Abstract
For over a century, muscle formation in the ascidian embryo has been representative of ‘mosaic’ development. The molecular basis of muscle-fate predetermination has been partly elucidated with the discovery of Macho1, a maternal zinc-finger transcription factor necessary and sufficient for primary muscle development, and of its transcriptional intermediaries Tbx6b and Tbx6c. However, the molecular mechanisms by which the maternal information is decoded by cis-regulatory modules (CRMs) associated with muscle transcription factor and structural genes, and the ways by which a seamless transition from maternal to zygotic transcription is ensured, are still mostly unclear. By combining misexpression assays with CRM analyses, we have identified the mechanisms through which Ciona Macho1 (Ci-Macho1) initiates expression of Ci-Tbx6b and Ci-Tbx6c, and we have unveiled the cross-regulatory interactions between the latter transcription factors. Knowledge acquired from the analysis of the Ci-Tbx6b CRM facilitated both the identification of a related CRM in the Ci-Tbx6c locus and the characterization of two CRMs associated with the structural muscle gene fibrillar collagen 1 (CiFCol1). We use these representative examples to reconstruct how compact CRMs orchestrate the muscle developmental program from pre-localized ooplasmic determinants to differentiated larval muscle in ascidian embryos.
Keywords: Ciona intestinalis, Ascidian, Brachyury, Tbx6, Collagen, Macho, Muscle, Notochord, T-box, Transcription
Introduction
Ascidians, or tunicates, provide an essential genetic and evolutionary reference for studies of chordate development because of their well-characterized modes of development, their experimental amenability, their phylogenetic proximity to vertebrates and the organization of their larval body plan (Davidson and Christiaen, 2006; Delsuc et al., 2006; Passamaneck and Di Gregorio, 2005). Ascidian larvae are characterized by the presence of an axial notochord flanked by two rows of muscle cells, classified as primary (B-lineage) and secondary (A- and b-lineage) (Meedel et al., 1987), and by a rudimentary dorsal nervous system (Satoh, 1994). As shown by classical embryological studies, the development of the primary larval muscle cells in solitary ascidians proceeds cell-autonomously from the first cleavages (Meedel et al., 1987; Nishida, 1992; Ortolani, 1955). Around the neurula stage, muscle precursors begin to position themselves paraxially (e.g. Rhee et al., 2005) and continue to divide synchronously until the early tailbud stage; after which they elongate ~fourfold, in the absence of cell division and evident positional reorganization, to allow tail extension (Passamaneck et al., 2007).
Recent molecular investigations have shown that primary muscle development is initially orchestrated by a maternally deposited muscle determinant, the zinc-finger transcription factor Macho-1 (Nishida, 2002; Nishida and Sawada, 2001). In Ciona intestinalis, overexpression of Ci-macho1 and its morpholino-mediated knockdown cause ectopic expression and silencing, respectively, of the T-box transcription factor genes Ci-Tbx6b and Ci-Tbx6c, indicating that Ci-Macho1 somehow activates their early expression (Yagi et al., 2004a). In turn, Ci-Tbx6b and Ci-Tbx6c, which are considered the result of a recent lineage-specific duplication event (Dehal et al., 2002), function as mediators of Ci-Macho1, because their morpholino-induced knockdown results in the downregulation of muscle-specific structural genes, such as those encoding muscle actin, myosin chains and creatine kinase, among others (Yagi et al., 2005). Similarly, in Halocynthia roretzi, another solitary ascidian distantly related to Ciona, Hr-Macho1 has been shown to activate the expression of Hr-Tbx6, as well as that of structural muscle genes, such as Hr-muscle actin (Sawada et al., 2005). In addition to controlling primary muscle formation, Ci-Macho1 acts cooperatively with β-catenin to induce the formation of the heart field by activating Ci-Mesp (Christiaen et al., 2009; Davidson et al., 2005; Satou et al., 2004); the function of Ci-Macho1 in heart specification is also mediated by the Ci-Tbx6-related transcription factors (Christiaen et al., 2009; Davidson et al., 2005).
Despite the wealth of information on the gene regulatory network that initiates and sustains muscle development in Ciona and other ascidian embryos (Hudson and Yasuo, 2008; Imai et al., 2006; Meedel et al., 2007), the cis-regulatory mechanisms that integrate maternal and zygotic information along this complex gene cascade, from egg to swimming larva, are largely unexplored, although some common logic has been identified in a limited subset of cis-regulatory modules (CRMs) (Erives, 2009). For example, it is still unclear how Ci-Macho1 activates its transcriptional intermediaries, and information on the structure and function of the CRMs controlling expression of direct Ci-Macho1 targets, and of their downstream structural muscle genes, is limited and fragmentary.
By combining misexpression assays and CRM analyses, we have begun to address these points and to gain a mechanistic understanding of how the activity of maternal and zygotic transcription factors is coordinated at the cis-regulatory level. Through misexpression assays, we have probed the transcriptional plasticity of the Ciona mesoderm to assess the ability of temporally and spatially misexpressed Ci-Macho1 to ectopically activate muscle genes in notochord cells, and we have identified a cross-regulatory interaction between Ci-Tbx6b and Ci-Tbx6c. Through a combination of sequence inspection, point-mutation analyses and electrophoretic mobility assays, we have characterized CRMs from the upstream genomic regions of these two genes, which occupy important nodes in the muscle gene regulatory network, as well as two muscle CRMs from a representative structural gene, Fibrillar Collagen-1 (CiFCol1), which is robustly expressed in muscle cells from late gastrulation until the late tailbud stages.
We present evidence that the expression patterns of Ci-Tbx6b and CiFCol1 are recapitulated by compact bipartite CRMs, consisting of ‘early’ and ‘late’ moieties that are essential for initiation and maintenance of gene expression, respectively, and that the coordinated activity of these CRMs ensures continuity in the expression of genes necessary for muscle development and differentiation.
Results
Effects of the ectopic expression of Ci-Macho1 on the transcription of Ci-Tbx6b and Ci-Tbx6c
To gain insights on the mechanisms used by Ci-Macho1 to control expression of Ci-Tbx6b and Ci-Tbx6c, we misexpressed this transcription factor spatially and temporally in notochord cells using the Ciona Brachyury (Ci-Bra) promoter region (Corbo et al., 1997). Ci-macho1 is normally expressed in germ-cell precursors in 110-cell-stage embryos, and in sensory vesicle, nerve cord and germ cells in mid-tailbud embryos (Fig. 1A,C,D) (Satou et al., 2002a); no detectable expression in notochord cells was seen at any stage. For this reason, the Ci-Bra promoter, which is active predominantly in notochord cells and mesenchyme, and only sporadically in a few muscle cells, was chosen for the misexpression experiments. When the misexpression experiments were attempted using the Ci-FoxA-a promoter, which encompasses a wider expression territory (Di Gregorio et al., 2001), the effects on embryogenesis were too extensive to allow meaningful interpretation of the results (data not shown).
Fig. 1.
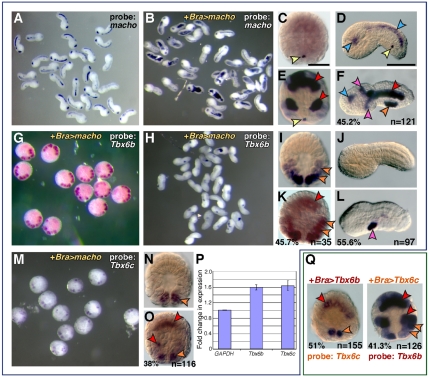
Transcriptional changes in response to ectopically expressed Ci-Macho1, Ci-Tbx6b and Ci-Tbx6c. (A-O,Q) Microphotographs of control and transgenic Ciona intestinalis embryos hybridized in situ with digoxigenin-labeled antisense RNA probes. Note that in some cases, because of mosaic incorporation of the transgene(s), not all the cells of the same lineage show staining. The percentages of transgenic embryos displaying the patterns shown, and the total number of embryos scored, are indicated at the bottom of each panel. (A,B,D,F,H,J,L) Mid-tailbud embryos; individual embryos are oriented with anterior to the left, dorsal up. (A,B,G,H,M) Low-magnification microphotographs of embryos from representative experiments. (C,E,G,I,K,M-O,Q) 110-cell-stage embryos, vegetal views. (A-P) Effects of the misexpression of Ci-macho1 on Ci-Tbx6b and Ci-Tbx6c expression. (A,C,D) Control embryos expressing zygotic Ci-macho1 in germ-line precursors (yellow arrowhead), sensory vesicle and nerve cord (blue arrowheads). (B,E,F) Bra>macho transgenics efficiently misexpress Ci-macho1 in notochord (red arrowheads) and mesenchyme cells (pink arrowheads) at early (E) and late stages (F). (G,H) Bra>macho transgenics hybridized with the Ci-Tbx6b probe. (I,J) Ci-Tbx6b is expressed at the 110-cell stage in muscle precursors (I; orange arrowheads) but is no longer expressed by the early tailbud stage (J; see also supplementary material Fig. S1A-C). (K) Ectopic expression of Ci-Tbx6b is detected in the notochord of 110-cell-stage Bra>macho transgenics (red arrowhead). (L) Mid-tailbud Bra>macho embryos show ectopic expression of Ci-Tbx6b only in mesenchyme (pink arrowhead). (M) Bra>macho transgenics hybridized with the Ci-Tbx6c probe. (N) Expression of Ci-Tbx6c in control embryos is detected in a subset of muscle precursors (orange arrowheads). (O) In 110-cell Bra>macho embryos, ectopic expression of Ci-Tbx6c is detected in notochord precursors (red arrowheads). (P) Changes in gene expression in 110-cell stage Bra>macho embryos versus control embryos, as monitored by qRT-PCR. (Q) Left panel shows a 110-cell-stage Bra>Tbx6b embryo hybridized with the Ci-Tbx6c probe. Right panel shows a 110-cell-stage Bra>Tbx6c embryo hybridized with the Ci-Tbx6b probe. Ectopic notochord staining is indicated by red arrowheads. Scale bars: 50 μm.
