Abstract
Background
Although pulsed electromagnetic fields (PEMFs) are used to treat delayed unions and nonunions, their mechanisms of action are not completely clear. However, PEMFs are known to affect the expression of certain genes.
Questions/purposes
We asked (1) whether PEMFs affect gene expression in human osteoblastlike cells (MG63) in vitro, and (2) whether and to what extent stimulation by PEMFs induce cell proliferation and differentiation in MG-63 cultures.
Methods
We cultured two groups of MG63 cells. One group was treated with PEMFs for 18 hours whereas the second was maintained in the same culture condition without PEMFs (control). Gene expression was evaluated throughout cDNA microarray analysis containing 19,000 genes spanning a substantial fraction of the human genome.
Results
PEMFs induced the upregulation of important genes related to bone formation (HOXA10, AKT1), genes at the transductional level (CALM1, P2RX7), genes for cytoskeletal components (FN1, VCL), and collagenous (COL1A2) and noncollagenous (SPARC) matrix components. However, PEMF induced downregulation of genes related to the degradation of extracellular matrix (MMP-11, DUSP4).
Conclusions and Clinical Relevance
PEMFs appear to induce cell proliferation and differentiation. Furthermore, PEMFs promote extracellular matrix production and mineralization while decreasing matrix degradation and absorption. Our data suggest specific mechanisms of the observed clinical effect of PEMFs, and thus specific approaches for use in regenerative medicine.
Introduction
PEMFs have been used for many years [44]. They reportedly are effective for treating nonunions [1, 7, 10], delayed unions [1, 42, 44], osteotomies [32], avascular necrosis of the femoral head [5, 34], bone grafts [11], and spinal fusion [36]. Although the therapeutic properties of PEMFs are well known, the sequence of events by which electromagnetic stimulation can bring about its desirable effects on bone healing is not completely understood. PEMFs modify some important physiologic parameters of cells, such as proliferation, transduction, transcription, synthesis, and secretion of growth factors [24]. PEMFs induce cell proliferation in mitogen-stimulated lymphocytes [10] and improve IL-2 receptor expression and IL-2 use in lymphocytes from aged donors, which are characterized by defective production and use of this growth factor [10]. PEMF exposure induces cell proliferation in human osteoblasts and chondrocytes cultured in vitro [18, 20, 38, 44, 45]. PEMFs determine signal transduction by means of intracellular release of Ca2+ leading to an increase in cytosolic Ca2+ and an increase in activated cytoskeletal calmodulin [9]. PEMFs induce a dose-dependent increase in bone [2] and cartilage differentiation [2–4, 33], and upregulation of mRNA expression of extracellular matrix molecules, proteoglycan, and Type II collagen [3]. The acceleration of chondrogenic differentiation is associated with increased expression of TGF-β1 mRNA and protein [4], suggesting the stimulation of TGF-β1 may be a mechanism through which PEMFs affect complex tissue behavior such as cell differentiation and through which the effects of PEMFs may be amplified [4]. PEMFs also are postulated to act at a membrane level influencing signal transduction of several hormones or growth factors such as parathyroid hormone, IGF 2, and adenosine A2a, producing the amplification of their transmembrane receptors [1, 19, 21, 23, 31, 46]. Studies of single genes using RT-PCR suggest activation of osteocalcin, osteopontin, and TGF-β transcription during osteogenesis [22] and inhibition of cyclooxygenase 2 in synovial fibroblasts stimulated with TNFα or lipopolysaccharide [21]. A wide analysis of gene expression in cells exposed to PEMFs has not been performed: most studies focus on a few aspects of cell activities or they have been performed using different types of signals in different experimental conditions.
We therefore asked (1) whether PEMFs affected a wide array of genes in human osteoblastlike cells (MG63), and (2) whether and to what extent PEMFs induce proliferation and differentiation of osteoblasts.
Materials and Methods
We treated osteoblastlike cell cultures (MG-63) with PEMFs for 18 hours, and maintained similar nontreated controls. Gene expression of both groups therefore was evaluated with cDNA microarray analysis, containing 19,000 genes spanning a substantial fraction of the human genome. All experiments were performed in triplicate in the same culture conditions for control and treated cells.
Osteoblastlike cells (MG63) were grown in sterile Falcon wells (Becton & Dickinson, Franklin Lakes, NJ) containing Eagle’s minimum essential medium supplemented with 10% fetal calf serum (Sigma-Aldrich, St Louis, MO) and antibiotics (penicillin 100 U/mL and streptomycin 100 μg/mL; Sigma-Aldrich). Cultures were maintained in a 5% CO2 humidified atmosphere at 37°C. For the assay, cells were collected and seeded at a density of 1 × 105 cells/mL in two multiwells (one for the control and one for the treated). Each multiwell was comprised of six wells, 9-cm2, in which 3-mL of complete medium was added.
After 24 hours, cells were exposed to PEMFs for 18 hours using a PEMF generator system (Igea, Carpi, Italy). The PEMF used in this study is used clinically to treat nonunions or delayed unions and avascular necrosis of the femoral head [32–34]. The solenoids were powered using a Biostim pulse generator (Igea), a PEMF generator. The electromagnetic bioreactor applied to the cells has the following characteristics: intensity of the magnetic field, 2 ± 0.2 mT; amplitude of the induced electric tension, 5 ± 1 mV; signal frequency, 75 ± 2 Hz; and pulse duration, 1.3 ms. The stimulated multiwell was placed parallel between the two solenoids of the PEMF generator. The solenoids were placed at a distance of 10 cm and the multiwell was located on an acrylic support exactly at the center of the two solenoids. Control cultures were placed in the same incubator; nevertheless, the presence of the electromagnetic field was checked and its value was less than 0.05 mT. This value was ineffective in previous studies [38–46]. After 18 hours, when cultures were subconfluent, cells were processed for RNA extraction.
For DNA microarray screening and analysis, we used the same protocol as described previously [12–16]. Briefly, RNA was extracted from cells by using RNAzol. Ten micrograms of total RNA was used for each sample. cDNA was synthesized by using Superscript II (Life Technologies, Invitrogen, Milano, Italy) and amino-allyl dUTP (Sigma-Aldrich). Monoreactive Cy3 and Cy5 esters (Amersham Pharmacia, Little Chalfont, UK) were used for indirect cDNA labeling. RNA extracted from untreated cells was labeled with Cy3 and used as control against the Cy5-labeled treated (PG) cDNA in the first experiment and then switched. For 20 K human DNA microarrays slides (MWG Biotech AG, Ebersberg, Germany), 100 μL of the sample and control cDNAs in DIG Easy hybridization solution (Roche, Basel, Switzerland) were used in a sandwich hybridization of the two slides, constituting the 20 K set at 37°C overnight. Washing was performed three times for 10 minutes with 1× saline sodium citrate (SSC) and 0.1% sodium dodecyl sulfate at 42°C and three times for 5 minutes with 0.1× SSC at room temperature. Slides were dried by centrifugation for 2 minutes at 2000 rpm. Hybridized arrays were scanned with a GenePix 4000 scanner (Axon Instruments) at variable photomultiplier tube (PMT) voltage to obtain maximal signal intensities with less than 1% probe saturation.
The Foreground Median intensity for Cy3 and Cy5, Background Median intensity for Cy3 and Cy5, spot size data were imported into BRB-ArrayTools software [43] using the Import wizard function. Global normalization was used to median center the log-ratios on each array r to adjust for differences in labeling intensities of the Cy3 and Cy5 dyes.
The normalized Log ratios also were imported to Significance Analysis of Microarray (SAM) [48] software to identify differentially expressed genes. SAM assigns a score to each gene on the basis of a change in gene expression relative to the standard deviation of repeated measurements. For genes with scores greater than an adjustable threshold, SAM uses permutations of the repeated measurements to estimate the percentage of genes identified by chance—the false discovery rate (FDR). Analysis parameters (Delta) were set to result in zero FDR.
Results
PEMF affected gene expression in MG-63 osteoblastlike cells (Fig. 1). The genes differentially expressed in cells treated with PEMFs were either upregulated (268 genes) (Table 1) or downregulated (277 genes) (Table 2). PEMF induced osteoblast proliferation and differentiation and regulated genes involved in bone formation in the direction of an enhancement of osteogenesis (Tables 3, 4).
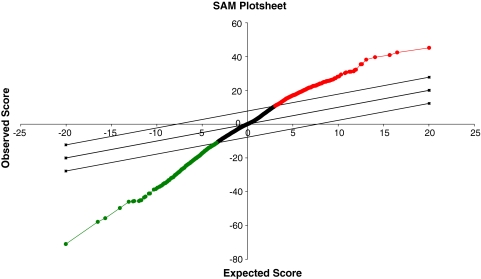
Fig. 1.
A microarray (SAM) plot of MG63 exposed to PEMFs versus control is shown. Expected differentially expressed genes are reported on the x axis, whereas observed differentially expressed genes are reported on the y axis. Downregulated genes (green) are located in the lower left of the graph; upregulated genes (red) are in the upper right; genes with different expression but statistically insignificant are shown in black. Parallel lines drawn from the lower left to upper right squares are the cutoff limits. The solid line indicates the equal value of observed and expected differentially expressed genes.
Table 1.
