Abstract
The hyaluronic acid receptor for endocytosis (HARE)/Stabilin-2 is the primary systemic scavenger receptor for 13 ligands including hyaluronan (HA), heparin and chondroitin sulfates. Most ligand-binding sites are within the 190 kDa isoform, which contains ∼25 kDa of N-glycans and is the C-terminal half of the full-length 315 kDa HARE. Glycoproteomic analyses of purified recombinant human 190-HARE ecto-domain identified a diverse population of glycans at 10 of 17 consensus sites. The most diversity (and the only sialylated structures) occurred at N2280, within the HA-binding Link domain. To determine if these N-glycans are required for HA binding, we created human Flp-In 293 cell lines expressing membrane-bound or soluble ecto-domain variants of 190-HARE(N2280A). Membrane-bound HARE lacking Link domain N-glycans mediated rapid HA endocytosis, but purified 190-HARE(N2280A) ecto-domain showed little or no HA binding in ELISA-like, HA-HARE pull-down assays or by surface plasmon resonance analysis (which detected very high apparent affinity for 190-HARE ecto-domain binding to HA; Kd = 5.2 nM). The results indicate that Link domain N-glycans stabilize interactions that facilitate HA binding to HARE.
Keywords: coated pit mediated, conformation, glycosaminoglycan turnover, HA binding affinity, Stabilin-2
Introduction
The hyaluronic acid (HA) receptor for endocytosis (HARE), also known as Stabilin-2 or FEEL-2, is a clearance/scavenger receptor that is highly expressed in the sinusoidal endothelial cells of liver, lymph node and spleen (Weigel and Yik 2002). HARE is also found in oviduct, corneal and lens epithelium, heart valve mesenchymal cells, ependymal cells lining ventricles in brain, macrophages and epithelial cells covering renal papillae (Falkowski et al. 2003). Human HARE is a 2551 aa, 315 kDa, type-1 transmembrane receptor expressed on the cell surface and in intracellular (e.g. endocytic) compartments. The receptor is composed of modular repeats of epidermal growth factor (EGF), EGF-like and Fasciclin domains. Native tissues express two HARE isoforms of different mass (190 kDa and 315 kDa) that are not splice variants (Zhou et al. 1999; Weigel et al. 2002; Zhou et al. 2003). In cells stably expressing cDNA encoding full-length 315-HARE, a small fraction of the 315 kDa protein is proteolytically cleaved to create the 190-HARE, the C-terminal 1416 aa of the 315-HARE protein (Harris et al. 2007). Both HARE isoforms are functional endocytic receptors, targeted to coated pits, with identical ligand-binding profiles (Harris et al. 2004; Harris et al. 2007; Harris and Weigel 2008). A defining feature of HARE is its Link domain, 168 aa from the membrane domain, which has a sequence similar to Link domains in other HA-binding proteins such as TSG-6, CD44 and aggrecan (Day and Prestwich 2002).
HARE recognizes at least 13 distinct ligands, including HA (Yannariello-Brown et al. 1997; Politz et al. 2002), collagen N-terminal propeptides (Hansen et al. 2005), advanced glycation end-products (Tamura et al. 2003), acetylated low density lipoprotein (Adachi and Tsujimoto 2002), chondroitin and chondroitin sulfates type A–E (Harris et al. 2004), αMβ2 integrin (Jung et al. 2007), heparin (Harris et al. 2008) and phosphatidylserine (Park et al. 2008a). The function of HARE/Stab2 in liver and lymph nodes to remove circulating HA and chondroitin sulfates has been known for over two decades. However, three recent findings indicate that HARE mediates additional important functions: i) HARE mediates intracellular ERK signaling in response to HA binding (Kyosseva et al. 2008); ii) HARE is the long-sought macrophage apoptotic receptor that recognizes exposed phosphatidylserine on dying cells and mediates their phagocytosis (Park et al. 2008a; Park et al. 2008b); and iii) HARE is the major liver clearance receptor for circulating heparin (Harris et al. 2008; Harris et al. 2009).
The large size of HARE and its recognition of multiple ligands present challenges in efforts to define subdomains within the protein that mediate specific ligand-binding events. Considerable additional complexity is contributed by earlier findings that the 190-HARE and 315-HARE glycoproteins contain >25 kDa in N-glycans (Weigel and Weigel 2003; Zhou et al. 2003). Within the 190-HARE, 13 consensus N-glycosylation sites are conserved among rat, mouse and human (including one NCTC site). In order to express and characterize stable HARE subdomains that retain a specific ligand-binding activity, it is important to know more about HARE N-glycans, the sites that are modified and if N-glycans at particular sites are important for ligand binding.
Our goals in this study were to identify the types of N-glycans at occupied Asn sites in 190-HARE and to determine if glycans at N2280 in the Link domain are required for HA binding by purified ecto-domain or membrane-bound receptor.
Results
Glycomics analysis of s190-HARE
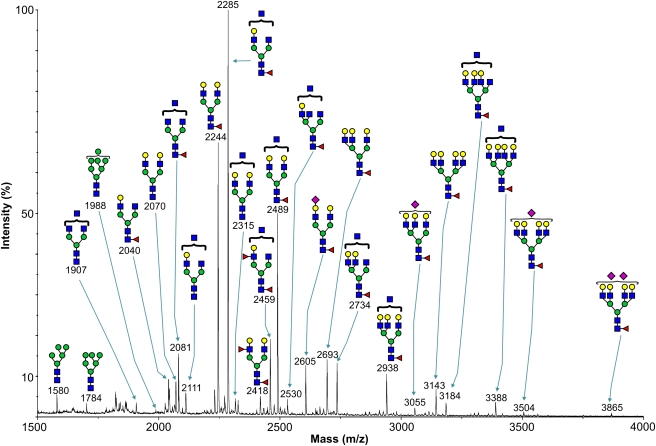
Recombinant soluble 190-HARE ecto-domain protein (s190-HARE), containing C-terminal His-6 and V5 tags, was expressed in human Flp-In 293 cells, purified from media and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The s190-HARE band was excised, digested with trypsin and then with PNGase-F. The released N-glycans were separated from peptides by Sep-Pak C18 purification, permethylated and analyzed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Figure 1). For each ion, putative structural assignments were made based on compositional information and knowledge of mammalian glycosylation biosynthetic pathways. Where possible, components observed in the MALDI-MS profile (Figure 1) were subjected to electrospray ionization tandem mass spectrometry (ESI-MS/MS) to assist sequence assignment (not shown).
Fig. 1.
Glycomic analysis of s190-HARE. MALDI-TOF mass spectrum of permethylated s190-HARE N-glycans. N-Glycans were derived from the 50% (v/v) acetonitrile fraction from a C18 Sep-Pak. All molecular ions are [M+-Na]+-, and nominal masses of the 12C isotope are shown. The putative structures shown are based on composition, tandem mass spectrometry analysis and knowledge of biosynthetic pathways. Symbol code: Gal (yellow circle), Man (green circle), GlcNAc (blue square), Fuc (red triangle) and NeuAc (purple diamond)
The mass spectrum indicates that s190-HARE N-glycans have compositions consistent with minor amounts of high mannose structures (Hex5–7HexNAc2) plus major complex type bi-, tri- and tetra-antennary glycans of compositions NeuAc0–2Fuc0–2Hex3–7HexNAc4–7. The most abundant ion (m/z 2285) has a composition (Fuc1Hex4HexNAc5) consistent with either a core fucosylated bisected bi-antennary glycan or a core fucosylated tri-antennary glycan, each having a single antenna terminated with galactose (Figure 1). The next two most abundant components (m/z 2244 and 2489; Figure 1), 70% and 50% of the base (highest) peak, respectively, have two antennae capped with galactose. Many components are of more modest abundance. Those showing signals greater than 5% of the base peak include glycans lacking fucose (for example m/z 2070 and 2111; Figure 1), and constituents whose compositions are consistent with the presence of a fucosylated antenna in addition to core fucose (m/z 2418 and 2459; Figure 1). Very few glycans are sialylated, and based on comparisons of peak heights, they constitute less than 5% of the total population. Most sialylated glycans have only a single sialic acid (m/z 2605, 3055 and 3504; Figure 1), although a trace amount of a disialyl component was identified at m/z 3865.
