Abstract
MPlot is a webserver that provides a quick and easy way for structural biologists to analyze, visualize and plot tertiary structure contacts of helical membrane proteins. As input, experimentally determined or computationally modeled protein structures in PDB format are required. The automatic analysis concatenates in house tools to calculate cut-off dependent van der Waals contacts or crossing angles of transmembrane helices with third party tools to compute main chain or side chain hydrogen bonds or membrane planes. Moreover, MPlot allows new features and tools to be added on a regular basis. For that purpose, MPlot was embedded in a framework that facilitates advanced users to compose new workflows from existing tools, or to substitute intermediate results with results from their (own) tools. The outputs can be viewed online in a Jmol based protein viewer, or via automatically generated scripts in PyMOL. For further illustration, the results can be downloaded as a 2D graph, representing the spatial arrangement of transmembrane helices true to scale. For analysis and statistics, all results can be downloaded as text files that may serve as inputs for or as standard data to validate the output of knowledge based tertiary structure prediction tools.
INTRODUCTION
Helical membrane proteins operate at the interface of the different cell compartments. They are involved in various clinical relevant cell-mediated processes such as immune response, signaling or homeostasis. Intra-membrane proteases are crucial for the pathogenesis of severe diseases such as cancer and Alzheimer’s disease (1). Human membrane proteins are therefore relevant drug targets (2) and consequently at the focus of many structural biologists (3). Knowing their tertiary structure is not only essential for protein-based virtual screenings of chemical databases (4), but also to gain detailed insights into the structure–function relationship of these proteins that account for about 30% of all proteins in the different genomes.
Despite recent progress in the crystallization of membrane proteins (3,5,6) still very few structures are known compared to water soluble proteins. At the moment 1.8% (Feb 2010 http://pdbtm.enzim.hu/) of the proteins deposited in the protein data bank (PDB) account for membrane proteins (7,8). For proteins sharing a sequence identity of at least 30–50% with a structural template, homology modeling is a well established method (9) to obtain valuable tertiary structure models. In other cases low resolution models are constructed using specialized knowledge based approaches (10–12). Most of these methods profit from sequence structure relationships derived from statistical analysis of known tertiary structures. However, specialized and easy to use tools to analyze helical membrane protein structures are still sparse. In the following we will shortly review some important structural features of helical membrane proteins together with available tools applicable for their analysis.
Most helix pairs in membrane proteins are not arranged parallel to each other, but cross at different right- or left-handed crossing angles (13). These packing motifs are relevant for the proteins’ functions. The right-handed packing mainly found in channels allows much greater flexibility than the left handed packing overrepresented in membrane-coils, that constitute a class of membrane proteins whose structures are expected to be more rigid (14). In right handed packing, the side chains point away from the packing interface (15). In left handed packing motifs, there is an interdigitation of side chains and consequently a preference for anti parallel ‘tightly packed’ arrangements. Detailed analysis of the sequence structure relationship has shown that right handed helix pairs are mainly arranged from octad repeat patterns of small and medium polar amino acids, while left handed helix pairs are arranged from heptad repeat patterns of bulky and polar residues (16). For example, the octad repeat GxxxGxxxG and related motifs are well known to promote right-handed helix–helix packings (17). These findings have been proven valuable for the prediction of structural features such as helix–helix and helix–membrane interactions (16,18). However, tools to quickly evaluate these packing features are still missing.