Upregulated genes
| GenBank | Name | Symbol | Cytoband | Score (d)* |
|---|---|---|---|---|
| W19447 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 | DDX51 | 12q24.33 | 45.15 |
| BM908669 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 12p13 | 42.40 |
| W33064 | Tubulin, alpha 4a | TUBA4A | 2q35 | 40.89 |
| BI258438 | Cofilin 1 (nonmuscle) | CFL1 | 11q13 | 39.61 |
| H80610 | Hypothetical protein LOC729176 | LOC729176 | 6q24.3 | 35.42 |
| R23641 | Vacuolar protein sorting 13 homolog A | VPS13A | 9q21 | 32.28 |
| BM006748 | Enolase 1 (alpha) | ENO1 | 1p36.3-p36.2 | 31.99 |
| W44826 | Major histocompatibility complex, class I, E | HLA-E | 6p21.3 | 31.04 |
| BG547115 | Ferritin, heavy polypeptide 1 | FTH1 | 11q13 | 30.55 |
| BG288116 | Integrin, alpha 2 | ITGA2 | 5q23-q31 | 27.14 |
| BQ014343 | Family with sequence similarity 62 | FAM62B | 7q36.3 | 26.05 |
| R89805 | ELOVL family member 7 | ELOVL7 | 5q12.1 | 25.81 |
| BQ108591 | Ribosomal protein S5 | RPS5 | 19q13.4 | 25.71 |
| H61302 | Hexose-6-phosphate dehydrogenase | H6PD | 1p36 | 25.10 |
| AA151568 | Testis enhanced gene transcript (BAX inhibitor 1) | TEGT | 12q12-q13 | 24.95 |
| H30300 | Small nuclear ribonucleoprotein polypeptide N | SNRPN | 15q11.2 | 24.75 |
| N72456 | Similar to RIKEN cDNA A730055C05 gene | LOC388335 | 17p13.1 | 24.38 |
| H25618 | Chromatin modifying protein 5 | CHMP5 | 9p13.3 | 24.12 |
| AA059376 | Similar to phosphodiesterase 4D interacting protein isoform 2 | LOC653513 | 1q21.1 | 24.01 |
| W30787 | DnaJ (Hsp40) homolog, subfamily C, member 15 | DNAJC15 | 13q14.1 | 24.00 |
| BM801770 | Solute carrier family 35, member E3 | SLC35E3 | 12q15 | 23.64 |
| AA099240 | NIPA-like domain containing 3 | NPAL3 | 1p36.12-p35.1 | 23.55 |
| W00391 | Solute carrier family 11 member 2 | SLC11A2 | 12q13 | 23.36 |
| H12528 | Annexin A5 | ANXA5 | 4q26-q28|4q28-q32 | 23.13 |
| N54759 | Prenylcysteine oxidase 1 | PCYOX1 | 2p13.3 | 23.08 |
| T89646 | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | ST3GAL2 | 16q22.1 | 22.82 |
| AA029517 | KCNQ1 overlapping transcript 1 | KCNQ1OT1 | 11p15 | 22.75 |
| W47664 | NAD(P)H dehydrogenase, quinone 1 | NQO1 | 16q22.1 | 22.73 |
| W02597 | PMS1 postmeiotic segregation increased 1 | PMS1 | 2q31-q33|2q31.1 | 22.61 |
| R11416 | Seryl-tRNA synthetase | SARS | 1p13.3-p13.1 | 22.55 |
| H67332 | GTP binding protein 1 | GTPBP1 | 22q13.1 | 22.43 |
| H86020 | NADH dehydrogenase | NDUFB5 | 3q26.33 | 22.21 |
| AA031564 | Chromosome 1 open reading frame 212 | C1orf212 | 1p34.3 | 21.90 |
| W67485 | Zinc finger protein 136 | ZNF136 | 19p13.2-p13.12 | 21.68 |
| W32906 | Zinc finger protein 702 | ZNF702 | 19q13.41 | 21.49 |
| BI492783 | Zinc finger protein 207 | ZNF207 | 17q11.2 | 21.43 |
| BE278092 | Ribosomal protein L10 | RPL10 | Xq28 | 21.42 |
| N90960 | Par-6 partitioning defective 6 homolog beta | PARD6B | 20q13.13 | 21.32 |
| BG565169 | Ferritin, light polypeptide | FTL | 19q13.3-q13.4 | 21.29 |
| H75902 | Complement component (3b/4b) receptor 1 | CR1 | 1q32 | 21.15 |
| W31736 | NADH dehydrogenase (ubiquinone) flavoprotein 1, 51 kDa | NDUFV1 | 11q13 | 20.96 |
| AA417686 | Casein kinase 1, gamma 3 | CSNK1G3 | 5q23 | 20.94 |
| R18627 | Amyloid beta precursor protein binding protein 2 | APPBP2 | 17q21-q23 | 20.81 |
| W38809 | Kelch-like 8 (Drosophila) | KLHL8 | 4q22.1 | 20.76 |
| BM456402 | Hypothetical gene LOC96610 | LOC96610 | 22q11.22 | 20.70 |
| AA044942 | Eukaryotic translation initiation factor 4 gamma, 1 | EIF4G1 | 3q27-qter | 20.44 |
| BM041235 | Actin, alpha 2, smooth muscle, aorta | ACTA2 | 10q23.3 | 20.42 |
| AI690073 | Glutamate-cysteine ligase, catalytic subunit | GCLC | 6p12 | 20.30 |
| AI734239 | Coiled-coil domain containing 120 | CCDC120 | Xp11.23 | 20.28 |
| N72922 | PDZ and LIM domain 5 | PDLIM5 | 4q22 | 20.23 |
| N50768 | Chromosome X open reading frame 57 | CXorf57 | Xq22.3 | 20.05 |
| N76504 | Hypothetical protein LOC257407 | LOC257407 | 2q37.1 | 19.69 |
| N45145 | Zinc finger, CCHC domain containing 4 | ZCCHC4 | 4p15.2 | 19.60 |
| BM922198 | Tubulin, beta 2C | TUBB2C | 9q34 | 19.57 |
| H65175 | Solute carrier family 31 (copper transporters), member 1 | SLC31A1 | 9q31-q32 | 19.50 |
| H83172 | Cytochrome b5 domain containing 2 | CYB5D2 | 17p13.2 | 19.28 |
| N73208 | Zinc finger protein 207 | ZNF207 | 17q11.2 | 19.26 |
| H95413 | Hydroxysteroid (17-beta) dehydrogenase 7 | HSD17B7 | 1q23 | 19.21 |
| N72546 | Cathepsin S | CTSS | 1q21 | 19.08 |
| BM705000 | Cold shock domain protein A | CSDA | 12p13.1 | 19.08 |
| W86495 | Coiled-coil-helix-coiled-coil-helix domain containing 7 | CHCHD7 | 8q12.1 | 19.07 |
| BI092679 | H19, imprinted maternally expressed untranslated mRNA | H19 | 11p15.5 | 18.99 |
| N45602 | Serine/threonine kinase 4 | STK4 | 20q11.2-q13.2 | 18.93 |
| H78769 | Interleukin-1 receptor-associated kinase 4 | IRAK4 | 12q12 | 18.88 |
| W35195 | Lethal giant larvae homolog 1 (Drosophila) | LLGL1 | 17p11.2 | 18.88 |
| AA062617 | Myotubularin related protein 9 | MTMR9 | 8p23-p22 | 18.85 |
| BE315195 | Ribosomal protein L8 | RPL8 | 8q24.3 | 18.84 |
| BQ067508 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 12p13 | 18.76 |
| R35530 | RAD23 homolog B (S cerevisiae) | RAD23B | 9q31.2 | 18.75 |
| H24644 | AlkB, alkylation repair homolog 5 (E coli) | ALKBH5 | 17p11.2 | 18.57 |
| BM010025 | Signal transducer and activator of transcription 3 | STAT3 | 17q21.31 | 18.53 |
| H08490 | Chloride channel 2 | CLCN2 | 3q27-q28 | 18.39 |
| H80175 | Radixin | RDX | 11q23 | 18.27 |
| H46045 | Tripartite motif-containing 46 | TRIM46 | 1q22 | 18.18 |
| N25456 | Mutated in colorectal cancers | MCC | 5q21 | 18.07 |
| AA047157 | CD82 molecule | CD82 | 11p11.2 | 18.01 |
| AA044701 | ADAMTS-like 5 | ADAMTSL5 | 19p13.3 | 17.82 |
| BM477950 | Ribosomal protein L8 | RPL8 | 8q24.3 | 17.79 |
| AI587328 | Radical S-adenosyl methionine domain containing 2 | RSAD2 | 2p25.2 | 17.68 |
| W03282 | Dihydrofolate reductase | DHFR | 5q11.2-q13.2 | 17.62 |
| BQ072807 | Ribosomal protein L13a | RPL13A | 19q13.3 | 17.52 |
| H01638 | Coiled-coil domain containing 82 | CCDC82 | 11q21 | 17.48 |
| BG529617 | Ribosomal protein, large, P1 | RPLP1 | 15q22 | 17.45 |
| H63198 | RAB interacting factor | RABIF | 1q32-q41 | 17.34 |
| BG397205 | Proteasome (prosome, macropain) subunit, beta type, 4 | PSMB4 | 1q21 | 17.33 |
| W31052 | Nephronophthisis 3 (adolescent) | NPHP3 | 3q22.1 | 17.26 |
| BM925268 | Chromosome 12 open reading frame 32 | C12orf32 | 12p13.33 | 17.25 |
| H83233 | Malate dehydrogenase 1, NAD (soluble) | MDH1 | 2p13.3 | 17.23 |
| W19108 | UBX domain containing 4 | UBXD4 | 2p23.3 | 17.22 |
| AA004532 | Fusion (involved in t(12;16) in malignant liposarcoma) | FUS | 16p11.2 | 17.21 |
| R50299 | SHANK-associated RH domain interactor | SHARPIN | 8q24.3 | 17.01 |
| R47837 | Zinc finger, RAN-binding domain containing 2 | ZRANB2 | 1p31 | 16.99 |
| H85307 | V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | KRAS | 12p12.1 | 16.89 |
| N49567 | Agmatine ureohydrolase (agmatinase) | AGMAT | 1p36.21 | 16.88 |
| N57076 | KIAA1909 protein | KIAA1909 | 5p15.33 | 16.84 |
| BI116974 | Ribosomal protein L18 | RPL18 | 19q13 | 16.83 |
| N40643 | Chromosome 10 open reading frame 18 | C10orf18 | 10p15.1 | 16.74 |
| R82575 | KIAA1704 | KIAA1704 | 13q13-q14 | 16.51 |
| BI196362 | Tubulin, alpha 1a | TUBA1A | 12q12-q14.3 | 16.36 |
| AA132192 | Pleckstrin homology domain containing, family H member 2 | PLEKHH2 | 2p21 | 16.34 |
| N31221 | Hypothetical protein DKFZp667M2411 | DKFZp667M2411 | 17q11.2 | 16.31 |
| H45243 | GDNF-inducible zinc finger protein 1 | GZF1 | 20p12.3-p11.21 | 16.29 |
| T86807 | Serine/threonine kinase 19 | STK19 | 6p21.3 | 16.28 |
| AA057270 | Choline kinase alpha | CHKA | 11q13.2 | 16.20 |
| R66209 | Synaptosomal-associated protein, 29 kDa | SNAP29 | 22q11.21 | 16.13 |
| R92306 | DnaJ (Hsp40) homolog, subfamily C, member 19 | DNAJC19 | 3q26.33 | 16.13 |
| T80698 | Glycine-N-acyltransferase-like 1 | GLYATL1 | 11q12.1 | 16.12 |
| N42722 | Guanine nucleotide binding protein (G protein), gamma 12 | GNG12 | 1p31.3 | 16.03 |
| BM911128 | Secreted protein, acidic, cysteine-rich (osteonectin) | SPARC | 5q31.3-q32 | 15.99 |
| N34619 | Coagulation factor II (thrombin) receptor-like 2 | F2RL2 | 5q13 | 15.97 |
| AA039528 | C-Maf-inducing protein | CMIP | 16q23 | 15.76 |
| N53715 | Neural precursor cell expressed | NEDD8 | 14q12 | 15.75 |
| BQ055308 | Ribosomal protein L4 | RPL4 | 15q22 | 15.73 |
| N44567 | Torsin A interacting protein 1 | TOR1AIP1 | 1q24.2 | 15.63 |
| AA046698 | Selenoprotein I | SELI | 2p23.3 | 15.62 |
| T75376 | Notch homolog 2 (Drosophila) | NOTCH2 | 1p13-p11 | 15.49 |
| N46675 | Unkempt homolog (Drosophila) | UNK | 17q25.1 | 15.44 |
| R93756 | Calmodulin 1 (phosphorylase kinase, delta) | CALM1 | 14q24-q31 | 15.42 |
| R67177 | Adenylate cyclase 1 (brain) | ADCY1 | 7p13-p12 | 15.39 |
| W24597 | Deoxyribonuclease II, lysosomal | DNASE2 | 19p13.2 | 15.28 |
| H43825 | HLA-B associated transcript 2 | BAT2 | 6p21.3 | 15.26 |
| N20577 | Leucine rich repeat containing 57 | LRRC57 | 15q15.1 | 15.24 |
| BI598074 | Neugrin, neurite outgrowth associated | NGRN | 15q26.1 | 15.23 |
| W79562 | Arginyltransferase 1 | ATE1 | 10q26.13 | 15.20 |
| R68004 | Poly(rC) binding protein 2 | PCBP2 | 12q13.12-q13.13 | 15.19 |
| AA040826 | Major histocompatibility complex, class I, C | HLA-C | 6p21.3 | 15.12 |
| H52744 | Abhydrolase domain containing 12 | ABHD12 | 20p11.21 | 15.11 |
| N78350 | RAN binding protein 1 | RANBP1 | 22q11.21 | 15.10 |
| BQ026918 | Collagen, type I, alpha 2 | COL1A2 | 7q22.1 | 15.05 |
| BG109286 | COX18 cytochrome c oxidase assembly homolog | COX18 | 4q13.3 | 15.02 |
| W47525 | Trans-golgi network protein 2 | TGOLN2 | 2p11.2 | 15.02 |
| BI494911 | Nck-associated protein 5 | NAP5 | 2q21.2 | 14.89 |
| N94192 | Glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | GNS | 12q14 | 14.84 |
| N28281 | Zinc finger protein 552 | ZNF552 | 19q13.43 | 14.61 |
| R84726 | Adenosine A1 receptor | ADORA1 | 1q32.1 | 14.59 |
