Abstract
The RosettaBackrub server (http://kortemmelab.ucsf.edu/backrub) implements the Backrub method, derived from observations of alternative conformations in high-resolution protein crystal structures, for flexible backbone protein modeling. Backrub modeling is applied to three related applications using the Rosetta program for structure prediction and design: (I) modeling of structures of point mutations, (II) generating protein conformational ensembles and designing sequences consistent with these conformations and (III) predicting tolerated sequences at protein–protein interfaces. The three protocols have been validated on experimental data. Starting from a user-provided single input protein structure in PDB format, the server generates near-native conformational ensembles. The predicted conformations and sequences can be used for different applications, such as to guide mutagenesis experiments, for ensemble-docking approaches or to generate sequence libraries for protein design.
INTRODUCTION
Ensembles of conformations are often a better representation of protein native states than a single static structure (1), and several computational methods have been developed to model conformational ensembles consistent with experimental data (2–4). It has moreover been suggested that modeled ensembles describing protein native state dynamics can encompass conformations similar to those a protein adopts in response to binding other molecules or to sequence mutations (2,3,5,6). Thus, ensemble representations have been found useful for improving applications of molecular modeling such as protein-small molecule (7) and protein–protein docking (8) as well as in protein design (9–14).
Methods to generate conformational ensembles are often computationally costly and require expert users. We have implemented a computationally efficient method for flexible backbone modeling called Backrub (5) into the modeling and design software program Rosetta (15,16) co-developed by our lab (2,6,17–22). The Backrub model, developed by the Richardsons’ lab (5) and also implemented by Donald and coworkers (23), is inspired by alternative conformations observed in high-resolution crystal structures of proteins, and has been suggested to capture a significant fraction of small conformational changes proteins undergo in solution (5). We have assessed the Rosetta Backrub model in various applications related to protein structure modeling and design (2,6,20,22). Here we make three common modeling and design applications using Backrub— (I) modeling of structures of point mutations, (II) generation of protein ensembles and computational sequence design on these conformational ensembles and (III) interface sequence plasticity prediction—available via a web server. RosettaBackrub provides a set of empirically determined default parameters and a common user interface for all three applications. Each method was validated and benchmarked using experimental data, as described in the referenced publications from our lab. Each application creates an ensemble of structures rather than a single output structure, to facilitate further analysis and computational method development.
SERVER APPLICATIONS
A common task in biological research is the mutagenesis of a protein. Application I: Modeling of Point Mutations (6) (Figure 1) aims to predict the structural effects of one or more point mutations to guide experimental analyses. In the past several years, a variety of different methods for estimating the energetic contributions of point mutations have been developed and made accessible via web servers [Fold-X (24), I-Mutant2.0 (25), CUPSAT (26), Eris (27), CC/PBSA (28) and Hunter (29)]. Our application builds on the method for computational alanine scanning we developed several years ago (17,30). This method has provided ∼9000 predictions since 2004. It estimates the energetic contribution of interface residues to the stability of a protein–protein interface and has been used for example in applications to receptor recognition in immunology (31–34) and for evaluating docked models of antibody–antigen interactions (35). Here we extend the modeling capabilities to simulate mutations other than to alanine and to include backbone conformational changes. Our method differs from others in that it is based on structural modeling of protein conformations instead of using machine learning approaches (25), takes into account backbone flexibility and has been validated through comparison of modeled and experimentally determined structures of mutants using the dataset described in (36). The RosettaBackrub server provides structural models of the mutated proteins, which can then be further analyzed by other scoring methods to evaluate energetic effects of sequence changes.
