Abstract
The mammalian gut is an attractive model for exploring the general question of how habitat impacts the evolution of gene content. Therefore, we have characterized the relationship between 16 S rRNA gene sequence similarity and overall levels of gene conservation in four groups of species: gut specialists and cosmopolitans, each of which can be divided into pathogens and non-pathogens. At short phylogenetic distances, specialist or cosmopolitan bacteria found in the gut share fewer genes than is typical for genomes that come from non-gut environments, but at longer phylogenetic distances gut bacteria are more similar to each other than are genomes at equivalent evolutionary distances from non-gut environments, suggesting a pattern of short-term specialization but long-term convergence. Moreover, this pattern is observed in both pathogens and non-pathogens, and can even be seen in the plasmids carried by gut bacteria. This observation is consistent with the finding that, despite considerable interpersonal variation in species content, there is surprising functional convergence in the microbiome of different humans. Finally, we observe that even within bacterial species or genera 16S rRNA divergence provides useful information about average conservation of gene content. The results described here should be useful for guiding strain selection to maximize novel gene discovery in large-scale genome sequencing projects, while the approach could be applied in studies seeking to understand the effects of habitat adaptation on genome evolution across other body habitats or environment types.
INTRODUCTION
The human gut harbors the largest collection of microbes in any of our body habitats; its microbiome is of great interest because the microbiota appears to have pervasive effects on health and disease, including the development of a functional immune system, vitamin synthesis and nutrient processing (1). Culture-independent methods for the discovery of novel microbial lineages using 16 S rRNA gene sequencing have revolutionized our understanding of microbial diversity (2–4). The 16 S rRNA gene is an excellent marker of average genomic evolution because it is a core gene that seldom undergoes horizontal gene transfer and has a phylogeny that matches other core genes, because it appears to evolve largely independently of ecological diversification, and because it contains both fast- and slow-evolving regions and can thus be used to resolve relationships among taxa at different phylogenetic depths [see (2,5–7) for reviews on the topic]. 16 S rRNA based-surveys indicate that bacterial communities of the mammalian gut differ more from non-gut communities, than even the most extreme free-living communities differ from one another (8). This observation suggests that life in the intestinal environment may have demanding and distinctive functional requirements. Understanding whether 16 S rRNA surveys that reveal which species (or higher taxa) are present relate directly to diversity in functional gene repertoires is critical for Human Microbiome Projects (1): these projects generally seek to relate variation in the phylogenetic composition of the microbiome, as profiled by 16S rRNA surveys, to health and disease (9–13). To begin addressing this question, we ask whether gut-dwelling species have converged on more closely related gene repertoires than we would expect from their phylogenetic relationship. In particular, is the degree of overlap in the gene repertoire of gut dwellers greater than that for non-gut dwellers after a given amount of evolutionary time?
Differences in 16S rRNA gene sequences between genomes are related to overall levels of gene conservation between those genomes and to the average nucleotide identity (ANI) of genes conserved between them (14), although whether the same trends hold true for very closely related genomes (e.g. those within the same bacterial species) is unknown. Several mechanisms alter genome content, including genome reduction, gene duplications and horizontal gene transfer. These have been extensively studied. However, the effect of differences in habitat on the rate of evolution of gene content has only been systematically studied using a small number of species, primarily from non-host-associated habitats (15). Substantial variation in gene content has been observed within individual bacterial species, whether isolated from many environments (such as Escherichia coli) (16) or highly habitat-restricted [such as Helicobacter pylori (17)].
These observations that bacterial species vary in their degree of gene conservation (15,18,19), raise the question of whether the differences are due to differences in population structure (17), diversity within and/or between habitats or ecological interactions with other organisms (16). For example, the rate at which gene content varies with phylogenetic distance (15) might be due to any of the mechanisms outlined above. Two well-characterized examples of associations between specific environments and mechanisms of genomic change are the extreme genome reduction observed in obligate intracellular symbionts and intracellular pathogens (20–22) as well as microbial adaptation to hypersaline environments through enrichment of proteins throughout the proteome with the acidic amino acids aspartate and glutamate (23,24). However, signatures of adaptation to specific environments have generally been difficult to obtain.
The mammalian gut provides an attractive model to explore these issues, because it harbors an especially restricted group of lineages (8). If this restriction results from a highly selective environment, we might expect that different species adapt to the gut by convergent evolution in gene content. More generally, there are several reasons why bacteria sharing a habitat may share more or fewer genes than phylogenetic distance alone would predict (15). For example, adaptation to a shared environment might enrich the same genes necessary for growth and survival in that environment, and horizontal gene transfer may increase in densely packed communities, leading to more shared genes (e.g. the distal mammalian gut can contain up to 1012 cells/ml lumenal contents). Alternatively, competition within a shared environment could produce niche specialization (25–27) as strains diversify their gene content and exploit underutilized resources. Thus, we reason that inferring the relationship between evolutionary distance, as measured by 16 S rRNA sequence divergence, and functional relatedness, at the level of overlap in gene repertoires, could assist in discriminating among these various mechanisms.
