Abstract
Yin Yang 1 (YY1) is a critical transcription factor controlling cell proliferation, development and DNA damage responses. Retrotranspositions have independently generated additional YY family members in multiple species. Although Drosophila YY1 [pleiohomeotic (Pho)] and its homolog [pleiohomeotic-like (Phol)] redundantly control homeotic gene expression, the regulatory contributions of YY1-homologs have not yet been examined in other species. Indeed, targets for the mammalian YY1 homolog YY2 are completely unknown. Using gene set enrichment analysis, we found that lentiviral constructs containing short hairpin loop inhibitory RNAs for human YY1 (shYY1) and its homolog YY2 (shYY2) caused significant changes in both shared and distinguishable gene sets in human cells. Ribosomal protein genes were the most significant gene set upregulated by both shYY1 and shYY2, although combined shYY1/2 knock downs were not additive. In contrast, shYY2 reversed the anti-proliferative effects of shYY1, and shYY2 particularly altered UV damage response, platelet-specific and mitochondrial function genes. We found that decreases in YY1 or YY2 caused inverse changes in UV sensitivity, and that their combined loss reversed their respective individual effects. Our studies show that human YY2 is not redundant to YY1, and YY2 is a significant regulator of genes previously identified as uniquely responding to YY1.
INTRODUCTION
Yin Yang 1 (YY1, δ, NF-E1, UCRBP or CF1) is a multifunctional zinc-finger transcription factor (1), and a member of the Polycomb protein Group (PcG). PcG proteins control the plasticity of the pluripotent state, stem-cell development and maintenance of lineage-specific gene-expression programs (2,3). They were first described in Drosophila as regulators of body segmentation patterns through their repression of homeotic (Hox) gene expression (4). The PcG complex includes several oncogenes including posterior sex combs (PSC—the Bmi oncogene) and EZH2 (5). YY1 is the major DNA-binding protein in the Polycomb complex (6), and its fly homolog is pleiohomeotic (Pho) (7). In addition to their functions in polycomb repression, both Pho and YY1 are the DNA-binding proteins in the INO80 DNA transcriptional-remodeling complex (8,9). Independent retrotranspositions have given rise to YY1 homologs in many species that include pleiohomeotic-like (Phol) in Drosophila (10) and Yin Yang 2 (YY2) in mammals (11). Of note, Drosophila Phol is redundant with pleiohomeotic in homeotic gene silencing since double mutants show more severe mis-expression of homeotic genes and more extreme developmental phenotypes (10). On the other hand, a genome-wide analysis of Pho and PhoL targets has not been performed, nor have gene targets of YY2 been compared to those of YY1.
In addition to its participation in polycomb and INO80 complexes, YY1 interacts with additional components of transcriptional control (1). It was simultaneously discovered as a regulator of ribosomal protein synthesis (12), and as a key repressor of adenoviral oncogenesis (13). In association with various cofactors, YY1 is involved in transcription activation and repression (1), histone modification (14), chromatin remodeling (15), DNA damage repair (9), as well as tumorigenesis (16–18). YY1 knock-out causes embryonic lethality (19,20), and development of several tissues is critically dependent on YY1 including B lymphocytes, the axial skeleton and neural progenitors (21–23). In addition, its expression is needed to support cell-cycle proliferation (20), and elements of this requirement have been proposed to occur through interactions with p53 or E2F (18,24,25). Likewise, YY1 contributes to genomic stability and DNA damage repair (9). YY1’s cis target sequence is the tenth most common mammalian DNA-binding motif, which is present in 2% of well annotated promoters (26). Although a large number of genes have been proposed to be regulated by YY1 (1), transcriptome analyses of YY1 knockout mice did not show significant changes in mRNA expression for most of these genes (20). This discrepancy suggests that another transcription factor can compensate for YY1 deficiency.
Two additional YY1-site-binding proteins have been identified in mammals—YY2 and zinc-finger protein 42 (ZFP42, Rex-1) (11,27). Both share 95% identity in their zinc-finger-binding regions with YY1, and their DNA-binding targets are nearly identical to those of YY1 (27,28). ZFP42 was described initially as Reduced Expression in retinoic acid-induced embryonal carcinoma cells (Rex-1) (29), which is an embryonic stem-cell marker that controls embryonic stem-cell self-renewal (28,30,31). It was actually the first YY family member to be cloned, but its expression levels are low in most cell lines (28). It has therefore received much less attention than YY1. YY2 was recently identified in a homology screen (11). It appears to be a recently transposed, but functional, transcription factor that was inserted into the intron of Mbtps2, which is undergoing rapid evolution in mammals (32). YY2 and Rex1 largely share DNA-binding motifs with YY1, although their affinities and specific target sequences differ somewhat (27). The only test of interaction between mammalian YY1 and YY2 suggests they antagonistically affect reporter genes carrying canonical YY1 cis target sequences (33). Almost nothing is known about YY2- or ZFP42-regulated genes, and their interactions with YY1 have not been explored. Given the importance of YY1 in mammalian development and carcinogenesis, we sought to determine whether YY2 or ZFP42 might contribute to the functional complexity of YY1’s various roles in cellular physiology.
