Abstract
The sterol regulatory element-binding protein (SREBP) transcription factor family is a critical regulator of lipid and sterol homeostasis in eukaryotes. In mammals, SREBPs are highly active in the fed state to promote the expression of lipogenic and cholesterogenic genes and facilitate fat storage. During fasting, SREBP-dependent lipid/cholesterol synthesis is rapidly diminished in the mouse liver; however, the mechanism has remained incompletely understood. Moreover, the evolutionary conservation of fasting regulation of SREBP-dependent programs of gene expression and control of lipid homeostasis has been unclear. We demonstrate here a conserved role for orthologs of the NAD+-dependent deacetylase SIRT1 in metazoans in down-regulation of SREBP orthologs during fasting, resulting in inhibition of lipid synthesis and fat storage. Our data reveal that SIRT1 can directly deacetylate SREBP, and modulation of SIRT1 activity results in changes in SREBP ubiquitination, protein stability, and target gene expression. In addition, chemical activators of SIRT1 inhibit SREBP target gene expression in vitro and in vivo, correlating with decreased hepatic lipid and cholesterol levels and attenuated liver steatosis in diet-induced and genetically obese mice. We conclude that SIRT1 orthologs play a critical role in controlling SREBP-dependent gene regulation governing lipid/cholesterol homeostasis in metazoans in response to fasting cues. These findings may have important biomedical implications for the treatment of metabolic disorders associated with aberrant lipid/cholesterol homeostasis, including metabolic syndrome and atherosclerosis.
Keywords: Cholesterol, fasting, lipid, SIRT1, SREBP
Lipids and sterols play key roles in diverse biological processes in eukaryotes, such as membrane biosynthesis, intra- and extracellular signaling, and energy storage. In humans, aberrant lipid and cholesterol homeostasis has been linked to a number of diseases prevalent in the developed world, including metabolic syndrome—a constellation of conditions and diseases that includes obesity, insulin resistance, liver steatosis, and hypertension, as well as type 2 diabetes, cardiovascular disease, and cancers (Cornier et al. 2008). An improved understanding of the molecular mechanisms governing lipid/cholesterol homeostasis might lead to novel therapeutic strategies to ameliorate such diseases.
Fasting (short-term food deprivation) produces a rapid metabolic shift from lipid/cholesterol synthesis and fat storage to mobilization of fat, and recent studies have suggested that fasting may improve conditions associated with metabolic syndrome (Varady and Hellerstein 2008; Fontana et al. 2010). There is thus keen interest in determining the mechanism of fasting-dependent regulation of lipid/cholesterol metabolism to facilitate the development of novel therapeutic strategies to treat disorders associated with aberrant lipid and cholesterol homeostasis.
The sterol regulatory element-binding protein (SREBP) transcription factors represent a highly conserved family of gene regulators controlling key genes involved in the biosynthesis and trafficking of lipids and sterols in eukaryotes from Schizosaccharomyces pombe to humans (Osborne and Espenshade 2009). In vertebrates, the SREBP-2 isoform primarily modulates intracellular cholesterol homeostasis by promoting the expression of the low-density lipoprotein (LDL) receptor gene and cholesterol biosynthesis genes (e.g., HMG-CoA reductase), whereas the SREBP-1 isoform preferentially controls lipid homeostasis by activating fatty acid and lipid biosynthesis genes (e.g., fatty acid synthase [FASN] and stearoyl-CoA desaturases) (Horton et al. 2002; Osborne and Espenshade 2009). In cholesterol auxotroph invertebrates such as Caenorhabditis elegans and Drosophila melanogaster, SREBP homologs appear to primarily regulate fatty acid/lipid homeostasis (Rawson 2003; Osborne and Espenshade 2009).
SREBP family members are controlled in a classic negative feedback manner by the downstream products of the metabolic pathways regulated by SREBPs, such as cholesterol/lipids (Brown and Goldstein 1997; Osborne and Espenshade 2009). SREBPs are synthesized as precursor proteins that are inserted into the endoplasmic reticular (ER) membrane in a hairpin fashion by two transmembrane domains. When intracellular cholesterol/lipid levels are high, SREBP precursors are tethered and retained in the ER by the sterol-sensing SCAP–Insig complex (Espenshade 2006). Upon cholesterol/lipid depletion, the SCAP–Insig interaction is disrupted and SCAP chaperones SREBPs to the Golgi, where they undergo two sequential intramembrane proteolytic cleavage steps, releasing the free mature transcription factor form, which then migrates to the nucleus to activate gene transcription (Espenshade 2006).
While the negative feedback mechanisms controlling the processing of the precursor forms of SREBPs are quite well understood, much less is known of how SREBPs are controlled by nutrient deprivation, such as during fasting. The mature nuclear forms of SREBP-1 and SREBP-2 are abundant in the mouse liver during feeding, and the expression of fatty acid and cholesterol biosynthesis genes is high, whereas, during fasting, SREBPs are markedly down-regulated, correlating with decreased lipogenic and cholesterogenic gene expression (Horton et al. 1998). The expression of the SREBP-1c isoform responds to changes in insulin signaling, and altered insulin levels may thus contribute to the effects of fasting on the levels of nuclear SREBP-1c in liver (Horton et al. 1998). However, the SREBP-2 family member is not transcriptionally regulated in response to insulin signaling, and the levels of the precursor form is not changed in response to fasting or refeeding in the mouse liver (Horton et al. 1998); hence, the mechanism of fasting-dependent down-regulation of nuclear SREBP-2 cannot be explained by decreased insulin signaling. Moreover, regardless of upstream regulatory events governing the expression and proteolytic processing of precursor SREBPs, the active, nuclear forms of SREBPs must be removed rapidly to terminate the gene activation signals promoting lipogenic and cholesterogenic transcriptional programs in response to fasting cues. However, the mechanism of fasting-dependent down-regulation of nuclear SREBPs has not been elucidated.
Nuclear SREBPs recruit coactivators, such as the histone acetyltransferases (HATs) CBP/p300 and the RNA polymerase II-binding ARC/Mediator complex, to improve chromatin accessibility and facilitate the assembly of the transcription machinery at promoters of regulated genes (Oliner et al. 1996; Ericsson and Edwards 1998; Näär et al. 1998, 1999). Interestingly, recent studies have shown that the nuclear mature form of the SREBP-1 isoform can be acetylated by the CBP/p300 acetyltransferases, and SREBP-1 acetylation correlated with decreased ubiquitination and proteasome-dependent turnover of SREBP-1 (Giandomenico et al. 2003). The resulting increased and sustained SREBP activity and elevated downstream target gene expression would be predicted to promote SREBP-dependent lipogenesis and cholesterol synthesis and uptake during feeding. Based on this notion, we speculated that fasting-regulated deacetylation of SREBPs might serve to initiate clearing of nuclear SREBPs by promoting ubiquitin/proteasome-dependent SREBP turnover, and to inhibit de novo synthesis of lipids/cholesterol.
One attractive candidate to mediate deacetylation of the SREBP family of transcription factors in response to fasting cues is the SIRT1 member of the class III NAD+-dependent family of protein deacetylases (Guarente 2006). First, SIRT1 orthologs not only remove acetyl groups from histone tails, they can also deacetylate transcription factors such as p53, NF-κB, and E2F (Haigis and Guarente 2006; Saunders and Verdin 2007; Feige and Auwerx 2008). In addition, a number of studies have shown that SIRT1 orthologs contribute to the effects of decreased caloric intake on life span from yeast to mammals, and sirtuins may thus act as energy sensors (Guarente 2006). Consistent with this notion, NAD+ is a critical cellular metabolite tightly linked to the energetic/metabolic status of cells, and altered levels of NAD+ or NAD+/NADH ratios in response to changes in caloric intake and/or energy consumption could have profound effects on SIRT1 activity (Haigis and Guarente 2006). Importantly, SIRT1 protein levels are elevated in the livers of fasted mice and decrease with refeeding (Rodgers and Puigserver 2007), a regulatory pattern converse to that of SREBPs. Several studies in rodents have shown that SIRT1 is indeed involved directly in modulating animal metabolism in response to fasting cues. For example, mouse SIRT1 deacetylates and potentiates the activities of the PPARα nuclear receptor as well as the PGC-1α and CRTC2/TORC2 transcription coactivators in the mouse liver during fasting, resulting in increased expression of gluconeogenic and fatty acid β-oxidation genes (Nemoto et al. 2005; Rodgers et al. 2005; Liu et al. 2008; Purushotham et al. 2009). SIRT1 also inhibits the expression of the lipogenic nuclear receptor PPARγ in adipose cells (Picard et al. 2004), and acts downstream from AMPK signaling, which also serves to inhibit lipogenesis and promote energy consumption (Hou et al. 2008; Canto et al. 2009). Moreover, small-molecule activators of sirtuins—such as the polyphenol resveratrol as well as newer, more potent SIRT1 activators—appear to act as fasting mimetics by promoting increased fat mobilization, fatty acid β-oxidation, and improved cholesterol homeostasis (Baur et al. 2006; Milne et al. 2007; Feige et al. 2008; Smith et al. 2009). Additionally, knockdown or knockout of SIRT1 in the mouse liver or transgenic overexpression of SIRT1 in mice results in altered lipid/cholesterol levels in serum and the liver (Li et al. 2007; Rodgers and Puigserver 2007; Pfluger et al. 2008; Erion et al. 2009; Purushotham et al. 2009). Altogether, these findings are consistent with an essential role for SIRT1 in controlling lipid/cholesterol homeostasis, at least in rodents (Yu and Auwerx 2009). While there is clear evidence for a role for SIRT1 in promoting fatty acid β-oxidation, it is not known whether SIRT1 orthologs participate directly in mediating the inhibitory effects of fasting on SREBP-dependent lipid/cholesterol biosynthesis. To understand how lipid/cholesterol synthesis is shut down during fasting in invertebrates and mammals, we investigated the functional and mechanistic interaction between SIRT1 and SREBP.