METHODS
Selection and classification of genomes
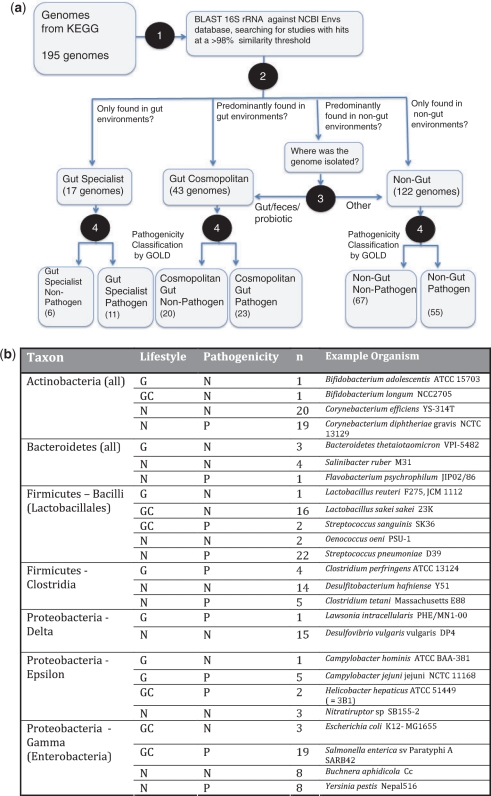
We sought to identify genomes representing abundant gut lineages that were specialist or cosmopolitan, and non-pathogenic or pathogenic. To do so, we downloaded 195 genomes from the KEGG database that were members of the Actinobacteria, Bacteroidetes, Firmicutes (separating the Clostridiales and the Lactobacillales), δ-Proteobacteria, ε-Proteobacteria and the γ-Proteobacteria (Enterobacteria). The bacteria from which these genomes were sequenced were then characterized according to their habitat and pathogenicity status (Figure 1) according to the following workflow: (i) To obtain information on the lifestyle of the isolates from which genome sequences were obtained, we determined which 16 S rRNA-based environmental surveys of microbial assemblages had deposited sequences in GenBank that were nearly identical to the 16 S rRNA sequence in the corresponding complete genome. We first downloaded the gbenv files from the NCBI ftp site on 31 December 2007 and used them to create a BLAST database. These files contain GenBank records for the ENV database, a component of the non-redundant nucleotide database (nt) where 16 S rRNA environmental survey data are deposited. GenBank records for hits with >98% sequence identity over 400 bp to the 16S rRNA sequence of each genome were parsed to obtain a list of study titles associated with the hits. (ii) These study titles were used to determine whether close relatives of each of the isolates had been found only in the gut (gut specialist), never in the gut (non-gut) or in the gut as well as a diversity of free-living communities (gut cosmopolitan). (iii) In ambiguous cases, where close relatives of the isolate were found in many environmental samples and only rarely in gut samples, isolation information from the GOLD database was used to decide how a genome should be categorized. In these ambiguous cases, strains annotated as probiotic or strains isolated from the distal gut or feces, were categorized as ‘gut cosmopolitan’ whereas others were categorized as non-gut. Thirteen genomes were removed from subsequent analysis because their isolation and phenotypic annotations from GOLD were ambiguous or conflicted. This classification process yielded 17 gut specialists, 43 gut cosmopolitan and 122 non-gut bacteria. (iv) Within each of these four categories, pathogens were identified using GOLD annotations downloaded 8 October 2009 (28).
Figure 1.
Classification of species by habitat and pathogenicity. (a) All genomes for the Actinobacteria, Bacteroidetes, Firmicutes (separating the Clostridiales and the Lactobacillales), δ-Proteobacteria, ε-Proteobacteria, and the γ-Proteobacteria (Enterobacteria) present in the KEGG database were downloaded (195 genomes total). The genomes were classified as follows (see ‘Materials and Methods’ section for detailed description): (i) BLAST was used to compare 16 S rRNA sequences for each genome against the NCBI Envs database to determine the environmental distribution of the species. (ii) Genomes were characterized by examination of the study titles of hits: genomes found exclusively in gut or fecal samples were labeled ‘gut specialist’, those found in several studies of the gut, but also in other environments were categorized as ‘gut cosmopolitan’, while those never found in the gut were labeled ‘non-gut’. (iii) In borderline cases where genomes were found in several environmental samples and only a small number of gut samples, isolation information from the GOLD database was used to determine whether the genome should be categorized as ‘gut cosmopolitan’ or ‘non-gut’. Probiotic bacteria, or those isolated from the gastrointestinal tract or feces in this abundance class were taken to be ‘gut cosmopolitan’. (iv) Finally, genomes in each category were categorized by pathogenicity using the GOLD (26) annotations for ‘phenotype’ and ‘disease’. Commensal microbes capable of only opportunistic infection were treated as non-pathogens in this analysis. Additionally, 13 genomes where annotation information was ambiguous or conflicted with observations from 16 S rRNA observations were removed from the analysis. (b) Example output of this annotation process, and numbers of genomes in each subcategory. Abbreviations are as follows: ‘G’, gut specialist, ‘GC’ cosmopolitan resident of the gut, ‘N’ non-gut. Pathogens are denoted ‘P’ and non-pathogens ‘N’.