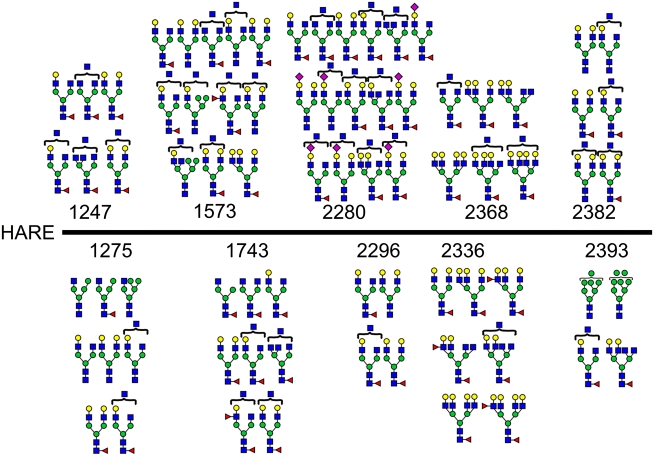
Glycoproteomic analysis of s190-HARE
We performed extensive glycoproteomic analyses on gel-purified s190-HARE protein using a variety of proteases, alone and in combinations, as well as chemical cleavage. Resulting peptides were analyzed by nano-liquid chromatography (LC)-ESI-MS/MS. As glycosidic bond fragmentation occurs during collisional activation in ESI-MS/MS and results in diagnostic fragment ions at m/z 204 (HexNAc) and 366 (HexHexNAc), MS/MS spectra were analyzed for these ions, and when present the spectra were examined manually to identify the glycopeptide. Using this technique, we identified the populations of N-glycans at 10 of the 17 consensus sites for N-glycosylation (Figure 2). Of the 10 sites, two are localized to regions of special biological interest. The first important site is at N2280KS within the HA-binding Link region. Although the HARE Link domain has not been independently expressed and assessed directly for HA-binding activity, cells expressing a 190-HARE(ΔLink) mutant endocytose <10% of the HA endocytosed by wild type (WT) 190-HARE cells (Kyosseva et al. 2008; Harris and Weigel 2008). The N2280 site is occupied by a very heterogeneous range of N-glycans with and without sialic acid. In fact, all the sialylated structures identified were found only at N2280. N-Glycans with compositions consistent with bisected structures are also a feature of the glycan population at N2280 (Figure 2). A second site of presumed importance is N2296, which is in the stem region between the Link and membrane domains. This site carries the most homogeneous range of N-glycans of all the identified sites, having a limited number of small neutral glycans with compositions Fuc0–1Hex4–5HexNAc4–5 (Figure 2). The N2296CT site is also unusual in that it overlaps a CTC sequence, both of which are conserved. In such cases, disulfide bond formation might preclude glycosylation and vice versa.
Fig. 2.
N-Glycosylation sites and N-glycans identified in s190-HARE. The glycans observed at the indicated sites are shown; numbering is based on the full-length 2551 aa HARE. The glycan structures are based on the composition of the glycan, as calculated from the mass of the glycopeptide minus the mass of the peptide, complemented by information from CAD-ESI-MS/MS data and from the glycomic profile shown in Figure 1. Symbol code: Gal (yellow circle), Man (green circle), GlcNAc (blue square), Fuc (red triangle) and NeuAc (purple diamond)
In addition to N-glycopeptides observed during the LC-ESI-MS/MS analysis of s190-HARE, ions consistent with two O-glycopeptide glycoforms were also detected. The data were attributable to the peptide CLPAYTGDGKVCTL1555 carrying O-glycans with compositions HexNAc and HexHexNAc. Peptide fragmentation enabled sequencing through most of the peptide and assignment of T1547 as the substituted residue (not shown). We do not know what other sites may also be O-glycosylated, but results shown below confirm that s190-HARE contains only a few O-glycans.
Estimation of HARE N-glycan content
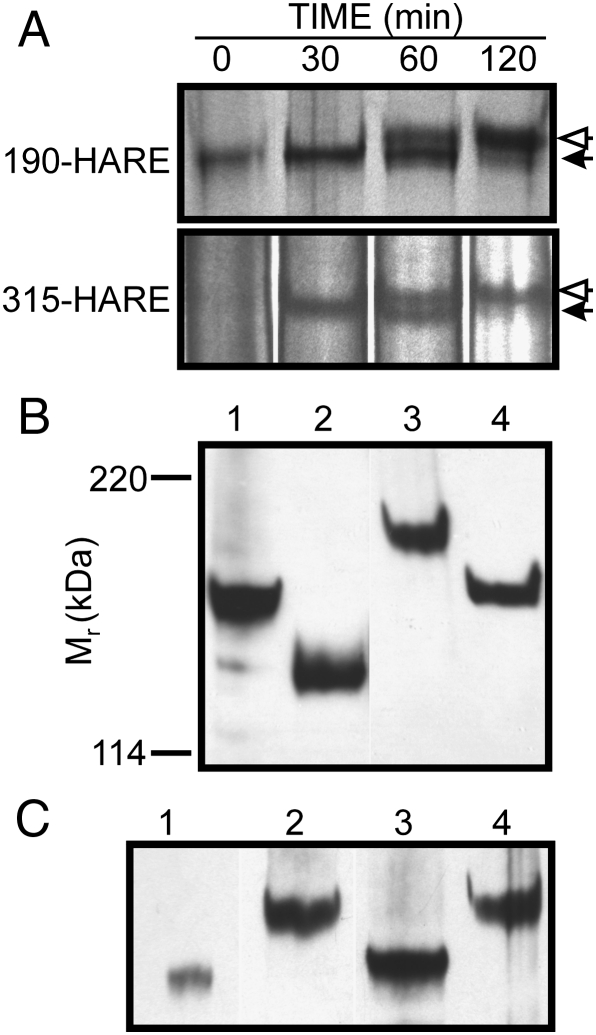
Based on treatment of nonreduced protein with PNGase-F, we concluded that the two isoforms of purified native rat and human HARE contain ∼25 kDa of N-glycans (Zhou et al. 2000; Weigel and Weigel 2003; Zhou et al. 2003), equivalent to ∼10 oligosaccharides. To complement the present glycomics analysis, we determined if similar results were obtained for recombinant WT 190-HARE and 315-HARE during biosynthesis. Metabolic pulse-chase labeling of human Flp-In 293 cells expressing 190-HARE or 315-HARE with 35S-Met/Cys revealed increased sizes of 190-HARE and 315-HARE between 60 and 120 min (Figure 3A), as expected for the addition and assembly of N-glycans in the ER/Golgi. After 2 h, most labeled protein was ∼25 kDa larger than the initial core protein.
Fig. 3.
Estimation of s190-HARE N-glycan content. A. Glycosylation of HARE during biosynthesis. Cells expressing either membrane-bound 190-HARE (top) or 315-HARE (bottom) were incubated with 35S-Met/Cys for 5 min, washed and then incubated (chased) for the indicated times with fresh DMEM as described in Materials and methods. HARE proteins were purified using mAb-30, separated by SDS–PAGE, and the gels were processed for autoradiography. The nascent HARE core proteins (solid arrows) are shifted to larger mass (open arrows) as they are glycosylated and move from the ER/Golgi to the cell surface. B. Effect of reduction on N-glycan release from s190-HARE. Purified s190-HARE was untreated (lanes 1 and 2) or reduced and alkylated (lanes 3 and 4) and then incubated with (lanes 2 and 4) or without (lanes 1 and 3) PNGase-F. Samples were separated by SDS–PAGE, electro-blotted to nitrocellulose, and HARE was detected using anti-V5 Ab. C. Effect of chemical or enzymatic deglycoslyation. Purified s190-HARE was reduced and alkylated and then either untreated (lane 2) or treated with TMFS (lane 1), PNGase-F (lane 3) or Endo-H (lane 4) and analyzed as above
PNGase-F treatment of purified and reduced s190-HARE also released ∼25 kDa of N-glycans (Figure 3B), indicating that the majority of N-glycans are in the C-terminal 190-HARE portion of 315-HARE. We also determined if some N-glycans were not released from the folded protein by comparing protein mass shifts after digestion of reduced or nonreduced HARE. Reduced and alkylated s190-HARE shifted from ∼150 kDa (Figure 3B, lane 1) to ∼170 kDa (Figure 3B, lane 3), and the mass loss of ∼25 kDa after PNGase-F treatment was similar for reduced and nonreduced receptor (Figure 3B, lanes 2 and 4). The mass of Endo-H-treated s190-HARE was identical to that of undigested protein (Figure 3C, lanes 2 and 4), indicating that essentially all WT s190-HARE N-linked glycans are mature complex structures with few high mannose or immature N-glycans (consistent with the glycomics analysis; Figure 1). To assess the amount of O-glycans or PNGase-F-resistant N-glycans present, we also treated the protein with trifluoromethanesulfonic acid (TFMS) to release all glycans (Edge et al. 1981). TFMS-treated and PNGase-F-treated s190-HARE proteins differed by only 4 kDa (Figure 3C, lanes 1 and 3), indicating that s190-HARE has little O-glycosylation.