The driving forces for tertiary structure folding of helical membrane proteins are still a matter of debate (19). Various forces like van der Waals interactions, hydrogen bonding or entropic effects contribute energetically to the stability of helical membrane proteins (20,21). The hydrophobic effect, namely the gain in entropy when residues are dissolved in water is the likely driving force of the folding of water soluble globular proteins. However, within the lipid bilayer, the hydrophobic effect is nearly absent. Therefore, other forces must energetically compensate for the absence of the hydrophobic effect within the membrane. The application of different mathematical methods to estimate the contribution of van der Waals forces to the stability of helical membrane proteins resulted in a conflict of statements (22,23). Accordant to the occluded surface method helices of membrane proteins have higher atomic packing densities than water soluble proteins (22). As a consequence, van der Waals forces would contribute significantly to their stability. Applying the Voronoi Cell method a contrary conclusion was made (23). For most structural or computational biologists it would be very laborious to reassess the outcome of these analyses, or to easily repeat the analysis for their own data. Therefore, we lately published Voronoia, an online version allowing recalculating, updating and reproducing the results mentioned above (24).
Hydrogen bonded networks are an invaluable source to elucidate the stability or the dynamics of biological macromolecules (14,25). The strength of hydrogen bonds depends on the distance, the chemistry, and the relative arrangement of donor and acceptor atoms and the nature of the surrounding milieu, e.g. its dielectric constant ε (26,27). Within the lipid bilayer ε is low and the strength of interhelical hydrogen bonds is considered to be high, so that in this milieu, even Cα-H—H-N main chain hydrogen bonds may contribute to the stability of the protein (17). Nevertheless, the exact value of ε in the protein interior is difficult to estimate. It is influenced by the exact position of the hydrogen donor and acceptor atoms relative to the lipid bilayer and by their accessibility to polar or lipophilic solvents. Only few tools for the calculation of hydrogen bonds as HBPlus (28) and Hbexplore (27) are available. But, there is currently no web tool accessible that combines these features with an up-to-date and intuitive user interface.
Generally, the landscape of modeling and structure analysis tools is quite cluttered. There are several free available applications for protein visualization and modeling as the Swiss-PDB viewer (29) or the extendible molecular viewers PyMOL (30) and Chimera (31). These tools also allow some very profound structural analyses i.e. the calculation of electrostatic surfaces or the generation of Ramachandran plots. Other web-based tools, as the Ligand Explorer build with the Molecular Biology Toolkit (32) provide details for hydrophilic and hydrophobic interactions between protein and ligand at different cutoff distances. The TMDET web server classifies membrane proteins and calculates membrane planes by determining their position relative to the position of atomic coordinates (33). However, at the moment, there is no specific program for a comprehensive analysis of structural features of helical membrane proteins.
Here we present MPlot, a framework for membrane protein structure analysis. It is designed based on the Galaxy framework that has originally been implemented for genomic data. This framework was successfully adapted for machine learning based tools for sequence and tiling array data analysis before (34). Here, we reconstructed it for MPlot, a web server that provides a completely integrated environment for the comprehensive analysis of hydrogen bonds, the calculation of helix crossing angles and the calculation of cut-off dependent van der Waals contacts. The results of these analyses are automatically computed from a membrane protein structure in PDB format. Results can either be downloaded as a table, viewed online in the Jmol (35) based protein viewer or in PyMOL on a local computer running a script. For illustration most results can be depicted by a 2D transmembrane helix interaction graph. It shows a clearly laid out view of the calculated interaction measures while retaining the relative helix positioning in the middle of the membrane.
DESCRIPTION/RESULTS
The framework
The galaxy framework is a web application for the analysis of genomic data that is heavily developed and used by a thriving community (36). The advantage of such a framework is that it integrates many different tools, so that they become available through a common user interface, which simplifies their utilization. To adapt it for MPlot, we extended it by several tools, data types and visualizations required for our protein structure analyses. Shortly, a data type for PDB files that allows the visualization of protein structures in a Jmol based protein viewer was created, which also includes a graphical interface to select and highlight chains, residues or individual atoms. In this viewer outputs from other tools as the membrane planes, membrane exposed versus buried residues or atoms and hydrogen bonds are also visualized.
