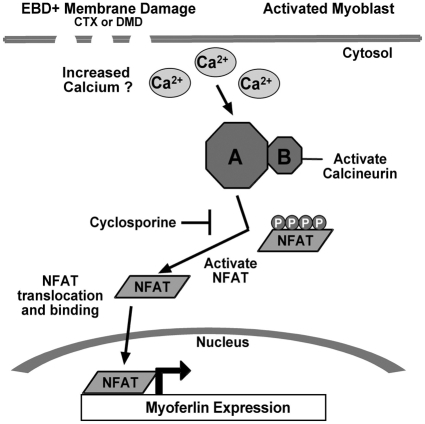
The expression pattern directed by the upstream region of the myoferlin promoter recapitulates the expression of endogenous myoferlin, including both its positive and negative regulation. Myoferlin expression is highest in myoblasts that are elongated and are preparing to fuse to myotubes (Davis et al., 2002). Following fusion, myoferlin expression is reduced in mature, undamaged muscle fibers. The 1543-bp myoferlin promoter demonstrated this same pattern of expression when introduced into cultured myoblasts and myotubes. When these myotubes were damaged by brief exposure to cardiotoxin, the levels of myoferlin expression in myotubes increased. This increase might represent a contribution from myoblasts that have recently fused or from myotube nuclei, or both. GFP expression was seen 48 hours after cardiotoxin treatment of myotubes, and this time frame is sufficient to support myoblast fusion in addition to gene expression from myotube nuclei. Similarly, the pattern of myoferlin expression in vivo was recapitulated using transgenesis. In vivo, we separated the mononuclear component from the multinucleate myotubes. Interestingly, the fibers that expressed the highest levels of GFP were the same fibers that also displayed high levels of Ca2+. Thus, we favor a model in which intracellular Ca2+ levels increase in damaged myofibers, activating both calcineurin and NFAT transcription factors. NFATs bind and activate the myoferlin promoter, increasing the expression of myoferlin mRNA and protein (Fig. 8). Furthermore, upon muscle damage, myoblasts are activated, myoferlin is expressed, and myoblasts are recruited to the site of damage for fusion to the injured fibers. Therefore, we believe that myoferlin protein expression is increased from myoferlin mRNA that derives from both myoblast as well as myotube nuclei.
Fig. 8.
Model for myoferlin expression in injured myofibers. Disruption of myofiber membranes leads to increased intracellular Ca2+. This increase in Ca2+ activates calcineurin, which results in NFAT dephosphorylation and nuclear translocation. Within the nucleus, NFAT is now positioned to activate myoferlin expression.
The 1543-bp–GFP transgenic mouse line is an ideal tool for studying muscle repair in general, because this model serves as a ‘damage sensor’. To date, transgenic GFP-tagged mouse models have been created for the key myogenic regulators, Pax7 and Pax3. During muscle repair, these proteins are transiently expressed in activated satellite cells and are downregulated in daughter myoblasts and myotubes (Buckingham et al., 2003; Seale et al., 2000). Thus, although the Pax3-GFP and Pax7-GFP mouse models are well suited for the study of satellite-cell activation, they do not allow the study of damaged myotubes or myoblasts participating in the repair process. Marking myotubes that are undergoing degeneration and/or regeneration and myoblasts, which actively mediate muscle repair, is crucial in furthering our understanding of the repair process both in heritable and environmentally induced muscle damage.
Materials and Methods
Animals and tissue harvest for laser microdissection, verification and muscle-injury assays
Mice lacking γ-sarcoglycan (Sgcg null) were generated by targeting exon 2 (Hack et al., 2000; Hack et al., 1998). mdx mice were obtained from Jackson Laboratories (Bar Harbor, ME). Myoferlin (mko) mice were generated previously by deleting the first exon (Doherty et al., 2005). For laser microdissection, two Sgcg-null mice, 13.5 weeks of age, were injected intraperitoneally with a 10 mg/ml solution of EBD at 1% of total body weight (Hack et al., 2000; Hamer et al., 2002). The mice were sacrificed 24 hours post-injection. Quadriceps muscles were dissected and frozen in cooled isopentane as described (Hack et al., 1998). For verification, EBD-injected and non-injected wild-type, Sgcg null and mdx muscle was harvested as described above. To induce muscle injury, cardiotoxin injections were done as 100 μl injections of 10 μM cardiotoxin in PBS or PBS alone into the quadriceps or gastrocnemius/soleus muscles from 14- to 20-week-old mice as described (Volonte et al., 2005). After 3 or 5 days, muscle from injected and control legs were harvested for immunoblot analysis and for immunofluorescence microscopy. For live-fiber analysis, cardiotoxin-damaged fibers were isolated by manual dissection, placed in PBS between a glass coverslip and slide, and immediately imaged using Ivision software and a Zeiss Axiophot microscope.