Cell lines expressing 190-HARE(N2280A)
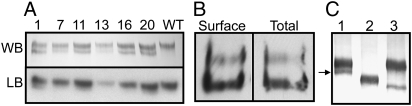
To determine if Link domain N-glycans are required for function, we made cDNA variants of ecto-domain or membrane-bound 190-HARE with a single N(2280)A mutation to eliminate N-glycosylation at N2280KS. We characterized multiple stably transfected Flp-In 293 cell lines expressing either 190-HARE variant. In all clones examined (Figure 4A, top), membrane 190-HARE(N2280A) was a doublet (separated by ∼10 kDa), unlike WT 190-HARE (Figure 4A, last lane); the major upper band migrated at the WT position. Both HARE bands bound 125I-HA in a ligand blot assay (Figure 4A, bottom).
Fig. 4.
190-HARE(N2280A) cells coexpress a smaller variant with altered glycosylation that is delivered to the surface. A. Although HARE expression levels varied among the indicated stable cell lines, each expressed two membrane-bound 190-HARE(N2280A) bands that bind 125I-HA in ligand blot (LB) assays. After exposure on film, western blot (WB) analysis with anti-V5 Ab visualized the HARE doublet. B. Both 190-HARE(N2280A) variants are on the cell surface. 190-HARE(N2280A) cells were incubated for 1 h at 4°C with HBSS containing 1 μg/mL mAb-30 with 0.055% digitonin (Weigel et al. 1983) [permeabilized to assess Total] or without [nonpermeabilized to assess Surface only]. Solubilized Ab-HARE complexes were captured using Protein A/G Sepharose. After centrifugation, the resin was eluted with 2× Laemmli buffer (Laemmli 1970), and the eluate was separated by SDS–PAGE, electro-blotted and HARE proteins detected by enhanced chemiluminescence using anti-V5 Ab. The ratios of the two HARE bands recovered in each lane were the same as assessed in digital images, and the Total signal was attenuated by ∼80% for comparison to the Surface signal (Harris et al. 2007; Harris et al. 2004). C. The smaller band has immature glycans. Immuno-purified nonreduced s190-HARE(N2280A) protein was subjected to SDS–PAGE after no treatment (lane 1) or digestion with PNGase-F (lane 2) or Endo-H (lane 3). The arrow indicates the minor smaller band, which was completely shifted by Endo-H, which did not affect the larger band
Since the smaller 190-HARE(N2280A) binds HA, it might also mediate HA endocytosis. Although this could not be tested directly (due to endocytosis mediated by the normal variant), we determined if the smaller variant is delivered to the cell surface. Cultured cells were incubated at 4°C with HARE-specific mAb-30 (Zhou et al. 2000), with or without digitonin, to monitor HARE recovery from intact or permeabilized cells (Weigel et al. 1983), respectively. HARE-antibody (Ab) complexes were then solubilized, captured using Protein A/G Sepharose and analyzed by SDS–PAGE and western blotting (Figure 4B). Since the intracellular receptor pool is 5–6-fold greater than the surface pool (Harris et al. 2004; Harris et al. 2007), the Total HARE signals (Figure 4B) were adjusted by this factor to compare the two doublet bands. The fraction of the smaller HARE band recovered by mAb bound to cell surfaces only (no digitionin) was identical to that for mAb bound to intracellular and surface HARE (digitonin-permeabilized). Thus, the smaller 190-HARE(N2280A) species is not a biosynthetic intermediate; it is a folded glycoform (different than WT) that traffics normally to the cell surface.
To test if the lack of N-glycans at N2280 alters the normal processing of 190-HARE(N2280A) proteins during biosynthesis (e.g. proteolysis or smaller N-glycans), we assessed the size of the two s190-HARE(N2280A) proteins after treating purified receptor with PNGase-F or Endo-H to release, respectively, almost all or only immature (e.g. high mannose) glycans. Digestion with PNGase-F collapsed the doublet into a single band (Figure 4C, lanes 1 and 2), demonstrating that proteolysis had not occurred. Endo-H digestion quantitatively shifted the mass of the smaller HARE band but, as expected, not the larger normal band (Figure 4C, lanes 1, 2 and 3). The broad PNGase-F treated band (lane 2) still contains N-glycans, whereas the Endo-H product (lane 3) does not, since PNGase-F digestion of reduced and alkylated doublet yielded one protein of the same size as the small band in lane 3 (not shown).
Cells expressing 190-HARE(N2280A) mediate HA endocytosis
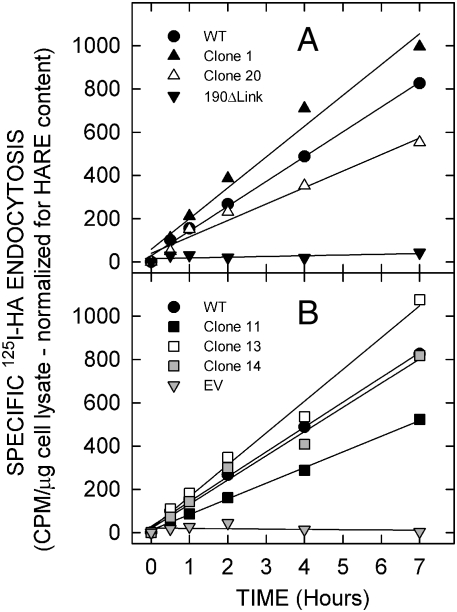
Previous studies showed that Flp-In 293 cells expressing either HARE isoform show robust binding and coated pit-mediated endocytosis of HA, whereas mock or empty vector (EV) transfected cells show essentially no specific HA binding or endocystosis, and HA uptake is reduced >90% in 190-HARE(ΔLink) cells compared to WT (Harris et al. 2004; Harris et al. 2007; Harris and Weigel 2008; Kyosseva et al. 2008; Pandey et al. 2008). Cells expressing 190-HARE lacking N2280-glycans also mediated specific binding and endocytosis of 125I-HA (Figure 5). Five cell lines expressing the N2280 variant each internalized HA rapidly and efficiently, with linear kinetics similar to WT and at similar rates (±30%), for at least 7 h (data were normalized for total HARE content relative to WT at 100%; Figure 5). During this time, the cell surface 190-HARE pool would be internalized and recycled back to the surface ∼46 times (Harris et al. 2004). In contrast, the specific HA uptake rate by EV and 190-HARE(ΔLink) cells was −1.2% and 2.8%, respectively, of WT. The variability in HA uptake rates by variant HARE cell lines may reflect unique clonal differences in the function of the complex cellular pathways for coated pit-mediated endocytosis and receptor recycling (Harris et al. 2004). The results indicate that the ability of membrane-bound HARE(N2280A) variant to bind and mediate endocytosis of HA is not affected by lack of the Link domain N-glycans.
Fig. 5.
Cell lines expressing 190-HARE(N2280A) endocytose HA normally. WT 190-HARE, EV, 190-HARE(ΔLink) cells and five 190-HARE(N2280A) cell lines, indicated by different numbers, were incubated for up to 7 h at 37°C with 125I-HA (plus or minus excess unlabeled HA) and processed at the indicated times to assess specific HA endocytosis as described in Materials and methods. Specific endocytosis values are presented as mean (n = 3) CPM/μg cell lysate protein, normalized for HARE expression relative to WT, which was 100%. SEM values for nearly all data points were within ± 4% and all SEM values were within < ± 6.3%
Purified soluble 190-HARE(N2280A) ecto-domain does not bind HA in three in vitro assays
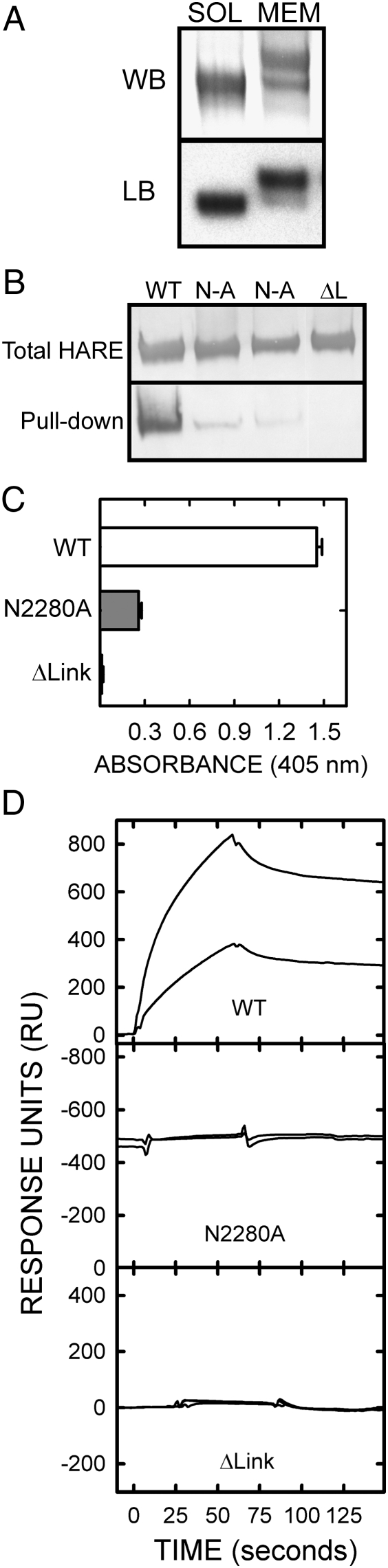
All s190-HARE(N2280A) cell lines tested expressed the ecto-domain as a single band that bound HA in a ligand blot assay (Figure 6A), and no differences were seen in overall glycosylation due to loss of a glycan at N2280 (not shown); the glycan mass at N2280 is <4 kDa. Membrane 190-HARE(N2280A) and WT 190-HARE bind HA similarly in ligand blot assays and internalize HA in a similar fashion (Figures 4A and 5). There were also no differences in HA-binding ability of the soluble and membrane 190-HARE(N2280A) proteins in ligand blot assays (Figure 6A).
