Abstract
iPARTS is an improved web server for aligning two RNA 3D structures based on a structural alphabet (SA)-based approach. In particular, we first derive a Ramachandran-like diagram of RNAs by plotting nucleotides on a 2D axis using their two pseudo-torsion angles η and θ. Next, we apply the affinity propagation clustering algorithm to this η-θ plot to obtain an SA of 23-nt conformations. We finally use this SA to transform RNA 3D structures into 1D sequences of SA letters and continue to utilize classical sequence alignment methods to compare these 1D SA-encoded sequences and determine their structural similarities. iPARTS takes as input two RNA 3D structures in the PDB format and outputs their global alignment (for determining overall structural similarity), semiglobal alignments (for detecting structural motifs or substructures), local alignments (for finding locally similar substructures) and normalized local structural alignments (for identifying more similar local substructures without non-similar internal fragments), with graphical display that allows the user to visually view, rotate and enlarge the superposition of aligned RNA 3D structures. iPARTS is now available online at http://bioalgorithm.life.nctu.edu.tw/iPARTS/.
INTRODUCTION
As both the number and the size of RNA tertiary 3D structures deposited in the database continue to grow, the techniques of RNA structure comparison have become an increasingly crucial bioinformatics tool because structures of molecules evolve more slowly than their sequences and, therefore, their structural comparison can bring more significant insights into their functions and even evolutionary relationships that would not be detected by analyzing sequence information alone. Basically, detecting structural similarities in two RNA 3D molecules is not an easy problem because it has been shown to be computationally intractable (1). Due to this reason, currently available software tools for comparing two RNA 3D structures, such as ARTS (2,3), DIAL (4), PARTS (5), SARA (6,7) and LaJolla (8), are all based on some heuristic approaches.
ARTS is a web server for detecting maximum common substructures between two given RNA 3D structures, which was implemented by Dror et al. (2,3) based on a heuristic algorithm of cubic running time. By representing each RNA 3D structure by a set of its phosphate atoms, ARTS identifies all structurally similar quadrats (i.e. four phosphate atoms located on two successive base pairs) between the two input RNA 3D structures and continues to extend them by using a greedy method for including additional coincident base pairs and unpaired nucleotides. ARTS is a good tool for detecting RNA structural motifs, but it is still time-consuming for ARTS to compare large RNA molecules (e.g. ribosomal RNAs) because of its cubic time complexity and, as was pointed out in (4), the structural alignments produced by ARTS may be incorrect sometimes. Later on, to overcome the inaccurate problems caused by ARTS, Ferré et al. (4) implemented DIAL, a web server for aligning two RNA 3D structures, by using a dynamic programming algorithm of quadratic running time based on a scoring function that combines similarities of nucleotide sequences, base pairs, pseudo-torsion (or pseudo-dihedral) and torsion (or dihedral) angles. DIAL is a versatile web server by providing the user three types of alignments: (i) global alignment, (ii) local alignment and (iii) an extension of global-semiglobal alignment [i.e. a global alignment of a motif A consisting of one or more contiguous segments is aligned to a contiguous sequence B; while gap penalties apply throughout for A (global alignment), gaps at the end of B as well as between portions aligned to contiguous segments of A are not penalized (so-called middle gaps)]. Next, we developed PARTS (5) for pairwise alignments of RNA tertiary structures based on a structural alphabet (SA)-based algorithm. Its basic idea is to reduce input RNA 3D structures to 1D sequences of SA letters using backbone torsion angles of constituent residues and continue to use algorithms of classical sequence alignments (including global, local, semiglobal and normalized local alignments) to compare these 1D SA-encoded sequences for determining their structural similarities. As was demonstrated in (5), the structural alignments by PARTS were comparable to those by DIAL, but the running time of PARTS was generally faster than that of DIAL. Recently, Capriotti and Marti-Renom (6) have proposed a new web server, called SARA, for globally aligning two RNA 3D structures based on the unit-vector approach and have further shown its ability in function assignment of RNA structures (7). For each input RNA 3D structure, SARA first identifies an atom trace that consists of all contiguous atoms of user-defined type and also calculates all unit-vectors between any two consecutive atoms along this trace. For each nucleotide of an input RNA structure, it then groups a set of k consecutive unit-vectors starting from this nucleotide and places these k unit-vectors at the origin of a unit-sphere, where k is a user-defined positive integer. Finally, SARA applies a dynamic programming algorithm without penalizing end gaps to the two sequences of unit-spheres to find an optimal semiglobal alignment between them. More recently, Bauer et al. (8) have used a hashing algorithm to develop a tool, called LaJolla, which can perform structural alignment of two RNA 3D structures. LaJolla first translates each of input RNA 3D structures into a 1D sequence of characters according to backbone pseudo-torsion angles of constituent residues, with one of these two 1D sequences being considered as query RNA and the other as target RNA. Next, it stores all n-grams (i.e. substrings of length n) of the target RNA in a hash table and searches each of all n-grams of the query RNA against the hash table for its occurrences in the target RNA. Finally, all corresponding n-grams between the query and target RNAs are aligned to determine their anchors and a superposition of these anchors are then performed.