Rapid immunofluorescence microscopy and laser microdissection
Skeletal muscle was collected in 6 μm longitudinal sections. One slide was processed for immunofluorescence microscopy with a rabbit polyclonal antibody to spectrin (catalog number AB993; Chemicon, Temecula, CA) using RNase-free technique, and staining was carried out as described previously (Bi et al., 2002). The anti-spectrin antibody was diluted (1:4) in PBS with 400 U/ml RNase inhibitor (Invitrogen, Carlsbad, CA) and applied to the slides for 4 minutes followed by a 30-second PBS wash. A goat anti-rabbit secondary antibody conjugated to Alexa Fluor 555 (Molecular Probes, Eugene, OR) was diluted in PBS (1:50) and applied to the slide for 2 minutes. The slide was washed three times, for 30 seconds each, dipped in ethanol for 3 seconds and then allowed to air dry for 2 minutes prior to laser microdissection.
A Leica AS LMD microscope was used to capture individual cells or cell clusters and regions of interest were selected manually. Upon command, an ultraviolet laser beam cut and released the cells into a PCR tube that contained lysis buffer from the RNeasy micro kit (Invitrogen, Carlsbad, CA). A Rhodamine filter set was used to detect both EBD and Cy3 fluorescence. For each group, a mean of 22.6×103±1.2×103 μm2 were collected (volume=135.5×103±7.3×103 μm3) representing approximately 27×103±1.5×103 cells (each 6 μm in thickness) from each group.
RNA isolation, amplification hybridization and analysis
Reverse transcriptase (RT)-PCR was performed to test the integrity of the RNA isolated using primers to dystrophin mRNA (data not shown). All samples within each group (four groups total) were pooled prior to RNA extraction. The RNeasy micro kit was used following the manufacturer's instructions except that 20 ng bacterial rRNA (Roche, Indianapolis, IA) was used as carrier RNA. The RiboAmp OA Amplification kit (Arcturus, Mountain View, CA) was used to perform one round of linear amplification prior to labeling, according to the manufacturer's instructions. A total of 200 ng Polyd(I-C) (Roche, Indianapolis, IA) was used as an RNA carrier. According to Affymetrix protocols, antisense RNA (aRNA) was streptavidin-labeled and each sample was hybridized to a Mouse 430A chip (Affymetrix, Santa Clara, CA) at the Functional Genomics Core Facility. Raw fluorescent data were imported into the Bioconductor platform (Gentleman et al., 2004). The background fluorescence was corrected, data were normalized and relative fluorescent values were determined by robust multichip analysis (RMA). Fluorescent values were then imported into Microsoft Excel for Significance Analysis of Microarray (SAM) to generate a differentially expressed gene list comparing the EBD-positive and EBD-negative myofibers (Tusher et al., 2001). The fluorescent data were analyzed by Student's t-test in the Bioconductor platform and by Rank products analysis by running a Windows script (rankproducts_FDR.exe) (Breitling et al., 2004).
Comparative sequence analysis
1543 bp 5′ of the translational start site of both human and mouse myoferlin was downloaded from http://genome.ucsc.edu and compared using the Vista program (http://genome.lbl.gov/vista) with a window size of 100 bp and a minimum sequence identity of 70%. Conserved NFAT-transcription-factor-binding sites were identified using the rVISTA program. Predictions were made based on the TRANSFAC Professional 9.2 library using the default core similarity value of 0.75 and the matrix similarity value of 0.70. Conserved transcription binding sites were defined as having greater than 80% similarity over a 24-bp window. Potential transcription-factor-binding sites were also determined using the Transcriptional Element Search System (TESS) (http://www.cbil.upenn.edu) using the default settings (Frazer et al., 2004; Loots et al., 2002; Mayor et al., 2000; Schug and Overton, 2007).
