Abstract
It is widely accepted that MAPK activation in budding and fission yeasts is often associated with negative effects on cell cycle progression, resulting in delay or arrest at a specific stage in the cell cycle, thereby enabling cells to adapt to changing environmental conditions. For instance, activation of the Cell Wall Integrity (CWI) pathway in the budding yeast Saccharomyces cerevisiae signals an increase in CDK inhibitory phosphorylation, which leads cells to remain in the G2 phase. Here we characterized the CWI pathway of Ustilago maydis, a fungus evolutionarily distant from budding and fission yeasts, and show that activation of the CWI pathway forces cells to escape from G2 phase. In spite of these disparate cell cycle responses in S. cerevisiae and U. maydis, the CWI pathway in both organisms appears to respond to the same class cell wall stressors. To understand the basis of such a difference, we studied the mechanism behind the U. maydis response. We found that activation of CWI pathway in U. maydis results in a decrease in CDK inhibitory phosphorylation, which depends on the mitotic phosphatase Cdc25. Moreover, in response to activation of the CWI pathway, Cdc25 accumulates in the nucleus, providing a likely explanation for the increase in the unphosphorylated form of CDK. We also found that the extended N-terminal domain of Cdc25, which is dispensable under normal growth conditions, is required for this G2 escape as well as for resistance to cell wall stressors. We propose that the process of cell cycle adaptation to cell stress evolved differently in these two divergent organisms so that each can move towards a cell cycle phase most appropriate for responding to the environmental signals encountered.
Author Summary
Signaling cascades connect changes in the extracellular environment with cellular responses. An obvious target for this signaling is cell cycle machinery, since cells have to integrate outside signals to decide whether to proliferate or not. Here we report that the response to cell wall damage in the fungus Ustilago maydis forces cells to escape from G2 phase. This finding contrasts with the response observed in Saccharomyces cerevisiae, in which cells are kept in G2 phase upon activation of the pathway responding to cell wall damage. We correlated this disparate response to differences in the growth pattern during cell cycle between both fungi, since during apical growth cells are more vulnerable to cell wall instability. While in S. cerevisiae polar growth is abrogated during G2 phase, in U. maydis pronounced polar growth occurs in G2 phase. Our results suggest that in response to stress stimuli (i. e. cell wall damage), cells move towards the cell cycle stage at which the cellular environment is most appropriate to respond to such stress (i. e. a phase where polar growth is minimal or absent). Depending on the individual organism, this cell cycle stage can differ, thus accounting for the observed differences in outcome.
Introduction
The mitogen-activated protein kinase (MAPK) family of kinases connects extracellular stimuli with diverse cellular responses [1]. Since progression through the cell cycle is critically dependent on the integration of external signals such as the presence of environmental growth factors or stress stimuli, it seems obvious that cell cycle regulation would be a prime target of MAPK-mediated signaling. In fact, patterns have begun to emerge from a large number of studies showing functions of MAPK subfamilies at different stages of the cell cycle [2]. However, in spite of these studies, a coherent role of MAPK-mediated signaling into cell cycle control has yet to be clarified. MAPK signaling produces both negative and positive effects in cell cycle regulation that depends not only on the stimulus or MAPK pathway implied in the signal transmission, but also on the extent of the MAPK activation and the cell type [3].
Studies carried out in fungal cells also support a role for MAPK pathways in regulating cell cycle [4], [5]. In Saccharomyces cerevisiae, which utilizes five separate MAPK cascades to respond appropriately to environmental changes [6], activation of MAPK pathway is often linked to a cell cycle arrest or delay. Activation of Fus3 MAPK cascade (in response to mating pheromone) imposes cell cycle arrest in the G1 phase via phosphorylation of Far1, a cyclin-dependent kinase inhibitor (CKI), which acts on Cdc28, the yeast Cdk1 [7]–[9]. The Hog1 MAPK pathway, in response to hyperosmotic stress also imposes cell cycle arrest both in the G1 and G2 phases via activation of the Sic1 CKI, or blocking the action of a protein kinase (Hsl1) that inhibits the protein kinase Swe1 (the Schizosaccharomyces pombe Wee1 ortholog that acts as a mitotic inhibitor), respectively [10], [11]. The Slt2/Mpk1 kinase, which is activated in response to cell wall stress, imposes G2 cell cycle arrest via inhibition (direct or indirect) of the yeast homologue of the Cdc25 phosphatase (Mih1), which is necessary to reverse the inhibitory tyrosine-specific phosphorylation conferred on Cdc28 by Swe1 [12]. Collectively, these observations lead to the general view that in fungi, MAPK activation negatively regulates cell cycle progression, by imposing delays or arrests at specific cell cycle stages to enable the cell to adapt to unfavorable stress conditions or to synchronize cell cycle progression before mating.
Our laboratory has been involved in the characterization of the connections between cell cycle and the induction of the virulence program in the phytopathogenic fungus Ustilago maydis [13]. This fungus has been used as a model system in fungal development for many years [14]. The virulence program in U. maydis is induced by a pheromone-dependent MAPK cascade [15]–[19]. We reported that activation of this cascade in U. maydis also resulted in cell cycle arrest, but in contrast to the well-known G1 cell cycle arrest described in S. cerevisae or S. pombe [7], [8], [20], [21], pheromone-induced cell cycle arrest in U. maydis takes place at the G2 phase [22]. These differences in the regulation of cell cycle arrest may reflect specific features of pheromone response in U. maydis or differences in the wiring between signal transduction and cell response in these organisms [23]. Therefore, we looked for cell cycle responses dependent on other MAPK cascades in U. maydis. We chose to characterize the Cell Wall Integrity (CWI) pathway, which utilizes MAPK cascades to facilitate the maintenance of the cell wall by mediating cell wall biosynthesis, actin organization and other events necessary to maintain cell wall integrity [24]. As fungal cell growth involves cell wall remodeling that must be coordinated with cell cycle, a connection between cell cycle and CWI pathway would be predicted. Furthermore, as this pathway is conserved among fungi including budding yeast, fission yeast and filamentous fungi [25], such a relationship between CWI pathway and cell cycle control could be of broad interest in fungal physiology.
In this work we report the surprising finding that activation of Mpk1, the MAPK central to CWI cascade, forces cells to escape from G2 phase in U. maydis, in contrast to the G2 cell cycle arrest that was reported for S. cerevisiae [12]. In both organisms it seems that the Cdc25 phosphatase is the target responsible of this disparate effect.
Results
Characterization of the central module of CWI pathway in U. maydis
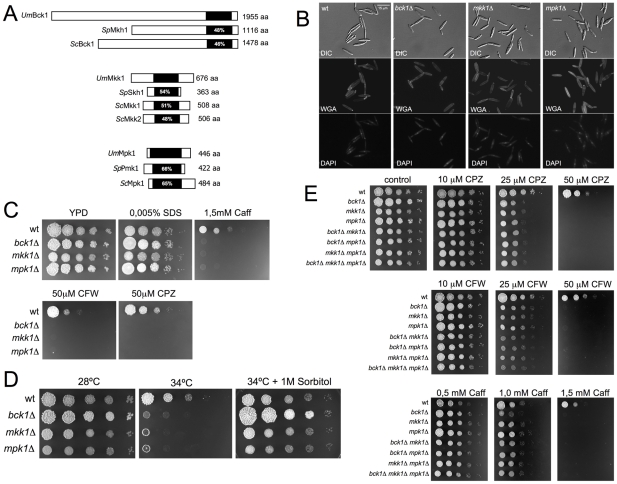
We were interested in characterizing the central core of kinases composing the Cell Wall Integrity (CWI) pathway of U. maydis. As putative proteins with sequence similarity to MEKK, MEK and MAPK were already annotated in U. maydis genome database [26], we performed a phylogenetic analysis to identify the closest homologues to CWI pathway components in U. maydis using the CWI core components from S. cerevisiae and S. pombe for comparison (Figure S1). From this analysis we identified putative homologues to S. cerevisiae MEKK (um01662), MEK (um10855), and MAPK (um10107) that were named Bck1, Mkk1 and Mpk1 respectively (Figure 1A).
Figure 1. Ustilago maydis cell wall integrity pathway.
(A) Scheme of the U. maydis MEKK, MEK and MAPK proteins in relation to homologous proteins from Saccharomyces cerevisiae and Schizosaccharomyces pombe. The catalytic domains are shown in black and were identified using the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de). The percentages inside each box represent the sequence identity in this domain when compared to the U. maydis sequence. (B) Morphology of wild-type (FB1) and bck1Δ, mkk1Δ and mpk1Δ cells growing in nutrient-rich medium, YPD. Cells were stained with 4',6-diamidino-2-phenylindole (DAPI) to visualize the nuclei and FITC-labeled wheat germ agglutinin (WGA), to detect cell wall. Bar 15 µm. (C) Sensitivity of CWI pathway mutant strains to cell wall inhibitors. Cultures of wild-type (FB1) and the indicated strains were grown overnight in rich medium, then diluted to an OD600 of 0.5. Tenfold serial dilutions were made and 2 µl of each was plated in solid YPD medium amended with the indicated compounds: Sodium dodecyl sulphate (SDS), caffeine (Caff), Calcofluor White (CFW) and Chlorpromazine (CPZ). The plates were grown for 4 days at 28°C. (D) Growth of FB1 (wt) and CWI pathway mutant strains at 28 and 34°C in solid medium (YPD). At 28°C the growth of control and mutant cells was similar. However, mutant cells showed severe growth defects at 34°C. These defects were alleviated when 1 M Sorbitol was added to culture media. (E) Sensitivity of CWI pathway single, double and triple mutant strains to cell wall inhibitors. Note that no enhancement in sensitivity was obtained when the individual mutations were combined.
To address whether these kinase homologues were the canonical components of CWI pathway in U. maydis, we generated complete deletions of the corresponding ORFs by PCR-based gene targeting [27]. Mutant strains in each gene as well as control wild-type cells were grown in nutrient rich-medium (YPD) and their duplication time was determined. Also, cells were stained with 4',6-diamidino-2-phenylindole (DAPI) to visualize the nuclei and FITC-labeled wheat germ agglutinin (WGA), a lectin that binds to chitin [28], to detect cell wall. We found no differences between mutant and wild-type cells with respect to doubling time (around 120 min in all cases, not shown) or morphology (Figure 1B). We tested for sensitivity to compounds that affect growth of strains with altered cell wall integrity. Cell dilutions of control and mutant cells were plated on YPD agar media containing different cell envelope stressors (Figure 1C): sodium dodecyl sulphate (SDS), which disrupts the plasma membrane and lyses cells with membrane defects; caffeine (Caff), which has been used extensively to probe signal transduction and cell integrity phenotypes in S. cerevisiae; calcofluor white (CFW), which binds to chitin and to a lesser extent glucan and disrupts the cell wall; and chlorpromazine (CPZ), an amphipathic molecule (and potent activator of the CWI pathway in S. cerevisiae) that inserts asymmetrically into the plasma membrane causing stretching [24], [29]–[31]. We observed that the U. maydis CWI mutant strains were more sensitive to all of these stressors except SDS. We also tested whether the mutant strains were more sensitive to growth at high temperature, as the CWI pathway is essential for the response to high temperature [30], and found this to be the case (Figure 1D). Moreover, we found that the temperature-sensitive growth of the mutant was rescued by 1 M sorbitol, an osmotic stabilizer that can rescue host defects in cell wall integrity [32].
We also constructed double mutant cells carrying different combinations of mutated alleles (bck1Δ, mkk1Δ; bck1Δ, mpk1Δ; and mkk1Δ, mpk1Δ) as well as the triple mutant (bck1Δ, mkk1Δ, mpk1Δ) and tested for enhanced sensitivity to cell wall stressors. The distinct double and triple mutant strains displayed sensitivities that were comparable to single mutants (Figure 1E) consistent with the notion that all these proteins function in a common pathway.