| W40304 | Apoptosis inhibitor 5 | API5 | 11p11.2 | 14.54 |
| W63760 | Coilin | COIL | 17q22-q23 | 14.52 |
| T97408 | BCL2-associated athanogene | BAG1 | 9p12 | 14.51 |
| BM923884 | Glutathione S-transferase pi | GSTP1 | 11q13 | 14.49 |
| N46186 | Glutaredoxin 5 homolog (S cerevisiae) | GLRX5 | 14q32.13 | 14.40 |
| N57438 | Vitamin K epoxide reductase complex, subunit 1-like 1 | VKORC1L1 | 7q11.21 | 14.38 |
| W19461 | Abl interactor 2 | ABI2 | 2q33 | 14.33 |
| R74572 | Serine incorporator 1 | SERINC1 | 6q22.31 | 14.24 |
| N28330 | Melanoma cell adhesion molecule | MCAM | 11q23.3 | 14.23 |
| N20611 | GTP-binding protein 10 (putative) | GTPBP10 | 7q21.13 | 14.23 |
| H74119 | Sec61 beta subunit | SEC61B | 9q22.32-q31.3 | 14.13 |
| N52748 | Zinc finger protein 536 | ZNF536 | 19q12 | 14.13 |
| R48809 | Hypothetical gene supported by AK123662 | LOC388692 | 1q21.1 | 14.13 |
| AA005393 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24 kDa | NDUFV2 | 18p11.31-p11.2 | 14.12 |
| R89913 | CD58 molecule | CD58 | 1p13 | 14.08 |
| W03395 | Elongation of very long chain fatty acids-like 1 | ELOVL1 | 1p34.2 | 13.97 |
| BM541374 | Peptidylprolyl isomerase H (cyclophilin H) | PPIH | 1p34.1 | 13.96 |
| AA046918 | Splicing factor 3b, subunit 2, 145 kDa | SF3B2 | 11q13.1 | 13.95 |
| R60604 | TAF5-like RNA polymerase II | TAF5L | 1q42.13 | 13.89 |
| H39844 | Small nuclear RNA activating complex, polypeptide 3 | SNAPC3 | 9p22.3 | 13.84 |
| N39630 | Purinergic receptor P2X, ligand-gated ion channel, 7 | P2RX7 | 12q24 | 13.82 |
| H57205 | Vinculin | VCL | 10q22.1-q23 | 13.77 |
| N39274 | Hook homolog 3 (Drosophila) | HOOK3 | 8p11.21 | 13.74 |
| H14054 | Beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) | B3GAT3 | 11q12.3 | 13.59 |
| BG676419 | Potassium channel tetramerisation domain containing 13 | KCTD13 | 16p11.2 | 13.59 |
| N31020 | Similar to Signal peptidase complex subunit 2 | LOC653566 | 1p35.3 | 13.57 |
| BG110260 | FK506 binding protein 14, 22 kDa | FKBP14 | 7p15.1 | 13.56 |
| H53224 | Transferrin receptor (p90, CD71) | TFRC | 3q29 | 13.54 |
| R31353 | Glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | GNS | 12q14 | 13.51 |
| AA128133 | Nexilin (F actin binding protein) | NEXN | 1p31.1 | 13.49 |
| BQ070812 | Proteasome (prosome, macropain) 26S subunit, ATPase, 3 | PSMC3 | 11p12-p13 | 13.48 |
| H94761 | Disrupted in schizophrenia 1 | DISC1 | 1q42.1 | 13.48 |
| BQ050099 | Ras homolog gene family, member A | RHOA | 3p21.3 | 13.39 |
| BG169474 | UTP14, U3 small nucleolar ribonucleoprotein | UTP14A | Xq25 | 13.35 |
| R69639 | Carbohydrate (chondroitin 4) sulfotransferase 11 | CHST11 | 12q | 13.35 |
| T77351 | Rotatin | RTTN | 18q22.2 | 13.33 |
| AA203284 | Basic transcription factor 3 | BTF3 | 5q13.2 | 13.33 |
| AA056664 | V-akt murine thymoma viral oncogene homolog 1 | AKT1 | 14q32.32|14q32.32 | 13.31 |
| BE385427 | Chromatin modifying protein 6 | CHMP6 | 17q25.3 | 13.26 |
| BI850411 | Calnexin | CANX | 5q35 | 13.19 |
| BG687243 | Similar to ribosomal protein S13 | LOC729236 | 1p32.3 | 13.16 |
| BE256276 | Ribosomal protein L32 | RPL32 | 3p25-p24 | 13.15 |
| W17368 | Hexose-6-phosphate dehydrogenase | H6PD | 1p36 | 13.04 |
| N56629 | Hypoxia upregulated 1 | HYOU1 | 11q23.1-q23.3 | 13.02 |
| R48663 | Nuclear factor of activated T-cells, cytoplasmic | NFATC2IP | 16p11.2 | 12.98 |
| BQ052715 | Pyruvate kinase, muscle | PKM2 | 15q22 | 12.97 |
| R02012 | Downstream neighbor of SON | DONSON | 21q22.1 | 12.97 |
| AA203750 | Dimethylglycine dehydrogenase | DMGDH | 5q14.1 | 12.96 |
| AA058399 | Zinc finger protein 720 | ZNF720 | 16p11.2 | 12.89 |
| H69509 | ATP-binding cassette, sub-family B (MDR/TAP) | ABCB10 | 1q42 | 12.86 |
| W20454 | Fibronectin 1 | FN1 | 2q34 | 12.85 |
| N80357 | NDRG family member 2 | NDRG2 | 14q11.2 | 12.84 |
| W16514 | Rho family GTPase 1 | RND1 | 12q12-q13 | 12.81 |
| AA021382 | Secreted protein, acidic, cysteine-rich (osteonectin) | SPARC | 5q31.3-q32 | 12.76 |
| H90355 | Ubiquitin protein ligase E3 component n-recognin 1 | UBR1 | 15q13 | 12.67 |
| N44935 | B-cell receptor-associated protein 31 | BCAP31 | Xq28 | 12.66 |
| AA054571 | Phosphatidylinositol glycan anchor biosynthesis, class V | PIGV | 1p36.11 | 12.65 |
| W61045 | Polymerase (DNA-directed), delta 4 | POLD4 | 11q13 | 12.65 |
| R25725 | Cylindromatosis (turban tumor syndrome) | CYLD | 16q12.1 | 12.63 |
| BM468576 | Chaperonin containing TCP1, subunit 6A (zeta 1) | CCT6A | 7p11.2 | 12.61 |
| R55158 | V-ral simian leukemia viral oncogene homolog B | RALB | 2cen-q13 | 12.58 |
| N77205 | RAN binding protein 2 | RANBP2 | 2q12.3 | 12.55 |
| AA121350 | DCN1, defective in cullin neddylation 1 | DCUN1D2 | 13q34 | 12.49 |
| H22871 | Peptidase D | PEPD | 19q12-q13.2 | 12.41 |
| H71235 | Sialic acid binding Ig-like lectin 5 | SIGLEC5 | 19q13.3 | 12.41 |
| W25557 | Tripartite motif-containing 28 | TRIM28 | 19q13.4 | 12.37 |
| H78781 | Absent in melanoma 1 | AIM1 | 6q21 | 12.37 |
| N51173 | Spastin | SPAST | 2p24-p21 | 12.36 |
| AA001324 | TIMP metallopeptidase inhibitor 1 | TIMP1 | Xp11.3-p11.23 | 12.34 |
| R16054 | HMG-box transcription factor 1 | HBP1 | 7q22-q31 | 12.34 |
| R88469 | Dipeptidyl-peptidase 6 | DPP6 | 7q36.2 | 12.34 |
| AA037249 | ATP synthase | ATP5C1 | 10p15.1 | 12.32 |
| T84763 | Cell division cycle associated 8 | CDCA8 | 1p34.3 | 12.30 |
| R69935 | Hypothetical protein FLJ10404 | FLJ10404 | 5q35.3 | 12.29 |
| H89836 | Phospholipase D1, phosphatidylcholine-specific | PLD1 | 3q26 | 12.28 |
| R48131 | SH3-domain binding protein 2 | SH3BP2 | 4p16.3 | 12.27 |
| AA007268 | Polyhomeotic homolog 2 (Drosophila) | PHC2 | 1p34.3 | 12.19 |
| H52288 | Metallothionein 1E | MT1E | 16q13 | 12.18 |
| AA044796 | Similar to BMS1-like, ribosome assembly protein | LOC729096 | 10q22.2 | 12.17 |
| T70535 | NUAK family, SNF1-like kinase, 1 | NUAK1 | 12q23.3 | 12.16 |
| BQ083501 | Ribosomal protein L12 | RPL12 | 9q34 | 12.16 |
| H72796 | Hexose-6-phosphate dehydrogenase | H6PD | 1p36 | 12.05 |
| N99693 | Chromosome 12 open reading frame 32 | C12orf32 | 12p13.33 | 12.05 |
| BQ063705 | Coiled-coil-helix-coiled-coil-helix domain containing 2 | CHCHD2 | 7p11.2 | 12.04 |
| AA040816 | Cleavage and polyadenylation specific factor 3, 73 kDa | CPSF3 | 2p25.1 | 12.03 |
| R50700 | Mercaptopyruvate sulfurtransferase | MPST | 22q13.1 | 11.95 |
| H38879 | Phosphoserine phosphatase | PSPH | 7p15.2-p15.1 | 11.93 |
| AA059211 | Male germ cell-associated kinase | MAK | 6p24 | 11.93 |
| W21187 | Thymidylate synthetase | TYMS | 18p11.32 | 11.92 |
| W49716 | GRAM domain containing 3 | GRAMD3 | 5q23.2 | 11.90 |
| W05242 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | DDX5 | 17q21 | 11.90 |
| N28562 | Exportin, tRNA (nuclear export receptor for tRNAs) | XPOT | 12q14.2 | 11.86 |
| AA128587 | Zinc finger protein 629 | ZNF629 | 16p11.2 | 11.83 |
| N76529 | Membrane metallo-endopeptidase | MME | 3q25.1-q25.2 | 11.72 |
| N44807 | NF-kappaB activating protein | NKAP | Xq24 | 11.67 |
| AA147560 | Hect domain and RLD 2 pseudogene | LOC440248 | 15q13.1 | 11.67 |
| H62176 | E1A binding protein p300 | EP300 | 22q13.2 | 11.65 |
| AA030048 | Protein kinase, cAMP-dependent, regulatory, type I, beta | PRKAR1B | 7p22 | 11.63 |
| R82429 | Alpha-methylacyl-CoA racemase | AMACR | 5p13 | 11.63 |
| BM457262 | Non-metastatic cells 1, protein (NM23A) expressed in | NME1 | 17q21.3 | 11.61 |
| W19413 | Cytoskeleton-associated protein 4 | CKAP4 | 12q23.3 | 11.59 |
| AA054778 | Homeobox A10 | HOXA10 | 7p15-p14 | 11.58 |
| N36197 | Proline-rich protein HaeIII subfamily 1 | PRH1 | 12p13.2 | 11.57 |
| W47528 | Overexpressed in colon carcinoma-1 | OCC-1 | 12q23.3 | 11.54 |
| BM559619 | MOB1, Mps One Binder kinase activator-like 1B (yeast) | MOBK1B | 2p13.1 | 11.53 |
| N46377 | Galactose-3-O-sulfotransferase 4 | GAL3ST4 | 7q22.1 | 11.53 |
| AA033651 | UDP-N-acetyl-alpha-D-galactosamine | GALNT6 | 12q13 | 11.50 |
| R97614 | Ribosomal protein L32 pseudogene 3 | RPL32P3 | 3q21.3 | 11.49 |
| H82707 | Protein phosphatase 2 (formerly 2A), regulatory subunit B’ | PPP2R3A | 3q22.1 | 11.48 |
| H16005 | Niemann-Pick disease, type C2 | NPC2 | 14q24.3 | 11.46 |
| R23610 | Zinc and ring finger 2 | ZNRF2 | 7p15.1 | 11.45 |
| H25541 | Ring finger protein 138 | RNF138 | 18q12.1 | 11.44 |
| AA114919 | Y box binding protein 1 | YBX1 | 1p34 | 11.43 |
| H57747 | Betaine-homocysteine methyltransferase | BHMT | 5q13.1-q15 | 11.42 |
| BM974828 | Ribosomal protein L18 | RPL18 | 19q13 | 11.41 |
| H97422 | NOL1/NOP2/Sun domain family, member 3 | NSUN3 | 3q11.2 | 11.40 |
| BG545342 | Synaptojanin 1 | SYNJ1 | 21q22.2 | 11.40 |
| BE790941 | Centromere protein O | CENPO | 2p23.3 | 11.36 |
| H66235 | Ataxin 2 | ATXN2 | 12q24.1 | 11.31 |
| H82010 | Transcription termination factor, RNA polymerase II | TTF2 | 1p22 | 11.30 |
| N40640 | WW domain binding protein 5 | WBP5 | Xq22.1-q22.2 | 11.27 |
| H21773 | Hypothetical protein LOC145758 | LOC145758 | 15q26.3 | 11.25 |
| T97204 | Interleukin 6 receptor | IL6R | 1q21 | 11.22 |
| BQ050102 | Proteasome (prosome, macropain) subunit, beta type, 2 | PSMB2 | 1p34.2 | 11.22 |
| W02584 | Lysosomal trafficking regulator | LYST | 1q42.1-q42.2 | 11.06 |
| H61357 | Tumor protein p53 (Li-Fraumeni syndrome) | TP53 | 17p13.1 | 11.03 |
| N38855 | Cyclin B1 interacting protein 1 | CCNB1IP1 | 14q11.2 | 11.03 |
| BM928663 | Chromodomain helicase DNA binding protein 4 | CHD4 | 12p13 | 11.01 |
| BM905720 | LSM12 homolog (S cerevisiae) | LSM12 | 17q21.31 | 10.99 |
| H64813 | Ribosomal protein S28 pseudogene | LOC646195 | 11q14.1 | 10.98 |
| AA058638 | ATPase, H + transporting, lysosomal 13 kDa, V1 subunit G1 | ATP6V1G1 | 9q32 | 10.98 |
| BG469305 | Keratin 18 | KRT18 | 12q13 | 10.96 |
| T95392 | Microfibrillar-associated protein 3-like | MFAP3L | 4q32.3 | 10.91 |
| AA037600 | Regulator of chromosome condensation 1 | RCC1 | 1p36.1 | 10.90 |
* SAM assigns a score to each gene on the basis of a change in gene expression relative to the standard deviation of repeated measurements.
