Abstract
Peptides perform many roles in cell–cell signaling; examples include neuropeptides, hormones, and growth factors. Although the vast majority of known neuropeptides are produced in the secretory pathway, a number of bioactive peptides are derived from cytosolic proteins. For example, the hemopressins are a family of peptides derived from alpha and beta hemoglobin which bind to the CB1 cannabinoid receptor, functioning as agonists or antagonists/inverse agonists depending on the size of the peptide. However, the finding that peptides derived from cytosolic proteins can affect receptors does not prove that these peptides are true endogenous signaling molecules. In order for the hemopressins and other peptides derived from cytosolic proteins to be considered neuropeptide-like signaling molecules, they must be synthesized in brain, they must be secreted in levels sufficient to produce effects, and either their synthesis or secretion should be regulated. If these criteria are met, we propose the name “non-classical neuropeptide” for this category of cytosolic bioactive peptide. This would be analogous to the non-classical neurotransmitters, such as nitric oxide and anandamide, which are not stored in secretory vesicles and released upon stimulation but are synthesized upon stimulation and constitutively released. We review some examples of cytosolic peptides from various protein precursors, describe potential mechanisms of their biosynthesis and secretion, and discuss the possibility that these peptides are signaling molecules in the brain, focusing on the criteria that these peptides would have to fill in order to be considered non-classical neuropeptides.
Key words: endocannabinoid, neurotransmitter, peptidase, peptide, protease, proteasome
INTRODUCTION
Cells communicate via a range of signaling molecules, including classical neurotransmitters, non-classical neurotransmitters, hormones, growth factors, and other molecules. A large number of signaling molecules have well-established roles, and many additional molecules have been proposed to function in signaling but are still being validated. For a molecule to be considered a bona fide cell–cell signaling molecule, it must fulfill several criteria. First, the molecule must be secreted from the signaling cell. Second, it must interact with a target cell to induce a change in cellular activity. Typically, this is accomplished by the binding of the molecule to a receptor on the target cell, although some signaling molecules interact with other targets such as enzymes or transcription factors. Third, the signaling molecule must be regulated in either a temporal or spatial way; temporal regulation is more common for neurotransmitters while spatial regulation is generally found during development. Finally, there must be a mechanism for the signaling molecule to be terminated, either via uptake into cells and/or degradation (1). This review is focused on signaling molecules in brain and specifically a class of peptide that we have termed “non-classical neuropeptides.” Major emphasis is placed on the criteria that need to be met for these molecules to be accepted as genuine signaling molecules.
CLASSICAL AND NON-CLASSICAL NEUROTRANSMITTERS
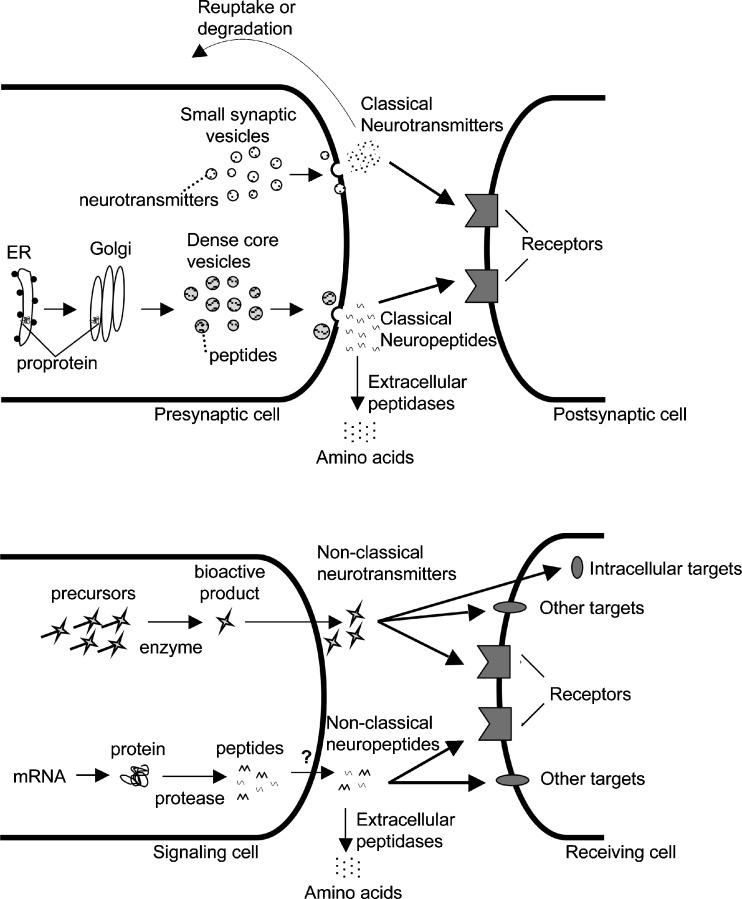
Classical neurotransmitters are released from neurons and act on neighboring cells (1). These neurotransmitters are often referred to as “small-molecule transmitters.” In general, they are synthesized in the presynaptic terminals of neurons and are packaged into synaptic vesicles. Upon stimulation of the neuron (usually by an action potential), these vesicles fuse with the presynaptic membrane and release neurotransmitters into the synaptic cleft, where neurotransmitters can travel to the postsynaptic cell and elicit their effect (Fig. 1). Termination of the signal is usually accomplished by reuptake of the neurotransmitter into the presynaptic cell where it is recycled back into the secretory vesicles. Examples of small-molecule transmitters include amino acids such as glutamate and monoamines such as dopamine and serotonin (1).
Fig. 1.
Schematic of classical and non-classical neurotransmitters and neuropeptides. Top panel Classical neurotransmitters are typically synthesized in the presynaptic cell and packaged into synaptic vesicles. Upon stimulation, vesicles fuse with the membrane and release the neurotransmitters into the synaptic cleft where the neurotransmitters can bind to postsynaptic receptors to elicit their effects. Neurotransmitters in the synaptic cleft are removed by reuptake or are degraded. Classical neuropeptides are derived from precursors synthesized in the rough endoplasmic reticulum (ER). Proteolytic processing begins in the trans-Golgi network or immature secretory vesicles and continues in the maturing vesicles. Classical neuropeptides are packaged into dense core vesicles, which can also contain classical neurotransmitters. Upon stimulation, vesicles fuse with the membrane to release their contents. Classical neuropeptides typically elicit their effects on G-protein coupled receptors and are eliminated by extracellular peptidases. Bottom panel Non-classical neurotransmitters and the proposed pathway of non-classical neuropeptides. Non-classical neurotransmitters are synthesized from their precursors upon stimulation of the signaling cell and are not stored in vesicles. Examples such as anandamide and 2-arachidonoylglycerol are derived from lipid precursors while nitric oxide is produced from arginine. Once produced, non-classical neurotransmitters are rapidly secreted and act upon receptors or other targets which can be membrane-bound or intracellular. Non-classical signaling can proceed in a retrograde manner as it does not require synaptic transmission. In our proposed model, non-classical neuropeptides are derived from intracellular proteins by the action of proteases. The peptides are then secreted by an unknown mechanism, and elicit their effects on receptors or other targets. Although some of these targets may be distinct from the targets of classical neurotransmitters, some may also be common. For example, the hemopressins, a family of putative non-classical neuropeptides, bind to CB1 receptors which are also the target of the non-classical neurotransmitters anandamide and 2-arachidonoylglycerol
In contrast to the classical neurotransmitters, the non-classical neurotransmitters are synthesized upon stimulation of the signaling cell. These molecules, also termed neuromodulators, are not packaged in vesicles or secreted via a regulated pathway (Fig. 1). Instead, non-classical neurotransmitters are rapidly synthesized in the cytosol and diffuse out of the cell, often in the absence of specific transporters (1). For example, nitric oxide is produced from arginine by nitric oxide synthase and diffuses out of the cell into neighboring cells where it has a number of functions including activation of guanylate cyclase (2). Other non-classical neurotransmitters include the CB1 cannabinoid receptor ligands anandamide and 2-arachidonoylglycerol; these are produced by the action of lipases on lipids present in brain membranes (3).