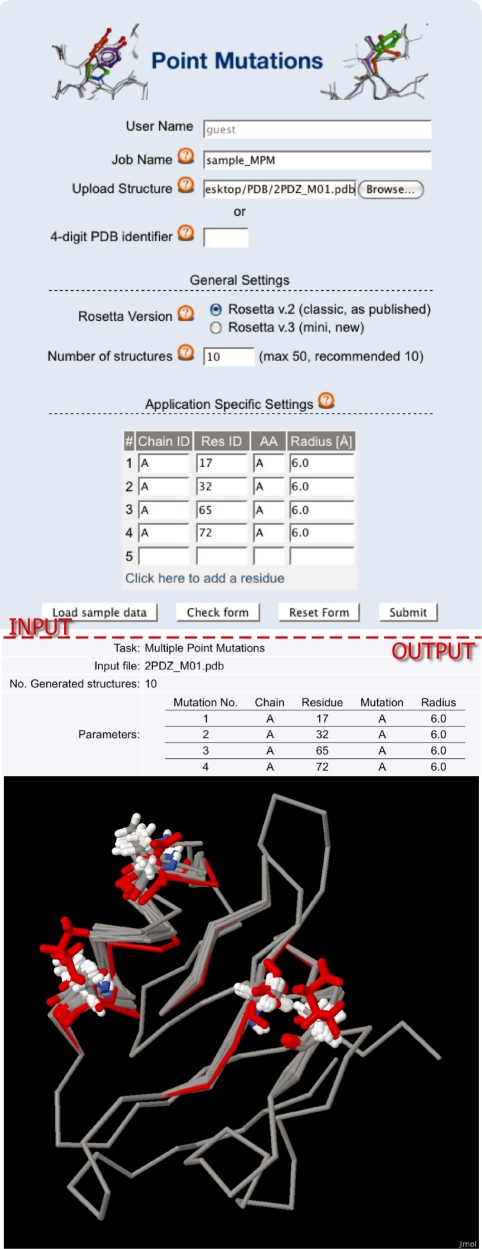
Figure 1.
Application (I) Modeling of Point Mutations. Shown are the input form for a prediction of multiple point mutations (top) and the corresponding output (bottom). The form contains fields to enter a job name to identify jobs later and fields to upload or provide a link to a PDB file. The general simulation settings are identical for all three applications, i.e. the Rosetta version (if applicable) and the number of structures generated. The application-specific settings are the chain identifier (Chain ID), the residue position of the mutation site (Res ID), the target amino acid (AA) and the Backrub radius in Ångströms. The output shows the table of all mutations made in a PDZ-domain from Mus musculus (PDB ID code: 2PDZ) and the Jmol plug-in with the resulting modeled structures. The initially uploaded structure is shown in red.
Application II: Generation of Protein Ensembles/Ensemble Design (2) (Figure 2) produces conformational ensembles based on a single crystal structure that are intended to represent aspects of the native state dynamics of a protein in solution. The application can also be used to generate conformations that may be adopted by other closely related members of the protein’s family. The method has been validated by showing that it generates conformational ensembles consistent with protein dynamical parameters measured by nuclear magnetic resonance (NMR) (2,20) and with the structural variation observed within a naturally occurring protein family (2). The ensembles are also used as a starting point for computational protein design, to predict sequences consistent with the conformations in the ensemble. Such computationally generated sequence families using Rosetta have been shown to resemble sequences observed in the evolutionary family of a protein (2,37). More generally, the RosettaDesign method has been experimentally validated through the design of protein structures (38,39), protein–protein interfaces (18,40,41), protein–DNA interfaces (42) and enzymes (43,44). Most of these applications have used a fixed backbone approximation. Our server adds the capability for an end-user to easily model backbone flexibility in RosettaDesign.