Table 2.
Downregulated genes
| GenBank | Name | Symbol | Cytoband | Score (d)* |
|---|---|---|---|---|
| BG700671 | Potassium inwardly rectifying channel | KCNJ13 | 2q37 | −57.88 |
| H88081 | Otoraplin | OTOR | 20p12.1-p11.23 | −55.73 |
| H81127 | Protein kinase, AMP-activated | PRKAB2 | 1q21.1 | −49.72 |
| H44375 | Myocyte enhancer factor 2B | MEF2B | 19p12 | −45.98 |
| N48215 | Solute carrier family 20 | SLC20A1 | 2q11-q14 | −45.74 |
| AA099522 | MORC family CW-type zinc finger 4 | MORC4 | Xq22.3 | −45.56 |
| BE904276 | Protein tyrosine phosphatase, non-receptor type 3 | PTPN3 | 9q31 | −45.11 |
| H03729 | Epidermal growth factor receptor | EGFR | 7p12 | −41.14 |
| T84537 | Fanconi anemia, complementation group D2 | FANCD2 | 3p26 | −41.10 |
| BG776239 | Wilms tumor 1 | WT1 | 11p13 | −38.77 |
| AA044149 | Methylmalonyl CoA epimerase | MCEE | 2p13.3 | −38.44 |
| BF437100 | Transmembrane protein 87B | TMEM87B | 2q13 | −37.68 |
| AA010608 | Parvalbumin | PVALB | 22q12-q13.1|22q13.1 | −36.84 |
| BG818724 | Solute carrier family 7 | SLC7A1 | 13q12-q14 | −36.44 |
| AA156812 | Collagen, Type XVIII, alpha 1 | COL18A1 | 21q22.3 | −35.48 |
| N76723 | Hypothetical protein LOC150166 | LOC150166 | 22q11.21 | −35.03 |
| N50000 | Methionine adenosyltransferase I, alpha | MAT1A | 10q22 | −34.96 |
| AA136950 | Plexin domain containing 2 | PLXDC2 | 10p12.32-p12.31 | −34.86 |
| BG620850 | Chorionic somatomammotropin hormone 2 | CSH2 | 17q24.2 | −34.04 |
| H79911 | Core-binding factor, runt domain | CBFA2T3 | 16q24 | −33.25 |
| N55596 | NOL1/NOP2/Sun domain family, member 7 | NSUN7 | 4p14 | −33.05 |
| W44535 | Neurochondrin | NCDN | 1p34.3 | −33.00 |
| N42329 | Suppressor of cytokine signaling 6 | SOCS6 | 18q22.2 | −32.03 |
| AA127799 | FYVE and coiled-coil domain containing 1 | FYCO1 | 3p21.31 | −31.87 |
| BG622452 | ADAM metallopeptidase domain 12 (meltrin alpha) | ADAM12 | 10q26.3 | −31.86 |
| BQ073808 | Proteasome (prosome, macropain) | PSMC4 | 19q13.11-q13.13 | −31.65 |
| BM466167 | Septin 6 | SEP6 | Xq24 | −31.55 |
| H52445 | Leucine rich repeat containing 31 | LRRC31 | 3q26.2 | −31.41 |
| W87840 | Helicase with zinc finger | HELZ | 17q24.2 | −29.81 |
| R53682 | SH2 domain containing 3C | SH2D3C | 9q34.11 | −29.56 |
| H69334 | Pirin (iron-binding nuclear protein) | PIR | Xp22.2 | −29.15 |
| W05657 | E74-like factor 1 (ets domain transcription factor) | ELF1 | 13q13 | −28.60 |
| W47223 | Mitochondrial trans-2-enoyl-CoA reductase | MECR | 1p36.1-p35.1 | −28.59 |
| AA053903 | FRY-like | FRYL | 4p12 | −28.54 |
| N44611 | Transmembrane protein 50B | TMEM50B | 21q22.11 | −28.28 |
| R99229 | Hydroxymethylbilane synthase | HMBS | 11q23.3 | −28.12 |
| BM857788 | Nuclear receptor co-repressor 2 | NCOR2 | 12q24 | −27.72 |
| N52672 | Nuclear receptor subfamily 1, group D, member 2 | NR1D2 | 3p24.2 | −27.52 |
| H17037 | Similar to CG4502-PA | FLJ25076 | 5p15.31 | −26.95 |
| BE779318 | Transcription elongation factor B (SIII) | TCEB3 | 1p36.1 | −26.77 |
| T90862 | Remodeling and spacing factor 1 | RSF1 | 11q14.1 | −26.74 |
| AA455435 | Chromosome 9 open reading frame 5 | C9orf5 | 9q31 | −26.67 |
| AI188464 | Matrix metallopeptidase 11 (stromelysin 3) | MMP11 | 22q11.2|22q11.23 | −26.36 |
| N54724 | Chromosome 14 open reading frame 24 | C14orf24 | 14q13.2 | −26.14 |
| W38932 | Heme oxygenase (decycling) 2 | HMOX2 | 16p13.3 | −25.94 |
| N51855 | Poly (ADP-ribose) polymerase family, member 2 | PARP2 | 14q11.2-q12 | −25.87 |
| R99225 | Keratin associated protein 4-7 | KRTAP4-7 | 17q12-q21 | −25.75 |
| T78280 | Histone acetyltransferase 1 | HAT1 | 2q31.2-q33.1 | −24.75 |
| R16431 | Chromosome 4 open reading frame 29 | C4orf29 | 4q28.2 | −24.61 |
| H91396 | Bile acid coenzyme A: amino acid N-acyltransferase | BAAT | 9q22.3 | −24.49 |
| T66756 | Sprouty homolog 3 (Drosophila) | SPRY3 | Xq28 and Yq12 | −24.16 |
| BG565707 | Fibrinogen gamma chain | FGG | 4q28 | −24.00 |
| AA031920 | Cytochrome b-245, alpha polypeptide | CYBA | 16q24 | −23.63 |
| H77390 | Golgi autoantigen, golgin subfamily a, 1 | GOLGA1 | 9q33.3 | −23.34 |
| R98300 | KIAA0286 protein | KIAA0286 | 12q13.3 | −23.28 |
| H25352 | Serum response factor binding protein 1 | SRFBP1 | 5q23.1 | −22.98 |
| H67225 | Solute carrier family 7 | SLC7A2 | 8p22-p21.3 | −22.56 |
| AA151360 | Rho GTPase activating protein 12 | ARHGAP12 | 10q11.1 | −22.36 |
| N34285 | Solute carrier family 26 | SLC26A2 | 5q31-q34 | −22.34 |
| H59530 | CHK1 checkpoint homolog (S pombe) | CHEK1 | 11q24-q24 | −22.25 |
| H68793 | Yip1 interacting factor homolog B (S cerevisiae) | YIF1B | 19q13.2 | −22.09 |
| H75715 | Membrane bound O-acyltransferase domain containing 2 | MBOAT2 | 2p25.1 | −21.77 |
| W19459 | Dipeptidyl-peptidase 8 | DPP8 | 15q22 | −21.59 |
| R61012 | CDC42 binding protein kinase alpha (DMPK-like) | CDC42BPA | 1q42.11 | −21.38 |
| H61387 | Reticulon 4 receptor | RTN4R | 22q11.21 | −21.33 |
| H28872 | Aspartyl-tRNA synthetase | DARS | 2q21.3 | −21.21 |
| W47361 | Folate receptor 3 (gamma) | FOLR3 | 11q13 | −21.16 |
| T92079 | Proteasome (prosome, macropain) activator subunit 2 | PSME2 | 14q11.2 | −21.15 |
| N55035 | Peroxisomal biogenesis factor 3 | PEX3 | 6q23-q24 | −21.03 |
| N48524 | TIA1 cytotoxic granule-associated RNA binding protein-like 1 | TIAL1 | 10q | −20.94 |
| BG770889 | RAB11 family interacting protein 2 (class I) | RAB11FIP2 | 10q26.11 | −20.75 |
| H97449 | Integrin, beta 5 | ITGB5 | 3q21.2 | −20.71 |
| T78739 | EPH receptor B2 | EPHB2 | 1p36.1-p35 | −20.62 |
| H10896 | Dual specificity phosphatase 4 | DUSP4 | 8p12-p11 | −20.42 |
| T87012 | CD79a molecule, immunoglobulin-associated alpha | CD79A | 19q13.2 | −20.31 |
| W16524 | CDC42 binding protein kinase alpha (DMPK-like) | CDC42BPA | 1q42.11 | −20.30 |
| R47766 | Transient receptor potential cation channel, subfamily C | TRPC4AP | 20q11.22 | −20.23 |
| T64848 | Period homolog 3 (Drosophila) | PER3 | 1p36.23 | −20.16 |
| N54874 | Chromosome 20 open reading frame 39 | C20orf39 | 20p11.21 | −20.16 |
| T95182 | Chromosome 6 open reading frame 86 | C6orf86 | 6p25.2 | −20.14 |
| BM994830 | UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase | B4GALT1 | 9p13 | −19.97 |
| N44094 | Cyclin J | CCNJ | 10pter-q26.12 | −19.77 |
| R02669 | Adaptor-related protein complex 3, beta 1 subunit | AP3B1 | 5q14.1 | −19.77 |
| H00518 | Multiple inositol polyphosphate histidine phosphatase, 1 | MINPP1 | 10q23 | −19.73 |
| R64061 | Pregnancy specific beta-1-glycoprotein 5 | PSG5 | 19q13.2 | −19.62 |
| T84786 | TRNA splicing endonuclease 2 homolog (S cerevisiae) | TSEN2 | 3p25.1 | −19.50 |
| BI818657 | Serine/threonine kinase 10 | STK10 | 5q35.1 | −19.49 |
| H79636 | KIAA1012 | KIAA1012 | 18q12.1 | −19.41 |
| AA055329 | Hypothetical locus LOC678655 | LOC678655 | 12p13.31 | −19.28 |
| H80810 | Formin-like 2 | FMNL2 | 2q23.3 | −19.26 |
| R91604 | Solute carrier family 38, member 2 | SLC38A2 | 12q | −19.21 |
| T77428 | ELOVL family member 5, elongation of long chain fatty acids | ELOVL5 | 6p21.1-p12.1 | −19.17 |
| H85608 | Protein phosphatase 1, regulatory (inhibitor) subunit 2 | PPP1R2 | 3q29 | −19.08 |
| T98709 | Major facilitator superfamily domain containing 11 | MFSD11 | 17q25 | −19.04 |
| AA033653 | Major histocompatibility complex, class II, DR beta 1 | HLA-DRB1 | 6p21.3 | −19.03 |
| W78787 | Complement component 5 | C5 | 9q33-q34 | −19.01 |
| AA001996 | MutS homolog 6 (E coli) | MSH6 | 2p16 | −18.87 |
| N32361 | PQ loop repeat containing 3 | PQLC3 | 2p25.1 | −18.85 |
| H45525 | Ras homolog gene family, member G (rho G) | RHOG | 11p15.5-p15.4 | −18.79 |
| AI927909 | Homogentisate 1,2-dioxygenase (homogentisate oxidase) | HGD | 3q13.33 | −18.75 |
| R05896 | Sodium channel modifier 1 | SCNM1 | 1q21.2 | −18.59 |