Differences in individual genes identified using standard DNA microarray analyses are often subtle; therefore studies of coordinated changes in groups of functionally related genes have attracted increasing interest (34). Since the previous microarray study that evaluated YY1 target genes in embryonic tissues did not use gene set enrichment analyses (GSEA) (20), we considered that GSEA might yield important information about sets of YY1 target genes and their potential overlaps with other YY family target gene sets (35–37). We therefore used lentiviral-expressed shRNAs from the RNAi consortium (38) to systematically reduce expression levels of YY1, YY2 and ZFP42 in a model cell line (11). We then used microarray and GSEA to identify relationships between their respective targets (35,37). Even though many gene sets were altered in a similar manner by shYY1 and shYY2, here we show that YY2 contributes to expression control of several important genes and gene sets that differ from YY1.
MATERIALS AND METHODS
Plasmids and primer sequences
Lentiviral pLKO-shRNAs were obtained from Sigma-Aldrich. The tracking numbers of the most effective shRNAs are TRCN0000019894 (YY1), TRCN0000016494 (YY2), TRCN0000107810 (ZFP42) and TRCN0000039642 (Myc). To replace the puromycin-resistance gene in pLKO plasmids with G418-resistance for combined knock down, an 827 bp fragment containing the neoR gene was amplified from plasmid pCNH (39) with primers 5′-TCA GGA TCC AGG ATC GTT TCG CAT GAT TGA AC-3′ and 5′-TCA GGT ACC GAT GCA TGA GTC CCG CTC AGA AGA ACT CG-3′, digested by Kpn I and BamH I, and ligated to Kpn I/BamH I-treated pLKO- shRNA plasmids. Plasmids pCEP4-FLAGYY1 and pCEP4-HAYY2 were derived from pcDNA-FLAGYY1 and pcDNA-FLAG/HAYY2 by PCR and ligation with vector pCEP4. These two plasmids were used as standards to measure endogenous YY1 and YY2 expression in quantitative real-time polymerase chain reaction (qRT-PCR). Double-stranded oligonucleotides used as electrophoresis mobility shift assay (EMSA) probes were based on an YY1-binding site in the eIF4E promoter (5′-CCA CAG TCG CCA TCT TAG A-3′).
Cell culture and shRNA transduction
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, maintained in 37°C incubators with 5% CO2. OV90 ovarian cells, MDA-MB-468 breast cells, MCF7 breast cells, T47D breast cells and Raji B cells were grown in conditions suggested by the ATCC. pLKO-shRNAs were packaged in 293T cells following the manufacturer’s standard protocol (Sigma-Aldrich). Spent medium of 293T cells was collected at 24 and 48 h post-transfection, pooled and cleared through a 0.45-µm syringe filter. To infect, HeLa cells were grown in six-well culture plates in 2 ml medium and 0.5 ml of lentivirus-containing medium was added. After 72 h, cells were selected and maintained in medium containing 2 µg/ml of puromycin. To make combined shRNA transfectants, this process was repeated but additionally selected using 500 µg/ml of Geneticin (G418) for dual selections. MCF7 and T47D cells were similarly treated to obtain stable transfectants to validate the HeLa cell results. Stable transfectants were split once a week and cells of <15 passages were used in all assays. To perform MTT, spent medium was aspirated and cells were incubated with 10 µl of 5 mg/ml thiazolyl blue tetrazolium bromide (Sigma) and 20 µl of 100 mM disodium succinic acid at 37°C for 3 h, then with 130 µl of DMSO for 20 min at room temperature. Absorbance was measured in a plate reader at 550 nm with a 620 nm reference filter.