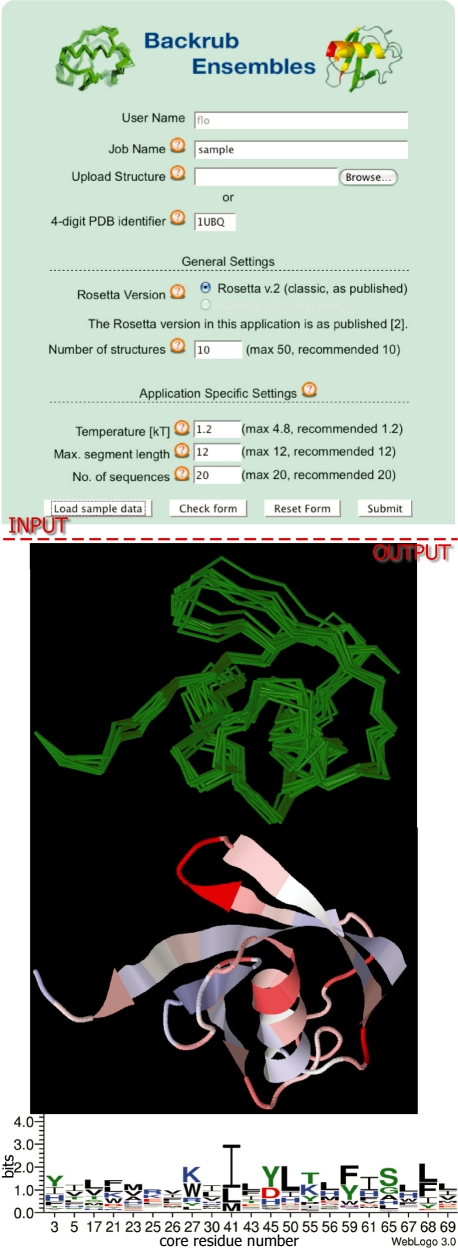
Figure 2.
Application (II) Backrub Ensemble Design. The input form (top) for Backrub Design contains the general settings (as in Figure 1) and in addition application-specific fields for the simulation temperature (controlling the amplitude of the structural variation in the modeled conformational ensemble), and the segment length for the Backrub simulation, as well as the number of sequences generated per structure. The output (bottom) features the backbone traces of 10 generated ensemble members in green and a representation of the dynamic regions of an Ubiquitin ensemble (from blue to white to red with increasing variability) mapped onto the ubiquitin starting structure (PDB ID code: 1UBQ). Furthermore a sequence motif of the designed core residues is shown.
Application III: Protein Interface Sequence Plasticity Prediction (22) predicts the sequence diversity (‘plasticity’) in protein–protein interfaces (Figure 3). The application seeks to find sequence variations in a protein–protein interface that can be tolerated without significantly compromising the stability of the complex and its partners by sampling mutations within the interface region. The method can be used to automatically generate libraries of sequences for protein interfaces that can then be screened experimentally for changes in protein interaction affinity or selectivity. The sequence plasticity prediction method has been validated through comparison of experimentally selected sequences from comprehensive phage display experiments (45) with modeled sequences (22).
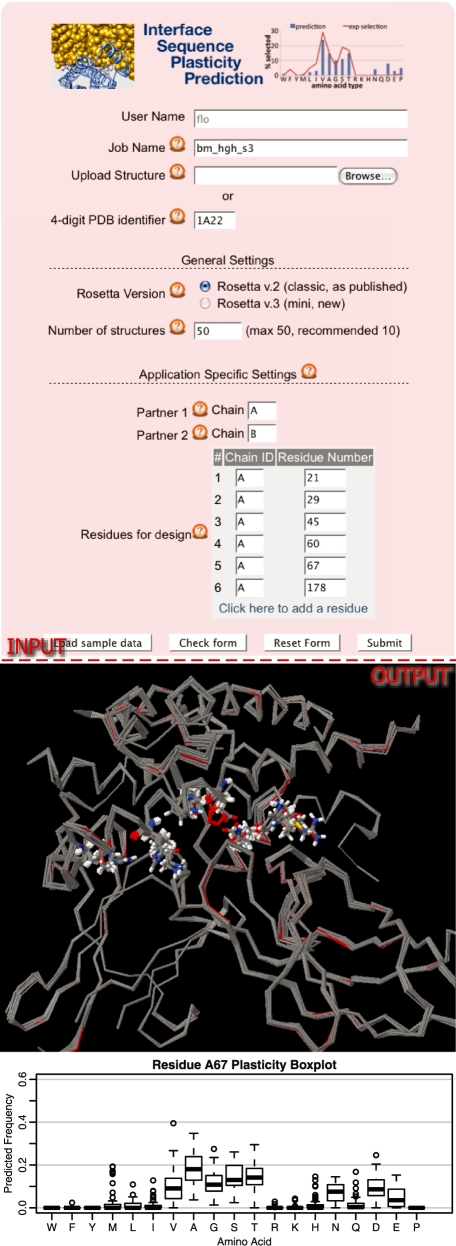
Figure 3.