| N80988 | GTP binding protein 2 | GTPBP2 | 6p21-p12 | −18.53 |
| N40600 | SUMO1/sentrin specific peptidase 7 | SENP7 | 3q12 | −18.47 |
| R23473 | PAK1 interacting protein 1 | PAK1IP1 | 6p24.2 | −18.40 |
| BQ063621 | Calsyntenin 1 | CLSTN1 | 1p36.22 | −18.36 |
| H58311 | Coagulation factor V (proaccelerin, labile factor) | F5 | 1q23 | −18.29 |
| H08311 | DTW domain containing 2 | DTWD2 | 5q23.1 | −18.26 |
| W02106 | Solute carrier family 26 (sulfate transporter), member 2 | SLC26A2 | 5q31-q34 | −18.22 |
| H39162 | 1-acylglycerol-3-phosphate O-acyltransferase 1 | AGPAT1 | 6p21.3 | −17.90 |
| AI368607 | Family with sequence similarity 13, member A1 | FAM13A1 | 4q22.1 | −17.89 |
| AA040364 | Hypothetical protein LOC284513 | LOC284513 | 1p36.13 | −17.84 |
| H38322 | SET binding factor 1 | SBF1 | 22q13.33 | −17.69 |
| R73417 | Peptidase inhibitor 16 | PI16 | 6p21.2 | −17.60 |
| T67154 | IMP2 inner mitochondrial membrane peptidase-like | IMMP2L | 7q31 | −17.45 |
| R96767 | Phospholipase C, beta 4 | PLCB4 | 20p12 | −17.42 |
| N35681 | Diablo homolog (Drosophila) | DIABLO | 12q24.31 | −17.29 |
| W25288 | SNAP-associated protein | SNAPAP | 1q21.3 | −17.27 |
| AA203442 | Chromosome 9 open reading frame 39 | C9orf39 | 9p22.2 | −17.21 |
| R73337 | Zinc finger protein 777 | ZNF777 | 7q36.1 | −17.08 |
| N73236 | Storkhead box 1 | STOX1 | 10q21.3 | −16.94 |
| H39156 | Myotubularin related protein 6 | MTMR6 | 13q12 | −16.93 |
| T77015 | GSG1-like | GSG1L | 16p11.2 | −16.66 |
| R23489 | Zinc finger protein 354A | ZNF354A | 5q35.3 | −16.62 |
| H64555 | S100 calcium binding protein A2 | S100A2 | 1q21 | −16.52 |
| W90519 | Zinc finger protein 652 | ZNF652 | 17q21.32 | −16.50 |
| W19130 | Plexin A2 | PLXNA2 | 1q32.2 | −16.39 |
| H78273 | Sperm associated antigen 9 | SPAG9 | 17q21.33 | −16.33 |
| AA156879 | Zinc finger protein 615 | ZNF615 | 19q13.33 | −16.30 |
| N53192 | Hypothetical protein MGC22014 | hCG_40738 | 2p13.1 | −16.00 |
| BG682138 | Secreted protein, acidic, cysteine-rich (osteonectin) | SPARC | 5q31.3-q32 | −15.87 |
| H41974 | Integrin, alpha 3 | ITGA3 | 17q21.33 | −15.84 |
| H79050 | Protein tyrosine phosphatase, receptor type, E | PTPRE | 10q26 | −15.83 |
| W48559 | Zinc finger, MYM-type 1 | ZMYM1 | 1p34.3 | −15.78 |
| H87048 | ADP-ribosylation factor GTPase activating protein 3 | ARFGAP3 | 22q13.2-q13.3 | −15.76 |
| T84174 | Eukaryotic translation initiation factor 3 | EIF3S9 | 7p22.2 | −15.76 |
| N40120 | Zinc finger protein 33B | ZNF33B | 10q11.2 | −15.74 |
| W35313 | Sterile alpha motif and leucine zipper containing kinase AZK | ZAK | 2q24.2 | −15.70 |
| AA040656 | Zinc finger protein 502 | ZNF502 | 3p21.31 | −15.70 |
| H18810 | Importin 8 | IPO8 | 12p11.21 | −15.68 |
| R12736 | Staufen, RNA binding protein, homolog 2 (Drosophila) | STAU2 | 8q13-q21.1 | −15.61 |
| N74741 | BTG family, member 3 | BTG3 | 21q21.1-q21.2 | −15.61 |
| AA069533 | Chromosome 7 open reading frame 42 | C7orf42 | 7q11.21 | −15.55 |
| N28267 | Integrin, alpha X | ITGAX | 16p11.2 | −15.47 |
| AA135718 | Neuropilin 1 | NRP1 | 10p12 | −15.46 |
| R50902 | Tubulin, gamma complex associated protein 6 | TUBGCP6 | 22q13.31-q13.33 | −15.46 |
| BQ020504 | Translocase of outer mitochondrial membrane 20 homolog | TOMM20 | 1q42 | −15.43 |
| BM545369 | Hect domain and RLD 2 pseudogene 2 | HERC2P2 | 15q11.2 | −15.39 |
| W58640 | SECIS binding protein 2 | SECISBP2 | 9q22.2 | −15.21 |
| W49512 | Bradykinin receptor B1 | BDKRB1 | 14q32.1-q32.2 | −15.19 |
| N46282 | PiggyBac transposable element derived 2 | PGBD2 | 1q44 | −15.12 |
| W31642 | Early B-cell factor 3 | EBF3 | 10q26.3 | −15.05 |
| BG623586 | ADAM metallopeptidase with thrombospondin type 1 motif | ADAMTS5 | 21q21.3 | −14.97 |
| W87709 | Kelch-like 23 (Drosophila) | KLHL23 | 2q31.1 | −14.97 |
| N44005 | EF-hand calcium binding domain 2 | EFCAB2 | 1q44 | −14.96 |
| W52509 | ARV1 homolog (S cerevisiae) | ARV1 | 1q42.2 | −14.95 |
| H79753 | Death-associated protein | DAP | 5p15.2 | −14.93 |
| BI905854 | Glycerophosphodiester phosphodiesterase domain containing 3 | GDPD3 | 16p11.2 | −14.71 |
| H53660 | HLA-B associated transcript 3 | BAT3 | 6p21.3 | −14.71 |
| AA037312 | ATP synthase mitochondrial F1 complex assembly factor 1 | ATPAF1 | 1p33-p32.3 | −14.68 |
| N57425 | DAZ associated protein 2 | DAZAP2 | 12q12 | −14.67 |
| BM545099 | Lectin, galactoside-binding, soluble, 9 (galectin 9) | LGALS9 | 17q11.1 | −14.66 |
| W16685 | N-glycanase 1 | NGLY1 | 3p24.2 | −14.64 |
| N76853 | Golgi autoantigen, golgin subfamily b, macrogolgin | GOLGB1 | 3q13 | −14.59 |
| AA001311 | Hypothetical protein LOC129293 | LOC129293 | 2p11.2 | −14.54 |
| W40439 | Forkhead box J1 | FOXJ1 | 17q22-q25 | −14.44 |
| H61030 | REX2, RNA exonuclease 2 homolog (S cerevisiae) | REXO2 | 11q23.1-q23.2 | −14.39 |
| H03728 | G1 to S phase transition 1 | GSPT1 | 16p13.1 | −14.37 |
| W48584 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase | P4HA2 | 5q31 | −14.30 |
| BI861012 | Mannosidase, alpha, class 1B, member 1 | MAN1B1 | 9q34 | −14.24 |
| AA203133 | DNA (cytosine-5-)-methyltransferase 1 | DNMT1 | 19p13.2 | −14.15 |
| AA481714 | Chromosome 6 open reading frame 62 | C6orf62 | 6p22.2 | −14.02 |
| H08319 | Zinc finger protein 783 | ZNF783 | 7q36.1 | −14.01 |
| N44142 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | HMGCR | 5q13.3-q14 | −13.97 |
| R15789 | Tumor suppressing subtransferable candidate 1 | TSSC1 | 2p25.3 | −13.97 |
| H79770 | Tripartite motif-containing 27 | TRIM27 | 6p22 | −13.93 |
| AI056197 | Amidohydrolase domain containing 2 | AMDHD2 | 16p13.3 | −13.92 |
| BE871226 | TAF6 RNA polymerase II | TAF6 | 7q22.1 | −13.87 |
| BM468475 | Keratin 8 pseudogene 12 | KRT8P12 | 3q26.1 | −13.84 |
| H10533 | Plasminogen activator, tissue | PLAT | 8p12 | −13.82 |
| H93653 | Collagen-like tail subunit of asymmetric acetylcholinesterase | COLQ | 3p25 | −13.82 |
| BG333273 | CD47 molecule | CD47 | 3q13.1-q13.2 | −13.80 |
| H44717 | Cytochrome c oxidase subunit 8A (ubiquitous) | COX8A | 11q12-q13 | −13.79 |
| W38526 | Exostoses (multiple)-like 2 | EXTL2 | 1p21 | −13.75 |
| R86053 | Nuclear factor of kappa light polypeptide gene enhancer | NFKB2 | 10q24 | −13.71 |
| AA203387 | Trophinin associated protein (tastin) | TROAP | 12q13.12 | −13.70 |
| H52351 | Transmembrane protein 150 | TMEM150 | 2p11.2 | −13.70 |
| W47015 | Ts translation elongation factor, mitochondrial | TSFM | 12q13-q14 | −13.60 |
| T89583 | Spermatogenesis associated 21 | SPATA21 | 1p36.13 | −13.59 |
| BM993318 | Ubiquitin specific peptidase 24 | USP24 | 1p32.3 | −13.58 |
| W88434 | Carboxypeptidase B2 (plasma) | CPB2 | 13q14.11 | −13.55 |
| N48417 | GA binding protein transcription factor, alpha subunit 60 kDa | GABPA | 21q21-q22.1|21q21.3 | −13.53 |
| H81801 | Phosphatidylinositol 3,4,5-trisphosphate-dependent RAC exchanger 1 | PREX1 | 20q13.13 | −13.52 |
| AI478910 | FERM domain containing 6 | FRMD6 | 14q22.1 | −13.45 |
| AA450143 | WD repeat domain 27 | WDR27 | 6q27 | −13.41 |
| H61757 | ELK4, ETS-domain protein (SRF accessory protein 1) | ELK4 | 1q32 | −13.31 |
| BM459914 | Serine/threonine kinase 4 | STK4 | 20q11.2-q13.2 | −13.31 |
| W60673 | CREB regulated transcription coactivator 3 | CRTC3 | 15q26.1 | −13.28 |
| R32668 | Component of oligomeric golgi complex 3 | COG3 | 13q14.12 | −13.27 |
| AA045300 | CDC42 small effector 2 | CDC42SE2 | 5q31.1 | −13.17 |
| W56454 | Furin (paired basic amino acid cleaving enzyme) | FURIN | 15q26.1 | −13.12 |
| W61099 | Chromosome X open reading frame 36 | CXorf36 | −13.06 | |
| AA037834 | Methylmalonic aciduria (cobalamin deficiency) cblB type | MMAB | 12q24 | −13.06 |