Plasmid construction
The 1543-bp fragment of the myoferlin promoter was amplified from genomic DNA using primers from supplementary material Table S2. All products were sequenced and verified. All promoter fragments were cloned into the pGL2-basic vector (Promega, E1641) using KpnI and Sac1 to generate the reporter vectors 1543-bp–pGL2, 918-bp–pGL2, 729-bp–pGL2 and 226-bp–pGL2. NFAT mutations were introduced into the 1543-bp–pGL2 reporter construct sequences listed in supplementary material Table S2. Using the restriction enzymes Sac1 and KpnI, the 1543-bp promoter was cloned into the pEGFP-N3 vector (Clontech, catalog number 6080-1). The CMV promoter was removed from the vector using AseI and BglII to generate the 1543-bp–GFP plasmid. The transgene construct was generated from the 1543-bp–GFP construct using the restriction enzymes SspI and BglII.
Cell culture and isolation
C2C12 cells were obtained from ATCC (catalog number CRL-1772). For all experiments, cells were grown to 95% confluency, the point at which C2C12 cells are prefusion myoblasts, in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in 7% CO2. DMEM supplemented with 2% horse serum and 1% penicillin/streptomycin was used as differentiation media. Myoblasts were isolated as described previously (Doherty et al., 2008). Myoblasts were lysed in 1 ml of lysis buffer [25 mM Tris (pH 7.4), 300 mM NaCl, 1 mM CaCl2, 1% Triton-X 100 with Complete Mini Protease Inhibitor cocktail (Roche Molecular Biochemicals)]. Cellular debris was removed, and the protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Protein Laboratories). A total of 50 μg of protein was separated on a 12% acrylamide gel. Equivalently loaded gels were stained with Gel Code Blue (Pierce) or transferred to PVDF Immobilon-P membrane (Millipore).
Transfections and luciferase reporter assays
For transfections, 50,000 C2C12 cells were plated into each well of a 12-well tissue-culture plate. After 24 hours, cells were transfected with 2.5 μg reporter plasmid, 10 μl Lipofectamine (Invitrogen), 10 μl Plus reagent (Invitrogen) and 50 ng of pRL-SV40 Renilla luciferase plasmid (Promega, E2231) in Opti-MEM. The pGL2-basic vector or the pGL2-SV40 control vector (Promega, E1631) replaced the reporter vector as a negative or positive control, respectively. For co-transfection assays, 0.5 μg/well of 1543-bp–pGL2 reporter vector was transfected with 0.5 mg/well of pcDL-Sra-ΔCnA, which was generously provided by Jeffery Molkentin (Oka et al., 2005), pREP-NFATc1 (AddGene) and pREP-NFATc3 (AddGene), alone or in combination. The total amount of DNA used per well was adjusted with the promoterless pGL-2-basic vector. Cells were incubated with the transfection mix for 3 hours at 37°C and then the mix was replaced with growth media. Forty-eight hours later, cells were lysed and analyzed for firefly and Renilla luciferase reporter gene activity using the Dual Luciferase Reporter Assay system as specified by the manufacturer (Promega, E1910). Data was normalized to Renilla luciferase activity. All experiments were performed in triplicate and repeated at least three times. For the CsA luciferase assay, cells were transfected with the 1543-bp–pGL2 construct as described above. At 18 hours post-transfection, 1 μM or 5 μM CsA (Invitrogen, PHZ1054) was added to the cells. Twenty-four hours later, cells were lysed and analyzed for luciferase activity as described above. For transfections with cardiotoxin exposure, 2.5 μg of the 1543-bp–GFP construct or CMV-GFP construct was transfected into 50,000 C2C12 cells plated on glass coverslips in six-well plates. Growth media was replaced with differentiation media after 24 hours. Cultures were allowed to differentiate for 3 days. GFP expression was analyzed using live-cell immunofluorescence microscopy. Cells were treated with 1 μM cardiotoxin (Calbiochem, catalog number 217504) after having differentiated for 6 days or cells were treated with 1 μM cardiotoxin and 1 μM CsA. GFP expression was analyzed using live-cell immunofluorescence microscopy 48 hours after the addition of cardiotoxin.