U. maydis Mpk1 is activated after Bck1-Mkk1–mediated phosphorylation
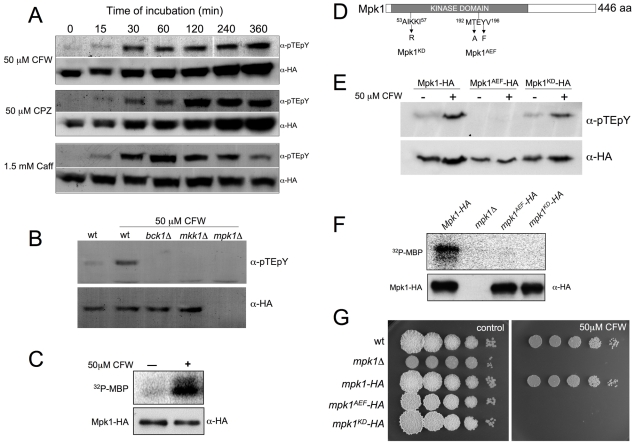
MAPKs are activated by phosphorylation at a pair of conserved Thr and Tyr residues, located in the so-called T-loop [33], [34]. To correlate the function and regulation of CWI pathway in U. maydis with the response to agents that stress the cell wall, we used a commercial specific antibody raised against dually phosphorylated (Thr202/Tyr204)-p44/42 MAPK to monitor the phosphorylation of Thr193 and Tyr195 of U. maydis Mpk1. This antibody has been successfully utilized to characterize CWI pathway activation in other fungi [35]–[38]. Previously, we constructed an N-terminal HA tagged mpk1 allele under the control of the native mpk1 promoter (mpk1-HA allele), and replaced the endogenous copy with this allele. When challenged with the different cell wall stressors the resulting strain was indistinguishable from wild-type cells with respect to sensitivity to these compounds (not shown, see also Figure 2G). We incubated cultures of the strain carrying the HA-tagged Mpk1 allele either with CFW, CPZ or Caff and analyzed the amount of phosphorylated signal at different times by immunoblotting. We observed that the signal corresponding to the phosphorylated form of Mpk1 increased upon the addition of the different cell wall stressors to culture medium (Figure 2A). As it has been reported in other organisms [35]–[37], we found that the kinetics of the response to cell wall stressors was slightly different depending on the compound added (Figure S2).
Figure 2. Phosphorylation of Mpk1 T-loop is required for in vivo activity.
(A) Time course of Mpk1 phosphorylation in response to Calcofluor White (CFW), Chlorpromazine (CPZ) and Caffeine (Caff). Mid log phase cells of a strain carrying a HA-tagged Mpk1 allele (UMA44) were treated with the indicated amount of cell wall stressors and aliquots of the same culture were withdrawn before (0) and at the indicated times (in minutes) following compound addition. Protein extracts were prepared and a similar amount of total protein of each time point were loaded in SDS-PAGE gels and submitted to Western blot assays. The same blot, after stripping, was incubated with anti-pTEpY to detect T-loop phosphorylation and with anti-HA to detect HA-tagged proteins. (B) T-loop phosphorylation of Mpk1 in vivo is dependent on upstream kinases. Cell extracts from CFW-treated cultures over a 2 h period were prepared from UMA44 (wt control), UMA44.2 (bck1Δ), UMA44.3 (mkk1Δ), and UMA3 (mpk1Δ), cells and analyzed as described above. (C) Catalytic activity of Mpk1 is enhanced after CFW treatment. HA-tagged proteins were immunoprecipitated from cells extracts prepared from UMA44 cells grown in the presence or absence of 50 µM CFW for 2 h. Protein kinase activity was measured by incubation of immunoprecipitates with purified Myelin Basic Protein (MBP) as substrate and [γ-32P] ATP. On the bottom, 8% SDS-PAGE and immunoblot with anti-HA was used to show comparable levels of Mpk1 proteins in the reaction mixtures. On the top, 12.5% SDS-PAGE and autorradiography was used to detect in vitro phosphorylated MBP. (D) Scheme of Mpk1 showing the mutated residues in Mpk1AEF and Mpk1KD alleles. (E) T-loop phosphorylation of Mpk1 mutants. Cell extracts from CFW-treated cultures over 2 h period were prepared from UMA44 (Mpk1-HA), UMA59 (Mpk1AEF-HA), and UMA60 (Mpk1KD-HA), cells and analyzed as described above. (F) Kinase activity of Mpk1 mutants. Immunoprecipitates from UMA44 (mpk1-HA), UMA3 (mpk1Δ), UMA59 (mpk1AEF-HA), and UMA60 (mpk1KD-HA) cells were analyzed as described above. (G) Sensitivity to CFW of Mpk1 mutants.
To address whether activation of Mpk1 was dependent on Bck1 or Mkk1, we examined the phosphorylation levels of Mpk1-HA in bck1Δ and mkk1Δ cells after treatment with 50 µM CFW. No activated form of Mpk1 in either bck1Δ or mkk1Δ cells was detected (Figure 2B). Thus, both Bck1 and Mkk1 appear to be required for appropriate phosphorylation of Mpk1 in vivo.
We also analyzed whether kinase activity of Mpk1 was increased after exposure to CFW. Catalytic activity of Mpk1 was directly monitored in an immunoprecipitation-kinase assay. Cell extracts from cultures treated with 50 µM CFW were prepared, HA-tagged Mpk1 was immunoprecipitated and activity in phosphorylating Myelin Basic Protein (MBP) was determined (Figure 2C). Immunoprecipitated kinase obtained from CFW-treated cells was more active in phosphorylating MBP than that from control non-treated cells.
To determine whether Mpk1 phosphorylation as well as kinase activity was required for in vivo activity, two Mpk1 mutant alleles were constructed. First, Mpk1AEF contains mutations (T193A, Y195F) in the conserved phosphoacceptor site and should therefore be unable to be phosphorylated at the conserved TEY motif. Second, Mpk1KD contains a single mutation (K55R) in the ATP-binding pocket and should therefore produce a kinase-dead mutant protein (Figure 2D). These mutant alleles were also engineered to produce HA-tagged variants. By immunoblotting we found no evidence for phosphorylation using the phospho-epitope specific MAPK antibody with proteins extracted from the strain expressing the Mpk1AEF variant. In contrast, the Mpk1KD mutant protein showed a phosphorylation signal that was comparable to wild-type levels (Figure 2E). When kinase activity was measured by immunoprecipitation, we found that both immunoprecipitated Mpk1KD and Mpk1AEF were strongly reduced in the capacity to phosphorylate MBP (Figure 2F). Finally, we asked whether cells carrying these mutant alleles were more sensitive to CFW and found this to be the case (Figure 2G). Taken together, these results indicate that kinase activity of Mpk1 is dependent on T-loop phosphorylation, which is required for appropriate response to cell wall damage.
In summary, these results support the notion that the Mpk1, Mkk1 and Bck1 kinases are bona fide core elements of the CWI pathway of U. maydis.
Over-activation of CWI pathway inhibits growth in U. maydis
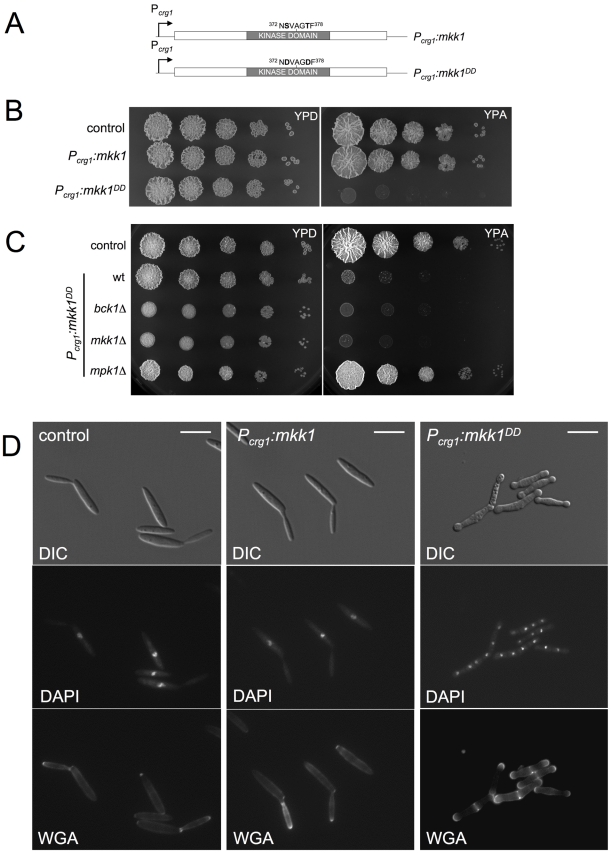
We wished to examine the consequences of over-activation of CWI pathway in U. maydis. To approach this issue and to avoid possible side effects on growth by the presence of cell wall stressors known to activate CWI pathway in U. maydis, we generated a conditionally activated allele of Mkk1. MEKs are activated by phosphorylation of two Ser/Thr residues located between the kinase subdomains VII and VIII [39], [40]; and previous studies have shown that mutation of these residues to Asp results in constitutive activation [41], [42]. Therefore we generated Mkk1DD in which the conserved Ser373 and Thr377 residues were both replaced with Asp. We constructed an integrative plasmid in which the mkk1DD allele was under the control of the crg1 promoter [43]. Thus, expression of mkk1DD can be achieved by inducing crg1 with the addition of arabinose to the culture medium. We also constructed a control plasmid carrying the wild-type allele under crg1 control (Figure 3A). A single copy of either construct or the empty vector was inserted at the ip locus in wild-type cells [44]. We found that under conditions that promoted expression of crg1 promoter (medium containing arabinose as carbon source, YPA) the growth of the strain carrying the activated allele was severely impaired on solid medium (Figure 3B). This inhibitory effect was rescued by loss of function of the downstream kinase mpk1 but not by the upstream kinase bck1 (Figure 3C).
Figure 3. Expression of an activated Mkk1 allele is toxic in U. maydis.
(A) mkk1 and mkk1DD alleles under the control of Pcrg1 are shown schematically. Residues mutated are shown at the top. (B) Growth ability of cells expressing an ectopic copy of mkk1 and mkk1DD alleles was examined by spotting serial dilutions of exponential cultures of UMA10 (control), UMA12 (Pcrg1:mkk1) and UMA7 (Pcrg1:mkk1DD) strains in rich medium with either 2% glucose (YPD, repressing conditions) or 2% arabinose (YPA, inducing conditions). Plates were incubated at 28°C. (C) Toxicity is suppressed by deletion of gene encoding downstream kinase but not by upstream kinase. Dilution of cell cultures from UMA10 (control), UMA7 (Pcrg1:mkk1DD wt), UMA57 (Pcrg1:mkk1DD bck1Δ), UMA9 (Pcrg1:mkk1DD mkk1Δ) and UMA13 (Pcrg1:mkk1DD mpk1Δ) were treated as above. (D) Morphology of cells expressing mkk1DD. YPA cultures of UMA10 (control), UMA12 (Pcrg1:mkk1) and UMA7 (Pcrg1:mkk1DD) were incubated at 22°C (to delay massive cell lysis, see text) for 8 h. Bar 15 µm.