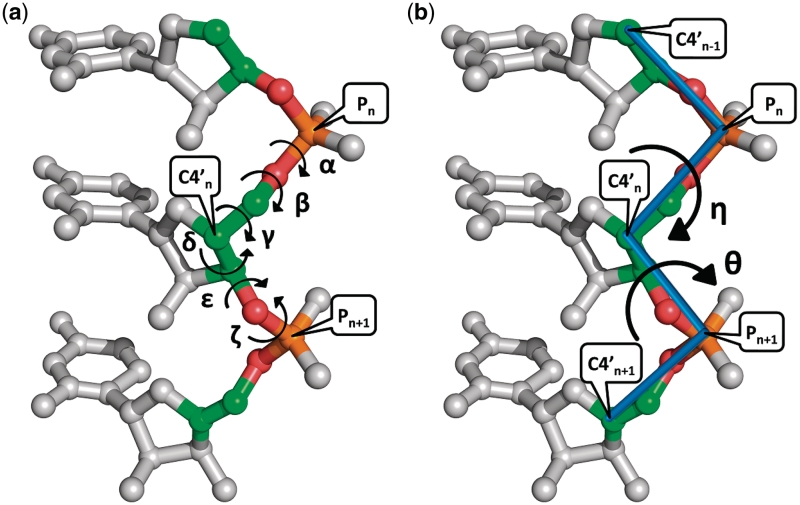
For proteins, two torsion angles (ϕ and ψ) are sufficient to describe the backbone conformation of each amino acid. In contrast, RNA molecules have much higher dimensionality, since six standard torsion angles (α, β, γ, δ, ε and ζ as shown in Figure 1a) are needed to specify the backbone conformation of a single nucleotide. This leads the analysis and classification of nucleotide conformation to be a high-dimensional problem that is computationally intractable and cannot be evaluated visually. In addition, it is difficult to use these standard torsion angles to distinguish important nucleotide conformations in RNA structural motifs, because the so-called ‘crankshaft effect’, in which large changes in individual torsion angles are compensated by changes in other torsion angles, usually leads to a result that different combinations of standard torsion angles can describe identical nucleotide conformations (9). In fact, as was suggested by Duarte and Pyle (10), the pseudo-torsion angles (η and θ as illustrated in Figure 1b) are at least as sensitive as standard torsion angles and even may be superior when specifying the backbone conformation of an individual nucleotide. Particularly, by representing the η and θ pseudo-torsion angles of nucleotides on a 2D plot, one can obtain a Ramachandran-like diagram in which clusters of nucleotides appear at discrete regions and nucleotides in the same cluster have similar conformation (9,10). Therefore, in this study, we aim to develop a novel SA for RNA 3D structures using their η-θ plot of pseudo-torsion angles, rather than using four standard torsion angles (α, γ, δ and ζ) as done in our previous work of PARTS (5) that was motivated from the works by Hershkovitz et al. (11,12). For this purpose, we utilize a recently introduced clustering algorithm, called affinity propagation (13), to classify the nucleotides in the 2D η-θ plot, instead of using the vector quantization (VQ) approach as used in PARTS (5). Like k-means clustering approaches, the VQ methods suffer from local optimality and are sensitive to outliers and noise (14). Moreover, for the VQ clustering methods, the identified centers in their clusters may be virtual nucleotides that cannot be evaluated visually. The so-called ‘affinity propagation’ (AP) algorithm, first proposed by Frey and Dueck (13), basically is an exemplar-based clustering method that considers all data points as potential exemplars (or centers) and exchanges messages (of how proper a data point serves as the exemplar of another one or of how proper a data point chooses another one as its exemplar) between data points until a good set of exemplars and clusters emerges (13). More importantly, Frey and Dueck (13) have shown that the AP algorithm can obtain better solutions than other frequently used methods, such as K-centers clustering and hierarchical agglomerative clustering algorithms.
Figure 1.