NEUROPEPTIDES
Peptides represent a large class of cell–cell signaling molecules. In the endocrine system, many hormones are peptides; examples include insulin and oxytocin. The term neuropeptide is usually reserved for peptides secreted from neurons that signal nearby cells, although the term has been used to refer to peptides secreted from a range of neuronal cells and not specifically neurons. Neuropeptides take part in a wide array of functions such as body weight regulation, pain, anxiety, memory, and more (4). They are produced by selective cleavage of precursors at specific well-defined sites, usually containing basic amino acids (5). The precursors, usually referred to as prohormones, generally have no biological activity until cleaved into the mature peptide forms. Initially, an endopeptidase such as prohormone convertase 1/3 or prohormone convertase 2 cleaves after basic residues (Lys, Arg) and then carboxypeptidase E removes the C-terminal basic amino acids (6,7). Neuropeptide precursors are synthesized in the rough endoplasmic reticulum and transported to the trans-Golgi network, where they are cleaved into intermediate neuropeptides, packaged into secretory vesicles, and further processed into the mature forms. Like the classical neurotransmitters, the neuropeptides are secreted in an activity-dependent manner upon depolarization of the cell (Fig. 1). However, unlike classical neurotransmitters, neuropeptides do not go through reuptake; rather, the secreted molecules are broken down by extracellular peptidases (1). Examples of neuropeptides include enkephalin, dynorphin, and neuropeptide Y. Because the well-known neuropeptides are analogous to classical neurotransmitters, we refer to them as “classical neuropeptides” (Fig. 2) to distinguish them from another category of bioactive peptides, the “non-classical neuropeptides,” which are described below.
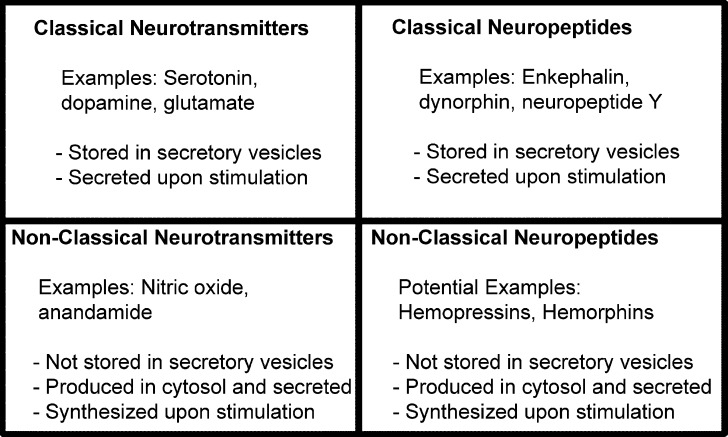
Fig. 2.
Summary of the key properties of classical and non-classical neurotransmitters and neuropeptides. Examples of molecules in each category are listed
BIOACTIVE PEPTIDES FROM CYTOSOLIC PROTEINS
Over the past several decades, numerous bioactive peptides have been reported that arise from cytosolic or mitochondrial protein precursors (8,9). In general, the scientific community has not accepted these as bona fide endogenous signaling molecules because of the dogma that neuropeptides are produced in the secretory pathway and released from cells upon stimulation. However, these intracellular protein fragments may represent a new non-classical type of neuropeptide that is synthesized and released from the cytosol, in an analogous fashion to the non-classical neurotransmitters (Figs. 1 and 2). There are examples of bioactive peptide hormones that are regulated via biosynthesis rather than regulated by secretion. For example, angiotensin II, a peptide that causes vasoconstriction, is derived from circulating angiotensinogen by the action of renin and angiotensin converting enzyme (10), and bradykinin, which causes vasodilation, is produced from circulating kininogen by plasma kallikrein and tissue kallikreins (11). Thus, there is precedent for the idea that bioactive peptides can be generated “on demand” and not just synthesized in advance and secreted on demand. Several bioactive peptides that are present in brain and derived from cytosolic or mitochondrial proteins are described in this section; these may represent non-classical neuropeptides.
Hemopressin and Other Hemoglobin-Derived Peptides
The two main chains of hemoglobin, α and β, are precursors of many peptides, some of which have been found to have biological activity. Hemoglobin is a well-known cytosolic protein. Although α and β hemoglobin are the main constituent of red blood cells, hemoglobin mRNA and/or protein have recently been found in many other cell types including activated macrophages, lens cells, lung epithelial type II and Clara cells, kidney messangeal cells, and endometrial epithelial and stromal cells (12–18). Most recently, α and β hemoglobin mRNA and protein have been found in brain, including nigral and mesencephalic dopaminergic, striatal GABAergic, and cortical pyramidal neurons and glial cells (19,20). Also, cultured neurons express hemoglobin mRNA (19,20). Because many of the hemoglobin-derived peptides found in brain are different from those found in blood or heart, it is likely that the brain hemoglobin peptides are produced in brain and not a reflection of blood contamination (21).
Hemopressin is a 9 amino acid peptide (Table I) derived from α hemoglobin that was originally identified using an enzyme-substrate capture approach with endopeptidase 24.15, a cytosolic enzyme (22,23). The 9-residue form of hemopressin is an inverse agonist of CB1 cannabinoid receptors. Hemopressin is orally active and is antinociceptive (24,25). Hemopressin has been shown to decrease blood pressure by lowering the systemic vascular resistance through the endogenous release of nitric oxide (26,27). Recently, longer forms of hemoglobin have been discovered (Table I); RVD-hemopressin-α and VD-hemopressin-α are both derived from the α chain of hemoglobin, and VD-hemopressin-β is from the β chain (28). All three of these hemopressins are agonists of the CB1 cannabinoid receptors, and VD-hemopressin-β is also an agonist of the CB2 cannabinoid receptors (28). Both RVD-hemopressin-α and VD-hemopressin-α were shown to be upregulated in mouse brain following ischemia (28) and in Cpefat/fat mice (21).
Table I.
Sequences of the Peptides
| Precursor | Peptide | Sequence | Species and Reference |
|---|---|---|---|
| Hemoglobin alpha | Hemopressin | PVNFKFLSH | Rat (22) |
| Hemoglobin alpha | VD-Hemopressin-α | VDPVNFKLLSH | Mouse (28) |
| Hemoglobin alpha | RVD-Hemopressin-α | RVDPVNFKLLSH | Mouse (28) |
| Hemoglobin alpha | Bradykinin-potentiating peptide | ASHLPSDFTPAVHASL | Bovine (45) |
| Hemoglobin alpha | Bradykinin-potentiating peptide | LANVST | Bovine (44) |
| Hemoglobin alpha | Neokyotorphin | TSKYR | Bovine (40) |
| Hemoglobin alpha | Kyotorphin | YR | Bovine (41) |
| Hemoglobin beta | VD-Hemopressin-β | VDPENFRLLGNM | Mouse (28) |
| Hemoglobin beta | Hemorphin-4 | YPWT | Bovine (31) |
| Hemoglobin beta | Hemorphin-5 | YPWTQ | Bovine (31) |
| Hemoglobin beta | Hemorphin-7 | YPWTQRF | Human and Bovine (30) |
| Hemoglobin beta | VV-hemorphin-7 | VVYPWTQRF | Human (32) |
| Hemoglobin beta | LVV-hemorphin-4 | LVVYPWTQRF | Bovine and Human (33) |
| Hemoglobin beta | VV-Hemorphin-5 (Valorphin) | VVYPWTQ | Bovine (34) |
| Cytochrome c oxidase subunit VIII | Mitocryptide-1 | LSFLIPAGWVLSHLDHYKRSS AA | Porcine (59) |
| Cytochrome b | Mitocryptide-2 | formyl-MTNIRKSHPLMKIIN | Porcine (61) |
| Prothymosin alpha | C-terminal peptide | TKKQKTDEDD | Human (67) |
| Prothymosin alpha | Thymosin Alpha 1 | SDAAVDTSSEITTKDLKEKKE VVEEAEN | Bovine (68) |
| Phosphatidylethanolamine-binding protein | HCNP | Acetyl-AADISQWAGPL | Rat (54) |
| DBI | ODN | QATVGDVNTDRPGLLDLK | Rat (47) |
| DBI | TTN | TQPTDEEMLFIYSHFKQA TVGDVNTDRPGLLDLK | Rat (49) |
The hemorphins are peptides derived from the β chain of hemoglobin which bind and stimulate opiate receptors (29,30). The first of the hemorphins, hemorphin-4 (YPWT), was isolated from enzyme-treated bovine blood (31). Hemorphins were then found to be endogenous in the brain, and more sequences of this group have been discovered using conventional approaches (30,32–34) and also by peptidomics analyses (Table I). The hemorphins inhibit the actions of angiotensin converting enzyme, thereby lowering arterial blood pressure and are antinociceptive (35,36). They also have bradykinin-potentiating activity (37) and can bind to and inhibit insulin-regulated aminopeptidase, which was previously named the angiotensin-4 receptor (38). LVV-hemorphin-7 has recently been shown to be secreted from stimulated synaptoneurosomes (39).