When the crg1 promoter was repressed, cells carrying mkk1DD were morphologically indistinguishable from control strains (not shown). When induced, cells expressing the ectopic copy of mkk1DD allele had a strong lysis phenotype as evidenced by the frequent presence of cell “ghosts” and cell debris (Figure S3). Lysis could be largely delayed by growing the cells at low temperature (22°C, unpublished observations). Therefore, we analyzed the morphological effects of expression of mkk1DD growing cells at 22°C. Under this condition, ectopic expression of the wild-type copy of mkk1 had no effect on cell morphology (Figure 3D, middle column). Expressing mkk1DD resulted in aberrant morphology, featuring cell aggregates containing several septa that delimited cell compartments, each carrying a single nucleus (Figure 3D, right column). In addition, there was swelling of the tips of these cell aggregates and these swollen areas stained strongly with FITC-WGA, indicating a continuous deposition of cell wall material at the tips (Figure 3D). These morphological defects were suppressed when mkk1DD was expressed in cells deleted of mpk1 (Figure S4).
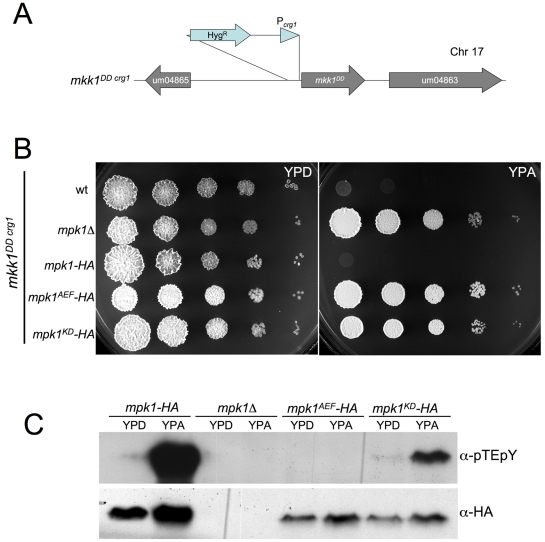
To obtain further support for the idea that it was the activation of the Mpk1 kinase activity that was responsible of these inhibitory effects, we replaced the endogenous mkk1 locus with the constitutively activated mkk1DD allele under the control of crg1 (mkk1DDcrg1 allele, Figure 4A) in strains harboring either the HA-tagged mpk1 allele or its derivates mpk1AEF-HA and mpk1KD-HA. Growth impairment after expression of mkk1DD allele was suppressed when the downstream Mpk1 kinase was either unphosphorylatable due to mutation at the conserved TEY motif or was disabled for kinase activity (Figure 4B). Moreover, with expression of the mkk1DD allele there was a dramatic increase in the level of Mpk1 phosphorylation (Figure 4C).
Figure 4. Expression of mkk1DD overactivates Mpk1.
(A) Scheme of the conditional allele mkk1DDcrg1. (B) Toxicity after mkk1DD expression is suppressed by mpk1 alleles defectives in T-loop phosphorylation or kinase activity. Dilution of cell cultures from UMA20 (control, wt), UMA42 (mpk1Δ), UMA43 (mpk1-HA), UMA64 (mpk1AEF-HA) and UMA65 (mpk1KD-HA) were treated as in Figure 3. Plates were incubated at 28°C. (C) T-loop phosphorylation of above strains. Cells were incubated in the indicated media for 8 h at 22°C.
These results are consistent with a model in which Mpk1 is activated by Mkk1, and its over-activation results in inhibitory effects on U. maydis cell growth.
Over-activation of Mpk1 forces cells to escape from G2 phase in U. maydis
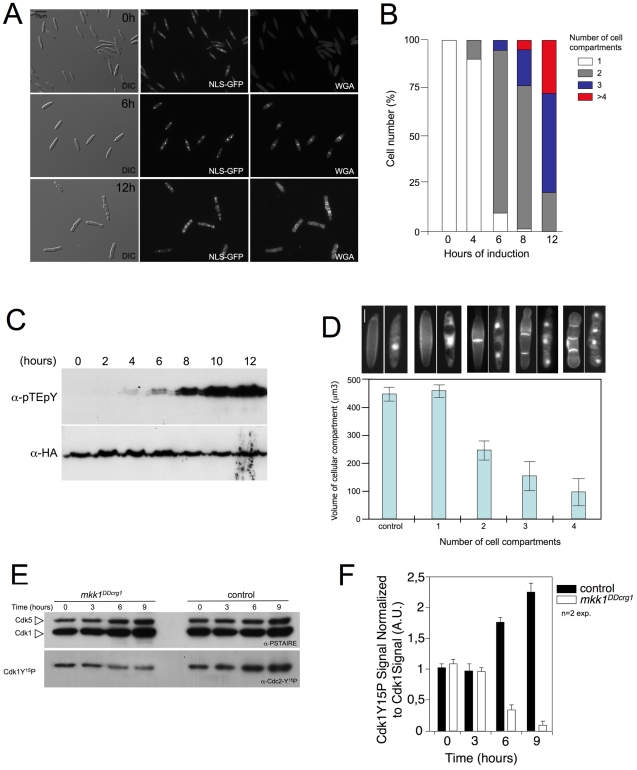
To characterize the morphogenetic defects caused by the over-activation of Mpk1 further, we monitored the morphological appearance of nuclei and cell septa. For nuclear staining we expressed GFP fused with a nuclear localization signal (NLS) and for septa, we used WGA-TRITC for visualization. Cells carrying the mkk1DDcrg1 allele, as well as the mpk1-HA allele and constitutively expressed NLS-GFP [45], were incubated at inducing conditions for 12 h. Samples were obtained at different time points and used for Western analysis (to asses the Mpk1 activation level, Figure 5C) as well as for microscopic observations (to asses nuclear number and the presence of septa after WGA-TRITC staining, Figure 5A). Four hours after induction, cells started to divide via a central septum, and this correlated with the presence of activated Mpk1 kinase. By 6 h induction more than 75% of the cells showed two cell compartments and at this time a clear activated Mpk1 signal was apparent. Further incubation (8 h) resulted in the appearance of cell aggregates (septa-mediated division produced cell aggregates due to the lack of cell separation) composed of three or more cell compartments, each carrying one nucleus. Finally, after 12 h of incubation more than 80% of the cells showed three or more cell compartments (Figure 5B).
Figure 5. Over-activation of Mpk1 resulted in shortened G2 phase.
(A) Time-course of morphological changes induced after mkk1DD expression. UMA54 cells carrying a NLS-GFP fusion to detect nuclei as well as a HA-tagged mpk1 allele, were incubated in YPA at 22°C along 12 h. Samples were withdrawn at different times and submitted, after WGA-TRITC staining, to microscopic observation. Bar 15 µm. (B) Quantification of the number of cell compartments per cell aggregate at different time points. At least 50 cell aggregates were counted at each time point (2 independent experiments). (C) Time-course of T-loop phosphorylation of Mpk1 after mkk1 DD induction. (D) Size of cell compartments decreases after each mitosis. Volume of each cell compartment (assuming each cell compartment was a cylinder) was obtained after measurement of length and wide of 30 cell compartments in each cell type (2 independent experiments). At the top, cell images at same magnification (Bar 5 µm) of the four cell types measured. Cells were stained with WGA-TRITC to detect septa. Mean and s.d. are shown. (E) Levels of Cdk1 inhibitory phosphorylation after mkk1DD expression. Protein extracts from FB1 wild-type cells (control) or UMA20 (mkk1DDcrg1) growing in inducing conditions (YPA) for the indicated time (hours) were separated by SDS-PAGE. Immunoblots were incubated successively with an antibody that recognizes the Cdk1 phosphorylated form (anti-Cdc2-Y15P) and anti-PSTAIRE, which recognizes Cdk1 as well as Cdk5. (F) Levels of Cdk1 phosphorylation were determined by quantifying the level of antibody signal using a ChemiDoc (Bio-Rad). Signal from the phosphopeptide-specific antibodies was normalized to the amount of phosphorylation of control strain at time zero. Differences in loading of samples were corrected by dividing each phosphopeptide-specific antibody signal by the Cdk1 (anti-PSTAIRE) antibody signal. Mean and s.d. are shown (n = 2 exp.).
An interesting observation was that the size of cell compartments appeared to decrease after each cell division. We quantified volume in each cell compartment by measuring length and width of each, and deduced that the volume of each cell compartments decreased as the number of cell compartments per aggregate increased (Figure 5D). In U. maydis, cell size in daughter cells is dependent on the length of G2 phase: the longer the G2 phase, the larger the daughter cell and vice versa [13]. Previous reports [46], [47] indicated that shortening of G2 phase by decreasing the levels of Cdk1 inhibitory phosphorylation at Tyr15 causes a decrease in the size of daughter cells as well as abolition of the budding program and the division by inserting septa. This change in the division mode is also accompanied by the inhibition of cell separation, resulting in cell aggregates. In fact, alteration of the levels of cell cycle regulators affecting Cdk1 inhibitory phosphorylation such as overproduction of Cdc25 phosphatase or down-regulation of Wee1 kinase [46], [47] resulted in the formation of cell aggregates that were reminiscent of those observed after over-activation of Mpk1. Therefore, we analyzed levels of Cdk1 inhibitory phosphorylation upon over-activation of Mpk1. For this we used a specific antibody raised against the Tyr15 phosphorylated human Cdc2 peptide (Y15P), which also recognizes the Tyr15-phosphorylated form of U. maydis Cdk1 [47]. We found a decrease of around 9-fold in the levels of Tyr15P-Cdk1 when the mkk1DDcrg1 allele was expressed (Figure 5E and 5F).
Since the observed decrease in inhibitory phosphorylation could promote the escape from G2 phase, and as a consequence a reduction in the size of daughter cells as well as production of cell aggregates, we sought to analyze what was the percentage of cells in G2 phase in conditions of mkk1DDcrg1 allele expression. For this we took advantage of previous studies showing that the mitotic cyclin Clb2 is synthesized during G2 phase and degraded at anaphase [48]. We constructed a strain carrying a clb2-GFP fusion at its native locus as well as the cut11-RFP fusion [49] to localize nuclear membrane and confirmed that the presence of GFP signal correlated with G2 phase (Figure S5). Cells carrying clb2-GFP marker and expressing mkk1DDcrg1 allele were scored as being in G2 phase or not, based on the presence or absence of a nuclear GFP signal. We observed that while in the control strain the percentage of cells being in G2 remained high during the incubation period, in the strain expressing mkk1DDcrg1 allele it was obvious a dramatic decrease in the number of cells showing clb2-GFP signal (Table 1). In summary these data indicated that overactivation of Mpk1 seems to force cells to escape from G2 phase.
Table 1. Cell population showing the G2 marker clb2-GFP.
| Induction time (h) | Cells with nuclear GFP signal* (%) | |
| control | mkk1DDcrg1 | |
| 0 | 30.4 | 29.2 |
| 3 | 28.6 | 18.4 |
| 6 | 32.1 | 6.7 |
| 9 | 26.4 | 4.4 |
| 12 | 22.3 | 1.8 |
* UMA88 (control) and UMA89 (mkk1DDcrg1) strains were incubated in YPA for the indicated time and around 100 cells in each time point (n = 2 experiments) were scored for the presence of Clb2-GFP signal. Numbers indicated percentage of cells in G2 phase.
