Introduction
PIPs are phosphorylated derivatives of phosphatidylinositol (PtdIns) that serve primary intracellular roles: (i) in the specification of dedicated membrane microdomains that organize signal transduction processes, (ii) as co-factors for the regulated activities of proteins, and (iii) as precursors for second messengers such as diacylglycerol and soluble inositol phosphates (Fruman et al., 1998; Martin, 1998; Waselle et al., 2005; Di Paolo and De Camilli, 2006). In part, this diversification of function reflects the chemical diversity afforded by the poly-hydroxylated inositol headgroup. Mammalian cells express seven distinct PIP species -- phosphatidylinositol 3-phosphate (PtdIns-3-P), PtdIns-4-P, PtdIns-5-P, phosphatidylinositol 3,5-bisphosphate (PtdIns-3,5-P2), PtdIns-4,5-P2, PtdIns-3,4-P2, and phosphatidylinositol 3,4,5-trisphosphate (PtdIns-3,4,5-P3) --each of which interfaces with specific downstream effectors. Tight regulation of each of these PIP levels is essential to cellular processes that include vesicular trafficking, apoptosis, metabolism, actin reorganization, cell proliferation and cell growth (Fruman et al., 1998; Martin, 1998). Yeast do not synthesize PtdIns-3,4-P2 or PtdIns-3,4,5-P3 and, indeed, 3-OH PIPs are nonessential for yeast viability although these play important homeostatic functions (Fruman et al., 1998; Martin, 1998).
PtdIns generally constitutes less than 15% of the total cellular phospholipids in eukaryotic cells and PIPs are usually less abundant in terms of mass by an order of magnitude. PtdIns-4-P and PtdIns-4,5-P2 are the major PIP species in mammalian cells --representing ∼90% of total PIP mass. By comparison, PtdIns-3-P and PtdIns-5-P represent only ∼0.25% of total PIP mass in mammalian cells (Rameh et al., 1997; Fruman et al., 1998; Martin, 1998; Waselle et al., 2005; Di Paolo and De Camilli, 2006).
Individual PIP species exhibit specific signaling capabilities and exhibit specific subcellular localizations that help define organelle identity. With regard to the latter set of functions, PtdIns-3-P and derived lipid species are mainly distributed in endosomal organelles, PtdIns-4-P is presented predominantly in Golgi membranes, PtdIns-4,5-P2 and PtdIns-3,4,5-P3, are minor phospholipids and enriched in the inner leaflet of the plasma membrane, and PtdIns-3,4,5-P3 may also accumulate on endomembranes following growth factor receptor activation. Moreover, the lateral distribution of specific PIPs within the two-dimensional space of a given membrane is also heterogeneous (Shisheva, 2001; De Matteis and Godi, 2004; Waselle et al., 2005; Di Paolo and De Camilli, 2006). The presence of nuclear PIPs has also been documented, and these may fuel an autonomous nuclear soluble inositol phosphate cycle that regulates gene transcription, mRNA processing and mRNA export from the nucleus (De Matteis and Godi, 2004; Waselle et al., 2005; Di Paolo and De Camilli, 2006).
PIP metabolism requires a highly coordinated balance between the activities of lipid kinases that generate PIPs and the activities of lipid phosphatases and phospholipases that degrade them. PIPs are subject to robust phosphatase-mediated turnover through via dephosphorylation at the 3-OH, 4-OH and 5-OH positions of the inositol ring. PtdIns-4-P and PtdIns-4,5-P2 are the major PIP species in mammalian cells, and represent 90% of total cellular phosphorylated PIPs. (Fruman et al., 1998; Martin, 1998; Whisstock et al., 2002; Waselle et al., 2005; Di Paolo and De Camilli, 2006). As will be discussed below, homeostasis of the 4-OH PIPs is strongly influenced by the action of Sac1 phosphatases. However, phosphatases of the Sac1 family play important roles in regulating the degradation of the 3-OH PIPs as well. Compromised activity of such phosphatases dedicated to the 3-OH arm of the PIP pathway (e.g. the tumor suppressor PTEN) has devastating consequences for higher eukaryotes (Li et al., 1997; Simpson and Parsons, 2001). PIP phosphatases, while still poorly studied as a group, are increasingly subjects of intense research effort – in part because of their newly recognized roles as tumor suppressors. The integral membrane protein, Sac1, is a prototypical member of a major class of such lipid phosphatases. Yeast Sac1 (ySac1) has been broadly studied and Sac1 loss of function (LOF) in yeast causes a wide array of phenotypes such as cold sensitivity, inositol auxotrophy, and “bypass Sec14p”. However, the mammalian homologs of Sac1 are not well explored. At present, mammalian cell systems and mouse knockout models provide powerful systems for analysis of the mammalian Sac1 functions in vivo, and the mechanisms by which these functions are executed. Herein, we review the functional involvement of Sac1 phosphatases in regulating PIP metabolism, the functional properties of ySac1, and recent progress in deciphering Sac1 function in mammals.
Materials and Methods
The experimental procedures and reagents employed herein are described in Cleves et al. (1989), Whitters et al., 1993; Guo et al., 1999; Rivas et al., 1999 ; Kochendörfer et al. (1999), Nemoto et al. (2000), Li et al (2002), Rohde et al. (2003), and Liu et al. (2008).
Results and Discussion
The Yeast Sac1 PIP Phosphatase
The Sac1 phosphatases are the prime subjects of this review. These enzymes not only represent a major class of PIP phosphatases, but these have the signature feature of being integral membrane proteins (Cleves et al., 1989; Whitters et al., 1993). The first member of the Sac1 family of phosphatases was identified in yeast by two independent genetic screens searching for modifiers of actin cytoskeleton defects and of trans-Golgi network exocytic failure caused by inactivation of the major yeast phosphatidylinositol (PtdIns)/phosphatidylcholine (PtdCho) transfer protein, respectively (Novick et al., 1989). Sac1 was demonstrated to be an integral membrane protein that localized to the endoplasmic reticulum (ER) and Golgi membranes in yeast and in mammalian cells (Xie et al., 1998; Nemoto et al., 2000). It consists of a 300-amino acid catalytic domain, designated the SAC1-like domain, and the catalytic domain is disposed to the cytosol (Cleves et al., 1989; Whitters et al., 1993). The catalytic SAC1 domain is common to other phosphoinositide phosphatases such as PTEN (Maehama et al., 2001), synaptojanins (Cremona et al., 1999) and yeast synaptojanin-like proteins (Srinivasan et al., 1997; Stolz et al., 1998). A highly conserved CX5R(T/S) motif almost certainly represents the core catalytic motif of the Sac1 domain – given that mutations in this motif eliminate catalytic activity (Nemoto et al., 2000, Rohde et al., 2003) and this motif is a signature of metal-independent phosphatases (Hughes, 2001). Sac1 is anchored to membranes by two C-terminal transmembrane domains such that the C-terminus of the protein is also disposed to the cytosol (Konrad et al., 2002).