Gene conservation
Gene conservation was measured as the proportion of genes in the query genome with at least one homolog conserved in the subject genome (see BLAST analysis, below). This measure is asymmetric because the query and subject genome can be of different sizes (e.g. if genome A contains 500 genes, genome B contains 5000 genes and they share 250 genes, B contains 50% of the genes in A, but A contains only 5% of the genes in B). The comparisons between genomes with large size differences was found to produce aberrant clusters of high or low gene conservation (see ‘Results’ section), therefore genomes were placed into three size categories ±1 SD from the mean genome size: these categories were small (<1783 genes), medium (1783–4964 genes) and large (>4964 genes). The comparisons between genomes in different size categories were then excluded from the analyses in Figures 3c, 3d, 5a and 5b, 6b, d and Supplementary Figure 1, as noted below. Since plasmids are subject to frequent horizontal gene transfer and the absence of plasmids in the strain chosen for genome sequencing does not indicate their absence in the corresponding natural populations, queries from plasmids were excluded from the analysis for comparisons of gene content to evolutionary distance. To assess the significance of correlations between evolutionary distance and gene content conservation, Mantel tests with 10 000 permutations were run on either the full matrix of comparisons for each taxon analyzed, as well as subsets of those matrices subdivided by environment, pathogenicity or chromosome type (chromosome or plasmid). Tests were performed using the Mantel test implementation in the PyCogent toolkit (29).
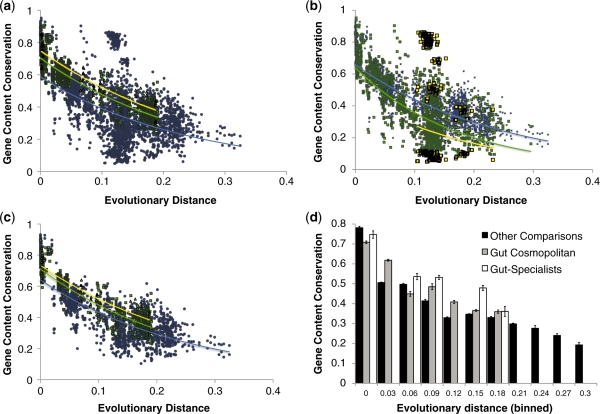
Figure 3.
Gene conservation in gut-adapted bacteria. Relationship between evolutionary distance in terms of 16 S rRNA divergence and gene content conservation. For these graphs, the x-axis shows evolutionary divergences in terms of nucleotide substitutions per site in the 16S rRNA gene, and the y-axis shows the fraction of genes in the first species that are found in the second species using BLASTP on the translated sequences. (a) Each point represents a comparison between two genomes. Yellow points are comparisons between two genomes that are both gut specialists, green points are comparisons between two genomes that are both cosmopolitan members of the gut microbiota, wheras all other comparisons are considered together and colored in blue. Although much variation in gene conservation is explained by phylogenetic distance, examples of genomes that vary little or greatly in gene conservation can be found at any given distance. r2 = 0.82 for gut specialists; 0.80 for gut cosmopolitans; and 0.22 for other comparisons. (b) Effects of relative genome size on conservation of gene content (size categories are defined in ‘Materials and Methods’ section above). Genome–genome comparisons were plotted separately for pairs of genomes where both are in the same size category (blue squares), where one genome is medium and the other is either large or small (green squares), or where one genome is large and the other is small (yellow squares). (c) Gene content conservation in pairs of gut-adapted bacteria with similar genome sizes. When only gut specialist or gut cosmpolitan genomes are considered, and when both genomes in each pair are similarly sized, phylogenetic distance is predictive of gene content conservation: r2 = 0.81 gut specialists; 0.78 gut cosmopolitan; and 0.57 for other comparisons. (d) Depicts the same data as in (c), but binned into increments of 0.03 corrected substitutions per site in the 16S rRNA, to clarify trends in conservation. Specialist (white bars) and cosmopolitan (gray bars) bacteria inhabiting the gut have somewhat lower levels of gene conservation at evolutionary distances below 0.03 substitutions per site than non-gut bacteria (black bars), but elevated levels between ∼0.06–0.18 substitutions per site. Error bars depict standard error.
Figure 5.
Gene conservation in plasmids borne by gut-adapted bacteria. (a) Gene conservation in bacterial chromosomes (red squares) or plasmids (blue squares). Plasmids show both lower average gene conservation than bacterial chromosomes, and, as would be expected given frequent conjugative exchange, a weaker relationship between evolutionary distance and gene conservation (r2 = 0.60 genomes; r2 = 0.06 plasmids). (b) Plasmids borne by specialist (white bars) or cosmopolitan (gray bars) bacteria tend to have higher gene conservation at evolutionary distances between 0.09 and 0.21 16 S rRNA substitutions per site than those borne by non-gut bacteria (black bars). These plasmids also exhibit markedly reduced gene conservation at distances under 0.03 substitutions per site.