Transgenic embryos carrying the Bra>macho construct (Fig. 1B) efficiently express Ci-macho1 in notochord cells and their precursors (Fig. 1E,F) and display a short, stubby tail (Fig. 1B,F,L). The 110-cell-stage transgenic Bra>macho embryos hybridized with a probe for Ci-Tbx6b (Fig. 1G), which is normally expressed in muscle precursors (Fig. 1I) (Takatori et al., 2004), showed ectopic expression of this gene in notochord precursors (Fig. 1K); however, at the early tailbud stage (Fig. 1H), when Ci-Tbx6b is normally undetectable (Fig. 1J), a consistent ectopic signal was observed only in a subpopulation of trunk mesenchyme cells (Fig. 1H,L). This result can be explained by previous observations of the activity of the Ci-Bra promoter region in mesenchyme cells (Corbo et al., 1997).
Ci-Tbx6c (Fig. 1M-O), which is normally expressed only in a subset of muscle precursors at the 110-cell stage (Fig. 1N), was ectopically activated in transgenic Bra>macho embryos in notochord precursors only at this stage (Fig. 1O), and was not detected at later stages (data not shown). The transcriptional changes induced by the misexpression of Ci-macho1 in 110-cell transgenic embryos were validated and quantified by qRT-PCR (Fig. 1P). Together, these results suggest that notochord precursors are competent to respond to Ci-Macho1, although only at early stages of their development.
Cross-regulatory interactions between Ci-Tbx6b and Ci-Tbx6c
We next sought to assess whether Ci-Tbx6b and Ci-Tbx6c, which possess identical binding affinities in vitro (Yagi et al., 2005), are each capable of activating transcription of the other gene. To test this hypothesis, we misexpressed Ci-Tbx6b and Ci-Tbx6c in the notochord by cloning the respective cDNAs downstream of the Ci-Bra promoter. As a first step, we ascertained whether these genes were efficiently transcribed in the notochord of transgenic embryos; a representative control experiment is presented in supplementary material Fig. S1. Subsequently, we hybridized embryos carrying the Bra>Tbx6b transgene with the Ci-Tbx6c probe, and embryos carrying the Bra>Tbx6c transgene with the Ci-Tbx6b probe (Fig. 1Q). We found ectopic expression of Ci-Tbx6c in notochord precursors in embryos bearing the Bra>Tbx6b transgene (Fig. 1Q, left panel; compare with Fig. 1N) and similarly, transcription of Ci-Tbx6b was ectopically activated in embryos carrying the Bra>Tbx6c transgene (Fig. 1Q, right panel; compare with Fig. 1I). Under the experimental conditions that we used, no cross-hybridization between the two probes was observed. These results indicate that these related transcription factors possess the potential for reciprocal activation.
A bipartite CRM recapitulates spatial and temporal muscle expression of Ci-Tbx6b
Beginning at the 16-cell stage, Ci-Tbx6b is specifically expressed in the precursors of the larval muscles derived from the B5.1 blastomere pair (Takatori et al., 2004). In 110-cell embryos, Ci-Tbx6b expression expands to include numerous muscle precursors (Fig. 1I), whereas by the neurula stage it becomes confined to the posterior-most muscle cells (Takatori et al., 2004). Ci-Tbx6b (gene model ci0100144249) is clustered in a tandem arrangement with Ci-Tbx6c (ci0100144293) on sequence scaffold 126 (JGI v2.0; http://genome.jgi-psf.org/Cioin2/Cioin2.home.html) (Dehal et al., 2002) (Fig. 2A,S; supplementary material Fig. S2A). A genomic fragment spanning 2.4 kb was PCR-amplified from the 5′-flanking region of Ci-Tbx6b and its various truncations were tested in vivo by parallel electroporations (e.g. Di Gregorio and Levine, 2002) to identify the minimal sequences necessary for its function. Truncated versions of the 2.4 kb fragment were cloned upstream of both the endogenous Ci-Tbx6b promoter and the Ci-FoxA-a basal promoter (supplementary material Fig. S2B). The 2.4 kb CRM directed strong expression in muscle and mesenchyme cells (Fig. 2B-D; supplementary material Fig. S2D-F and Table S1); occasional staining was also observed in trunk ventral cells, which are the heart precursors (data not shown). The use of the Ci-FoxA-a basal promoter (Oda-Ishii and Di Gregorio, 2007) allowed the identification of a minimal 112 bp fragment sufficient for activity in a heterologous context (shaded area in supplementary material Fig. S2B).
Fig. 2.
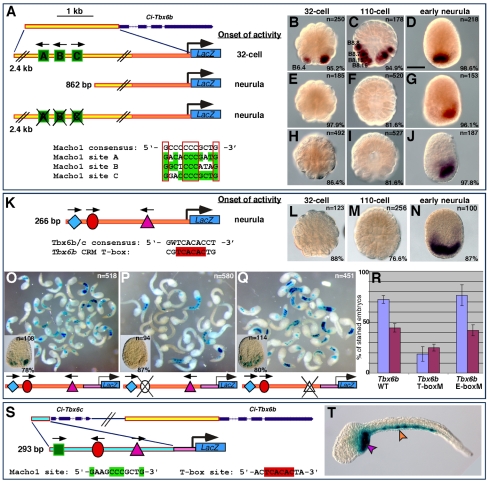
Identification of the early and late Ci-Tbx6b CRMs and prediction of a related muscle CRM in the Ci-Tbx6c locus. (A) Schematic representation of the Ci-Tbx6b locus as shown in the JGI Ciona intestinalis genome browser (Dehal et al., 2002) (http://genome.jgi-psf.org/Cioin2/Cioin2.home.html). Dark blue boxes and dashed lines indicate exons and introns, respectively. A yellow rectangle represents the 2.4 kb fragment that was analyzed. Green boxes represent putative Ci-Macho1-binding sites; black arrows indicate their orientations. The orange rectangle depicts the proximal region of the 2.4 kb fragment. Bottom panel shows alignment of the Ci-Macho1 sites with the consensus sequence identified by Yagi and colleagues (Yagi et al., 2004a). Highly conserved nucleotides are boxed in red. (B-J) Embryos electroporated with either the 2.4 kb Ci-Tbx6b CRM (B-D), the 862 bp fragment (E-G) or the 2.4 kb CRM carrying mutations in all Ci-Macho1-binding sites (H-J) were fixed at the 32-cell (B,E,H), 110-cell (C,F,I), or early neurula (D,G,J) stage and hybridized in situ with a lacZ probe. Stained blastomeres are labeled only on one side of the embryos (Conklin, 1905; Meinertzhagen and Okamura, 2001). (K) The 266 bp proximal region of the Ci-Tbx6b CRM (orange in A) contains putative binding sites for CREB (blue diamond), T-box (red oval) and bHLH (‘AC’-core E-box; pink triangle) transcription factors. (L-N) Embryos carrying the 266 bp Ci-Tbx6b transgene in K, hybridized in situ with a lacZ probe. (O-Q) Mutation analysis of the Ci-Tbx6b proximal CRM. A 112 bp subfragment of the 266 bp CRM, containing the three putative binding sites shown in K, was cloned upstream of the Ci-FoxA-a basal promoter (pink box) and subjected to truncation (data not shown) and mutation analysis. (O) Low-magnification image of mid-tailbud embryos electroporated with the wild-type 112 bp Ci-Tbx6b CRM. (P) Mid-tailbud embryos electroporated with the 112 bp Ci-Tbx6b CRM carrying a mutation in the Ci-Tbx6b/c core binding site (TCACAC>TCAaca). (Q) Mid-tailbud embryos electroporated with the 112 bp Ci-Tbx6b CRM carrying a mutation in the E-box sequence (CAGTTG>actggt). Insets in (O-Q) show transgenic embryos fixed at the neurula stage. (R) Graph showing the percentage (mean ± s.d.) of stained mid-tailbud embryos in O-Q; blue columns, muscle staining; purple, mesenchyme. The number of embryos scored for each construct is shown in white in O-Q. (S) Organization of the Ci-Tbx6c and Ci-Tbx6b genomic locus. The predicted Ci-Tbx6c CRM (blue rectangle) contains one Ci-Macho1-binding site, one T-box site and an AC-core E-box. (T) Late-tailbud embryo electroporated with the Ci-Tbx6c CRM, showing activity in muscle (orange arrowhead) and mesenchyme (pink arrowhead). B,E,H,L, vegetal views; C,F,I,M, dorsal-vegetal views; D,G,J,N, dorsal views, with anterior to the top. Scale bar: 50 μm.
