Abstract
All but five of the N-terminal 23 residues of the HA2 domain of the influenza virus glycoprotein hemagglutinin (HA) are strictly conserved across all 16 serotypes of HA genes. The structure and function of this HA2 fusion peptide (HAfp) continues to be the focus of extensive biophysical, computational, and functional analysis, but most of these analyses are of peptides that do not include the strictly conserved residues Trp21-Tyr22-Gly23. The heteronuclear triple resonance NMR study reported here of full length HAfp of sero subtype H1, solubilized in dodecylphosphatidyl choline, reveals a remarkably tight helical hairpin structure, with its N-terminal α-helix (Gly1-Gly12) packed tightly against its second α-helix (Trp14-Gly23), with six of the seven conserved Gly residues at the interhelical interface. The seventh conserved Gly residue in position 13 adopts a positive ϕ angle, enabling the hairpin turn that links the two helices. The structure is stabilized by multiple interhelical CαH to C = O hydrogen bonds, characterized by strong interhelical HN-Hα and Hα-Hα NOE contacts. Many of the previously identified mutations that make HA2 nonfusogenic are also incompatible with the tight antiparallel hairpin arrangement of the HAfp helices.15N relaxation analysis indicates the structure to be highly ordered on the nanosecond time scale, and NOE analysis indicates HAfp is located at the water-lipid interface, with its hydrophobic surface facing the lipid environment, and the Gly-rich side of the helix-helix interface exposed to solvent.
Keywords: aliphatic hydrogen bond, fusion peptide, helix-helix interaction, NMR, relaxation
As exemplified by the 2009 outbreak of the swine flu pandemic, influenza remains a high priority health concern. Of the three different types of RNA influenza virus, the influenza A type is the most common and virulent human pathogen. Based on the antibody response to its viral surface proteins, hemagglutinin and neuraminidase, influenza type A is further subdivided into serotypes, ranging from subtypes H1-16 for hemagglutinin, and N1-9 for neuraminidase. Subtypes H1, H2, H3, N1, and N2 are the most common in human disease (1). Whereas neuraminidase is an enzyme responsible for cleavage of cellular sialic acid carbohydrates, a key step in viral exit, HA mediates cellular entry through binding to carbohydrates on epithelial cells and subsequent delivery of the viral contents to the cellular interior (2, 3).
Prior to infection of human tissue, the homotrimeric hemagglutinin, HA0, requires cleavage by one of several human proteases (4), resulting in a trimer of heterodimeric proteins, HA1 and HA2, covalently linked to one another by a single disulfide bridge. The atomic structures of the precursor and processed HA have been solved by X-ray crystallography (5–7) and reveal that the main rearrangement taking place upon cleavage is the relocation of the newly formed N-terminal HA2 tail from the surface of HA0 to the helical core of the HA2 ecto domain. Upon lowering the pH, the N-terminal HA2 fusion domain is extruded from the HA2 helical core to a position where it can target the endosomal membrane, while the C-terminal transmembrane helix keeps HA2 anchored to the viral membrane. These two membrane anchors are separated by the homotrimeric rod-shaped ecto domain which subsequently undergoes a drastic structural rearrangement, bringing the N- and C-terminal membrane anchors in close proximity (2, 3). During this rearrangement, the C-terminal transmembrane anchor of HA2 remains embedded in the viral membrane, but the structural rearrangement brings the N and C termini of HA2, and thereby their associated endosomal and viral membranes, in proximity of one another—a process preceding viral fusion (6, 8). Fusion then proceeds through a hemi-fusion intermediate (9), with mixing of the outer leaflets of the two membranes, followed by formation of a fusion pore (10–12).
Despite many years of intense study, the detailed atomic-level mechanism of membrane fusion remains elusive and challenges our understanding of protein-lipid interactions. The role of the fusion peptide has been investigated extensively by mutagenesis, carried out on the intact protein (13–19) and also on isolated HAfp peptides, which exhibit fusogenic activity. The latter studies investigated lipid-bound, fusion-active structures (20, 21), how they interact with the host membrane to promote fusion (22–25), and how conservative mutations have fatal consequences on fusion activity. Many of these studies focused on a truncated form of the HAfp subtype H3 peptide (20) and its various mutants in dodecylphosphatidyl choline (DPC) micelles at both neutral and fusogenic pH values (26, 27). This truncated form only contains the N-terminal 20 residues of HA2, followed by a Lys-rich heptapeptide for residues 21–27, enhancing its water solubility. Use of this so-called P20H7 host-guest system not only facilitated peptide synthesis and purification, it also enabled straightforward thermodynamic studies of its interaction energy with various lipid environments (28). However, the use of P20H7 is not without shortcomings. In particular, the absence of the strictly conserved residues, Trp21-Tyr22-Gly23, is a concern. Trp and Tyr often play a pivotal role in anchoring peptides and proteins at the membrane-water interface (29, 30), and conserved Gly residues in membrane proteins, in particular when part of a GXXXG or GXXG motif, as is Gly23 in HAfp, can be important in helix stabilization and interhelical interaction (31, 32).
The P20H7 wild-type peptide was reported to have a boomerang shaped structure (20, 33), with its apex exposed to solvent, and with its positively charged N-terminal amino group (34) and negatively charged Asp19 side chain, more than 20 Å away from the N terminus, deeply embedded in the hydrophobic interior. Without any clear charge compensation, this raises the question of what drives the burial of these charged groups. A second question not resolved by the P20H7 structure relates to the impact of mutation of Gly8 to Ala, which abolishes fusion activity, without requiring disruption of the reported P20H7 backbone structure. A third question is whether the absence of the highly conserved residues 21–23 impacts the structure of the remainder of HAfp.