Based on genetic data, Sac1 was proposed to negatively regulate PIP signaling (Cleves et al., 1991; Whitters et al., 1993), and this was demonstrated to be directly true when Shuling Guo in John York’s laboratory demonstrated Sac1 domains are PIP phosphatase domains (Guo et al., 1999). Biochemically, ySac1 catalyzes dephosphorylation of PtdIns-3-P, PtdIns-4-P, and PtdIns-3,5-P2 to PtdIns in vitro and in vivo but, interestingly, is unable to utilize PtdIns-4,5-P2 as substrate in either context (Guo et al., 1999; Rivas et al., 1999; Hughes et al., 2000). Moreover, ySac1 represents a major pathway for PtdIns-4-P degradation in vivo. Genetic ablation of ySac1 activity results in a nearly 10-fold increase in the steady-state levels of PtdIns-4-P with little effect on PtdIns-4,5-P2 and far more modest increases in the steady-state levels of the 3-OH PIP species (Guo et al., 1999; Rivas et al., 1999; Hughes et al., 2000). Paradoxically, and a point of discussion revisited below, ySac1 degrades a PtdIns-4-P pool that is produced apparently exclusively by the plasma membrane-localized Stt4 PtdIns 4-OH kinase – one of three PtdIns 4-OH kinases in this organism (Nemoto et al., 2000; Foti et al., 2001).
The single SAC1 gene in yeast is not essential for cell viability. Rather, sac1 nullizygous alleles (sac1Δ) evoke a wide array of phenotypes, such as alterations in the actin cytoskeleton, cold sensitivity for growth, a curious inositol auxotrophy independent of the ability of Sac1-deficient yeast to produce their own inositol de novo, a ‘bypass Sec14’phenotype where Sac1-insufficient yeast are able to survive the normally lethal consequences of loss of function of the major PtdIns/PtdCho transfer protein of this organism (Sec14), compromised cell integrity at alkaline pH, and deranged neutral lipid metabolism (Cleves et al., 1989; Novick et al., 1989; Whitters et al., 1993; Boyum and Guidotti, 1997; Kearns et al., 1997; Rivas et al., 1999). Moreover, ySac1 deficiencies somehow interfere with ATP uptake into the ER lumen resulting in compromised ER protein quality control systems (Kochendorfer et al., 1999).
ySac1 is proposed to be retained in the yeast ER through a direct interaction of its COOH-terminal region with a very abundant ER-localized integral membrane protein dolicholphosphate-mannose synthase (Dpm1; Faulhammer et al., 2005). Its interaction with Dpm1 is regulated by cell growth conditions. The interaction is detected only during exponential cell division – it is apparently lost when cells are challenged with limited nutrient conditions. Under such suboptimal growth conditions, ySac1 accumulates in Golgi membranes (Faulhammer et al., 2005). This cell-growth controlled switch of ySac1 between the ER and the Golgi provides reciprocal control of PtdIns-4-P levels at these organelles (Faulhammer et al., 2005). As PtdIns-4-P helps promote anterograde transport of secretory proteins from the Golgi system to the plasma membrane (Walch-Solimena and Novick, 1999; Wang et al., 2003; Godi et al., 2004), and yeast exocytic capacity is maximized during exponential growth (Finger and Novick, 1998), such a redistribution of Sac1 may coordinate secretory capacity with larger aspects of yeast cell physiology.
Biochemical and Cell Biological Features of Mammalian Sac1 Proteins
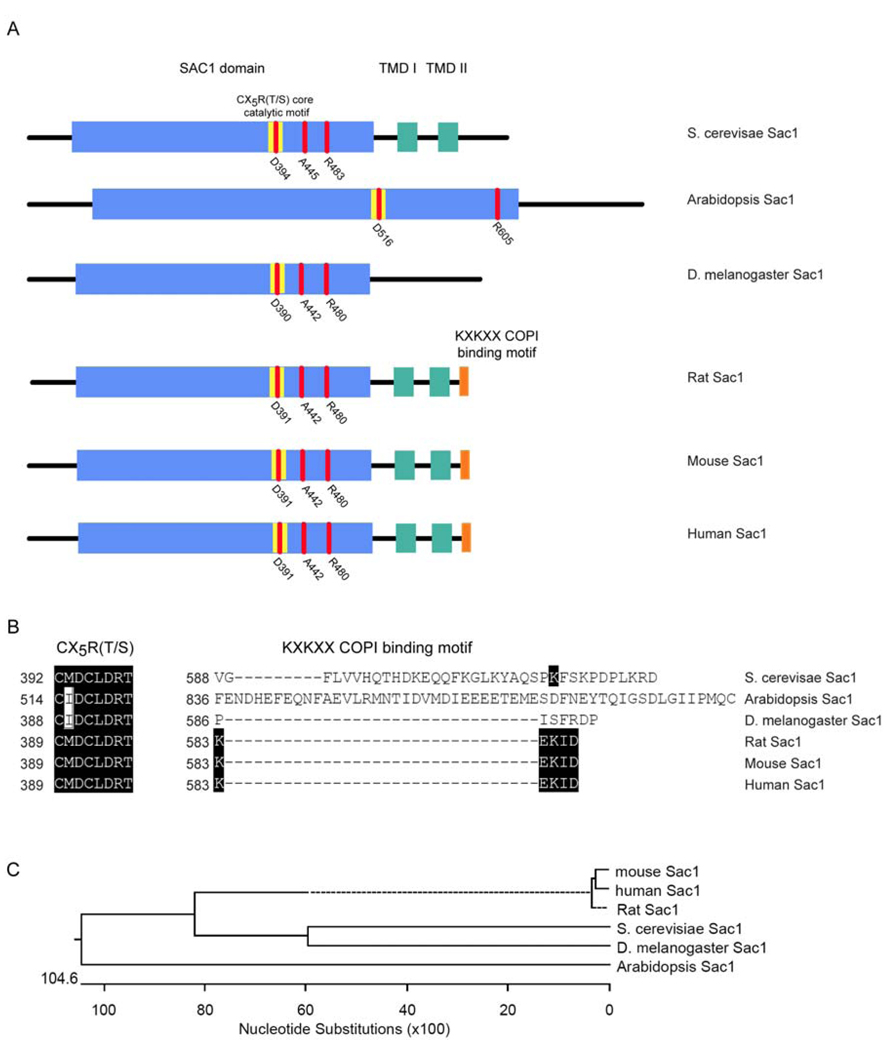
Homologs of the ySac1 PIP phosphatase are disseminated throughout the eukaryotic kingdom as evidenced by inspection of insect, plant, and mammalian genomes (Figure 2). Interestingly, mammalian genomes encode only a single protein that closely resembles ySac1 in its molecular architecture. As expected, the rat Sac1 (rSac1), human Sac1 (hSac1) and murine Sac1 (mSac1) all exhibit the expected PIP phosphatase activities --with PtdIns-3-P, PtdIns-4-P, and PtdIns-3,5-P2 as primary substrates – and these mammalian Sac1 proteins share the CX5R(T/S) catalytic motif with ySac1 (Nemoto et al., 2000). The mammalian proteins exhibit a molecular mass of ca. 65-kDa, similar to ySac1, are integral membrane proteins of the ER and Golgi, and exhibit the same membrane topology as ySac1 (Figure 2). Moreover, heterologous complementation analyses demonstrate that individual expression of each of these mammalian proteins rescues the pleiotropic phenotypes that are signatures of sac1Δ alleles.