Microarray and gene set enrichment analysis
Total RNA was prepared by Trizol reagent (Invitrogen). Ten micrograms of RNA were used to prepare probes to hybridize to Affymetrix U133 Plus 2.0 human genome arrays. Probes were synthesized using Affymetrix One-Cycle target labeling and control reagents and the Enzo BioArray high yield RNA transcript labeling kit. Raw data were generated by the GeneChip Scanner 3000 and its bundled software. Each analysis was performed using triplicate samples. Raw data were normalized using the GeneChip Operating System algorithm and the resulting matrix table was filtered to remove absent data points. While this filter may decrease our discovery of more robust changes, it should significantly decrease false-positive discoveries. The resulting matrix tables were uploaded using the desktop module for Gene Set Enrichment Analysis (Broad Institute) (35,37). We analyzed curated, motif and computational data sets using 1000 gene set permutations. The results were downloaded and compiled to compare the effects of shYY1, shYY2 and the combination of shYY1 and shYY2 to cells expressing the LKO vector control.
cDNA synthesis and qRT-PCR
Two micrograms of total RNA were used to make cDNA. Prior to reverse transcription, samples were first treated with one unit of DNase I for 15 min at room temperature to remove residual genomic DNA. Following a 5-min 65°C incubation to inactivate DNase I, samples were annealed with oligo(dT) and reverse-transcribed using a Superscript III First-strand cDNA Synthesis Kit (Invitrogen) following the manufacturer’s standard protocol. The resulting cDNAs were diluted 25-fold and 10 µl samples were mixed with SYBR-Green Supermix (Bio-Rad) and 0.4 µM of each primer in a 25-µl PCR reaction and thermocycled in the Bio-Rad MyiQ single color real time PCR detection system. We designed primer sets for 57 candidate genes chosen for their responsiveness to shYY1, shYY2 or their combination (Supplementary Table S1). We then compared the average of every individual microarray set for the 57 genes to the average of the qRT-PCR results (n = 3) for each gene in a Pearson correlation analysis (Sigmastat software).
Western blot and immunoprecipitation
For immunoblotting, cells were lysed using NP40 lysis buffer (50 mM Tris, pH 8.0, 0.42 M NaCl, 0.5% NP40, 2 mM EDTA) supplemented with fresh complete protease inhibitor cocktail tablets (Roche) for 20 min on ice. Lysates were cleared by centrifugation at 14 000 rpm for 10 min and protein concentrations determined by Bio-Rad protein assays. Equal amounts of lysates were fractionated on SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotted with YY1 antibody (Santa Cruz sc-281) at 1 : 1000 dilution, or 1 : 1000 thrombospodin-1 (sc-59887), 1 : 100 CD-36 (sc-9154) and 1 : 5000 actin antibodies (Chemicon). To perform immunoprecipitation, cells were grown in 10-cm tissue culture dishes. Following incubation of the monolayer in methionine- and cysteine- free medium (Cellgro) plus 5% dialyzed FBS (Hyclone) for 3 h, 500 µCi 35S-methionine/cysteine (Perkin Elmer) was added and incubated for 3 h at 37°C, 5% CO2. Cells were rinsed once with cold PBS and RIPA buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris, pH 7.4, 0.05% NP-40, 1% deoxycholic acid, 1% Triton X-100, fresh complete protease inhibitor cocktail tablets) was added for 10 min on ice. The lysates were pre-cleared by incubating with 5 µg non-specific goat serum (Santa Cruz) plus 20 µl Protein-G Agarose (Santa Cruz) for 30 min at 4°C. The beads were removed by centrifugation at 2500 rpm 5 min at 4°C, and the supernatant treated with 10 µg of each anti-YY2 antibody (sc-47637 and sc-47635). Protein-G Agarose was added to the preparation and agitated overnight at 4°C. Beads were then rinsed with RIPA buffer, and boiled in 1× Laemmli electrophoresis buffer for 2 min to isolate the immunoprecipitated proteins. The labeled-immunoprecipitated proteins were then separated using standard 10% SDS–PAGE, and autoradiographed.
Eletrophoretic mobility shift assay
To prepare nuclear extracts, cells were washed with PBS, lifted from dishes by scraping, and incubated in hypotonic buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT). Nuclei were isolated by Dounce-homogenization and collected by centrifugation at 14 000 r.p.m. for 20 min. Proteins were extracted from pellets by high-salt buffer (20 mM HEPES, pH 7.9, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT), and dialyzed with storage buffer (20 mM HEPES, pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT). All buffers were supplemented with fresh protease inhibitors and all procedures were performed either on ice or at 4°C. Double-stranded oligonucleotides were end-labeled with 32P. To perform DNA binding, 5 µg nuclear extracts were incubated with 5 × 104 cpm of probes in the presence of 10 µg/ml poly(dIdC) in binding buffer (10 mM Tris, pH 7.5, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 5 mM MgCl2, 5% glycerol). After 5 min of incubation, mixtures were fractionated at 4% polyacrylamide/0.5 × TBE at 280 V, 4°C for 1 h.