A graphical designer assists in creating workflows from individual tools, given that the output format of a tool is a valid input format of another tool. Such a workflow could include i.e. the analysis of interhelical hydrogen bonds between residues lying within the membrane, or those being positioned outside. Importantly, the framework also keeps track of every analysis and dataset produced, so that they can be inspected anytime later. Therefore, the framework enables functions to facilitate collaborative analysis. All datasets and workflows can be shared among users. This way the workflow of an analysis and all its associated datasets can be provided in a completely transparent way to colleagues or as a supplement alongside of a publication (36).
To use this data management and collaboration functionality a log in is required. For simple access the MPlot pipeline is provided separately as a ready to use tool that constitutes the membrane protein analysis without need to create a special workflow. A detailed introduction on the main functions of our membrane protein analysis web server is available at http://proteinformatics.charite.de/mplot/static/howto.html. From a developers perspective it is worth noting, that the framework greatly simplifies the process of adding new tools for helical membrane protein analysis and visualization. Of course, all programs part of the MPlot pipeline are also available as separate tools. This means that i.e. hydrogen bonds in water soluble proteins can also be calculated and visualized.
The MPlot pipeline
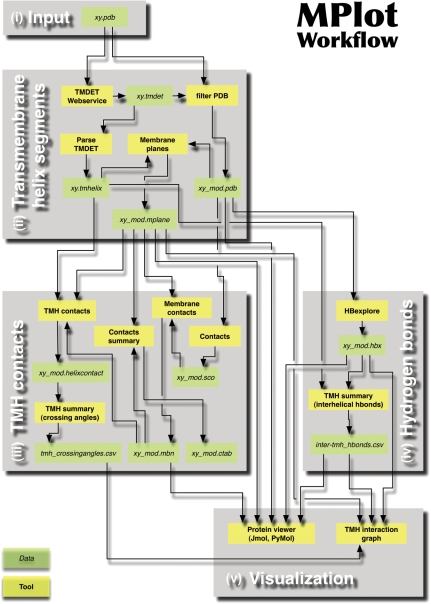
For simple access of the membrane protein analysis framework the pipeline MPlot is provided as a ready to use compact tool. It integrates all presently available tools in the framework that could as well be combined to a workflow by the user. Therefore, the MPlot pipeline is an appropriate and fast way to access the full range of the present functionalities given by our web server. Given the appropriate input, the following steps are done automatically (Figure 1).
Figure 1.
The MPlot workflow presented as a chart providing an overview of all tools (yellow boxes), their outputs (green boxes) and interdependencies (arrows). A more detailed description is provided in the running text. (i) With the PDB file as input, MPlot performs the following calculations automatically: (ii) First transmembrane segments, membrane planes and the biological unit are predicted using the TMDET web service (33) and are pre-processed for further use. (iii) Then atomic contacts between helices, the membrane and other structure elements are specified. If at least two atomic contacts between a helix pair are observed, the crossing angle is calculated. (iv) Next, interhelical hydrogen bonds are extracted from the complete list of potential hydrogen bonds identified by HBexplore (27). (v) For visualization the location and distances of the transmembrane helices, hydrogen bonds and crossing angles are depicted by a TMH–2D interaction graph. The 3D-structure of the uploaded protein can be visualized in conjunction with the interhelical hydrogen bonds, the membrane exposed residues and the membrane planes by using the Jmol based protein viewer or the script for PyMOL.
Input
MPlot requires a file in PDB format as input, to be uploaded by the user. Alternatively a PDB file can automatically be directly obtained from the PDB data base by using the ‘get PDB’ tool and by specification of a four-character PDB code (8).
Transmembrane helix segments determination
The TMDET server is queried by the ‘TMDET’ tool to determine the membrane spanning regions and the biological unit (33). The results file (.tmdet) specifies the biological unit that is used for further calculations. It also contains the coordinates of the membrane planes and the structural elements that lie inside and outside of the membrane bilayer. From the latter information the membrane planes are recalculated with ‘Membrane planes’. This tool basically places two parallel planes in such a way in the protein structure, that the distance from beginnings and ends of the membrane spanning regions (extracted by ‘Parse TMDET’) to the planes is minimized. The helical sections lying between these two planes are defined as transmembrane spanning Helices (TMH).