G2 escape after over-activation of Mpk1 is dependent on Cdc25 phosphatase
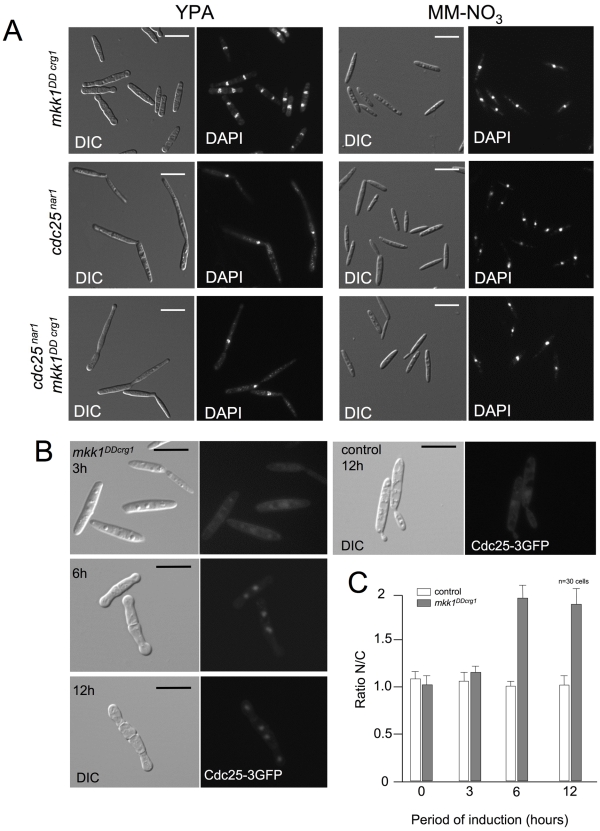
Levels of Cdk1 inhibitory phosphorylation in U. maydis are dependent on the activity of Wee1 kinase [47]. We found that over-activation of Mpk1 can overcome high levels of Wee1 produced ectopically (Figure S6). Two possible explanations could account for this result: either Mpk1 over-activation inhibits Wee1 activity or Mpk1 over-activation leads to an increase of some phosphatase activity able to remove inhibitory phosphorylation from Cdk1 -including conditions of high levels of Wee1. In U. maydis, the levels of Cdk1 inhibitory phosphorylation also depends on the activity of the Cdc25 phosphatase, which reverses the CDK inhibition [46]. To determine whether the Mpk1-induced escape from G2 was dependent on Cdc25, we introduced the conditional allele cdc25nar1 [46] into a strain carrying the mkk1DDcrg1 allele. This conditional allele enables the expression of cdc25 in cells incubated in minimal medium plus nitrate as nitrogen source (MM-NO3) as well as the down-regulation of cdc25 expression in rich medium. When cells carrying the cdc25nar1 conditional allele were grown in rich medium (repressive conditions) and in the presence of arabinose (inducing conditions for mkk1DDcrg1), they arrested in G2 phase (large buds, single nucleus) whether or not the mkk1DDcrg1 allele was expressed, indicating that Cdc25 was necessary for Mpk1-induced G2 escape (Figure 6A). This result indicates that no “alternative” phosphatase, putatively activated by Mpk1, is supplanting the Cdc25 role to promote the observed G2 escape; and also that it is unlikely that Wee1 is the target (direct or indirect) of Mpk1 overactivation: If the activity of Wee1 had been strongly inhibited, then the observed escape from G2 would not require Cdc25 activity. However, we found the opposite result: Cdc25 is required for Mpk1-mediated G2 escape. Therefore, the most plausible explanation is that Cdc25 is activated (direct or indirectly) by the CWI pathway over-activation.
Figure 6. Cdc25 is required for Mpk1-activated premature G2/M transition.
(A) Cdc25 is required for Mpk1-induced premature G2/M transition. DIC and DAPI images of UMA20 (mkk1DDcrg1), UMC27 (cdc25nar1) and UM56 (cdc25nar1 mkk1DDcrg1) cells grown for 6 h in YPA at 22°C (induction conditions for crg1, repressive for nar1) or minimal medium amended with nitrate and glucose (MM-NO3; repressing conditions for crg1 and induction for nar1). Bar 15 µm. (B) Cdc25 concentrates in the nucleus upon Mpk1 over-activation. DIC and epifluorescence images of MUM72 cells (control) carrying an endogenous Cdc25-3GFP as well as its derivated UMP184, expressing mkk1DD (mkk1DDcrg1), at different time points (hours) of incubation in YPA at 22°C. Bar 10 µm. (C) Graph indicates the quantification of nucleus/cytoplasm ratio of GFP fluorescence signal. Quantification of fluorescence signal was performed by measuring pixel intensity in the nuclei and in an equivalent area of the cytoplasm. Mean and s.d. are shown. 30 cells were measured in each time point (2 independent experiments).
We tried to address the organizational level at which Cdc25 is regulated after Mpk1 over-activation. Western blot analysis of Cdc25 levels in a strain carrying a myc-tagged Cdc25 [46] as well as the mkk1DDcrg1 allele showed no changes in Cdc25 levels after Mpk1 over-activation (not shown). Previous work done in our laboratory established that conditions resulting in nuclear accumulation of Cdc25 correlated with accelerated G2/M transition in U. maydis [50], [51]. Therefore, we also analyzed whether Mpk1 over-activation correlated with nuclear accumulation of Cdc25. To address this possibility we monitored the Cdc25 nucleus/cytoplasm distribution in cells carrying the mkk1DDcrg1 allele and expressing GFP-tagged Cdc25 (Figure 6B). To assess changes in localization quantitatively, fluorescence intensities of the Cdc25 signal were determined for a circular region corresponding to the nucleus (N) and a comparable area in the cytoplasm (C) as described [50]. We found under induction conditions (YPA) the ratio N/C in control cells was about 1, while the ratio in cell expressing the mkk1DDcrg1 allele was around 2 (Figure 6C). This correlation would suggest that the nuclear accumulation of Cdc25 is one possible cause for G2 escape when Mpk1 is over-activated.
N-terminal extension of Cdc25 is important for the response to cell wall damage
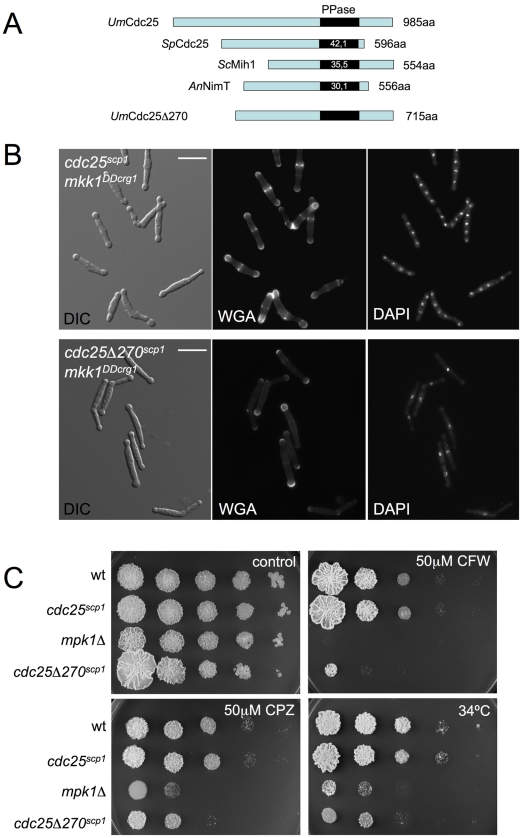
In comparison with other fungal Cdc25-like phosphatases, U. maydis Cdc25 carries an unusually long N-terminal extension of around 300 amino acids with no similarity to orthologues in the database (Figure 7A). Previous attempts to ascribe a role to this domain were not definitive because its deletion seemed not to affect Cdc25 function in U. maydis growing under normal conditions [46]. We wondered whether this region might be related to the G2 escape in response to Mpk1 over-activation. To address this question we constructed a N-terminal deletion that removed the first 270 amino acids, and we cloned the resulting deletion allele as well as a full-length allele under the control of the constitutively expressed scp1 promoter, that transcribes at a similar rate as the native cdc25 promoter (unpublished observations). The alleles cdc25scp1 and cdc25Δ270scp1 were exchanged with cdc25 locus in cells carrying mkk1DDcrg1. We found that deletion of the extra N-terminal domain of Cdc25 precludes the morphological effects of G2 escape. After 12 h of incubation under inducing conditions the majority of cells carried a single nucleus and these were able to divide by budding. Other effects of Mpk1 over-activation such as the swelling at the tips (Figure 7B) or the inability to form colonies in solid medium (not shown) were unaffected.
Figure 7. The N-terminal end of Cdc25 is required for Mpk1-induced premature G2/M transition.
(A) Representation of the U. maydis Cdc25 protein in relation to other fungal Cdc25-like proteins. The percentages inside each box represent the sequence identity of catalytic phosphatase domain when compared to the U. maydis sequence. Note the extended N-terminal end of UmCdc25. Derivate lacking the first 270 amino acids is also shown. (B) Cells carrying a cdc25 allele lacking the first 270 amino acids do not accelerate G2/M transition upon Mpk1 activation. Images show DIC as well as DAPI and WGA-FITC stained cells from UMA62 (cdc25scp1 mkkDDcrg1) and UMA63 (cdc25Δ270scp1 mkkDDcrg1) cultures in YPA for 8 h at 22°C. Bar 15 µm. (C) Cells lacking the N-terminal extension of Cdc25 were less resistant to cell wall stressors. Tenfold serial dilutions of cultures of FB1 (wt), UMA61 (cdc25scp1), UMA3 (mpk1Δ) and UMA79 (cdc25Δ270scp1) were spotted in solid YPD medium amended with Calcofluor White (CFW) and Chlorpromazine (CPZ). Plates were grown for 4 days at 28°C (control, CFW and CPZ) or 34°C.
To investigate the relationship between an impaired escape from G2 and cell resistance to cell wall stressors, we introduced the cdc25Δ270scp1 allele in wild-type cells and measured growth in the presence of cell wall stressors (Figure 7C). We found that cells carrying the cdc25Δ270scp1 allele were more sensitive than wild-type cells and control (cdc25scp1) cells, although to a lesser degree than mpk1Δ mutants, suggesting that cell cycle adjustment is one part of the adaptation of cells to these harsh conditions.
Cell cycle was not accelerated upon CWI pathway activation
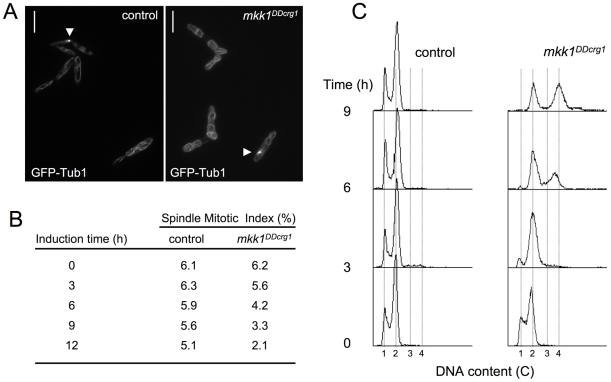
The above results indicated that CWI pathway over-activation resulted in an escape from G2 phase as a consequence of the decrease in Cdk1 inhibitory phosphorylation, most likely upon Cdc25 activation. Strikingly, this cell cycle adjustment seems to be necessary for appropriate response of U. maydis cells to cell wall stress. The general view is that in fungi, response to stress stimuli often involves a cell cycle arrest in order for cells to adapt to unfavorable stress conditions. However, escape from G2 -without any compensatory change in other phases of cell cycle- would result in a substantial decrease in the generation time (i. e., cell cycle acceleration). To address whether cell cycle was accelerated in response to CWI pathway over-activation, we estimated the doubling time of cells after Mpk1 over-activation. In control wild-type cells growing in YPA the doubling time changed from 2.2 to 2.5 h during the measured interval (12 h of incubation). However, in same conditions cells expressing the mkk1DDcrg1 allele showed a doubling time of around 3.2 h during the first cycle (cells carrying one single nucleus to cell aggregates showing two nuclei), while the second cycle (cell aggregates carrying two nuclei to cell aggregates carrying 4 nuclei) was roughly 6.4 h. As this result suggests that upon Mpk1 over-activation the cell cycle is slowing, we analyzed the proportion of cells entering into mitosis. Cells carrying a GFP-a tubulin fusion were scored as being in mitosis or not being in mitosis based on the presence or absence of a mitotic spindle (Spindle Mitotic Index, SMI; Figure 8A). With the observed increase in the duplication time, we consistently found a lower SMI in cells expressing the mkk1DDcrg1 allele (Figure 8B). In summary these data indicated that in spite of the apparent shorter G2 phase cells expressing the mkk1DDcrg1 allele showed a more lengthy cell cycle than control cells.