Results
The C. elegans SIRT1 ortholog SIR-2.1 mediates fasting-dependent down-regulation of the SREBP ortholog SBP-1, and inhibits lipid synthesis and fat storage in response to fasting cues
We hypothesized that increased sirtuin activity during the fasting response promotes loss of nuclear SREBP, resulting in down-regulation of SREBP-responsive genes and decreased potential to store lipids. To test this hypothesis, we first used invertebrate models containing single SREBP orthologs. The nematode C. elegans represents a powerful and facile model system for investigating conserved mechanisms governing lipid homeostasis (Ashrafi 2007; Watts 2009). The SREBP ortholog in C. elegans, SBP-1, is necessary for lipid production and for expression of multiple lipogenic genes (McKay et al. 2003; Yang et al. 2006).
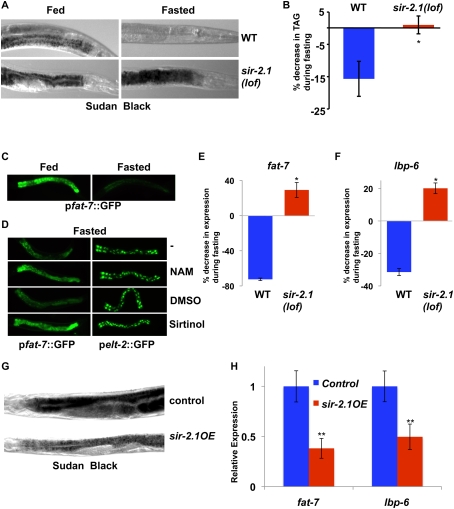
To investigate the role of sirtuins in the invertebrate fasting response, we assessed lipid levels in the intestine (liver/adipose equivalent) by staining with the lipophilic dye Sudan Black. Fasting of wild-type C. elegans results in strongly decreased levels of lipids in the intestines (Fig. 1A). Strikingly, nematodes null for the SIRT1 ortholog sir-2.1 exhibit high levels of lipids under both fed and fasted conditions (Fig. 1A; Supplemental Fig. 1A), indicating that SIR-2.1 is required for the decreased lipid synthesis and/or storage in response to fasting cues. Thin-layer chromatography and gas chromatography analyses confirmed that total levels of triglycerides decrease during fasting in wild-type animals, but not in sir-2.1-null animals (Fig. 1B). Previous studies of fasting-dependent transcriptional responses in C. elegans revealed marked down-regulation of the expression of several genes involved in lipid homeostasis, including fat-7, a gene encoding a stearoyl-CoA desaturase that plays a key and conserved role in lipid/triglyceride biosynthesis and fat storage (Van Gilst et al. 2005; Flowers and Ntambi 2008), which is also an important regulatory target of SBP-1 (Yang et al. 2006). Using a C. elegans strain harboring a fat-7 promoter∷GFP reporter, we confirmed that fasting elicits a strong decrease in the intestinal GFP expression directed by the fat-7 promoter (Fig. 1C). Treatment of nematodes with the sirtuin inhibitors nicotinamide and sirtinol results in markedly increased expression of the fat-7p∷GFP reporter in the intestine under fasting conditions, while intestinal GFP expression driven by the elt-2 promoter was unaffected by these treatments, revealing a gene-selective effect of the sirtuin inhibitors (Fig. 1D). Accordingly, deletion of sir-2.1 largely abrogates the fasting-dependent decline in expression of the endogenous fat-7 gene (Fig. 1E). Additionally, we found that the expression of lipid-binding protein 6 (lbp-6, a novel SBP-1 target gene) (AK Walker and AM Näär, unpubl.), is also down-regulated by fasting in wild-type animals, but not in sir-2.1 loss-of-function (lof) animals (Fig. 1F). To further investigate the regulation of SBP-1 target genes by SIR-2.1, we used a C. elegans strain overexpressing SIR-2.1 (sir-2.1OE) (Tissenbaum and Guarente 2001). Converse to the findings with the sir-2.1(lof) strain, the SIR-2.1OE strain exhibits decreased transcription of fat-7 and lbp-6 under both fed and fasted conditions, and has markedly lower intestinal lipid storage as compared with control animals (Fig. 1G,H; Supplemental Fig. 1B; data not shown). These results reveal an essential role for the SIRT1 ortholog SIR-2.1 in down-regulating expression of SBP-1 lipogenic target genes and lipid/triglyceride biosynthesis and storage in C. elegans in response to fasting cues.
Figure 1.
SIR-2.1 is essential for proper fasting-dependent down-regulation of lipid synthesis and fat storage in C. elegans (A) Sudan Black staining reveals that fat storage is decreased in wild-type animals upon fasting, but is maintained in fasted sir-2.1(lof) animals. (B) Thin-liquid chromatography/gas chromatography analysis shows that triacylglycerols (TAGs) normally reduced during fasting are still present in sir-2.1(lof) animals. (C) Intestinal expression of fat-7p∷GFP is strongly decreased during fasting. (D) Chemical interference with sirtuin activity (nicotinamide [NAM], 12.5 mM; sirtinol, 0.1 mM) causes a retention of fat-7p∷GFP expression in fasted animals. (E) Fasting-dependent decreases in expression of endogenous fat-7 do not occur in sir-2.1(lof) animals. fat-7 expression is normalized to act-1. (F) The SBP-1 target gene lbp-6 is abnormally regulated in the fasting response of sir-2.1(lof) animals. Gene expression was measured by qRT–PCR normalized to act-1. Error bars represent standard deviations between parallel reactions. (G) Increased SIR-2.1 activity in nematodes results in lower levels of stored fat, as measured by Sudan Black staining. (H) C. elegans overexpressing sir-2.1 exhibit lower levels of SBP-1 target gene expression. Relative mRNA amounts of the SBP-1 target genes fat-7 or lbp-6 from fed animals were measured by qRT–PCR. Error bars represent standard deviations from parallel reactions. (*) P < 0.05; (**) P < 0.01.
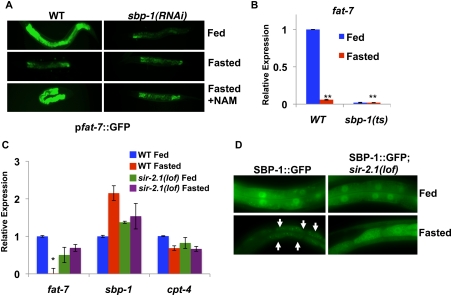
We next examined whether the alterations in fat-7 expression and lipid storage during fasting or after manipulating SIR-2.1 levels or activity were a consequence of changes in SBP-1 activity. Using sbp-1 RNAi, we found that the elevated intestinal GFP expression driven by the fat-7 promoter∷GFP reporter observed in fasted animals in response to nicotinamide treatment is dependent on SBP-1 (Fig. 2A). The expression of the endogenous fat-7 gene in fed or fasted sir-2.1-null animals is also dependent on SBP-1, as judged by sbp-1 RNAi experiments (data not shown). Using animals harboring a hypomorphic allele of sbp-1 [sbp-1(ep79)], we also show that SBP-1 is required for the expression of endogenous fat-7 in fed and fasted animals, and that the accumulation of lipids in fasted sir-2.1-null animals is dependent on SBP-1 (Fig. 2B; Supplemental Fig. 2A,B). A previous study revealed that the C. elegans orphan nuclear receptor NHR-49 is also involved in fasting-dependent regulation of lipid homeostasis genes (Van Gilst et al. 2005); however, the expression of several NHR-49 target genes (e.g., cpt-4) is unaffected by sir-2.1 deletion during either feeding or fasting (Fig. 2C; data not shown). Furthermore, nhr-49(lof);sir-2.1(lof) double mutants have similar lipid storage levels during feeding or fasting when stained by Sudan Black (Supplemental Fig. 3), suggesting that NHR-49 is dispensable for lipid storage in sir-2.1 animals. Together, these results are consistent with a specific role for SIR-2.1 inhibition of SBP-1 function in down-regulation of lipid synthesis and storage in C. elegans during fasting.
Figure 2.
SIR-2.1 is essential for proper fasting-dependent down-regulation of SBP-1-dependent gene expression and localization in C. elegans. (A) fat-7p∷GFP expressed during fasting in response to sirtuin inactivation is also dependent on sbp-1. (B) fat-7 expression is decreased in sbp-1(ep79) animals at the nonpermissive temperature in both fed and fasted conditions. (C) In contrast with fat-7 expression, mRNA expression of sbp-1 or the NHR-49 target cpt-4 was very modestly altered in sir-2.1(lof) animals. C. elegans were placed in fed or fasted conditions for 7 h. Gene expression was measured by qRT–PCR normalized to act-1. Error bars represent standard deviations between parallel reactions. (*) P < 0.05; (**) P < 0.01. (D) Nuclear levels of SBP-1∷GFP are robust in fasted SBP-1∷GFP;sir-2.1(lof) intestines. White arrows represent positions of nuclei as visualized by light microscopy.