TMH contacts
Knowing the membrane spanning regions, lipid accessibility and crossing angles between TMHs can be calculated. For the calculation of atomic contacts between structural elements several issues have to be addressed. First, secondary structures have to be assigned. We applied the DSSP algorithm (33) to assign alpha helices and beta sheets but also atomic contacts to water molecules and other hetero atoms. Second, the atomic contacts between TMHs or with the membrane have to be defined using ‘Contacts’, a tool that was applied for some analyses before (11,13,32). The membrane accessible surface (MAS) is determined through a 1.4 Å radius probe rolled along the van der Waals spheres of atoms lying at the protein surface enclosed by the membrane planes. The MAS is then defined by all atoms touched by that probe. Atomic contacts between the van der Waals surfaces of two neighboring structure elements (TMHs, MAS, coils or hetero atoms) are defined dependent on the chosen distance cutoff value. Instead of limiting this measure to a single value, the contacts are calculated and stored for cutoff values ranging from −0.5 to 2.8 Å. This allows the user to specify the distance cutoff values when creating statistics or during visualization (see v). The raw data are stored in the .sco file and the .mbn file together with the atomic coordinates of the PDB file. The former output file is also available for water soluble globular proteins. The latter additionally includes the 3D coordinates of the membrane planes and information on contacts with the MAS. The raw data are further processed using ‘Contacts summary’. The output .ctab file contains the frequency of atomic contact types per residue in a tabular format.
For the evaluation of helix–helix crossing angles, only TMH pairs that are connected by at least two residues (interhelical distance <1.5 Å) are considered. Since transmembrane helices are regularly kinked or curved, the crossing angle is identified using local instead of global helix axes. The local axis is defined by the line that minimizes the distances to the coordinates of the backbone atoms of the TMH segment actually involved in the contact plus one additional turn at both termini. Thus helices are only considered in their entirety for pairs of straight helices with virtually parallel arrangements. For each pair of axes (‘a’,’b’), axis ‘a’ is projected along the vector representing the shortest distance between the lines ‘a’ and ‘b’ onto axis ‘b’ yielding a new line ‘ap’. The crossing angle is then defined as the angle between the intersecting lines ‘ap’ and ‘b’. These calculations are done by ‘TMH contacts’ written to the file .helixcontact, which is further processed by ‘TMH summary’ to extract the list of helix pairs and their crossing angles. This list is then available in the ‘.hcsummary’ dataset and visualized in the TMH–2D interaction graph together with interhelical hydrogen bonds.
Hydrogen bonds
A set of potential hydrogen bonds is identified with the HBexplore program, which selects all potential hydrogen bonds according to geometrical criteria (27). The integrated version of HBexplore gives access to all configuration options of the original command line program. Accordingly, the maximum distance and the angle between donor and acceptor atoms can be specified. The calculation of Cα-H—H-N main chain hydrogen bonds can additionally be integrated into the analysis. Inter helical TMH hydrogen bonds are extracted from the complete list by removing all hydrogen bonds with donor or acceptor atoms outside of the predicted lipid bilayer or within the same TMH.
Visualization
The TMH–2D interaction graph (Figures 2A and 3A) provides a clearly laid out view of the calculated TMH interaction measures while retaining the relative helix positioning inside the membrane. It provides a cross-section of the membrane protein, where the cutting plane is placed in the center of the membrane. The graphs’ nodes represent the membrane helices and directly correspond to their positions in the center of the membrane. The edges between the nodes (helices) are labeled with information about the crossing angles between two TMHs and the number of interhelical hydrogen bonds. All labels can be switched on or off. The graph can also be modified so that only certain protein subunits, chains or TMHs are depicted. Since it is available in SVG format (http://www.w3.org/Graphics/SVG/About.html) the graph also provides a useful template for the user to add additional information.