In order to address the above questions, we have isolated and characterized the full length HAfp for the H1 sero subtype, followed by a Lys-rich heptapeptide similar to that used for the shorter sequence, again solubilized in DPC micelles. The peptide was obtained using a bacterially expressed HAfp fusion protein, followed by proteolytic cleavage and purification. This approach allowed us to include 13C, 15N and 2H labeling, enabling use of the full battery of modern multidimensional isotope-based NMR methods, including the measurement of very extensive NOE, residual dipolar coupling (RDC), and J coupling restraints. The isotope labeling approach also facilitates evaluation of the pH dependence of the structure, its position at the water-lipid interface, and the protonation state of Gly1. Our results show that the three conserved residues absent in P20H7 but present in our full length HAfp construct play a key role in stabilizing a highly ordered structure, where its N- and C-terminal helices pack tightly together in a hairpin-like structure. One side of the helical hairpin is completely hydrophobic and shows NOE interactions to the alkyl chains in the interior of the DPC micelle, whereas the other surface includes the polar side chains of Glu11, Thr15, and Asp19, and the Hα2 hydrogens of five Gly residues located at the interhelical interface.
Results
Comparison of the 1H-15N heteronuclear single quantum correlation (HSQC) NMR spectra of a truncated HAfp peptide containing only the first 20 residues, HAfp1–20, with that of the full length HAfp1–23 shows remarkably large shifts in resonance positions, suggestive of substantial structural differences (Fig. S1). Whereas narrower 15N line widths towards the amino- and carboxy-terminal ends of HAfp1–20 are indicative of increased disorder, HAfp1–23 shows highly homogeneous line widths with residues 21–23 protected from rapid exchange with solvent, even at the relatively high pH value of 7.4, indicative of stable hydrogen bonding. Comparison of the HAfp1–23 HSQC spectra recorded at pH 7.4 and pH 4.0 shows only small changes in peak positions, with the exception of the backbone amides of Glu11 and Asp19, which alter their side chain protonation state (Fig. S2). The similarity in the full length HAfp1–23 HSQC spectra, recorded at high and low pH values, points to the absence of a major structural rearrangement. This conclusion is confirmed by the measurement of RDCs and NOE spectra (vide infra).
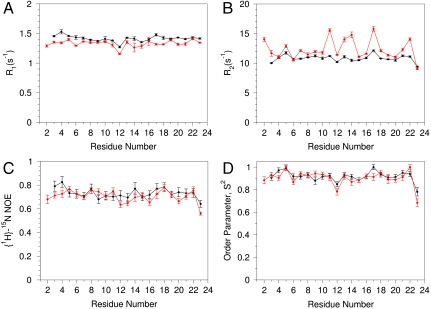
Quantitative evaluation of the HAfp1–23 backbone rigidity was carried out by analysis of amide 15N relaxation rates (35, 36). Both the longitudinal (R1) and transverse (R2) 15N relaxation rates at pH 7.4, as well as the heteronuclear 15N-{1H} NOE, are remarkably uniform over the entire HAfp region, from residue Leu2 to Gly23 (Fig. 1). Note that the amino-terminal 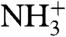 resonance of Gly1 is not detectable due to the intrinsic fast exchange of
resonance of Gly1 is not detectable due to the intrinsic fast exchange of 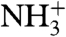 hydrogens with solvent. Analysis of the amide 15N relaxation rates in terms of the standard model-free Lipari-Szabo formalism (37) yields rotational correlation times of 8.4 and 9.1 ns at pH 7.4 and 4.0, respectively, for the micelle-attached peptide, comparable to values measured for other peptides embedded in DPC micelles (38). Slightly elevated 15N R2 rates for a subset of the backbone amides at pH 4.0 (Glu11, Trp14, and Met17) suggest the presence of an exchange contribution to these rates at acidic pH. However, considering that the corresponding resonances remain sharp, with RDC and 1H-1H NOE values remaining fully consistent with those observed at high pH (Fig. 2D), the alternate conformation sampled by HAfp1–23 must either have a very low population or be structurally very similar to the major conformer.
hydrogens with solvent. Analysis of the amide 15N relaxation rates in terms of the standard model-free Lipari-Szabo formalism (37) yields rotational correlation times of 8.4 and 9.1 ns at pH 7.4 and 4.0, respectively, for the micelle-attached peptide, comparable to values measured for other peptides embedded in DPC micelles (38). Slightly elevated 15N R2 rates for a subset of the backbone amides at pH 4.0 (Glu11, Trp14, and Met17) suggest the presence of an exchange contribution to these rates at acidic pH. However, considering that the corresponding resonances remain sharp, with RDC and 1H-1H NOE values remaining fully consistent with those observed at high pH (Fig. 2D), the alternate conformation sampled by HAfp1–23 must either have a very low population or be structurally very similar to the major conformer.
Fig. 1.
HAfp1–23 backbone dynamics extracted from 15N relaxation rates at a magnetic field strength of 14.1 Tesla, pH 7.4 (black) and 4.0 (red). (A) Longitudinal 15N relaxation rates, R1; (B) Transverse 15N relaxation rates, R2; (C) 15N-{1H} NOE; (D) Model-free generalized order parameters, S2, derived from these data for a librationally corrected 1.04 Å bond length and an axially symmetric 15N Δσ value of -173 ppm applicable for α-helices (57), using an isotropic diffusion model.
Fig. 2.