To explore the mechanism by which SIR-2.1 inhibits SBP-1-dependent gene regulation and lipid synthesis/storage in C. elegans, we examined whether SIR-2.1 affects SBP-1 levels. Nematodes expressing a SBP-1∷GFP fusion protein driven by the sbp-1 promoter exhibit strong nuclear fluorescence in the intestinal cells of fed animals, whereas fasted animals show markedly reduced nuclear SBP-1∷GFP levels, consistent with previously published results in mammals (Fig. 2D; Horton et al. 1998). In contrast, nuclear SBP-1∷GFP expression remains high in fasted nematodes carrying a deletion in sir-2.1, or in animals treated with the sirtuin inhibitors nicotinamide and sirtinol (Fig. 2D; Supplemental Fig. 4A; data not shown). Transcript levels of sbp-1 are only very modestly altered by feeding/fasting or loss of sir-2.1 (Fig. 2C), suggesting that SIR-2.1 mediates fasting-dependent regulation of SBP-1 protein levels by a post-transcriptional mechanism. Together, these results reveal a critical role for the C. elegans SIRT1 ortholog SIR-2.1 in fasting-dependent inhibition of lipid synthesis and storage, an effect at least in part due to post-transcriptional down-regulation of the key lipogenic gene regulator and SREBP ortholog SBP-1.
We next wished to establish whether the role of SIRT1 orthologs in negative regulation of lipogenic genes in response to fasting is conserved among metazoans. We examined whether the Drosophila SIRT1 ortholog dSIR2 is required for fasting-dependent inhibition of lipogenic genes that are known to be controlled by the Drosophila SREBP ortholog dSREBP (Kunte et al. 2006). The dSREBP-regulated genes encoding the lipogenic enzymes acetyl-CoA synthase, acetyl-CoA carboxylase, and fatty acid synthase are all significantly down-regulated during fasting of Drosophila larvae (Supplemental Fig. 4B). Importantly, Drosophila larvae homozygously deleted for dSir2 exhibit strongly increased transcription of the three dSREBP target genes, in particular during fasting (Supplemental Fig. 4B). These results, together with the C. elegans findings, are consistent with an essential and conserved role for SIRT1 orthologs in negatively regulating SREBP-dependent transactivation of lipogenic genes and fat storage in invertebrate metazoans during fasting.
Mammalian SIRT1 represses SREBP-1 and SREBP-2 target gene expression
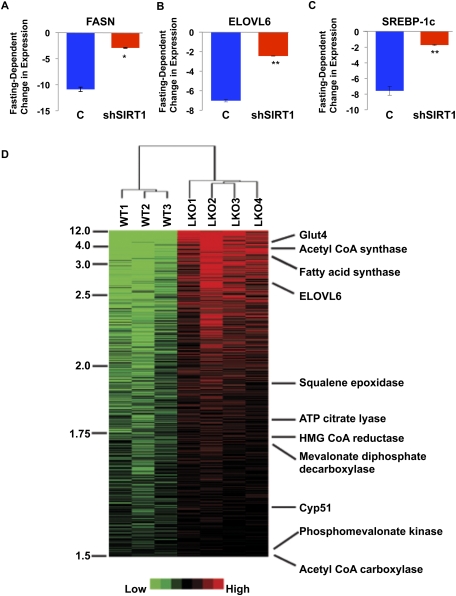
Next, we investigated whether mammalian SIRT1 also regulates SREBPs and their target genes. Consistent with our findings suggesting a central role for SIRT1 orthologs in regulating the lipogenic SREBP-1 orthologs in C. elegans and Drosophila, we show here that shRNA-mediated knockdown of SIRT1 in the mouse liver results in significantly decreased fasting-dependent down-regulation of lipogenic gene expression—including FASN, ELOVL6 (fatty acid elongase), and SREBP-1c—suggesting that SIRT1 mediates fasting-dependent regulation of multiple SREBP-1 target genes in mice (Fig. 3A–C; Rodgers and Puigserver 2007).
Figure 3.
SIRT1 regulation of SREBP target gene expression during fasting in the mouse liver. qRT–PCR was used to analyze the mRNA levels of SREBP target genes FASN](A), ELOVL6 (B), and SREBP-1c (C) in the mouse liver. The fasting-dependent changes of gene expression are compared between livers of mice subjected to tail-vein injection of adenoviruses programmed to express control shRNA (C; n = 7) and SIRT1 shRNA (shSIRT1; n = 7) (fasted for 20 h). All data are normalized to 18s rRNA expression. Error bars represent SEM. (*) P < 0.05; (**) P < 0.01. (D) Hierarchical clustering of mouse liver transcriptomes with or without SIRT1 knockout. The heat map demonstrates that SREBP target genes are among the most significantly up-regulated genes in SIRT1 knockout livers.
A SIRT1 liver-specific knockout (SIRT1 LKO) mouse model has been developed recently (Purushotham et al. 2009). This study showed that, in addition to effects on PPARα target genes involved in fatty acid β-oxidation, livers from the SIRT1 LKO mice became steatotic after being fed a high-fat diet and exhibited high levels of cholesterol, and several SREBP target genes examined (FASN, ACC1, and SREBP-1c) were increased on both normal chow and high-fat diets (Purushotham et al. 2009). We therefore performed DNA microarray analysis on isolated livers from wild-type and SIRT1 LKO animals to more comprehensively evaluate the possible effects on expression of SREBP target genes in response to SIRT1 deletion in the liver. Indeed, the DNA microarray studies, together with quantitative RT–PCR (qRT–PCR) analysis and unsupervised hierarchical clustering analysis, revealed a broad spectrum of SREBP-1 and SREBP-2 targets among the genes significantly up-regulated in the SIRT1 LKO livers (Fig. 3D; Supplemental Fig. 5; data not shown), including genes in the lipogenic programs (e.g., FASN, acetyl-CoA synthase and carboxylase, ATP citrate lyase, and ELOVL6), as well as cholesterol biosynthesis pathways (e.g., squalene epoxidase, mevalonate diphosphate decarboxylase, HMG-CoA reductase, and phosphomevalonate kinase). In contrast, we did not observe any significant changes in the expression of SREBP-1a/c or SREBP-2 themselves in response to SIRT1 deletion in the liver, consistent with our model of SIRT1-dependent post-transcriptional of SREBPs, with ensuing effects on SREBP target gene expression. These in vivo data together suggest that loss of SIRT1 in the mouse liver broadly and significantly impacts SREBP-dependent gene regulation.
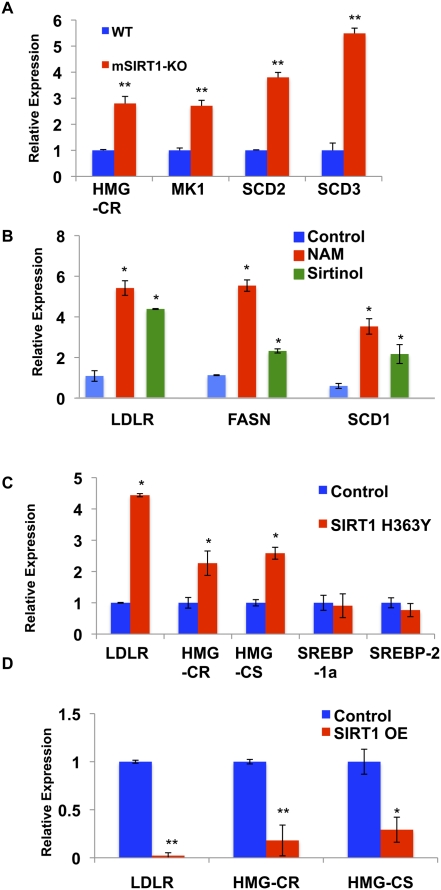
In accord with the in vivo findings, mouse embryo fibroblasts (MEFs) isolated from SIRT1 knockout mice (Cheng et al. 2003) exhibit increased expression of several SREBP target genes, confirming an important role for SIRT1 in regulating SREBP-dependent gene expression (Fig. 4A). Additionally, we found that manipulation of sirtuin activity or SIRT1 levels in different human cell lines by treatment with the sirtuin inhibitors nicotinamide and sirtinol (Fig. 4B), or overexpression of a dominant-negative, catalytically inactive SIRT1 mutant protein (H363Y) (Fig. 4C), all result in significantly increased expression of SREBP-regulated genes. Conversely, overexpression of wild-type SIRT1 causes strong repression of SREBP target genes in human cells (Fig. 4D). These regulatory changes encompass genes from lipogenic programs such as stearoyl-CoA desaturases and fatty acid synthase, primarily regulated by SREBP-1a, and cholesterol biosynthesis genes (HMG-CoA reductase [HMG-CR] and the low-density lipoprotein receptor [LDLR]), which are likely to be under the control of SREBP-2 in these cell culture models. Consistent with the SIRT1 LKO data, we do not observe changes in the expression of the SREBPs themselves upon manipulations of SIRT1 function (Fig. 4C; data not shown). Taken together, our findings reveal that SIRT1 plays a central and conserved role in fasting-dependent down-regulation of SREBP target gene expression in mammals.
Figure 4.