Figure 2.
Visualization of the MPlot analysis’ results of the Mechanosensitive Channel of Large Conductance (PDB id 1msl). (A) TMH–2D interaction graph. The blue circles represent the membrane helix positions in the middle of the membrane; the green lines hold the crossing angle between two helices; the second number denotes the number of interhelical hydrogen bonds. (B) The protein viewer, showing the membrane interface (yellow), the membrane planes (blue) and potential hydrogen bonds (blue, red).
Figure 3.
Visualization of the MPlot analysis’ results of Bacteriorhodopsin in purple membrane (PDB id 2brd). The lipids were temporarily removed during the analysis. Instead they were simulated as described in the running text. All potential hydrogen bonds (blue, red) are shown. For more details see Figure 2.
The TMH–2D graph is constructed as follows: A plane ‘mp’ in the middle of the membrane is derived from the two membrane planes representing the membranes cytosolic and extracellular border respectively. Next, the intersection points of the plane ‘mp’ and the axes of the transmembrane helix segments are calculated. This set ‘s’ of intersection points is then transformed into a new 3D cartesian coordinate system where the plane ‘mp’ is parallel to a plane spanned by two coordinate axes of the new system. The set ‘s’ of intersection points is then orthogonally projected onto the middle plane ‘mp’. This projection represents a view perpendicular to the membrane borders and the set of intersection points is now interpreted as the locations of the nodes for the TMH interaction graph.
For 3D visualization two well-known protein viewers can be used: (i) a PyMOL script is supplied to download and visualize the results in the PyMOL molecular viewer (Figures 2B and 3B). The script encapsulates all MPlot results data in a self contained file that can be executed in PyMOL via the ‘run’ command. (ii) the Jmol based protein viewer is a web application with an intuitive user interface specifically designed for the MPlot data. It allows i.e. selecting inter TMH hydrogen bonds, main chain hydrogen bonds or specifying the cut-off dependent atomic contacts between TMHs and MAS. The viewer facilitates the exploration of the MPlot analyses results without any need to install other programs or to download the data. To quickly find and highlight specific residues, a tree-like graphical interface to select and highlight chains, residues or individual atoms known from other visualization tools as the DeepView—Swiss-PdbViewer or the DS Viewer (Accelrys Software, Inc.) is also included. For more advanced users the Jmol scripting interface is exposed through a convenient console. To minimize waiting times the analysis data are only dynamically loaded upon request.
CONCLUSION
The MPlot membrane protein analysis framework integrates tools for the analysis and visualization in a web based, easy to use workbench that also provides functionality for sharing data, analyses and workflows. Analyses and associated datasets can be supplied in a completely transparent way to others enhancing reproducibility or updating of results. The analyses of tertiary structure contacts and geometrical features of helical membrane proteins, however is at the present stage, by no means exhaustive. More tools will be integrated to address other issues dealing with membrane protein structures or to simply broaden the analysis by adding alternative tools for existing analyses. The framework in which MPlot is integrated facilitates such extensions. By reducing the complexity of installing and maintaining programs, MPlot allows us and also other researchers to instantly deal with their tasks at hand and less with the administrative problems around them. Ideas for new tools and requests to integrate existing tools are therefore most welcome.
MPlot targets not only computer scientists but all structural biologists dealing with membrane protein structures. Therefore it was constructed as an easy to use and automated tool. However, it is important to note that not all kinds of analyses may work properly without user interference. Some helical membrane proteins such as the Photosystem II contain a large number of hetero atoms that must be included in the calculation of the lipid accessible surface, because otherwise parts of the protein would be misleadingly identified as lipid accessible. While this is possible to do with MPlot, it requires user intervention, e.g. a custom workflow must be created with the appropriate configuration option set and the user must make sure the PDB file does not contain lipids bound to the protein surface that could be falsely seen as hetero atoms belonging to the protein complex.