Neokyotorphin (Table I) is derived from the C terminus of the α hemoglobin chain (40). Neokyotorphin binds to angiotensin receptors and has been shown to exhibit analgesic activity (40). Kyotorphin, the two amino acid peptide YR, has also been shown to have analgesic activity (41). Neokyotorphin is secreted from erythrocytes and stimulates proliferation of tumor cells and fibroblasts (42,43).
Other peptides derived from hemoglobin have also been shown to have biological activities. For example, peptides corresponding to residues 110–125 and 129–134 of the α hemoglobin sequence have been shown to exhibit bradykinin potentiating activity (Table I) (44,45).
Diazepam-Binding Inhibitor
Diazepam-binding inhibitor (DBI), also known as acyl-CoA-binding protein, is the precursor of endozepines, compounds that are able to displace diazepam from its binding sites (46). Two of these peptides, octadecaneuropeptide (ODN) and triakontatetraneuropeptide (TTN), have greater biological activity than the parent protein (Table I). DBI and its products ODN and TTN are capable of modulating a cell’s response to γ-aminobutyric acid (GABA) (47–49). The peptides displace diazepam binding to GABAA receptors and thereby inhibit GABAergic transmission (46,50). ODN acts as a ligand for central-type benzodiazepine receptors, and TTN acts through the peripheral-type benzodiazepine receptors to stimulate the biosynthesis of neurosteroids (51). Both ODN and TTN also act in vivo to elicit proconflict action (47,49).
Hippocampal Cholinergic Neurostimulating Peptide
Hippocampal cholinergic neurostimulating peptide (HCNP) is the N-terminal fragment of phosphatidylethanolamine-binding protein (Table I). It enhances the differentiation of hippocampal neurons and can also modulate cardiac response (52–55) HCNP also plays a role in activating acetylcholine release by stimulating choline acetyltransferase secretion (56). Recently, it has been suggested that HCNP plays a role in Alzheimer’s disease and hypoxia (56,57). Both HCNP and its precursor protein have been found to be secreted, but it is not yet known how this secretion occurs (58).
Mitocryptides
Mitocryptide-1 is a fragment of cytochrome C oxidase subunit 8. It activates neutrophils via Gi2-type G proteins and stimulates β-hexosaminidase secretion and chemotaxis (59,60). Mitocryptide-2 is derived from mitochondrial cytochrome b and also activates neutrophils, leading to chemotaxis and β-hexosaminidase release (61). Sequences of the peptides are shown in Table I.
Thymosins
Beta thymosins are actin sequestering peptides (62). Thymosin β4 inhibits inflammation and stimulates wound healing by promoting angiogenesis (63,64).
Thymosin β4 and smaller peptides are secreted from tissues and have been implicated in controlling apoptosis (62,65,66). The 10 amino acid C-terminal fragment of prothymosin α (Table I) is generated by caspase cleavage and stimulates lymphocytes (67). Thymosin alpha 1, a 28 amino acid peptide derived from prothymosin α (Table I), plays a role in stimulation of the immune response and enhances the antitumor response (68–70).
CRITERIA FOR NEUROPEPTIDES
Many of the peptides described above have been studied for years by a number of investigators but are not universally accepted as signaling molecules because they are produced from cytosolic proteins. However, it is possible that they represent non-classical neuropeptides. Based on established criteria for the acceptance of classical neurotransmitters and neuropeptides, we propose the following criteria for non-classical neuropeptides:
The peptide is synthesized in brain cells.
The peptide is secreted from brain cells in physiologically relevant levels.
The peptide is either synthesized or secreted in a regulated fashion.
The peptide is able to influence the function of another cell.
Another criterion for neurotransmitters is the demonstration of a mechanism for termination of action, often uptake. However, neuropeptides are thought to be degraded by extracellular peptidases with broad specificities; therefore, selective peptidases or other mechanisms of elimination are not needed for secreted peptides.
All of the peptides described above have met the 4th criteria, and some such as LVV-hemorphin-7 have been shown to be present in brain and secreted in response to stimulation. Further proof of the proposed role as non-classical neuropeptides requires demonstration of a physiological effect when the peptide is removed from a biological system. For classical neuropeptides, this can be achieved by gene knockout or mRNA knockdown approaches. However, for peptides derived from cytosolic proteins, these techniques will be difficult to interpret because the proteins have additional roles. Alternative approaches, such as blocking the biosynthesis and/or secretion, can be used to help demonstrate the proposed biological role of these peptides. For this, it is essential to understand the mechanism of synthesis and secretion. The rest of this review will focus on the possible mechanisms for synthesis and secretion of non-classical neuropeptides, and consideration of how these processes can be regulated.
BIOSYNTHESIS OF PEPTIDES
There are three different ways that cells make peptides. One is to simply link amino acids together using cellular ligases, without involving ribosomes and protein synthesis; this is the route used to make the cellular antioxidant peptide glutathione, a tripeptide. It is likely that this approach is also used to make N-acetylaspartylglutamate (commonly known as NAAG), a dipeptide that has been called a neuropeptide although it is possible that its mechanism of action is after hydrolysis into glutamate and not as a functional peptide. Another approach that cells may use to make peptides is by translation of short open reading frames within RNAs. In this case, the peptide is made directly using protein synthetic machinery. A possible example of this is the peptide named humanin (from human mitochondrial 16s ribosomal RNA) or the rat homolog, rattin (from rat mitochondrial 16s ribosomal RNA). However, it is not clear if these peptides are truly found in vivo or if the immunoreactive peptides detected in many studies represent another peptide. The most common method for the generation of peptides is by enzymatic cleavage of proteins. Because protein precursors exist for bioactive peptides listed in Table I, proteolytic processing is clearly the route for the production of these peptides.
PROTEASES
Many intracellular proteases are well studied, and it is possible that one or more of these are responsible for the generation of the cytosolic/mitochondrial peptides described above. Because most intracellular proteases are highly regulated, knowledge of the enzymatic pathway will be important in understanding the regulation of the peptides. Below, we discuss some of the well-known proteases and address the possibility of their involvement in the production of the bioactive intracellular peptides.
Proteasome
The ubiquitin-proteasome system is found in all cell types and is involved in the turnover of many intracellular proteins. This system is a likely candidate for the formation of the observed cytosolic peptides because many of these peptides are produced by cleavage at sites favored by the proteasome: hydrophobic residues (except for Ile) and basic residues. Proteasomes are large protein complexes which include a core particle (20S proteasome) and two regulatory particles (19S cap). The core subunit has different activity for substrates, chymotrypsin-like, trypsin-like, and peptidyl-glutamyl peptide-hydrolyzing activity (71). Together, this comprises the 26S proteasome which degrades proteins in a ubiquitin-dependent manner and produces peptides 4–25 amino acids in length. Some proteasome-generated peptides are used for presentation on the cell surface by major histocompatibility complex class I molecules (72). Proteasome activity levels can be altered in response to some physiological conditions and oxidative stress (73). Inhibition of the proteasome can lead to cell arrest and apoptosis (74).
Calpains
Calpains are broadly expressed calcium-dependent cysteine proteases which have 15 isoforms (75). Well-studied calpains are µ-calpain, which is activated by low calcium concentrations (µM range) and m-calpain, which is activated by higher, mM concentrations of calcium (76). Calpastatin is an endogenous protein inhibitor of calpain (77). Both µ- and m-calpains have pH optima of 7.2–8.2 and are found intracellularly, mostly associated with subcellular organelles. Calpains are not known to cleave small peptides but rather produce large polypeptides by cleaving at specific sites within proteins (78). The specificity of calpain cleavage is not well defined and likely represents a combination of conformation (79) and sequence such as hydrophobic amino acids (80). Cleavage specificity can be affected by phosphorylation, which can also change the rate of calpain actions (75).
Calpains may be involved in the formation of the observed cytosolic peptides based on their intracellular location and preference for cleaving at hydrophobic residues. But, because the known calpain products are generally large polypeptides while the majority of the cytosolic peptides are small, a role for calpains in the production of the cytosolic peptides would represent a novel role for these enzymes.