Application (III) Interface Sequence Plasticity Prediction. The input form (top) holds the general settings (Figure 1) and parameters for the definition of the interface partners and the designed residues. The output (bottom) shows an overlay of the structures of the best scoring sequence for each of 10 initial backrub structures of the human growth hormone receptor complex (PDB ID code: 1A22) and a box-plot of the predicted frequencies of designed amino acid residues at a given designed sequence position (position 67 in chain A).
All applications use the Rosetta software suite (http://www.rosettacommons.org) for predicting and designing protein structures and interactions (15). Today, there are five web servers in use that provide access to applications implemented in Rosetta. Robetta (46) includes ab initio protein-structure prediction, peptide-fragment library generation, DNA interface amino acid affinity/specificity scan (47) and the previous computational alanine scanning protocol for protein–protein interfaces (17,30). RosettaDesign (48) identifies low-energy sequences for specified fixed protein backbones. RosettaDock (49) predicts the structure of a protein complex from individual components and an approximate orientation. The Antibody FV Region Prediction Server (50) uses homology modeling to predict antibody FV structures. Finally, FunHunt (51) is a classifier of correct protein–protein complex orientations. Our server, RosettaBackrub, adds three more applications. Each of the three applications utilizes the Backrub method (5) for flexible protein-backbone modeling and design. This is the first server that generates near-native protein conformational ensembles using Rosetta to model protein mutations, small protein conformational changes upon binding and flexible backbone design of protein sequence libraries. The following section gives a more detailed description of the methods used in each of these applications.
METHODS
Backrub modeling
RosettaBackrub creates ensembles of structures by utilizing the Backrub method (5) for flexible backbone modeling. This method has been derived from observations of alternative conformations in high-resolution crystal structures and involves local backbone rotations about axes between Cα atoms of protein segments. First, a segment of typically 2–12 residues is randomly selected. Then, all atoms of this fragment are rotated as a rigid body by an angle of up to 11°–40° around the axis between the two Cα pivot atoms (see the web server online documentation section for details). Backrub moves, interleaved with rearrangements of the surrounding side chains, are sampled by a Monte Carlo algorithm using the Rosetta all-atom force field (39). Backrub and side chains moves are made 10 000 times, with each new conformation being scored with the Rosetta scoring function. The resulting score is used to determine whether the new conformation is accepted or not according to the Metropolis criterion. In order to create an ensemble of size N this algorithm is independently applied N times to the input structure. Side-chain sampling and scoring in sequence design are as described in (39).
Input structure files
The input to all RosettaBackrub applications is a structure file, the ensemble size to create and a set of application-dependent parameters. The structure file must be provided in PDB format (http://www.wwpdb.org/docs.html). There may be gaps in the structure, but each residue must have a complete set of backbone atoms (N, Cα, C and O); side chain atoms may be missing. Rosetta automatically picks the first conformation if multiple conformations for a single residue are found in the PDB file. Every polypeptide chain needs to have a unique identifier. This identifier is one of the input parameters, and must be identical to the identifier present in the input file (a capital letter in most cases). If the file contains coordinates for hydrogen atoms, those are ignored and automatically rebuilt according to the Rosetta force field. Water and other heteroatoms (e.g. ligands) are not considered in the simulations in the present version. Non-standard amino acids are currently removed and a gap is introduced. While it is possible to upload structures determined by NMR methods, only the first model in the file is used. If a specific structure should be used, the other models need to be deleted from the file.