Sequence analysis of the 2.4 kb Ci-Tbx6b CRM identified three bona fide binding sites for Ci-Macho1 (green squares in Fig. 2A), all of which partially matched the consensus sequence previously identified (Yagi et al., 2004a) (Fig. 2A, bottom). We performed an electrophoretic mobility-shift assay (EMSA) to verify the functionality of these sites, and found that all three sequences were bound in vitro by a Histidine-tagged Ci-Macho1 fusion protein (supplementary material Fig. S3A), as well as by a GST-tagged Ci-Macho1 protein (data not shown), with affinities that reflected their different homologies with the published consensus sequence (Fig. 2A, bottom). The three Ci-Macho1-binding sites were found within a 463 bp region. To verify the involvement of the Ci-Macho1-binding sites in the temporal control of the CRM activity, we compared the developmental windows of transcriptional activity of the three Ci-Tbx6b constructs shown in Fig. 2A. The first construct contains the 2.4 kb wild-type Ci-Tbx6b CRM fused to lacZ, the second construct contains an 862 bp 5′-truncated version of the previous fragment, which lacks all Ci-Macho1-binding sites, and the third construct consists of the 2.4 kb Ci-Tbx6b CRM carrying mutations in all three Ci-Macho1-binding sites. To accurately determine the respective windows of activity, one-cell stage Ciona embryos were electroporated separately with each construct, cultured until the 32-cell (Fig. 2B,E,H), 110-cell (Fig. 2C,F,I) and early neurula stages (Fig. 2D,G,J), then fixed and hybridized in situ with a lacZ RNA probe. In embryos electroporated with the 2.4 kb wild-type Ci-Tbx6b CRM, a strong hybridization signal was detected in the B6.4 pair of muscle precursors starting from the 32-cell stage (Fig. 2B). At the 110-cell stage, lacZ expression was detected in descendants of B6.4 blastomeres and in the B8.7 and B8.8 pairs, which derive from the B6.2 pair (Fig. 2C). In early neurulae, lacZ expression was detected only in the posterior-most muscle cells (Fig. 2D). This pattern faithfully recapitulates the expression of Ci-Tbx6b (Takatori et al., 2004). Embryos electroporated with the 862 bp CRM, which does not contain the Ci-Macho1-binding sites, showed no detectable signal at either the 32-cell stage (Fig. 2E) or at the 110-cell stage (Fig. 2F); lacZ expression became detectable by the early neurula stage in only the posterior-most muscle cells (Fig. 2G; note that the asymmetrical expression is due to mosaic incorporation of the transgene). Finally, the vast majority of embryos carrying the 2.4 kb CRM with mutations in all Ci-Macho1-binding sites showed no expression of the reporter at either the 32-cell (Fig. 2H) or the 110-cell stage (Fig. 2I), and displayed active transcription in the posterior-most muscle cells only at the neurula stage (Fig. 2J). These results indicate that mutations in the Ci-Macho1-binding sites do not affect the spatial (i.e. lineage-specific) activity of the 2.4 kb Ci-Tbx6b CRM, but are sufficient to cause a considerable delay in the onset of transcription driven by this fragment.
Together, these observations suggest that: (1) the temporal information required for the early activity of the 2.4 kb Ci-Tbx6b CRM is stored in its distal region; (2) this temporal information is encoded by the Ci-Macho1-binding sites; and (3) the 862 bp proximal region contains the cis-regulatory elements responsible for the late muscle activity observed in neurulae.
The proximal region of the Ci-Tbx6b CRM recapitulates the late muscle expression of Ci-Tbx6b
The proximal 112 bp region of the Ci-Tbx6b CRM (shaded area in supplementary material Fig. S2B), which was first identified using the Ci-FoxA-a basal promoter, was further tested using the endogenous 154 bp Ci-Tbx6b promoter. The resultant 266 bp construct (Fig. 2K; dark orange area in the schematics in Fig. 2A) was used for lacZ time-course experiments. The majority of the embryos electroporated with the 266 bp construct did not show staining at the 32-cell (Fig. 2L) or at the 110-cell stage (Fig. 2M); however, by the early neurula stage, 87% of the embryos showed strong staining in muscle cells (Fig. 2N). These results suggest that this 266 bp region is sufficient to recapitulate the late activity of the 862 bp fragment previously analyzed (Fig. 2H-J).
Sequence inspection identified at least three putative binding sites within this region: a distal sequence showing an incomplete match with the CREB-binding sites previously identified in other Ciona muscle CRMs (TGACG core; blue diamond in Fig. 2K) (Kusakabe et al., 2004; Brown et al., 2007), a T-box-binding site matching the core consensus sequence identified for Ci-Tbx6b and Ci-Tbx6c via SELEX assays (7 out of 10 matches; red oval in Fig. 2K) (Yagi et al., 2005), and an ‘AC’-core E-box (pink triangle in Fig. 2K) (Erives et al., 1998). In vivo analysis of progressive truncations and specific point mutations showed that removal of the imperfect CREB-binding site did not noticeably affect either the intensity of the muscle staining or the percentage of embryos showing activity (data not shown). However, when the T-box-binding site was mutated, a considerable reduction was observed not only in the intensity of the muscle staining but also in the percentage of embryos showing activation of the reporter gene (Fig. 2O,P,R). Finally, a mutation of the ‘AC’-core E-box left the muscle staining unaffected (Fig. 2Q,R). The results of three independent experiments are quantified in the graph in Fig. 2R. Comparable results were obtained when transgenic embryos were fixed at the neurula stage (insets in Fig. 2O-Q).
Together, these findings suggest that the proximal region of the Ci-Tbx6b CRM is mainly controlled by a T-box transcription factor. To further investigate this point, we carried out EMSA using bacterially expressed Ci-Tbx6b and Ci-Tbx6c proteins and a radiolabeled oligonucleotide probe containing the putative T-box-binding site (supplementary material Fig. S3B). For these experiments we synthesized full-length proteins to identify possible differences in binding affinities (see Materials and Methods); nevertheless, we found that this sequence was bound with similar intensity by both GST-Ci-Tbx6b and GST-Ci-Tbx6c fusion proteins (supplementary material Fig. S3B). These results suggest that the late-acting Ci-Tbx6b CRM functions as an autoregulatory and/or a cross-regulatory enhancer sequence.
Lastly, we used the information gathered from the analysis of the Ci-Tbx6b CRM to predict the location of a related muscle CRM in the Ci-Tbx6c locus. Ci-Tbx6c is clustered with Ci-Tbx6b within a 8 kb genomic region (Fig. 2S; supplementary material Fig. S2A). Within the 2 kb sequence directly upstream of Ci-Tbx6c, we identified a region spanning 293 bp which contained bona fide binding sites for Ci-Macho1 and Ci-Tbx6b/c, as well as an ‘AC’-core E-box (Fig. 2S). When we tested this fragment in vivo we found that, as expected, it was able to direct expression in most muscle cells (Fig. 2T), recapitulating the expanded expression pattern seen for Ci-Tbx6c at stages later than 110-cell (Takatori et al., 2004). This result suggests that the muscle activity of the highly related Ciona Tbx6 genes is controlled by the same basic set of structural cis-regulatory elements.
The composite cis-regulatory region of CiFcol1 harbors two muscle CRMs with different temporal onsets
We next sought to investigate whether the structural and functional criteria that we had identified for the Tbx6-related transcription factor genes applied to the cis-regulatory region of a structural muscle gene. The Ciona Fibrillar Collagen-1 gene (CiFCol1; JGI gene model ci0100150759) (Wada et al., 2006) encodes a member of the fibrillar collagen family related to the vertebrate clade A collagen genes (Wada et al., 2006). During Ciona early embryogenesis, this gene is first expressed mostly in muscle precursors and by the neural plate stage its expression has expanded to mesenchyme, notochord, endoderm and CNS (Satou et al., 2001) (our unpublished results). Expression of CiFCol1 is maintained in all these tissues at very high levels throughout the mid-tailbud stage (Fig. 3A) until hatching (Satou et al., 2001) (our unpublished results). The sustained expression of CiFCol1 in muscle cells suggested that transcription of this gene might rely upon a combination of early and late activators. Sequence inspection of its 5′-flanking region indicated that CiFCol1 might be controlled, at least at early developmental stages, by Ci-Macho1 and/or its intermediaries. To verify this hypothesis, we first used the misexpression assays described in Fig. 1. Since CiFCol1 is normally expressed in notochord precursors starting from the late gastrula stage, and is not present at detectable levels in these cells in early gastrulae (Fig. 3B), we harvested embryos at this developmental stage which had been electroporated with either Bra>macho, Bra>Tbx6b or Bra>Tbx6c and monitored the effects on the activation of CiFCol1 transcription in notochord precursors by WMISH (Fig. 3C-E). These experiments show that Ci-Macho1, Ci-Tbx6b and Ci-Tbx6c are all able to induce precocious onset of CiFCol1 expression in the notochord lineage.
