Abstract
The PiRaNhA web server is a publicly available online resource that automatically predicts the location of RNA-binding residues (RBRs) in protein sequences. The goal of functional annotation of sequences in the field of RNA binding is to provide predictions of high accuracy that require only small numbers of targeted mutations for verification. The PiRaNhA server uses a support vector machine (SVM), with position-specific scoring matrices, residue interface propensity, predicted residue accessibility and residue hydrophobicity as features. The server allows the submission of up to 10 protein sequences, and the predictions for each sequence are provided on a web page and via email. The prediction results are provided in sequence format with predicted RBRs highlighted, in text format with the SVM threshold score indicated and as a graph which enables users to quickly identify those residues above any specific SVM threshold. The graph effectively enables the increase or decrease of the false positive rate. When tested on a non-redundant data set of 42 protein sequences not used in training, the PiRaNhA server achieved an accuracy of 85%, specificity of 90% and a Matthews correlation coefficient of 0.41 and outperformed other publicly available servers. The PiRaNhA prediction server is freely available at http://www.bioinformatics.sussex.ac.uk/PIRANHA.
INTRODUCTION
RNA-binding proteins (RBPs) play key roles in many cellular processes, including gene expression regulation. The fundamental role of RBPs within the cell is reflected in the wide range of human diseases, including neurological disorders and cancer, to which they have been linked (1). However, proteomic sequence data includes a significant percentage of RBPs in which the location of RNA-binding residues (RBRs) is unknown. Hence, computational methods to predict the location of the RBRs in such proteins are of great significance.
The availability of the structures of an increasing number of protein–RNA complexes has allowed RBRs to be characterized using features such as hydrophobicity, solvent accessibility, evolutionary conservation and charge (2–4). Such analysis of RBRs has led to the development of methods to predict RBRs from protein sequence information, using machine learning techniques (5–10). The identification of RNA-binding sites from protein sequence information alone is critically important for understanding the function of RBPs when the structure of the protein–RNA complex is not known. Three of the prediction methods are currently available as publicly available web servers: RNABindR (6), BindN (7) and PPRInt (9). The majority of these methods use inclusive definitions of RBRs, based on lenient distance constraints between the RNA and the protein entities (between 5 Å and 6 Å). These methods also use either position-specific scoring matrices (PSSMs) (to estimate evolutionary conservation) or residue parameters as features to differentiate RBRs from non-RBRs. These two separate approaches give results with varying levels of accuracy, and combined with their inclusive definition of RBRs can give large numbers of false positive predictions (11).
This article presents a new publicly available online server (PiRaNhA, capitals denote Protein-RNA; pronounced ‘piranha’) for the prediction of RBRs from protein sequence information alone. This server differs from other prediction servers in three important criteria: (i) the server is based on a support vector machine (SVM) model that has been trained on known structures of protein–RNA complexes from the Protein Data Bank (PDB) (12). The RBRs in these structures are defined using a restrictive distance constraint based on intermolecular interaction data calculated using the HBPLUS software (13). RBRs are defined as those residues making an intermolecular hydrogen bond or van der Waals contact, with a distance of ≤3.9 Å. (ii) The SVM model integrates PSSMs with three physicochemical residue parameters to identify RBRs. The three parameters are residue interface propensity, predicted residue accessibility and residue hydrophobicity. (iii) The server provides the unique facility for users to make predictions for multiple sequences. The PiRaNhA server achieves a Matthews correlation coefficient (MCC, a balanced performance measure that includes the numbers of true and false, positive and negative, predictions) of 0.41 for a data set of 42 known RNA-binding protein sequences not used in training. Thus, PiRaNhA outperforms other web server prediction tools; RNABindR (6), BindN (7) and PPRInt (9), which achieve an MCC of 0.36, 0.29 and 0.34, respectively, on the same data set (11). The PiRaNhA server allows researchers to upload the sequence of one or more proteins and obtain a set of RBR predictions. The accurate nature of the predictions means fewer site-directed mutagenesis experiments are required to verify the RNA-binding function of residues. The PiRaNhA web server is freely available at http://www.bioinformatics.sussex.ac.uk/PIRANHA.
METHODS
Server input
The PiRaNhA server is designed to be easy to use and to provide results that are easily interpreted. It allows the submission of single or multiple (up to a maximum of 10) protein sequences in FASTA format, by cutting and pasting to the submission page or by file upload. Protein sequences of unlimited length are accepted, but the calculation time required for the PSI-BLAST alignments required for the PSSM generation is related to the sequence length. Hence, for sequences longer than 150 residues selection of the email option is recommended (as is the case with batch submissions), so results can be emailed when the calculations are complete.