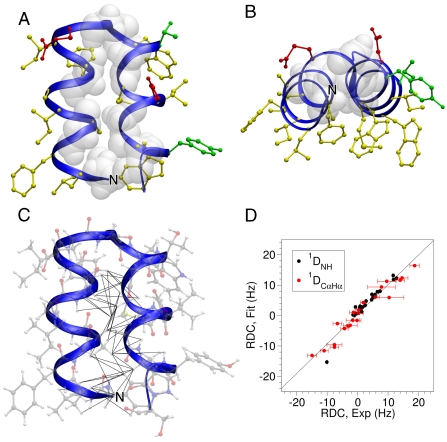
Structure of HAfp1–23. (A) Longitudinal view with the hydrophobic side chains drawn in yellow, polar side chains in green, acidic side chains in red, and Gly residues shown in white van der Waals representation. The interhelical angle between helix A (residues 2–12) and helix B (residues 14–22) is 158 °, and the interhelical distance calculated using the program interhlx (K. Yap, University of Toronto) is 6.3 Å. The amino terminus is marked N.(B) Lateral view. (C) Longitudinal view showing all atoms, and interhelical NOEs below 5 Å as black lines. (D) Singular value decomposition fit of the pH 4.0 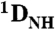 and
and 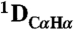 RDCs to the refined pH 7.4 structure. The
RDCs to the refined pH 7.4 structure. The 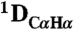 couplings have been scaled by -0.484 relative to
couplings have been scaled by -0.484 relative to 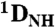 to account for the relative internuclear dipolar interaction strength. The Q-factor of the pH 4.0 data to the pH 7.4 structure is 22.3%.
to account for the relative internuclear dipolar interaction strength. The Q-factor of the pH 4.0 data to the pH 7.4 structure is 22.3%.
The high degree of backbone order, as revealed by near-unity values of the generalized order parameter, S2, across the entire length of HAfp1–23 is comparable to that found in well-structured globular proteins. For such well-structured systems, interpretation of RDCs in terms of orientations of the internuclear vector is straightforward (39), and a large number of such couplings were therefore measured, including all 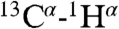 and
and 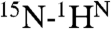 couplings, the geminal Gly
couplings, the geminal Gly 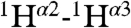 values, and the intrinsically much smaller backbone
values, and the intrinsically much smaller backbone 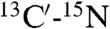 and
and 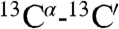 RDCs. In addition, more than 450 1H-1H distance restraints were derived from recording two- and three-dimensional NMR spectra (Fig. S3). Both the short and medium range NOE data, the NMR chemical shifts, and the RDC data are fully compatible with α-helical structures extending from Gly1 to Gly12, and Trp14-Gly23. A remarkable feature in these NOE spectra, however, is the presence of relatively strong
RDCs. In addition, more than 450 1H-1H distance restraints were derived from recording two- and three-dimensional NMR spectra (Fig. S3). Both the short and medium range NOE data, the NMR chemical shifts, and the RDC data are fully compatible with α-helical structures extending from Gly1 to Gly12, and Trp14-Gly23. A remarkable feature in these NOE spectra, however, is the presence of relatively strong 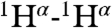 ,
, 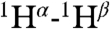 and
and 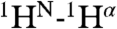 NOE interactions between residues located in these separate α-helices, including interactions between Met17 and Phe9, Trp21 and Ala5, and Trp21 and Gly1 (Fig. S3). Use of mixtures of 13C-labeled and unlabeled peptides combined with isotope editing and filtering procedures (40, 41), proved all these interhelical NOEs to be intramolecular, thereby excluding antiparallel homodimeric arrangements of fully α-helical HAfp1–23 molecules.
NOE interactions between residues located in these separate α-helices, including interactions between Met17 and Phe9, Trp21 and Ala5, and Trp21 and Gly1 (Fig. S3). Use of mixtures of 13C-labeled and unlabeled peptides combined with isotope editing and filtering procedures (40, 41), proved all these interhelical NOEs to be intramolecular, thereby excluding antiparallel homodimeric arrangements of fully α-helical HAfp1–23 molecules.
Close contacts between Hα protons which are distant in the amino acid sequence are commonly observed in β-sheet structures (42). The presence of such contacts between residues in neighboring α-helices is rare, but not unprecedented. For example, short (≤ 3 Å) interhelical Hα-Hα distances are seen in the classic glycophorin A (GpA) dimer structure (43), even though the corresponding NOEs were not reported at that time, and in a substantial number of X-ray structures (44). Multiple interhelical Hα-Hα NOEs are reported in a recent NMR study of the BNIP3 transmembrane domain (45). Both GpA and BNIP3 structures are homodimeric and stabilized by intermolecular H-bonds between CαH and backbone carbonyl oxygens, with the H-bond-linked transmembrane helices in a parallel orientation. Our NMR structure, derived from the NOE, RDC, and three-bond 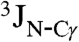 and
and 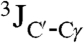 coupling restraints, shows a tightly packed helical hairpin (Fig. 2), where the helices are antiparallel to one another but also exhibit the characteristic interhelical hydrogen bonds between CαH and backbone carbonyl oxygens. For each of these four H-bonds (Trp21 CαH to Gly1 C = O; Met17 CαH to Ala5 C = O; Ala5 CαH to Met17 C = O; Phe9 CαH to Gly13 C = O), the carbonyl oxygen of residue i accepts the regular helical H-bond from its
coupling restraints, shows a tightly packed helical hairpin (Fig. 2), where the helices are antiparallel to one another but also exhibit the characteristic interhelical hydrogen bonds between CαH and backbone carbonyl oxygens. For each of these four H-bonds (Trp21 CαH to Gly1 C = O; Met17 CαH to Ala5 C = O; Ala5 CαH to Met17 C = O; Phe9 CαH to Gly13 C = O), the carbonyl oxygen of residue i accepts the regular helical H-bond from its 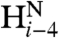 as well as from the
as well as from the 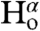 atom of the other helix, putting in close proximity these hydrogens from separate helices. Therefore, such interhelical H-bonds predict strong interhelical NOE interactions between
atom of the other helix, putting in close proximity these hydrogens from separate helices. Therefore, such interhelical H-bonds predict strong interhelical NOE interactions between 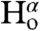 and
and 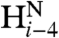 , as indeed observed for all four above listed H-bonds (Fig. S3D). Furthermore, the proximity and geometry between the Gly1
, as indeed observed for all four above listed H-bonds (Fig. S3D). Furthermore, the proximity and geometry between the Gly1 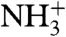 protons and the carbonyl of Gly20 produces another hydrogen bond, as identified by the potential-of-mean-force hydrogen bond database (HBDB) module (46) of the XPLOR-NIH structure calculation program (47). Inclusion of these hydrogen bonds as fixed restraints in the refinement did not adversely affect structural statistics and improved the quality validation factors,
protons and the carbonyl of Gly20 produces another hydrogen bond, as identified by the potential-of-mean-force hydrogen bond database (HBDB) module (46) of the XPLOR-NIH structure calculation program (47). Inclusion of these hydrogen bonds as fixed restraints in the refinement did not adversely affect structural statistics and improved the quality validation factors,  and
and 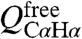 (Table S1).