Figure 8. Cell cycle was not accelerated upon CWI pathway activation.
(A) Cell images of control (UMP61) and mkk1DDcrg1-derived (UMA84) cells carrying GFP-Tub1 to detect the microtubule cytoskeleton after 9 h of growth in YPA. Cells in interphase showed microtubules forming a cytoplasmic network, while in mitotic cells the microtubules are concentrated in the spindle (arrowhead). Bar: 15 µm. (B) Spindle Mitotic Index of control cells as well as cells with over-activated Mpk1. Cells were incubated in YPA for the indicated time and around 300 cells in each time point (n = 3 experiments) were scored for the presence of mitotic spindle. Numbers indicated percentage of cells showing mitotic spindle. (C) FACS analysis of the DNA content of FB1 (control) and UMA20 (mkk1DDcrg1) strains growing in YPA. The period of incubation in testing media is indicated (hours).
We believe that a plausible explanation for the above apparently contradictory results is that escape from G2 phase is compensated by the lengthening at another cell cycle phase (i.e. G1 or S-phase). To analyze whether G1 or S-phase was affected by over-activation of CWI pathway, we analyzed the DNA content of cells at different time points upon induction of mkk1DDcrg1 allele using Fluorescence/Activated Cell Sorter (FACS) (Figure 8C). We observed that upon over-activation of Mpk1 kinase, cells (or cell aggregates) accumulated with a 2C DNA content and over time even higher (3-4C). Since during that period the majority of cell aggregates carried two nuclei (for instance, after 6 h upon induction around 70% of cells showed a 2C DNA content and also showed 2 cell compartments, Figure 5B) we can deduce that these nuclei have a 1C DNA content each and therefore most likely are in G1 phase.
Taken together, these results suggest that upon over-activation of the CWI pathway, the cell cycle is affected in at least two different ways. Cells seem to escape from G2 phase and G1-S transition appears to be dramatically delayed.
Cell wall stressors affect cell cycle in wild-type cells
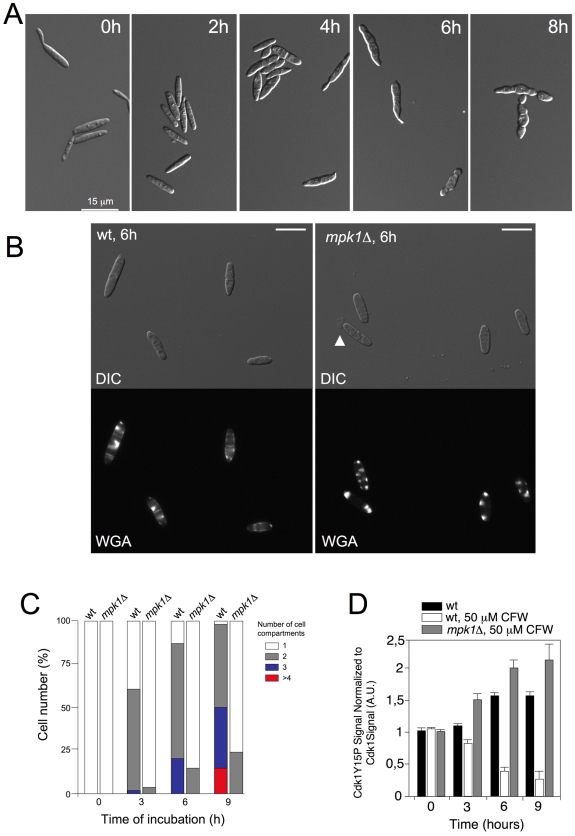
One concern in our approach is the use of a constitutive activated form of MEK. We reasoned that if our above results reflect a physiological response we should to be able to observe a similar response in wild-type cells under cell wall stress and that response should be abolished in a CWI pathway-defective mutant. Consistently, we observed a morphological response reminiscent of that observed with the mkk1DDcrg1 allele when wild-type cells were treated with 50 µM CFW: Cell aggregates resulted from septation without cell separation (Figure 9A–9C). When mpk1Δ cells were treated with CFW, cells remained undivided, WGA-FITC accumulation was observed around cell surface, and eventually these cells lysed near the cell tip (Figure 9B, arrowhead). In wild-type cells these results also correlated with a decrease in the levels of Cdk1 inhibitory phosphorylation after treatment with CFW, and again this response was dependent on an active CWI pathway (Figure 9D). We also found that the Spindle Mitotic Index of wild-type cells decreased to 3.5 after 6 hours of CFW while in untreated cells it was of 5.6 (no measurements were taken in mpk1Δ mutants because microtubules were severely disordered in these cells after CFW treatment). In summary, these results mirrored those found when a constitutively activated mkk1DDcrg1 allele was used.
Figure 9. Cell cycle changes after CFW treatment.
(A) DIC images of wild-type (FB1) cells treated treated with 50 µM CFW for the time indicated. Bar: 15 µm. (B) Aspect of wild-type and mpk1Δ cells grown in the presence of 50 µM CFW for 6 hours. WGA-FITC staining shows the presence of cell compartments in wild-type cells as well as its absence in mutant cells. Arrowhead indicates a cell leaking cytoplasmic material at the tip. Bar: 10 µm. (C) Quantification of the number of cell compartments per cell aggregate at different time points in wild-type and mpk1Δ cells. At least 50 cell aggregates were counted at each time point (2 independent experiments). (D) Levels of Cdk1 inhibitory phosphorylation after CFW treatment in control (FB1) and mpk1Δ (UMA3) cells. Cdk1 inhibitory phosphorylation was measured as described in Figure 5. Mean and s.d. are shown (2 independent experiments).
Discussion
The main conclusion of this study is that in U. maydis activation of the CWI pathway promotes escape from G2 phase. This result contrasts with the G2 cell cycle arrest that occurs in S. cerevisiae in response to CWI pathway activation [12]. Since our results strongly suggest that the pathway described in this study is the bona fide CWI pathway in U. maydis, it is of interest to understand what might explain the disparate responses in these organisms. This distinct pattern of cell cycle arrest likely reflects different cellular tactics used to pursue a common strategic mechanism. Cell wall is remodeled throughout the cell cycle. During periods of polarized cell growth, the wall is loosened by digestive enzymes, such a chitinases and glucanases, and expanded at these specific regions on the cell surface. During these periods, cells experience the greatest wall stress and are most vulnerable to lysis. Several reports support the view that apical growth is exquisitely sensitive to loss of integrity –tip lysis occurs in response to osmotic stress or treatment with drugs targeting chitin and glucan synthases such as nikkomycin and caspofungin [52], [53]. It follows that phases where polar growth is most active are the phases in which cells are most vulnerable to cell wall instability.
Under these conditions, if for some reason cell wall biosynthesis is compromised, a survival strategy might be to escape from cell cycle phases where polar growth is higher. Interestingly, the cell cycle stage at which polar growth is maximum is different in each organism. While in S. cerevisiae polar growth reaches its maximal at late G1 and switches to isotropic growth during G2 [54], in U. maydis the maximum of polar growth occurs at the G2 phase where the characteristic elongated bud is made [55]. Consistently, activation of CWI pathway in S. cerevisiae drives the cell cycle to remain in G2 phase, where cell growth is isotropic, while in U. maydis activation of CWI drives the cell cycle to escape from G2, where polar growth is more pronounced.
Our results also suggest that the promoted escape from G2 does not necessarily imply a faster cell cycle. In fact, we observed a pronounced increase in the generation time upon CWI pathway over-activation, suggesting that shortening of the G2 phase is compensated by the lengthening of another cell cycle phase. We believe that this delay occurs on G1/S transition, although we do not understand the mechanism through which this delay takes place. At this point we do not know whether this G1/S delay is part of the cell cycle response to over-activation of CWI pathway or is simply a consequence of the observed growth defect. Previously reported U. maydis G1 regulators such as the APC activator Cru1 [56] or the G1 cyclin Cln1 [57] seem not to affect this delay (NC, unpublished observations). We investigated in more detail the mechanism responsible for the observed G2 escape upon CWI pathway over-activation. It appears that activation of the CWI pathway results in the accumulation of the Tyr15-unphosphorylated form of Cdk1 and most likely it results in the observed escape from G2 towards mitosis. We found that Cdc25 was the sole mitotic-inducer phosphatase in U. maydis required for this phenotype. In response to activation of CWI pathway, Cdc25 accumulates in the nucleus, providing a likely explanation for the increase of the unphosphorylated form of Cdk1. We also found that a mutant allele of Cdc25 lacking the first 270 residues ameliorates the premature G2/M transition upon activation of CWI pathway. In addition, these mutant cells showed a higher sensitivity to the presence of cell wall stressors. We hypothesize that Mpk1 directly targets Cdc25 at its extended N-terminal domain promoting nuclear import. Recently, it has been reported in Xenopus that p42 MAPK, the Xenopus ortholog of mammalian ERK2, is able to directly phosphorylate and activate Cdc25C [58]. To date, however, we have obtained no experimental support for this notion in U. maydis. We did not detect any physical interaction between Cdc25 and Mpk1 by co-immunoprecipitation (not shown), nor did we observe any differential migration of Cdc25 in SDS-PAGE gels after response to Mpk1 activation that might suggest direct phosphorylation. We considered alternative explanations. A three-amino acid motif (S/TPS/T) was described that is phosphorylated upon MAPK-associated stimulation to induce Importin 7-dependent nuclear translocation [59]. We were intrigued to note the N-terminal extension of U. maydis Cdc25 contains several S/TPS/T motifs. However, deletion of the putative Importin 7 (um01606) had no effect on the MAPK-mediated nuclear translocation of Cdc25 (not shown). We are now investigating other factors that are able to specifically interact with the N-terminal domain of Cdc25, such as 14-3-3 protein [50].
One cautionary aspect of our approach is whether the cell cycle effects observed are a consequence of the toxicity resulting from forced expression of mkk1DD. We believe that our results provide strong evidence that at least, G2 escape seems to be a result of CWI pathway activation unrelated from the toxicity. First, we were able to observe similar effects on the cell cycle upon treatment of wild-type cells with cell wall stressors and these effects were dependent on the presence of an active CWI pathway. Second, the correlation between inability to escape from G2 and enhanced sensitivity to cell wall stressors observed with the Cdc25 mutant lacking the N-terminal extension, strongly supports the idea that cell cycle modification is part of the cell response to cell wall insults in U. maydis. Third, the toxicity effects we observed are unlikely to be related to the cell cycle effects. Cells expressing mkk1DD had a strong lysis phenotype and the Mkk1DD-induced toxicity can be alleviated growing the cells in the presence of an osmotic stabilizer or at low temperature (not shown). However, these conditions do not preclude the premature G2/M transition observed when CWI pathway is activated. Furthermore in a strain carrying the cdc25Δ270 allele, the expression of mkk1DD was also toxic. We propose that this toxicity could be a consequence of unbalanced cell wall synthesis because of the sustained high activation of the CWI pathway. Concomitant, but independent of G2 escape, was the dramatic swelling of cell tips that stained strongly with WGA, indicating a continuous deposition of new cell wall material occurring at the tip. It is worth noting that toxicity was also reported in S. cerevisiae upon expression of an activated MKK1 allele [60], in spite of the different effects on cell cycle that activation of CWI pathway has in these species.