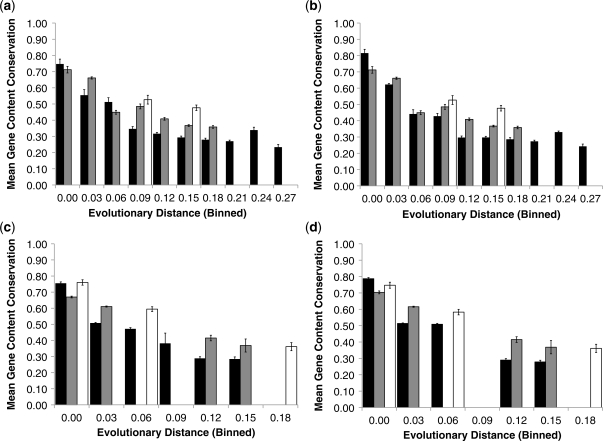
Figure 6.
Gut pathogens, like gut commensals, exhibit different patterns of gene content conservation from non-gut genomes. Each panel depicts average levels of gene content conservation, binned in ranges of 0.03 16 S rRNA substitutions per site. Values for comparisons between pairs of non-gut bacteria are shown in black, pairs of gut cosmopolitan bacteria in gray and pairs of gut specialists in white. (a) Gene conservation in non-pathogens, including comparison between pairs in all size categories. (b) As in (a), but showing only comparisons between pairs of genomes in the same size category. (c) As in (a), but for pathogenic bacteria. (d) As in (b), but for pathogens. Error bars depict the standard error of the mean.
BLAST analysis
BLASTp analyses were conducted using a custom python script based on PyCogent (29) to run NCBI BLAST (30). Analyses were run using the BLOSUM62 matrix (-M BLOSUM62) with maximum hits was set to 1 (-m 1). Hits were then filtered to an e-value threshold of 10−10 (analyses using alternative e-value thresholds altered the slope of results but not the qualitative outcome, data not shown), and hits with alignable regions <75% of the length of both query and subject were rejected.
Tree construction
16S rRNA sequences for each of the genomes under study were identified by BLASTing the E. coli rrsG gene against the nucleotide (nuc) file from KEGG, (http://www.genome.ad.jp/), for each genome with an e-value threshold of 1e–20 and word length of 11. Some genomes contain multiple 16S rRNA sequences. We verified manually that the BLAST settings used identified all 16 S rRNA sequences from several such genomes (and no others) that had been identified in a previous study (31). 16 S rRNA sequences identified in this manner were then aligned using NAST (32).
In cases where multiple 16 S rRNA sequences in a single genome passed the NAST screen, sequences were selected randomly. The Lane mask (33) from GreenGenes (34) was applied to the selected NAST-aligned sequences. Phylogenetic trees were constructed in ClearCut (35) using traditional neighbor-joining and the Kimura two-parameter distance correction. In order to determine whether short reads such as those generated by pyrosequencing would suffice for analyses of gene content and evolutionary distance, trees were also constructed using simulated pyrosequencing reads. In this case, trees were also constructed by the same procedure, but instead using only the regions of the 16 S rRNA corresponding to 250 bases of the regions amplified by V2, V4 and V6 primers (36). These were generated by taking only the corresponding regions from the full-length 16 S rRNA sequences. The gaps were then removed and the sequences realigned. The coordinates in the GreenGenes 7682 bp format for these regions were: V2, 1869–2353; V4, 2310–4100; and V6, 4625–5877.
RESULTS
A scale relates gene content to 16S rRNA evolutionary distance
We calculated gene conservation for all pairs of bacterial genomes in the KEGG database from within the Actinobacteria, Bacteroidetes, Firmicutes (separating the Clostridiales and the Lactobacillales), δ-Proteobacteria, ε-Proteobacteria and γ-Proteobacteria (Enterobacteria). These taxa were selected because they contain prominent members of the mammalian gut microbiota (37). Plotting proportions of shared genes against tip-to-tip distances on a 16S rRNA neighbor-joining tree for the resulting 5737 intra-taxon genome-to-genome comparisons allowed us to infer a model for the relationship between 16S rRNA distances and protein conservation. The proportion of shared genes was determined by performing protein BLAST queries for each gene in that genome against a database composed of all genes in each other genome within the taxon at an e-value threshold of 10−10. The proportions of genes with homologs below the e-value threshold were then plotted against the tip-to-tip distance between the two genomes on a neighbor-joining tree. Initial studies indicated that the BLAST stringency varied only the steepness of the slope but not the overall patterns; therefore only data for the 10−10 threshold is shown although 10−4 and 10−7 were also used. Gene conservation as measured by protein BLAST was found to decrease exponentially with 16S rRNA distance, in agreement with previous observations (14,38). Exponential regression of 16S rRNA distance alone explained only 29% of the overall variance in gene conservation levels. This regression also suggested that gene conservation falls at a rate of 0.62e–4.326d where d is the corrected tip-to-tip distance on a 16S rRNA neighbor-joining phylogeny.