Chromatin immunoprecipitation
ChIP assays were performed in triplicate using the Upstate Chip Assay Kit according to the manufacturer's instructions (Upstate, catalog number 17-295) with Proteinase K (Puregene, catalog number D50K5) and glycogen (Roche, catalog number 901393). 106 confluent C2C12 cells were sonicated to shear chromatin to 200-500 bp. For immunoprecipitation, 2 μg of the antibodies anti-NFATc1, anti-NFATc2 or anti-NFATc3 (Santa Cruz Biotechnology, catalog numbers sc-7294, sc-7296 and sc-8321, respectively) were incubated with the cell lysates overnight at 4°C with rotation. Immunoprecipitation with Immunopure IgG (Pierce, catalog number 31160) was performed as a control. Myoferlin-promoter NFAT-binding sequences were amplified using the primers in supplementary material Table S2.
Electrophoretic mobility shift assay
EMSAs were performed using a double-stranded oligonucleotide containing a hypothetical NFAT-binding site from the 1543-bp myoferlin promoter (see supplementary material Table S1). Twenty-five μg of C2C12 nuclear extracts were incubated with 30,000 cpm of 32P-labeled double-stranded oligonucleotide, 1 μg of poly(dI-dC)-poly(dI-dC) and 1.5 μl of 10× Ficoll binding buffer [20% Ficoll, 50 mM Tris (pH 7.5), 50 mM KCl, 5 mM EDTA, 5 mM dithiothreitol, 375 mM KCL] on ice for 30 minutes in a 30 μl reaction. For super shift assays, 2 μg of anti-NFATc1 (Santa Cruz Biotechnology, catalog number sc-7294) or anti-NFATc3 (Santa Cruz Biotechnology, catalog number sc-8321) was incubated with the reaction mixture. Protein complexes were resolved from the free 32P-labeled probe using a 5% acrylamide gel. Primers are shown in supplementary material Table S1.
Immunoblotting, immunofluorescence microscopy and antibodies
The anti-myoferlin antibody (MYOF3, 1:2000) (Doherty et al., 2005) and an anti-GFP antibody (1:1000, Santa Cruz, sc-9996) were used. The secondary antibodies goat anti-rabbit or goat anti-mouse conjugated to horseradish peroxidase (Jackson ImmunoResearch) were used at a dilution of 1:5000. Blocking and antibody incubations were done in Starting Block T20 (Invitrogen) for all antibodies. ECL-Plus chemiluminescence (Amersham-Pharmacia) and Kodak Biomax MS film was used for detection. For microscopy, MYOF3 was used at 1:100 or 1:500 dilution (Doherty et al., 2005). Goat anti-rabbit conjugated to Alexa Fluor 594 was used at 1:2000 dilution or to Alexa Fluor 488 at 1:5000 dilution (Molecular Probes). Phalloidin conjugated to Alexa Fluor 488 (Molecular Probes, A12379) was used at 1:2000. Mac-1 (BD Bioscience, 557395) was used at 1:100 and a strepavidin–Alexa-Fluor-488 secondary (Molecular Probes, S-11223) was used at 1:2500. Blocking and antibody incubations were in 1×TBS and 5% fetal bovine serum. Coverslips were mounted using Vectashield with DAPI. Images were captured using a Zeiss Axiophot microscope and AxioVision software (Carl Zeiss).
FACS analysis
Mononuclear cells analyzed by flow cytometry were prepared as above. Cells were washed and resuspended in PBS for analysis on a FACS Canto (BD Biosciences). After gating to remove dead cells, aggregated cells and debris, a minimum of 10,000 events was scored per culture. Cells were derived from independent animals per genotype. Fluorescent gates were set to include 0% wild-type cells. The percentage of fluorescent cells was determined by the number of cells remaining in this gate. FACS data was analyzed using FlowJo software (Tree Star).
Mdx 1543-bp–GFP transgenic mouse tissue analysis
The 1543-bp–GFP mouse line 68 was crossed to an mdx female mouse (Jackson Laboratories). Diaphragm muscles from male transgene-positive mdx mice and male transgene-negative littermates were analyzed using 7-μm serial sections. Unfixed tissue sections were subjected to hematoxylin and eosin, 2% Alizarin Red (Chiu et al., 2009) for 2 minutes then rinsed in acetone to visualize calcification, or 10 ng/ml EBD for 30 minutes then three 5-minute PBS washes to identify damaged myofibers. Tissues were fixed in ethanol for endogenous GFP expression. Coverslips were mounted with permamount or Vectashield with DAPI. Images were captured using a Zeiss Axiophot microscope and Ivision software.