Figure 2.
Domain and primary structure of Sac1 protein family. A Diagrams of the domain organization of Sac1 proteins. Domains and motifs of interest are highlighted by colored boxes. Blue boxes show the Sac1 catalytic domain, yellow boxes show the CX5R(T/S) core catalytic motif, green boxes show the two transmembrane domains (TMD I and TMD II respectively), and the orange boxes show the KXKXX COPI binding motif. Three key amino acids within the Sac1 domain known to affect catalytic activity (Rivas et al., 1999; Liu et al., 2008) are indicated in red. B Sequence alignment of the core catalytic domains and the KXKXX COPI binding motifs of the Sac1 protein family. The CX5R(T/S) core catalytic domain of Sac1 is highly conserved in yeast, higher plants, Drosophila, and mammals, whereas the KXKXX COPI binding motif is unique to mammalian Sac1 proteins. Identical amino acids are highlighted by black boxes. C. Phylogenetic tree of the Sac1 protein family.
The intracellular distribution of the Sac1 PIP phosphatase in the ER and Golgi of mammalian cells recapitulates aspects of what is observed in yeast, but with some unanticipated twists. The first interesting aspect is that hSac1 (and other mammalian versions – see below) contain a C-terminal KXKXX motif that serves as binding site for the coatomer (COP1) complex. This COPI interaction motif is a unique feature of mammalian Sac1s as it is not conserved in yeast, plant or Drosophila Sac1 proteins (Figure 2). This divergence reflects the differing mechanisms for ER retention of mammalian Sac1 enzymes. Compromise of the KXKXX motif abolishes retrieval of hSac1 from the Golgi back to the ER, and provokes accumulation of hSac1 in Golgi membranes (Rohde et al., 2003). Interestingly, the phosphatase activity of hSac1 is also apparently required for its interaction with the COPI complex. A “catalytic-dead” hSac1-C/S mutant (where Cys289 residue of the CX5R(T/S) core catalytic motif is replaced with Ser) is incapable of interacting with the COPI complex, and is incompetent for retrieval back to the ER – an intact KXKXX motif notwithstanding (Rohde et al., 2003). The mechanism by which the core catalytic motif regulates interaction with COPI is unclear, and this result suggests that the phosphatase activity of hSac1 may function as a switch to adjust its interaction with the COPI complex, thereby influencing hSac1 distribution in the ER and Golgi as a function of catalytic cycle.
Why the strategic adjustment in how mammalian Sac1 is retrieved from the Golgi system back to the ER relative to the Sac1 enzymes of other eukaryotes? Blagoveshchenskaya et al. (2008) report that, in quiescent mammalian cells, hSac1 is mainly localized in the Golgi complex – perhaps to downregulate Golgi PtdIns-4-P levels. Escape to the Golgi complex from the ER is achieved via oligomerization of hSac1 and subsequent recruitment of the coat protein II (COPII) complex (Blagoveshchenskaya et al., 2008). In the presence of growth factors, the p38 mitogen-activated kinase (MAPK) pathway induces dissociation of Sac1 oligomers -- thereby ‘activating’ the KXKXX complex to render hSac1 as competent cargo for COPI-mediated retrograde traffic from the Golgi to the ER. The proposed consequence of this ER retrieval is to increase the Golgi PhtdIns-4-P pool which helps promote protein export from the Golgi to the plasma membrane (Blagoveshchenskaya et al., 2008). This study suggests a link between growth factor signaling and lipid signaling at the Golgi complex. Thus, although yeast and mammalian cells use distinct mechanisms to control Sac1 PIP phosphatase distribution between ER and Golgi membranes, these widely divergent organisms appear to use similar strategies for coupling Sac1 localization to the regulation of anterograde transport activities from the Golgi system.
Functional Analyses of Mammalian Sac1
Present interpretations of the work of Rohde et al (2003) and Blagoveshchenskaya et al (2008) hold, as principle thesis, that ER retention of Sac1 archives the enzyme in the ER and restrict its effects on Golgi function. Yet, the observed Sac1 redistribution between ER and Golgi membranes (and the corresponding interpretation of those data), is not coupled to any direct functional readout. One is left with the principal question of what do these redistribution phenomena truly mean for the mammalian cell? In that regard, mammalian Sac1 proteins remain poorly studied from the intracellular and organismal perspectives. This lack of information is made more striking by the fact that mammalian Sac1 proteins, while ubiquitously expressed (also in embryonic stem cells, Figure 3), nonetheless show interesting tissue distributions – e.g. particularly high expression is detected in cerebellum, hippocampus, and heart (Figure 3, Nemoto et al., 2000). From the perspective of the vertebrate organism, does mammalian Sac1 play any interesting tissue-specific functions? From the standpoint of individual cells, does Sac1 play a significant role in integrating growth factor signals with Golgi exocytic activity? From that perspective, a difficulty with the simple Golgi PtdIns-4-P control model is presented by existing information regarding ySac1. This enzyme only degrades the PtdIns-4-P pool generated by the plasma membrane-localized ySac1 – not the PtdIns-4-P pool generated by the Pik1 PtdIns 4-OH kinase which is generally though to be the enzyme responsible for the PtdIns-4-P pool relevant to membrane trafficking from the Golgi complex (see above). If such a PtdIns-4-P pool specificity also holds true in mammalian cells (and it is not known whether this is in fact the case), then a revision of how we think Stt4- and Pik1-kinases act on the Golgi complex is in order. Finally, if the “ER-archive” model is generally true, this level of control likely represents a fine-tuning mechanism. This interpretation is based on the demonstration that a chimeric Sac1 protein, where the catalytic domain is tethered to an ER resident protein (Sec61), and is unable to shuttle between ER and Golgi membranes like the native Sac1 enzyme, is nonetheless able to functionally substitute for ySac1 in vivo (Rivas et al., 1999). The cumulative data raise the question: do Sac1 enzymes play more direct roles in ER or nuclear envelope function? As discussed below, emerging evidence suggests this is likely so – at least in the case of the mammalian Sac1 enzymes.