Mammalian SIRT1 represses SREBP-1 and SREBP-2 target gene expression. (A) SREBP target genes are expressed at higher levels in SIRT1 knockout MEFs when compared with the wild-type cells, as judged by qRT–PCR; β-actin served as the control of total RNA. (MK1) Mevalonate kinase-1; (SCD2) stearoyl-CoA desaturase-2; (SCD3) stearoyl-CoA desaturase-3. (B) Treatment of human IMR-90 fibroblasts with the sirtuin inhibitors nicotinamide (NAM; 20 mM) or sirtinol (0.1 mM) for 5 h resulted in increased expression of SREBP target genes by qRT–PCR. Data were first normalized by β-actin, then normalized by the control (0.1% DMSO), and expressed as mean ± SD. (SCD1) Stearoyl-CoA desaturase-1. (C) Expression of a dominant-negative form of SIRT1 (H363Y) in HEK293T cells increased the transcription of the LDLR, HMG-CR, HMG-CS, and SCD1 genes as assessed by qRT–PCR. SREBP-1a or SREBP-2 transcriptional levels were unchanged. GAPDH served as a loading control of total RNA. (D) Overexpression of SIRT1 decreases expression of SREBP target genes in HEK293T cells. SIRT1 and SIRT1(H363Y) (Fig. 4C) were expressed at similar levels. Error bars represent standard deviations between cDNA made from independently treated cells. (*) P < 0.05; (**) P < 0.01.
SIRT1 directly deacetylates SREBP and controls SREBP protein stability
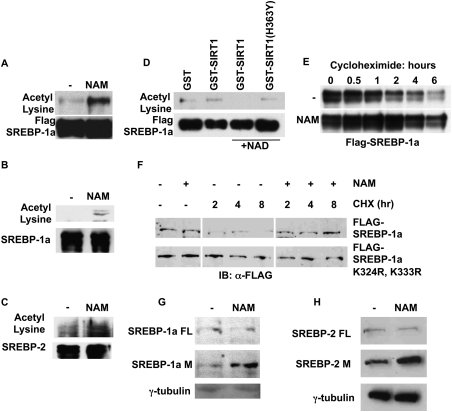
Based on our findings of regulation of SREBP target gene expression by SIRT1 orthologs in metazoans during fasting, we speculated that SIRT1 might also directly deacetylate SREBPs and inhibit SREBP-dependent transactivation functions. Previous studies have shown that SREBPs interact with the CBP/p300 acetyltransferases, which acetylate SREBPs on lysine residues located in the DNA-binding domain (Oliner et al. 1996; Ericsson and Edwards 1998; Näär et al. 1998; Giandomenico et al. 2003). To examine whether SREBP acetylation might be regulated by SIRT1, we initially treated human cells with the sirtuin inhibitor nicotinamide, and then examined acetylation of immunopurified SREBPs. Nicotinamide treatment caused a significant increase in acetylation of both overexpressed and endogenous mature forms of SREBPs, consistent with sirtuin regulation of the SREBP acetylation state (Fig. 5A–C). To test whether SIRT1 is capable of directly deacetylating SREBP in vitro, we immunopurified Flag-SREBP-1a from cells treated with nicotinamide to enhance SREBP-1a acetylation, and then performed deacetylation reactions with bacterially expressed and purified SIRT1 proteins fused to GST in the presence or absence of the essential cofactor NAD+. These experiments revealed that wild-type SIRT1 deacetylates SREBP-1a in an NAD+-dependent manner, whereas the catalytically inactive mutant SIRT1(H363Y) does not (Fig. 5D). Interestingly, acetylation has been suggested to result in SREBP protein stabilization (Giandomenico et al. 2003), and deacetylation of SREBPs by SIRT1 would thus be predicted to increase SREBP protein turnover. Consistent with this hypothesis, we found that nicotinamide treatment of human cells expressing Flag-SREBP-1a results in markedly elevated SREBP stability (half-life increased from ∼1 h to ∼6 h), as judged by treatment with the translation inhibitor cycloheximide (Fig. 5E). Nicotinamide had little effect on the transcription of the Flag-SREBP-1a construct (data not shown). To determine if sirtuin-dependent deacetylation and regulation of stability of SREBP-1a depended on the lysine residues in the DNA-binding domain shown previously to be acetylated by p300 (K324 and K333) (Giandomenico et al. 2003), we compared the half-lives of wild-type Flag-SREBP-1a with Flag-SREBP-1a K324R/K333R in the presence of sirtuin inhibitors. As demonstratedpreviously (Giandomenico et al. 2003), mutations of Lys 324 and Lys 333 increased the stability of SREBP-1a (Fig. 5F). Importantly, however, nicotinamide did not further increase the stability of Flag-SREBP-1a K324R/K333R, suggesting that sirtuin-dependent deacetylation of SREBP-1a indeed occurs at these residues (Fig. 5F). We also found that nicotinamide treatment of normal human fibroblasts (IMR-90) resulted in increased levels of endogenous, mature (nuclear) SREBP-1 and SREBP-2 proteins, while levels of the SREBP-1 and SREBP-2 precursor proteins were unaffected (Fig. 5G, H). Collectively, these results suggest that SIRT1 can control the stability of mature, nuclear SREBPs by direct deacetylation.
Figure 5.
SIRT1 directly deacetylates SREBP and controls SREBP protein stability. (A) HeLa cells were transfected with Flag-tagged SREBP-1a, and then were treated with (+) or without (−) 20 mM NAM for 20 h. Flag-SREBP-1a was purified by immunoprecipitation using anti-Flag antibodies, and was eluted with Flag peptide. The presence of acetylated SREBP-1a was detected by immunoblotting using an acetyl lysine-specific antibody. Flag-SREBP-1a served as the loading control. (B) HeLa cells were treated with proteasome inhibitors alone or with proteasome inhibitors and 25 mM NAM for 6 h. Endogenous SREBP-1a was immunoprecipitated by antibodies specific to SREBP-1a. The presence of acetylated SREBP-1a was detected by immunoblotting using an acetyl lysine-specific antibody. (C) HeLa cells were treated with proteasome inhibitors alone or with proteasome inhibitors and 25 mM NAM for 6 h. Endogenous SREBP-2 was immunoprecipitated. The presence of acetylated SREBP-2 was detected by immunoblotting using an acetyl lysine-specific antibody. (D) Overexpressed Flag-SREBP-1a proteins were purified from HeLa cells treated with NAM, and were incubated with GST fusion proteins as indicated in the in vitro deacetylation assay. The degree of SREBP-1a acetylation was detected by immunoblotting using an acetyl-lysine-specific antibody. Flag-SREBP-1a served as the loading control. (E) HeLa cells were transfected with Flag-tagged SREBP-1a, and then were treated with (+) or without (−) 20 mM NAM for 20 h. After the indicated time of cycloheximide (0.1 mM) treatment, whole-cell extracts were prepared, and Flag-SREBP-1a was detected by immunoblotting using anti-Flag antibodies. Equal amounts of proteins were loaded in each lane. (F) Overexpressed Flag-SREBP-1a or Flag-SREBP-1a K324R, K333R were treated with nicotinamide and cycloheximide as in E. (G) IMR-90 cells were placed in 1% lipid-depleted serum with or without NAM for 16 h. Nuclear extracts were prepared and detected by immunoblotting with an antibody to SREBP-1. (FL) Full-length SREBP-1 precursor; (M) mature, nuclear SREBP-1. (H) Endogenous SREBP-2 in IMR-90 cells was detected as in G.
Sirtuin inhibition results in increased nuclear SREBP levels
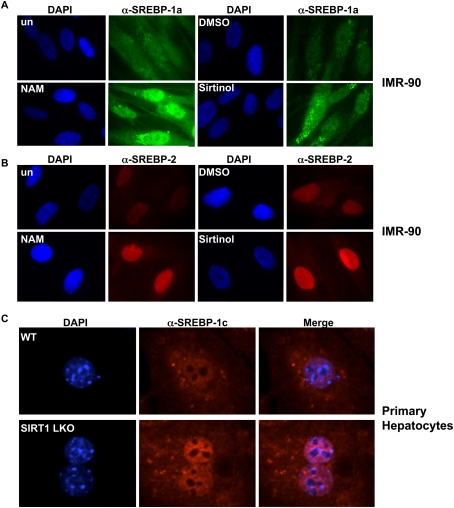
Our gene expression studies suggest that transcriptional targets of multiple SREBP isoforms are changed in response to altered SIRT1 activity. To more fully investigate the role of SIRT1 in affecting distinct SREBP isoforms, we examined the levels of nuclear SREBP-1a and SREBP-2 in human cells treated with sirtuin inhibitors and SIRT1 siRNA. We found that nicotinamide, sirtinol, and siRNA-mediated ablation of SIRT1 all increased the nuclear abundance of SREBP-1a and SREBP-2 in primary human fibroblasts (IMR-90) and cancer cells (HeLa S) (Fig. 6A,B; Supplemental Figs. 7, 8). To examine the response of the SREBP-1c isoform to alterations of SIRT1, we performed immunofluorescence studies on primary hepatocytes isolated from wild-type or SIRT1 LKO mice (Purushotham et al. 2009), and found that, similar to SREBP-1a and SREBP-2, nuclear SREBP-1c levels increased when SIRT1 was absent (Fig. 6C). These experiments are consistent with the notion that all three SREBP isoforms can be regulated by SIRT1.
Figure 6.
Sirtuin inhibitors regulate nuclear abundance of SREBP-1a, SREBP-2, and SREBP-1c in primary cells. IMR-90 cells were placed in 1% lipid-depleted serum and proteasome inhibitors and then treated with NAM or sirtinol for 6 h. Cells were stained with antibodies to SREBP-1a (A) or SREBP-2 (B) along with DAPI, and were visualized by immunofluorescence. (C) Hepatocytes from wild-type or SIRT1 LKO mice were stained with antibodies specific for SREBP-1c, and were stained with DAPI to visualize nuclei.