Caspases
Caspases are cysteine proteases that are key factors in the processes of apoptosis and inflammation. They are synthesized as inactive precursors which are activated in response to apoptosis and in some cases of disease (81,82). The substrate specificity of caspases includes a requirement for aspartic acid in the P1 position of the substrate. Also, they have selectivity for glutamic acid in the P3 position and do not accept charged residues in the P1′ position (83). It is unlikely that caspases are responsible for the formation of the cytosolic peptides because all of the bioactive peptides listed in Table I, and most other cytosolic peptides that have been detected, are not the products of cleavage at Asp residues.
Cathepsins
Cathepsins are lysosomal proteases. There are 15 known cathepsins, 11 of which are cysteine proteases, two are serine proteases, and two are aspartyl proteases (84–87) Cathepsins are typically activated at low pH, as found in lysosomes, but some are active at neutral pH and secreted out of the lysosome into the cytoplasm (87–90).
There is evidence that some cytosolic peptides result from cleavage by the cathepsins. Neokyotorphin and VV-hemorphin-7 have been shown to be generated by incubation of hemoglobin with cathepsin D, an aspartyl protease (91,92). The cathepsins are, therefore, candidates for production of the hemoglobin-derived peptides and other cytosolic/mitochondrial peptides. For the cathepsins to perform this function in vivo, either the cytosolic molecules need to be transported into lysosomes or the cathepsins need to get out of the lysosomes into the cytosol; both of these processes are known to occur (90,93).
Intramembrane Proteases
Since the peptides discussed in this review do not come from membrane bound proteins, it is not likely that they are formed by intramembrane proteases. However, intramembrane proteases contribute to the production of other brain peptides. For example, beta and gamma secretases are transmembrane proteases which cleave amyloid precursor protein into amyloid beta peptide (94). Beta secretase (memapsin 2) is an aspartic acid protease which consists of a C-terminal cytosolic domain, a transmembrane domain, and a catalytic N-terminal ectodomain (95). Gamma secretase is a protease complex composed of nicastrin, PEN-2, APH-1, and the catalytic subunit, presenilin (96). Gamma secretase is involved in processing of amyloid precursor protein and notch protein (97). Rhomboids are intramembrane serine proteases which cleave transmembrane proteins resulting in release of a part of the protein that was tethered to the membrane (98,99). Signal peptide peptidases are transmembrane aspartyl proteases which cleave in the transmembrane region of their substrates, signal peptides (100,101).
Mitochondrial Proteases
A variety of proteases are present in the mitochondria, and although most of these are thought to function within the mitochondria, some have been shown to be released into the cytosol in an active form (102). An example is the serine protease HtrA2/Omi, which plays a role in apoptosis (103). HtrA2/Omi has protease activity within mitochondria and also in the cytosol after it is released (103). Since it is secreted to the cytosol, HtrA2/Omi may cleave cytosolic proteins to form bioactive peptides. Furthermore, HtrA2/Omi cleaves the amyloid precursor protein in the mitochondria to produce the C161 fragment, which is then released into the cytosol (104). A mechanism such as this could also be responsible for the bioactive peptides described above, especially for the microcryptides which are likely to be formed in the mitochondria and then released to the cytosol.
Oligopeptidases
A number of intracellular oligopeptidases have been described which process peptides of ∼5–20 or ∼5–30 residues into smaller peptides (depending on the enzyme). Although these enzymes cannot convert proteins into bioactive peptides, they may contribute to the biosynthesis of the observed peptides following an endoprotease step. Alternatively, these enzymes may function in the degradation of the bioactive peptides. Thimet-oligopeptidase (endopeptidase 24.15) and neurolysin (endopeptidase 24.16) are intracellular peptidases which are located primarily in the cytosol in most cell types (105,106). Endopeptidase 24.15 is known to play a role in the processing of peptides produced by the proteasome (107). Insulin-degrading enzyme is also active in the cytosol and can process a range of peptides, in addition to insulin (108). Prolyl-oligopeptidase catalyzes the cleavage of several bioactive peptides that contain proline residues (109). Tripeptidyl peptidase II works downstream of the proteasome and cleaves N-terminal tripeptides from oligopeptides (110). Although some of these enzymes can be secreted and function outside of the cell, all are active within the intracellular compartments and are, therefore, candidates for cytosolic peptide formation.
Other Intracellular Proteases
In addition to the well-known proteases described above, there are a number of additional proteases that are present within the cytosol. Autophagins are cysteine proteinases which are activated during autophagy (111,112). Alpha secretases are zinc metalloproteinases which cleave amyloid precursor protein at the Lys16-Leu17 bond in a nonamyloidogenic processing pathway (94,113). Separase is a cysteine protease involved in separation of sister chromatids by cleaving the cohesion molecule during the transition from metaphase to anaphase in the process of mitosis (114,115). These enzymes are limited in their specificity of substrates thus making them unlikely candidates in the formation of the cytosolic peptides.
SECRETION OF CYTOSOLIC PROTEINS/PEPTIDES
Secretory proteins have N-terminal signal peptides that are removed by a signal peptidase (116). The signal peptide is a hydrophobic sequence which is required for proteins to be transported out of the cell. The signal peptide promotes translocation of the protein to the endoplasmic reticulum, and the proteins are secreted via vesicular transport. The question arises, how then would the cytosolic peptides be secreted if they do not have a signal peptide? Without a signal peptide, a protein cannot go through the usual endoplasmic reticulum secretory pathway.
There are many examples of unconventionally secreted proteins, and this pathway has been referred to as ER/Golgi independent protein secretion. The annexins are calcium-dependent phospholipid-binding proteins. Although annexins lack signal peptides, some members of the annexin family have been found to be secreted and function in anti-inflammation (117). For example, annexin A1 is synthesized and rapidly secreted in response to a stimulus or treatment with glucocorticoids; the mechanism of secretion is unknown (118). Annexin A2 is another intracellular protein that is secreted and functions outside of the cell (119). Plasminogen activator inhibitor-2 is a serine protease inhibitor which has been shown to be secreted without cleavage of a signal peptide (120). Some of the other well-known examples of cytosolic proteins that are secreted include interleukins, fibroblast growth factors, phosphatidylethanolamine-binding protein, galectin-1, glycosylation-inhibiting factor, and macrophage migration inhibitory factor (121–126). These are just a few of the many examples of cytosolic proteins that are secreted from intact cells.
There are several possible mechanisms by which cytosolic proteins and peptides can be secreted (126). Translocation may occur directly across the plasma membrane or through vesicle mediators such as secretory lysosomes, microvesicles, or endosomal internal vesicles that are subsequently released as exosomes (126). Fibroblast growth factor 2 is secreted via translocation across the plasma membrane (127). Interleukin-1β secretion is induced with monocyte activation, and the secretion of this cytokine may involve exocytosis of endocytic vesicles (128). Recently, P2X7 receptor activation has been shown to be crucial for the secretion of interleukin-1β; the mechanism is thought to involve P2X7 receptor-regulated formation of multivesicular bodies and exosome-mediated secretion of the interleukin (129). It is possible that these mechanisms are used for the secretion of cytosolic peptides or that a combination of mechanisms exist for the different peptides.
SUMMARY
Although a number of biologically active peptides are produced from cytosolic/mitochondrial proteins, additional studies are needed to firmly establish these as non-classical neuropeptides. The mechanisms for their synthesis and secretion need to be identified. Regulation of synthesis and/or secretion has been shown for some of the cytosolic peptides and needs to be demonstrated for the others. Because the cytosolic peptides do not go through the classical ER/Golgi-mediated secretory pathway and do not get packaged into conventional secretory vesicles, the synthesis of the peptides is a likely point of regulation, as is the case with non-classical neurotransmitters such as nitric oxide. In addition to the handful of peptides discussed in this review, there are a very large number of additional intracellular peptides that have been detected in peptidomic studies, raising the possibility that these peptides represent a large and diverse group of cell–cell signaling molecules.
Acknowledgments
This work was supported in part by National Institutes of Health grant DA-04494 (L.D.F.). Thanks to Prof. Lakshmi Devi and Emer Ferro for helpful discussions.