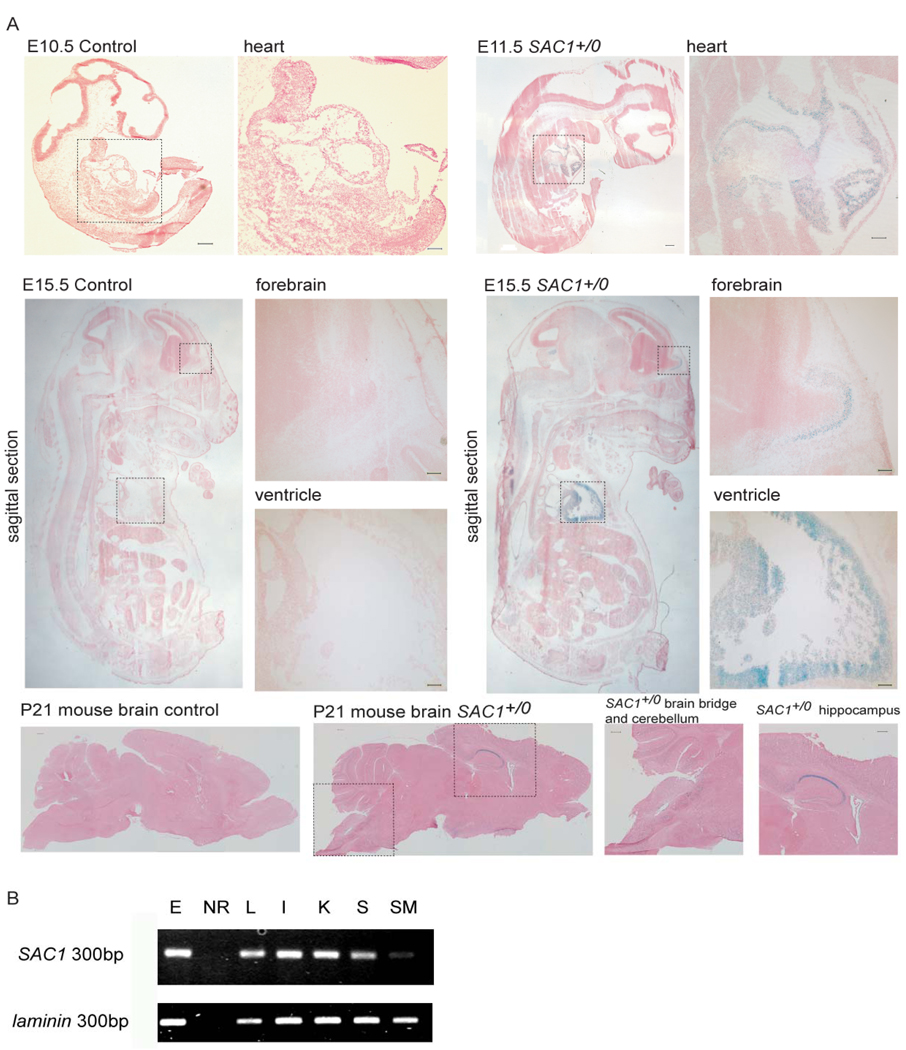
Figure 3.
Expression of mSAC1 in the developing mouse. (A) mSAC1 transcriptional expression patterns are indicated by X-gal staining in E10.5 and E11.5 Sac1+/0 embryos. Moderate mSAC1 expression was detected in the heart. In E15.5 Sac1+/0 embryos, ubiquitous mSAC1 expression was detected with particularly robust expression in the ventricle and the forebrain. p21 brains exhibited ubiquitous mSAC1 expression that was particularly robust in the hippocampus, the cerebellum, and the brain stem. Scale bar, 100 µm. (B) Total mRNA from various mouse tissues and ES cells was purified, and RT-PCR amplification was used to estimate mSAC1 expression. E, ES cells; NR, no RNA control; L, liver; I, intestine; K, kidney; S, stomach; SM, skeletal muscle.
Meaningful study of mammalian Sac1 function requires transgenic mouse approaches, and such approaches have recently provided new insights. Gene trap experiments demonstrate that genetic ablation of the single murine SAC1 gene results in a recessive (and fully penetrant) pre-implantation lethality -- most sac10/0 progeny failing to progress past E3.5 – and sac10/0 cells cannot be generated by selection from heterozygous ES cells (Liu et al., 2008). These collective data indicate the Sac1 PIP phosphatase plays an essential housekeeping role in mammalian cells. No obvious phenotypes derive from SAC1+/0 haplo-insufficiency, however, nor are sac10/0 sperm or eggs deficient in any obvious way. Thus, 50% reductions in intracellular Sac1 load do not threaten obvious functional thresholds (Liu et al., 2008). As described below, the mechanisms for how Sac1 insufficiencies cause cell death are now being clarified, and functional rescue experiments demonstrate essential Sac1 functions require both PIP phosphatase activity and proper COP1-dependent retrieval of the enzyme back to the mammalian ER.