Supplementary Material
Acknowledgments
This work was supported by NIH NS4776, HL61322 and T32 HL 7381, and the Muscular Dystrophy Association. We acknowledge the Jain Foundation. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/14/2413/DC1
References
- Abbott K. L., Friday B. B., Thaloor D., Murphy T. J., Pavlath G. K. (1998). Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol. Biol. Cell 9, 2905-2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio J., Gonyea W. J. (1993). Skeletal muscle fiber hyperplasia. Med. Sci. Sports Exerc. 25, 1333-1345 [PubMed] [Google Scholar]
- Ault K. A., Springer T.A. (1981). Cross reaction of a rant-anti-mouse phagocyte specific monoclonal antibody (anti Mac-1) with human monocytes and natural killer cells. J. Immunol. 126, 359-364 [PubMed] [Google Scholar]
- Bansal D., Campbell K. P. (2004). Dysferlin and the plasma membrane repair in muscular dystrophy. Trends Cell. Biol. 14, 206-213 [DOI] [PubMed] [Google Scholar]
- Bashir R., Britton S., Strachan T., Keers S., Vafiadaki E., Lako M., Richard I., Marchand S., Bourg N., Argov Z., et al. (1998). A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 20, 37-42 [DOI] [PubMed] [Google Scholar]
- Bi W. L., Keller-McGandy C., Standaert D. G., Augood S. J. (2002). Identification of nitric oxide synthase neurons for laser capture microdissection and mRNA quantification. Biotechniques 33, 1274-1283 [DOI] [PubMed] [Google Scholar]
- Breitling R., Armengaud P., Amtmann A., Herzyk P. (2004). Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83-92 [DOI] [PubMed] [Google Scholar]
- Buckingham M., Bajard L., Chang T., Daubas P., Hadchouel J., Meilhac S., Montarras D., Rocancourt D., Relaix F. (2003). The formation of skeletal muscle: from somite to limb. J. Anat. 202, 59-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabria E., Ciciliot S., Moretti I., Garcia M., Picard A., Dyar K. A., Pallafacchina G., Tothova J., Schiaffino S., Murgia M. (2009). NFAT isoforms control activity-dependent muscle fiber type specification. Proc. Natl. Acad. Sci. USA 106, 13335-13340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y. H., Hornsey M. A., Klinge L., Jorgensen L. H., Laval S. H., Charlton R., Barresi R., Straub V., Lochmuller H., Bushby K. (2009). Attenuated muscle regeneration is a key factor in dysferlin-deficient muscular dystrophy. Hum. Mol. Genet. 18, 1976-1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. Y., Yao K., Bode A. M., Bergen H. R., 3rd, Madden B. J., Oh S. M., Ermakova S., Kang B. S., Choi H. S., Shim J. H., et al. (2007). RSK2 mediates muscle cell differentiation through regulation of NFAT3. J. Biol. Chem. 282, 8380-8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D. B., Delmonte A. J., Ly C. T., McNally E. M. (2000). Myoferlin, a candidate gene and potential modifier of muscular dystrophy. Hum. Mol. Genet. 9, 217-226 [DOI] [PubMed] [Google Scholar]
- Davis D. B., Doherty K. R., Delmonte A. J., McNally E. M. (2002). Calcium-sensitive phospholipid binding properties of normal and mutant ferlin C2 domains. J. Biol. Chem. 277, 22883-22888 [DOI] [PubMed] [Google Scholar]
- de la Pompa J. L., Timmerman L. A., Takimoto H., Yoshida H., Elia A. J., Samper E., Potter J., Wakeham A., Marengere L., Langille B. L., et al. (1998). Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 392, 182-186 [DOI] [PubMed] [Google Scholar]
- Delling U., Tureckova J., Lim H. W., De Windt L. J., Rotwein P., Molkentin J. D. (2000). A calcineurin-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol. Cell. Biol. 20, 6600-6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun A. R., Posey A. D., Heretis K., Swaggart K. A., Earley J. U., Pytel P., McNally E. M. (2009). Myoferlin is required for insulin-like growth factor response and muscle growth. FASEB J. 24, 1284-1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty K. R., Cave A., Davis D. B., Delmonte A. J., Posey A., Earley J. U., Hadhazy M., McNally E. M. (2005). Normal myoblast