Fig. 3.
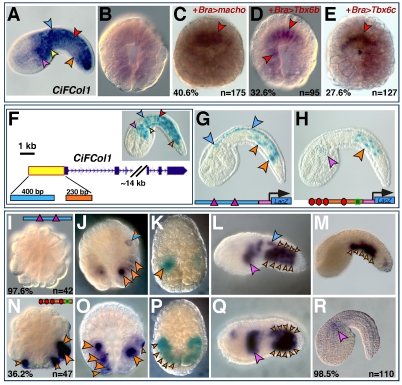
The Ciona CiFCol1 gene and its CRMs. (A) Mid-tailbud Ciona embryo hybridized in situ with the CiFCol1 probe. Arrowhead color code: blue, CNS; pink, mesenchyme; red, notochord; orange, muscle; yellow: endoderm. Expression at other stages is available online at http://ghost.zool.kyoto-u.ac.jp/cgi-bin3/photoget2.cgi?CLSTR00093 (Satou et al., 2001). (B-E) 110-cell-stage embryos hybridized with the CiFCol1 probe. (B) Control embryo. (C-E) Embryos carrying the transgenes indicated on the top right. Ectopic notochord staining is highlighted by red arrowheads. (F) Partial representation of the CiFCol1 gene locus. The yellow box depicts the 2.2 kb cis-regulatory region that fully recapitulates the CiFCol1 expression pattern; a blue and an orange box indicate the distal and proximal muscle CRMs, respectively. Inset shows a mid-tailbud embryo electroporated with the 2.2 kb region. (G,H) Microphotographs of X-Gal-stained Ciona embryos electroporated with the 400 bp and 230 bp regions shown in F, cloned upstream of the FoxA-a basal promoter (pink box in the schematics). The distal region (G) contains two E-boxes (pink triangles); the proximal region (H) contains four putative Ci-Tbx6b/c-binding sites (red ovals) and one putative Ci-Macho1 site (green square). (I-R) Embryos electroporated with either the distal (I-M), or the proximal muscle CRM (N-R), fixed at the 32-cell (I,N) 110-cell (J,O), early tailbud (L,Q) and mid-tailbud (M,R) stage and hybridized with a lacZ probe. (K,P) Early neurula embryos electroporated with either the distal (K) or the proximal CRM (P), stained with X-Gal.
We then investigated the molecular mechanisms mediating the response of this structural gene to the misexpression of the three muscle transcription factors by characterizing the CiFCol1 cis-regulatory region. We found that a 2.2 kb fragment from the 5′-flanking region of CiFCol1 (yellow rectangle in Fig. 3F) was able to fully recapitulate its expression pattern (inset in Fig. 3F). Within this main fragment, three different CRMs were identified, including one 400 bp distal muscle CRM (blue rectangle in Fig. 3F), and two adjacent proximal CRMs, one directing expression in notochord (data not shown) and the other, 230 bp long, which was active in muscle cells (orange rectangle in Fig. 3F). The distal muscle CRM contains two generic E-boxes (Fig. 3G, pink triangles) and directs expression in muscle cells (Fig. 3G, orange arrowheads; see lineage map in supplementary material Fig. S4A) and in the two rows of lateral ependymal cells of the nerve cord (Fig. 3G, blue arrowheads). The 230 bp proximal muscle CRM contains four putative T-box-binding sites (generic sequence: TNNCAC; Fig. 3H, red ovals) and one putative low-affinity Ci-Macho1-binding site (Yagi et al., 2004a) (Fig. 3H, green rectangle) and directs expression in most of the primary muscle cells and in trunk mesenchyme.
To accurately define the window of activity of each muscle CRM, we performed time-course experiments using the lacZ probe (Fig. 3I,J,L-O,Q,R). In addition, we monitored the accumulation of β-galactosidase in the muscle cells where each CRM was active by performing X-Gal staining (Fig. 3K,P). Embryos carrying the distal 400 bp muscle CRM began accumulating lacZ transcripts between the 32-cell and the 110-cell stage (Fig. 3I,J). In the majority of the embryos analyzed, high levels of lacZ transcripts were predominantly detected in only one pair of muscle precursors (supplementary material Fig. S4A), although a few embryos also showed a faint signal in additional muscle precursors (orange arrowheads in Fig. 3J), including the mixed-lineage A8.16 blastomeres (orange and blue arrowhead in Fig. 3J). Consistent with this early pattern, at the early neurula stage β-galactosidase accumulation was detected only in a small subset of muscle and mesenchyme precursors (Fig. 3K). By the early tailbud stage (Fig. 3L), the activity of the CRM had considerably expanded, to encompass virtually all muscle cells (18 per side; supplementary material Fig. S4B), as well as part of the nerve cord and mesenchyme cells. The CRM remained active in most muscle cells through the mid-tailbud stage (Fig. 3M).
Starting at the 32-cell stage (Fig. 3N), over one third of the embryos electroporated with the proximal 230 bp CiFCol1 muscle CRM showed lacZ expression in primary muscle precursors; by the 110-cell stage, high levels of lacZ transcripts were observed in most pairs of muscle precursors, except for the A8.16 pair (supplementary material Fig. S4A). At the early neurula stage, this pattern translated into a homogeneous accumulation of β-galactosidase in the majority of muscle cells (Fig. 3P). At the early tailbud stage, this CRM was still active in muscle and mesenchyme (Fig. 3Q). However, at the mid-tailbud stage, transcriptional activity had faded almost completely from the muscle in the majority of the embryos, whereas a residual signal was occasionally seen in mesenchyme cells (Fig. 3R). The activity of each CRM in muscle cells is plotted in the graph in supplementary material Fig. S4C and compared with the number of muscle cells per side of the embryo at the stages analyzed.
From these observations, we conclude that of the two muscle CRMs that we have identified in the CiFCol1 5′-flanking region, the proximal one is activated earlier, beginning at the 32-cell stage, and ceases its function around the mid-tailbud stage, whereas the distal one is activated two to three cell divisions later, but persists beyond the mid-tailbud stage.
Selective activation of the CiFCol1 CRMs by ectopically expressed Tbx6 transcription factors
To test whether the muscle CRMs identified within the CiFCol1 locus were mediating the response of the endogenous gene to the ectopic expression of Ci-Tbx6b and/or Ci-Tbx6c, we co-electroporated each CRM with either Bra>Tbx6b or Bra>Tbx6c.
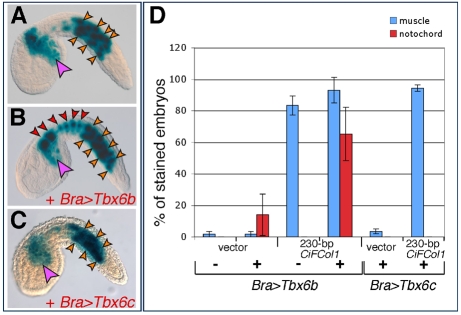
As shown in Fig. 4, misexpression of Ci-Tbx6b was sufficient to induce ectopic expression of the CiFCol1 proximal 230 bp muscle CRM in notochord cells (Fig. 4A,B), consistent with the possibility that Ci-Tbx6b activates this enhancer. While performing controls for these experiments, we noticed that in some batches of embryos the pFBΔSP6 vector was itself slightly responsive to the Bra>Tbx6b construct; however, the ectopic activation observed in the notochord when the 230 bp CiFCol1 CRM was used in these co-electroporations instead of the empty vector was at least 3.5-times higher (Fig. 4D). No ectopic activation in the notochord was observed when the 230 bp CiFCol1 CRM was co-electroporated with Bra>Tbx6c (Fig. 4C,D). In addition, we tested the response of these CRMs to another Tbx6-related transcription factor, Ci-Tbx6a (Takatori et al., 2004), and to a more distantly related T-box factor, Ci-Tbx15/18/22 (Erives and Levine, 2000; Takatori et al., 2004), and we found no response above background to either factor (data not shown). Finally, no ectopic activation was observed when the distal CiFCol1 CRM was co-electroporated with either Bra>Tbx6b or Bra>Tbx6c (data not shown); this result is consistent with the lack of evident Tbx6b/c-binding sites in this sequence (Fig. 3G).
Fig. 4.
Effects of the ectopic expression of Ci-Tbx6b and Ci-Tbx6c on CiFCol1 muscle CRMs. (A-C) Ciona embryos electroporated with the CiFCol1 proximal CRM, alone (A) or in combination with either Bra>Tbx6b (B) or Bra>Tbx6c (C). Arrowhead color code: red, notochord; orange, muscle; pink, mesenchyme. (D) Graph showing the percentages (mean ± s.d.) of embryos displaying staining in muscle (blue columns) and in notochord (red columns) in the experiments shown in (A-C), as well as the baseline staining attributable to the vector. ‘+’ and ‘−’ signs indicate the presence or absence, respectively, of either misexpression construct. Note that the height of the error bars on the red columns reflects the sporadic occurrence of the activation of the empty vector in notochord cells, which is observed only in some batches of embryos.