To test whether patterns of gene conservation over evolutionary distance were universal or varied by bacterial taxon, the results were broken down by taxonomy (Figure 2). For all taxa in the analysis, the negative correlation between evolutionary distance and gene content conservation was statistically significant by Mantel Test (P < 0.05; see Supplementary Table 1). However, the explanatory power of 16S rRNA gene distance varied greatly between the taxa studied, explaining as little as 28% (Enterobacteria) to as much as 70% (Bacteroidetes) of the variance in gene conservation levels (Figure 2). This heterogeneity could arise from several mechanisms, including different rates of horizontal gene transfer, genome reduction or habitat specialization in different taxa, which we investigate below.
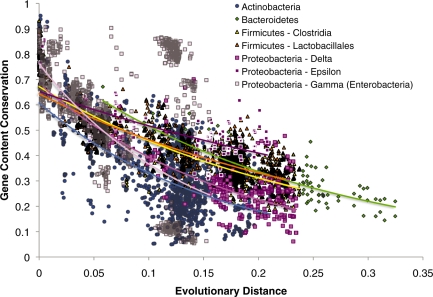
Figure 2.
Gene conservation by evolutionary distance. Gene content conservation at the protein level. Each point represents a BLAST comparison between two genomes at an E-value threshold cutoff of 10−10. The x-axis represents the 16 S distance between the two genomes, while the y-axis represents the proportion of proteins from the query genome that matches proteins from the subject genome. Genome–genome comparisons are subdivided by taxonomic group. Comparisons between members of the same taxonomic group are represented by the same shape and similar colors. Each colored line represents the exponential regression of the points within a single taxon. r2 values for exponential regression of each taxon were: Actinobacteria, r2 = 0.28; Bacteroidetes, r2 = 0.70; Clostridia, r2 = 0.57; Lactobacillales, r2 = 0.70; δ-Proteobacteria, r2 = 0.38; ε-Proteobacteria r2 = 0.48; γ-Proteobacteria r2 = 0.24.
Habitat adaptation and genome size alter aggregate gene conservation
In order to test whether the shared lifestyle of gut-adapted bacteria altered the relationship between gene conservation and evolutionary distance, the genomes in this analysis were categorized based on how often they have been observed in the gut relative to other environments in 16S rRNA studies, combined with information about isolation sources and pathogenicity status derived from the GOLD database (28) (see ‘Materials and Methods’ section and Figure 1). Species found exclusively in the gut were labeled ‘gut specialist’, while those frequently found in both the gut and other environments were labeled ‘gut cosmopolitan’ and those rarely or never observed in the gut but plentiful in other environments were labeled ‘non-gut’, with isolation information being used to decide borderline cases (28).
Gene content fell exponentially with increasing evolutionary distance for both specialist, cosmopolitan and non-gut species (Figure 3a). In each taxon and each habitat category, the correlation between gene content conservation and evolutionary distance was statistically significant (P < 0.05, Mantel test), except in subcategories for which very few (n < 5) genomes were available (Supplementary Table 2). Differences in gene content were well explained by evolutionary distance for gut-adapted bacteria (specialists: r2 = 0.82; cosmopolitan: r2 = 0.80), but poorly explained for other comparisons (r2 = 0.22). Importantly, regression analysis indicated that, for a broad range of phylogenetic distances, gut-adapted bacteria possess higher levels of gene conservation than their non-gut relatives, with cosmopolitan members of the gut community being intermediate between gut specialists and other species.
The measure of similarity in gene content (i.e. conservation) used was asymmetric (see ‘Materials and Methods’ section), therefore averages of pairwise comparisons among genomes of different sizes can be misleading. Differences in gene conservation attributable to genome reduction are captured in Figures 2 and 3a. Clusters of very high gene conservation were found when comparing reduced genomes to large genomes, and conversely clusters of very low levels of gene conservation were found when comparing large genomes to their reduced relatives.
To investigate the effect of relative genome size on the relationship between evolutionary distance and gene content, the genome–genome comparisons in Figure 3a were re-plotted according to relative genome size (Figure 3b). Each genome was categorized as small, medium or large according to the criteria defined in ‘Materials and Methods’ section. The results from Figure 3a were then re-plotted according to whether the genomes being compared belonged to the same size category (Figure 3b).
Comparisons between genomes with very unequal sizes explain many of the outliers from the overall trend in gene conservation over phylogenetic distance reported in the analyses above. While phylogenetic distance explained ∼60% of the variance in gene conservation between genome pairs within the same size category, it explained only 27% of the variance between genome pairs that differed by one size category and only 1% of the variance in genome pairs that differed by two size categories. This result suggests that controlling for genome size is critical for prediction of gene conservation from phylogenetic distance. Moreover, this is a difference that would be missed if gene conservation were calculated symmetrically. Recalculating the results from Figure 2 to include only genome–genome comparisons (Supplementary Figure S1) within the same size category yields an r2 of 0.60, ∼2-fold improvement in the degree to which variance in gene content can be explained by phylogenetic distance. This improvement applies only to lineages where variation in genome size is substantial. For example, the enterobacteria, rather than appearing as an outlier to the overall trend appear entirely typical, once differences in genome size are corrected for (γ-Proteobacteria r2 = 0.60; see Supplementary Figure S1).