Fig. 6.
The 190-HARE(N2280A) ecto-domain does not bind HA in most assays. A. Ligand blot assay. Soluble (SOL) s190-HARE(N2280A) is a single band by western blot (WB) that is smaller than membrane-bound (MEM) 190-HARE(N2280A). Both bind 125I-HA in ligand blot (LB) assays. B. HARE pull-down assay. SA-agarose with bound b-HA was incubated with 1 μg of purified s190-HARE [WT], s190-HARE(ΔLink) [ΔL] or s190-HARE(N2280A) [N-A; two different preparations] C. ELISA-like assay. Binding of the three purified ecto-domains to HA immobilized on plate wells was quantified by A405 as described in Materials and methods. D. Surface plasmon resonance assay. The three purified ecto-domains at 12.5 nM (lower lines) and 50 nM (upper lines) were each analyzed for binding to immobilized HA and then, after 60–90 s, for dissociation in buffer without ecto-domain as described in Materials and methods
In contrast, three other independent methods to evaluate HA-binding activity showed that the s190-HARE(N2280A) ecto-domain binds much less or no HA compared to WT ecto-domain (Figure 6B–D). The negative control was s190-HARE(Δlink), which does not bind HA. In a HARE pull-down assay, using biotin (b)-HA bound to SA-Agarose, two independent purified s190-HARE(N2280A) preparations showed only a trace of HA binding, whereas all the WT s190-HARE present, but no s190-HARE(Δlink), bound to HA (Figure 6B). Similarly, in an ELISA-like assay, s190-HARE(N2280A) binding to immobilized HA was only 18% of the WT value (Figure 6C). Finally, we used surface plasmon resonance (SPR) with b-HA as the ligand, attached to a SA chip, to quantify HA binding to each of the three ecto-domains used as the analyte (Figure 6D). No detectable binding to HA was observed for either the s190-HARE(N2280A) or the s190-HARE(ΔLink) ecto-domains (Figure 6D, middle and bottom panels). In contrast, WT s190-HARE bound HA in a reversible manner (Figure 6D, top panel) with exceptionally high apparent affinity (Kd = 5.2 nM) calculated from the measured kinetic values for kon (1.58 × 105 s−1 mol−1) and koff (8.22 × 10−4 s−1). This affinity of HARE binding to HA is the highest of all known hyalectins.
Discussion
We wanted to determine if the HARE Link domain is N-glycosylated and, if so, whether these N-glycans are required for HA binding. Two of the present findings indicate an unexpected complexity and importance for the physiological functions of HARE Link module N-glycans. First, of 10 glycosylated sites within the 190-HARE identified to contain multiple N-glycoforms, N2280 in the Link domain site showed the greatest structural diversity and was the only site with sialylated glycans. Second, although membrane-bound 190-HARE(N2280A) binds and endocytoses HA normally, the ecto-domain shows impaired or no HA binding in three of four in vitro assays.
HARE has the highest apparent affinity for binding to HA (Kd = 5–23 nM; Figure 6D and Harris et al. 2004; Harris et al. 2007) of all hyalectins, surpassing CD44 [Kd = 5–150 µM (Skelton et al. 1998)], TSG-6 [Kd = 200–500 nM (Kahmann et al. 2000)], aggrecan [Kd = 226 nM (Watanabe et al. 1997)] and Link protein [Kd = 82 nM (Watanabe et al. 1997)]. These values are all apparent (van der Merwe et al. 1993) since the presence of dimers and oligomers in the purified proteins is generally unknown and the HA ligand is inherently multivalent. Nonetheless, the Kd value of 5 nM determined here from SPR data (Figure 6D) is in excellent agreement with previously determined Kd values of 10 nM and 22 nM for HA binding to purified s315-HARE and s190-HARE, respectively, in ELISA-like assays (Harris et al. 2007), and of 7 nM for HA binding to membrane-bound 190-HARE in cells (Harris et al. 2004).
Both HARE isoforms have identical affinities for binding to heparin or HA and identical binding profiles for at least six other ligands (Harris and Weigel 2008). More than 90% of the HA-binding activity of HARE is lost in the HARE(ΔLink) variant (Kyosseva et al. 2008). CS-A, CS-C and CS-D also bind within the HA-binding Link domain of HARE (Harris et al. 2004; Harris et al. 2007; Harris and Weigel 2008). In contrast, s190-HARE(ΔLink) still binds to acetylated low density lipoprotein, heparin, dermatan sulfate (CS-B) and CS-E. Thus, HARE specifically binds to HA and heparin at two independent and nonoverlapping sites. Not all Link modules that bind both HA and heparin show independent binding. For example, unlike HARE (Harris and Weigel 2008; Harris et al. 2008), the TSG-6 Link module binds each ligand in separate grooves, but HA binding prevents heparin binding or vice versa; HA and heparin do not bind simultaneously (Mahoney et al. 2005).
Human 315-HARE has 28 predicted N-glycosylation sites, of which 17 are within the 190-HARE. Our glycoproteomic results identified multiple types of glycans on 10 of these 17 sites. Of these 10 human glycosylation sites, nine are conserved in rat and mouse HARE (Zhou et al. 2002; Zhou et al. 2003), indicating their importance to the functions of HARE. PNGase-F treatment releases ∼25 kDa of glycans from either isoform and from either rat or human native or recombinant HARE (Zhou et al. 1999; Zhou et al. 2002; Zhou et al. 2003). The same result was obtained with membrane-bound or soluble 190-HARE or 315-HARE. Glycans identified at the 10 sites (Figure 1) ranged in mass from 1580 Da to 3866 Da (7 to 16 sugars) corresponding to a total average mass of ∼25 kDa. Thus, most of the N-glycans attached to the full-length protein may be within its C-terminal 190-HARE region.
Several groups discovered that N- or O-glycans can influence the HA-binding function of proteins containing a Link module (Bennett et al. 1995; Katoh et al. 1995; Lesley et al. 1995; Bartolazzi et al. 1996; Dasgupta et al. 1996; English et al. 1998; Skelton et al. 1998). Although initially controversial, it is now generally accepted that N-glycans on the Link module of the HA receptor CD44 influence HA binding. In most cases, the presence of an N-glycan inhibits HA binding. The HA-binding ability of LYVE-1, a lymphatic HA receptor, is inhibited by sialylation of O-glycans (Nightingale et al. 2009), and HA binding could be unmasked in sialylated native or recombinant LYVE-1 by neuraminidase treatment. Locations of the sialylated O-glycans are unknown, but thought not to be on the LYVE-1 Link module. Inhibition due to the presence of a glycan can sometimes be attributed to steric occlusion; HA cannot physically interact with key binding site residues. Another attractive and inherently more responsive mechanism would involve reversible interactions between an oligosaccharide and domains within the protein to create an unfavorable conformation for HA binding (i.e. a temporary self-occluding steric effect). Reversing or disrupting this oligosaccharide interaction to allow a conformation favorable for HA binding might occur by multiple mechanisms, such as an allosteric-like response upon binding another protein or small ligand or by removing sialic acid residues needed to maintain the unfavorable conformation.