To address such difficulties we plan to create a curated dataset of MPlot results for all helical transmembrane proteins that will be carefully checked for the problems described above. While some problems can in time be overcome adding additional tools and new methods, there will always be unforeseen complexities that cannot be overcome in an automatic way, but need a human curator. The tight integration of analysis and visualization in MPlot will provide support for this task, by automating repetitive, non-analysis related burdens.
Technical notes
To use MPlot a fairly recent web browser is needed. We have tested the web application to work with Safari 3 & 4, Firefox 3 to 3.6 and Internet Explorer 7 & 8. However, compared to the other browsers, the Internet Explorer has a poor JavaScript performance, which results in a slow user interface, so that we recommend using one of the available alternatives.
The web application uses the Galaxy framework (http://galaxy.psu.edu/) for tool and data integration. The protein viewer uses Jmol to provide the molecular visualization and the Jmol JavaScript library is used for communication between the Jmol applet and the rest of the website. The Jmol applet requires a Java JRE that is freely available from http://java.net. The user interface makes extensive use of the jQuery JavaScript library (http://jquery.com/), which is also used for the Ajax based communication between the website and the server to retrieve the MPlot results’ data for visualization.
FUNDING
Deutsche Forschungsgemeinschaft [DFG HI 1502/1-1, SFB 740]; European Research Council [ERC Advanced Grant TUDOR]. Funding for open access charge: Deutsche Forschungsgemeinschaft [DFG HI 1502/1-1, SFB 740]; European Research Council [ERC Advanced Grant TUDOR].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Juergen Suehnel for kindly providing the HBexplore algorithm. Without the use of free and/or open source software this work would not have been possible.
REFERENCES
- 1.Strooper BD, Vassar R, Golde T. The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 2010;6:99–107. doi: 10.1038/nrneurol.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker OM, Marantz Y, Shacham S, Inbal B, Heifetz A, Kalid O, Bar-Haim S, Warshaviak D, Fichman M, Noiman S. G protein-coupled receptors: in silico drug discovery in 3D. Proc. Natl Acad. Sci. USA. 2004;101:11304–11309. doi: 10.1073/pnas.0401862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 4.Bissantz C, Bernard P, Hibert M, Rognan D. Protein-based virtual screening of chemical databases. II. Are homology models of G-protein coupled receptors suitable targets? Proteins. 2003;50:5–25. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 5.Privé GG. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Katzen F, Peterson TC, Kudlicki W. Membrane protein expression: no cells required. Trends Biotechnol. 2009;27:455–460. doi: 10.1016/j.tibtech.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Tusnady G, Dosztanyi Z, Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forrest L, Tang C, Honig B. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys. J. 2006;91:508–517. doi: 10.1529/biophysj.106.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehnert U, Xia Y, Royce T, Goh C, Liu Y, Senes A, Yu H, Zhang Z, Engelman D, Gerstein M. Computational analysis of membrane proteins: genomic occurrence, structure prediction and helix interactions. Quart. Rev. Biophys. 2004;37:121–146. doi: 10.1017/s003358350400397x. [DOI] [PubMed] [Google Scholar]
- 11.Punta M, Forrest L, Bigelow H, Kernytsky A, Liu J, Rost B. Membrane protein prediction methods. Methods. 2007;41:460–474. doi: 10.1016/j.ymeth.2006.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleishman S, Harrington SE, Friesner R, Honig B, Ben-Tal N. An automatic method for predicting transmembrane protein structures using cryo-EM and evolutionary data. Biophys. J. 2004;87:3448–3459. doi: 10.1529/biophysj.104.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walters RFS, DeGrado WF. Helix-packing motifs in membrane proteins. Proc. Natl Acad. Sci. USA. 2006;103:13658–13663. doi: 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildebrand PW, Gunther S, Goede A, Forrest L, Frommel C, Preissner R. Hydrogen-bonding and packing features of membrane proteins: functional implications. Biophys. J. 2008;94:1945–1953. doi: 10.1529/biophysj.107.110395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walther D, Eisenhaber F, Argos P. Principles of helix-helix packing in proteins: the helical lattice superposition model. J. Mol. Biol. 1996;255:536–553. doi: 10.1006/jmbi.1996.0044. [DOI] [PubMed] [Google Scholar]