(a) Six standard backbone torsion angles of α, β, γ, δ, ε and ζ and (b) two backbone pseudo-torsion angles of η and θ for a nucleotide (denoted by n), where η is defined by the atoms C4′n−1, Pn, C4′n and Pn+1, while θ is defined by Pn, C4′n, Pn+1 and C4′n+1.
In this study, we have derived a new SA of RNA nucleotide conformations using their η and θ pseudo-torsion angles and the AP algorithm. Based on this newly designed SA, we have re-implemented our previous tool PARTS as iPARTS (short for improved PARTS) to make its structural alignments of two RNA molecules more accurate. Our experimental results on some data sets have finally shown that our iPARTS outperforms its previous version PARTS, as well as ARTS and LaJolla, on accuracy of aligning two RNA 3D structures without compromising the computational efficiency and also outperforms SARA on the function assignment of RNA structures. Basically, the main differences between iPARTS and PARTS are 2-fold. First, iPARTS uses the AP algorithm to construct the SA according to two pseudo-torsion angles of η and θ, while PARTS uses the VQ method to construct it based on four standard torsion angles of α, γ, δ and ζ. Second, iPARTS uses two data sets, one of highly identical RNA structure pairs from the DARTS database (15) and the other of structurally similar RNA motif pairs from the SCOR database (16,17), to construct the BLOSUM-like substitution matrix, while PARTS uses only a data set of structurally similar RNA motif pairs from the SCOR database to construct it.
METHODS
The basic idea of our iPARTS algorithm is to reduce input RNA 3D structures to 1D sequences of SA letters and continue to use algorithms of classical sequence alignments to compare these 1D SA-encoded sequences and determine their structural similarities. As mentioned before, the 2D η-θ plot is a Ramachandran-like diagram that can provide us a graphic representation of quantitatively distinct structural features for analyzing and modeling RNA 3D structures (9,10). To depict this η-θ plot, we prepared a data set that includes non-redundant crystal structures with minimum resolution of 3.0 Å from the PDB database (18). This data set finally contains 117 crystal RNA structures with 9527 nt in total. Next, we used the AMIGOS program developed by Duarte and Pyle (10) to calculate the η and θ pseudo-torsion angles for all non-terminal nucleotides (9267 nt in total) from all RNA molecules in the above data set and plotted these calculated pseudo-torsion angles on the axes of a 2D plot (refer to Supplementary Data for the derived η-θ plot). We then continued to use the AP clustering algorithm (13) to classify all the non-terminal nucleotides in the η-θ plot into 23 conformation clusters, each of which was further assigned a letter. We used the set of these 23 letters as an SA and encoded RNA 3D structures as 1D sequences of SA letters using the so-called ‘nearest neighbor rule’, by which each nucleotide in an RNA molecule is assigned with the letter whose corresponding exemplar (or center) is nearest to the nucleotide to be encoded. Next, we derived a log-odds matrix for SA-letter substitutions using the statistical method that was proposed by Henikoff and Henikoff (19). Finally, we utilized classical sequence alignment algorithms, such as global (20), semiglobal (21), local (22) and normalized local (23) alignments, to compare two 1D SA-encoded sequences for determining the similarities of their corresponding RNA 3D structures. Notice that a grid-like search procedure was performed to optimize the parameters of open and extension gap penalties by varying the open gap penalty from −15 to −1 in steps of 1 and the extension gap penalty from −3 to −0.5 in steps of 0.5. The reader is referred to Supplementary Data for the details of above procedures. It is worth mentioning here that the Smith–Waterman algorithm (22) for the local alignment was originally designed to remove non-similar initial and terminal fragments but not non-similar internal fragments in a sequence alignment, resulting in a so-called ‘mosaic effect’ by including poor internal fragments in a local alignment (23). Such a mosaic effect still can be observed in local alignments of two RNA 3D structures, as demonstrated on the help page of our iPARTS server. To eliminate this mosaic effect, we implemented the algorithm proposed by Arslan et al. (23) to solve the so-called ‘normalized local alignment problem’, which aims to find the subsequences, say I and J, of two given sequences that maximizes S(I, J)/(|I| + |J|) among all subsequences I and J with |I| + |J| ≥ T, where S(I, J) is the alignment score between I and J, and T is a threshold for the minimal overall length of I and J. Usually, an alignment should be sufficiently long to be biologically meaningful. Therefore, the above length constraint of |I| + |J| ≥ T is necessary, since length normalization in the normalized local alignment problem favors short local alignments. The user can vary the value of T to control the result of optimal normalized local alignment. If T is small, the optimal normalized local alignment tends to be short; otherwise, it tends to be a long local alignment that may contain some non-similar internal fragments.