In summary, our results show that lack of an N-glycan on the HARE Link domain: (i) compromises HA binding to purified ecto-domain in three of four in vitro assays and (ii) does not decrease HA binding or endocytosis mediated by the membrane-bound protein. These effects indicate that N2280 oligosaccharides are important for the soluble truncated ecto-domain to maintain an HA binding conformation. Despite the ability of 190-HARE(N2280A) cells to internalize HA normally, preliminary results indicate that this variant HARE mediates little or no HA-dependent intracellular ERK activation (M.S. Pandey and P.H. Weigel unpublished). As noted above, the HARE Link glycans may directly interact (or indirectly facilitate interaction) with an adjacent HARE domain near the membrane to create an active HA binding site and ERK signaling competence. Supporting the idea that these activities depend on specific conformations, we previously found that the ability of mAb-174 to block HA binding by HARE in rat liver sinusoidal endothelial cells was lost at lower temperature (Weigel et al. 2003). We concluded that HARE undergoes a conformation change between 37°C and 4°C that does not prevent HA binding to HARE but that alters the epitope recognized by mAb-174 so that it does not block HA binding. Based on the present results, this conformational change is likely dependent on the Link domain glycan.
Materials and methods
Reagents and buffers
Flp-In 293 cells, culture medium, transfection reagents, cDNA purification and amplification reagents and plasmids were from Invitrogen-Gibco (Carlsbad, CA). pfu ultra HF was from Stratagene (La Jolla, CA). EasyTag Expre35S35S Labeling Mix (35S-Methionine/Cysteine) was from Perkin Elmer (Waltham, MA). Classic Blue BX film was from MidSci (St. Louis, MO). Streptavidin (SA), biotin-LC-hydrazide, sulfo-NHS-SS-biotin, SnakeSkin dialysis tubing (MWCO 3500) and luminol and peroxide solutions were from Pierce (Rockford, IL). RNA purification kits were from Qiagen, and plasmid DNA purification and cloning kits were from Fermentas (Glen Burnie, MD). Polysorp well strips were from Nunc (Roskilde, Denmark), and p-nitrophenylphosphate was from Kirkegaard & Perry Laboratories, Inc. (Gaithersburg, MD). HA from Genzyme (Framingham, MA) was acid-hydrolyzed under mild conditions (Raja et al. 1988), neutralized, size fractionated and weight-average mass was determined by light scattering (Baggenstoss and Weigel 2006). Biotinylated HA (b-HA; 108 kDa) was prepared by the method of Yu and Toole (1995), with slight modifications (Harris et al. 2007). Sodium 125I (100 mCi/mL; specific activity of >0.6 TBq/mg) in NaOH and Sepharose 6 Fast Flow (Nickel NTA) resin were from GE/Amersham Biosciences (Piscataway, NJ). 125I-SA and 125I-HA were prepared as described previously (Weigel 1980; Raja et al. 1984; McGary et al. 2003). Concentrator/desalting Centricon devices were from Amicon (Bedford, MA). Endo-H (Streptomyces plicatus) was from Calbiochem (San Diego, CA). SA-alkaline phosphatase conjugate, TFMS, trypsin and other salts and reagents were from Sigma-Aldrich (St. Louis, MO). PNGase-F (Flavobacterium meningosepticum), formic acid and sequence grade chymotrypsin, Asp-N, chymotrypsin/trypsin and Asp-N/Glu-C were from Roche Applied Science (San Francisco, CA). TFMS was from Acros Chemicals (Geel, Belgium). TBST contains 20 mM Tris–HCl, pH 7.0, 150 mM NaCl and 0.1% (v/v) Tween-20. Hank's balanced salt solution (HBSS) contains 5 mM KCl, 0.4 mM KH2PO4, pH 7.2, 0.8 mM MgSO4, 137 mM NaCl, 0.3 mM Na2HPO4, 5.5 mM glucose, 1.26 mM CaCl2, 0.5 mM MgCl2 and 28 μM phenol red.
Purification of 190-HARE ecto-domains
Recombinant s190-HARE, s190-HARE(N2280A) or s190-HARE(ΔLink) was purified from conditioned cell culture medium by immobilized Ni-chelate affinity chromatography as described previously (Harris et al. 2007). Protein content, here and in general, was determined by A280 or by a dye-binding method (Bradford 1976). Eluted fractions (pooled and concentrated using a 30 MWCO concentrator) were 10–20% pure, since some bovine serum proteins (e.g. α-2 macroglobulin and H factor 1/complement) copurified. Impurities were removed after 5% SDS–PAGE (Laemmli 1970) by excising s190-HARE bands. After overnight electroelution (35 V, 4°C) and subsequent concentration (Harris et al. 2007), the recovered s190-HARE was >98% pure, as assessed by SDS–PAGE and silver staining.
Trypsin digestion of s190-HARE prior to N-glycan determination
Partially purified s190-HARE was fractionated by SDS–PAGE (Novex NuPAGE Tris-Acetate gel system; Invitrogen) and the gel stained by colloidal Coomassie. The s190-HARE band, identified by parallel western blotting (Burnette 1981), was excised and reduced in-gel in 600 mM Tris–HCl, pH 8.4, containing 2 mg/mL dithiothreitol (37°C for 45 min) and then carboxymethylated by addition of 12 mg/mL iodoacetic acid (22°C for 1 h). Carboxymethylation was terminated by dialysis against 50 mM NH4HCO3, pH 8.5, at 4°C for 48 h, followed by lyophilization. Alkylated s190-HARE was incubated with trypsin (50:1 ratio, w/w) in 50 mM (NH4)HCO3, pH 8.4, for 16 h at 37°C. The digestion was terminated by heating at 100°C for 3 min, followed by C18 Sep-Pak chromatography (Waters Corp., Milford, MA, USA). Bound peptides were eluted with either 20% (v/v) or 40% (v/v) propanol in 5% aqueous acetic acid, pooled and lyophilized.
Preparation of N-glycans
PNGase-F (3 Roche units) digestion of tryptic peptides prepared above or from the whole s190 was carried out in 50 mM ammonium bicarbonate, pH 8.5, for 16 h at 37°C. Released N-glycans were separated from peptides using Sep-Pak C18 cartridges (Waters Corp.) and permethylated using a sodium hydroxide procedure (Dell et al. 1993).
Digestion of s190-HARE for glycoproteomic studies
Gel-purified 190-HARE was reduced and alkylated as described above and digested with trypsin, chymotrypsin, Asp-N, Asp-N/Glu-C, chymotrypsin/trypsin or chymotrypsin/trypsin/Asp-N as described previously (Dell et al. 1993; Dell et al. 1994). For CNBr cleavage, an aliquot of tryptically digested s190-HARE was dried down and resuspended in 100 μL of a 1 mL solution of 8 CNBr crystals dissolved in 70% formic acid. Samples were incubated at 22°C overnight and reactions then stopped by adding 4 volumes of water and lyophilization.
MS for glycan determination
MALDI-TOF MS data were acquired on a Voyager-DE sSTR mass spectrometer (PerSeptive Biosystems, Framingham, MA) in the reflectron mode with delayed extraction. Permethylated samples were dissolved in 10 µL of 80% (v/v) methanol in water, and 1 µL of dissolved sample was premixed with 1 µL of matrix (10 mg/mL 2,5-dihydroxybenzoic acid in 80% (v/v) aqueous methanol) before loading onto a metal plate. MALDI-TOF/TOF experiments were performed on a 4800 Proteomics Analyzer (Applied Biosystems, Framingham, MA) in reflectron positive-ion mode. Both 2,5-dihydroxybenzoic acid and α-cyano-4-hydroxycinnamic acid matrices (10 mg/mL in 50% (v/v) acetonitrile in 0.1% (v/v) aqueous trifluoroacetic acid) were used in conjunction with setting the potential difference between the source acceleration voltage and the collision cell at 1 kV to obtain different degrees and patterns of fragmentation.
MS for peptide mapping
Digests were analyzed by nano-LC-ESI-MS/MS using a reverse-phase nano-high-performance liquid chromatography (HPLC) system (Dionex, Sunnyvale, CA) connected to a quadrupole TOF mass spectrometer (Q-STAR Pulsar I, MDS Sciex). Components were separated by using a binary nano-HPLC gradient generated by an Ultimate pump fitted with a Famos autosampler and a Switchos microcolumn switching module (LC Packings, Amsterdam, The Netherlands). An analytical C18 nanocapillary (75 µm inside diameter × 15 cm, PepMap) and a micro precolumn C18 cartridge were employed for on-line peptide separation. The digest was first loaded onto the precolumn and eluted with 0.1% formic acid (Sigma) in water for 4 min. The eluent was then transferred onto an analytical C18 nanocapillary HPLC column and eluted at a flow rate of 150 nL/min using the following gradient of solvent A [0.05%, v/v formic acid in a 95:5, v/v water/acetonitrile mixture] and solvent B [0.04% formic acid in a 95:5, v/v acetonitrile/water mixture]: 99% A from 0 to 5 min, 99–90% A from 5 to 10 min, 90–60% A from 10 to 70 min, 60–50% A from 70 to 71 min, 50–5% A from 71 to 75 min, 5% A from 75 to 85 min, 5–95% A from 85 to 86 min and 95% A from 86 to 90 min. Data acquisition was performed using Analyst QS software with an automatic information-dependent acquisition function.