SIRT1 activators promote inhibition of SREBP target genes, increased SREBP ubiquitination, and decreased SREBP protein stability, and ameliorate aberrant lipogenesis in vivo
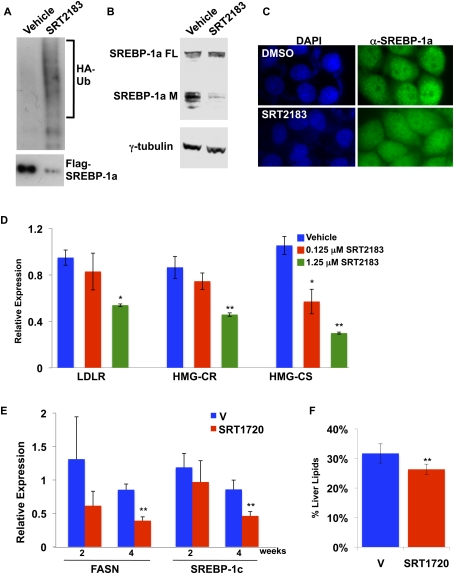
Recent studies have reported the identification of potent SIRT1-activating compounds, and showed that such SIRT1 activators could significantly ameliorate insulin resistance and improve energy homeostasis in mouse diet-induced obesity models (Milne et al. 2007). Additional studies showed a significant reduction of hepatic lipids in wild-type mice fed a high-fat diet and treated with the bioavailable SIRT1 activator SRT1720 (Feige et al. 2008). We investigated whether SIRT1-selective activators could also affect SREBP-dependent gene activation in vitro and in vivo, and whether they might influence hepatic steatosis in a mouse obesity model. First, our model predicts that, in the presence of sirtuin activators, the proportion of nuclear SREBP targeted for ubiquitination and degradation would increase, resulting in a decrease in the nuclear abundance of SREBP. Indeed, treatment with the potent SIRT1 activator SRT2183 causes strongly increased ubiquitination of Flag-SREBP-1a (Fig. 7A). Furthermore, SRT2183 treatment decreased the levels of endogenous mature, nuclear SREBP-1 in cultured cells, but had no effect on the SREBP-1 precursor (Fig. 7B,C). We also found that treatment of human cell lines with SRT2183 results in significantly decreased expression of SREBP target genes in a dose-dependent manner, consistent with our model (Fig. 7D; data not shown). CBP/p300 have been reported to be regulated by sirtuins, including SIRT1 and SIRT2 (Liu et al. 2008), and the effects of SIRT1 manipulations that we observe could therefore be indirect. However, while treatment of cells with the SIRT1 activator SRT2183 causes decreased nuclear SREBP levels and inhibition of SREBP target genes, immunopurified p300 from cells treated with SRT2183 does not exhibit decreased activity in terms of acetylation of histones, suggesting that impaired CBP/p300 activity is unlikely to provide an explanation for the observed effects on SREBPs and their target genes (Supplemental Fig. 8).
Figure 7.
SIRT1 activators promote increased SREBP ubiquitination, decreased SREBP nuclear levels, and inhibition of SREBP target genes, and ameliorate aberrant lipogenesis in vivo. (A) HeLa cells were transfected with Flag-SREBP-1a and HA-Ubiquitin, and were treated with DMSO (vehicle) or the SIRT1 activator SRT2183. Extracts were immunoprecipitated with Flag antibodies, and then were probed by immunoblotting with antibodies to Flag or HA. (B) HeLa cells were treated with the SIRT1 activator SRT2183, and extracts were immunoblotted with an antibody to SREBP-1a. (FL) Full-length SREBP-1a precursor; (M) mature, nuclear SREBP-1a. (C) HeLa cells were placed in medium containing 1% lipid-depleted serum, low glucose, and proteasome inhibitors. Cells were treated with DMSO or SRT2183 for 6 h, fixed, and stained with an antibody to SREBP-1a. (D) SIRT1 activation decreases SREBP-dependent gene activity in human cells. HeLa cells were placed in low-glucose DMEM, supplemented with 1% fetal bovine serum with or without SRT2183. Target gene expression was analyzed by qRT–PCR normalized to β-actin levels, and error bars represent standard deviations from three independently treated wells. (*) P < 0.05; (**) P < 0.01. (E) Livers from ob/ob mice fed a high-fat diet and treated daily with vehicle or the SIRT1 activator SRT1720 for 2 or 4 wk were analyzed by qRT–PCR. Expression of SREBP-1c target genes was analyzed. Expression was normalized to 18s rRNA. Error bars show standard error for five mice. (F) Livers from ob/ob mice fed a high-fat diet and treated with vehicle or SRT1720 for 4 wk were analyzed for lipid content. Total lipid content was normalized to body weight, and error bars represent standard error. Significance was determined by Student's t-test. (**) P < 0.01.
Next, we analyzed in vivo effects of the bioavailable SIRT1 activator SRT1720 (Milne et al. 2007; Feige et al. 2008) on SREBP target gene expression and steatosis in the mouse liver. Genetically (ob/ob) obese mice fed a high-fat diet and treated with SRT1720 daily for 2 and 4 wk exhibit significantly lower expression of the SREBP target gene FASN in the liver at 4 wk as compared with vehicle-treated mice (Fig. 7E). We also found that the SREBP-1c gene itself is significantly repressed in response to SRT1720 treatment for 4 wk in these mice, consistent with the fact that SREBP-1c controls its own expression (Fig. 7E; Amemiya-Kudo et al. 2000). Importantly, ob/ob mice treated with SRT1720 for 4 wk have significantly improved hepatosteatosis, as judged by decreased liver weight and hepatic lipids (Fig. 7F; data not shown). These data show that sirtuin activators are capable of reducing hepatosteatosis in several mouse models, and that, while sirtuins may affect multiple metabolic processes, down-regulation of SREBP activity appears to be a significant contributor to decreases in stored lipids.
Discussion
We revealed here that metazoan SIRT1 orthologs play key and conserved roles in regulating SREBP-dependent lipogenic gene expression and lipogenesis/lipid storage in response to fasting cues in metazoans. Sirtuins are NAD+-dependent deacetylases with multiple targets in metabolic pathways, including transcription factors and coactivators in the PPAR, FOXO, and PGC families (Yu and Auwerx 2009). Because sirtuin activators mimicking the fasting response are being actively developed for treatment of human metabolic syndrome (Guarente 2006), it is critical to understand mechanisms of sirtuin-induced changes in lipogenesis. We examined functional relationships between SIRT1 orthologs and SREBP family members in several metazoan models, and found consistent evidence that SIRT1 is important for fasting-dependent attenuation of SREBP function, and that this down-regulation contributes to the decrease in lipid stores during fasting (Supplemental Fig. 9). Indeed, by using animals that lack SIRT1 orthologs or employing RNAi, in addition to chemical sirtuin inhibitors, we show that SIRT1 orthologs are necessary for inhibition of SREBP target genes in response to fasting cues, and that these effects can be replicated in wild-type animals. We also provide critical mechanistic evidence that SIRT1 can directly deacetylate SREBP-1a, that chemical sirtuin inhibitors increase SREBP protein stability and nuclear abundance, and that mutations that prevent acetylation of lysine residues in the DNA-binding domain render SREBP-1a independent of sirtuin regulation. Finally, we show that recently developed SIRT1 activators, which are effective in improving energy homeostasis and decreasing hepatosteatosis in mouse models (Milne et al. 2007; Feige et al. 2008), also increase ubiquitination and proteasomal targeting of SREBP-1a, resulting in decreased SREBP-1 nuclear levels and target gene expression. These results suggest that the prolipogenic program driven by SREBP can be curtailed rapidly by increases in SIRT1 activity, and represents an important aspect of SIRT1 function in metabolic control.
In order to cope with periods of short-term food deprivation (fasting), animals dramatically alter their metabolism, shifting cellular programs to using energy stores rather than promoting lipid biogenesis. This process not only requires the coordination of multiple metabolic pathways, but also may vary in different animals depending on their ecology or life cycles. Conversely, fasting-dependent metabolic shifts toward fatty acid β-oxidation and away from lipogenesis must be reversible rapidly so that energy storage can be restarted when feeding resumes. The cholesterogenic and lipogenic potential of SREBP is tightly controlled at transcriptional and post-transcriptional levels, which may act preferentially on individual isoforms. SREBP-1c, which supports fatty acid synthesis in the liver and adipose tissue, has the most extensively characterized regulation at the transcriptional level (Eberle et al. 2004). SREBP-1c expression can be up-regulated by insulin signaling in response to carbohydrate uptake (Azzout-Marniche et al. 2000) or LXRα-mediated induction, which may coordinate fatty acid and aspects of cholesterol homeostasis (Repa et al. 2000; Schultz et al. 2000; Laffitte et al. 2003), or be subject to autoactivation (Amemiya-Kudo et al. 2000). SREBP-1a and SREBP-2 appear to be regulated primarily on the post-transcriptional level, where processing of the membrane-bound inactive form through the ER and Golgi can be repressed by high cholesterol (Brown and Goldstein 1997), addition of palmitate (Drosophila) (Seegmiller et al. 2002), or other physiological processes such as ER stress (Kammoun et al. 2009). Additionally, post-translational modifications of SREBPs have been shown to affect levels and activity. For example, the activity of nuclear SREBPs may be augmented by phosphorylation or acetylation (Eberle et al. 2004). Moreover, SUMOylation has been reported to decrease transcriptional activity (Hirano et al. 2003), and acetylation of lysines in the DNA-binding domain in active SREBP mask residues that may be ubiquitinated as a prelude to proteasomal degradation (Hirano et al. 2001; Sundqvist and Ericsson 2003). Our studies showing SIRT1-dependent regulation of nuclear SREBPs provided a biological context for control of SREBP stability by demonstrating a mechanism for rapid reductions in cellular potential for fatty acid or cholesterol production. Post-translational effects on the nuclear, active form of SREBPs are consistent with the rapid reactivation of lipogenic and cholesterogenic programs (Horton et al. 1998) that occur upon refeeding. While SIRT1/SIR-2.1 effects are most pronounced during fasting, it is also possible that deacetylation plays a minor role in SREBP turnover in nonfasted situations, as SREBP target genes respond to changes in sirtuin activity in fed animals, or cells in normal culture media (Figs. 1H, 2C, 4; data not shown). It is also notable that the effects of SIRT1 orthologs on SREBPs are conserved in multiple metazoan organisms with diverse feeding/fasting cycles, suggesting the biological importance of the regulatory mechanism. The regulation of SREBP proteins by other sirtuin family members has been demonstrated recently in a neurodegeneration model (Luthi-Carter et al. 2010). However, in this instance, SIRT2-mediated regulatory events occurred outside the nucleus and resulted in decreases in SREBP-2-dependent gene expression, rather than increases, as in our model of SIRT1-dependent regulation. This study raises important additional support for our assertion that pharmacological inhibition of sirtuins in our system, which mimics genetic SIRT1 manipulation in our gene expression and SREBP localization studies, is primarily acting through SIRT1.