Application I: Modeling of Point Mutations
This application computationally mutates one or more amino acid residues at position(s) defined by the user, and models backbone and side chain conformational changes in response to the mutations by applying Backrub moves to all residues that have at least one heavy atom within a certain radius of any heavy atom of the residue to be mutated before the mutation is made. This method is described and assessed in (6). The web server implements two flavors of the protocol: single point mutation allows only for one mutation at a time and uses a fixed radius of 6 Å. This protocol was used to benchmark the Backrub method on a set of ∼2000 single point mutations where predictions of modeled side chain conformations could be compared with crystal structures of pairs of wild-type and mutant protein structures (36). The application multiple point mutations allows the user to define more than one point mutation in a given structure. Furthermore, the Backrub radius for each mutation can be set independently. The number of mutations per simulation is limited to 30. The prediction of multiple mutations has not been validated extensively with experimental data (also, available data sets are much smaller). For single point mutations, the accuracy was found to be somewhat dependent on the sampling radius, with a radius of 6 Å for Backrub modeling around the site of the mutation giving optimal performance [see Figures 9–11 in the Supplementary Materials to (6)].
Application (I) input
A single PDB file, definition of the mutation by chain, residue number, amino acid type to be mutated to, and Backrub modeling radius.
Application (I) output
A set of structures containing the mutation(s) in PDB file format, along with the corresponding Rosetta force field scores. The force field scores are not parameterized to estimate changes in protein stability, an application to be added in the future.
Application II: Generation of Protein Ensembles and Ensemble Design
This application is described and assessed in (2). Backrub conformational ensembles are created by applying the Backrub method repeatedly to the entire uploaded structure without any spatial restrictions (no additional parameters are required). The second protocol, Ensemble Design, creates a Backrub ensemble first, for which it requires a simulation temperature in kT and the maximum size of the Backrub segment (see Backrub moves above and the online documentation) from the user. Then, this ensemble is used to predict low-energy sequences consistent with the ensemble structures, using a Monte Carlo simulated annealing protocol as described in (2,52). For each structure of the ensemble, a user-defined number of sequences are designed. These methods were shown to produce conformational ensembles consistent with protein dynamics in solution and sequence ensembles consistent with the structural and sequence variation observed within a naturally occurring protein family (2).
Application (II) input
A single PDB structure, the simulation temperature, the maximum size of a segment (length 3–12) rotated in a Backrub move, and the number of predicted sequences.
Application (II) output
A single PDB file with all structures of the generated ensemble, as well as different representations describing the conformational variability of the ensemble. These include plots and files with the mean Cα root-mean-square deviations (RMSD) of the ensemble, average Cα difference distances matrices, and average Cα difference distance values mapped onto the structure using the B-factor field. Sequences and sequence motifs for the designed ensemble are given as files and logos, respectively.
Application III: Interface Sequence Plasticity Prediction
This application models the tolerated sequence space for interface positions in a protein complex (22). First, Backrub is applied to the two interaction partners in a protein–protein complex to create a conformational ensemble. As in application (II), backbone flexibility is modeled at all positions in each complex partner. Each of the resulting complex structures is then subject to the sequence plasticity protocol. This protocol uses a genetic algorithm to sample amino acid changes at interface positions specified by the user. Modeled residues are scored according to their contributions to the stability of the protein partners as well as to the stability of the protein–protein interface. Interface sequences are recorded and kept if their score is within a threshold from the interface and complex scores of the sequence in the input file, as described in (22). The number of designable interface positions is limited to 10.
Application (III) input
A single PDB structure, a definition of the interaction partners (i.e. the chains that form the interface), and the interface positions that are subject to design.
Application (III) output
The PDB files of the generated ensemble, as well as the sequences and frequencies of amino acids of the designed ensemble.
For each application, relevant information from the Rosetta output is extracted and presented on the web server. The resulting PDB structures are loaded into the Jmol plug-in (http://www.jmol.org/) for immediate inspection. The user can download data such as the generated ensemble conformations, sequences, Cα difference distances and designed amino acid sequences as flat files for further analysis. Depending on the application, the files provide Rosetta force-field scores for each residue. Furthermore, most of this information is also plotted and presented on the website as downloadable image files, created with WebLogo (53), Matplotlib (http://matplotlib.sourceforge.net) and R (http://www.r-project.org/). All files can be downloaded individually or as an archive file.