In summary, our findings support the view that in response to environmental input, the cell has to choose which cell cycle stage is the most appropriate cellular environment to respond to such stimulus and then moves towards such a cell cycle phase. The appropriate cell cycle stage could well differ from one organism to other, and so the observed response.
Materials and Methods
Strains and growth conditions
Ustilago maydis strains are listed in Table 2 and are derived from FB1 background [61]. Media were prepared as described [62]. Controlled expression of genes under the crg1 and nar1 promoter was performed as described previously [48], [63]. FACS analyses were described previously [22]. Drug sensitivity assays were performed as described [64].
Table 2. U. maydis strains used in this study.
| Strain | Relevant genotype | Reference |
| FB1 | a1 b1 | [61] |
| UMA1 | a1 b1 bck1Δ | This work |
| UMA2 | a1 b1 mkk1Δ | This work |
| UMA3 | a1 b1 mpk1Δ | This work |
| UMA4 | a1 b1 bck1Δ mkk1Δ | This work |
| UMA5 | a1 b1 bck1Δ mpk1Δ | This work |
| UMA6 | a1 b1 mkk1Δ mpk1Δ | This work |
| UMA8 | a1 b1 bck1Δ mkk1Δ mpk1Δ | This work |
| UMA44 | a1 b1 mpk1-3HA | This work |
| UMA44.2 | a1 b1 mpk1-3HA bck1Δ | This work |
| UMA44.3 | a1 b1 mpk1-3HA mkk1Δ | This work |
| UMA59 | a1 b1 mpk1AEF-3HA | This work |
| UMA60 | a1 b1 mpk1KD-3HA | This work |
| UMA10 | a1 b1 Pcrg1 | This work |
| UMA12 | a1 b1 Pcrg1:mkk1 | This work |
| UMA7 | a1 b1 Pcrg1:mkk1DD | This work |
| UMA57 | a1 b1 Pcrg1:mkk1DD bck1Δ | This work |
| UMA9 | a1 b1 Pcrg1:mkk1DD mkk1Δ | This work |
| UMA13 | a1 b1 Pcrg1:mkk1DD mpk1Δ | This work |
| UMA20 | a1 b1 mkk1DD crg1 | This work |
| UMA42 | a1 b1 mkk1DD crg1 mpk1Δ | This work |
| UMA43 | a1 b1 mkk1DD crg1 mpk1-3HA | This work |
| UMA64 | a1 b1 mkk1DD crg1 mpk1AEF-3HA | This work |
| UMA65 | a1 b1 mkk1DD crg1 mpk1KD-3HA | This work |
| UMA54 | a1 b1 mkk1DD crg1 mpk1-3HA NLS-GFP | This work |
| UMC48 | a1 b1 Pcrg1:wee1 | [47] |
| UMA79 | a1 b1 Pcrg1:wee1 mkk1DD crg1 | This work |
| UMC27 | a1 b1 cdc25nar1 | [46] |
| UMA56 | a1 b1 cdc25nar1 mkk1DD crg1 | This work |
| MUM72 | a1 b1 cdc25-3GFP | [48] |
| UMP184 | a1 b1 cdc25-3GFP mkk1DD crg1 | This work |
| UMA62 | a1 b1 cdc25scp mkk1DD crg1 | This work |
| UMA63 | a1 b1 cdc25Δ270scp mkk1DD crg1 | This work |
| UMA61 | a1 b1 cdc25scp | This work |
| UMA79 | a1 b1 cdc25Δ270scp | This work |
| UMP61 | a1 b1 GFP-tub1 | [66] |
| UMA84 | a1 b1 GFP-tub1 mkk1DD crg1 | This work |
| UMA88 | a1 b1 clb2-GFP cut11-RFP | This work |
| UMA89 | a1 b1 clb2-GFP cut11-RFP mkk1DD crg1 | This work |
Protein analysis
Protein extracts and Western blotting were performed as described previously [65]. To purify Mpk1 complexes and analyze their kinase activity previously described protocols were followed [65]. All quantification was done using a Phosphorimager (Molecular Dynamics). To detect the phosphorylated and non-phosphorylated forms of Cdk1 and Mpk1 commercial antibodies were used as described [47], [48], [65]. Primary antibody was followed by a secondary antibody conjugated to horseradish peroxidase and immunoreactive proteins were visualized using a chemiluminescent susbtrate. The chemiluminescent signal was analyzed using ChemiDoc XCS+ (Molecular Imager, Bio-Rad).
Plasmid and strain constructions
Plasmid pGEM-T easy (Promega) was used for cloning, subcloning and sequencing of genomic fragments and fragments generated by PCR. Sequence analysis of fragments generated by PCR was performed with an automated sequencer (ABI 373A) and standard bioinformatic tools. To construct the different strains, transformation of U. maydis protoplasts with the indicated constructions was performed as described previously [66]. Gene replacement into the corresponding loci was verified by diagnostic PCR and subsequent Southern blot analysis. U. maydis DNA isolation was performed as previously described [66]. Fluorescent protein fusions were described previously: Cdc25-3GFP [50], NLS-GFP [45], GFP-Tub1 [55], Cut11-RFP [49], Clb2-GFP (JPM, unpublished).
Deletion of each kinase gene was performed using PCR-based gene targeting [27]. Briefly, a pair of DNA fragments flanking the corresponding kinase ORF were amplified and ligated to antibiotic resistance cassettes via SfiI sites. The 5′ and 3′ fragments were amplified using specific oligonucleotide pairs (Table 3). Each fragment was about 1 kbp in length. For bck1 we used primer pairs Bck1-2/Bck1-3 (5′ end) and Bck1-4/Bck1-5 (3′ end) and we ligated to a nourseotricin resistance cassette. For mkk1 we used primer pairs Mkk1-2/Mkk1-3 (5′ end) and Mkk1-4/Mkk1-5 (3′ end) and we ligated to a hygromycin resistance cassette. For mpk1 we used primer pairs Slt2-4/Slt2-r (5′ end) and Slt2-5/Slt2-f (3′ end) and we ligated to carboxin as well as hygromycin resistance cassettes. Double and triple mutants were obtained after cross the respective parental strains and selection of haploid progeny resistant to the respective markers [62].
Table 3. Oligonucleotides used in this study.
| Name | Sequence |
| BCK1-2 | 5′ACGCACCACGCACGACCACAGACTCGT3′ |
| BCK1-3 | 5′GTGGGCCATCTAGGCCAACGGTTGGATTGGCTTGA3′ |
| BCK1-4 | 5′CACGGCCTGAGTGGCCATTGCTTCATGCGGTTGC3′ |
| BCK1-5 | 5′TCTATGTCCACCAACATGCTCATGGGT3′ |
| MKK1-2 | 5′GCCTGATGCAAGCAAATCAGTGTCAGT3′ |
| MKK1-3 | 5′GTGGGCCATCTAGGCCAAGCAGCAGCACATACAGTC3′ |
| MKK1-4 | 5′CACGGCCTGAGTGGCCCTCACGTGCAATCTTAGTATG3′ |
| MKK1-5 | 5′CTCAAGCTTTTGTCGTAACGATGGTGT3′ |
| MKK1-E1 | 5′CATATGGCCTCGCTCATTCCACCGAAG3′ |
| MKK1-E2 | 5′GAGCTGATCAACGACGTTGCCGGCGATTTTACGGGCACA3′ |
| MKK1-E3 | 5′TGTGCCCGTAAAAGTACCGGCAACCGAGTTGATCAGCTC3′ |
| MKK1-E4 | 5′GAATTCAGCAACATAATCAGATTGCAC3′ |
| MKK1-8E | 5′GAATTCCGGCGTATACTCATACGCCAT3′ |
| MKK1-9B | 5′GGATCCGCCTTTTGACCATCAATCTC3′ |
| SLT2-4 | 5′AGTCACAACGTGTTGGTTCAAAGTTTA3′ |
| SLT2-r | 5′GTGGGCCATCTAGGCCTTGTCTACGTCAAGATCA3′ |
| SLT2-5 | 5′TTTCATGTGGGAATGGTATACCACCAA3′ |
| SLT2-f | 5′CACGGCCTGAGTGGCCACAGCTCACCTCGCGGTC3′ |
| SLT2-KF | 5′GGTACCACTCCCGACTGTACAGTGCAGTA3′ |
| SLT2-10 | 5′GCTGATCTTGACGTAGACCATATGTCGGCACATCCATCAAAC3′ |
| SLT2-11 | 5′GTTTGATGGATGTGCCGACATATGGCTTACGTCAAGATCAGC3′ |
| SLT2-KR | 5′GGTACCGATTTTGACCGCGAGGTGAGCTG3′ |
| SLT2-A1 | 5′GGCGCAGGTTTCATGGCAGAATTCGTAGCAACTCGATGG3′ |
| SLT2-A2 | 5′CCATCGAGTTGCTACGAATTCTGCCATGAAACCTGCGCC3′ |
| SLT2-B1 | 5′GAAAGCGTAGCTATCAGAAAGATCACCAACGTC3′ |
| SLT2-B2 | 5′GACGTTGGTGATCTTTCTGATAGCTACGCTTTC3′ |
For N-terminal fusion of Mpk1 with HA, a NdeI site was introduced at the ATG of mpk1 locus using PCR-based two-step mutagenesis protocol (amplification was performed using the primer pairs: Slt2-KF/Slt2-11 and Slt2-10/Slt2-KR). A stop-less NdeI 3xHA cassette was introduced at the respective NdeI site and the HA-Mpk1 fusion integrated by homologous recombination under their respective native mpk1 promoter.
The mutant mpk1AEF allele was constructed by the assembly of two fragments carrying the T193A and Y195F mutations generated by PCR with the primer pairs Slt2-KF/Slt2-A2 and Slt2-A1/Slt2-KR. As template we used the mpk1-HA allele.
The mutant mpk1KD allele was constructed by the assembly of two fragments carrying the K55R mutation generated by PCR with the primer pairs Slt2-KF/Slt2-B2 and Slt2-B1/Slt2-KR. As template we used the mpk1-HA allele.
The mutant mkk1DD allele was constructed by the assembly of two fragments carrying the S373D and T377D mutations generated by PCR with the primer pairs Mkk1-E1/Mkk1-E3 and Mkk1-E2/Mkk1-E4. The wild-type mkk1 allele was amplified using the primer pair Mkk1-E1/Mkk1-E4. In both cases they produced a 2.1 kb fragment flanked by NdeI and EcoRI sites that were cloned into pRU11 an integrative U. maydis vector that contains the crg1 promoter [63] and the resulting plasmid was linearized and integrated into the ip locus. To integrate the mkk1DDcrg1 allele into its native locus, the 2.1 kb NdeI-EcoRI fragment carrying the mkk1DD allele was ligated together a 0.68 kb EcoRI-BamHI fragment carrying the 3′ region of mkk1 (generated by PCR using the primer pair Mkk1-8E and Mkk1-9B) into the plasmid pRU11-Hyg [67] digested with NdeI-EcoRI. The resulting plasmid after linearization with EcoRI was integrated at the mkk1 locus.
To produce the alleles cdc25scp1 and cdc25Δ270scp1, we exchanged in the pCdc25nar1 and pCDC25Δ270nar1 plasmids [46] the nar1 promoter, which is flanked by NotI-NdeI sites, with a NotI-NdeI fragment carrying the scp1 promoter, a weak U. maydis constitutive promoter [47].
Microscopy
Samples were mounted on microscope slides and visualized in a Nikon Eclipse 90i microscope equipped with a Hamamatsu ORCA-ER CCD camera. All the images in this study are single planes. Standard DAPI, rhodamine and GFP filter sets were used for epifluorescence analysis. The software used with the microscope was MetaMorph 7.1 (Universal Imaging, Downingtown, PA). Images were further processed with Adobe Photoshop 8.0.