USAGE OF iPARTS
The kernel programs of iPARTS, as well as its web interface, were written in PHP. The server of iPARTS is currently installed on IBM PC with 2.8 GHz processor and 3 GB RAM under Linux system. iPARTS provides an intuitive and easy-to-operate interface that can be freely accessed at http://bioalgorithm.life.nctu.edu.tw/iPARTS/. It provides four types of alignments to compare two RNA 3D structures: (i) global alignment that is suitable to align two RNA 3D structures that have overall structural similarity, (ii) semiglobal alignment that can be used to detect known structural motifs with a single contiguous segment or substructures in an RNA 3D structure, (iii) local alignment that is to find common similar substructures between two RNA 3D structures, and (iv) normalized local alignment that can identify more similar local substructures without non-similar internal fragments. iPARTS takes as input two RNA 3D structures, each of which can be either a PDB/NDB ID or a PDB file uploaded by the user, and their chain IDs if they have multiple chains and optionally the starting and ending residue numbers of substructures to be aligned. If needed, the default settings of all the parameters can be modified by the user, including alignment method (whose default is semiglobal alignment), gap open and extension penalties (whose default values are −6 and −1, respectively), number of suboptimal semiglobal, local or normalized local alignments (at least one) and threshold of T (whose default value is 8) for controlling normalization degree of normalized local alignments. Notice that iPARTS currently is limited to align RNA 3D structures of length up to 1900 nt due to limited memory availability. In the output page, iPARTS first displays the information about input RNA molecules and user-specified parameters. In this display, the user can further review the details of the input RNA molecules annotated in the PDB database, as well as standard torsion and pseudo-torsion angles of nucleotides calculated by iPARTS, just by clicking their associated hyperlinks. Next, iPARTS shows the result of its pairwise RNA structural alignments in detail, including alignment score, number of aligned residues, RMSD (root mean square deviation), and resulting alignment of SA-encoded sequences and its corresponding RNA sequence alignment and RNA structural superposition. In the display of RNA structural superposition, the user can visually view, rotate and enlarge 3D structures of input RNA molecules and their structural superposition in a Jmol (an open-source Java viewer for chemical structures in 3D whose web site is at http://www.jmol.org/) window. Notably, in the top panel of this Jmol window, iPARTS provides the user some useful functions for displaying RNA molecules. For example, the user can choose either black or white (default) as window background color, spin RNA molecules or not (default no), display RNA molecules in a scheme of either ribbon, cartoon (default), wirefare or trace, and determine whether to display nucleotide IDs or not (default no). In addition, the user can click the hyperlink of PDB file to download a PDB file containing the superposition of aligned RNA 3D structures. Notice that if the number of suboptimal alignments is set to >1, then the user needs to click the associated hyperlink to display the structural superposition of each suboptimal alignment. We refer the user to the help page of iPARTS for the step-by-step guide of its detailed usage and examples to illustrate the applicability of its provided structural alignments.
EXPERIMENTAL RESULTS
To demonstrate the accuracy improvement of iPARTS over PARTS, as well as other tools, we conducted an experiment on a filtered and non-redundant data set (named data set #1, consisting of 34 families of 100 RNA structures) we newly prepared in this study as follows. Initially, we collected a total of 544 PDB files with 869 RNA chains from the SCOR database (16,17). We then prepared a temporary data set from this collection by removing sequence redundancy at 95% identity. Finally, we obtained data set #1 by further partitioning each RNA family in the temporary data set into several sub-families according to the structural similarity of its constituent RNAs. For the purpose of comparison, we calculated the receiver operating characteristic (ROC) curves of PARTS and iPARTS, as well as stand-alone programs of other tools at the time (i.e. ARTS and LaJolla), on data set #1 based on native alignment score and a geometric match measure, called structural alignment score (SAS) (24,25), where SAS = 100× RMSD/(number of aligned residues). The ROC curve is to depict the trade-off between true-positive rate (i.e. sensitivity) and false-positive rate (i.e. 1 − specificity). The ROC curve for each experiment in this study was obtained as follows. First, the alignments of all pairs of RNA structures are sorted by their native alignment or SAS score. A threshold is then varied between the maximum and minimum of the sorted alignment/SAS scores for producing the points of the ROC curve. For a fixed threshold, all pairs of aligned RNA structures whose alignment/SAS scores are above the threshold are assumed positive and all below it negative. Moreover, the pairs assumed positive are counted as true positives (TP) if they belong to the same family (i.e. they are structurally similar) and false positives (FP) otherwise (i.e. they are not structurally similar); the pairs assume negative are counted as true negatives (TN) if they do not belong to the same family and false negatives (FN) otherwise. Then a point of the ROC curve corresponding to this fixed threshold is produced by plotting its true positive rate on the y-axis and its false positive rate on the x-axis, where the ‘true positive rate’ is defined as TP/(TP+FN) and the ‘false positive rate’ as FP/(FP+TN).