- 16.Hildebrand PW, Lorenzen S, Goede A, Preissner R. Analysis and prediction of helix-helix interactions in membrane channels and transporters. Proteins. 2006;64:253–262. doi: 10.1002/prot.20959. [DOI] [PubMed] [Google Scholar]
- 17.Senes A, Ubarretxena-Belandia I, Engelman D. The Calpha —H … O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc. Natl Acad. Sci. USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rose A, Lorenzen S, Goede A, Gruening B, Hildebrand PW. RHYTHM–a server to predict the orientation of transmembrane helices in channels and membrane-coils. Nucleic Acids Res. 2009;37:W575–W580. doi: 10.1093/nar/gkp418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowie J. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 20.Harrington SE, Ben-Tal N. Structural determinants of transmembrane helical proteins. Structure. 2009;17:1092–1103. doi: 10.1016/j.str.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Helms V. Attraction within the membrane. Forces behind transmembrane protein folding and supramolecular complex assembly. EMBO Rep. 2002;3:1133–1138. doi: 10.1093/embo-reports/kvf245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eilers M, Shekar S, Shieh T, Smith S, Fleming P. Internal packing of helical membrane proteins. Proc. Natl Acad. Sci. USA. 2000;97:5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hildebrand PW, Rother K, Goede A, Preissner R, Frommel C. Molecular packing and packing defects in helical membrane proteins. Biophys. J. 2005;88:1970–1977. doi: 10.1529/biophysj.104.049585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rother K, Hildebrand PW, Goede A, Gruening B, Preissner R. Voronoia: analyzing packing in protein structures. Nucleic Acids Res. 2009;37:D393–D395. doi: 10.1093/nar/gkn769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adamian L, Liang J. Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins. 2002;47:209–218. doi: 10.1002/prot.10071. [DOI] [PubMed] [Google Scholar]
- 26.Baker EN, Hubbard RE. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 1984;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 27.Lindauer K, Bendic C, Suhnel J. HBexplore–a new tool for identifying and analysing hydrogen bonding patterns in biological macromolecules. Comput. Appl. Biosci. 1996;12:281–289. doi: 10.1093/bioinformatics/12.4.281. [DOI] [PubMed] [Google Scholar]
- 28.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 29.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl. 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 30.The PyMOL Molecular Graphics System DeLano Scientific, Palo Alto, CA, USA. Available at: http://www.pymol.org. [Google Scholar]
- 31.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE. The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics. 2005;6:21. doi: 10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tusnady G, Dosztanyi Z, Simon I. TMDET: web server for detecting transmembrane regions of proteins by using their 3D coordinates. Bioinformatics. 2004;21:1276–1277. doi: 10.1093/bioinformatics/bti121. [DOI] [PubMed] [Google Scholar]
- 34.Sonnenburg S, Zien A, Philips P, Rätsch G. POIMs: positional oligomer importance matrices–understanding support vector machine-based signal detectors. Bioinformatics. 2008;24:i6–i14. doi: 10.1093/bioinformatics/btn170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herráez A. Biomolecules in the computer: Jmol to the rescue. Biochem. Mol. Biol. Education. 2006;344:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]
- 36.Pond SK, Wadhawan S, Chiaromonte F, Ananda G, Chung W, Taylor J, Nekrutenko A, Team G. Windshield splatter analysis with the Galaxy metagenomic pipeline. Genome Res. 2009;19:2144–2153. doi: 10.1101/gr.094508.109. [DOI] [PMC free article] [PubMed] [Google Scholar]