To quantify the ratio of the nuclear intensity (N) to the cytoplasmic intensity (C) of Cdc25-GFP signal already described procedures were followed [50]. Briefly, the intensity of the nuclear and cytoplasmic signal was determined by measuring pixel intensity in the nucleus and of an equivalent area in the cytoplasm and the ratio was determined. 30 cells were quantified for each experiment.
Supporting Information
Dendrogram analysis of MEKK, MEK and MAPK extracted from U. maydis database and compared with S. cerevisiae and S. pombe. Branches were named after the S. cerevisiae MAPK pathway.
(0.30 MB TIF)
Levels of Thr193 and Tyr195 Mpk1 phosphorylation after treatment with cell wall stressors. Protein extracts from the indicated strains growing in the presence of cell wall stressors for the indicated time (minutes) were separated by SDS-PAGE. Immunoblots were incubated successively with an antibody that recognizes the Mpk1 phosphorylated form (anti-pTEpY) and anti-HA. The levels of phosphorylation were determined by quantifying the level of antibody signal using a ChemiDoc (Bio-Rad). Signal from the phosphopeptide-specific antibodies was normalized to the amount of phosphorylation of at time zero. Differences in loading of samples were corrected by dividing each phosphopeptide-specific antibody signal by the anti-HA antibody signal. Mean and s.d. are shown (n = 3 experiments).
(0.03 MB PDF)
Expression of mkk1DD resulted in cell lysis. Strain UMA7 was grown at 28°C in YPA for 8 h. Weakness of cell wall at the tips was apparent by the leakage of cell material.
(0.09 MB PDF)
Deletion of mpk1 gene abolishes the morphological defects observed upon expression of mkk1DD. Strains FB1 (wt), UMA7 (Pcrg1: mkk1DD) and UMA13 (Pcrg1: mkk1DD mpk1Δ) were grown in YPA at 28°C for 8 h.
(1.14 MB PDF)
Clb2-GFP is a marker of G2 phase in U. maydis. A. Scheme of cell cycle in U. maydis where different phases are associated to specific morphological markers: microtubules, nuclear membrane, and presence or absence of buds (based in results from different sources: [Perez-Martin, et al; Steinberg, et al; Straube, et al]). Presence of Clb2-GFP fluorescence is also included from our data. Cells without buds are in G1 and S phase. Cells with buds, one single nucleus surrounded by an intact nuclear membrane, and a cytoplasmic array of MT are G2 cells. Cells with large buds, mitotic spindle and nuclear membrane partially disassembled are in mitosis and finally cells with large buds, two nuclei (one in the bud and other in mother cell), cytoplasmic array of microtubules and in the process of cell separation are early G1 cells. B. Correlation between presence of GFP signal (GFP+) and G2 phase. A strain carrying Clb2-GFP and Cut11-RFP fusions was grown in YPD until mid-log phase. 200 cells were counted and sorted in 4 different groups depending on presence or absence of bud, number of nuclei and integrity of nuclear membrane. C. Cell images of cells carrying Clb2-GFP and Cut11-RFP fusion showing different cell cycle phases. [Perez-Martin J, Castillo-Lluva S, Sgarlata C, Flor-Parra I, Mielnichuk N, et al. (2006) Pathocycles: Ustilago maydis as a model to study the relationships between cell cycle and virulence in pathogenic fungi. Mol Genet Genomics 276: 211-229. Steinberg G, Wedlich-Soldner R, Brill M, Schulz I (2001) Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J Cell Sci 114: 609-622. Straube A, Weber I, Steinberg G (2005) A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J 24: 1674-1685.]
(2.32 MB TIF)
Ectopic expression of wee1 does not preclude accelerated G2/M transition upon over-activation of Mpk1. (A) DIC and DAPI images of UMA20 (mkk1DDcrg1), UMC48 (Pcrg1:wee1) and UMA79 (Pcrg1:wee1 mkk1DDcrg1) cells grown for 8 h in YPA at 22°C. Bar 15 µm. In this experiment, we wondered whether high levels of wee1 expression were able to preclude the G2 shortening observed after over-activation of Mpk1. To address this question we introduced an ectopic copy of a MYC-tagged wee1 allele under the control of crg1 promoter in a strain carrying the mkk1DDcrg1 allele. By this manipulation, over-activation of Mpk1 (upon expression of the mkk1DDcrg1 allele) will occur concomitantly with over-expression of wee1. We found that high levels of Wee1 in a control strain produced elongated buds with a single nucleus (A, middle column), concomitant with a G2 cell cycle arrest. However, when wee1 was overexpressed in cells carrying the mkk1DDcrg1 allele, the presence of high levels of Wee1 did not prevent the G2 shortening. The observed cell compartments in wee1-overexpressing strain were slightly larger than those observed in the strain not expressing wee1 (182 µm3 in cells overexpressing wee1 versus 104 µm3 in cells not overexpressing wee1 on average for cell aggregates carrying at least four cell compartments). (B) Western blot showing the levels of Wee1-myc in the above strains, after grown for 8 h under inducing (YPA) and repressing (YPD) conditions for crg1. As loading control Cdk1 and Cdk5 levels were analyzed using anti-PSTAIRE antibodies.
(1.15 MB TIF)
Acknowledgments
We are indebted to Prof. William K. Holloman (Cornell University, USA) for editing, critical reading, and suggestions of this manuscript. We also thank Prof. N. A. R. Gow (University of Aberdeen, UK) for critical reading and suggestions. Also we want to thank to members of Signalpath Consortium for helpful discussions. The helpful suggestions from the anonymous reviewers were also appreciated.
Footnotes
The authors have declared that no competing interests exist.
This work was funded by Grants from the Spanish government (BIO2008-04054), local government (S-SAL-246-2006), and the EU (MRTN-CT-2005-019277). NC was supported by the CSIC/BSCH contract B104BSCH02. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson MG, Millar JB. Control of the eukaryotic cell cycle by MAP kinase signaling pathways. FASEB J. 2000;14:2147–2157. doi: 10.1096/fj.00-0102rev. [DOI] [PubMed] [Google Scholar]
- 3.MacCorkle RA, Tan TH. Mitogen-activated protein kinases in cell-cycle control. Cell Biochem Biophys. 2005;43:451–461. doi: 10.1385/CBB:43:3:451. [DOI] [PubMed] [Google Scholar]
- 4.Clotet J, Posas F. Control of cell cycle in response to osmostress: lessons from yeast. Methods Enzymol. 2007;428:63–76. doi: 10.1016/S0076-6879(07)28004-8. [DOI] [PubMed] [Google Scholar]
- 5.Peter M, Herskowitz I. Joining the complex: cyclin-dependent kinase inhibitory proteins and the cell cycle. Cell. 1994;79:181–184. doi: 10.1016/0092-8674(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 8.Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 9.Tyers M, Futcher B. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol Cell Biol. 1993;13:5659–5669. doi: 10.1128/mcb.13.9.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clotet J, Escote X, Adrover MA, Yaakov G, Gari E, et al. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J. 2006;25:2338–2346. doi: 10.1038/sj.emboj.7601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escote X, Zapater M, Clotet J, Posas F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat Cell Biol. 2004;6:997–1002. doi: 10.1038/ncb1174. [DOI] [PubMed] [Google Scholar]
- 12.Harrison JC, Bardes ES, Ohya Y, Lew DJ. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat Cell Biol. 2001;3:417–420. doi: 10.1038/35070104. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Martin J, Castillo-Lluva S, Sgarlata C, Flor-Parra I, Mielnichuk N, et al. Pathocycles: Ustilago maydis as a model to study the relationships between cell cycle and virulence in pathogenic fungi. Mol Genet Genomics. 2006;276:211–229. doi: 10.1007/s00438-006-0152-6. [DOI] [PubMed] [Google Scholar]
- 14.Steinberg G, Perez-Martin J. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 2008;18:61–67. doi: 10.1016/j.tcb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Andrews DL, Egan JD, Mayorga ME, Gold SE. The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol Plant Microbe Interact. 2000;13:781–786. doi: 10.1094/MPMI.2000.13.7.781. [DOI] [PubMed] [Google Scholar]
- 16.Banuett F, Herskowitz I. Identification of Fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 1994;8:1367–1378. doi: 10.1101/gad.8.12.1367. [DOI] [PubMed] [Google Scholar]
- 17.Mayorga ME, Gold SE. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol Microbiol. 1999;34:485–497. doi: 10.1046/j.1365-2958.1999.01610.x. [DOI] [PubMed] [Google Scholar]
- 18.Muller P, Aichinger C, Feldbrugge M, Kahmann R. The MAP kinase Kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol Microbiol. 1999;34:1007–1017. doi: 10.1046/j.1365-2958.1999.01661.x. [DOI] [PubMed] [Google Scholar]
- 19.Muller P, Weinzierl G, Brachmann A, Feldbrugge M, Kahmann R. Mating and pathogenic development of the Smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot Cell. 2003;2:1187–1199. doi: 10.1128/EC.2.6.1187-1199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stern B, Nurse P. Fission yeast pheromone blocks S-phase by inhibiting the G1 cyclin B-p34cdc2 kinase. EMBO J. 1997;16:534–544. doi: 10.1093/emboj/16.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stern B, Nurse P. Cyclin B proteolysis and the cyclin-dependent kinase inhibitor rum1p are required for pheromone-induced G1 arrest in fission yeast. Mol Biol Cell. 1998;9:1309–1321. doi: 10.1091/mbc.9.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Muse T, Steinberg G, Perez-Martin J. Pheromone-induced G2 arrest in the phytopathogenic fungus Ustilago maydis. Eukaryot Cell. 2003;2:494–500. doi: 10.1128/EC.2.3.494-500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez-Martinez C, Perez-Martin J. Dimorphism in fungal pathogens: Candida albicans and Ustilago maydis–similar inputs, different outputs. Curr Opin Microbiol. 2001;4:214–221. doi: 10.1016/s1369-5274(00)00191-0. [DOI] [PubMed] [Google Scholar]
- 24.Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rispail N, Soanes DM, Ant C, Czajkowski R, Grunler A, et al. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol. 2009;46:287–298. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- 27.Brachmann A, Konig J, Julius C, Feldbrugge M. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol Genet Genomics. 2004;272:216–226. doi: 10.1007/s00438-004-1047-z. [DOI] [PubMed] [Google Scholar]
- 28.Nagata Y, Burger MM. Wheat germ agglutinin. Molecular characteristics and specificity for sugar binding. J Biol Chem. 1974;249:3116–3122. [PubMed] [Google Scholar]
- 29.Elorza MV, Rico H, Sentandreu R. Calcofluor white alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J Gen Microbiol. 1983;129:1577–1582. doi: 10.1099/00221287-129-5-1577. [DOI] [PubMed] [Google Scholar]
- 30.Kamada Y, Jung US, Piotrowski J, Levin DE. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 31.Levin DE, Bowers B, Chen CY, Kamada Y, Watanabe M. Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae. Cell Mol Biol Res. 1994;40:229–239. [PubMed] [Google Scholar]
- 32.Torres L, Martin H, Garcia-Saez MI, Arroyo J, Molina M, et al. A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2 mutants. Mol Microbiol. 1991;5:2845–2854. doi: 10.1111/j.1365-2958.1991.tb01993.x. [DOI] [PubMed] [Google Scholar]