References
- 1.Schwartz JH. Neurotransmitters. In: Kandel ER, Schwartz JH, Jessell TM, editors. Principles of Neural Science. 4. New York: McGraw-Hill; 2000. pp. 280–297. [Google Scholar]
- 2.Jaffrey SR, Snyder SH. Nitric oxide: a neural messenger. Annu Rev Cell Dev Biol. 1995;11:417–440. doi: 10.1146/annurev.cb.11.110195.002221. [DOI] [PubMed] [Google Scholar]
- 3.Boyd ST. The endocannabinoid system. Pharmacotherapy. 2006;26(12 Pt 2):218S–221S. doi: 10.1592/phco.26.12part2.218S. [DOI] [PubMed] [Google Scholar]
- 4.Fricker LD. Neuropeptide-processing enzymes: applications for drug discovery. Aaps J. 2005;7(2):E449–E455. doi: 10.1208/aapsj070244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eipper BA, Mains RE, Herbert E. Peptides in the nervous system. Trends Neurosci. 1986;9:463–468. doi: 10.1016/0166-2236(86)90149-9. [DOI] [Google Scholar]
- 6.Zhou A, Webb G, Zhu X, Steiner DF. Proteolytic processing in the secretory pathway. J Biol Chem. 1999;274(30):20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 7.Fricker LD. Carboxypeptidase E. Annu Rev Physiol. 1988;50:309–321. doi: 10.1146/annurev.ph.50.030188.001521. [DOI] [PubMed] [Google Scholar]
- 8.Skold K, Svensson M, Kaplan A, Bjorkesten L, Astrom J, Andren PE. A neuroproteomic approach to targeting neuropeptides in the brain. Proteomics. 2002;2(4):447–454. doi: 10.1002/1615-9861(200204)2:4<447::AID-PROT447>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 9.Che FY, Zhang X, Berezniuk I, Callaway M, Lim J, Fricker LD. Optimization of neuropeptide extraction from the mouse hypothalamus. J Proteome Res. 2007;6(12):4667–4676. doi: 10.1021/pr060690r. [DOI] [PubMed] [Google Scholar]
- 10.Leung PS, Carlsson PO. Tissue renin-angiotensin system: its expression, localization, regulation and potential role in the pancreas. J Mol Endocrinol. 2001;26(3):155–164. doi: 10.1677/jme.0.0260155. [DOI] [PubMed] [Google Scholar]
- 11.Yousef GM, Diamandis EP. The new human tissue kallikrein gene family: structure, function, and association to disease. Endocr Rev. 2001;22(2):184–204. doi: 10.1210/er.22.2.184. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Zeng M, Stamler JS. Hemoglobin induction in mouse macrophages. Proc Natl Acad Sci USA. 1999;96(12):6643–6647. doi: 10.1073/pnas.96.12.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wride MA, Mansergh FC, Adams S, Everitt R, Minnema SE, Rancourt DE, et al. Expression profiling and gene discovery in the mouse lens. Mol Vis. 2003;9:360–396. [PubMed] [Google Scholar]
- 14.Bhaskaran M, Chen H, Chen Z, Liu L. Hemoglobin is expressed in alveolar epithelial type II cells. Biochem Biophys Res Commun. 2005;333(4):1348–1352. doi: 10.1016/j.bbrc.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dassen H, Kamps R, Punyadeera C, Dijcks F, de Goeij A, Ederveen A, et al. Haemoglobin expression in human endometrium. Hum Reprod. 2008;23(3):635–641. doi: 10.1093/humrep/dem430. [DOI] [PubMed] [Google Scholar]
- 16.Newton DA, Rao KM, Dluhy RA, Baatz JE. Hemoglobin is expressed by alveolar epithelial cells. J Biol Chem. 2006;281(9):5668–5676. doi: 10.1074/jbc.M509314200. [DOI] [PubMed] [Google Scholar]
- 17.Nishi H, Inagi R, Kato H, Tanemoto M, Kojima I, Son D, et al. Hemoglobin is expressed by mesangial cells and reduces oxidant stress. J Am Soc Nephrol. 2008;19(8):1500–1508. doi: 10.1681/ASN.2007101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullal AJ, Litaker RW, Noga EJ. Antimicrobial peptides derived from hemoglobin are expressed in epithelium of channel catfish (Ictalurus punctatus, Rafinesque) Dev Comp Immunol. 2008;32(11):1301–1312. doi: 10.1016/j.dci.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Biagioli M, Pinto M, Cesselli D, Zaninello M, Lazarevic D, Roncaglia P, et al. Unexpected expression of alpha- and beta-globin in mesencephalic dopaminergic neurons and glial cells. Proc Natl Acad Sci USA. 2009;106(36):15454–15459. doi: 10.1073/pnas.0813216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter F, Meurers BH, Zhu C, Medvedeva VP, Chesselet MF. Neurons express hemoglobin alpha- and beta-chains in rat and human brains. J Comp Neurol. 2009;515(5):538–547. doi: 10.1002/cne.22062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelman JS, Sironi J, Castro LM, Ferro ES, Fricker LD. Hemopressins and other hemoglobin-derived peptides in mouse brain: Comparison between brain, blood, and heart peptidome and regulation in Cpe(fat/fat) mice. J Neurochem. 2010 (in press) [DOI] [PMC free article] [PubMed]
- 22.Rioli V, Gozzo FC, Heimann AS, Linardi A, Krieger JE, Shida CS, et al. Novel natural peptide substrates for endopeptidase 24.15, neurolysin, and angiotensin-converting enzyme. J Biol Chem. 2003;278(10):8547–8555. doi: 10.1074/jbc.M212030200. [DOI] [PubMed] [Google Scholar]
- 23.Dale CS, Pagano Rde L, Rioli V. Hemopressin: a novel bioactive peptide derived from the alpha1-chain of hemoglobin. Mem Inst Oswaldo Cruz. 2005;100(Suppl 1):105–106. doi: 10.1590/s0074-02762005000900017. [DOI] [PubMed] [Google Scholar]
- 24.Heimann AS, Gomes I, Dale CS, Pagano RL, Gupta A, de Souza LL, et al. Hemopressin is an inverse agonist of CB1 cannabinoid receptors. Proc Natl Acad Sci USA. 2007;104(51):20588–20593. doi: 10.1073/pnas.0706980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dale CS, Pagano Rde L, Rioli V, Hyslop S, Giorgi R, Ferro ES. Antinociceptive action of hemopressin in experimental hyperalgesia. Peptides. 2005;26(3):431–436. doi: 10.1016/j.peptides.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 26.Blais PA, Cote J, Morin J, Larouche A, Gendron G, Fortier A, et al. Hypotensive effects of hemopressin and bradykinin in rabbits, rats and mice. A comparative study. Peptides. 2005;26(8):1317–1322. doi: 10.1016/j.peptides.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 27.Lippton H, Lin B, Gumusel B, Witriol N, Wasserman A, Knight M. Hemopressin, a hemoglobin fragment, dilates the rat systemic vascular bed through release of nitric oxide. Peptides. 2006;27(9):2284–2288. doi: 10.1016/j.peptides.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Gomes I, Grushko JS, Golebiewska U, Hoogendoorn S, Gupta A, Heimann AS, et al. Novel endogenous peptide agonists of cannabinoid receptors. Faseb J. 2009;23(9):3020–3029. doi: 10.1096/fj.09-132142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Q, Garreau I, Sannier F, Piot JM. Opioid peptides derived from hemoglobin: hemorphins. Biopolymers. 1997;43(2):75–98. doi: 10.1002/(SICI)1097-0282(1997)43:2<75::AID-BIP2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 30.Nyberg F, Sanderson K, Glamsta EL. The hemorphins: a new class of opioid peptides derived from the blood protein hemoglobin. Biopolymers. 1997;43(2):147–156. doi: 10.1002/(SICI)1097-0282(1997)43:2<147::AID-BIP8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 31.Brantl V, Gramsch C, Lottspeich F, Mertz R, Jaeger KH, Herz A. Novel opioid peptides derived from hemoglobin: hemorphins. Eur J Pharmacol. 1986;125(2):309–310. doi: 10.1016/0014-2999(86)90044-0. [DOI] [PubMed] [Google Scholar]
- 32.Glamsta EL, Meyerson B, Silberring J, Terenius L, Nyberg F. Isolation of a hemoglobin- derived opioid peptide from cerebrospinal fluid of patients with cerebrovascular bleedings. Biochem Biophys Res Commun. 1992;184(2):1060–1066. doi: 10.1016/0006-291X(92)90699-L. [DOI] [PubMed] [Google Scholar]