To test whether the elevated gene conservation in gut-adapted genomes seen in Figure 3a is an artifact caused by wide variation in genome sizes amongst non-gut genomes, we repeated the analysis in Figure 3a excluding genome–genome comparisons from different size categories. Similar patterns emerged to those observed in the full dataset (Figure 3c), indicating that differences in the evolution of gene content between gut and non-gut genomes were not simply attributable to trends in genome size. In order to quantify the effects of adaptation to the gut habitat on gene conservation at various phylogenetic distances, and to test whether this difference was significant, genome–genome comparisons were binned into increments of 0.03 corrected substitutions/site in the 16S rRNA (Figure 3d). This analysis revealed that gut specialist and gut cosmopolitan lineages have greater gene conservation for evolutionary distances between 0.06 and 0.18 substitutions/site. However, at distances of <0.03 16 S rRNA substitutions per site (roughly corresponding to the traditional bacterial species boundary, see Supplementary Figure S2), gut genomes tended to have much lower gene conservation than is present at greater distances. This could reflect increased niche specialization in very closely related gut genomes or increased convergence in other environments.
16S rRNA distance predicts genomic diversity within bacterial species
Patterns of niche specialization within and between bacterial species may operate according to different principles, which could provide insight into the ecological mechanisms which underlie them within a given habitat. To follow up on this question of niche specialization, we next examined the ability of 16 S rRNA distances to predict gene content within bacterial species. This analysis is interesting for two reasons. First, because barriers to horizontal gene transfer are believed to be lower between closely related genomes (39), it might be expected that the phylogenetic signal would have little effect on gene content within bacterial species. Second, although genome sequencing is increasingly affordable, criteria for choosing strains that maximize divergence in genome content so as to maximize the discovery of new components of the pan-genome are essential. If 16S rRNA distance had little effect on gene conservation within bacterial species, then it would be preferable to select strains based on other criteria or at random to maximize statistical power.
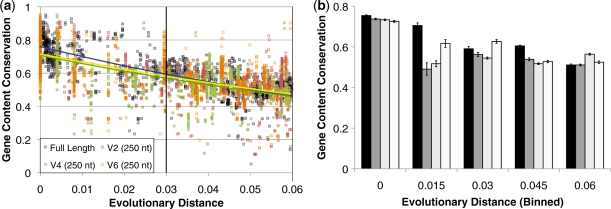
Even when examining gene conservation at scales that correspond to the most commonly used cut-off for bacterial species (16 S rRNA distances below 3% divergence), we found that 16 S rRNA gene distance is an important predictor of gene conservation. Gene conservation between strains of the same species fell as evolutionary distances approached 0.03 nucleotide substitutions per site (Figure 4a and b). These results are consistent with those of Konstantinidis and Tiedje (15), who found a relationship between 16 S rRNA divergence, overall gene content, ANI in orthologous genes and DNA rehybridization kinetics. In addition, these trends can be recovered using not just full-length 16S rRNA, but also using 250 nucleotide reads from the V2, V4 or V6 regions of this gene. This result reveals that even short 16S rRNA gene reads, such as those produced with pyrosequencing, are associated with genomic differences (Figure 4). On an average, selecting a strain with 16 S rRNA distance between 0.015 and 0.03 from the nearest known strain will produce ∼9% fewer conserved genes (and, conversely, greater gene novelty) than selecting a random genome within the species; whereas a similar criterion applied to phylogenies constructed from 250 nucleotide reads from V2, V4 or V6 primers will yield an average 17, 16 or 4% reduction in conserved genes, respectively (Figure 4b). A similar concept applies when selecting species within the same genus (using the >94% rRNA percent identity threshold). Selecting the most divergent strains within a genus (i.e. those with 94–95% identity in the 16 S rRNA) provides an average 8–12% reduction in gene conservation relative to randomly chosen species belonging to the same genus, depending on the primers used. It should be noted, however, that variation is sufficiently high in either case that this technique is most useful when sequencing a large number of genomes; although choosing divergent lineages at the genus or species level provides access to a pool of strains or species with reduced gene conservation, it is not the case that gene conservation for every genome pair will be reduced.
Figure 4.
Greater 16 S rRNA divergence implies greater divergence in gene content within bacterial species. (a) Trees constructed from either the full length 16 S rRNA or 250 nucleotide stretches of its V2, V4 or V6 regions. The vertical bar corresponds to the species boundary, using the traditional bacterial species definition of >97% 16 S rRNA identity. (This boundary was determined by regressing the corrected 16 S rRNA distances displayed here against 16 S rRNA percent identity. See Supplementary Figure S2). The results demonstrate that even within the same bacterial species, the average gene conservation of a genome pair falls as phylogenetic distance increases. (b) Binning the results from (a) to bins of 0.015 16 S rRNA substitutions per site allows quantification of the effects of phylogenetic distance on gene conservation. Black bars represent average gene conservation at a given distance when distances are calculated using the full-length 16 S rRNA gene sequence, while progressively lighter gray bars represent gene conservation when calculating distance with fragments of the V2, V4 or V6 regions, respectively.