Sequence feature vectors
For all residues in the submitted protein sequence(s), four sequence features are calculated and encoded into feature vectors in the SVM model. These features are: (i) a PSSM created using PSI-BLAST (14) with an E-value threshold of 0.001 for three iterations, and the NCBI nr sequence database. This feature describes the evolutionary conservation of the residue positions. (ii) A residue interface propensity that describes the likelihood of a residue of a specific type being found in an RNA-binding site. The interface propensity values are taken from our previous analysis of known RNA-binding sites (3). (iii) A predicted residue accessibility value that quantifies the estimated solvent exposure of each residue and is calculated using SABLE (15). (iv) A residue hydrophobicity score based on the Kyte and Doolittle hydropathy scale (16). These four properties are integrated into a single feature vector based on a sequence window of 23 residues, with the residue being described in the centre. The calculation of the PSSMs and the prediction of accessible surface area are computationally intensive, and it is for this reason that users may want to obtain results via email when making a batch submission of more than one protein sequence.
The SVM model
The SVM model used in the PiRaNhA server is based on LIBSVM (Chang C-C and Lin C-J: LIBSVM: a library for SVMs, 2001. www.csie.ntu.edu.tw/∼cjlin/libsvm, version 2.8) and uses the Radial Basis Function kernel. In order to distinguish between RBRs and non-RBRs in protein sequences, the SVM model was trained on a non-redundant set of 81 known RNA-binding protein sequences (RNAset81) whose complexed structures are known. The model was tested on a non-redundant data set of 42 known RNA-binding protein sequences not used in training (RNAtestset42); of the 8554 residues in these sequences, 14.8% were known RBRs. The SVM model achieved an MCC of 0.41, a sensitivity of 53%, a specificity of 90% and a precision of 48% and outperforms the other publicly available servers [RNABindR (6), BindN (7) and PPRInt (9)] tested on the same data set (11).
RESULTS
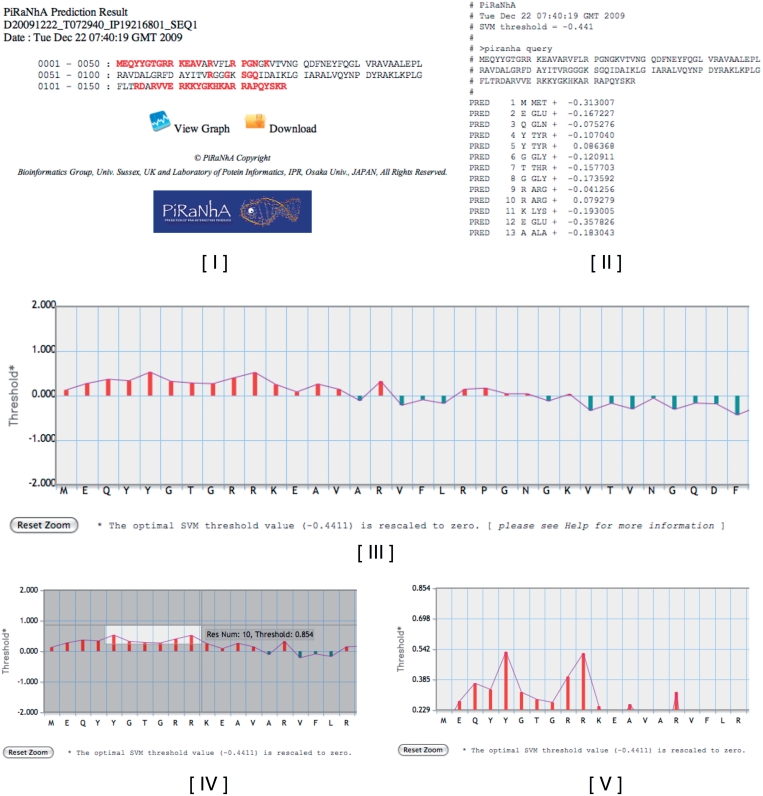
The PiRaNhA server provides predictions for each submitted sequence in a separate web page and, if requested, by email with links to URLs and attached files. The results are provided in three formats: (i) the user’s submitted sequence shown with residues involved in protein–RNA interactions highlighted in red (Figure 1I), (ii) a text file of raw prediction results with SVM scores that can be downloaded (Figure 1II) and (iii) a graph where the submitted sequence is plotted against SVM threshold values (Figure 1III). On the graph, the x-axis shows the submitted sequence and the y-axis shows the threshold for the prediction. The optimal threshold value (–0.4411) [which achieves an MCC of 0.50, and an area under ROC curve (AUC) of 0.86 in a 5-fold cross-validation on the training data set (11)] is rescaled to zero for the ease of interpretation. The graph has a built in ‘click and drag’ zoom function to enable users to highlight and select residues above a desired threshold (Figure 1IV and V). By increasing the threshold value, a user can effectively decrease the false positive rate of predictions. This graph function enables users to quickly determine which residues are most likely to bind to RNA, and hence be the initial targets for site-directed mutagenesis.