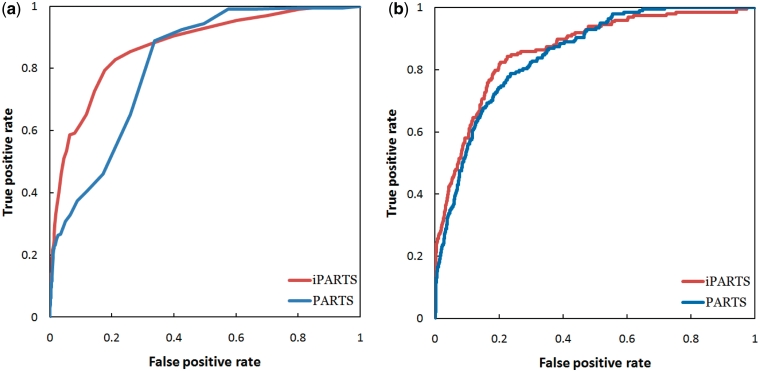
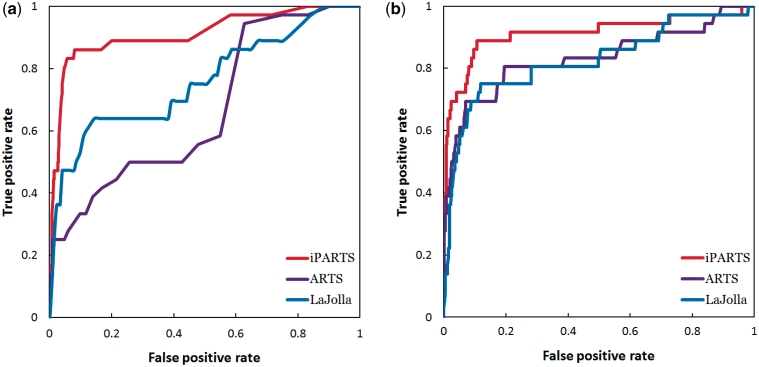
In the experimental results obtained by testing iPARTS and PARTS on data set #1, as shown in Figure 2a, the semiglobal alignment of iPARTS performed much better than that of PARTS, because the AUC (area under ROC curve) of the former ROC curve based on native alignment score is 0.87, while the AUC of the latter is just 0.81. On the other hand, the ROC curve of iPARTS based on SAS score is still better than that of PARTS, because the AUC values of iPARTS and PARTS are 0.86 and 0.85, respectively, as illustrated in Figure 2b. However, when testing ARTS and LaJolla on data set #1, we found that not all pairs of RNA structures can be aligned to successfully yield their common substructures. Two RNA structures can be aligned by ARTS only when each of them possess at least two successive base pairs. For instance, ARTS cannot find a common substructure between two RNA loop structures even though they are similar structurally. LaJolla may also fail to identify a common substructure shared by two RNA structures, when the size of the used n-gram is too large so that no exactly matching n-gram can be found between query and target RNAs. Among 4950 possible pairs of RNA structures within data set #1, there are only 613 and 4251 pairs whose common substructures can be successfully identified by ARTS and LaJolla (using a n-gram size of 3 bp), respectively. To fairly compare the alignment results from ARTS, LaJolla and iPARTS, we calculated their ROC curves based on native alignment score and SAS score using the 613 pairs of RNA structures that can be aligned by ARTS. When sorting the structural alignments by their native alignment score, the comparison of ROC curves in Figure 3a suggest that iPARTS is the best tool. When sorting the structural alignments by their SAS score, iPARTS is still best, as illustrated in Figure 3b. Notice that if we use the 4251 pairs of RNA structures that can be aligned by LaJolla to calculate the ROC curves of LaJolla, then its AUC values based on native alignment and SAS scores are 0.82 and 0.79, respectively.
Figure 2.
The ROC curves of iPARTS and PARTS on data set #1 based on (a) native alignment score, where the AUC values of iPARTS and PARTS are 0.87 and 0.81, respectively, and (b) based on SAS score, where the AUC values of iPARTS and PARTS are 0.86 and 0.85, respectively.
Figure 3.