| W90717 | Solute carrier family 24 (sodium/potassium/calcium exchanger) | SLC24A4 | 14q32.12 | −13.05 |
| N44262 | Pecanex homolog (Drosophila) | PCNX | 14q24.2 | −12.99 |
| T70417 | REC8 homolog (yeast) | REC8 | 14q11.2-q12 | −12.98 |
| BE899110 | Family with sequence similarity 105, member B | FAM105B | 5p15.2 | −12.94 |
| R06564 | UDP-galactose-4-epimerase | GALE | 1p36-p35 | −12.92 |
| R50922 | Neuroligin 4, X-linked | NLGN4X | Xp22.32-p22.31 | −12.89 |
| H15612 | COX10 homolog, cytochrome c oxidase assembly protein | COX10 | 17p12-p11.2 | −12.88 |
| R18433 | Opioid binding protein/cell adhesion molecule-like | OPCML | 11q25 | −12.85 |
| AA143060 | Melanoma associated antigen (mutated) 1 | MUM1 | 19p13.3 | −12.81 |
| BF969700 | Chromosome 12 open reading frame 35 | C12orf35 | 12p11.21 | −12.79 |
| N43949 | Mitogen-activated protein kinase kinase kinase kinase 4 | MAP4K4 | 2q11.2-q12 | −12.77 |
| N71526 | Inhibitor of kappa light polypeptide gene enhancer in B-cells | IKBKB | 8p11.2 | −12.73 |
| W78799 | Nudix (nucleoside diphosphate linked moiety X)-type motif 13 | NUDT13 | 10q22.1 | −12.69 |
| W20458 | Tripartite motif-containing 59 | TRIM59 | 3q26.1 | −12.69 |
| H89618 | WNK lysine deficient protein kinase 1 | WNK1 | 12p13.3 | −12.67 |
| R65820 | SLC2A4 regulator | SLC2A4RG | 20q13.33 | −12.61 |
| R87913 | Potassium voltage-gated channel, delayed-rectifier, subfamily S | KCNS1 | 20q12 | −12.60 |
| N57603 | Solute carrier organic anion transporter family, member 1C1 | SLCO1C1 | 12p12.2 | −12.55 |
| AA045905 | Forkhead box P1 | FOXP1 | 3p14.1 | −12.55 |
| H29349 | Abelson helper integration site 1 | AHI1 | 6q23.3 | −12.54 |
| H08988 | Ubiquitin specific peptidase 7 (herpes virus-associated) | USP7 | 16p13.3 | −12.43 |
| R23677 | Nucleolar protein 4 | NOL4 | 18q12 | −12.43 |
| H46899 | Adenosine deaminase, RNA-specific, B2 (RED2 homolog rat) | ADARB2 | 10p15.3 | −12.41 |
| BI667959 | Reticulon 1 | RTN1 | 14q23.1 | −12.39 |
| BG323782 | Coiled-coil domain containing 14 | CCDC14 | 3q21.1 | −12.36 |
| R38905 | Dihydropyrimidinase-like 5 | DPYSL5 | 2p23.3 | −12.35 |
| N23456 | Cytoglobin | CYGB | 17q25.3 | −12.34 |
| R11685 | COP9 constitutive photomorphogenic homolog subunit 5 | COPS5 | 8q13.2 | −12.33 |
| R06410 | O-6-methylguanine-DNA methyltransferase | MGMT | 10q26 | −12.31 |
| H63698 | N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D | NAPE-PLD | 7q22.1 | −12.27 |
| H93191 | Ubiquitin specific peptidase 3 | USP3 | 15q22.3 | −12.26 |
| AA069502 | Hypothetical protein DKFZp434H1419 | DKFZp434H1419 | 2q35 | −12.24 |
| AA043530 | MORN repeat containing 2 | MORN2 | 2p22.1 | −12.21 |
| N57339 | LMBR1 domain containing 1 | LMBRD1 | 6q13 | −12.20 |
| H40732 | PPAR binding protein | PPARBP | 17q12-q21.1 | −12.19 |
| AI822112 | Similar to SR protein related family member (rsr-1) | LOC728676 | 1q42.13 | −12.18 |
| R12743 | HECT domain containing 1 | HECTD1 | 14q12 | −12.14 |
| N48445 | RAB33A, member RAS oncogene family | RAB33A | Xq25 | −12.13 |
| H03305 | Bromodomain containing 1 | BRD1 | 22q13.33 | −12.13 |
| AA010089 | Hypothetical protein LOC157860 | LOC157860 | 8p11.23 | −12.09 |
| H93176 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | PFKFB3 | 10p14-p15 | −12.07 |
| R12649 | Solute carrier family 13 (sodium-dependent dicarboxylate transporter) | SLC13A3 | 20q12-q13.1 | −12.05 |
| R14154 | Hypothetical protein LOC286063 | LOC286063 | 8q11.21 | −12.03 |
| R61444 | Thyroid adenoma associated | THADA | 2p21 | −12.02 |
| W17278 | Solute carrier family 25 (mitochondrial oxodicarboxylate carrier) | SLC25A21 | 14q11.2 | −12.00 |
| T77303 | Leucine-rich repeat LGI family, member 2 | LGI2 | 4p15.2 | −11.98 |
| AA043837 | Coiled-coil domain containing 45 | CCDC45 | 17q24.1 | −11.93 |
| R52735 | THAP domain containing 8 | THAP8 | 19q13.12 | −11.92 |
| W46207 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 | B4GALT1 | 9p13 | −11.79 |
| BI914695 | Immunoglobulin superfamily containing leucine-rich repeat 2 | ISLR2 | 15q24.1 | −11.78 |
| BQ000722 | Deoxyribonuclease I-like 1 | DNASE1L1 | Xq28 | −11.73 |
| N52657 | MYC binding protein 2 | MYCBP2 | 13q22 | −11.68 |
| AI340082 | Sulfotransferase family 1E, estrogen-preferring, member 1 | SULT1E1 | 4q13.1 | −11.67 |
| N54717 | Acyl-coenzyme A binding domain containing 5 | ACBD5 | 10p12.1 | −11.67 |
| R16400 | Elongation factor, RNA polymerase II, 2 | ELL2 | 5q15 | −11.63 |
| AA151264 | ALS2 C-terminal like | ALS2CL | 3p21.31 | −11.59 |
| T80372 | Calcium channel, voltage-dependent, alpha 2/delta subunit 2 | CACNA2D2 | 3p21.3 | −11.58 |
| AW872398 | Amphiphysin | AMPH | 7p14-p13 | −11.53 |
| H08101 | Glutaminase 2 (liver, mitochondrial) | GLS2 | 12q13 | −11.51 |
| AA134026 | Ubiquitin-conjugating enzyme E2A (RAD6 homolog) | UBE2A | Xq24-q25 | −11.49 |
| R72472 | HLA-B associated transcript 1 | BAT1 | 6p21.3 | −11.47 |
| BM450631 | Heat shock protein 90 kDa alpha (cytosolic), class A member 2 | HSP90AA2 | 11p14.1 | −11.45 |
| AA534429 | Chromosome 1 open reading frame 38 | C1orf38 | 1p35.3 | −11.44 |
| BI753390 | Amyloid beta (A4) precursor protein-binding, family A | APBA2 | 15q11-q12 | −11.44 |
| BF112255 | Tousled-like kinase 2 | TLK2 | 17q23 | −11.42 |
| T78737 | KIAA2026 | KIAA2026 | 9p24.1 | −11.42 |
| R39428 | Protein tyrosine phosphatase, receptor type, G | PTPRG | 3p21-p14 | −11.40 |
| N42943 | PHD finger protein 17 | PHF17 | 4q26-q27 | −11.38 |
| H86918 | Pleckstrin homology domain containing, family B (evectins) | PLEKHB1 | 11q13.5-q14.1 | −11.38 |
| R23434 | Membrane-bound transcription factor peptidase, site 2 | MBTPS2 | Xp22.1-p22.2 | −11.37 |
| R17293 | GLE1 RNA export mediator-like (yeast) | GLE1L | 9q34.11 | −11.33 |
| BI522504 | Zinc finger protein 605 | ZNF605 | 12q24.33 | −11.29 |
| AA056151 | SEC24 related gene family, member C (S. cerevisiae) | SEC24C | 10q22.2 | −11.28 |
| AA055164 | Low density lipoprotein receptor-related protein 6 | LRP6 | 12p11-p13 | −11.21 |
| R27647 | KIAA1333 | KIAA1333 | 14q12 | −11.18 |
* SAM assigns a score to each gene on the basis of a change in gene expression relative to the standard deviation of repeated measurements.
Table 3.
Main upregulated genes related to bone formation
| Gene | Function | Effect | Reference |
|---|---|---|---|
| STAT 3 | Transcriptional factor, activation of the MAP kinase | Bone turnover | Itoh et al. [27] |
| HOXA 10 | Activation of Runx2, alkaline phosphatase, osteocalcin, and bone sialoprotein | Osteogenic response | Hassan et al. [25] |
| AKT1 | Suppression of osteoblasts apoptosis through inhibition of Fox03a and Bim; Mediation of the osteoblastic bone formation by IGF-1 and insulin. | Bone formation | Kawamura et al. [29] |
| CALM1 | Signal transduction, stimuli to proliferation | Bone formation | Rhymer et al. [40] |
| P2RX7 | Activation of P2RX7 receptors stimulates expression of osteoblast markers and enhances mineralization in cultures cells | Promote osteogenesis | Ohlendorff et al. [37] |
| FN1 | Adhesion and migration cellular process | Extracellular matrix stability; tissue healing | Potts and Campbell [39] |
| COL1A2 | Collagen 1α2, chain of the most abundant collagen in the human organism | Extracellular matrix stability | Antoniv et al. [6] |
| SPARC | The most abundant noncollagenous protein in the bone tissue. | Modulation of the cell-matrix interaction and production of the matrix | Yan and Sage [47] |
| VCL | Associated with the intercellular junctions between the cells and the matrix | Anchorages the actin to the cellular membrane | Ziegler et al. [49] |
| TIMP1 | Inhibits collagen and other components of extracellular matrix degradation operated by the metalloproteinase | Decrease matrix degradation | Hatori et al. [26] |
Table 4.