Habitat adaptation in bacterial plasmids
Bacterial plasmids are frequently subject to horizontal transfer. Because plasmids supplement an existing bacterial genome, they are not constrained to contain genes essential for cellular life. The 132 plasmids sequenced with the genomes included in this analysis thus provide a window into gene conservation amongst frequently transferred genes. We compared the genes carried on each plasmid with the combined pool of genes carried on the chromosomes and plasmids of each other isolate in the analysis (Figure 5a). Both overall gene conservation and the ability to predict gene conservation from phylogenetic distance were dramatically reduced in plasmids. This contrast between conservation of plasmid-borne genes and those located on bacterial chromosomes suggests that horizontal gene transfer in genomes is not so frequent that phylogeny and gene conservation are uncoupled (in which case the ability of phylogenetic distance to predict gene conservation would be similar for both plasmids and chromosomes). Instead, once we account for differences in overall genome size, the gene content of chromosomes is substantially more predictable than that of plasmids (r2 = 0.60 chromosomes; r2 = 0.06 plasmids). Surprisingly, despite explaining little of the variation in gene content conservation, the correlation between evolutionary distance and gene content conservation is still statistically significant for the taxa in the analysis (P < 0.05, Mantel test), except in cases where the number of plasmids is very small (n < 5; see Supplementary Table S3).
Given the observation that the dense bacterial community of the mammalian gut presents ample opportunities for horizontal gene transfer, and horizontal gene transfer is thought to be a process promoting habitat adaptation, we tested whether the effect of environmental adaptation on gene conservation observed in bacterial chromosomes also occurs on plasmids. The plasmids of gut cosmopolitan genomes clearly show a similar effect of habitat on gene content to that observed in bacterial chromosomes (Figure 5b). That is, at short phylogenetic distances gene content conservation is reduced for comparisons within the same environment, whereas at longer phylogenetic distances gene conservation is enriched, suggesting that the same pattern of short range specialization and long range convergence observed for bacterial chromosomes may be acting on plasmids. For gut-specialist plasmids the dataset is limited to a small number of examples, but overall the results appear consistent with the patterns observed for the full chromosomes. Indeed, the effect of habitat on gene content conservation over short phylogenetic distances appears to be even more dramatic in plasmids than in bacterial chromosomes (Figure 5b).
The effects of habitat adaptation on gene conservation occur in both pathogens and non-pathogens
Finally, we tested whether the effects of shared habitat, phylogenetic distance and genome content were common across commensal and pathogenic genomes. When we divide the genomes into more categories, the statistical power is reduced, but in cases where data are available gut-adapted commensal (Figure 6a) and pathogenic (Figure 6b) genomes generally display the same elevated levels of gene conservation at intermediate phylogenetic distances relative to non-gut genomes. This effect persists when also limiting the data to comparisons between genomes of similar size (Figures 6c and d).
DISCUSSION
This study reveals that gut-adapted genomes are more similar in gene content at a given evolutionary distance than non-gut genomes. Thus, common functional requirements or increased horizontal gene transfer cause similarities in gene content within the gut habitat. This trend holds over a broad range of phylogenetic distances. However, niche specialization at short phylogenetic distances (e.g. of strains within the same bacterial species) is also important in the mammalian gut. The well-known result that genome content can vary radically for genomes with identical 16 S rRNA sequences (14,40), and studies that report high levels of horizontal gene transfer (41,42) have raised doubts about our ability to understand genome and community functions based on phylogeny. The results presented here, together with the demonstration from GEBA (http://www.jgi.doe.gov/programs/GEBA/) that phylogenetically chosen genomes maximize novel gene lineage discovery, suggest that these effects, while important, do not obscure the overall trend that evolutionarily related organisms tend to share genomic features and, presumably, ecological niches.
The finding that gene conservation between gut-adapted bacteria is reduced over very short phylogenetic distances but elevated at greater distances suggests that gene content filters the persistent lineages of microbes in the gut (43). The reduced gene conservation at short phylogentic distances might thus indicate that competitive exclusion amongst bacteria with very similar functional profiles dominates amongst closely related bacteria, while the gene content of more divergently related gut bacteria is more strongly influenced by the shared selective pressures imposed by life in the gut. This interpretation is further supported by the convergence of very different species assemblages on similar functional repertoires in the human gut, as revealed by metagenomic studies (44).
A survey of microbial communities across 27 body habitats in healthy individuals has emphasized the importance of body habitat in determining community composition relative to interpersonal or temporal variation (45). If there is more convergence in function in the gut due to extreme selective pressure and/or horizontal gene transfer, would this be mirrored by more consistent metagenomic profiles and/or more divergence at fine phylogenetic scales in the gut than in other body habitats? Although difficulties with low sample biomass currently preclude metagenomic studies of these other body habitats, large-scale sequencing of strains associated with other body habitats could address these important questions by allowing the application of the techniques introduced here.