UV irradiation
Two thousand cells per well were seeded in 96-well tissue culture plates and grown overnight. The four cell lines to be tested (LKO vector, LKO-shYY1, LKO-shYY2 and LKO-shYY1 and two combined) were seeded within individual plates at interval rows to avoid exposure and measurement variations. Plates were placed on the glass plate of a UVP Bioimaging System. A stack of microplate films (MJ Research) was inserted under the 96-well plates. These films reduced the UV permeability so that exposure time could be precisely controlled. Cells were irradiated from 10 to 90 s. Cells were incubated for 6 days after irradiation and measured by MTT assays. All readings were normalized to an unexposed plate.
RESULTS
YY1 and YY2 expression
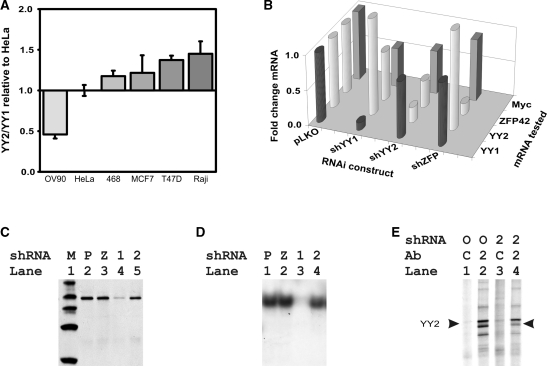
Studies of the YY family proteins have to date failed to address their interactions, despite strong homology in their DNA-binding domains. Interestingly, the genes for both YY2 and ZFP42 are expressed retrotransposons that have recently appeared in the mammalian genome. Since YY2 was initially described in HeLa cells, we first compared its expression levels to YY1 levels in those cells. YY1 was present at 751 ± 0.1 mRNA copies and YY2 was present at 10.3 ± 0.7 copies per nanogram total RNA. ZFP42 levels were at the border of detectability in HeLa cells. We then compared YY1 and YY2 mRNA copy numbers in five additional cell lines to assess their relative expression of YY1 and YY2 (Figure 1A). Notably, YY1 and YY2 levels varied little between cell lines and we consequently used HeLa cells for further studies.
Figure 1.
Altered expression of YY1 and YY2. (A) Copy numbers of YY1 and YY2 mRNAs in total RNA extracted from HeLa cells were compared with those in five other cell lines. Copy numbers were derived from qRT-PCR using plasmids pCEP4-FLAGYY1 and pCEP4-HAYY2 to provide standards. The YY2/YY1 copy number ratio was compared to that of HeLa and is plotted for each cell line. Standard deviations (SD) are shown, n = 3. The indicated cells are OV90, HeLa, MDA-MB-468 (468), MCF7, T47D and Raji cells. (B) shRNAs decreased YY family and Myc gene expression. The z-axis plots fold change between LKO-vector expressing control cells and the indicated shRNA identified along the x-axis. The y-axis identifies the mRNA assayed using qRT-PCR. (C) Western blots revealed reduced expression of YY1 in shYY1-treated cells (lane 4), but not in pLKO-, shZFP42- and shYY2-treated cells (lanes 2, 3, 5). Lane 1, MagicMark protein ladder (Invitrogen). (D) Electrophoretic mobility shift assay indicated loss of binding activity to the YY1-binding element in shYY1 treatment. Lane 1, pLKO vector treatment; Lane 2, shZFP42 treatment; Lane 3. shYY1 treatment; Lane 4. shYY2 treatment. (E) Immunoprecipitation of YY2 from 35S-methionine incorporated cells. Lanes 1 and 2, HeLa cells treated with control pLKO vectors; Lanes 3 and 4, HeLa cells treated with shYY2. Lanes 1 and 3, lysates were incubated with non-specific goat serum as antibody controls. Lanes 2 and 4, lysates were incubated with anti-YY2 antibodies. The indicated YY2-specific band migrates just under 60 kD, as expected.