The flow of maternal and zygotic developmental information in the muscle cells of Ciona
Our findings are summarized by the model in Fig. 5, which reconstructs part of the cis-regulatory hierarchy underlying the cell-autonomous developmental program that characterizes primary muscle formation in Ciona. Arrows in Fig. 5 indicate the interactions between transcription factors and cis-regulatory sequences, either demonstrated by the work presented here (black) or inferred (gray).
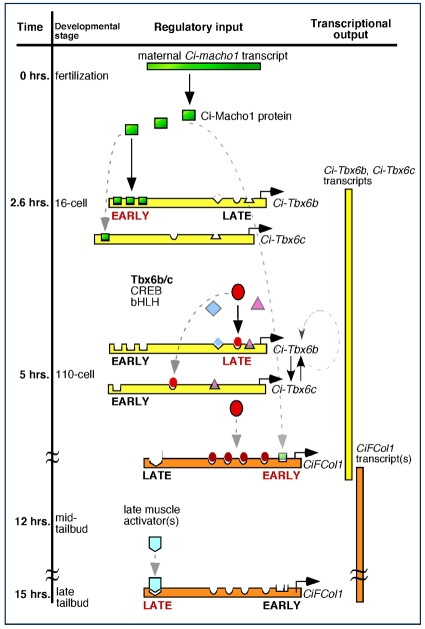
Fig. 5.
A working model for the muscle cis-regulatory hierarchy in Ciona. The regulatory interactions identified by this study are summarized and plotted against the developmental time-course of Ciona embryogenesis at 18°C (black vertical bar on left). Horizontal green bar represents the maternally-deposited Ci-macho1 transcripts, green squares depict the Ci-Macho1 protein. Horizontal bars symbolize the cis-regulatory regions of Ci-Tbx6b and Ci-Tbx6c (yellow) and CiFCol1 (orange); transcription start sites are indicated by horizontal arrows. Transcripts are shown in scheme as vertical bars on the right. Transcription factors are depicted as described in Fig. 2.
After fertilization, binding of maternally encoded Ci-Macho1 protein to the early Ci-Tbx6b CRM activates transcription of this gene in muscle precursors. By the 16-cell stage, when maternal Ci-macho1 transcripts starts declining (Satou et al., 2002a), transcription of Ci-Tbx6b begins. At later stages, it is likely that the Ci-Tbx6b early CRM is progressively vacated and is first aided and then progressively replaced by the late CRM, which is activated by Ci-Tbx6b and/or Ci-Tbx6c. Given the similarities that we have identified in the Ci-Tbx6b and Ci-Tbx6c cis-regulatory sequences, it is plausible that similar mechanisms activate the Ci-Tbx6c CRM. Ci-Tbx6b in turn activates the proximal CRM of CiFCol1, which recapitulates the early transcription of this gene. By the neurula stage, as expression of Ci-Tbx6b fades, different muscle activators begin binding the distal, late-acting CiFCol1 CRM and allow transcription of this gene to proceed without interruptions until the late tailbud stage.
Discussion
Spatial and temporal heterogeneity of the embryonic territories responsive to Ci-Macho1
For over a century, it has been known that upon fertilization and first cleavages of the ascidian egg, maternally loaded cytoplasmic determinants are differentially segregated into the resulting blastomeres, thus giving rise to the early determination and invariance that characterize their developmental fates (Conklin, 1905; Deno et al., 1984; Ortolani, 1955). Although recent molecular evidence has highlighted the major role of inductive processes in the formation of most embryonic tissues (reviewed by Lemaire, 2009), the cell-autonomous development of the primary muscle of the ascidian larva is largely attributable to the maternal determinant Ci-Macho1 (Nishida and Sawada, 2001). Ci-macho1 postplasmic mRNA is relocated after fertilization by the cortical centrosome-attracting body (CAB) (Sardet et al., 2003). As cleavage proceeds, in both Halocynthia (Nishida and Sawada, 2001) and Ciona (Satou et al., 2002a) Ci-macho1 mRNA becomes progressively restricted to a narrow region of the embryo, the B7.6 blastomeres; however, the Macho1 protein is generally believed to persist in an unlocalized form, and to be distributed to all descendants of the B4.1 cells (Kondoh et al., 2003). Studies in Halocynthia show that for the proper formation of other lineages that also derive from the B4.1 cells, such as mesenchyme and endoderm, the function of Macho1 needs to be actively suppressed by FGF and BMP signaling pathways (Kim et al., 2000; Kim and Nishida, 1999; Kondoh et al., 2003). Similar mechanisms are also likely responsible for the functional suppression of zygotically expressed Ci-Macho1 in the Ciona CNS, considering that Ci-FGF16/19/20 is expressed in the Ciona CNS through tailbud stages and is required for neural development (Imai et al., 2004; Bertrand et al., 2004).
The misexpression experiments described here suggest that no such restraining mechanism is present in notochord cells before the early tailbud stage. In fact, at early developmental stages the ectopic activation of both Ci-Tbx6b and Ci-Tbx6c was seen in notochord precursors of both lineages in Bra>macho embryos, whereas at the mid-tailbud stage only the ectopic activation of Ci-Tbx6b was observed, and it was confined to a subset of mesenchyme cells. These cells are most likely descendants of the B7.3 blastomere, a 64-cell stage precursor of both secondary notochord and mesenchyme cells (Satoh, 1994). The differential competence of the notochord to respond to Ci-Macho1 might be explained by the requirement for temporally and spatially localized co-factors and/or transcriptional intermediaries. Alternatively, as in the case of the CNS, Ci-Macho1 might be functionally suppressed in the notochord of tailbud embryos by the activation of the FGF signaling pathway, as suggested by the observation that Ci-FGFR is expressed in the notochord beginning at the early tailbud stage (Imai et al., 2004; Shi et al., 2009). These mechanisms might also account for the relatively mild phenotype that we observed in embryos carrying the Bra>macho transgene, whereby the notochord is still able to form, even in transgenic embryos where mosaic incorporation is minimal.
The spatio-temporal expression pattern of Ci-Tbx6b is recapitulated by early and late cis-regulatory sequences
Using in vivo transient transgenic assays, we have identified a 2.4 kb CRM upstream of Ci-Tbx6b that is able to faithfully recapitulate the muscle expression of this gene. The temporal muscle activity of the 2.4 kb CRM represents the composite read-out of early- and late-acting cis-regulatory sequences, which interpret maternal and zygotic information. The Ci-Tbx6b CRM contains a distal region which functions as the repository of the temporal information necessary to recapitulate the early expression pattern previously reported for Ci-Tbx6b (Takatori et al., 2004). When this distal region is deleted, muscle activity is not lost, but its onset is considerably delayed. Sequence inspection and point-mutation analyses suggested that this early-acting distal region might be controlled by maternal Ci-Macho1, because three putative binding sites for this factor are present in this sequence. We found that these sites are bound in vitro by Macho1 and that their concomitant mutation is sufficient to cause the same delay in the onset of transcriptional activity that was observed when the entire fragment encompassing them was deleted. Together, these observations provide a mechanistic cis-regulatory explanation to the results of our misexpression assay, as well as to previous results showing that overexpression of Ci-Macho1 is sufficient to induce ectopic expression of Ci-Tbx6b (Yagi et al., 2004a) and that, likewise, Hr-Macho-1 is able to ectopically induce Hr-Tbx6, among other muscle genes (Sawada et al., 2005). It is noteworthy that in Ciona, Ci-ZicL cooperates with Ci-Macho1 to promote muscle development (Yagi et al., 2005); this zygotic zinc-finger transcription factor is related to Ci-Macho1 and recognizes a similar consensus binding site in vitro (Yagi et al., 2004b). Interestingly, one of the three Ci-Macho1-binding sites that we have characterized in the Ci-Tbx6b CRM, namely site ‘C’, contains permutations of the published ZicL consensus site that are compatible with binding in vitro (Yagi et al., 2004b). If this site is bound in vivo by either transcription factor, then this would explain the observation that Ci-Tbx6b is still weakly expressed in Ci-Macho1 morphant embryos, whereas its expression is no longer detectable in Ci-Macho1 and Ci-ZicL double-morphants (Yagi et al., 2005).
Within the 2.4 kb CRM, a 266 bp proximal region is able to direct transcription only from neurulation onwards, thus acting as a late muscle enhancer. Sequence analysis of this region revealed the presence of an imperfect CREB-binding site, a T-box-binding site (generic sequence: TNNCAC) partly matching the core consensus sequence previously reported for Ci-Tbx6b/c (Yagi et al., 2005), and an ‘AC’-core E-box. Both CREB-binding sites and AC-core E-boxes have been previously shown to be necessary for muscle activity of other muscle CRMs (Brown et al., 2007; Erives et al., 1998; Kusakabe et al., 2004); however, in this case, only the T-box site substantially contributes to the muscle activity, qualitatively and quantitatively. Through EMSA, we have shown that this T-box site is bound in vitro by both Ci-Tbx6b and Ci-Tbx6c.
Modular organization and temporal properties of the cis-regulatory region of CiFCol1
Originally isolated in a subtractive screen aimed to identify genes downstream of Ci-Bra (Takahashi et al., 1999), the CiFCol1 gene attracted our interest because of its sustained muscle expression, which begins around mid-gastrulation, and because its upstream region is enriched in T-box-binding sites.