- 33.Cobb MH, Goldsmith EJ. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 34.Marshall CJ. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 35.Kraus PR, Fox DS, Cox GM, Heitman J. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol Microbiol. 2003;48:1377–1387. doi: 10.1046/j.1365-2958.2003.03508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madrid M, Soto T, Khong HK, Franco A, Vicente J, et al. Stress-induced response, localization, and regulation of the Pmk1 cell integrity pathway in Schizosaccharomyces pombe. J Biol Chem. 2006;281:2033–2043. doi: 10.1074/jbc.M506467200. [DOI] [PubMed] [Google Scholar]
- 37.Martin H, Rodriguez-Pachon JM, Ruiz C, Nombela C, Molina M. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J Biol Chem. 2000;275:1511–1519. doi: 10.1074/jbc.275.2.1511. [DOI] [PubMed] [Google Scholar]
- 38.Verna J, Lodder A, Lee K, Vagts A, Ballester R. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 40.Zheng CF, Guan KL. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J. 1994;13:1123–1131. doi: 10.1002/j.1460-2075.1994.tb06361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pages G, Brunet A, L'Allemain G, Pouyssegur J. Constitutive mutant and putative regulatory serine phosphorylation site of mammalian MAP kinase kinase (MEK1). EMBO J. 1994;13:3003–3010. doi: 10.1002/j.1460-2075.1994.tb06599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiozaki K, Shiozaki M, Russell P. Heat stress activates fission yeast Spc1/StyI MAPK by a MEKK-independent mechanism. Mol Biol Cell. 1998;9:1339–1349. doi: 10.1091/mbc.9.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bottin A, Kamper J, Kahmann R. Isolation of a carbon source-regulated gene from Ustilago maydis. Mol Gen Genet. 1996;253:342–352. doi: 10.1007/pl00008601. [DOI] [PubMed] [Google Scholar]
- 44.Loubradou G, Brachmann A, Feldbrugge M, Kahmann R. A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol Microbiol. 2001;40:719–730. doi: 10.1046/j.1365-2958.2001.02424.x. [DOI] [PubMed] [Google Scholar]
- 45.Straube A, Enard W, Berner A, Wedlich-Soldner R, Kahmann R, et al. A split motor domain in a cytoplasmic dynein. EMBO J. 2001;20:5091–5100. doi: 10.1093/emboj/20.18.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sgarlata C, Perez-Martin J. The Cdc25 phosphatase is essential for the G2/M phase transition in the basidiomycete yeast Ustilago maydis. Mol Microbiol. 2005;58:1482–1496. doi: 10.1111/j.1365-2958.2005.04925.x. [DOI] [PubMed] [Google Scholar]
- 47.Sgarlata C, Perez-Martin J. Inhibitory phosphorylation of a mitotic cyclin-dependent kinase regulates the morphogenesis, cell size and virulence of the smut fungus Ustilago maydis. J Cell Sci. 2005;118:3607–3622. doi: 10.1242/jcs.02499. [DOI] [PubMed] [Google Scholar]
- 48.Garcia-Muse T, Steinberg G, Perez-Martin J. Characterization of B-type cyclins in the smut fungus Ustilago maydis: roles in morphogenesis and pathogenicity. J Cell Sci. 2004;117:487–506. doi: 10.1242/jcs.00877. [DOI] [PubMed] [Google Scholar]
- 49.Perez-Martin J. DNA-damage response in the basidiomycete fungus Ustilago maydis relies in a sole Chk1-like kinase. DNA Repair (Amst) 2009;8:720–731. doi: 10.1016/j.dnarep.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 50.Mielnichuk N, Perez-Martin J. 14-3-3 regulates the G2/M transition in the basidiomycete Ustilago maydis. Fungal Genet Biol. 2008;45:1206–1215. doi: 10.1016/j.fgb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 51.Mielnichuk N, Sgarlata C, Perez-Martin J. A role for the DNA-damage checkpoint kinase Chk1 in the virulence program of the fungus Ustilago maydis. J Cell Sci. 2009;122:4130–4140. doi: 10.1242/jcs.052233. [DOI] [PubMed] [Google Scholar]
- 52.Bartnicki-Garcia S, Lippman E. The bursting tendency of hyphal tips of fungi: presumptive evidence for a delicate balance between wall synthesis and wall lysis in apical growth. J Gen Microbiol. 1975;73:487–500. [Google Scholar]
- 53.Gooday GW. The dynamics of hyphal growth. Mycol Res. 1995;99:385–394. [Google Scholar]
- 54.Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinberg G, Wedlich-Soldner R, Brill M, Schulz I. Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J Cell Sci. 2001;114:609–622. doi: 10.1242/jcs.114.3.609. [DOI] [PubMed] [Google Scholar]
- 56.Castillo-Lluva S, Garcia-Muse T, Perez-Martin J. A member of the Fizzy-related family of APC activators is regulated by cAMP and is required at different stages of plant infection by Ustilago maydis. J Cell Sci. 2004;117:4143–4156. doi: 10.1242/jcs.01304. [DOI] [PubMed] [Google Scholar]
- 57.Castillo-Lluva S, Perez-Martin J. The induction of the mating program in the phytopathogen Ustilago maydis is controlled by a G1 cyclin. Plant Cell. 2005;17:3544–3560. doi: 10.1105/tpc.105.036319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang R, He G, Nelman-Gonzalez M, Ashorn CL, Gallick GE, et al. Regulation of Cdc25C by ERK-MAP kinases during the G2/M transition. Cell. 2007;128:1119–1132. doi: 10.1016/j.cell.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 59.Chuderland D, Konson A, Seger R. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol Cell. 2008;31:850–861. doi: 10.1016/j.molcel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe Y, Irie K, Matsumoto K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banuett F, Herskowitz I. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci U S A. 1989;86:5878–5882. doi: 10.1073/pnas.86.15.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holliday R. Ustilago maydis. In: King RC, editor. Handbook of Genetics. New York: Plenum Press; 1974. pp. 575–595. [Google Scholar]
- 63.Brachmann A, Weinzierl G, Kamper J, Kahmann R. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol Microbiol. 2001;42:1047–1063. doi: 10.1046/j.1365-2958.2001.02699.x. [DOI] [PubMed] [Google Scholar]
- 64.Castillo-Lluva S, Alvarez-Tabares I, Weber I, Steinberg G, Perez-Martin J. Sustained cell polarity and virulence in the phytopathogenic fungus Ustilago maydis depends on an essential cyclin-dependent kinase from the Cdk5/Pho85 family. J Cell Sci. 2007;120:1584–1595. doi: 10.1242/jcs.005314. [DOI] [PubMed] [Google Scholar]
- 65.Garrido E, Voss U, Muller P, Castillo-Lluva S, Kahmann R, et al. The induction of sexual development and virulence in the smut fungus Ustilago maydis depends on Crk1, a novel MAPK protein. Genes Dev. 2004;18:3117–3130. doi: 10.1101/gad.314904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsukuda T, Carleton S, Fotheringham S, Holloman WK. Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol Cell Biol. 1988;8:3703–3709. doi: 10.1128/mcb.8.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carbo N, Perez-Martin J. Spa2 is required for morphogenesis but it is dispensable for pathogenicity in the phytopathogenic fungus Ustilago maydis. Fungal Genet Biol. 2008;45:1315–1327. doi: 10.1016/j.fgb.2008.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dendrogram analysis of MEKK, MEK and MAPK extracted from U. maydis database and compared with S. cerevisiae and S. pombe. Branches were named after the S. cerevisiae MAPK pathway.
(0.30 MB TIF)
Levels of Thr193 and Tyr195 Mpk1 phosphorylation after treatment with cell wall stressors. Protein extracts from the indicated strains growing in the presence of cell wall stressors for the indicated time (minutes) were separated by SDS-PAGE. Immunoblots were incubated successively with an antibody that recognizes the Mpk1 phosphorylated form (anti-pTEpY) and anti-HA. The levels of phosphorylation were determined by quantifying the level of antibody signal using a ChemiDoc (Bio-Rad). Signal from the phosphopeptide-specific antibodies was normalized to the amount of phosphorylation of at time zero. Differences in loading of samples were corrected by dividing each phosphopeptide-specific antibody signal by the anti-HA antibody signal. Mean and s.d. are shown (n = 3 experiments).
(0.03 MB PDF)
Expression of mkk1DD resulted in cell lysis. Strain UMA7 was grown at 28°C in YPA for 8 h. Weakness of cell wall at the tips was apparent by the leakage of cell material.
(0.09 MB PDF)
Deletion of mpk1 gene abolishes the morphological defects observed upon expression of mkk1DD. Strains FB1 (wt), UMA7 (Pcrg1: mkk1DD) and UMA13 (Pcrg1: mkk1DD mpk1Δ) were grown in YPA at 28°C for 8 h.
(1.14 MB PDF)
Clb2-GFP is a marker of G2 phase in U. maydis. A. Scheme of cell cycle in U. maydis where different phases are associated to specific morphological markers: microtubules, nuclear membrane, and presence or absence of buds (based in results from different sources: [Perez-Martin, et al; Steinberg, et al; Straube, et al]). Presence of Clb2-GFP fluorescence is also included from our data. Cells without buds are in G1 and S phase. Cells with buds, one single nucleus surrounded by an intact nuclear membrane, and a cytoplasmic array of MT are G2 cells. Cells with large buds, mitotic spindle and nuclear membrane partially disassembled are in mitosis and finally cells with large buds, two nuclei (one in the bud and other in mother cell), cytoplasmic array of microtubules and in the process of cell separation are early G1 cells. B. Correlation between presence of GFP signal (GFP+) and G2 phase. A strain carrying Clb2-GFP and Cut11-RFP fusions was grown in YPD until mid-log phase. 200 cells were counted and sorted in 4 different groups depending on presence or absence of bud, number of nuclei and integrity of nuclear membrane. C. Cell images of cells carrying Clb2-GFP and Cut11-RFP fusion showing different cell cycle phases. [Perez-Martin J, Castillo-Lluva S, Sgarlata C, Flor-Parra I, Mielnichuk N, et al. (2006) Pathocycles: Ustilago maydis as a model to study the relationships between cell cycle and virulence in pathogenic fungi. Mol Genet Genomics 276: 211-229. Steinberg G, Wedlich-Soldner R, Brill M, Schulz I (2001) Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J Cell Sci 114: 609-622. Straube A, Weber I, Steinberg G (2005) A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J 24: 1674-1685.]
(2.32 MB TIF)
Ectopic expression of wee1 does not preclude accelerated G2/M transition upon over-activation of Mpk1. (A) DIC and DAPI images of UMA20 (mkk1DDcrg1), UMC48 (Pcrg1:wee1) and UMA79 (Pcrg1:wee1 mkk1DDcrg1) cells grown for 8 h in YPA at 22°C. Bar 15 µm. In this experiment, we wondered whether high levels of wee1 expression were able to preclude the G2 shortening observed after over-activation of Mpk1. To address this question we introduced an ectopic copy of a MYC-tagged wee1 allele under the control of crg1 promoter in a strain carrying the mkk1DDcrg1 allele. By this manipulation, over-activation of Mpk1 (upon expression of the mkk1DDcrg1 allele) will occur concomitantly with over-expression of wee1. We found that high levels of Wee1 in a control strain produced elongated buds with a single nucleus (A, middle column), concomitant with a G2 cell cycle arrest. However, when wee1 was overexpressed in cells carrying the mkk1DDcrg1 allele, the presence of high levels of Wee1 did not prevent the G2 shortening. The observed cell compartments in wee1-overexpressing strain were slightly larger than those observed in the strain not expressing wee1 (182 µm3 in cells overexpressing wee1 versus 104 µm3 in cells not overexpressing wee1 on average for cell aggregates carrying at least four cell compartments). (B) Western blot showing the levels of Wee1-myc in the above strains, after grown for 8 h under inducing (YPA) and repressing (YPD) conditions for crg1. As loading control Cdk1 and Cdk5 levels were analyzed using anti-PSTAIRE antibodies.
(1.15 MB TIF)