Glycosidase treatments
Cells were lysed in phosphate buffered saline (PBS) with 0.5% NP-40 containing protease inhibitors, debris was removed by centrifugation and membrane-bound HARE was collected by incubation for 2 h at 22°C with 20 μL of a 1:1 suspension of mAb-30 (Zhou et al. 2000) coupled to CNBr-activated Sepharose 4B (1 mg/mL resin). After centrifugation, the supernatant was removed and PBS (20 μL) and 0.5% SDS (5 μL) were added to the resin, which was incubated at 22°C for 10 min to dissociate HARE. The sample was centrifuged, the supernatant containing HARE was placed in a 0.2 mL screw cap microfuge tube and NP-40 was added to a final concentration of 2%. PNGase-F or Endo-H (1.25 mU) was added and the sample incubated for 16 h at 37°C. For purified soluble HARE proteins, 0.5 μg of protein in 2% NP-40 in PBS was incubated with enzyme under the same conditions.
Chemical deglycosylation
Five micrograms of s190-HARE was lyophilized in a 1.5 mL tube and placed on ice. TFMS (70 μL) was mixed with 7.5 μL anisole, cooled on ice and added to the sample. The tube was shaken gently (not vortexed) until the protein was dissolved and the mixture incubated for 3 h on ice with occasional mixing. Pyridine was chilled to −20°C in an ethanol/dry ice bath, and 2 μL of 0.2% bromophenol blue in ethanol (as a pH indicator) was added to the protein mixture. The red TFMS protein solution was cooled to −20°C and cold pyridine added until the solution turned yellow. To prevent solidification, 10 μL of water was added. More pyridine was added slowly, to minimize exothermic heating, until the solution was purple/blue. Deglycosylated HARE was then dialyzed in three changes of 500 mL PBS over 10 h and stored at −80°C.
Preparation of s190-HARE(N2280A) and 190-HARE(N2280A) constructs
To make a N2280A mutant, two primers (forward: 5′-CTATGGACCTAGACCCGCCAAGAGTGAAATGTGGG-3′ and reverse: 5′-CCCACATTTCACTCTTGGCGGGTCTAGGTCCATAG-3′) (altered codon is underlined) were used in individual mutagenic reactions with the WT 190-HARE cDNA (in pSecTag/FRT/V5/His) in an Ericomp thermocycler (18 cycles: 94°C, 20 s; 62°C, 20 s; 71°C, 1 min/plasmid kb) using pfu Ultra HF. Plasmids were ethanol precipitated, resuspended in 17 μL H2O, 2 μL NEB4 buffer and 2.5 U of DpnI to cut template plasmids while retaining intact mutant plasmids. After overnight incubation at 37°C, the digestion mixtures were heated to 95°C for 10 min and immediately transformed into E. cloni 10G super-competent E. coli cells (Lucigen, Middleton, WI). Bacterial cells were screened by a mini-prep procedure, PCR and DNA sequencing to confirm the mutation. Plasmids with correct mutations, open reading frames and promoter regions were used to make stable cell lines. A cDNA construct encoding the secreted ecto-domain was then produced from a validated 190-HARE(N2280A) cDNA using the primer (5′-GTGACCTTGACCCACACTGGATCCGAAGGTAAGCCTATC-3′) to delete the transmembrane and cytoplasmic domains, while retaining C-terminal V5 and His6 epitopes (Harris et al. 2007).
Creation of Flp-In 293 cell lines expressing soluble or membrane-bound 190-HARE(N2280A)
Cell lines with a single identical chromosome insertion site were created using recombinase-mediated integration in human Flp-In 293 cells (embryonic kidney derived) as described previously (Harris et al. 2004; Harris et al. 2007). Multiple clones were tested for HARE expression by SDS–PAGE (5% gels) and western analysis (Burnette 1981) with anti-V5 Ab. Each positive clone was then tested for correct insertion of the pSecTag(190HARE) cDNA into the unique chromosomal Flp-In recombination site, mediated by the Flp-In recombinase encoded by pOG44, using the assays described earlier (Harris et al. 2004; Harris et al. 2007).
HA endocytosis assays
WT 190-HARE, EV, 190-HARE(ΔLink) and 190-HARE(N2280A) cells were grown in 24-well plates to ∼90% confluence, washed and incubated in serum-free medium at 37°C for 1 h. Cells were washed with 1 mL HBSS and incubated at 37°C for the indicated times in 0.4 mL of Dulbecco's modified Eagle's medium (DMEM) containing 0.05% bovine serum albumin and 1.0 µg/mL 125I-HA with or without a 100-fold excess of unlabeled HA to assess nonspecific or total uptake, respectively. Medium was aspirated and cells were washed three times with 1 mL HBSS, solubilized in 1 mL 0.3 N NaOH, and radioactivity (using a gamma counter) and protein were determined. Multiple experiments were performed with different combinations of control and N2280A cell lines. Since receptor expression often differs among cell lines, HARE content was quantified by western analysis using 10 μg of cell lysate (in triplicate) and enhanced chemiluminescence according to the manufacturer's instructions using Classic Blue BX film. Digital images were captured using a FluoroChem 8000 imaging system (Alpha Innotech Corporation, San Leandro, CA), and band densities were quantified as integrated densitometry values (i.e. the sum of all pixel values minus background correction). Values for specific endocytosis (total minus nonspecific) are presented as CPM/μg cell lysate protein (normalized to the mean integrated digital value of each mutant compared to WT, which was 100%).
35S Pulse-chase metabolic labeling
190-HARE and 315-HARE cells were grown in DMEM with 8% FBS and 100 μg/mL hygromycin B in 6-well dishes for 2 days. Confluent cells were then washed twice with PBS, incubated in DMEM without Met/Cys for 45 min, washed and 1.0 mL of 37°C DMEM with 200 mCi/mL 35S-Met/Cys (no unlabeled Met/Cys) was added to each well. After 10 min at 37°C, the media were aspirated, and the cells were washed twice in PBS and then incubated in fresh DMEM with 8% FBS at 37°C. At the indicated chase times, 0.5 mL of 4°C DMEM with 1.5% NP-40 and protease inhibitors was added, lysates were centrifuged to clear debris and HARE was removed by incubation overnight at 4°C with CNBr-activated Sepharose 4B coupled to three anti-HARE mAbs (30, 154 and 159). The resin was washed, bound proteins released with SDS, separated by 5% SDS–PAGE, and the gel was dried on filter paper and subjected to autoradiography using BioMax MS film (Kodak) for 2–7 days at −80°C.
In vitro assays for HA binding to HARE ecto-domains
ELISA-Like Assay. HA (142 kDa) and T-10 dextran (as a control) were mixed separately in PBS at 50 μg/mL and incubated overnight in heparin-binding plates (BD Biosciences, San Jose, CA, USA). After blocking with 0.2% (w/v) gelatin in 0.2% (v/v) Tween-20, 100 mM NaCl, 50 mM sodium acetate, pH 7.2, the wells were washed and incubated with 1.0 μg/mL purified soluble WT, N2280A or ΔLink 190-HARE ecto-domains for 1.5 h at 37°C. The wells were washed with 100 mM NaCl, 50 mM sodium acetate, 0.2% (v/v) Tween-20, pH 7.2 three times, and bound HARE was detected with anti-V5 Ab and alkaline phosphatase-anti goat IgG and color development using p-nitrophenylphosphate was quantified at A405 over 30 min.
Ligand Blot Assay. Cell pellets were resuspended in 0.5% NP-40 with protease inhibitors, and 20 mg of total protein was separated by 5% (w/v) SDS–PAGE. Gels were electro-transferred to nitrocellulose and incubated with 125I-HA as described previously (Yannariello-Brown et al. 1996). After washing, bound 125I-HA was detected by autoradiography using Kodak BioMax MS film. The blots were then processed for western analysis to identify HARE as noted above.
HA-HARE Pull-Down Assay. SA-Agarose CL-4B (25 μL, 50% slurry; Fluka, Sigma-Aldrich) was mixed by rotation with 100 nM b-HA in TBST for 1 h at 22°C in a 1.5 mL tube. The resin was washed three times by centrifugation in TBST and incubated for 1 h at 37°C in 1 mL of TBST with 1 μg of s190-HARE, s190-HARE(N2280A) or s190-HARE(ΔLink). The resin was then washed three times with TBST, resuspended in 20 μL of 4× Laemmli buffer (Laemmli 1970) and subjected to 5% SDS–PAGE and western blotting using anti-V5 Ab. HARE was visualized as above, and the bands were digitized and compared to the total protein used to assess the fraction bound to HA.