Dissection of a 2.2 kb genomic fragment located upstream of the transcription start site of CiFCol1 revealed the presence of discrete CRMs active in all the tissues where CiFCol1 is expressed. In particular, this 2.2 kb fragment harbors two distinct muscle CRMs: a distal CRM containing two generic E-boxes and depleted of T-box-binding sites and Ci-Macho1-binding sites, and a proximal CRM containing four clustered T-box-binding sites, some of which are bound weakly in vitro by the Ci-Tbx6b protein (data not shown), and a low-affinity Ci-Macho1-binding site. The heterogeneity of these sequences is reflected by the temporal activity of the two CRMs, because the distal one, which does not contain any apparent T-box-binding sites, is activated later than the proximal one, which is enriched in these motifs. In particular, the distal CiFCol1 muscle CRM is active in a small subset of muscle precursors from the 110-cell stage to the neurula stage, and only by the early tailbud stage does its territory expand to encompass all muscle cells. Afterwards, it remains active in the majority of muscle cells. Therefore, the spatial range of action of this CRM in the muscle seems to be controlled by an activator(s) functioning from neurulation onwards. The presence of two E-boxes in this sequence prompted us to investigate the possible involvement of transcription factors of the bHLH family in the regulation of this CRM. We found that neither mutation of the E-boxes nor misexpression, individual or combined, of two bHLH transcription factors, Ci-MRF (Meedel et al., 2007) and Ci-paraxis (Erives, 2009; Imai et al., 2004) had any detectable effect (data not shown), thus leaving the identification of the late activator(s) to future investigations.
Conversely, the proximal CiFCol1 muscle CRM is ignited early in most muscle cell precursors, starting from the 32-cell stage, but its activity fades by the mid-tailbud stage. We conclude that the additive activity of the two CRMs is probably responsible for the sustained expression of CiFCol1 in muscle cells.
Interestingly, misexpression of Ci-Macho1, Ci-Tbx6b or Ci-Tbx6c in notochord cells all result in ectopic activation of CiFCol1 in this territory. Although we cannot rule out that this might be attributable to the low-affinity Ci-Macho1-binding site in the CiFCol1 early CRM, given the late onset of CiFCol1 muscle expression it seems more likely that Ci-Macho1 activates expression of CiFCol1 indirectly, through Ci-Tbx6b. To test this hypothesis we monitored the response of the CiFCol1 proximal muscle CRM to the misexpression of Ci-Tbx6b in notochord cells. We found that misexpression of Ci-Tbx6b caused the ectopic activation of the CiFCol1 proximal muscle CRM in the notochord, whereas misexpression of Ci-Tbx6c did not have any effect. We conclude that the ectopic activation of CiFCol1 seen in notochord cells of embryos carrying the Bra>Tbx6c construct might occur indirectly, via the activation of Ci-Tbx6b expression by Ci-Tbx6c.
Finally, no ectopic activation was observed when the distal CiFCol1 muscle CRM was co-electroporated with either construct (data not shown), consistent with the lack of Tbx6b/c-binding sites in its sequence.
Muscle gene regulation in ascidians versus vertebrates: lineage-specific innovations and evolutionarily conserved mechanisms
By analyzing the cis-regulatory sequences that mediate the response to Ci-Macho1 and its mediators, this study has begun to provide sharper insights into the molecular mechanisms controlling cell-autonomous muscle development in the ascidian embryo. Given the large number of genes that respond to Ci-Tbx6b and Ci-Tbx6c (Yagi et al., 2005) (this study), it is conceivable that the mechanisms of transcriptional regulation that control the CRMs presented here might be shared by several other muscle genes. This hypothesis is supported by the abundance of putative Tbx6b/c-binding sites in muscle CRMs identified by other groups (Brown et al., 2007; Kusakabe et al., 2004) and by our laboratory (data not shown).
Although the early cell-fate determination mediated by Macho-like proteins in muscle cells has been described so far as an ascidian-specific mechanism, transcription factors of the Zic family, of which Macho, ZicL and related proteins represent a diverged branch (Aruga et al., 2006), are known to be required for shaping the body plan of widely different animals (Merzdorf, 2007). In addition, Tbx6-related proteins in Ciona appear to be part of an evolutionarily conserved kernel that is employed for the specification and differentiation of paraxial mesoderm in several other chordates, including mouse (Chapman and Papaioannou, 1998; White et al., 2003), Xenopus (Tazumi et al., 2008; Uchiyama et al., 2001) and zebrafish (Goering et al., 2003). Hence, the elucidation of the cis-regulatory mechanisms used by these transcription factors to modulate expression of their target genes should provide insights on the inner workings of other model systems in which cis-regulatory elements are less tractable, including higher chordates.
Materials and Methods
Ascidians and electroporation
Adult Ciona intestinalis were purchased from Marine Research and Educational Products (M-REP; Carlsbad, CA). Fertilization, dechorionation, electroporation and X-Gal staining were carried out as described (Corbo et al., 1997). Whenever necessary, embryos were fixed in 0.2% glutaraldehyde and stained at 37°C for 2-12 hours to enhance the signal. Each construct was tested on several different batches of embryos; graphs and error bars were obtained as previously described (Dunn and Di Gregorio, 2009).
Plasmid construction
The Bra>macho fusion was constructed by removing the coding region of Ciona Brachyury (Ci-Bra) from the −3.5 kb Ci-Bra>eGFP plasmid (Corbo et al., 1997) and by replacing eGFP with the coding region and 3′-UTR from the Ci-macho1 cDNA. The Ci-Bra promoter was amplified from Ci-Bra>eGFP using the primers: 5′-ATAAGAATTCGGCTTATGACGAAATAATGT-3′ and 5′-ATATGCGGCCGCTATAGGTTTGTAACTCGCACT-3′ and the resulting PCR product was digested with EcoRI and NotI. The Ci-macho1 coding sequence and 3′-UTR were amplified from a cDNA clone kindly provided by Yutaka Satou (Kyoto University, Kyoto, Japan), using primers 5′-GTAGGCGGCCGCATGGCCTTTACTGGTACGATGGGA-3′ and 5′-CAGGTGGCTCAGCTAATACGACTCACTATAGGGCG-3′ and the resulting PCR product was digested with NotI and BlpI. The two PCR products were cloned into Ci-Bra>eGFP digested with EcoRI and BlpI through a triple ligation.
To construct the Bra>Tbx6b fusion plasmid, the Ci-Tbx6b coding region was excised from the Ci-Tbx6b cDNA and cloned into the NotI and KpnI sites of Bra>eGFP. To create the Bra>Tbx6c plasmid, the full-length Ci-Tbx6c ORF was reconstructed from two separate PCR-amplified fragments. The 5′-most fragment, containing the region encoding amino acids 9-269 of Ci-Tbx6c, was PCR-amplified using as a template the published GST-Tbx6c fusion construct kindly provided by Michael Levine, UC Berkeley, CA (Yagi et al., 2005), with primer Ci-Tbx6c-top, 5′-TGAAGCGGCCGCATGGCGACAGACATGAGAAGCCCAACCTTTGAACCGAAAGTTCATCTTCAGG-3′, which adds to the GST-Tbx6c construct the first eight codons of Ci-Tbx6c, and primer 5′-CAGTTTAGTGATCTGTCCGTTTTGGC-3′. The remainder of the ORF was amplified using as a template clone GC43g03 from the Ciona cDNA collection release 1 (Satou et al., 2002b), with primer 5′-GGCACGAGGCCAAAACGGACAGATC-3′ and primer Ci-Tbx6c-ORF-bot, 5′-TTCCAAGCTAAGCTTTTATTCACTATAGGACACAATTACTAAC-3′. The two PCR products resulting from these reactions were then used as templates for a final PCR in the presence of primers Ci-Tbx6c-top and Ci-Tbx6c-ORF-bot. The resulting product, encoding the full-length Ci-Tbx6c protein, which spans 388 amino acids, was digested with NotI and BlpI and ligated into the pCi-Bra-Linker vector (Dunn and Di Gregorio, 2009).
The Ciona Tbx6b (Ci-Tbx6b) 5′-flanking region was PCR-amplified from Ciona intestinalis genomic DNA using the following primers: 5′-AATGTAGCGTCGCTTCACAACCAGTCG-3′ and 5′-GAGACTCGTTTTCGATGCCACTTTG-3′. The resulting ~2.4 kb fragment was cloned into the pFBΔSP6 vector (Oda-Ishii and Di Gregorio, 2007). The Ci-Tbx6b basal promoter was PCR-amplified using primers 5′-CGAGCCATGGGGCATCGAAAACGAGTCTCGC-3′ and 5′-ACTAGCGGCCGCCCATAGTCTTGTCTGGTCCAA-3′, which yielded a 154 bp fragment encompassing the TATA box and nearby start of the longest Ci-Tbx6b EST, which we refer to as the transcription start site. The PCR-amplified fragment was cloned into the NcoI and NotI sites of the pFBΔSP6 vector.