Main downregulated genes related to bone formation
| Gene | Function | Effect | Reference |
|---|---|---|---|
| MMP-11 | Metallopeptidase with substrate specificity, including proteoglycans, lamini, and fibronectin | Degradation of extracellular matrix | Matziari et al. [35] |
| DUSP4 | Inactivates the superfamily of MAP kinase | Inhibition of proliferation and differentiation | Caunt et al. [17] |
In particular, PEMFs induced upregulation of several genes at the transcriptional level like STAT3, homeobox A10 (HOXA10), and V-akt murine thymoma viral oncogene homolog 1 (AKT1). Some genes acting at the transductional level also are upregulated including calmodulin (CALM1), activator protein 1 (AP-1), Nuclear factor kappaB (NF-KB), cAMP response element binding (CREB), and P2RX7 (Table 3). Several interesting overexpressed genes are components of cytoskeleton and involved in cell adhesion (Table 3). Examples are fibronectin (FN1) and vinculin (VCL). PEMF also increased the expression of genes encoding for collagenous and noncollagenous extracellular matrix proteins including collagen Type 1α2 (COL1A2), osteonectin (SPARC), and metallopeptidase inhibitor 1 (TIMP1) (Table 3).
Some genes downregulated by PEMFs are related to degradation of extracellular matrix (ECM) (Table 4), specifically, matrix metallopeptidase 11 (MMP11), or stromelysin 3 and dual specificity phosphatase 4 (DUSP4).
Discussion
The improvement of osteogenesis is important because of the wide clinical applications it may have. PEMFs reportedly restart osteogenesis in disorders in which it has stopped [34] and in disorders in which osteogenesis needs to be enhanced [32]. Although considerable basic and clinical research on PEMFs has been reported, their mechanism of action is not completely clear. Moreover, studies in the existing literature have so far focused only on a few aspects of cell activities [9, 10, 46], or they have been performed by using different types of signals in different experimental conditions [1, 9, 22, 23]. To address these limitations in the literature, we asked (1) whether PEMFs affected a wide array of genes in human osteoblastlike cells (MG63), and (2) whether and to what extent PEMFs induce proliferation and differentiation of osteoblasts.
We acknowledge several limitations. First, the experiment was performed using a human osteosarcoma cell line (MG63), whereas the use of a primary human osteoblast cell culture might better replicate what happens in humans in vivo. We chose the MG63 cell line because these cells show a phenotype similar to that of normal human osteoblasts, while also providing a reproducible experimental model suitable for the microarray analysis. Second, as it is still difficult to explain the roles of all genes, whose expression was modified, we focused on the role of genes with well-known functions related to osteogenesis. Third, although microarray technology is widely accepted as a valid approach to describe changes induced by a factor on cell environment, additional research using, for example RT-PCR, might be useful to provide supplementary support for the results obtained. Fourth, we studied responses at only one time. We chose 18 hours exposure time on the basis of a previous time experiment, in which a peak in DNA synthesis was seen after 18 hours of stimulation in MG63 cultures maintained in the presence of 10% FCS [45]. In contrast, Lohmann et al. reported PEMFs enhanced cell differentiation in MG63 cultures and reduced cell proliferation [30]. The differences existing between the two sets of data regarding cell proliferation could be related to the different experimental conditions used. Lohmann et al. exposed MG63 cultures when they reached confluence. When cultures are confluent they stop to proliferate. We exposed cells to PEMF when cultures were subconfluent, therefore, they responded with an enhancement of proliferation. We cannot extrapolate our findings to shorter or longer exposures to PEMFs.
PEMFs appear to act on bone formation by inducing upregulation of several genes related to osteoblast proliferation and differentiation. Among those genes, HOXA10, a transcriptional factor that acts positively on RUNX2, is the main transcriptional regulator of osteoblast differentiation [25]. HOXA10 controls osteoblastogenesis via RUNX2-promoted osteoprogenitor cell differentiation in immature osteoblasts [25]. This protein also is believed to be involved in activation of alkaline phosphatase, osteocalcin, and sialoprotein genes [25]. We also observed STAT3, P2RX7, and AKT1 upregulation. It has been suggested that osteoblast-specific disruption of STAT3 results in an osteopenic phenotype [27, 41]. STAT3, involved in bone turnover [27], regulates the transcription of various genes that modulate cell proliferation and differentiation in a cell-specific manner [27]. P2RX7 is a purinergic receptor, which is correlated with calcium channels and interacts with the calmodulin-dependent protein [37]. Activation of P2RX7 receptors by exogenous nucleotides stimulates expression of osteoblast markers and enhances mineralization in cultures of rat calvarial cells promoting osteogenesis [37]. V-akt murine thymoma viral oncogene homolog 1 (AKT1), is a phosphoinositide-dependent serine-threonine protein kinase, and one of the key players in the signaling of potent bone anabolic factors [29]. The disruption of AKT1 in mice led to low-turnover osteopenia through dysfunction [29]. AKT1 deficiency causes decreased bone mass and formation [29], impairs RUNX2-dependent differentiation and function of osteoblasts [29], and impairs bone resorption via dysfunction of osteoblasts and osteoclasts [29]. AKT1 suppresses osteoblasts apoptosis through inhibition of FOXO3a and Bim [29], and may mediate the osteoblastic bone formation by IGF-1 [29]. The IGF-1/AKT1 pathway might be a common pathway for bone anabolic action of parathyroid, thyroid, and growth hormone [29].
We also observed upregulation of genes involved in connective and bone tissue formation (COL1A2) and noncollagenous extracellular matrix (ECM) synthesis (SPARC, FN1, VCL). COL1A2 encodes for collagen Type 1α2. Collagen Type 1 is the most represented collagen in the human organism and is important for ECM stability [6]. Osteonectin (SPARC), the most abundant noncollagenous protein in bone tissue, modulates cell-matrix interaction and is involved in the tissue-remodeling process [47]. FN1 is important for ECM stability and involved in adhesion and migration cellular processes such as tissue healing [39]. VCL is a cytoskeletal protein associated with the intercellular junctions between the cells and the matrix [49].
The effect of TIMP1 upregulation and of MMP-11 and DUSP4 downregulation can be interpreted as a decrease in the degradation process. TIMP1 promotes apposition of ECM by inhibiting collagen and other components of ECM degradation operated by the metalloproteinase [26]. DUSP4 inactivates the superfamily of MAP kinase, which is involved with proliferation and differentiation. DUSP4 downregulation, then, stimulates proliferation [17]. MMPs potentially can degrade almost all components of the periprosthetic ECM and contribute to prosthetic loosening and osteolysis through pathologic ECM degradation and bone remodeling around prostheses [28, 35]. The stromelysins especially have broad substrate specificity, including proteoglycans, laminin, and fibronectin [35]. Stromelysin-1 determines the release and activation ECM-bound latent TGF-ß1 and is involved with ECM turnover [8]. Upregulation of CALM1 promotes enhancement of calmodulin1, a protein involved in proliferative cell activation [40]. Calmodulin also is involved in the transduction mechanism of PEMFs [9].
Our data suggest many effects of PEMFs on human osteoblastlike cells in vitro. PEMFs seem to exert an anabolic effect on cells. In particular, they are consistent with abundant preclinical and clinical findings showing a positive effect of PEMFs on osteogenesis. Stimulation by PEMFs induces bone healing in patients, shortens the time of healing processes, and stimulates healing of nonunions. Exposure to PEMFs acts on cell behavior in different ways. More specifically, PEMFs stimulate cell proliferation and induce osteoblastogenesis and differentiation of osteoblasts. Moreover, PEMFs promote ECM apposition and mineralization, and decrease degradation and absorption processes of ECM. These data suggest a more comprehensive explanation of the observed clinical effect of PEMFs on the induction of osteogenesis. Given their broad effects, PEMFs might be useful in other fields such as regenerative medicine.
Footnotes
Each author certifies that he or she has no commercial associations (eg, consultancies, stock ownership, equity interest, patent/licensing arrangements, etc) that might pose a conflict of interest in connection with the submitted article.
One of more of the authors received (VS, FC, LM) grants from Regione Emilia Romagna (Istituto Ortopedico Rizzoli, Bologna) for the study of bone regeneration and from the University of Ferrara.
This work was performed at Istituto di Clinica Ortopedica Università di Ferrara.