fusion requires myoferlin. Development 132, 5565-5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty K. R., Demonbreun A. R., Wallace G. Q., Cave A., Posey A. D., Heretis K., Pytel P., McNally E. M. (2008). The endocytic recycling protein EHD2 interacts with myoferlin to regulate myoblast fusion. J. Biol. Chem. 283, 20252-20260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273-W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friday B. B., Horsley V., Pavlath G. K. (2000). Calcineurin activity is required for the initiation of skeletal muscle differentiation. J. Cell Biol. 149, 657-666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack A. A., Ly C. T., Jiang F., Clendenin C. J., Sigrist K. S., Wollmann R. L., McNally E. M. (1998). Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J. Cell Biol. 142, 1279-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack A. A., Lam M. Y., Cordier L., Shoturma D. I., Ly C. T., Hadhazy M. A., Hadhazy M. R., Sweeney H. L., McNally E. M. (2000). Differential requirement for individual sarcoglycans and dystrophin in the assembly and function of the dystrophin-glycoprotein complex. J. Cell Sci. 113, 2535-2544 [DOI] [PubMed] [Google Scholar]
- Hamer P. W., McGeachie J. M., Davies M. J., Grounds M. D. (2002). Evans Blue Dye as an in vivo marker of myofibre damage: optimising parameters for detecting initial myofibre membrane permeability. J. Anat. 200, 69-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslett J. N., Sanoudou D., Kho A. T., Bennett R. R., Greenberg S. A., Kohane I. S., Beggs A. H., Kunkel L. M. (2002). Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc. Natl. Acad. Sci. USA 99, 15000-15005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslett J. N., Sanoudou D., Kho A. T., Han M., Bennett R. R., Kohane I. S., Beggs A. H., Kunkel L. M. (2003). Gene expression profiling of Duchenne muscular dystrophy skeletal muscle. Neurogenetics 4, 163-171 [DOI] [PubMed] [Google Scholar]
- Horsley V., Friday B. B., Matteson S., Kegley K. M., Gephart J., Pavlath G. K. (2001). Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J. Cell Biol. 153, 329-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V., Jansen K. M., Mills S. T., Pavlath G. K. (2003). IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell 113, 483-494 [DOI] [PubMed] [Google Scholar]
- Kegley K. M., Gephart J., Warren G. L., Pavlath G. K. (2001). Altered primary myogenesis in NFATC3(−/−) mice leads to decreased muscle size in the adult. Dev. Biol. 232, 115-126 [DOI] [PubMed] [Google Scholar]
- Kelley G. (1996). Mechanical overload and skeletal muscle fiber hyperplasia: a meta-analysis. J. Appl. Physiol. 81, 1584-1588 [DOI] [PubMed] [Google Scholar]
- Klamut H. J., Lin C. H., Strickland K. P. (1986). Normal and dystrophic hamster myoblast and fibroblast growth in culture. Muscle Nerve 9, 597-605 [DOI] [PubMed] [Google Scholar]
- Lennon N. J., Kho A., Bacskai B. J., Perlmutter S. L., Hyman B. T., Brown R. H., Jr (2003). Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J. Biol. Chem. 278, 50466-50473 [DOI] [PubMed] [Google Scholar]
- Liu J., Aoki M., Illa I., Wu C., Fardeau M., Angelini C., Serrano C., Urtizberea J. A., Hentati F., Hamida M. B., et al. (1998). Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 20, 31-36 [DOI] [PubMed] [Google Scholar]
- Loots G. G., Ovcharenko I., Pachter L., Dubchak I., Rubin E. M. (2002). rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 12, 832-839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rodriguez C., Aramburu J., Rakeman A. S., Rao A. (1999). NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc. Natl. Acad. Sci. USA 96, 7214-7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda R., Nishikawa A., Tanaka H. (1995). Visualization of dystrophic muscle fibers in mdx mouse by vital staining with Evans blue: evidence of apoptosis in dystrophin-deficient muscle. J. Biochem. (Tokyo) 118, 959-964 [DOI] [PubMed] [Google Scholar]