Mutations in the core sequence (CCC>TTT) of the three Ci-Macho1-binding sites of the 2.4 kb Ci-Tbx6b CRM were introduced sequentially by PCR amplification. The Ciona Fibrillar Collagen-1 (CiFCol1) CRMs were identified through the analysis of a 2.2 kb fragment originally cloned from a Ciona genomic library. The Ci-Tbx6c CRM was PCR-amplified from Ciona genomic DNA using the primers 5′-ATCTCGAGGATTCTTTAAGAATATTTTTTGATAATG-3′ and 5′-CATCTAGACGTAACGCATGATCAAAGTTAAATTAAAC-3′.
In all the PCR amplifications, ~125 ng of genomic DNA extracted from the sperm of a single Ciona intestinalis adult were used as a template, in the presence of either Hi-Fi Taq (Invitrogen, Carlsbad, CA, USA) or Turbo Pfu (Stratagene, La Jolla, CA) DNA polymerase. All plasmids were checked for accuracy either manually (Sambrook et al., 1989) or through automated sequencing facilities.
Whole-mount in situ hybridization (WMISH)
This was performed as previously described (Oda-Ishii and Di Gregorio, 2007) using digoxigenin-labeled antisense RNA probes. The CiFCol1probe was prepared from EST GC01e23, the Ci-macho1 probe from EST GC18h07, the Ci-Tbx6b probe from EST GC09i19, and the Ci-Tbx6c probe from EST GC43g03 (Satou et al., 2002b). The lacZ probe was kindly provided by Yutaka Nibu (Weill Cornell Medical College, New York, NY).
Quantitative RT-PCR (qRT-PCR)
Ciona zygotes from a single round of fertilization were divided into two groups and were either electroporated with Ci-Bra>eGFP or co-electroporated with Ci-Bra>eGFP and Bra>macho constructs. Once they reached the 110-cell stage, only the embryos showing fluorescence in the notochord were selected for RNA extraction using an epifluorescence microscope. RNA was extracted using the RNeasy Protect Mini kit (Qiagen, Valencia, CA), and treated with RNase-free DNase (Qiagen) using the on-column digestion protocol to remove possible genomic DNA contamination. cDNA was synthesized using the Superscript III kit (Invitrogen). This process was repeated twice on different batches of embryos to generate two independent samples of Bra>macho cDNA and corresponding wild-type control cDNA.
qRT-PCR samples were prepared using 2× SYBR Green Hotstart Mix (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The resulting data were analyzed using the SDS2.3 software (Applied Biosystems). Both sets of cDNAs were loaded with the appropriate primers on a single plate, to eliminate variation between PCR amplifications. Samples were run in triplicate, using Ci-GAPDH as a control. Error bars represent the variation seen between the two sets of cDNAs. The relative standard curve method was used to calculate the fold change in gene expression, as described by the manufacturer (http://www3.appliedbiosystems.com/AB_Home/index.htm). We considered the 1.6-fold increase in gene expression relevant in consideration of the low number of notochord precursors at the 110-cell stage (ten), the electroporation efficiency, and the high variability in the number of cells that efficiently incorporate the construct(s) within each transgenic embryo.
Recombinant protein synthesis and purification
The full-length Ci-macho1 coding region was PCR-amplified from the Bra>macho plasmid described above and cloned into the BamHI and SacI sites of the pRSET-B vector (Invitrogen). Full-length Ci-Tbx6b and Ci-Tbx6c coding regions were PCR-amplified and cloned into the pGEX-KG vector (GE Healthcare, Piscataway, NJ) in frame with the GST tag. The resulting plasmids were transformed into E. coli BL21 (DE3) cells harboring the pJY2 plasmid (Affiniti Research Products, Exeter, UK) and the fusion proteins were purified as previously described (Gazdoiu et al., 2005). The 6×His-tagged Ci-Macho1 protein was expressed at 15°C in the presence of 0.1 mM IPTG.
Electrophoretic mobility shift assays (EMSA)
The following double-stranded oligonucleotides were used (only the 5′-3′ strand is reported, mutations are underlined): Macho site-A-wt, CAAACTATGCATCGGGTGTCAGGGAGACA; Macho site-A-MUT, CAAACTATGCATCtttTGTCAGGGAGACA; Macho site-B-wt, GACAACGATCTATGGGAGCCGTGAGGATA; Macho site-B-MUT, GACAACGATCTATtttAGCCGTGAGGATA; Macho site-C-wt, AAGTAATGAGGACCCCGCTGCGGTAACCT; Macho site-C-MUT, AAGTAATGAGGACtttGCTGCGGTAACCT; T-box site wt, 5′-CGGTATGCGTCACACTGAGTTTTG-3′; T-box site MUT, 5′-CGGTATGCGTCAacaTGAGTTTTG-3′. Radioactive labeling and subsequent procedures were performed as previously described (Dunn and Di Gregorio, 2009).
Supplementary Material
Acknowledgments
We thank Yutaka Nibu and the members of the Di Gregorio and Nibu labs for valuable comments on the manuscript and Hiroki Takahashi (NIBB, Japan) for the Ci-Tbx6b cDNA clone. We are grateful to Mami Takeda and Gary Esses for technical help. This work was supported by grant 1-FY08-430 from the March of Dimes Birth Defects Foundation and by NIH/NICHD grant R01HD050704 to A.D.G. J.E.K. was supported in part by a Jacques Cohenca pre-doctoral fellowship from the Weill Graduate School of Medical Sciences; I.O.-I. was supported in part by a post-doctoral fellowship from the Uehara Memorial Foundation (Japan). A.D.G. is an Irma T. Hirschl Scholar. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/14/2453/DC1
References
- Aruga J., Kamiya A., Takahashi H., Fujimi T. J., Shimizu Y., Ohkawa K., Yazawa S., Umesono Y., Noguchi H., Shimizu T., et al. (2006). A wide-range phylogenetic analysis of Zic proteins: implications for correlations between protein structure conservation and body plan complexity. Genomics 87, 783-792 [DOI] [PubMed] [Google Scholar]
- Bertrand V., Hudson C., Caillol D., Popovici C., Lemaire P. (2004). Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell 115, 615-627 [DOI] [PubMed] [Google Scholar]
- Brown C. D., Johnson D. S., Sidow A. (2007). Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science 317, 1557-1560 [DOI] [PubMed] [Google Scholar]
- Chapman D. L., Papaioannou V. E. (1998). Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature 391, 695-697 [DOI] [PubMed] [Google Scholar]
- Christiaen L., Stolfi A., Davidson B., Levine M. (2009). Spatio-temporal intersection of Lhx3 and Tbx6 defines the cardiac field through synergistic activation of Mesp. Dev. Biol. 328, 552-560 [DOI] [PubMed] [Google Scholar]
- Conklin E. G. (1905). The organization and cell-lineage of the ascidian egg. J. Acad. Natl. Sci. Philadelphia 13, 1-119 [Google Scholar]
- Corbo J. C., Levine M., Zeller R. W. (1997). Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124, 589-602 [DOI] [PubMed] [Google Scholar]
- Davidson B., Christiaen L. (2006). Linking chordate gene networks to cellular behavior in ascidians. Cell 124, 247-250 [DOI] [PubMed] [Google Scholar]
- Davidson B., Shi W., Levine M. (2005). Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development 132, 4811-4818 [DOI] [PubMed] [Google Scholar]
- Dehal P., Satou Y., Campbell R. K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D., et al. (2002). The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298, 2157-2167 [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Chourrout D., Philippe H. (2006). Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965-968 [DOI] [PubMed] [Google Scholar]
- Deno T., Nishida H., Satoh N. (1984). Autonomous muscle cell differentiation in partial ascidian embryos according to the newly verified cell lineages. Dev. Biol. 104, 322-328 [DOI] [PubMed] [Google Scholar]
- Di Gregorio A., Levine M. (2002). Analyzing gene regulation in ascidian embryos: new tools for new perspectives. Differentiation 70, 132-139 [DOI] [PubMed] [Google Scholar]
- Di Gregorio A., Corbo J. C., Levine M. (2001). The regulation of forkhead/HNF-3beta expression in the Ciona embryo. Dev. Biol. 229, 31-43 [DOI] [PubMed] [Google Scholar]
- Dunn M. P., Di Gregorio A. (2009). The evolutionarily conserved leprecan gene: its regulation by Brachyury and its role in the developing Ciona notochord. Dev. Biol. 328, 561-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erives A. (2009). Non-homologous structured CRMs from the Ciona genome. J. Comput. Biol. 16, 369-377 [DOI] [PubMed] [Google Scholar]
- Erives A., Levine M. (2000). Characterization of a maternal T-Box gene in Ciona intestinalis. Dev. Biol. 225, 169-178 [DOI] [PubMed] [Google Scholar]