(Table S1).
In the resulting structures, one side of the helical hairpin is completely covered by hydrophobic side chains (Fig. 2B), whereas the other side exposes the polar side chains of Glu11, Thr15, and Asp19, as well as the Hα2 protons of the α-helical Gly residues. The side chains of the majority of hydrophobic residues are locked into a single χ1 conformer, as judged by 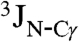 and
and 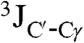 values (Table S2), and confirmed by intraresidue and sequential NOEs. A positive ϕ angle on Gly13 reverses the backbone chain, leading into the second helix, and a total of 66 long-range NOEs (Fig. 2C) lock in the tertiary contacts between the two helices. Small regions taken from the two-dimensional and three-dimensional NOESY spectra illustrate the unambiguous nature of such interactions when spectra are recorded at high field (900 MHz) and at high spectral resolution (Fig. S3).
values (Table S2), and confirmed by intraresidue and sequential NOEs. A positive ϕ angle on Gly13 reverses the backbone chain, leading into the second helix, and a total of 66 long-range NOEs (Fig. 2C) lock in the tertiary contacts between the two helices. Small regions taken from the two-dimensional and three-dimensional NOESY spectra illustrate the unambiguous nature of such interactions when spectra are recorded at high field (900 MHz) and at high spectral resolution (Fig. S3).
The close similarity between the HSQC spectra recorded at pH 7.4 and 4.0 (Fig. S2) strongly suggests the absence of major structural rearrangement between the two pH values. Indeed, the interhelical contacts found at pH 7.4 remain present at pH 4.0 (Fig. S3 B, C). The RDC values recorded at pH 4.0 differ substantially from those at pH 7.4 (Fig. S4), which could be caused by a change in structure or by a different orientation of HAfp1–23 relative to the magnetic field. The finding that the RDCs measured at pH 4.0 fit well to the structure calculated at pH 7.4 (Fig. 2D) indicates that it is the change in alignment, presumably reflecting the change in electrostatic repulsion between the alignment medium (negatively charged dGpG (48), or negatively charged polyacrylamide gel (49, 50)) and the HAfp1–23 upon protonation of Glu11 and Asp19, which dominates the change in RDCs, and not a change in HAfp1–23 structure.
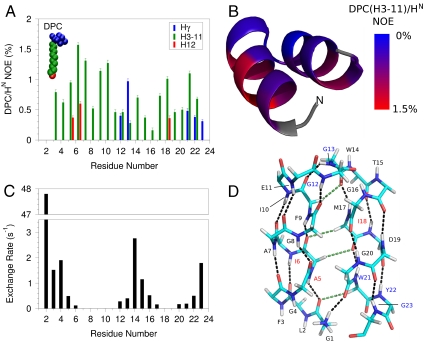
Implicit in the segregated distribution of the side chains according to hydrophobicity (Fig. 2B) is the orientation of HAfp1–23 relative to a phospholipid surface, with the aromatic and hydrophobic side chains pointing downward, towards the hydrophobic interior, and the top of the molecule exposed to solvent. The intermolecular NOE contacts between the micelle and HAfp1–23 provide additional support for this interfacial location (Fig. 3). NOEs to the alkyl chains of DPC are more than twofold weaker for the amide hydrogens of Gly4, Gly8, Gly16, and Gly20, located at the top of the molecule, than for amides on the opposing face, despite the Gly amide protons not being shielded from close intermolecular contacts by the side chains of the perdeuterated HAfp1–23, used for these measurements. Moreover, the amides of Ala5, Ile6, and Ile18 on the hydrophobic face are the only ones showing detectable NOE interactions with the terminal methyl group of DPC. Intermolecular NOEs between the DPC choline methyl resonance and the Trp14 and Trp21 Hε1 indole protons are also observed (Fig. S5). Trp Hε1 and Tyr OH groups in membrane proteins frequently form H-bonding interactions to the polar atoms at the water-lipid interface of membrane lipids (29, 51), and the observation of these interactions is therefore not surprising.
Fig. 3.