Figure 1.
Example PiRaNhA server prediction results for 30S ribosomal protein S9 (PDB-ID 2J00, chain I). (I) The sequence format webpage where the predicted RBRs are highlighted in red. (II) The text format results, which includes the sequence and the SVM values that can be downloaded. (III) The graphical interpretation of the results in which the submitted sequence is plotted against SVM threshold values. The x-axis shows the submitted sequence and the y-axis the threshold for the prediction. The optimal SVM threshold value (0.4411) is rescaled to zero, for ease of interpretation. The graph has a ‘click and drag’ zoom function to enable the easy highlighting of residues (IV) above a desired threshold to produce a finer grained graph (V).
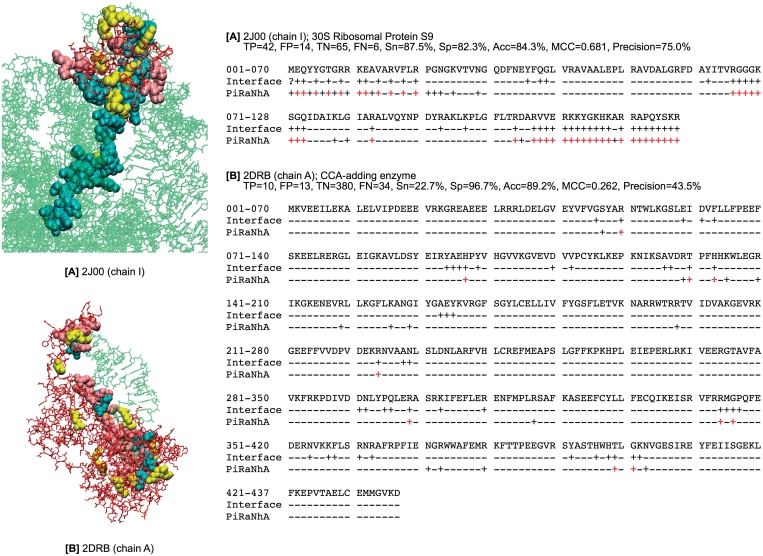
Figure 2 shows examples of two PiRaNhA predictions taken from the RNAtestset42 data set; (Figure 2A) 30S ribosomal protein S9 (PDB-ID 2J00, chain I), (Figure 2B) CCA-adding enzyme (PDB-ID 2DRB, chain A), in which the overlap between the predictions and the known RBRs from the protein–RNA complex is detailed. This clearly shows the accuracy of the PiRaNhA server, but also highlights the fact that predictions can include false positives (FPs), i.e. residues predicted to be RNA binding that, when compared to the RNA–protein structure in the PDB, are not defined in the known RNA-binding site. The relatively high number of FPs for some predictions is reflected in a mean precision value of 48%. However, to address this issue the PiRaNhA server allows the user to increase the SVM threshold value above the default (–0.4411). Increasing the threshold value decreases the number of false positives; a strategy that would be recommended if potential RBRs are being selected for mutagenesis. For example, when RBR predictions are made for the RNAtestset42 data set with increasing SVM threshold levels, the precision and the specificity increase: precision and specificity at a threshold of −0.44 are 90.0 and 47.9%, respectively, at a SVM threshold of −0.20 they rise to 97.4 and 64.7% and at a threshold of 0.00 they rise to 99.1 and 76.4%.
Figure 2.
Two example PiRaNhA predictions; (A) 30S ribosomal protein S9 (PDB-ID 2J00, chain I), (B) CCA-adding enzyme (PDB-ID 2DRB, chain A). In the left panel, the three-dimensional structures of the protein–RNA complexes are shown, where TP, FP and FN are highlighted as cyan, yellow and pink CPK spheres, respectively, and the remaining protein residues and RNA nucleotides are represented as red and green sticks, respectively. In the right panel, the protein sequence is shown. Non-RBRs are indicated with the symbol ‘−’ and RBRs with ‘+’. The TPs are highlighted in red. The prediction performance: TP (true positives), FP (false positives), TN (true negatives), FN (false negatives), Sn (sensitivity), Sp (specificity), Acc (accuracy), MCC (Mathews Correlation Coefficient) and precision are listed for each example.