- 33.Barkhudaryan N, Gambarov S, Gyulbayazyan T, Nahapetyan K. LVV-hemorphin-4 modulates Ca2+/calmodulin-dependent pathways in the immune system by the same mechanism as in the brain. J Mol Neurosci. 2002;18(3):203–210. doi: 10.1385/JMN:18:3:203. [DOI] [PubMed] [Google Scholar]
- 34.Erchegyi J, Kastin AJ, Zadina JE, Qiu XD. Isolation of a heptapeptide Val-Val-Tyr-Pro- Trp-Thr-Gln (valorphin) with some opiate activity. Int J Pept Protein Res. 1992;39(6):477–484. doi: 10.1111/j.1399-3011.1992.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 35.Davis TP, Gillespie TJ, Porreca F. Peptide fragments derived from the beta-chain of hemoglobin (hemorphins) are centrally active in vivo. Peptides. 1989;10(4):747–751. doi: 10.1016/0196-9781(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 36.Lantz I, Glamsta EL, Talback L, Nyberg F. Hemorphins derived from hemoglobin have an inhibitory action on angiotensin converting enzyme activity. FEBS Lett. 1991;287(1–2):39–41. doi: 10.1016/0014-5793(91)80011-Q. [DOI] [PubMed] [Google Scholar]
- 37.Ianzer D, Konno K, Xavier CH, Stocklin R, Santos RA, de Camargo AC, et al. Hemorphin and hemorphin-like peptides isolated from dog pancreas and sheep brain are able to potentiate bradykinin activity in vivo. Peptides. 2006;27(11):2957–2966. doi: 10.1016/j.peptides.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Lee J, Mustafa T, McDowall SG, Mendelsohn FA, Brennan M, Lew RA, et al. Structure- activity study of LVV-hemorphin-7: angiotensin AT4 receptor ligand and inhibitor of insulin- regulated aminopeptidase. J Pharmacol Exp Ther. 2003;305(1):205–211. doi: 10.1124/jpet.102.045492. [DOI] [PubMed] [Google Scholar]
- 39.Annangudi SP, Luszpak AE, Kim SH, Ren S, Hatcher NG, Weiler IJ, et al. Neuropeptide Release Is Impaired in a Mouse Model of Fragile X Mental Retardation Syndrome. ACS Chemical Neuroscience. 2010 (in press) [DOI] [PMC free article] [PubMed]
- 40.Takagi H, Shiomi H, Fukui K, Hayashi K, Kiso Y, Kitagawa K. Isolation of a novel analgesic pentapeptide, neo-kyotorphin, from bovine brain. Life Sci. 1982;31(16-17):1733–1736. doi: 10.1016/0024-3205(82)90197-7. [DOI] [PubMed] [Google Scholar]
- 41.Takagi H, Shiomi H, Ueda H, Amano H. A novel analgesic dipeptide from bovine brain is a possible Met-enkephalin releaser. Nature. 1979;282(5737):410–412. doi: 10.1038/282410a0. [DOI] [PubMed] [Google Scholar]
- 42.Blishchenko EY, Mernenko OA, Yatskin ON, Ziganshin RH, Philippova MM, Karelin AA, et al. Neokyotorphin and neokyotorphin (1-4): secretion by erythrocytes and regulation of tumor cell growth. FEBS Lett. 1997;414(1):125–128. doi: 10.1016/S0014-5793(97)00991-5. [DOI] [PubMed] [Google Scholar]
- 43.Sazonova OV, Blishchenko EY, Tolmazova AG, Khachin DP, Leontiev KV, Karelin AA, et al. Stimulation of fibroblast proliferation by neokyotorphin requires Ca influx and activation of PKA, CaMK II and MAPK/ERK. FEBS J. 2007;274(2):474–484. doi: 10.1111/j.1742-4658.2006.05594.x. [DOI] [PubMed] [Google Scholar]
- 44.Piot JM, Zhao Q, Guillochon D, Ricart G, Thomas D. Isolation and characterization of a bradykinin-potentiating peptide from a bovine peptic hemoglobin hydrolysate. FEBS Lett. 1992;299(1):75–79. doi: 10.1016/0014-5793(92)80104-O. [DOI] [PubMed] [Google Scholar]
- 45.Ivanov VT, Karelin AA, Philippova MM, Nazimov IV, Pletnev VZ. Hemoglobin as a source of endogenous bioactive peptides: the concept of tissue-specific peptide pool. Biopolymers. 1997;43(2):171–188. doi: 10.1002/(SICI)1097-0282(1997)43:2<171::AID-BIP10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 46.Costa E, Guidotti A. Diazepam binding inhibitor (DBI): a peptide with multiple biological actions. Life Sci. 1991;49(5):325–344. doi: 10.1016/0024-3205(91)90440-M. [DOI] [PubMed] [Google Scholar]
- 47.Ferrero P, Santi MR, Conti-Tronconi B, Costa E, Guidotti A. Study of an octadecaneuropeptide derived from diazepam binding inhibitor (DBI): biological activity and presence in rat brain. Proc Natl Acad Sci USA. 1986;83(3):827–831. doi: 10.1073/pnas.83.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbaccia ML, Berkovich A, Guarneri P, Slobodyansky E. DBI (diazepam binding inhibitor): the precursor of a family of endogenous modulators of GABAA receptor function. History, perspectives, and clinical implications. Neurochem Res. 1990;15(2):161–168. doi: 10.1007/BF00972206. [DOI] [PubMed] [Google Scholar]
- 49.Slobodyansky E, Guidotti A, Wambebe C, Berkovich A, Costa E. Isolation and characterization of a rat brain triakontatetraneuropeptide, a posttranslational product of diazepam binding inhibitor: specific action at the Ro 5-4864 recognition site. J Neurochem. 1989;53(4):1276–1284. doi: 10.1111/j.1471-4159.1989.tb07425.x. [DOI] [PubMed] [Google Scholar]
- 50.Alho H, Costa E, Ferrero P, Fujimoto M, Cosenza-Murphy D, Guidotti A. Diazepam- binding inhibitor: a neuropeptide located in selected neuronal populations of rat brain. Science. 1985;229(4709):179–182. doi: 10.1126/science.3892688. [DOI] [PubMed] [Google Scholar]
- 51.Rego JL, Leprince J, Luu-The V, Pelletier G, Tonon MC, Vaudry H. Structure-activity relationships of a series of analogs of the endozepine octadecaneuropeptide (ODN(11)(-)(18)) on neurosteroid biosynthesis by hypothalamic explants. J Med Chem. 2007;50(13):3070–3076. doi: 10.1021/jm0610548. [DOI] [PubMed] [Google Scholar]
- 52.Angelone T, Goumon Y, Cerra MC, Metz-Boutigue MH, Aunis D, Tota B. The emerging cardioinhibitory role of the hippocampal cholinergic neurostimulating peptide. J Pharmacol Exp Ther. 2006;318(1):336–344. doi: 10.1124/jpet.106.102103. [DOI] [PubMed] [Google Scholar]
- 53.Ojika K, Mitake S, Tohdoh N, Appel SH, Otsuka Y, Katada E, et al. Hippocampal cholinergic neurostimulating peptides (HCNP) Prog Neurobiol. 2000;60(1):37–83. doi: 10.1016/S0301-0082(99)00021-0. [DOI] [PubMed] [Google Scholar]
- 54.Ojika K, Kojima S, Ueki Y, Fukushima N, Hayashi K, Yamamoto M. Purification and structural analysis of hippocampal cholinergic neurostimulating peptide. Brain Res. 1992;572(1–2):164–171. doi: 10.1016/0006-8993(92)90465-L. [DOI] [PubMed] [Google Scholar]
- 55.Kim HG, Kim KL. Decreased hippocampal cholinergic neurostimulating peptide precursor protein associated with stress exposure in rat brain by proteomic analysis. J Neurosci Res. 2007;85(13):2898–2908. doi: 10.1002/jnr.21407. [DOI] [PubMed] [Google Scholar]
- 56.Burgula S, Medisetty R, Jammulamadaka N, Musturi S, Ilavazhagan G, Singh SS. Downregulation of PEBP1 in Rat Brain Cortex in Hypoxia. J Mol Neurosci. 2010;41:36–47. doi: 10.1007/s12031-009-9275-7. [DOI] [PubMed] [Google Scholar]
- 57.George AJ, Holsinger RM, McLean CA, Tan SS, Scott HS, Cardamone T, et al. Decreased phosphatidylethanolamine binding protein expression correlates with Abeta accumulation in the Tg2576 mouse model of Alzheimer's disease. Neurobiol Aging. 2006;27(4):614–623. doi: 10.1016/j.neurobiolaging.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 58.Goumon Y, Angelone T, Schoentgen F, Chasserot-Golaz S, Almas B, Fukami MM, et al. The hippocampal cholinergic neurostimulating peptide, the N-terminal fragment of the secreted phosphatidylethanolamine-binding protein, possesses a new biological activity on cardiac physiology. J Biol Chem. 2004;279(13):13054–13064. doi: 10.1074/jbc.M308533200. [DOI] [PubMed] [Google Scholar]
- 59.Mukai H, Hokari Y, Seki T, Takao T, Kubota M, Matsuo Y, et al. Discovery of mitocryptide-1, a neutrophil-activating cryptide from healthy porcine heart. J Biol Chem. 2008;283(45):30596–30605. doi: 10.1074/jbc.M803913200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueki N, Someya K, Matsuo Y, Wakamatsu K, Mukai H. Cryptides: functional cryptic peptides hidden in protein structures. Biopolymers. 2007;88(2):190–198. doi: 10.1002/bip.20687. [DOI] [PubMed] [Google Scholar]