A more detailed description of the methods can be found in the documentation section of the website and in the publications referenced for each application (2,6,22).
SYSTEM ARCHITECTURE
The web server consists of three parts: a user interface (front-end), a MySQL database and a daemon that controls simulations (back-end). The front-end is implemented in Python and runs as a CGI-script in an Apache web server environment. It allows the user to add a new job to the job-queue and access the resulting data for each simulation. The front-end accesses a MySQL database that stores the initial PDB file, the parameters and the directory where the results can be found. The back-end daemon watches the status of the jobs in the database and starts them, checks whether they are done, and updates the database accordingly. This modular design allows distributing the load onto different computers. A possible scenario is to add the computationally inexpensive processes, such as the web server and the database, to an existing web server, while the actual simulations are distributed onto the nodes of a cluster. Furthermore the back-end is designed in a modular fashion that makes it easy to extend it when new applications become available.
Documentation
The documentation of RosettaBackrub is available online and can be accessed using the ‘Documentation’ link in the menu at the top of every server page. The documentation contains descriptions of the methods, tutorials and benchmarks. The online version enables us to specifically address issues a user might have and share it with others, as well as update benchmark information when new Rosetta versions are implemented.
Registration
This website is free and open to all users and there is no login requirement. It can be used at any time via ‘guest access’. However, registration is possible and provides the advantage for users to receive a notification via email after a simulation finishes.
SERVER PERFORMANCE
Jobs on the web server are processed on a first-come first-served basis. As long simulations are not run on all available processor cores, processor cores are left open to allow short, newly submitted jobs to be processed more quickly. The runtime of all simulations is most dependent on the size of the uploaded structure, the size of the computed ensembles and the number of designed sequence positions for Interface Sequence Plasticity Predictions. Predictions that use design algorithms generally take much longer than simulations of point mutations and ensemble creation. During tests we experienced computing times between 6 and 96 h for Ensemble Design and Interface Sequence Plasticity Prediction. More detailed information on compute times dependent on application type and parameters is available via the web server online documentation to allow for updates when the server setup changes.
Rosetta versions
The Backrub algorithm was first implemented in Rosetta version 2.2 and benchmarked (6). It was shown that the flexible backbone model improves the quality of the prediction of side chains conformations significantly compared to the fixed backbone model. In 2009, a new version of Rosetta (3.0) was released. Rosetta 3.0 was almost entirely rewritten and is the basis for current developments by the Rosetta community. For the basic Backrub applications, i.e. Modeling of Point Mutations and Backrub Conformational Ensembles, RosettaBackrub thus provides two options: first, the original published implementation and second, a current version based on Rosetta 3.x. Since this software is in active development, we plan to upgrade the server as needed. Benchmarks for the respective most recent version can be found on our online documentation. Both design applications Ensemble Design and Interface Sequence Plasticity Prediction are currently provided as described in (2) and (22), respectively.
FUNDING
National Science Foundation (EF-0849400, MCB-0744541); National Institute of Health Roadmap Initiative in Nanomedicine (2PN2EY016525; 2PN2EY016546); DOD NDSEG fellowship program and the Genentech Scholars program (to C.A.S). Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Cristina Melero, Greg Kapp, Alexandra Schnoes, Alan Barber, Daniel Almonacid and members of the Kortemme lab for discussion and testing of the web server. We would also like to thank the entire Rosetta Developers’ community for many discussions and contributions to shared development of Rosetta.