A further point to consider is that some FP residues could in fact be true positives (i.e. be present in the RNA-binding site) in RNA–protein complexes where the PDB structure does not represent the complete functional complex. One example of this is Pop7 recently deposited in the PDB (PDB-ID 3IAB) (17). The PDB structure comprises Pop7 and Pop6 RNase MRP proteins bound to the P3 RNA domain, whereas the complete functional complex in Saccharomyces cerevisiae comprises more than 10 RNA secondary structure domains and many additional proteins (17). Hence, predictions made for proteins in incomplete complexes such as this, may give rise to FP residues, which are in fact novel RBRs. Such sites will only be validated when complete structures are determined.
CONCLUSION
The PiRaNhA server predicts potential RBRs based on protein sequence information alone. Four sequence-based features, such as PSSM, amino acid interface propensity, predicted residue accessibility and hydrophobicity, are integrated in a feature vector and used for predicting RBRs using an SVM model. PiRaNhA outperforms other publicly available RBR prediction servers [RNABindR (6), BindN (7) and PPRInt (9)] in a benchmark test (11). The accuracy of the PiRaNhA predictions could be improved further by the inclusion of (i) additional non-homologous RNA-binding protein structures and (ii) the structures of complete functional complexes of RNA–protein moieties into the SVM training data set. Such structures are starting to be determined as part of the many structural genomics projects (18). The aim is for the PiRaNhA server to be retrained on an updated set of non-homologous RNA-binding proteins on an annual basis, thus increasing its predictive potential.
The PiRaNhA server is of use to experimental biologists studying RNA-binding proteins in specific systems in which the structure of the protein–RNA complex is as yet unknown. The predictions made by the server allow for fewer and more targeted mutations to be made to verify RNA binding. In addition, the server is of interest to theoretical researchers wishing to analyse and compare functional residues in multiple protein data sets.
FUNDING
Strategic International Cooperative Program, Japan Science and Technology (to Y.M. and H.N.); Medical Research Council (grant number 70760 to R.V.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, Daley DT, Luscombe NM, Berman HM, Thornton JM. Protein-RNA interactions: a structural analysis. Nucleic Acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis JJ, Broom M, Jones S. Protein-RNA interactions: structural analysis and functional classes. Proteins. 2007;66:903–911. doi: 10.1002/prot.21211. [DOI] [PubMed] [Google Scholar]
- 4.Bahadur RP, Zacharias M, Janin J. Dissecting protein-RNA recognition sites. Nucleic Acids Res. 2008;36:2705–2716. doi: 10.1093/nar/gkn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeong E, Chung IF, Miyano S. A neural network method for identification of RNA-interacting residues in protein. Genome Inform. 2004;15:105–116. [PubMed] [Google Scholar]
- 6.Terribilini M, Sander JD, Lee JH, Zaback P, Jernigan RL, Honavar V, Dobbs D. RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res. 2007;35:W578–W584. doi: 10.1093/nar/gkm294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Xue Z, Shen G, Xu J. PRINTR: prediction of RNA binding sites in proteins using SVM and profiles. Amino Acids. 2008;35:295–302. doi: 10.1007/s00726-007-0634-9. [DOI] [PubMed] [Google Scholar]
- 9.Kumar M, Gromiha MM, Raghava GP. Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins. 2008;71:189–194. doi: 10.1002/prot.21677. [DOI] [PubMed] [Google Scholar]
- 10.Cheng CW, Su EC, Hwang JK, Sung TY, Hsu WL. Predicting RNA-binding sites of proteins using support vector machines and evolutionary information. BMC Bioinformatics. 2008;9(Suppl. 12):S6. doi: 10.1186/1471-2105-9-S12-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spriggs RV, Murakami Y, Nakamura H, Jones S. Protein function annotation from sequence: prediction of residues interacting with RNA. Bioinformatics. 2009;25:1492–1497. doi: 10.1093/bioinformatics/btp257. [DOI] [PubMed] [Google Scholar]
- 12.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner M, Adamczak R, Porollo A, Meller J. Linear regression models for solvent accessibility prediction in proteins. J. Comput. Biol. 2005;12:355–369. doi: 10.1089/cmb.2005.12.355. [DOI] [PubMed] [Google Scholar]
- 15.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 16.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS. Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO J. 2010;29:761–769. doi: 10.1038/emboj.2009.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair R, Liu J, Soong TT, Acton TB, Everett JK, Kouranov A, Fiser A, Godzik A, Jaroszewski L, Orengo C, et al. Structural genomics is the largest contributor of novel structural leverage. J. Struct. Funct. Genomics. 2009;10:181–191. doi: 10.1007/s10969-008-9055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]