- 61.Mukai H, Seki T, Nakano H, Hokari Y, Takao T, Kawanami M, et al. Mitocryptide-2: purification, identification, and characterization of a novel cryptide that activates neutrophils. J Immunol. 2009;182(8):5072–5080. doi: 10.4049/jimmunol.0802965. [DOI] [PubMed] [Google Scholar]
- 62.Huff T, Muller CS, Otto AM, Netzker R, Hannappel E. beta-Thymosins, small acidic peptides with multiple functions. Int J Biochem Cell Biol. 2001;33(3):205–220. doi: 10.1016/S1357-2725(00)00087-X. [DOI] [PubMed] [Google Scholar]
- 63.Goldstein AL, Hannappel E, Kleinman HK. Thymosin beta4: actin-sequestering protein moonlights to repair injured tissues. Trends Mol Med. 2005;11(9):421–429. doi: 10.1016/j.molmed.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Low TL, Goldstein AL. Chemical characterization of thymosin beta 4. J Biol Chem. 1982;257(2):1000–1006. [PubMed] [Google Scholar]
- 65.Choi SY, Noh MR, Kim DK, Sun W, Kim H. Neuroprotective function of thymosin-beta and its derivative peptides on the programmed cell death of chick and rat neurons. Biochem Biophys Res Commun. 2007;362(3):587–593. doi: 10.1016/j.bbrc.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 66.Huang CM, Wang CC, Barnes S, Elmets CA. In vivo detection of secreted proteins from wounded skin using capillary ultrafiltration probes and mass spectrometric proteomics. Proteomics. 2006;6(21):5805–5814. doi: 10.1002/pmic.200600163. [DOI] [PubMed] [Google Scholar]
- 67.Skopeliti M, Iconomidou VA, Derhovanessian E, Pawelec G, Voelter W, Kalbacher H, et al. Prothymosin alpha immunoactive carboxyl-terminal peptide TKKQKTDEDD stimulates lymphocyte reactions, induces dendritic cell maturation and adopts a beta-sheet conformation in a sequence-specific manner. Mol Immunol. 2009;46(5):784–792. doi: 10.1016/j.molimm.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 68.Goldstein AL, Low TL, McAdoo M, McClure J, Thurman GB, Rossio J, et al. Thymosin alpha1: isolation and sequence analysis of an immunologically active thymic polypeptide. Proc Natl Acad Sci USA. 1977;74(2):725–729. doi: 10.1073/pnas.74.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romani L, Bistoni F, Montagnoli C, Gaziano R, Bozza S, Bonifazi P, et al. Thymosin alpha1: an endogenous regulator of inflammation, immunity, and tolerance. Ann N Y Acad Sci. 2007;1112:326–338. doi: 10.1196/annals.1415.002. [DOI] [PubMed] [Google Scholar]
- 70.Garbin F, Eckert K, Immenschuh P, Kreuser ED, Maurer HR. Prothymosin alpha 1 effects, in vitro, on the antitumor activity and cytokine production of blood monocytes from colorectal tumor patients. Int J Immunopharmacol. 1997;19(6):323–332. doi: 10.1016/S0192-0561(97)00024-6. [DOI] [PubMed] [Google Scholar]
- 71.Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J Biol Chem. 1997;272(40):25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- 72.Voges D, Zwickl P, Baumeister W. The 26 S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 73.Breusing N, Grune T. Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol Chem. 2008;389(3):203–209. doi: 10.1515/BC.2008.029. [DOI] [PubMed] [Google Scholar]
- 74.Naujokat C, Fuchs D, Berges C. Adaptive modification and flexibility of the proteasome system in response to proteasome inhibition. Biochim Biophys Acta. 2007;1773(9):1389–1397. doi: 10.1016/j.bbamcr.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 75.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83(3):731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 76.Vosler PS, Brennan CS, Chen J. Calpain-mediated signaling mechanisms in neuronal injury and neurodegeneration. Mol Neurobiol. 2008;38(1):78–100. doi: 10.1007/s12035-008-8036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pontremoli S, Viotti PL, Michetti M, Salamino F, Sparatore B, Melloni E. Modulation of inhibitory efficiency of rat skeletal muscle calpastatin by phosphorylation. Biochem Biophys Res Commun. 1992;187(2):751–759. doi: 10.1016/0006-291X(92)91259-S. [DOI] [PubMed] [Google Scholar]
- 78.Goll DE, Thompson VF, Taylor RG, Zalewska T. Is calpain activity regulated by membranes and autolysis or by calcium and calpastatin? Bioessays. 1992;14(8):549–556. doi: 10.1002/bies.950140810. [DOI] [PubMed] [Google Scholar]
- 79.Stabach PR, Cianci CD, Glantz SB, Zhang Z, Morrow JS. Site-directed mutagenesis of alpha II spectrin at codon 1175 modulates its mu-calpain susceptibility. Biochemistry. 1997;36(1):57–65. doi: 10.1021/bi962034i. [DOI] [PubMed] [Google Scholar]
- 80.Cuerrier D, Moldoveanu T, Davies PL. Determination of peptide substrate specificity for mu-calpain by a peptide library-based approach: the importance of primed side interactions. J Biol Chem. 2005;280(49):40632–40641. doi: 10.1074/jbc.M506870200. [DOI] [PubMed] [Google Scholar]
- 81.Yuan J, Horvitz HR. A first insight into the molecular mechanisms of apoptosis. Cell. 2004;116(2 Suppl):S53–S56. doi: 10.1016/S0092-8674(04)00028-5. [DOI] [PubMed] [Google Scholar]
- 82.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5(11):897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 83.Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, et al. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272(15):9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 84.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6(10):764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 85.Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci. 2008;29(1):22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 86.Turk V, Kos J, Turk B. Cysteine cathepsins (proteases)–on the main stage of cancer? Cancer Cell. 2004;5(5):409–410. doi: 10.1016/S1535-6108(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 87.Turk B, Turk D, Turk V. Lysosomal cysteine proteases: more than scavengers. Biochim Biophys Acta. 2000;1477(1–2):98–111. doi: 10.1016/s0167-4838(99)00263-0. [DOI] [PubMed] [Google Scholar]
- 88.Oorni K, Sneck M, Bromme D, Pentikainen MO, Lindstedt KA, Mayranpaa M, et al. Cysteine protease cathepsin F is expressed in human atherosclerotic lesions, is secreted by cultured macrophages, and modifies low density lipoprotein particles in vitro. J Biol Chem. 2004;279(33):34776–34784. doi: 10.1074/jbc.M310814200. [DOI] [PubMed] [Google Scholar]
- 89.Mort JS, Recklies AD, Poole AR. Extracellular presence of the lysosomal proteinase cathepsin B in rheumatoid synovium and its activity at neutral pH. Arthritis Rheum. 1984;27(5):509–515. doi: 10.1002/art.1780270505. [DOI] [PubMed] [Google Scholar]
- 90.Lou X, Xiao T, Zhao K, Wang H, Zheng H, Lin D, et al. Cathepsin D is secreted from M-BE cells: its potential role as a biomarker of lung cancer. J Proteome Res. 2007;6(3):1083–1092. doi: 10.1021/pr060422t. [DOI] [PubMed] [Google Scholar]
- 91.Dagouassat N, Garreau I, Sannier F, Zhao Q, Piot JM. Generation of VV-hemorphin-7 from globin by peritoneal macrophages. FEBS Lett. 1996;382(1–2):37–42. doi: 10.1016/0014-5793(96)00144-5. [DOI] [PubMed] [Google Scholar]
- 92.Zhao Q, Piot JM. Neokyotorphin formation and quantitative evolution following human hemoglobin hydrolysis with cathepsin D. Peptides. 1998;19(4):759–766. doi: 10.1016/S0196-9781(98)00002-3. [DOI] [PubMed] [Google Scholar]