Mammalian Sac1 and Golgi Membrane Organization
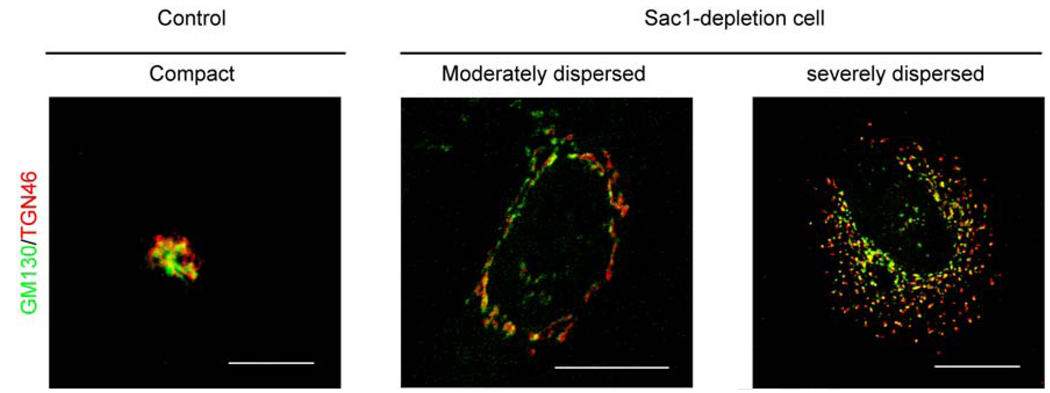
The intracellular localization of mammalian Sac1 enzymes to the ER and Golgi systems suggested these as likely points of failure under conditions of Sac1 insufficiency. Indeed, one striking intracellular phenotype associated with hSac1 depletion (by siRNA methods) in HeLa or HEK293 cells is a dramatic structural derangement of the Golgi membranes that is registered across the entire Golgi stack (Liu et al., 2008). A consistent dispersal of cis-, medial-, and trans-Golgi compartments from the typical compact structure to dispersed arrangements is observed. These phenotypes are further classified as ‘moderately dispersed’ or ‘severely dispersed’ as a function of magnitude of disorganization (Figure 4), and these phenotypic gradations presumably reflect the extent to which Sac1 levels have been diminished in those cells. Because Sac1-deficient cells exhibit significant reductions in viability, it was formally possible that disorganization of the Golgi system was an indirect consequence of activating apoptotic pathways. This is not the case, however, as evidenced by the fact that treatment with the pan-caspase inhibitor Z-VAD-fmk does not eliminate either the Golgi membrane derangements, or the reduced cell viability, evoked by hSac1-depletion (Liu et al., 2008).
Figure 4.
hSac1 deficiency results in defective Golgi morphology in human cells. The TGN marker TGN46 is stained red, and the cis-Golgi marker GM130 is in green. The left panel shows a representative Golgi system of HeLa cells challenged with an irrelevant siRNA. The middle and right panels show the two types of Golgi dispersion in human Sac1-deficient cells. Based on the severity of dispersion, these are classified as moderately and severely dispersed, respectively. Moderately dispersed Golgi are defined by reticulation and relaxation of Golgi membranes, and severely dispersed Golgi are defined by extensive fragmentation of the Golgi system. Bar, 20 µm.
With regard to intracellular membrane disorganization, the effect is apparently limited to the Golgi system as the gross morphologies of lysosomes, endosomes, mitochondria, and ER are not affected in hSac1-depleted cells. As discussed in more detail below, these Golgi structural phenotypes reflect ‘on-target’ effects that report the consequences of Sac1-deficiency as evidenced by the demonstration that the compromised viability and Golgi disorganization phenotypes are both rescued by expression of silencing-resistant mSac1 transgenes in these human cell lines.
Structural compromise of Golgi membranes is often associated with defects in Golgi secretory function. For example, depletion of the peripheral Golgi protein Nir2 compromises Golgi structure, and Nir2 depletion also inhibits protein export from the TGN (Litvak et al., 2005). Considering that: (i) Sac1 is a major PtdIns-4-P phosphatase in cells that plays an important role in PtdIns-4-P turnover (Nemoto et al., 2000; Foti et al., 2001; Schorr et al., 2001), and (ii) PtdIns-4-P is an essential regulator for anterograde transport of secretory proteins from the Golgi system, it seemed likely that mammalian Sac1 deficiency compromises protein trafficking through this organelle. However, several lines of evidence indicate that both rate and efficiency of transport of a variety of cargo to, through, and from the disorganized Golgi system is not impaired in Sac1-depletion cells (Liu et al., 2008).
What are the effects of Sac1 depletion on PIP homeostasis in mammalian cells? Analyses of [3H]-inositol labeled PIP species report modest (30%) increases in PtdIns-4-P in hSac1-depleted cells – an effect that is almost certainly an underestimate because it is an averaging measurement and only partial Sac1 deficiencies are being scored. In imaging experiments where individual cells with severely dispersed Golgi membranes can be analyzed individually, localization of endogenous TGN-associated PtdIns-4-P binding proteins (such as FAPP1, FAPP2 and Orp9), or other peripheral Golgi proteins (βCOP and PITPβ), is not disturbed under conditions of hSac1 deficiency that lead to severe Golgi dispersal phenotypes (Liu et al., 2008). Taken together, these data indicate that, unlike the case in yeast (Li et al., 2002), mammalian Sac1 deficiencies do not evoke redistribution of Golgi-associated PIP binding proteins to inappropriate intracellular locations (presumably as a result of ectopic accumulation of PtdIns-4-P in those locations). With the caveat that the effects of genuine Sac1 nullizygosity are not monitored in these experiments, the data suggest mammalian Sac1 may not represent as major a pathway for PtdIns-4-P degradation in mammalian cells ySac1 is in yeast. How Sac1 deficiency influences Golgi membrane organization remains to be elucidated, and represents an interesting question for future work.
Mammalian Sac1 and Mitotic Spindle Organization
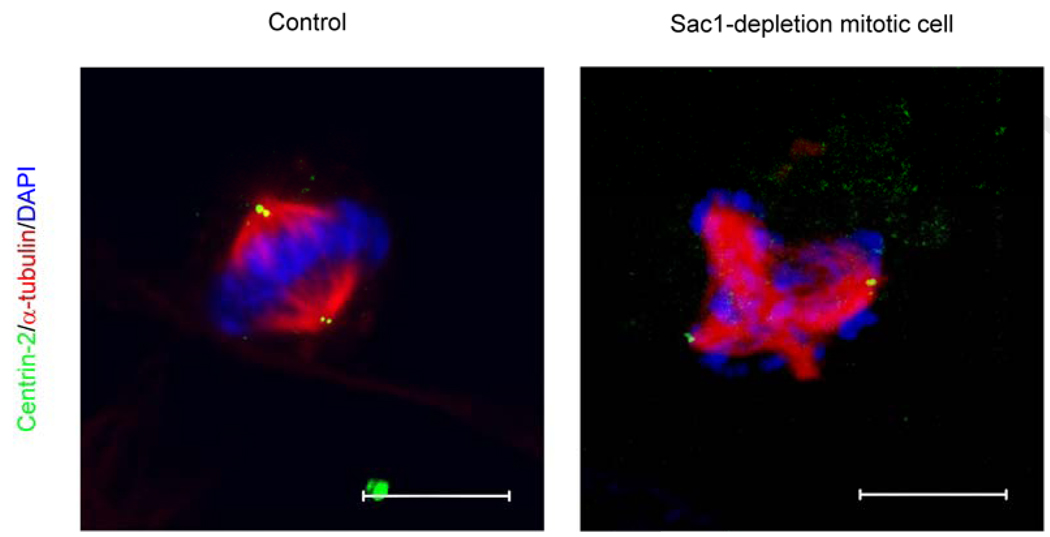
That mammalian Sac1 executes essential housekeeping functions is demonstrated not only by the pre-implantation lethality associated with sac1 nullizygosity in mice, but also by the compromised viability of mammalian cells depleted for Sac1 protein (Liu et al., 2008). Fluorescence-activated cell sorting analyses demonstrate Sac1 depletion results in difficulties in progression through the G2/M phase of the cell cycle. This defect is accompanied by a significant (10-fold) increase, as assessed by α-tubulin staining, in the incidence of abnormal multipolar spindles in mitotic hSac1-depleted cells (Liu et al., 2008). Interestingly, the ectopic spindles are mechanically active and generate sufficient force to drive aberrant segregation of chromosomal material, as indicated by 4,6- diamidino-2-phenylindole (DAPI) staining of DNA at each spindle (Figure 5, Liu et al., 2008). Again, this mitotic phenotype is not rescued by challenge of Sac1-deficient mammalian cells with the pan-caspase inhibitor Z-VAD-fmk.