HAfp interactions with the micelle and with water at pH 7.4. (A) Strength of NOE interactions between perdeuterated, amide-protonated HAfp1–23 and DPC micelle protons. Amides with cross peaks to choline methyl protons (Hγ) are at the lipid-water interface. Residues with stronger intensity to the DPC methylene (H3-H11) and methyl (H12) protons are closer to the lipid core of the micelle. A NOE mixing time of 100 ms was used, and the plotted intensities have been scaled by the intensity of the diagonal amide resonance. DPC/HN NOEs at pH 7.4 and 4.0 are compared in Fig. S5C. (B) NOE intensity between the DPC H3-H11 methylene protons and the backbone HN protons of the fusion peptide mapped on the structure, with red corresponding to strongest NOEs. Residues in gray could not be detected due to fast exchange of the amide/amino protons with water. The amino terminus is marked N. (C) Solvent hydrogen exchange rates of the backbone amide protons. (D) Hydrogen bonding pattern in the HAfp1–23 structure. Residues Gly1 to Gly12 and Trp14 to Gly23 have well satisfied αhelical hydrogen bonds, marked by black dashed lines. CαH-O = C hydrogen bonds are shown as green dashed lines. Hydrogen bonding between the 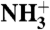 of the N-terminal Gly1 and the carbonyl oxygen of Gly20 is also suggested by the structure. Inclusion of the N-terminal hydrogen bond and the CαH-O = C hydrogen bonds does not adversely affect experimental statistics of the refined structure (SI Text). Blue residue labels indicate proximity to the DPC choline; red labels are for residues with NOEs to the DPC methyl groups.
of the N-terminal Gly1 and the carbonyl oxygen of Gly20 is also suggested by the structure. Inclusion of the N-terminal hydrogen bond and the CαH-O = C hydrogen bonds does not adversely affect experimental statistics of the refined structure (SI Text). Blue residue labels indicate proximity to the DPC choline; red labels are for residues with NOEs to the DPC methyl groups.
Amide hydrogen exchange rates indicate that all backbone amides engaged in α-helical hydrogen bonds are strongly protected from exchange with solvent (Fig. 3C), but the amides at the N-terminal ends of the two helices, which lack their C = O H-bond partner (preceding it by four residues) show less protection. Nevertheless, these hydrogen exchange rates are significantly (> 10-fold) slower than expected for random coil peptides dissolved in water at pH 7.4, 33 °C (52). This latter observation suggests that these amides are not fully accessible to solvent and/or engaged in hydrogen bonds with phospholipid headgroups. For the two Gly residues in the turn region of the hairpin, Gly12-NH makes a slightly distorted H-bond to the Gly8-C = O, while Gly13-NH donates a H-bond to the C = O of Phe9, resulting in relatively high hydrogen exchange protection factors of > 100. The structures calculated also suggest the presence of a hydrogen bond, with O-H distance of ≤ 2.5 Å and C-O-H angle of ≥120°, between the 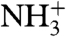 of Gly1 and the carbonyl oxygen of Gly20, and simultaneously for a subset of the calculated structures, between
of Gly1 and the carbonyl oxygen of Gly20, and simultaneously for a subset of the calculated structures, between 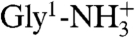 and either Trp21 C = O or Gly23 C = O. The N-terminal Gly1 amino group of HAfp1–23 is protonated, as indicated by its 15N chemical shift of 27.4 ppm. Despite the long-range H-bond interactions of the Gly1 amino hydrogens, their exchange with solvent remains too rapid for direct observation of their 1H NMR signal. Similarly, the amide signal of Leu2 at pH 7.4 also exchanges too rapidly with solvent to permit observation of its 1H-15N correlation in the two-dimensional HSQC spectrum, although it yields a visible resonance at pH 4.0 (Fig. S2), where hydrogen exchange is intrinsically much slower.
and either Trp21 C = O or Gly23 C = O. The N-terminal Gly1 amino group of HAfp1–23 is protonated, as indicated by its 15N chemical shift of 27.4 ppm. Despite the long-range H-bond interactions of the Gly1 amino hydrogens, their exchange with solvent remains too rapid for direct observation of their 1H NMR signal. Similarly, the amide signal of Leu2 at pH 7.4 also exchanges too rapidly with solvent to permit observation of its 1H-15N correlation in the two-dimensional HSQC spectrum, although it yields a visible resonance at pH 4.0 (Fig. S2), where hydrogen exchange is intrinsically much slower.
Discussion
The structure reported here for full length HAfp1–23 differs substantially from results reported for the construct that lacks the strictly conserved residues Trp21-Gly23. Nevertheless, initial NMR measurements by us on these shorter versions of the fusion peptides also exhibited long-range Hα-Hα NOE contacts between Phe9 and Met17, and Ala5 and Ser21 (with Ser21 being the first residue of the carboxy-terminal solubilization tag). However, these NOE interactions were quite weak, and 15N relaxation measurements indicated significant disorder at both the N- and C-terminal ends of this HAfp1–20 fusion peptide. The presence of these weak long-range NOEs suggest that HAfp1–20 also populates the helical hairpin conformation, but only transiently. Indeed, we were unable to fit the NOEs and RDCs observed for HAfp1–20 to a single static structure. These observations prompted us to study constructs containing all strictly conserved residues, encompassing residues 1–23.