- Erives A., Corbo J. C., Levine M. (1998). Lineage-specific regulation of the Ciona snail gene in the embryonic mesoderm and neuroectoderm. Dev. Biol. 194, 213-225 [DOI] [PubMed] [Google Scholar]
- Gazdoiu S., Yamoah K., Wu K., Escalante C. R., Tappin I., Bermudez V., Aggarwal A. K., Hurwitz J., Pan Z. Q. (2005). Proximity-induced activation of human Cdc34 through heterologous dimerization. Proc. Natl. Acad. Sci. USA 102, 15053-15058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering L. M., Hoshijima K., Hug B., Bisgrove B., Kispert A., Grunwald D. J. (2003). An interacting network of T-box genes directs gene expression and fate in the zebrafish mesoderm. Proc. Natl. Acad. Sci. USA 100, 9410-9415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C., Yasuo H. (2008). Similarity and diversity in mechanisms of muscle fate induction between ascidian species. Biol. Cell 100, 265-277 [DOI] [PubMed] [Google Scholar]
- Imai K. S., Hino K., Yagi K., Satoh N., Satou Y. (2004). Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131, 4047-4058 [DOI] [PubMed] [Google Scholar]
- Imai K. S., Levine M., Satoh N., Satou Y. (2006). Regulatory blueprint for a chordate embryo. Science 312, 1183-1187 [DOI] [PubMed] [Google Scholar]
- Kim G. J., Nishida H. (1999). Suppression of muscle fate by cellular interaction is required for mesenchyme formation during ascidian embryogenesis. Dev. Biol. 214, 9-22 [DOI] [PubMed] [Google Scholar]
- Kim G. J., Yamada A., Nishida H. (2000). An FGF signal from endoderm and localized factors in the posterior-vegetal egg cytoplasm pattern the mesodermal tissues in the ascidian embryo. Development 127, 2853-2862 [DOI] [PubMed] [Google Scholar]
- Kondoh K., Kobayashi K., Nishida H. (2003). Suppression of macho-1-directed muscle fate by FGF and BMP is required for formation of posterior endoderm in ascidian embryos. Development 130, 3205-3216 [DOI] [PubMed] [Google Scholar]
- Kusakabe T., Yoshida R., Ikeda Y., Tsuda M. (2004). Computational discovery of DNA motifs associated with cell type-specific gene expression in Ciona. Dev. Biol. 276, 563-580 [DOI] [PubMed] [Google Scholar]
- Lemaire P. (2009). Unfolding a chordate developmental program, one cell at a time: invariant cell lineages, short-range inductions and evolutionary plasticity in ascidians. Dev. Biol. 332, 48-60 [DOI] [PubMed] [Google Scholar]
- Meedel T. H., Crowther R. J., Whittaker J. R. (1987). Determinative properties of muscle lineages in ascidian embryos. Development 100, 245-260 [DOI] [PubMed] [Google Scholar]
- Meedel T. H., Chang P., Yasuo H. (2007). Muscle development in Ciona intestinalis requires the b-HLH myogenic regulatory factor gene Ci-MRF. Dev. Biol. 302, 333-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinertzhagen I. A., Okamura Y. (2001). The larval ascidian nervous system: the chordate brain from its small beginnings. Trends Neurosci. 24, 401-410 [DOI] [PubMed] [Google Scholar]
- Merzdorf C. S. (2007). Emerging roles for zic genes in early development. Dev. Dyn. 236, 922-940 [DOI] [PubMed] [Google Scholar]
- Nishida H. (1992). Developmental potential for tissue differentiation of fully dissociated cells of the ascidian embryo. Roux's Arch. Dev. Biol. 201, 81-87 [DOI] [PubMed] [Google Scholar]
- Nishida H. (2002). Patterning the marginal zone of early ascidian embryos: localized maternal mRNA and inductive interactions. BioEssays 24, 613-624 [DOI] [PubMed] [Google Scholar]
- Nishida H., Sawada K. (2001). macho-1 encodes a localized mRNA in ascidian eggs that specifies muscle fate during embryogenesis. Nature 409, 724-729 [DOI] [PubMed] [Google Scholar]
- Oda-Ishii I., Di Gregorio A. (2007). Lineage-independent mosaic expression and regulation of the Ciona multidom gene in the ancestral notochord. Dev. Dyn. 236, 1806-1819 [DOI] [PubMed] [Google Scholar]
- Ortolani G. (1955). The presumptive territory of the mesoderm in the ascidian germ. Experientia 11, 445-446 [Google Scholar]
- Passamaneck Y. J., Di Gregorio A. (2005). Ciona intestinalis: chordate development made simple. Dev. Dyn. 233, 1-19 [DOI] [PubMed] [Google Scholar]
- Passamaneck Y. J., Hadjantonakis A.-K., Di Gregorio A. (2007). Dynamic and polarized muscle cell behaviors accompany tail morphogenesis in the ascidian Ciona intestinalis. PLoS ONE 2, e714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J. M., Oda-Ishii I., Passamaneck Y. J., Hadjantonakis A.-K., Di Gregorio A. (2005). Live imaging and morphometric analysis of embryonic development in the ascidian Ciona intestinalis. Genesis 43, 136-147 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. G., Maniatis T. (1989). Molecular Cloning: a Laboratory Manual Plainview, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Sardet C., Nishida H., Prodon F., Sawada K. (2003). Maternal mRNAs of PEM and macho 1, the ascidian muscle determinant, associate and move with a rough endoplasmic reticulum network in the egg cortex. Development 130, 5839-5849 [DOI] [PubMed] [Google Scholar]
- Satoh N. (1994). Developmental Biology of Ascidians New York: Cambridge University Press; [Google Scholar]
- Satou Y., Takatori N., Yamada L., Mochizuki Y., Hamaguchi M., Ishikawa H., Chiba S., Imai K., Kano S., Murakami S. D., et al. (2001). Gene expression profiles in Ciona intestinalis tailbud embryos. Development 128, 2893-2904 [DOI] [PubMed] [Google Scholar]
- Satou Y., Yagi K., Imai K. S., Yamada L., Nishida H., Satoh N. (2002a). macho-1-related genes in Ciona embryos. Dev. Genes Evol. 212, 87-92 [DOI] [PubMed] [Google Scholar]
- Satou Y., Yamada L., Mochizuki Y., Takatori N., Kawashima T., Sasaki A., Hamaguchi M., Awazu S., Yagi K., Sasakura Y., et al. (2002b). A cDNA resource from the basal chordate Ciona intestinalis. Genesis 33, 153-154 [DOI] [PubMed] [Google Scholar]
- Satou Y., Imai K. S., Satoh N. (2004). The ascidian Mesp gene specifies heart precursor cells. Development 131, 2533-2541 [DOI] [PubMed] [Google Scholar]
- Sawada K., Fukushima Y., Nishida H. (2005). Macho-1 functions as transcriptional activator for muscle formation in embryos of the ascidian Halocynthia roretzi. Gene Expr. Patterns 5, 429-437 [DOI] [PubMed] [Google Scholar]
- Shi W., Peyrot S. M., Munro E., Levine M. (2009). FGF3 in the floor plate directs notochord convergent extension in the Ciona tadpole. Development 136, 23-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Hotta K., Erives A., Di Gregorio A., Zeller R. W., Levine M., Satoh N. (1999). Brachyury downstream notochord differentiation in the ascidian embryo. Genes Dev. 13, 1519-1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatori N., Hotta K., Mochizuki Y., Satoh G., Mitani Y., Satoh N., Satou Y., Takahashi H. (2004). T-box genes in the ascidian Ciona intestinalis: characterization of cDNAs and spatial expression. Dev. Dyn. 230, 743-753 [DOI] [PubMed] [Google Scholar]
- Tazumi S., Yabe S., Yokoyama J., Aihara Y., Uchiyama H. (2008). PMesogenin1 and 2 function directly downstream of Xtbx6 in Xenopus somitogenesis and myogenesis. Dev. Dyn. 237, 3749-3761 [DOI] [PubMed] [Google Scholar]
- Uchiyama H., Kobayashi T., Yamashita A., Ohno S., Yabe S. (2001). Cloning and characterization of the T-box gene Tbx6 in Xenopus laevis. Dev. Growth Differ. 43, 657-669 [DOI] [PubMed] [Google Scholar]
- Wada H., Okuyama M., Satoh N., Zhang S. (2006). Molecular evolution of fibrillar collagen in chordates, with implications for the evolution of vertebrate skeletons and chordate phylogeny. Evol. Dev. 8, 370-377 [DOI] [PubMed] [Google Scholar]
- White P. H., Farkas D. R., McFadden E. E., Chapman D. L. (2003). Defective somite patterning in mouse embryos with reduced levels of Tbx6. Development 130, 1681-1690 [DOI] [PubMed] [Google Scholar]
- Yagi K., Satoh N., Satou Y. (2004a). Identification of downstream genes of the ascidian muscle determinant gene Ci-macho1. Dev. Biol. 274, 478-489 [DOI] [PubMed] [Google Scholar]
- Yagi K., Satou Y., Satoh N. (2004b). A zinc finger transcription factor, ZicL, is a direct activator of Brachyury in the notochord specification of Ciona intestinalis. Development 131, 1279-1288 [DOI] [PubMed] [Google Scholar]
- Yagi K., Takatori N., Satou Y., Satoh N. (2005). Ci-Tbx6b and Ci-Tbx6c are key mediators of the maternal effect gene Ci-macho1 in muscle cell differentiation in Ciona intestinalis embryos. Dev. Biol. 282, 535-549 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.