(a) ROC curves based on native alignment score, where the AUC values of ARTS, LaJolla and iPARTS are 0.65, 0.74 and 0.91, respectively. (b) ROC curves based on SAS score, where the AUC values of ARTS, LaJolla and iPARTS are 0.83, 0.83 and 0.91, respectively.
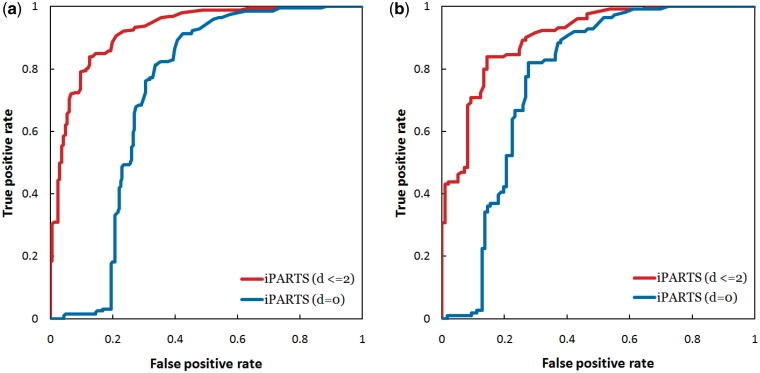
Next, we tested our iPARTS for its capability of RNA function assignment on three data sets (called FSCOR, R-FSCOR and T-FSCOR, respectively) that were prepared by Capriotti and Marti-Renom (7) from the SCOR database on their recent study of SARA. We, here, did not evaluate the accuracies of ARTS and LaJolla in the RNA function assignment because, as explained previously, they were not able to successfully return the common substructures between all pairs of RNA structures in these data sets and hence it is impossible now to make a fair comparison of these two tools with SARA and iPARTS. The FSCOR data set includes 419 RNA chains that were classified into 192 classes, the R-FSCOR data set contains the representative structures of 192 classes in the FSCOR data set and the T-FSCOR data set has all structures of the FSCOR data set not present in the R-SCOR data set. In the study by Capriotti and Marti-Renom (7), two RNA structures have a ‘geodesic distance’ d = 0 if they were annotated with the same function in the SCOR database, and d ≤ 2 if the number of edges between their SCOR function annotations, which are organized in a directed acyclic graph, is ≤2. The evaluation of structure-based function assignment was usually done by searching with a query RNA structure against a representative data set of annotated RNA structures and predicting the function of the query as the annotated function of the top hit RNA structure. For this purpose, Capriotti and Marti-Renom (7) performed two different tests using their SARA tool: (i) a leave-one-out test using the FSCOR data set and (ii) a test using each structure in the T-SCOR data set as the query and searching it against the R-FSCOR data set. As described in (7), SARA resulted in an AUC of 0.61 and 0.83 for d = 0 and d ≤ 2, respectively, on the leave-one-out test and an AUC of 0.58 and 0.85 for d = 0 and d ≤ 2, respectively, on the other test. Here, we repeated these two experiments using our iPARTS tool. Consequently, the AUC values obtained by iPARTS on the leave-one-out test are 0.72 and 0.92 for d = 0 and d ≤ 2, respectively (see Figure 4a for their ROC curves) and 0.77 and 0.90 for d = 0 and d ≤ 2, respectively, on the second test (Figure 4b), suggesting that our iPARTS performs better than SARA on the function assignment of RNA 3D structures.
Figure 4.
The ROC curves when testing our iPARTS for its capability of function assignment using (a) the FSCOR data set, where the AUC values for d = 0 and d ≤ 2 are 0.72 and 0.92, respectively, and (b) the R-FSCOR and T-FSCOR data sets, where the AUC values for d = 0 and d ≤ 2 are 0.77 and 0.90, respectively.