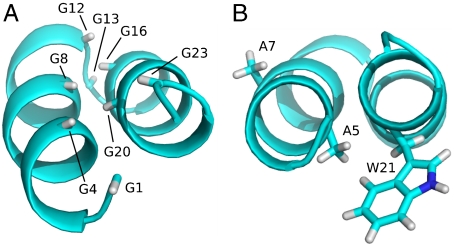
The helical hairpin structure observed in our study is stabilized by interactions between residues Trp21-Gly23 and the N-terminal residues of HAfp, explaining the high degree of backbone order observed for these residues (Fig. 1D). In retrospect, the tight antiparallel helical packing of the two helices in HAfp1–23 is perhaps not surprising, considering that both the N- and C-terminal helix contain the classic GXXXG motifs that are the hallmark of many tight transmembrane helix-helix associations (31, 32). In most, but not all of these associations, the helical orientations are parallel, not antiparallel. However, the Protein Data Bank does not contain a single case where the antiparallel, aliphatic hydrogen-bond linked helices are connected by a tight hairpin turn, as found here for HAfp. The close packing is permitted by Gly residues that line the inner faces of the two helices (Fig. 4A), consisting of Gly1, Gly4, and Gly8 on one side, and Gly16, Gly20, and Gly23 on the other, all of which are completely conserved (8, 53). Sequence conservation of the HA2 fusion domain has been shown to be important for burial of the HAfp in the hydrophobic interior of the postcleavage but prefusion state of HA, playing a critical role in tuning the pH where membrane fusion occurs (8, 15, 54). Our structural results on HAfp1–23 after the peptide has been extruded from the hydrophobic core highlight a second, structural role for this high degree of sequence conservation. Positioning of the hydrophobic and hydrophilic residues relative to the hairpin surface appears key for defining its location at the water-lipid interface (Fig. 2, 3). Strict sequence conservation of Gly13 permits the positive ϕ angle, allowing formation of the exceptionally tight helical hairpin arrangement. The Gly8 Hα3 atom is fully buried at the interhelical interface (Fig. 4A), and modeling indicates that even a Gly → Ala mutation at this position is incompatible with the hairpin structure. This potentially explains the strict conservation of this residue, and the finding that a G8A mutation abrogates fusogenicity (14). In fact, six Gly residues in HAfp are found at the helical interface, and the remaining two are at the center of the hairpin turn. The only HAfp residue that lacks strong sequence conservation is Thr15, for which mutations to Glu, Gln, Ser, and even Pro are observed in the various HA sero subtypes. Residue 15 is located on the solvent-side of the hairpin. The position of the molecule at the N-terminal end of helix 2 causes it to lack the intrahelical hydrogen bond for its amide proton, and mutation to Pro is therefore less disruptive. Indeed, Pro is commonly found as one of the first residues in an α-helix.
Fig. 4.
Ribbon structures showing the orientation of Gly residues and various side chains. (A) The Gly Hα3 protons, which would be replaced by a side chain Cβ atom if mutated, are displayed. (B) Side chains of Ala5, Ala7, and Trp21. Mutation of Ala5 to Val would require rearrangement of the Trp21 side chain.
The two Hα protons of Gly1 are highly nonequivalent and differ in 1H chemical shift by 0.7 ppm, confirming the high degree of ordering for this residue. Its position is defined both by NOEs and by 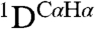 and
and 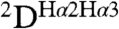 RDCs, orienting it such that its amino group donates a H-bond to the carbonyl of Gly20, while its carbonyl accepts an α-helical H-bond from Ala5. The N-terminal Gly is highly conserved, and it has long been known that fusion activity is sensitive to its mutation. Not unexpectedly, deletions of G1 and L2 abolish fusion (14) as removal of Gly1 or Leu2 abrogates these interactions, which appear key in stabilizing the hairpin configuration. In contrast to the bulky G1V mutation, substitutions of Gly1 by a non-β-branched polar residue (G1E, G1Q, and G1K) appear possible without disruption of the backbone or its H-bond interactions, but these mutations have been reported to lack fusion activity (19). Instead, the more conservative mutation to a small hydrophobic side chain, G1A, does not abolish fusogenic activity (14, 19). Interestingly, the G1S mutation is fusogenic, but activity is arrested at the hemi-fusion state (19), highlighting the critical role of the N-terminal residue in transitioning from the hemi-fused state to channel formation.
RDCs, orienting it such that its amino group donates a H-bond to the carbonyl of Gly20, while its carbonyl accepts an α-helical H-bond from Ala5. The N-terminal Gly is highly conserved, and it has long been known that fusion activity is sensitive to its mutation. Not unexpectedly, deletions of G1 and L2 abolish fusion (14) as removal of Gly1 or Leu2 abrogates these interactions, which appear key in stabilizing the hairpin configuration. In contrast to the bulky G1V mutation, substitutions of Gly1 by a non-β-branched polar residue (G1E, G1Q, and G1K) appear possible without disruption of the backbone or its H-bond interactions, but these mutations have been reported to lack fusion activity (19). Instead, the more conservative mutation to a small hydrophobic side chain, G1A, does not abolish fusogenic activity (14, 19). Interestingly, the G1S mutation is fusogenic, but activity is arrested at the hemi-fusion state (19), highlighting the critical role of the N-terminal residue in transitioning from the hemi-fused state to channel formation.
Strict sequence conservation is found for the two Trp residues in HAfp, at positions 14 and 21. Even though mutations to residues with smaller side chains are structurally feasible without disrupting the backbone, Trp residues are known to be key in positioning proteins at the membrane-water interface. It therefore is likely that the conserved nature of these two residues relates to their function in interacting with the membrane, rather than being dictated by conserving the backbone structure. Interestingly, the inactive A5V mutation (24) would cause a steric clash with the Trp21 side chain, requiring its rearrangement (Fig. 4B). In contrast, the A7V mutation is fully active and fusogenic (24). When retaining the backbone structure of HAfp1–23, the Val7 side chain is located in the plane of the hairpin, pointing sideways at the water-lipid interface, without causing any steric interference with the interhelical interaction.