- 93.Salvador N, Aguado C, Horst M, Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem. 2000;275(35):27447–27456. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- 94.Walter J, Kaether C, Steiner H, Haass C. The cell biology of Alzheimer's disease: uncovering the secrets of secretases. Curr Opin Neurobiol. 2001;11(5):585–590. doi: 10.1016/S0959-4388(00)00253-1. [DOI] [PubMed] [Google Scholar]
- 95.Hong L, He X, Huang X, Chang W, Tang J. Structural features of human memapsin 2 (beta-secretase) and their biological and pathological implications. Acta Biochim Biophys Sin (Shanghai) 2004;36(12):787–792. doi: 10.1093/abbs/36.12.787. [DOI] [PubMed] [Google Scholar]
- 96.Kaether C, Haass C, Steiner H. Assembly, trafficking and function of gamma-secretase. Neurodegener Dis. 2006;3(4–5):275–283. doi: 10.1159/000095267. [DOI] [PubMed] [Google Scholar]
- 97.Ikeuchi T, Sisodia SS. The Notch ligands, Delta1 and Jagged2, are substrates for presenilin-dependent “gamma-secretase” cleavage. J Biol Chem. 2003;278(10):7751–7754. doi: 10.1074/jbc.C200711200. [DOI] [PubMed] [Google Scholar]
- 98.Freeman M. Rhomboid proteases and their biological functions. Annu Rev Genet. 2008;42:191–210. doi: 10.1146/annurev.genet.42.110807.091628. [DOI] [PubMed] [Google Scholar]
- 99.Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17(11):1634–1646. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J Immunol. 2001;167(11):6441–6446. doi: 10.4049/jimmunol.167.11.6441. [DOI] [PubMed] [Google Scholar]
- 101.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296(5576):2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 102.Koppen M, Langer T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol. 2007;42(3):221–242. doi: 10.1080/10409230701380452. [DOI] [PubMed] [Google Scholar]
- 103.Vande Walle L, Lamkanfi M, Vandenabeele P. The mitochondrial serine protease HtrA2/Omi: an overview. Cell Death Differ. 2008;15(3):453–460. doi: 10.1038/sj.cdd.4402291. [DOI] [PubMed] [Google Scholar]
- 104.Park HJ, Kim SS, Seong YM, Kim KH, Goo HG, Yoon EJ, et al. Beta-amyloid precursor protein is a direct cleavage target of HtrA2 serine protease. Implications for the physiological function of HtrA2 in the mitochondria. J Biol Chem. 2006;281(45):34277–34287. doi: 10.1074/jbc.M603443200. [DOI] [PubMed] [Google Scholar]
- 105.Fontenele-Neto JD, Massarelli EE, Gurgel Garrido PA, Beaudet A, Ferro ES. Comparative fine structural distribution of endopeptidase 24.15 (EC3.4.24.15) and 24.16 (EC3.4.24.16) in rat brain. J Comp Neurol. 2001;438(4):399–410. doi: 10.1002/cne.1323. [DOI] [PubMed] [Google Scholar]
- 106.Ferro ES, Carreno FR, Goni C, Garrido PA, Guimaraes AO, Castro LM, et al. The intracellular distribution and secretion of endopeptidases 24.15 (EC 3.4.24.15) and 24.16 (EC 3.4.24.16) Protein Pept Lett. 2004;11(5):415–421. doi: 10.2174/0929866043406706. [DOI] [PubMed] [Google Scholar]
- 107.Ferro ES, Hyslop S, Camargo AC. Intracellullar peptides as putative natural regulators of protein interactions. J Neurochem. 2004;91(4):769–777. doi: 10.1111/j.1471-4159.2004.02757.x. [DOI] [PubMed] [Google Scholar]
- 108.Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998;19(5):608–624. doi: 10.1210/er.19.5.608. [DOI] [PubMed] [Google Scholar]
- 109.Fulop V, Bocskei Z, Polgar L. Prolyl oligopeptidase: an unusual beta-propeller domain regulates proteolysis. Cell. 1998;94(2):161–170. doi: 10.1016/S0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 110.Rockel B, Baumeister W. A tale of two giant proteases. Ernst Schering Found Symp Proc. 2008;2008(1):17–40. doi: 10.1007/2789_2008_099. [DOI] [PubMed] [Google Scholar]
- 111.Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278(6):3671–3678. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 112.Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282(25):18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 113.Hooper NM, Turner AJ. The search for alpha-secretase and its potential as a therapeutic approach to Alzheimer s disease. Curr Med Chem. 2002;9(11):1107–1119. doi: 10.2174/0929867023370121. [DOI] [PubMed] [Google Scholar]
- 114.Uhlmann F. Separase regulation during mitosis. Biochem Soc Symp. 2003;2003(70):243–251. doi: 10.1042/bss0700243. [DOI] [PubMed] [Google Scholar]
- 115.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103(3):375–386. doi: 10.1016/S0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 116.Wickner WT, Lodish HF. Multiple mechanisms of protein insertion into and across membranes. Science. 1985;230(4724):400–407. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- 117.Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002;82(2):331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 118.D'Acquisto F, Perretti M, Flower RJ. Annexin-A1: a pivotal regulator of the innate and adaptive immune systems. Br J Pharmacol. 2008;155(2):152–169. doi: 10.1038/bjp.2008.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Siever DA, Erickson HP. Extracellular annexin II. Int J Biochem Cell Biol. 1997;29(11):1219–1223. doi: 10.1016/S1357-2725(97)00057-5. [DOI] [PubMed] [Google Scholar]
- 120.Ye RD, Wun TC, Sadler JE. Mammalian protein secretion without signal peptide removal. Biosynthesis of plasminogen activator inhibitor-2 in U-937 cells. J Biol Chem. 1988;263:4869–4875. [PubMed] [Google Scholar]
- 121.Flieger O, Engling A, Bucala R, Lue H, Nickel W, Bernhagen J. Regulated secretion of macrophage migration inhibitory factor is mediated by a non-classical pathway involving an ABC transporter. FEBS Lett. 2003;551(1–3):78–86. doi: 10.1016/S0014-5793(03)00900-1. [DOI] [PubMed] [Google Scholar]
- 122.Engling A, Backhaus R, Stegmayer C, Zehe C, Seelenmeyer C, Kehlenbach A, et al. Biosynthetic FGF-2 is targeted to non-lipid raft microdomains following translocation to the extracellular surface of CHO cells. J Cell Sci. 2002;115(Pt 18):3619–3631. doi: 10.1242/jcs.00036. [DOI] [PubMed] [Google Scholar]
- 123.Hicks KK, Shin JT, Opalenik SR, Thompson JA. Molecular mechanisms of angiogenesis: experimental models define cellular trafficking of FGF-1. P R Health Sci J. 1996;15(3):179–186. [PubMed] [Google Scholar]
- 124.Seelenmeyer C, Wegehingel S, Tews I, Kunzler M, Aebi M, Nickel W. Cell surface counter receptors are essential components of the unconventional export machinery of galectin-1. J Cell Biol. 2005;171(2):373–381. doi: 10.1083/jcb.200506026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hengst U, Albrecht H, Hess D, Monard D. The phosphatidylethanolamine-binding protein is the prototype of a novel family of serine protease inhibitors. J Biol Chem. 2001;276(1):535–540. doi: 10.1074/jbc.M002524200. [DOI] [PubMed] [Google Scholar]
- 126.Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10(2):148–155. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- 127.Schafer T, Zentgraf H, Zehe C, Brugger B, Bernhagen J, Nickel W. Unconventional secretion of fibroblast growth factor 2 is mediated by direct translocation across the plasma membrane of mammalian cells. J Biol Chem. 2004;279(8):6244–6251. doi: 10.1074/jbc.M310500200. [DOI] [PubMed] [Google Scholar]
- 128.Andrei C, Dazzi C, Lotti L, Torrisi MR, Chimini G, Rubartelli A. The secretory route of the leaderless protein interleukin 1beta involves exocytosis of endolysosome-related vesicles. Mol Biol Cell. 1999;10(5):1463–1475. doi: 10.1091/mbc.10.5.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Qu Y, Franchi L, Nunez G, Dubyak GR. Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J Immunol. 2007;179(3):1913–1925. doi: 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]