- Mayor C., Brudno M., Schwartz J. R., Poliakov A., Rubin E. M., Frazer K. A., Pachter L. S., Dubchak I. (2000). VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046-1047 [DOI] [PubMed] [Google Scholar]
- O'Connor R. S., Mills S. T., Jones K. A., Ho S. N., Pavlath G. K. (2007). A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J. Cell Sci. 120, 149-159 [DOI] [PubMed] [Google Scholar]
- Oka T., Dai Y. S., Molkentin J. D. (2005). Regulation of calcineurin through transcriptional induction of the calcineurin A beta promoter in vitro and in vivo. Mol. Cell. Biol. 25, 6649-6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontell M., Hughes D., Bourke D. (1988). Morphometric analysis of the developing mouse soleus muscle. Am. J. Anat. 181, 279-288 [DOI] [PubMed] [Google Scholar]
- Pena J., Jimena I., Luque E., Vaamonde R. (1995). New fiber formation in rat soleus muscle following administration of denervated muscle extract. J. Neurol. Sci. 128, 14-21 [DOI] [PubMed] [Google Scholar]
- Rando T. A., Blau H. M. (1994). Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125, 1275-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger A. M., Grusby M. J., Hodge M. R., Gravallese E. M., de la Brousse F. C., Hoey T., Mickanin C., Baldwin H. S., Glimcher L. H. (1998). The transcription factor NF-ATc is essential for cardiac valve formation. Nature 392, 186-190 [DOI] [PubMed] [Google Scholar]
- Rao A., Luo C., Hogan P. G. (1997). Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15, 707-747 [DOI] [PubMed] [Google Scholar]
- Sakuma K., Nishikawa J., Nakao R., Watanabe K., Totsuka T., Nakano H., Sano M., Yasuhara M. (2003). Calcineurin is a potent regulator for skeletal muscle regeneration by association with NFATc1 and GATA-2. Acta Neuropathol. 105, 271-280 [DOI] [PubMed] [Google Scholar]
- Schreiber S. L., Crabtree G. R. (1992). The mechanism of action of cyclosporin A and FK506. Immunol. Today 13, 136-142 [DOI] [PubMed] [Google Scholar]
- Schug J., Overton G. C. (2007). TESS: Transcription Element Search Software on the WWW: Computational Biology and Informatics Laboratory School of Medicine University of Pennsylvania, 1997
- Seale P., Sabourin L. A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M. A. (2000). Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777-786 [DOI] [PubMed] [Google Scholar]
- Straub V., Rafael J. A., Chamberlain J. S., Campbell K. P. (1997). Animal models for muscular dystrophy show different patterns of sarcolemmal disruption. J. Cell Biol. 139, 375-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupka N., Gregorevic P., Plant D. R., Lynch G. S. (2004). The calcineurin signal transduction pathway is essential for successful muscle regeneration in mdx dystrophic mice. Acta Neuropathol. 107, 299-310 [DOI] [PubMed] [Google Scholar]
- Stupka N., Michell B. J., Kemp B. E., Lynch G. S. (2006a). Differential calcineurin signalling activity and regeneration efficacy in diaphragm and limb muscles of dystrophic mdx mice. Neuromuscular Disord. 16, 337-346 [DOI] [PubMed] [Google Scholar]
- Stupka N., Plant D. R., Schertzer J. D., Emerson T. M., Bassel-Duby R., Olson E. N., Lynch G. S. (2006b). Activated calcineurin ameliorates contraction-induced injury to skeletal muscles of mdx dystrophic mice. J. Physiol. 575, 645-656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupka N., Schertzer J. D., Bassel-Duby R., Olson E. N., Lynch G. S. (2007). Calcineurin-A alpha activation enhances the structure and function of regenerating muscles after myotoxic injury. Am. J. Physiol. Regul. Integr. Comp. Physiol. 293, R686-R694 [DOI] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R., Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116-5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volonte D., Liu Y., Galbiati F. (2005). The modulation of caveolin-1 expression controls satellite cell activation during muscle repair. FASEB J. 19, 237-239 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.