SPR Assay. Experiments were performed at 37°C on a Biacore 3000 (Biacore AB [GE Healthcare] Uppsala, Sweden). b-HA was captured on research grade SA-coated sensor chips (Sensor Chip SA, Biacore Inc.) pretreated according to the manufacturer's instructions. A solution of b-HA (1.0 μg/mL) was injected at 30 μL/min in PBS, pH 7, containing 0.005% Tween 20 until a saturating amount of HA was captured on the surface (600 RU). The control surface contained only pretreated SA without b-HA on the same sensor chip. The specific HA binding of purified s190-HARE WT (positive control), ΔLink (negative control) and N2280A variant ecto-domains were measured using the Biacore 3000 in-line reference subtraction feature. Two concentrations (12.5 and 50 nM) of each analyte were used to evaluate the kinetics of association (kon) and dissociation (koff, assessed in the same buffer without ecto-domain) at a flow rate of 5 μL/min over both surfaces of the sensor chip. Regeneration of the chip by stripping the analyte from the HA was performed by washing all surfaces with 5 M urea containing 0.2% SDS. Association and dissociation rate constants were calculated using 1:1 Langmuir modeling by the Biaevaluation software v3.2.
Acknowledgments
We thank Jennifer Washburn for the purification of s190HARE(N2280A) and s190-HARE, and Jennifer and Amy Padgett-McCue for general laboratory and technical assistance.
Conflict of interest statement
None declared.
Glossary
Abbreviations
- 190-HARE
the 190 kDa HA receptor for endocytosis
- 315-HARE
the 315 kDa HA receptor for endocytosis
- Ab
antibody
- b
biotin
- DMEM
Dulbecco's modified Eagle's medium
- EGF
epidermal growth factor
- ESI
electrospray ionization
- EV
empty vector
- HA
hyaluronic acid, hyaluronate, hyaluronan
- HARE
HA receptor for endocytosis
- HBSS
Hank's balanced salt solution
- HPLC
high-performance liquid chromatography
- LC
liquid chromatography
- MALDI
matrix-assisted laser desorption ionization
- MS/MS
tandem mass spectrometry
- PBS
phosphate buffered saline
- s190-HARE
soluble 190 kDa HARE ecto-domain
- SA
streptavidin
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SPR
surface plasmon resonance
- TFMS
trifluoromethanesulfonic acid
- Tris
tris(hydroxymethyl)aminomethane
- TBST
Tris-buffered saline containing Tween-20
- TOF
time of flight
- WT
wild type
Funding
This research was supported by National Institutes of Health/National Institute of General Medical Sciences grant GM69961 (to P.H.W.) and Biotechnology and Biological Sciences Research Council grants BBF0083091 and B19088 (to A.D., S.M.H. and H.R.M.).
References
- Adachi H, Tsujimoto M. FEEL-1, a novel scavenger receptor with in vitro bacteria-binding and angiogenesis-modulating activities. J Biol Chem. 2002;277:34264–34270. doi: 10.1074/jbc.M204277200. [DOI] [PubMed] [Google Scholar]
- Baggenstoss BA, Weigel PH. SEC-MALLS analysis of hyaluronan size distributions made by membrane-bound hyaluronan synthase. Anal Biochem. 2006;352:243–251. doi: 10.1016/j.ab.2006.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolazzi A, Nocks A, Aruffo A, Spring F, Stamenkovic I. Glycosylation of CD44 is implicated in CD44-mediated cell adhesion to hyaluronan. J Cell Biol. 1996;132:1199–1208. doi: 10.1083/jcb.132.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett KL, Modrell B, Greenfield B, Bartolazzi A, Stamenkovic I, Peach R, Jackson DG, Spring F, Aruffo A. Regulation of CD44 binding to hyaluronan by glycosylation of variably spliced exons. J Cell Biol. 1995;131:1623–1633. doi: 10.1083/jcb.131.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Burnette WN. “Western blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Dasgupta A, Takahashi K, Cutler M, Tanabe KK. O-linked glycosylation modifies CD44 adhesion to hyaluronate in colon carcinoma cells. Biochem Biophys Res Commun. 1996;227:110–117. doi: 10.1006/bbrc.1996.1475. [DOI] [PubMed] [Google Scholar]
- Day AJ, Prestwich GD. Hyaluronan-binding proteins: Tying up the giant. J Biol Chem. 2002;277:4585–4588. doi: 10.1074/jbc.R100036200. [DOI] [PubMed] [Google Scholar]
- Dell A, Khoo KH, Panico M, McDowell RA, Etienne AT, Reason AJ, Morris HR. In: Glycobiology: A Practical Approach. Fukuda M, Kobata A, editors. Oxford: Oxford University Press; 1993. pp. 187–222. [Google Scholar]
- Dell A, Reason AJ, Khoo KH, Panico M, McDowell RA, Morris HR. Mass spectrometry of carbohydrate-containing biopolymers. Methods Enzymol. 1994;230:108–132. doi: 10.1016/0076-6879(94)30010-0. [DOI] [PubMed] [Google Scholar]
- Edge AS, Faltynek CR, Hof L, Reichert LE, Jr, Weber P. Deglycosylation of glycoproteins by trifluoromethanesulfonic acid. Anal Biochem. 1981;118:131–137. doi: 10.1016/0003-2697(81)90168-8. [DOI] [PubMed] [Google Scholar]
- English NM, Lesley JF, Hyman R. Site-specific de-N-glycosylation of CD44 can activate hyaluronan binding, and CD44 activation states show distinct threshold densities for hyaluronan binding. Cancer Res. 1998;58:3736–3742. [PubMed] [Google Scholar]
- Falkowski M, Schledzewski K, Hansen B, Goerdt S. Expression of stabilin-2, a novel fasciclin-like hyaluronan receptor protein, in murine sinusoidal endothelia, avascular, tissues, and at solid/liquid interfaces. Histochem Cell Biol. 2003;120:361–369. doi: 10.1007/s00418-003-0585-5. [DOI] [PubMed] [Google Scholar]
- Hansen B, Longati P, Elvevold K, Nedredal GI, Schledzewski K, Olsen R, Falkowski M, Kzhyshkowska J, Carlsson F, Johansson S, et al. Stabilin-1 and stabilin-2 are both directed into the early endocytic pathway in hepatic sinusoidal endothelium via interactions with clathrin/AP-2, independent of ligand binding. Exp Cell Res. 2005;303:160–173. doi: 10.1016/j.yexcr.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Harris EN, Baggenstoss BA, Weigel PH. Rat and human HARE/Stabilin-2 are clearance receptors for high- and low-molecular-weight heparins. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1191–G1199. doi: 10.1152/ajpgi.90717.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EN, Kyosseva SV, Weigel JA, Weigel PH. Expression, processing and glycosaminoglycan binding activity of the recombinant human 315-kDa HA receptor for endocytosis (HARE) J Biol Chem. 2007;282:2785–2797. doi: 10.1074/jbc.M607787200. [DOI] [PubMed] [Google Scholar]
- Harris EN, Weigel PH. The ligand-binding profile of HARE/Stabilin-2: Hyaluronan and chondroitin sulfates A, C, and D bind to overlapping sites distinct from the sites for heparin, acetylated low-density lipoprotein and dermatan sulfate. Glycobiology. 2008;18:638–648. doi: 10.1093/glycob/cwn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EN, Weigel JA, Weigel PH. Endocytic function, glycosaminoglycan specificity, and antibody sensitivity of the recombinant human 190 kDa HA receptor for endocytosis (HARE) J Biol Chem. 2004;279:36201–36209. doi: 10.1074/jbc.M405322200. [DOI] [PubMed] [Google Scholar]
- Harris EN, Weigel JA, Weigel PH. The human hyaluronan receptor for endocytosis (HARE/Stabilin-2) is a systemic clearance receptor for heparin. J Biol Chem. 2008;283:17341–17350. doi: 10.1074/jbc.M710360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung MY, Park SY, Kim IS. Stabilin-2 is involved in lymphocyte adhesion to the hepatic sinusoidal endothelium via the interaction with alphaMbeta2 integrin. J Leukoc Biol. 2007;82:1156–1165. doi: 10.1189/jlb.0107052. [DOI] [PubMed] [Google Scholar]