In mammals, SIRT1 appears to be key to shifts in energetic equilibrium occurring as nutritional states change. SIRT1 levels increase in the liver in response to fasting (Rodgers and Puigserver 2007), and become more active as cellular NAD+ levels rise; thus, SIRT1 is poised to modify and activate regulators of lipid mobilization. Indeed, SIRT1 has a well-documented role in stimulation of lipid breakdown in multiple tissues and through several distinct pathways (Yu and Auwerx 2009). In adipose tissue, SIRT1 can repress PPARγ, limiting fatty acid uptake and triacylglycerol (TAG) synthesis (Tontonoz and Spiegelman 2008), while activating PPARα/PGC-1α-dependent programs of fatty acid β-oxidation in the liver (Rodgers et al. 2005; Gerhart-Hines et al. 2007; Purushotham et al. 2009; Smith et al. 2009). Several recent reports have also linked SIRT1 to changes in lipogenic programs. First, resveratrol, an activator of SIRT1, can increase the levels of SREBP in livers of alcohol-treated mice (Ajmo et al. 2008). Second, the SIRT1 activator SRT1720 was shown to decrease lipogenic gene expression in a mouse model of hepatosteatosis (Yamazaki et al. 2009). Together with our data demonstrating biological and mechanistic links, this strengthens the observation that SIRT1 can suppress factors such as SREBP that promote lipid storage, and highlights the multilayered control sirtuins can exert on metabolic circuits.
SIRT1 function also appears to be promoted by AMPK, which is activated upon energy depletion and plays key roles in modulating lipid and energy homeostasis (Ruderman et al. 2010), at least in mammalian contexts (Hou et al. 2008; Canto et al. 2009), and may thus act as an upstream fasting-regulated signal transducer to coordinately regulate multiple facets of lipogenesis, including SIRT1-dependent control of SREBPs. It is presently unclear whether AMPK also plays a role in regulating SREBP in lower metazoans in response to fasting cues.
Recent work has demonstrated that sirtuins employ multiple mechanisms allowing cells to access stored energy during fasting, depleting lipid deposits (Yu and Auwerx 2009). The therapeutic benefits of treating disorders such as type 2 diabetes, metabolic syndrome, and hepatosteatosis have driven the development of sirtuin activators that may act as fasting mimetics (Guarente 2006). Resveratrol treatment attenuates the effects of a high-fat diet in mice (Lagouge et al. 2006; Bordone et al. 2007), and, recently, more potent SIRT1 activators act to decrease serum triglyceride levels and protect against obesity in mice (Milne et al. 2007; Feige et al. 2008). While the precise mechanistic details of how these SIRT1 activators function is still being debated (Ledford 2008; Pacholec et al. 2010), they do exert potent effects on a number of metabolic regulators, with important physiological consequences. For example, SIRT1 activators increase SIRT1-dependent activation of PGC-1α, FOXO1, and p53, promoting the expression of fatty acid β-oxidation genes (Feige et al. 2008; Smith et al. 2009). Our work shows that the SIRT1 activator SRT2183 impinges on transcriptional programs governing de novo cholesterol/lipid biosynthesis in cultured cells by rendering SREBP susceptible to ubiquitination; the resulting proteasomal processing decreases the pool of active SREBP, and, consequently, SREBP-dependent gene expression (Fig. 7A–D). This link is likely to hold true in vivo, as the bioavailable SIRT1 activator SRT1720 also decreases expression of SREBP target genes and stored fats in livers from ob/ob mice fed a high-fat diet (Fig. 7 E,F). Our in vitro and in vivo results thus provide important insights into the mechanistic and physiological effects of SIRT1 activators; understanding how such potential treatments affect the breadth of SIRT1 function is critical to their success as therapeutics in humans.
Materials and methods
Antibodies
Anti-Flag M2 was purchased from Sigma. Anti-HA was purchased from Covance Research Products. Anti-SIRT1 and anti-pan-acetyl were purchased from Cell Signaling. SREBP antibodies were as follows: SREBP-1a antibody (2121) was purchased from Upstate Biotechnologies, and SREBP-2 was purchased from Cell Signaling. Anti-β-tubulin antibodies were obtained from Santa Cruz Biotechnology, and anti-γ-tubulin antibodies were procured from Sigma.
Tissue culture
HeLa cells, 293T cells, and MEFs were cultured at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Sigma), supplemented with 100 mg/mL penicillin–streptomycin (GIBCO-BRL), 10% (or otherwise indicated) fetal bovine serum (Hyclone), and 20 mM glutamine. Treatment of HeLa cells with SRT2183 was performed in low-glucose DMEM with 1% fetal bovine serum. IMR-90 cells were cultured in MEM, supplemented as other cells with the addition of 0.1 mM sodium pyruvate (GIBCO-BRL). For activator and inhibitor studies, cells were plated and allowed to recover overnight before addition of drug. Nicotinamide (25 mM) treatment was performed for 6–20 h; as noted, this concentration is within the range of physiological effect for SIRT1 in cell culture (Rodgers et al. 2005). Sirtinol (0.1 mM) treatment occurred for 20 h, and SRT2183 was added for 16 h. Lipid-depleted bovine calf serum was obtained from Biomedical Technologies, Inc.
Plasmids
Full-length human SIRT1 cDNA (wild-type or H363Y mutated) was subcloned into pcDNA4/TO-Flag or pGEX-2TKN. Human SREBP-1a cDNA encoding amino acids 1–487 was subcloned into pcDNA4/TO with an N-terminal Flag tag. shRNA constructs of human SIRT1 were purchased from Open Biosystems. Adenoviruses expressing control and SIRT1 shRNA were described previously (Rodgers and Puigserver 2007). Flag-SREBP-1a K324R, K333R was kindly provided by J. Ericsson (Giandomenico et al. 2003).
Transfection and siRNA
To overexpress epitope-tagged proteins in HeLa and 293T cells, 2 μg of plasmid DNA was transfected by Lipofectamine 2000 (Invitrogen) into each well (4 × 105 cells) of six-well plates. Whole-cell extracts were prepared after 24 h of culture. Smart pool of double-stranded siRNA oligonucleotides (control or SIRT1) were synthesized by Dharmacon Research, Inc. (a Thermo-Fisher Company). siRNA oligonucleotides (1 μg per well) were transfected into HeLa cells in six-well plates by Lipofectamine 2000. After 48 h, cells were replated into 24-well plates, and were transfected with reporter vectors. Cell extracts for immunodetection of SIRT1 and β-tubulin were generated 65 h after siRNA transfection.
Immunoprecipitation, immunoblotting, and immunofluorescence
For immunoprecipitation, 5 μL of anti-Flag, 20 μL of SREBP-1a (m2121, Upstate Biotechnologies), or 20 μL of SREBP-2 was incubated with 20 μL of protein A/G beads (Pharmacia) for 1 h, nutating at room temperature. After extensive washes, 1 mL of HeLa or 293T whole-cell lysates was incubated for 3 h, nutating at 4°C, with the antibody-coupled protein A/G beads. For immunoprecipitations of endogenous SREBP isoforms from HeLa cells, cultures were pretreated with proteasome inhibitor and nicotinamide for 6 h. The beads were washed five times with 1 mL of wash buffer containing 20 mM HEPES (pH 7.6), 250 mM KCl, 0.1 mM EDTA, 10% glycerol, 0.1% NP-40, 1 mM DTT, 1 mM benzamidine, 0.25 mM PMSF, and 2 μg/mL aprotinin. The interacting proteins were then eluted with binding buffer containing 10 mM Flag peptide (for Flag-tagged proteins) for 1 h at 4°C. For immunoblotting, protein samples were resolved by SDS-PAGE and transferred to PVDF membranes. The membranes were blocked by 0.5% nonfat milk for 1 h at room temperature and were incubated with primary antibodies overnight at 4°C. After several washes, the membranes were incubated with HRP-conjugated secondary antibody for 1 h at room temperature. For detection of endogenous acetylated SREBP-1a and SREBP-2, secondary anti-light chain-conjugated HRP antibodies (Jackson Immunologicals) were used. For detection of endogenous SREBP-1 in immunoblots, IMR-90 cells were cultured in low-glucose DMEM with 1% lipid-depleted serum (LDS) with vehicle (DMSO) or SRT2183. ALLN (25 mg/mL; Peptide Biosciences) was added to the culture media 1 h before harvesting. Nuclear extracts were prepared as in Andrews and Faller (1991). For immunofluorescence, IMR-90 cells were plated overnight in 1% LDS, and HeLa cells were treated with 1% LDS/low glucose. Six hours before harvest, ALLN was added at 1 mM, along with nicotinamide (25 mM) or sirtinol (0.1 mM). The cells were then washed with PBS and fixed with paraformaldehyde. Primary antibody incubations were performed overnight. Cells for photography were selected at random by DAPI staining, and three independent fields were photographed.