Even with the current structure of HAfp in hand, the mechanism by which the peptide mediates viral fusion is not immediately obvious. Single cell fusion studies indicate the formation of fusion pores, forming narrow aqueous channels, as an early intermediate in the fusion process (11, 12), with models suggesting six HA trimers, i.e., 18 HAfp peptides participating in the formation of a single pore (55). The length of the HAfp hairpin structure appears too short to span a lipid bilayer, and it is conceivable that during the process of pore formation the peptide rearranges from the helical hairpin to a contiguous α-helical state upon forming oligomeric pores. Although highly speculative, it is possible that the small degree of conformational exchange broadening observed in HAfp1–23 at low pH, but absent at high pH (Fig. 1C), is related to such a structural transition. In this respect, it is interesting to note that another class I viral protein fusion domain, gp41 of HIV-1, adopts a contiguous α-helical conformation when studied in detergent micelles (56).
Materials and Methods
HAfp1–23 of sero type H1 was expressed as a fusion protein, flanked by the residues SGKKKKD at its C terminus and by the Igg-binding domain B1 of streptococcal protein G (GB1; PDB entry 3GB1) at its N terminus. Cleavage by factor Xa protease resulted in the peptide with sequence GLFGAIAGFI EGGWTGMIDG WYGSGKKKKD (Fig. S6). Details regarding expression and purification are included as SI Text.
NMR measurements were carried out at 600 and 900 MHz on uniformly 15N/13C- and 2H/15N/13C-enriched samples at peptide concentrations of ca 0.6 mM, in 130–180 mM DPC. NMR structures were calculated using semiquantitative NOE distance restraints (Fig. S7), starting from a fully extended chain, using the XPLOR-NIH software (47). For details, see SI Text.
Supplementary Material
Acknowledgments.
We thank Annie Aniana for help with protein expression and purification, and Alex Grishaev, Dan Garrett, Charles Schwieters, Marius Clore, and Dennis Torchia for many helpful suggestions. This work was funded by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH); Intramural AIDS-Targeted Antiviral Program of the Office of the Director, NIH.
Footnotes
The authors declare no conflict of interest.
Data deposition: The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 16907). The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2KXA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006142107/-/DCSupplemental.
References
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza-A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 3.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinhauer DA. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 5.Wilson IA, Skehel JJ, Wiley DC. Structure of the hemagglutinin membrane glycoprotein of influenze virus at 3-A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 6.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 7.Bizebard T, et al. Structure of influenza virus hemagglutinin complexed with a neutralizing antibody. Nature. 1995;376:92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 8.Cross KJ, Langley WA, Russell RJ, Skehel JJ, Steinhauer DA. Composition and functions of the influenza fusion peptide. Protein Peptide Lett. 2009;16:766–778. doi: 10.2174/092986609788681715. [DOI] [PubMed] [Google Scholar]
- 9.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 10.Tse FW, Iwata A, Almers W. Membrane flux through the pore formed by a fusogenic viral envelope protein during cell fusion. J Cell Biol. 1993;121:543–552. doi: 10.1083/jcb.121.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoch C, Blumenthal R. Role of the fusion peptide sequence in intial stages of influenza hemagglutinin-induced cell fusion. J Biol Chem. 1993;268:9267–9274. [PubMed] [Google Scholar]
- 12.Spruce AE, Iwata A, Almers W. The first milliseconds of the pore formed by a fusogenic viral envelope protein during membrane fusion. Proc Nat'l Acad Sci USA. 1991;88:3623–3627. doi: 10.1073/pnas.88.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gething MJ, Doms RW, York D, White J. Studies on the mechanism of membrane-fusion. Site-specific mutagenesis of the hemagglutinin of influenza virus. J Cell Biol. 1986;102:11–23. doi: 10.1083/jcb.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinhauer DA, Wharton SA, Skehel JJ, Wiley DC. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cross KJ, Wharton SA, Skehel JJ, Wiley DC, Steinhauer DA. Studies on influenza haemagglutinin fusion peptide mutants generated by reverse genetics. EMBO J. 2001;20:4432–4442. doi: 10.1093/emboj/20.16.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinhauer DA, et al. Studies using double mutants of the conformational transitions in influenza hemagglutinin required for its membrane fusion activity. Proc Nat'l Acad Sci USA. 1996;93:12873–12878. doi: 10.1073/pnas.93.23.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orlich M, Rott R. Thermolysin activation mutants with changes in the fusogenic region of an influenza virus hemagglutinin. J Virol. 1994;68:7537–7539. doi: 10.1128/jvi.68.11.7537-7539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yewdell JW, et al. Mutations in or near the fusion peptide of the influenze virus hemagglutinin affect an antigenic site in the globular region. J Virol. 1993;67:933–942. doi: 10.1128/jvi.67.2.933-942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao H, Armstrong RT, Melikyan GB, Cohen FS, White JM. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol Biol Cell. 1999;10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han X, Bushweller JH, Cafiso DS, Tamm LK. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat Struct Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 21.Li YL, et al. Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion. J Virol. 2005;79:12065–12076. doi: 10.1128/JVI.79.18.12065-12076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durrer P, et al. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J Biol Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 23.Tsurudome M, et al. Lipid interactions of the hemagglutinin HA2 NH2-terminal segment during influenze virus-induced membrane fusion. J Biol Chem. 1992;267:20225–20232. [PubMed] [Google Scholar]