- Kahmann JD, O'Brien R, Werner JM, Heinegard D, Ladbury JE, Campbell ID, Day AJ. Localization and characterization of the hyaluronan-binding site on the link module from human TSG-6. Structure Fold Des. 2000;8:763–774. doi: 10.1016/s0969-2126(00)00163-5. [DOI] [PubMed] [Google Scholar]
- Katoh S, Zheng Z, Oritani K, Shimozato T, Kincade PW. Glycosylation of CD44 negatively regulates its recognition of hyaluronan. J Exp Med. 1995;182:419–429. doi: 10.1084/jem.182.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyosseva SV, Harris EN, Weigel PH. The hyaluronan receptor for endocytosis (HARE) mediates hyaluronan-dependent signal transduction via extracellular signal-regulated kinases (ERK) J Biol Chem. 2008;283:15047–15055. doi: 10.1074/jbc.M709921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lesley J, English N, Perschl A, Gregoroff J, Hyman R. Variant cell lines selected for alterations in the function of the hyaluronan receptor CD44 show differences in glycosylation. J Exp Med. 1995;182:431–437. doi: 10.1084/jem.182.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney DJ, Mulloy B, Forster MJ, Blundell CD, Fries E, Milner CM, Day AJ. Characterization of the interaction between tumor necrosis factor-stimulated Gene-6 and heparin: Implications for the inhibition of plasmin in extracellular matrix microenvironments. J Biol Chem. 2005;280:27044–27055. doi: 10.1074/jbc.M502068200. [DOI] [PubMed] [Google Scholar]
- McGary CT, Weigel JA, Weigel PH. Study of hyaluronan-binding proteins and receptors using iodinated hyaluronan derivatives. Methods Enzymol. 2003;363:354–366. doi: 10.1016/S0076-6879(03)01064-4. [DOI] [PubMed] [Google Scholar]
- Nightingale TD, Frayne ME, Clasper S, Banerji S, Jackson DG. A mechanism of sialylation functionally silences the hyaluronan receptor LYVE-1 in lymphatic endothelium. J Biol Chem. 2009;284:3935–3945. doi: 10.1074/jbc.M805105200. [DOI] [PubMed] [Google Scholar]
- Pandey MS, Harris EN, Weigel JA, Weigel PH. The cytoplasmic domain of the hyaluronan receptor for endocytosis (HARE) contains multiple endocytic motifs targeting coated pit-mediated internalization. J Biol Chem. 2008;283:21453–21461. doi: 10.1074/jbc.M800886200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Jung MY, Kim HJ, Lee SJ, Kim SY, Lee BH, Kwon TH, Park RW, Kim IS. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ. 2008;15:192–201. doi: 10.1038/sj.cdd.4402242. [DOI] [PubMed] [Google Scholar]
- Park SY, Kim SY, Jung MY, Bae DJ, Kim IS. Epidermal growth factor-like domain repeat of stabilin-2 recognizes phosphatidylserine during cell corpse clearance. Mol Cell Biol. 2008;28:5288–5298. doi: 10.1128/MCB.01993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz O, Gratchev A, McCourt PAG, Schledzewski K, Guillot P, Johansson S, Svineng G, Franke P, Kannicht C, Kzhyshkowska J, et al. Stabilin-1 and-2 constitute a novel family of fasciclin-like hyaluronan receptor homologues. Biochem J. 2002;362:155–164. doi: 10.1042/0264-6021:3620155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja RH, LeBoeuf RD, Stone GW, Weigel PH. Preparation of alkylamine and 125I-radiolabeled derivates of hyaluronic acid uniquely modified at the reducing end. Anal Biochem. 1984;139:168–177. doi: 10.1016/0003-2697(84)90402-0. [DOI] [PubMed] [Google Scholar]
- Raja RH, McGary CT, Weigel PH. Affinity and distribution of surface and intracellular hyaluronic acid receptors in isolated rat liver endothelial cells. J Biol Chem. 1988;263:16661–16668. [PubMed] [Google Scholar]
- Skelton TP, Zeng CX, Nocks A, Stamenkovic I. Glycosylation provides both stimulatory and inhibitory effects on cell surface and soluble CD44 binding to hyaluronan. J Cell Biol. 1998;140:431–446. doi: 10.1083/jcb.140.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Adachi H, Osuga J, Ohashi K, Yahagi N, Sekiya M, Okazaki H, Tomita S, Iizuka Y, Shimano H, et al. FEEL-1 and FEEL-2 are endocytic receptors for advanced glycation end products. J Biol Chem. 2003;278:12613–12617. doi: 10.1074/jbc.M210211200. [DOI] [PubMed] [Google Scholar]
- van der Merwe PA, Brown MH, Davis SJ, Barclay AN. Affinity and kinetic analysis of the interaction of the cell adhesion molecules rat CD2 and CD48. EMBO J. 1993;12:4945–4954. doi: 10.1002/j.1460-2075.1993.tb06188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Cheung SC, Itano N, Kimata K, Yamada Y. Identification of hyaluronan-binding domains of aggrecan. J Biol Chem. 1997;272:28057–28065. doi: 10.1074/jbc.272.44.28057. [DOI] [PubMed] [Google Scholar]
- Weigel PH. Characterization of the asialoglycoprotein receptor on isolated rat hepatocytes. J Biol Chem. 1980;255:6111–6120. [PubMed] [Google Scholar]
- Weigel PH, McGary CT, Zhou B, Weigel JA. Purification and characterization of the hyaluronan receptor for endocytosis (HARE) In: Kennedy JF, Philips GO, Williams PA, editors. Hyaluronan 2000. Vol. 1. Wales, England: Woodhead Publishing Ltd; 2002. pp. 401–410. [Google Scholar]
- Weigel PH, Ray DA, Oka JA. Quantitation of intracellular membrane-bound enzymes and receptors in digitonin-permeabilized cells. Anal Biochem. 1983;133:437–449. doi: 10.1016/0003-2697(83)90106-9. [DOI] [PubMed] [Google Scholar]
- Weigel JA, Raymond RC, McGary CT, Singh A, Weigel PH. A blocking antibody to the hyaluronan receptor for endocytosis (HARE) inhibits HA clearance by perfused liver. J Biol Chem. 2003;278:9808–9812. doi: 10.1074/jbc.m211462200. [DOI] [PubMed] [Google Scholar]
- Weigel JA, Weigel PH. Characterization of the recombinant rat 175-kDa hyaluronan receptor for endocytosis (HARE) J Biol Chem. 2003;278:42802–42811. doi: 10.1074/jbc.M307201200. [DOI] [PubMed] [Google Scholar]
- Weigel PH, Yik JHN. Glycans as endocytosis signals: The cases of the asialoglycoprotein and hyaluronan/chondroitin sulfate receptors. Biochim Biophys Acta. 2002;1572:341–363. doi: 10.1016/s0304-4165(02)00318-5. [DOI] [PubMed] [Google Scholar]
- Yannariello-Brown J, Zhou B, Ritchie D, Oka JA, Weigel PH. A novel ligand blot assay detects different hyaluronan-binding proteins in rat liver hepatocytes and sinusoidal endothelial cells. Biochem Biophys Res Commun. 1996;218:314–319. doi: 10.1006/bbrc.1996.0055. [DOI] [PubMed] [Google Scholar]
- Yannariello-Brown J, Zhou B, Weigel PH. Identification of a 175 kDa protein as the ligand-binding subunit of the rat liver sinusoidal endothelial cell hyaluronan receptor. Glycobiology. 1997;7:15–21. doi: 10.1093/glycob/7.1.15. [DOI] [PubMed] [Google Scholar]
- Yu Q, Toole BP. Biotinylated hyaluronan as a probe for detection of binding proteins in cells and tissues. Biotechniques. 1995;19:122–129. [PubMed] [Google Scholar]
- Zhou B, Oka JA, Singh A, Weigel PH. Purification and subunit characterization of the rat liver endocytic hyaluronan receptor. J Biol Chem. 1999;274:33831–33834. doi: 10.1074/jbc.274.48.33831. [DOI] [PubMed] [Google Scholar]
- Zhou B, McGary CT, Weigel JA, Saxena A, Weigel PH. Purification and molecular identification of the human hyaluronan receptor for endocytosis. Glycobiology. 2003;13:339–349. doi: 10.1093/glycob/cwg029. [DOI] [PubMed] [Google Scholar]
- Zhou B, Weigel JA, Fauss LA, Weigel PH. Identification of the hyaluronan receptor for endocytosis (HARE) J Biol Chem. 2000;275:37733–37741. doi: 10.1074/jbc.M003030200. [DOI] [PubMed] [Google Scholar]
- Zhou B, Weigel JA, Saxena A, Weigel PH. Molecular cloning and functional expression of the rat 175-kDa hyaluronan receptor for endocytosis. Mol Biol Cell. 2002;13:2853–2868. doi: 10.1091/mbc.02-03-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]