In vitro deacetylation assays
GST fusion proteins [GST alone, GST-SIRT1WT, and GST-SIRT1(H363Y)] were expressed in Escherichia coli (BL21; DE3) and purified from the lysates by glutathione-sepharose beads (Pharmacia). The amount of GST proteins was estimated using SDS-PAGE followed by Coomassie staining. Flag-tagged SREBP-1a proteins were expressed in HeLa cells in the presence of 25 mM nicotinamide, purified by immunoprecipitation from whole-cell extract, and eluted with Flag peptide. The purified Flag-SREBP-1a was incubated with purified GST fusion proteins for 1 h at 30°C in either the presence or absence of 5 mM NAD+. The reactions were performed in a buffer containing 50 mM Tris-HCl (pH 9.0), 50 mM NaCl, 4 mM MgCl2, 1 mM ZnCl2, 0.5 mM DTT, 0.2 mM PMSF, 0.02% NP-40, and 5% glycerol. The reactions were resolved on SDS-PAGE, and were analyzed by immunoblotting.
C. elegans
Nematodes were cultured using standard C. elegans methods (Brenner 1974) and were fed E. coli OP50, unless otherwise noted. Strains used in the analysis were N2, sir-2.1(ok434), fat-7p∷GFP(BC15777), elt-2∷GFP(rtIS26); CE500 Ex[SBP-1∷GFP, rol-6]; sbp-1(ep79); HA1894 sbp-1(ep79);sir-2.1(ok434); HA316 Is[rol-6]; and LG100 Is[sir-2.1, rol-6]. For fasting assays, animals were collected at the L4/young adult stage, washed extensively in M9 with 100 mg/mL kanamycin, divided into fed and fasted groups, and placed on nematode growth medium (NGM) plates unless otherwise noted. Sudan Black staining was carried out as described in Ogg and Ruvkun (1998). More than 30 images from each genotype were scored blind for level of staining in multiple independent experiments. Animals for Sudan Black staining or fat-7p∷GFP analysis were fasted for 18 h; when analyzing sbp-1(ts), fed and fasted animals were placed at 25°C. For experiments involving fat-7p∷GFP and SBP-1∷GFP, the assay was carried out in liquid culture in 24-well plates with constant rotation. Kanamycin (100 mg/mL) was added to both fed and fasted animals to control bacterial growth. For sbp-1(RNAi), nematodes were fed E. coli HT115 bacteria expressing clone III-6C01 from the Ahringer RNAi library (Kamath et al. 2003). Both fat-7 expression and Sudan Black staining levels were lower in wild-type animals fed HT115 bacteria. Fluorescence microscopy was carried out on a Zeiss Axioplan equipped with an Axiocam camera. Images were converted into Adobe Photoshop and pseudocolored. All alterations of levels were applied equally to all elements within a panel. qRT–PCR on C. elegans samples was performed following RNA extraction (Tri-Reagent, Sigma) and cDNA synthesis (Transcriptor, Roche) on an LC480 (Roche) with SYBR Green PCR kit (Roche). Values were normalized to act-1. For gene expression studies, fasting was carried out for 7 h. Primer sequences are available on request. For thin-layer chromatography and gas chromatography analysis of C. elegans TAG levels, please see Ashrafi et al. (2003) and Watts and Browse (2006).
Drosophila treatment and culture
All flies were cultured on standard cornmeal–agar–molasses medium, and w1118 strain was used as the wild-type control. A Sir2-null allele (Sir22A-7-11) deletes the entire coding sequence of the Sir2 gene, and was generated by targeted knockout (Xie and Golic 2004). The homozygous mutant animals of this Sir2-null allele are viable, allowing us to collect third instar mutant larvae for qRT–PCR analyses. Early third instar larvae of Sir2 mutants (w1118; Sir22A-7-11; +) or control (w1118; +; +) were maintained in vials containing either normal food or 1% agarose gel in PBS for fasting treatment, for 24 h at 25°C.
SIRT1 shRNA knockdown in the mouse liver
Experiments were performed in 6- to 8-wk-old male BALB/c mice, purchased from Harlan Laboratories. Animals were fed a standard rodent chow in a controlled environment with 14- to 10-h light–dark cycle. Control shRNA, SIRT1 shRNA, and recombinant adenoviruses were delivered by tail-vein injection into mice. Tolerance tests were performed 4–5 d after transduction. Mice were killed 7–9 d following adenoviral transduction. Livers were extracted and immediately snap-frozen in liquid nitrogen, and were stored at −80°C until analysis. Detailed methods are found in Rodgers and Puigserver (2007).
SIRT1 activator treatment of mice, and liver lipid and mRNA analyses
Six-week-old ob/ob mice were purchased from Jackson Laboratories, fed a high-fat diet (60% energy; Research Diets), acclimated for 2 wk before the study, and dosed with SRT1720 (100 mg/kg) or vehicle (2% HPMC + 0.2% DOSS) by oral gavage once a day for 2 or 4 wk as described (n = 5 per group) (Milne et al. 2007). All handling and dosing was performed according to the guidelines set out by the Institutional Animal Care and Use Committee (IACUC) at Sirtris. Animals were weighed immediately before sacrifice. Euthanasia was performed with CO2 2–3 h after the last oral gavage (with compound or vehicle). The weight of the whole liver was recorded, and the liver was frozen in N2(l). The frozen tissue was ground into a powder, from which lipid content and mRNA gene expression were measured. Roughly 500 mg of liver (powder) was weighed, and total lipids were extracted using the Folch method (Folch et al. 1957). The lipid content of the liver is represented as percentage lipids by weight. First, 0.5 mL of dH2O was added to livers and homogenized using a handheld tissue homogenizer. HPLC-grade chloroform (Fluka), methanol (Sigma), and water (Invitrogen) were added to the tissue homogenate in 8:4:3 volume, respectively. For most tissues, 3 mL of HPLC-grade chloroform was added, and sample was vortexed. Then, 1.5 mL of methanol was added, and the sample was vortexed. One milliliter of dH2O was added, and then the sample was vortexed. Tissue solution was allowed to sit for 20 min to allow phase separation. Using negative pressure, a plastic pipette was used to transfer the bottom aqueous layer to a preweighed new tube. The top supernatant layer was discarded. The extraction protocol was repeated with the middle layer, and the bottom was transferred to its respective tube. Tubes were placed overnight in a 70°C oven. When all of the liquid dissolved and the samples remained unchanged over several hours, a small drop of oil remained. This was weighed, and the percentage of fat in the original samples was extrapolated. Total RNA was extracted from the liver (powdered and stored at −80°C) with Trizol (Invitrogen), and quantified with an Agilent 2100 Bioanalyzer (Agilent Technologies). For mRNA quantification, 2 μg of RNA was converted to cDNA with a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems), and transcript levels of relevant genes were determined by real-time, quantitative PCR with an ABI 7300 Real-Time PCR machine according to the manufacturer's instructions. TaqMan probes and primers were designed using Primer Express 3.0 (Applied Biosystems).
qRT–PCR and semiquantitative RT–PCR assay
The total mRNA was extracted using Trizol Reagent (Invitrogen) following the manufacturer's protocols. For Drosophila samples, five larvae of each genotype per treatment were ground with a pellet mixer. The first strand of cDNA was generated using a first-strand cDNA synthesis kit (Promega). PCR primers are available on request.
Microarray experiment and data analysis
Total RNA extraction and NimbleGen Mus musculus 385K microarray experiments were performed according to the manufacturers' instructions. The absence of systemic bias among samples was confirmed by the box plot analysis supported by the R statistical programming language. Raw intensity values of 389,307 features were subjected to t-test (P < 0.05, two-tail) to extract 13,258 features whose expression significantly differed between the SIRT1 knockout and control animals. Genes detected with redundant features were then represented by features with the largest fold change between the knockout and control animals, while other features were removed. This resulted in a nonredundant list of 8573 genes. Fold changes between averages of the knockout and control animals were calculated for each of the 6252 RefSeq genes in the nonredundant list, and the fold change values were determined for genes involved in lipid metabolism.
p300 HAT assays
Nuclear extracts were prepared from IMR-90 cells treated with either SRT2183 SIRT1 activator or nicotinamide or vehicle control, and were incubated with Protein A sepharose beads (GE Healthcare) prebound with either anti-p300 (Santa Cruz Biotechnology) or control IgG. After 4 h, beads were collected and washed four times with 500 mM KCl immunoprecipitation wash buffer (20 mM Tris at pH 7.9, 10% glycerol, 0.05% NP-40, 1 mM MgCl2, 1 mM EDTA at pH 8, 1 mM DTT, 1 mM PMSF), followed by two washes with 100 mM KCl immunoprecipitation wash buffer, and then two washes with HAT assay buffer (50 mM Tris at pH 8, 10% glycerol, 10 mM butyric acid, 1 mM PMSF, 1 mM DTT). HAT assays were then performed using Calf Thymus IIA histones (Sigma) and 14C-acetyl-CoA (Amersham) substrates as described previously (Mizzen et al. 1999). SDS-PAGE and fluorography were used to detect acetylated histone products.