References
- 1.Aaron RK, Boyan BD, Ciombor DM, Schwartz Z, Simon BJ. Stimulation of growth factor synthesis by electric and electromagnetic fields. Clin Orthop Relat Res. 2004;419:30–37. doi: 10.1097/00003086-200402000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Aaron RK, Ciombor DM. Acceleration of experimental endochondral ossification by biophysical stimulation of the progenitor cell pool. J Orthop Res. 1996;14:582–589. doi: 10.1002/jor.1100140412. [DOI] [PubMed] [Google Scholar]
- 3.Aaron RK, Ciombor DM, Jolly G. Stimulation of experimental endochondral ossification by low-energy pulsing electromagnetic fields. J Bone Miner Res. 1989;4:227–233. doi: 10.1002/jbmr.5650040215. [DOI] [PubMed] [Google Scholar]
- 4.Aaron RK, Ciombor DM, Keeping H, Wang S, Capuano A, Polk C. Power frequency fields promote cell differentiation coincident with an increase in transforming growth factor-beta(1) expression. Bioelectromagnetics. 1999;20:453–458. doi: 10.1002/(SICI)1521-186X(199910)20:7<453::AID-BEM7>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 5.Aaron RK, Lennox D, Bunce GE, Ebert T. The conservative treatment of osteonecrosis of the femoral head: a comparison of core decompression and pulsing electromagnetic fields. Clin Orthop Relat Res. 1989;249:209–218. [PubMed] [Google Scholar]
- 6.Antoniv TT, Tanaka S, Sudan B, Val S, Liu K, Wang L, Wells DJ, Bou-Gharios G, Ramirez F. Identification of a repressor in the first intron of the human alpha2(I) collagen gene (COL1A2) J Biol Chem. 2005;280:35417–35423. doi: 10.1074/jbc.M502681200. [DOI] [PubMed] [Google Scholar]
- 7.Bassett CA, Mitchell SN, Gaston SR. Treatment of ununited tibial diaphyseal fractures with pulsing electromagnetic fields. J Bone Joint Surg Am. 1981;63:511–523. [PubMed] [Google Scholar]
- 8.Boyan BD, Schwartz Z. 1,25-Dihydroxy vitamin D3 is an autocrine regulator of extracellular matrix turnover and growth factor release via ERp60-activated matrix vesicle matrix metalloproteinases. Cells Tissues Organs. 2009;189:70–74. doi: 10.1159/000152916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brighton CT, Wang W, Seldes R, Zhang G, Pollack SR. Signal transduction in electrically stimulated bone cells. J Bone Joint Surg Am. 2001;83:1514–1523. doi: 10.2106/00004623-200110000-00009. [DOI] [PubMed] [Google Scholar]
- 10.Cadossi R, Bersani F, Cossarizza A, Zucchini P, Emilia G, Torelli G, Franceschi C. Lymphocytes and low-frequency electromagnetic fields. FASEB J. 1992;6:2667–2674. doi: 10.1096/fasebj.6.9.1612290. [DOI] [PubMed] [Google Scholar]
- 11.Capanna R, Donati D, Masetti C, Manfrini M, Panozzo A, Cadossi R, Campanacci M. Effect of electromagnetic fields on patients undergoing massive bone graft following bone tumor resection: a double blind study. Clin Orthop Relat Res. 1994;306:213–221. [PubMed] [Google Scholar]
- 12.Carinci F, Pezzetti F, Volinia S, Francioso F, Arcelli D, Farina E, Piattelli A. Zirconium oxide: analysis of MG63 osteoblast-like cell response by means of a microarray technology. Biomaterials. 2004;25:215–228. doi: 10.1016/S0142-9612(03)00486-1. [DOI] [PubMed] [Google Scholar]
- 13.Carinci F, Pezzetti F, Volinia S, Francioso F, Arcelli D, Marchesini J, Caramelli E, Piattelli A. Analysis of MG63 osteoblastic-cell response to a new nanoporous implant surface by means of a microarray technology. Clin Oral Implants Res. 2004;15:180–186. doi: 10.1111/j.1600-0501.2004.00997.x. [DOI] [PubMed] [Google Scholar]
- 14.Carinci F, Pezzetti F, Volinia S, Laino G, Arcelli D, Caramelli E, Degidi M, Piattelli A. P-15 cell-binding domain derived from collagen: analysis of MG63 osteoblastic-cell response by means of a microarray technology. J Periodontol. 2004;75:66–83. doi: 10.1902/jop.2004.75.1.66. [DOI] [PubMed] [Google Scholar]
- 15.Carinci F, Piattelli A, Stabellini G, Palmieri A, Scapoli L, Laino G, Caputi S, Pezzetti F. Calcium sulfate: analysis of MG63 osteoblast-like cell response by means of a microarray technology. J Biomed Mater Res B Appl Biomater. 2004;71:260–267. doi: 10.1002/jbm.b.30133. [DOI] [PubMed] [Google Scholar]
- 16.Carinci F, Volinia S, Pezzetti F, Francioso F, Tosi L, Piattelli A. Titanium-cell interaction: analysis of gene expression profiling. J Biomed Mater Res B Appl Biomater. 2003;66:341–346. doi: 10.1002/jbm.b.10021. [DOI] [PubMed] [Google Scholar]
- 17.Caunt CJ, Rivers CA, Conway-Campbell BL, Norman MR, McArdle CA. Epidermal growth factor receptor and protein kinase C signaling to ERK2: spatiotemporal regulation of ERK2 by dual specificity phosphatases. J Biol Chem. 2008;283:6241–6252. doi: 10.1074/jbc.M706624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciombor DM, Aaron RK, Wang S, Simon B. Modification of osteoarthritis by pulsed electromagnetic field: a morphological study. Osteoarthritis Cartilage. 2003;11:455–462. doi: 10.1016/S1063-4584(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 19.Clark AN, Youkey R, Liu X, Jia L, Blatt R, Day YJ, Sullivan GW, Linden J, Tucker AL. A1 adenosine receptor activation promotes angiogenesis and release of VEGF from monocytes. Circ Res. 2007;101:1130–1138. doi: 10.1161/CIRCRESAHA.107.150110. [DOI] [PubMed] [Google Scholar]
- 20.Mattei M, Caruso A, Traina GC, Pezzetti F, Baroni T, Sollazzo V. Correlation between pulsed electromagnetic fields exposure time and cell proliferation increase in human osteosarcoma cell lines and human normal osteoblast cells in vitro. Bioelectromagnetics. 1999;20:177–182. doi: 10.1002/(SICI)1521-186X(1999)20:3<177::AID-BEM4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 21.Mattei M, Varani K, Masieri FF, Pellati A, Ongaro A, Fini M, Cadossi R, Vincenzi F, Borea PA, Caruso A. Adenosine analogs and electromagnetic fields inhibit prostaglandin E2 release in bovine synovial fibroblasts. Osteoarthritis Cartilage. 2009;17:252–262. doi: 10.1016/j.joca.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Fassina A, Vasai L, Benazzo F, Benedetti L, Calligaro A, Angelis MG, Farina A, Maliardi V, Magenes G. Effects of electromagnetic stimulation on calcified matrix production by SAOS-2 cells over a polyurethane porous scaffold. Tissue Eng. 2006;12:1985–1999. doi: 10.1089/ten.2006.12.1985. [DOI] [PubMed] [Google Scholar]
- 23.Fitzsimmons RG, Ryaby JT, Magee FP, Baylink DJ. IGF-II receptor number is increased in TE-85 osteosarcoma cells by combined magnetic fields. J Bone Miner Res. 1995;10:812–819. doi: 10.1002/jbmr.5650100519. [DOI] [PubMed] [Google Scholar]
- 24.Goodman EM, Greenebaum B, Marron MT. Effects of electromagnetic fields on molecules and cells. Int Rev Cytol. 1995;158:279–338. doi: 10.1016/S0074-7696(08)62489-4. [DOI] [PubMed] [Google Scholar]
- 25.Hassan MQ, Tare R, Lee SH, Mandeville M, Weiner B, Montecino M, Wijnen AJ, Stein JL, Stein GS, Lian JB. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol Cell Biol. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatori K, Sasano Y, Takahashi I, Kamakura S, Kagayama M, Sasaki K. Osteoblasts and osteocytes express MMP2 and -8 and TIMP1, -2, and -3 along with extracellular matrix molecules during appositional bone formation. Anat Rec A Discov Mol Cell Evol Biol. 2004;277:262–271. doi: 10.1002/ar.a.20007. [DOI] [PubMed] [Google Scholar]
- 27.Itoh S, Udagawa N, Takahashi N, Yoshitake F, Narita H, Ebisu S, Ishihara K. A critical role for interleukin-6 family-mediated Stat3 activation in osteoblast differentiation and bone formation. Bone. 2006;39:505–512. doi: 10.1016/j.bone.2006.02.074. [DOI] [PubMed] [Google Scholar]
- 28.Jones GC, Riley GP. ADAMTS proteinases: a multi-domain, multi-functional family with roles in extracellular matrix turnover and arthritis. Arthritis Res Ther. 2005;7:160–169. doi: 10.1186/ar1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawamura N, Kugimiya F, Oshima Y, Ohba S, Ikeda T, Saito T, Shinoda Y, Kawasaki Y, Ogata N, Hoshi K, Akiyama T, Chen WS, Hay N, Tobe K, Kadowaki T, Azuma Y, Tanaka S, Nakamura K, Chung UI, Kawaguchi H. Akt1 in osteoblasts and osteoclasts controls bone remodelling. PloS ONE. 2007;2:e1058. doi: 10.1371/journal.pone.0001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmann CH, Schwartz Z, Liu Y, Guerkov H, Dean DD, Simon B, Boyan BD. Pulsed electromagnetic field stimulation of MG63 osteoblast-like cells affects differentiation and local factor production. J Orthop Res. 2000;18:637–646. doi: 10.1002/jor.1100180417. [DOI] [PubMed] [Google Scholar]
- 31.Luben RA, Cain CD, Chen MC, Rosen DM, Adey WR. Effects of electromagnetic stimuli on bone and bone cells in vitro: inhibition of responses to parathyroid hormone by low-energy, low-frequency fields. Proc Natl Acad Sci USA. 1982;79:4180–4184. doi: 10.1073/pnas.79.13.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mammi GI, Rocchi R, Cadossi R, Massari L, Traina GC. The electrical stimulation of tibial osteotomies: double-blind study. Clin Orthop Relat Res. 1993;288:246–253. [PubMed] [Google Scholar]
- 33.Massari L, Benazzo F, Mattei M, Setti S, Fini M, CRES Study Group Effects of electrical physical stimuli on articular cartilage. J Bone Joint Surg Am. 2007;899((suppl 3)):152–161. doi: 10.2106/JBJS.G.00581. [DOI] [PubMed] [Google Scholar]
- 34.Massari L, Fini M, Cadossi R, Setti S, Traina GC. Biophysical stimulation with pulsed electromagnetic fields in osteonecrosis of the femoral head. J Bone Joint Surg Am. 2006;88(suppl 3):56–60. doi: 10.2106/JBJS.F.00536. [DOI] [PubMed] [Google Scholar]
- 35.Matziari M, Dive V, Yiotakis A. Matrix metalloproteinase 11 (MMP-11; stromelysin-3) and synthetic inhibitors. Med Res Rev. 2007;27:528–552. doi: 10.1002/med.20066. [DOI] [PubMed] [Google Scholar]
- 36.Mooney V. A randomized double-blind prospective study of the efficacy of pulsed electromagnetic fields for interbody lumbar fusions. Spine (Phila PA 1976) 1990;15:708–712. doi: 10.1097/00007632-199007000-00016. [DOI] [PubMed] [Google Scholar]
- 37.Ohlendorff SD, Tofteng CL, Jensen JE, Petersen S, Civitelli R, Fenger M, Abrahamsen B, Hermann AP, Eiken P, Jørgensen NR. Single nucleotide polymorphisms in the P2X7 gene are associated to fracture risk and to effect of estrogen treatment. Pharmacogenet Genomics. 2007;17:555–567. doi: 10.1097/FPC.0b013e3280951625. [DOI] [PubMed] [Google Scholar]
- 38.Pezzetti F, Mattei M, Caruso A, Cadossi R, Zucchini P, Carinci F, Traina GC, Sollazzo V. Effects of pulsed electromagnetic fields on human chondrocytes: an in vitro study. Calcif Tissue Int. 1999;65:396–401. doi: 10.1007/s002239900720. [DOI] [PubMed] [Google Scholar]
- 39.Potts JR, Campbell ID. Structure and function of fibronectin modules. Matrix Biol. 1996;15:313–320. doi: 10.1016/S0945-053X(96)90133-X. [DOI] [PubMed] [Google Scholar]
- 40.Rhymer JA, Ottiger M, Wicki R, Greenwood TM, Strehler EE. Structure of the human CALM1 calmodulin gene and identification of two CALM1-related pseudogenes CALM1P1 and CALM1P2. Eur J Biochem. 1994;225:71–82. doi: 10.1111/j.1432-1033.1994.00071.x. [DOI] [PubMed] [Google Scholar]
- 41.Scott MJ, Godshall CJ, Cheadle WG. Jaks, STATs, cytokines, and sepsis. Clin Diagn Lab Immunol. 2002;9:1153–1159. doi: 10.1128/CDLI.9.6.1153-1159.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharrard WJ. A double-blind trial of pulsed electromagnetic fields for delayed union of tibial fractures. J Bone Joint Surg Br. 1990;72:347–355. doi: 10.1302/0301-620X.72B3.2187877. [DOI] [PubMed] [Google Scholar]
- 43.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-array tools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 44.Sollazzo V, Massari L, Caruso C, Mattei M, Pezzetti P. Effects of low-frequency pulsed electromagnetic fields on human osteoblast-like cells in vitro. Electro- and Magnetobiology. 1996;15:75–83. [Google Scholar]
- 45.Sollazzo V, Traina GC, DeMattei M, Pellati A, Pezzetti F, Caruso A. Responses of human MG-63 osteosarcoma cell line and human osteoblast-like cells to pulsed electromagnetic fields. Bioelectromagnetics. 1997;18:541–547. doi: 10.1002/(SICI)1521-186X(1997)18:8<541::AID-BEM2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 46.Varani K, Gessi S, Merighi S, Iannotta V, Cattabriga E, Spisani S, Cadossi R, Borea PA. Effect of low frequency electromagnetic fields on A2A adenosine receptors in human neutrophils. Br J Pharmacol. 2002;136:57–66. doi: 10.1038/sj.bjp.0704695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan Q, Sage EH. SPARC, a matricellular glycoprotein with important biological functions. J Histochem Cytochem. 1999;47:1495–1506. doi: 10.1177/002215549904701201. [DOI] [PubMed] [Google Scholar]
- 48.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]