Figure 5.
Disorganization of mitotic spindles and aberrant distribution of chromosomal material in Sac1 deficient mammalian cells. Immunofluorescence images of control and a Sac1-depeleted mitotic cells are shown. The spindle marker α-tubulin, the centrosome marker centrin-2, and DNA (stained with DAPI) are rendered red, green and blue, respectively). The control mitotic cell in the left panel presents the typical two spindle poles, each containing a centrosome, with the chromosomes evenly distributed by the force of those two spindles. The right panel shows a SAC1 siRNA treated cell presenting multiple ectopic spindle asters so that four spindle poles are evident. Each spindle pole generates sufficient force to pull chromosomes in an inappropriate manner. Only two centrosomes are observed, however. Bar, 10 µm.
One plausible mechanism for generating multipolar spindles is the deregulation of centrosome duplication which further induces MT nucleation at ectopic centrosomes (Khodjakov and Rieder, 1999; Hinchcliffe and Sluder, 2001). However, Sac1-depleted mitotic cells present only two spindle poles which contain the centrosome marker, Centrin-2. This fidelity of centrosome duplication is observed even in cases where as many as five ectopic spindle poles are observed. Immunofluorescence analyses further demonstrate that γ-tubulin foci, which identify the γ-tubulin ring complex (γTuRC) that nucleates spindle MT assembly (Moudjou et al., 1996), mark each of the ectopic spindle asters in cells with multipolar spindles. These data indicate centrosome duplication occurs normally in hSac1-deficient cells with multipolar spindles, and that the γTuRC is responsible for assembly of extra spindle poles in these cells. These findings are consistent with previous in vitro studies that demonstrate γTuRC is required, but not sufficient, for the formation of a functional centrosome (Martin et al. 1998; Moritz et al. 1998). Time-lapse video microscopy indicates the multipolar spindles of Sac1-deficient HeLa cells fail to resolve into bipolar spindles, and that such cells effectively fail to exit mitosis (Liu et al., 2008). Such catastrophic failure of organizing normal mechanically active spindles can cause chromosomal nondisjunction events (Wong and Stearns, 2003). This suggests an attractive mechanism for why Sac1-depleted mammalian cells fail to efficiently progress through G2/M and ultimately expire.
Functional Properties of Mammalian Sac1
The Golgi morphology and mitotic spindle phenotypes provide robust readouts for functional complementation experiments where the effects of expressing silencing-resistant versions of mammalian Sac1 on these phenotypes can be monitored. These experiments employ Sac1-depleted HeLa cells expressing murine Sac1 variants whose expression is recalcitrant to the siRNAs used to silence hSAC1 expression (Liu et al., 2008). Both the Golgi and multipolar spindle phenotypes are rescued by expression of wild-type mSac1, but not by the “PIP phosphatase-dead” mSac1D391N. In a genetic version of a biochemical dose-response experiment, expression of mSac1 variants with partial PIP phosphatase activity (e.g mSac1R480H) results in partial rescue of both phenotypes. Finally, expression of the mSac1AEAID mutant that is incompetent for COPI binding and retrieval from the Golgi complex to the ER fails to rescue the Golgi morphological phenotype or the defects in mitotic spindle organization (Liu et al., 2008). These complementation experiments demonstrate that proper Golgi membrane and mitotic spindle organization require Sac1 PIP phosphatase activities and, in both cases, localization of Sac1 PIP phosphatase activity to the ER is a functional requirement.
Sac1, The Golgi Complex, and Mitotic Spindle Organization
What is the relationship, if any, between the Golgi and spindle pole phenotypes of Sac1-deficient mammalian cells? The physical proximity of the Golgi apparatus with the centrosome is consistent with a functional linkage between these two organelles (Sütterlin et al., 2005; Kodani and Sütterlin, 2008). Indeed, several recent reports show that Golgi-associated proteins are involved in regulation of cell cycle progression related events. Specifically, a Golgi associated protein GRASP-65 plays an as yet undefined role in spindle dynamics (Sütterlin et al., 2005), and another (GM130) is involved in regulation of centrosome morphology, position and function during interphase (Kodani and Sütterlin, 2008). Are the mitotic spindle defects of Sac1-deficient mammalian cells a secondary effect of Golgi membrane disorganization? That remains a viable possibility. However, the Sac1-depletion effects on organization of the mitotic apparatus are not the result of secondary depletion of GRASP-65 or GM130 as both proteins are present in apparently normal amounts in Sac1-insufficent cells. Moreover, ectopic spindles in GRASP-65-deficient cells are not mechanically active, in contrast to those of Sac1-depleted mammalian cells (Liu et al., 2008). Thus, the mechanisms appear different.
Given that Sac1 is an integral membrane protein of the ER and the Golgi, and that ER localization is essential for Sac1 function in mammalian cells, possibilities for ER/nuclear functions for the Sac1 PIP phosphatase seem most likely. In this regard, PtdIns and PIP levels influence DNA synthesis, cell proliferation, and cell cycle progression (York and Majerus, 1994; Rubbini et al., 1997; Albi and Viola Magni, 2004). Although a direct connection between PIP species and regulation of γTRC organization during cell cycle progression remains to be demonstrated, these data suggest this to be an interesting avenue for future research. Novel functions for Sac1-mediated regulation of nuclear PIP metabolism must also be considered. The nuclear envelope is continuous with the ER, and it is possible that Sac1 phosphatases may localize in the ER to regulate nuclear PIP signal during cell cycle progression. It is now an interesting question as to what intracellular locations are compatible with Sac1 function and which ones are not.