Acknowledgments
We thank Kent Golic and the Bloomington Drosophila Stock Center for dSir2-null flies, and some nematode strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We thank J. Ericsson for the mutated SREBP-1a plasmid, David J. Gagne and Nekeya Meade for help with the ob/ob mice, Luisa DeStefano for Drosophila experiments, and Jill C. Milne and Michael R. Jirousek for helpful discussions. This work was supported by The Paul F. Glenn Laboratories for the Biological Mechanisms of Aging at Harvard Medical School and the following grants from NIH: R01DK07833 and R01GM071449 to A.M.N., and R01GM53203 to N.J.D.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1901210.
Supplemental material is available at http://www.genesdev.org.
References
- Ajmo JM, Liang X, Rogers CQ, Pennock B, You M 2008. Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol 295: G833–G842 doi: 10.1152/ajpgi.90358.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, et al. 2000. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem 275: 31078–31085 [DOI] [PubMed] [Google Scholar]
- Andrews NC, Faller DV 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res 19: 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K 2007. Obesity and the regulation of fat metabolism. In WormBook (ed. The C. elegans Research Community, WormBook). doi: 10.1895/wormbook.1.130.1. http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268–272 [DOI] [PubMed] [Google Scholar]
- Azzout-Marniche D, Becard D, Guichard C, Foretz M, Ferre P, Foufelle F 2000. Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem J 350: 389–393 [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. 2006. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, et al. 2007. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell 6: 759–767 [DOI] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL 1997. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89: 331–340 [DOI] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF 2003. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci 100: 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornier MA, Dabelea D, Hernandez TL, Lindstrom RC, Steig AJ, Stob NR, Van Pelt RE, Wang H, Eckel RH 2008. The metabolic syndrome. Endocr Rev 29: 777–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F 2004. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie 86: 839–848 [DOI] [PubMed] [Google Scholar]
- Ericsson J, Edwards PA 1998. CBP is required for sterol-regulated and sterol regulatory element-binding protein-regulated transcription. J Biol Chem 273: 17865–17870 [DOI] [PubMed] [Google Scholar]
- Erion DM, Yonemitsu S, Nie Y, Nagai Y, Gillum MP, Hsiao JJ, Iwasaki T, Stark R, Weismann D, Yu XX, et al. 2009. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc Natl Acad Sci 106: 11288–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade PJ 2006. SREBPs: Sterol-regulated transcription factors. J Cell Sci 119: 973–976 [DOI] [PubMed] [Google Scholar]
- Feige JN, Auwerx J 2008. Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr Opin Cell Biol 20: 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J 2008. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab 8: 347–358 [DOI] [PubMed] [Google Scholar]
- Flowers MT, Ntambi JM 2008. Role of stearoyl-coenzyme A desaturase in regulating lipid metabolism. Curr Opin Lipidol 19: 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH 1957. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226: 497–509 [PubMed] [Google Scholar]
- Fontana L, Klein S, Holloszy JO 2010. Effects of long-term calorie restriction and endurance exercise on glucose tolerance, insulin action, and adipokine production. Age 32: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P 2007. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J 26: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giandomenico V, Simonsson M, Gronroos E, Ericsson J 2003. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol Cell Biol 23: 2587–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L 2006. Sirtuins as potential targets for metabolic syndrome. Nature 444: 868–874 [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP 2006. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev 20: 2913–2921 [DOI] [PubMed] [Google Scholar]
- Hirano Y, Yoshida M, Shimizu M, Sato R 2001. Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin-proteasome pathway. J Biol Chem 276: 36431–36437 [DOI] [PubMed] [Google Scholar]
- Hirano Y, Murata S, Tanaka K, Shimizu M, Sato R 2003. Sterol regulatory element-binding proteins are negatively regulated through SUMO-1 modification independent of the ubiquitin/26 S proteasome pathway. J Biol Chem 278: 16809–16819 [DOI] [PubMed] [Google Scholar]
- Horton JD, Bashmakov Y, Shimomura I, Shimano H 1998. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci 95: 5987–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS 2002. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109: 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ, et al. 2008. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem 283: 20015–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kammoun HL, Hainault I, Ferre P, Foufelle F 2009. Nutritional related liver disease: Targeting the endoplasmic reticulum stress. Curr Opin Clin Nutr Metab Care 12: 575–582 [DOI] [PubMed] [Google Scholar]
- Kunte AS, Matthews KA, Rawson RB 2006. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab 3: 439–448 [DOI] [PubMed] [Google Scholar]
- Laffitte BA, Chao LC, Li J, Walczak R, Hummasti S, Joseph SB, Castrillo A, Wilpitz DC, Mangelsdorf DJ, Collins JL, et al. 2003. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc Natl Acad Sci 100: 5419–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. 2006. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell 127: 1109–1122 [DOI] [PubMed] [Google Scholar]
- Ledford H 2008. Mice share yeast's ageing system. Nature 456: 433. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L 2007. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell 28: 91–106 [DOI] [PubMed] [Google Scholar]
- Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J 3rd, et al. 2008. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456: 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi-Carter R, Taylor DM, Pallos J, Lambert E, Amore A, Parker A, Moffitt H, Smith DL, Runne H, Gokce O, et al. 2010. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc Natl Acad Sci 107: 7927–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay RM, McKay JP, Avery L, Graff JM 2003. C. elegans: A model for exploring the genetics of fat storage. Dev Cell 4: 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, et al. 2007. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450: 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen CA, Brownell JE, Cook RG, Allis CD 1999. Histone acetyltransferases: Preparation of substrates and assay procedures. Methods Enzymol 304: 675–696 [DOI] [PubMed] [Google Scholar]
- Näär AM, Beaurang PA, Robinson KM, Oliner JD, Avizonis D, Scheek S, Zwicker J, Kadonaga JT, Tjian R 1998. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev 12: 3020–3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näär AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398: 828–832 [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T 2005. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem 280: 16456–16460 [DOI] [PubMed] [Google Scholar]
- Ogg S, Ruvkun G 1998. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell 2: 887–893 [DOI] [PubMed] [Google Scholar]
- Oliner JD, Andresen JM, Hansen SK, Zhou S, Tjian R 1996. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev 10: 2903–2911 [DOI] [PubMed] [Google Scholar]
- Osborne TF, Espenshade PJ 2009. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: What a long, strange tRIP it's been. Genes Dev 23: 2578–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, et al. 2010. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem 285: 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH 2008. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci 105: 9793–9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L 2004. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429: 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X 2009. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab 9: 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson RB 2003. The SREBP pathway—Insights from Insigs and insects. Nat Rev Mol Cell Biol 4: 631–640 [DOI] [PubMed] [Google Scholar]
- Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev 14: 2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P 2007. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci 104: 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P 2005. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434: 113–118 [DOI] [PubMed] [Google Scholar]
- Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y 2010. AMPK and SIRT1: A long-standing partnership? Am J Physiol Endocrinol Metab 298: E751–E760 doi: 10.1152/ajpendo.00745.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Verdin E 2007. Sirtuins: Critical regulators at the crossroads between cancer and aging. Oncogene 26: 5489–5504 [DOI] [PubMed] [Google Scholar]
- Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev 14: 2831–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegmiller AC, Dobrosotskaya I, Goldstein JL, Ho YK, Brown MS, Rawson RB 2002. The SREBP pathway in Drosophila: Regulation by palmitate, not sterols. Dev Cell 2: 229–238 [DOI] [PubMed] [Google Scholar]
- Smith JJ, Kenney RD, Gagne DJ, Frushour BP, Ladd W, Galonek HL, Israelian K, Song J, Razvadauskaite G, Lynch AV, et al. 2009. Small molecule activators of SIRT1 replicate signaling pathways triggered by calorie restriction in vivo. BMC Syst Biol 3: 31 doi: 10.1186/1752-0509-3-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A, Ericsson J 2003. Transcription-dependent degradation controls the stability of the SREBP family of transcription factors. Proc Natl Acad Sci 100: 13833–13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410: 227–230 [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM 2008. Fat and beyond: The diverse biology of PPARγ. Annu Rev Biochem 77: 289–312 [DOI] [PubMed] [Google Scholar]
- Van Gilst MR, Hadjivassiliou H, Yamamoto KR 2005. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci 102: 13496–13501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varady KA, Hellerstein MK 2008. Do calorie restriction or alternate-day fasting regimens modulate adipose tissue physiology in a way that reduces chronic disease risk? Nutr Rev 66: 333–342 [DOI] [PubMed] [Google Scholar]
- Watts JL 2009. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol Metab 20: 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Browse J 2006. Dietary manipulation implicates lipid signaling in the regulation of germ cell maintenance in C. elegans. Dev Biol 292: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HB, Golic KG 2004. Gene deletions by ends-in targeting in Drosophila melanogaster. Genetics 168: 1477–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Usui I, Kanatani Y, Matsuya Y, Tsuneyama K, Fujisaka S, Bukhari A, Suzuki H, Senda S, Imanishi S, et al. 2009. Treatment with SRT1720, a SIRT1 activator, ameliorates fatty liver with reduced expression of lipogenic enzymes in MSG mice. Am J Physiol Endocrinol Metab 1: 1. [DOI] [PubMed] [Google Scholar]
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, et al. 2006. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442: 700–704 [DOI] [PubMed] [Google Scholar]
- Yu J, Auwerx J 2009. The role of sirtuins in the control of metabolic homeostasis. Ann NY Acad Sci 1173: E10–E19 doi: 10.1111/j.1749-6632.2009.04952.x [DOI] [PMC free article] [PubMed] [Google Scholar]