A key and pressing challenge is to understand how, if the gut is such a selective environment, some species are able to establish and maintain a broadly cosmopolitan lifestyle. To that end, it would be profitable to deliberately choose closely related gut and non-gut strains both for sequencing and for careful experiments to test survival across a broad set of conditions and environments where common metabolic themes such as fermentation may be represented. Ideally these would be newly isolated from well-characterized environments, sidestepping the issue of dubious provenance of many existing strains. As these species are being sequenced, our ability to gain insight will improve as annotations converge on improved standards such as Minimal Information about a Genome Sequence [MIGS (46)] and Minimal Information about an Environmental Sequence (MIENS; http://darwin.nerc- oxford.ac.uk/gc_wiki/index.php/MIENS). This combination of data and metadata will enable more general tests of the effects of environmental adaptation on genome composition and evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health predoctoral training (grant T32 GM08759 to J.Z.); National Institutes of Health (grant numbers P01DK078669, R01HG004872); Crohn’s and Colitis Foundation of America and Howard Hughes Medical Institute (HHMI). Funding for open access charge: National Institutes of Health; HHMI.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Justin Kuczynski, Elizabeth Costello, Tony Walters, Daniel McDonald and Sara Nakielny for helpful comments on the manuscript. J.Z. would also like to thank his classmates in “Genome Databases: Mining and Management”, MCDB 5621, where this analysis was initiated as a class project, for their valuable insight and support.
REFERENCES
- 1.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 3.Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl Acad. Sci. USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woese CR. Interpreting the universal phylogenetic tree. Proc. Natl Acad. Sci. USA. 2000;97:8392–8396. doi: 10.1073/pnas.97.15.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woese CR. Bacterial evolution. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olsen GJ, Woese CR. Ribosomal RNA: a key to phylogeny. FASEB J. 1993;7:113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- 7.Doolittle WF, Brown JR. Tempo, mode, the progenote, and the universal root. Proc. Natl Acad. Sci. USA. 1994;91:6721–6728. doi: 10.1073/pnas.91.15.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2008;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Wei H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl Acad. Sci. USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol. 2007;10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc. Natl Acad. Sci. USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welch RA, Burland V, Plunkett G, 3rd, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M, et al. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1:e43. doi: 10.1371/journal.pgen.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, Whittam TS, Musser JM. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl Acad. Sci. USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl Acad. Sci. USA. 2004;101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 21.Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 22.Sallstrom B, Andersson SG. Genome reduction in the alpha-Proteobacteria. Curr. Opin. Microbiol. 2005;8:579–585. doi: 10.1016/j.mib.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Fukuchi S, Yoshimune K, Wakayama M, Moriguchi M, Nishikawa K. Unique amino acid composition of proteins in halophilic bacteria. J. Mol. Biol. 2003;327:347–357. doi: 10.1016/s0022-2836(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 24.Paul S, Bag SK, Das S, Harvill ET, Dutta C. Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 2008;9:R70. doi: 10.1186/gb-2008-9-4-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchinson GE. Homage to Santa Rosalia, or why are there so many kinds of animals? Am. Nat. 1959;93:145–149. [Google Scholar]
- 26.Sokurenko EV, Chesnokova V, Dykhuizen DE, Ofek I, Wu XR, Krogfelt KA, Struve C, Schembri MA, Hasty DL. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl Acad. Sci. USA. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sokurenko EV, Feldgarden M, Trintchina E, Weissman SJ, Avagyan S, Chattopadhyay S, Johnson JR, Dykhuizen DE. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol. Biol. Evol. 2004;21:1373–1383. doi: 10.1093/molbev/msh136. [DOI] [PubMed] [Google Scholar]
- 28.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–D479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight R, Maxwell P, Birmingham A, Carnes J, Caporaso JG, Easton BC, Eaton M, Hamady M, Lindsay H, Liu Z, et al. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Coenye T, Vandamme P. Intragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomes. FEMS Microbiol. Lett. 2003;228:45–49. doi: 10.1016/S0378-1097(03)00717-1. [DOI] [PubMed] [Google Scholar]
- 32.DeSantis TZ, Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York: Wiley; 1991. [Google Scholar]
- 34.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheneman L, Evans J, Foster JA. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics. 2006;22:2823–2824. doi: 10.1093/bioinformatics/btl478. [DOI] [PubMed] [Google Scholar]
- 36.Liu Z, DeSantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 2008;36:e120. doi: 10.1093/nar/gkn491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 38.Tamames J. Evolution of gene order conservation in prokaryotes. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-research0020. RESEARCH0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 40.Jaspers E, Overmann J. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl. Environ. Microbiol. 2004;70:4831–4839. doi: 10.1128/AEM.70.8.4831-4839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 42.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 43.Green JL, Bohannan BJ, Whitaker RJ. Microbial biogeography: from taxonomy to traits. Science. 2008;320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- 44.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.