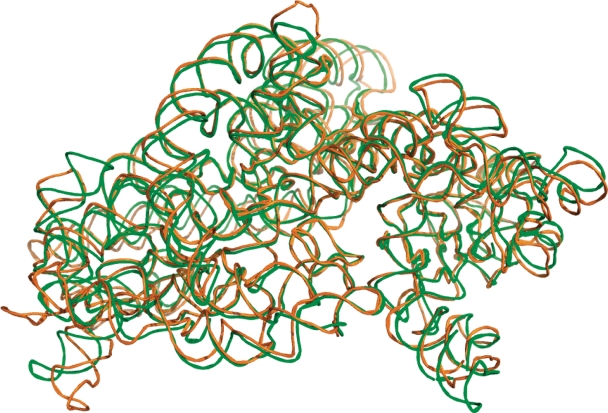
Finally, we tested iPARTS on two 16S rRNA 3D structures of Thermus thermophilus (PDB ID: 1J5E; NDB ID: RR0052, chain ID: A, length: 1513 bp) and Escherichia coli (PDB ID: 2AVY; NDB ID: RR0123, chain ID: A, length: 1530 bp) to demonstrate its capability for aligning large RNAs, which still remains a challenge to date due to their large size. Consequently, as shown in Figure 5, our iPARTS returned the global alignment of these two 16S rRNAs in 172.5 s with an RMSD of 7.491 Å. In Table 1, we show a comparison of average CPU time for RNA structural alignment tools of ARTS, PARTS, SARA, LaJolla and iPARTS. For the purpose of this comparison, we chose four data sets that contain RNA 3D structures at different scale of length: (i) five tRNAs (1EHZ:A, 1H3E:B, 1I9V:A, 2TRA:A and 1YFG:A) with an average structure length of 76 bp, (ii) three ribozyme P4-P6 domains (1GID:A, 1HR2:A and 1L8V:A) with an average structure length of 157 bp, (iii) two domains V of 23S rRNA (1FFZ:A and 1FG0:A) with an average structure length of 496 bp, and (iv) two 16S rRNAs (1J5E:A and 2AVY:A) with an average structure length of 1522 bp. An all-against-all comparison within each data set was then performed using all the tools mentioned above with their default parameters. Notice that LaJolla was performed using its stand-alone program with a default n-gram size of 7 bp (because it currently provides no web server for public access), while other tools were performed via their web servers. As indicated in Table 1, for the RNA structures with moderate length of <160 bp, all tools, except ARTS, can finish their job within several seconds. However, for the RNA structures with length >490 bp, our iPARTS and PARTS are the fastest tools.
Figure 5.
Superposition display of iPARTS global alignment between two 16S rRNA 3D structures of T. thermophilus (PDB ID: 1J5E, chain ID: A, length: 1513 bp) colored green and E. coli (PDB ID: 2AVY, chain ID: A, length: 1530 bp) colored orange with an RMSD of 7.491 Å.
Table 1.
Comparison of average CPU time for various RNA structural alignment tools
| Data set | ARTS | PARTS | SARA | LaJolla | iPARTS |
|---|---|---|---|---|---|
| tRNA | 38.7 s | 2.8 s | 5 s | 1.1 s | 1 s |
| Ribozyme P4-P6 domain | 52.4 s | 5.5 s | 12.9 s | 5.2 s | 2.8 s |
| Domain V of 23S rRNA | 79.8 s | 22.1 s | N/A | 119 s | 17.7 s |
| 16S rRNA | 3.4 min | 2.3 min | N/A | 5 h 9 min | 2.9 min |
Notice that LaJolla was performed using its stand-alone program (because it currently provides no web server for public access), while other tools were performed via their web servers. All these tools were tested using their default parameters. At the time of our testing, the SARA web server cannot deal with domain V of 23S rRNA as well as 16S rRNA.
In (26), Murray et al. proposed the concept of sugar-to-sugar suite unit and used it to define 42 RNA backbone rotamers, each of which is represented by two letters, according to the distributions of multi-dimensional backbone torsion angles. Recently, Richardson et al. (27) further refined and updated this work by proposing 46 RNA backbone rotamers. It will be interesting to further study whether the SA consisting of such 46 RNA backbone rotamers can be used to produce more accurate alignments between RNA 3D structures, when compared to the one we used in this study.