REFERENCES
- 1.Hartmann H, Parak F, Steigemann W, Petsko GA, Ponzi DR, Frauenfelder H. Conformational substates in a protein: structure and dynamics of metmyoglobin at 80 K. Proc. Natl Acad. Sci. USA. 1982;79:4967–4971. doi: 10.1073/pnas.79.16.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedland GD, Lakomek NA, Griesinger C, Meiler J, Kortemme T. A correspondence between solution-state dynamics of an individual protein and the sequence and conformational diversity of its family. PLoS Comput. Biol. 2009;5:e1000393. doi: 10.1371/journal.pcbi.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lange OF, Lakomek N.-A, Farès C, Schröder GF, Walter K.FA, Becker S, Meiler J, Grubmüller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 4.Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 5.Davis IW, Arendall WB, Richardson DC, Richardson JS. The backrub motion: how protein backbone shrugs when a sidechain dances. Structure. 2006;14:265–274. doi: 10.1016/j.str.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Smith CA, Kortemme T. Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J. Mol. Biol. 2008;380:742–756. doi: 10.1016/j.jmb.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei BQ, Weaver LH, Ferrari AM, Matthews BW, Shoichet BK. Testing a flexible-receptor docking algorithm in a model binding site. J. Mol. Biol. 2004;337:1161–1182. doi: 10.1016/j.jmb.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhury S, Gray JJ. Conformer selection and induced fit in flexible backbone protein-protein docking using computational and NMR ensembles. J. Mol. Biol. 2008;381:1068–1087. doi: 10.1016/j.jmb.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Comput. Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu X, Apgar JR, Keating AE. Modeling backbone flexibility to achieve sequence diversity: the design of novel alpha-helical ligands for Bcl-xL. J. Mol. Biol. 2007;371:1099–1117. doi: 10.1016/j.jmb.2007.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kono H, Saven JG. Statistical theory for protein combinatorial libraries. Packing interactions, backbone flexibility, and the sequence variability of a main-chain structure. J. Mol. Biol. 2001;306:607–628. doi: 10.1006/jmbi.2000.4422. [DOI] [PubMed] [Google Scholar]
- 12.Kraemer-Pecore CM, Lecomte J.TJ, Desjarlais JR. A de novo redesign of the WW domain. Protein Sci. 2003;12:2194–2205. doi: 10.1110/ps.03190903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larson SM, England JL, Desjarlais JR, Pande VS. Thoroughly sampling sequence space: large-scale protein design of structural ensembles. Protein Sci. 2002;11:2804–2813. doi: 10.1110/ps.0203902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang S, Li L, Hsu W.-L, Pilcher MN, Uversky V, Zhou Y, Dunker AK, Meroueh SO. Exploring the molecular design of protein interaction sites with molecular dynamics simulations and free energy calculations. Biochemistry. 2009;48:399–414. doi: 10.1021/bi8017043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das R, Baker D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 16.Rohl CA, Strauss C.EM, Misura K.MS, Baker D. Protein structure prediction using Rosetta. Meth. Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 17.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl Acad. Sci. USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kortemme T, Joachimiak LA, Bullock AN, Schuler AD, Stoddard BL, Baker D. Computational redesign of protein-protein interaction specificity. Nat. Struct. Mol. Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 19.Kortemme T, Morozov AV, Baker D. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J. Mol. Biol. 2003;326:1239–1259. doi: 10.1016/s0022-2836(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 20.Friedland GD, Linares AJ, Smith CA, Kortemme T. A simple model of backbone flexibility improves modeling of side-chain conformational variability. J. Mol. Biol. 2008;380:757–774. doi: 10.1016/j.jmb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphris EL, Kortemme T. Design of multi-specificity in protein interfaces. PLoS Comput. Biol. 2007;3:e164. doi: 10.1371/journal.pcbi.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphris EL, Kortemme T. Prediction of protein-protein interface sequence diversity using flexible backbone computational protein design. Structure. 2008;16:1777–1788. doi: 10.1016/j.str.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Georgiev I, Keedy D, Richardson JS, Richardson DC, Donald BR. Algorithm for backrub motions in protein design. Bioinformatics. 2008;24:i196–i204. doi: 10.1093/bioinformatics/btn169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat. Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 28.Benedix A, Becker CM, de Groot BL, Caflisch A, Bockmann RA. Predicting free energy changes using structural ensembles. Nat. Methods. 2009;6:3–4. doi: 10.1038/nmeth0109-3. [DOI] [PubMed] [Google Scholar]