- 24.Han X, Steinhauer DA, Wharton SA, Tamm LK. Interaction of mutant influenza virus hemagglutinin fusion peptides with lipid bilayers: probing the role of hydrophobic residue size in the central region of the fusion peptide. Biochemistry. 1999;38:15052–15059. doi: 10.1021/bi991232h. [DOI] [PubMed] [Google Scholar]
- 25.Ge M, Freed JH. Fusion peptide from influenza hemagglutinin increases membrane surface order: an electron-spin resonance study. Biophys J. 2009;96:4925–4934. doi: 10.1016/j.bpj.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai AL, Tamm LK. Locking the kink in the influenza hemagglutinin fusion domain structure. J Biol Chem. 2007;282:23946–23956. doi: 10.1074/jbc.M704008200. [DOI] [PubMed] [Google Scholar]
- 27.Lai AL, Park H, White JM, Tamm LK. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J Biol Chem. 2006;281:5760–5770. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- 28.Han X, Tamm LK. A host-guest system to study structure-function relationships of membrane fusion peptides. Proc Nat'l Acad Sci USA. 2000;97:13097–13102. doi: 10.1073/pnas.230212097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiffer M, Chang CH, Stevens FJ. The functions of tryptophan residues in membrane proteins. Protein Eng. 1992;5:213–214. doi: 10.1093/protein/5.3.213. [DOI] [PubMed] [Google Scholar]
- 30.Stopar D, Spruijt RB, Hemminga MA. Anchoring mechanisms of membrane-associated M13 major coat protein. Chem Phys Lipids. 2006;141:83–93. doi: 10.1016/j.chemphyslip.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 31.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 32.Kleiger G, Grothe R, Mallick P, Eisenberg D. GXXXG and AXXXA: common alpha-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry. 2002;41:5990–5997. doi: 10.1021/bi0200763. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y, Weliky DP. C-13-C-13 correlation spectroscopy of membrane-associated influenza virus fusion peptide strongly supports a helix-turn-helix motif and two turn conformations. J Am Chem Soc. 2009;131:13228–13229. doi: 10.1021/ja905198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z, et al. N-15 NMR study of the ionization properties of the influenza virus fusion peptide in zwitterionic phospholipid dispersions. Biophys J. 2000;78:2418–2425. doi: 10.1016/S0006-3495(00)76785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 36.Mandel AM, Akke M, Palmer AG. Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure nad function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 37.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 38.Krueger-Koplin RD, et al. An evaluation of detergents for NMR structural studies of membrane proteins. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 39.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 40.Wider G, Weber C, Wuthrich K. Proton-proton Overhauser effects of receptor-bound cyclosporin A observed with the use of a heteronuclear-resolved half-filter experiment. J Am Chem Soc. 1991;113:4676–4678. [Google Scholar]
- 41.Ikura M, Bax A. Isotope-filtered two-dimensional NMR of a protein-peptide complex: study of a skeletal muscle myosin light chain kinase fragment bound to calmodulin. J Am Chem Soc. 1992;114:2433–2440. [Google Scholar]
- 42.Wüthrich K. NMR of proteins and nucleic acids. New York: John Wiley & Sons; 1986. [Google Scholar]
- 43.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 44.Senes A, Ubarretxena-Belandia I, Engelman DM. The C alpha-H center dot center dot center dot O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc Nat'l Acad Sci USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sulistijo ES, MacKenzie KR. Structural basis for dimerization of the BNIP3 transmembrane domain. Biochemistry. 2009;48:5106–5120. doi: 10.1021/bi802245u. [DOI] [PubMed] [Google Scholar]
- 46.Grishaev A, Bax A. An empirical backbone-backbone hydrogen-bonding potential in proteins and its applications to NMR structure refinement and validation. J Am Chem Soc. 2004;126:7281–7292. doi: 10.1021/ja0319994. [DOI] [PubMed] [Google Scholar]
- 47.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 48.Lorieau J, Yao LS, Bax A. Liquid crystalline phase of G-tetrad DNA for NMR study of detergent-solubilized proteins. J Am Chem Soc. 2008;130:7536–7537. doi: 10.1021/ja801729f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tycko R, Blanco FJ, Ishii Y. Alignment of biopolymers in strained gels: a new way to create detectable dipole-dipole couplings in high-resolution biomolecular NMR. J Am Chem Soc. 2000;122:9340–9341. [Google Scholar]
- 50.Meier S, Haussinger D, Grzesiek S. Charged acrylamide copolymer gels as media for weak alignment. J Biomol NMR. 2002;24:351–356. doi: 10.1023/a:1021609207024. [DOI] [PubMed] [Google Scholar]
- 51.Kachel K, Asuncionpunzalan E, London E. Anchoring of tryptophan and tyrosine analogs at the hydrocarbon polar boundary in model membrane vesicles. Biochemistry. 1995;34:15475–15479. doi: 10.1021/bi00047a012. [DOI] [PubMed] [Google Scholar]
- 52.Bai Y, Milne JS, Englander SW. Primary structure effects on peptide group exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nobusawa E, et al. Comparison of complete amino acid sequences and receptor binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 54.Daniels RS, et al. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell. 1985;40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 55.Blumenthal R, Sarkar DP, Durell S, Howard DE, Morris SJ. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J Cell Biol. 1996;135:63–71. doi: 10.1083/jcb.135.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaroniec CP, et al. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 57.Yao L, Grishaev A, Cornilescu G, Bax A. Site-specific backbone amide 15N chemical shift anisotropy tensors in a small protein from liquid crystal and cross-correlated relaxation measurements. J Am Chem Soc. 2010;132:4295–4309. doi: 10.1021/ja910186u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.