Summary
The Sac1 PIP phosphatase is an enigmatic enzyme in that it occupies an intracellular location (ER) that is not normally associated with PIP signaling. Yet, genetic experiments in mice and silencing experiments in cultured cells report an essential housekeeping function for this protein. Detailed cellular analyses report maintenance of proper organization of the Golgi system, and of the mitotic spindle apparatus, are compromised when Sac1 functional thresholds are breached. While the Golgi derangements do not obviously affect protein transport through the organelle, the mitotic defects result in defects in progression though the G2/M stage of the cell cycle. Finally, both the catalytic PIP phosphatase activity, and its ability to be recycled back to the ER, represent essential functional features of the Sac1 enzyme.
We expect that current insights for Sac1 will set the blueprint for future analyses of its functions. Many questions remain to be answered in this field: does Sac1 have important roles in ER and plasma membranes connections since ySac1 only degrades the plasma membrane localized PtdIns 4-OH kinase Stt4 generated PtdIns-4-P? Does the ER-localized Sac1 play critical roles in regulating nuclear PIP signaling since ER is continuous with the nuclear envelope, and does it specifically happen in certain stage during cell cycle progression? Does anchoring Sac1-catalytic domain to other cellular membranes affect its function and what effects will be generated by mislocalizing the Sac1 catalytic domain to exotic membrane locations? Does the Golgi dispersion phenotype herald a crosstalk of the organelle with the mitotic apparatus (i.e. does Golgi disorganization provide ectopic nucleation sites for the γ-tubulin ring complex)? Clearly, there is much to be learned regarding the biological functions of Sac1-like lipid phosphatases, and we anticipate the discoveries yet to come will rival those derived from studies of the kinases – both in impact and in scope.
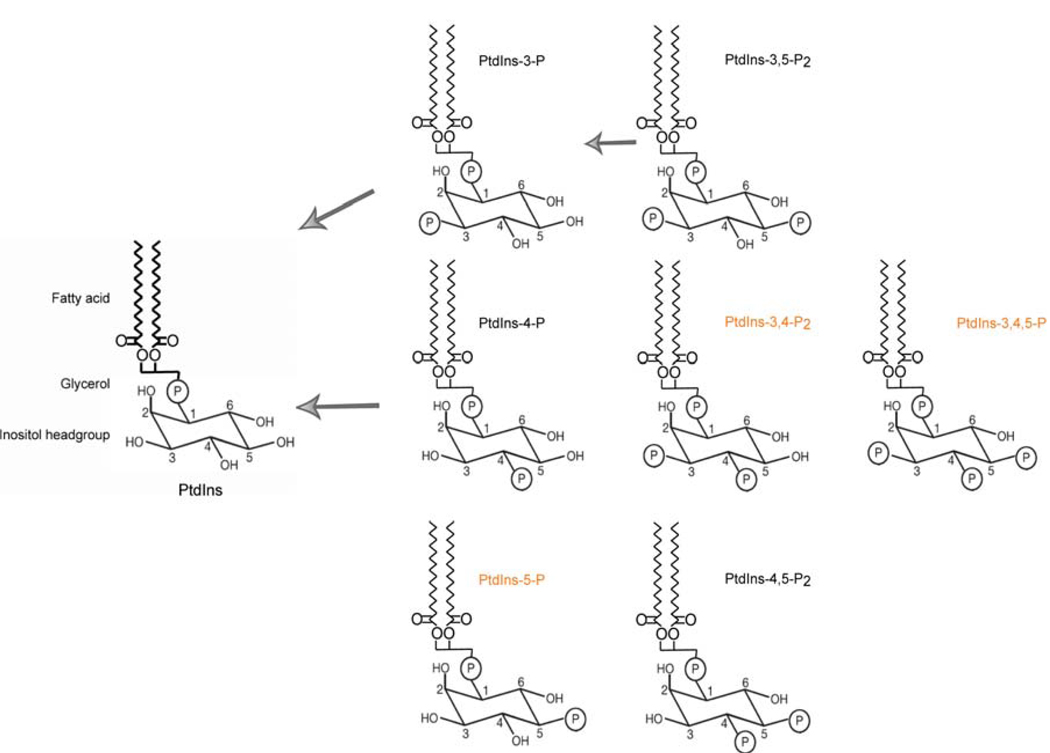
Figure 1.
Cellular PIP species and Sac1-mediated dephosphorylation. The chemical structures of PtdIns and its seven phosphorylated derivatives are shown. The PtdIns inositol headgroup, glycerol backbone and fatty acyl chains are illustrated. Mammals synthesize all seven phosphoinositides, although PtdIns-5-P is undetectable in several mammalian cell types (Kent, 1995; Fruman et al., 1998; Shisheva, 2001; Di Paolo and De Camilli, 2006), and those shown in red are not present in yeast. Black arrows indicate Sac1 PIP phosphatase-catalyzed dephosphorylation reactions.
Acknowledgments
This work was supported by National Institutes of Health grant RO1 NS-37723 to VAB. We acknowledge our University of North Carolina colleague Kristina Ile for critical comments on the manuscript. We thank Peter Mayinger (University of Oregon Health Sciences Center) for helpful discussions and for communicating his laboratory’s results prior to publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albi E, Viola-Magni MP. The role of intranuclear lipids. Biol Cell. 2004;96:657–667. doi: 10.1016/j.biolcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Malehorn DE, Emr SD, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya A, Cheong FY, Rohde HM, Glover G, Knodler A, Nicolson T, Boehmelt G, Mayinger P. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J Cell Biol. 2008;180:803–812. doi: 10.1083/jcb.200708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyum R, Guidotti G. Sac1p of Saccharomyces cerevisiae is not involved in ATP release to the extracellular fluid. Biochem Biophys Res Commun. 1997;236 doi: 10.1006/bbrc.1997.6805. 50-Z3. [DOI] [PubMed] [Google Scholar]
- Cleves AE, Novick PJ, Bankaitis VA. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J Cell Biol. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona O, Di Paolo G, Wenk MR, Luthi A, Kim WT, Takei K, Daniell L, Nemoto Y, Shears SB, Flavell RA, McCormick DA, De Camilli P. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 1999;99:179–188. doi: 10.1016/s0092-8674(00)81649-9. [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Godi A. PI-loting membrane traffic. Nat Cell Biol. 2004;6:487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Lippincott-Schwartz J. Sorting and signaling at the Golgi complex. Cell. 2000;101:693–696. doi: 10.1016/s0092-8674(00)80881-8. [DOI] [PubMed] [Google Scholar]
- Faulhammer F, Konrad G, Brankatschk B, Tahirovic S, Knodler A, Mayinger P. Cell growth-dependent coordination of lipid signaling and glycosylation is mediated by interactions between Sac1p and Dpm1p. J Cell Biol. 2005;168:185–191. doi: 10.1083/jcb.200407118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger FP, Novick P. Spatial regulation of exocytosis: lessons from yeast. J Cell Biol. 1998;142:609–612. doi: 10.1083/jcb.142.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. It takes two to tango: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- Hughes WE. The Sac phosphatase domain. Curr Biol. 2001;11:R249. doi: 10.1016/s0960-9822(01)00129-4. [DOI] [PubMed] [Google Scholar]
- Hughes WE, Woscholski R, Cooke FT, Patrick RS, Dove SK, McDonald NQ, Parker PJ. SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologous Inp52p and Inp53p phosphatases. J Biol Chem. 2000;275:801–808. doi: 10.1074/jbc.275.2.801. [DOI] [PubMed] [Google Scholar]