- 29.Potapov V, Cohen M, Schreiber G. Assessing computational methods for predicting protein stability upon mutation: good on average but not in the details. Protein Eng. Des. Sel. 2009;22:553–560. doi: 10.1093/protein/gzp030. [DOI] [PubMed] [Google Scholar]
- 30.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci. STKE. 2004;2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 31.Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol. Cell. 2003;12:577–589. doi: 10.1016/s1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- 32.Lengyel CSE, Willis LJ, Mann P, Baker D, Kortemme T, Strong RK, McFarland BJ. Mutations designed to destabilize the receptor-bound conformation increase MICA-NKG2D association rate and affinity. J. Biol. Chem. 2007;282:30658–30666. doi: 10.1074/jbc.M704513200. [DOI] [PubMed] [Google Scholar]
- 33.McBeth C, Seamons A, Pizarro JC, Fleishman SJ, Baker D, Kortemme T, Goverman JM, Strong RK. A new twist in TCR diversity revealed by a forbidden alphabeta TCR. J. Mol. Biol. 2008;375:1306–1319. doi: 10.1016/j.jmb.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McFarland BJ, Kortemme T, Yu SF, Baker D, Strong RK. Symmetry recognizing asymmetry: analysis of the interactions between the C-type lectin-like immunoreceptor NKG2D and MHC class I-like ligands. Structure. 2003;11:411–422. doi: 10.1016/s0969-2126(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 35.Sivasubramanian A, Chao G, Pressler HM, Wittrup KD, Gray JJ. Structural model of the mAb 806-EGFR complex using computational docking followed by computational and experimental mutagenesis. Structure. 2006;14:401–414. doi: 10.1016/j.str.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 36.Bordner AJ, Abagyan RA. Large-scale prediction of protein geometry and stability changes for arbitrary single point mutations. Proteins. 2004;57:400–413. doi: 10.1002/prot.20185. [DOI] [PubMed] [Google Scholar]
- 37.Saunders CT, Baker D. Recapitulation of protein family divergence using flexible backbone protein design. J. Mol. Biol. 2005;346:631–644. doi: 10.1016/j.jmb.2004.11.062. [DOI] [PubMed] [Google Scholar]
- 38.Dantas G, Kuhlman B, Callender D, Wong M, Baker D. A large scale test of computational protein design: folding and stability of nine completely redesigned globular proteins. J. Mol. Biol. 2003;332:449–460. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- 39.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 40.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 41.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein-protein interface. J. Mol. Biol. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 42.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–9. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas C.F., 3rd, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–91. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Ghallaher JL, Althoff EA, Zanghellini A, Dym O, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 45.Pál G, Kouadio J.-LK, Artis DR, Kossiakoff AA, Sidhu SS. Comprehensive and quantitative mapping of energy landscapes for protein-protein interactions by rapid combinatorial scanning. J. Biol. Chem. 2006;281:22378–22385. doi: 10.1074/jbc.M603826200. [DOI] [PubMed] [Google Scholar]
- 46.Chivian D, Kim DE, Malmstrom L, Schonbrun J, Rohl CA, Baker D. Prediction of CASP6 structures using automated Robetta protocols. Proteins. 2005;61(Suppl 7):157–166. doi: 10.1002/prot.20733. [DOI] [PubMed] [Google Scholar]
- 47.Ashworth J, Baker D. Assessment of the optimization of affinity and specificity at protein-DNA interfaces. Nucleic Acids Res. 2009;37:e73. doi: 10.1093/nar/gkp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Kuhlman B. RosettaDesign server for protein design. Nucleic Acids Res. 2006;34:W235–W238. doi: 10.1093/nar/gkl163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–W238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sircar A, Kim ET, Gray JJ. RosettaAntibody: antibody variable region homology modeling server. Nucleic Acids Res. 2009;37:W474–W479. doi: 10.1093/nar/gkp387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.London N, Schueler-Furman O. FunHunt: model selection based on energy landscape characteristics. Biochem. Soc. Trans. 2008;36:1418–1421. doi: 10.1042/BST0361418. [DOI] [PubMed] [Google Scholar]
- 52.Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proc. Natl Acad. Sci. USA. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]