SUMMARY
In this study, we have developed a web-based tool iPARTS that allows the user quickly and accurately to perform global, semiglobal, local and normalized local alignments of two (large-scale) RNA 3D structures. We have also demonstrated that iPARTS outperforms PARTS, as well as ARTS and LaJolla, on the pairwise RNA structural alignments and also outperforms SARA on the function assignment of RNA tertiary structures. Therefore, we believe that iPARTS can serve as a useful tool in the study of structural and functional biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council of Republic of China grant NSC97-2221-E-009-081-MY3. Funding for open access charge: National Science Council of Republic of China.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kolodny R, Linial N. Approximate protein structural alignment in polynomial time. Proc. Natl Acad Sci. USA. 2004;101:12201–12206. doi: 10.1073/pnas.0404383101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dror O, Nussinov R, Wolfson HJ. ARTS: alignment of RNA tertiary structures. Bioinformatics. 2005;21:47–53. doi: 10.1093/bioinformatics/bti1108. [DOI] [PubMed] [Google Scholar]
- 3.Dror O, Nussinov R, Wolfson HJ. The ARTS web server for aligning RNA tertiary structures. Nucleic Acids Res. 2006;34:W412–W415. doi: 10.1093/nar/gkl312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrè F, Ponty Y, Lorenz WA, Clote P. DIAL: a web server for the pairwise alignment of two RNA three-dimensional structures using nucleotide, dihedral angle and base-pairing similarities. Nucleic Acids Res. 2007;35:W659–W668. doi: 10.1093/nar/gkm334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang Y-F, Huang Y-L, Lu CL. SARSA: a web tool for structural alignment of RNA using a structural alphabet. Nucleic Acids Res. 2008;36:W19–W24. doi: 10.1093/nar/gkn327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capriotti E, Marti-Renom MA. RNA structure alignment by a unit-vector approach. Bioinformatics. 2008;24:i112–i118. doi: 10.1093/bioinformatics/btn288. [DOI] [PubMed] [Google Scholar]
- 7.Capriotti E, Marti-Renom MA. SARA: a server for function annotation of RNA structures. Nucleic Acids Res. 2009;37:W260–W265. doi: 10.1093/nar/gkp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer RA, Rother K, Moor P, Reinert K, Steinke T, Bujnicki JM, Preissner R. Fast structural alignment of biomolecules using a hash table, n-grams and string descriptors. Algorithms. 2009;2:692–709. [Google Scholar]
- 9.Wadley LM, Keating KS, Duarte CM, Pyle AM. Evaluating and learning from RNA pseudotorsional space: quantitative validation of a reduced representation for RNA structure. J. Mol. Biol. 2007;372:942–957. doi: 10.1016/j.jmb.2007.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duarte CM, Pyle AM. Stepping through an RNA structure: a novel approach to conformational analysis. J. Mol. Biol. 1998;284:1465–1478. doi: 10.1006/jmbi.1998.2233. [DOI] [PubMed] [Google Scholar]
- 11.Hershkovitz E, Tannenbaum E, Howerton SB, Sheth A, Tannenbaum A, Williams LD. Automated identification of RNA conformational motifs: theory and application to the HM LSU 23S rRNA. Nucleic Acids Res. 2003;31:6249–6257. doi: 10.1093/nar/gkg835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hershkovitz E, Sapiro G, Tannenbaum A, Williams LD. Statistical analysis of RNA backbone. IEEE/ACM Transaction on Computational Biology and Bioinformatics. 2006;3:33–46. doi: 10.1109/TCBB.2006.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 14.Xu R, Wunsch DI. Survey of clustering algorithms. IEEE Transactions on Neural Networks. 2005;16:645–678. doi: 10.1109/TNN.2005.845141. [DOI] [PubMed] [Google Scholar]
- 15.Abraham M, Dror O, Nussinov R, Wolfson HJ. Analysis and classification of RNA tertiary structures. RNA. 2008;14:2274–2289. doi: 10.1261/rna.853208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klosterman PS, Tamura M, Holbrook SR, Brenner SE. SCOR: a structural classification of RNA database. Nucleic Acids Res. 2002;30:392–394. doi: 10.1093/nar/30.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura M, Hendrix DK, Klosterman PS, Schimmelman NR, Brenner SE, Holbrook SR. SCOR: structural classification of RNA, version 2.0. Nucleic Acids Res. 2004;32:D182–D184. doi: 10.1093/nar/gkh080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Needleman S, Wunsch C. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Evol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 21.Setubal J, Meidanis J. Introduction to Computational Molecular Biology. Boston: PWS Publishing Company; 1997. [Google Scholar]
- 22.Smith T, Waterman M. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 23.Arslan AN, Eğecioğlu O, Pevzner PA. A new approach to sequence comparison: normalized sequence alignment. Bioinformatics. 2001;17:327–337. doi: 10.1093/bioinformatics/17.4.327. [DOI] [PubMed] [Google Scholar]
- 24.Subbiah S, Laurents DV, Levitt M. Structural similarity of DNA-binding domains of bacteriophage repressors and the globin core. Curr. Biol. 1993;3:141–148. doi: 10.1016/0960-9822(93)90255-m. [DOI] [PubMed] [Google Scholar]
- 25.Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J. Mol. Biol. 2005;346:1173–1188. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray LJ, Arendall WB, Richardson DC, Richardson JS. RNA backbone is rotameric. Proc. Natl Acad. Sci. USA. 2003;100:13904–13909. doi: 10.1073/pnas.1835769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, et al. RNA backbone: consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.