- Kent C. Eukaryotic phospholipid biosynthesis. Annu Rev Biochem. 1995;64:315–343. doi: 10.1146/annurev.bi.64.070195.001531. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochendorfer KU, Then AR, Kearns BG, Bankaitis VA, Mayinger P. Sac1p plays a crucial role in microsomal ATP transport, which is distinct from its function in Golgi phospholipid metabolism. EMBO J. 1999;18:1506–1515. doi: 10.1093/emboj/18.6.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Sütterlin C. The Golgi Protein GM130 Regulates Centrosome Morphology and Function. Mol Biol Cell. 2008;19:745–753. doi: 10.1091/mbc.E07-08-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad G, Schlecker T, Faulhammer F, Mayinger P. Retention of the yeast Sac1p phosphatase in the endoplasmic reticulum causes distinct changes in cellular phosphoinositide levels and stimulates microsomal ATP transport. J Biol Chem. 2002;277:10547–10554. doi: 10.1074/jbc.M200090200. [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH. Parsons R PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Li X, Rivas MP, Fang M, Marchena J, Mehrotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Dahan N, Ramachandran S, Sabanay H, Lev S. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat Cell Biol. 2005;7:225–234. doi: 10.1038/ncb1221. [DOI] [PubMed] [Google Scholar]
- Liu Y, Boukhelifa M, Tribble E, Morin-Kensicki E, Uetrecht A, Bear JE, Bankaitis VA. The Sac1 phosphoinositide phosphatase regulates Golgi membrane morphology and mitotic spindle organization in mammals. Mol Biol Cell. 2008;19:3080–3096. doi: 10.1091/mbc.E07-12-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Taylor GS, Dixon JE. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu Rev Biochem. 2001;70:247–279. doi: 10.1146/annurev.biochem.70.1.247. [DOI] [PubMed] [Google Scholar]
- Martin OC, Gunawardane R, Iwamatsu A, Zheng Y. Xgrip109: A γ-tubulin–associated protein with an essential role in γ-tubulin ring complex (γTuRC) assembly and centrosome function. J. Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TF. Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu Rev Cell Dev Biol. 1998;14:231–264. doi: 10.1146/annurev.cellbio.14.1.231. [DOI] [PubMed] [Google Scholar]
- Moritz M, Zheng Y, Alberts B, Oegema K. Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudjou M, Bordes N, Paintrand M, Bornens M. gamma-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J Cell Sci. 1996;109:875–887. doi: 10.1242/jcs.109.4.875. [DOI] [PubMed] [Google Scholar]
- Nemoto Y, Kearns BG, Wenk MR, Chen H, Mori K, Alb JG, Jr, De Camilli P, Bankaitis VA. Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J Biol Chem. 2000;275:34293–34305. doi: 10.1074/jbc.M003923200. [DOI] [PubMed] [Google Scholar]
- Novick P, Osmond BC, Botstein D. Suppressors of yeast actin mutations. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- Rivas MP, Kearns BG, Xie Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p” and inositol auxotrophy. Mol Biol Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde HM, Cheong FY, Konrad G, Paiha K, Mayinger P, Boehmelt G. The human phosphatidylinositol phosphatase SAC1 interacts with the coatomer I complex. J Biol Chem. 2003;278:52689–52699. doi: 10.1074/jbc.M307983200. [DOI] [PubMed] [Google Scholar]
- Rubbini S, Cocco L, Manzoli L, Lutterman J, Billi AM, Matteucci A, Wirtz KW. Phosphoinositide signalling in nuclei of Friend cells: DMSO-induced differentiation reduces the association of phosphatidylinositol-transfer protein with the nucleus. Biochem Biophys Res Commun. 1997;230:302–305. doi: 10.1006/bbrc.1996.5950. [DOI] [PubMed] [Google Scholar]
- Schorr M, Then A, Tahirovic S, Hug N, Mayinger P. The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr Biol. 2001;11:1421–1426. doi: 10.1016/s0960-9822(01)00449-3. [DOI] [PubMed] [Google Scholar]
- Shisheva A. PIKfyve: the road to PtdIns 5-P and PtdIns 3,5-P(2) Cell Biol Int. 2001;25:1201–1206. doi: 10.1006/cbir.2001.0803. [DOI] [PubMed] [Google Scholar]
- Simpson L, Parsons R. PTEN: life as a tumor suppressor. Exp Cell Res. 2001;26:29–41. doi: 10.1006/excr.2000.5130. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Seaman M, Nemoto Y, Daniell L, Suchy SF, Emr S, De Camilli P, Nussbaum R. Disruption of three phosphatidylinositol-polyphosphate 5-phosphatase genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis. Eur J Cell Biol. 1997;74:350–360. [PubMed] [Google Scholar]
- Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD. INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem. 1998;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- Sütterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- Waselle L, Gerona RR, Vitale N, Martin TF, Bader MF, Regazzi R. Role of phosphoinositide signaling in the control of insulin exocytosis. Mol Endocrinol. 2005;19:3097–3106. doi: 10.1210/me.2004-0530. [DOI] [PubMed] [Google Scholar]
- Whisstock JC, Wiradjaja F, Waters JE, Gurung R. The structure and function of catalytic domains within inositol polyphosphate 5-phosphatases. IUBMB Life. 2002;53:15–23. doi: 10.1080/15216540210814. [DOI] [PubMed] [Google Scholar]
- Whitters EA, Cleves AE, McGee TP, Skinner HB, Bankaitis VA. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J Cell Biol. 1993;122:79–94. doi: 10.1083/jcb.122.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- Xie Z, Fang M, Rivas MP, Faulkner AJ, Sternweis PC, Engebrecht JA, Bankaitis VA. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc Natl Acad Sci U S A. 1998;95:12346–12351. doi: 10.1073/pnas.95.21.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JD, Majerus PW. Nuclear phosphatidylinositols decrease during S-phase of the cell cycle in HeLa cells. J Biol Chem. 1994;269:7847–7850. [PubMed] [Google Scholar]