We expressed four to five independent lentiviral shRNA constructs for each of the three YY family members and identified the transduced cells exhibiting the strongest knock down effect for each gene. We compared the knock down effects of the strongest shRNA on its targeted mRNA and on each of the other YY family members. The three shRNAs had minimal effects on other YY family members (Figure 1B). We also assessed their effects on c-myc since it is a known YY1-regulated gene (40). Of note, both the 90% knock down of YY1 and the 50% knock down of YY2 had similar net effects on Myc gene expression, which decreased by 44% in shYY1-expressing cells, and 38% in shYY2-expressing cells. Thus, despite differing mRNA levels of YY1 and YY2, and the 2-fold difference in knock down effects, myc mRNA decreases were very similar in both knock downs, a strong argument for a significant contribution of YY2 to transcriptional regulation. The decreases in myc mRNA are consistent with YY1’s reported role as an initiator-binding transcription factor for the cognate site on the Myc promoter and suggest that YY2 shares the same function.
Downregulation of YY1 and YY2 proteins were also observed in response to the shRNA constructs. Immunoblots showed significantly reduced YY1 protein in shYY1-treated cells, but no change in pLKO vector-, shYY2- or shZFP42-expressing cells (Figure 1C). Correspondingly, nuclear extracts from shYY1-treated cells showed loss of binding activity to the YY1-cognate site (Figure 1D). Endogenous YY2 was not detectable by western blot due to either low protein levels and/or weak YY2 antibody activities. Two Santa Cruz antibodies recognizing the over-expressed HA-tagged YY2 protein were used to immunoprecipitate 35S-radiolabeled cells (Figure 1E). We identified reduced amounts of a protein migrating just under 60 kDa corresponding to YY2 protein in shYY2-treated cells, along with multiple co-precipitated bands.
Gene set enrichment analysis and quantitative real-time polymerase chain assessment of array performance
We performed gene-expression profiling to gain perspective on the relationships between genes regulated by the YY family members. RNA was harvested for profiling and analyzed using Affymetrix human genome U133 Plus 2.0 chips as described in methods. While knock down of YY1 and YY2 caused significant expression changes as assessed by statistical analysis of microarrays (SAM) (41), knock down of ZFP42 did not (Supplementary Figure S1). We therefore focused the remainder of our analyses on YY1 and YY2 since the lack of significance in array data for the ZFP42 knock downs was likely due to its borderline expression in the parental cells.
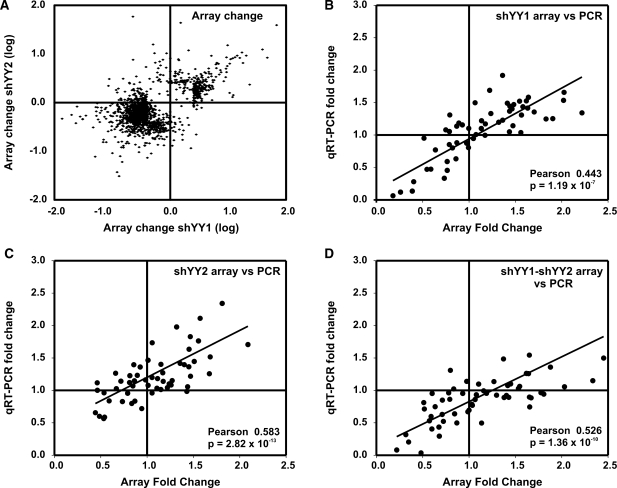
In general, genes responded similarly to YY1 and YY2 knock down in expression microarrays (Figure 2A). To identify the sets of genes that responded to decreased YY1 expression and/or decreased YY2, we applied gene set enrichment analysis (GSEA) using compiled gene sets from the Broad Institute (34,35,37,42) (Table 1–3). GSEA tests for the enrichment of genes in rank-ordered gene lists based on their relatedness to observed changes in test samples. Enrichment scores (ES) are calculated as a running sum statistic, which increases for every gene change that matches an equivalent change in a compiled gene set and decreases for every gene change absent from those sets (35,37). The statistical significance of the ES compares the measured ES to a calculated score assuming a null distribution. False determination rates (FDRs) calculate the probability that a given ES represents a false positive in multiple hypothesis testing. As shown for the individual genes, similar gene sets were affected by shYY1 and shYY2. Consequently, we sought to determine whether combined decreases in both YY1 and YY2 would have additive effects by expressing both shRNAs together using combined G418 and puromycin selection. We identified cells that expressed both the shYY1 and the shYY2 at the same levels when they were combined as when they were individually expressed (data not shown).
Figure 2.
Comparison of array versus qRT-PCR measurements of mRNA changes in cells expressing shYY1, shYY2 and shYY1/2. (A) Shown is a scatter plot comparing the effects of shYY1 to those of shYY2 in microarray experiments. We show mRNA signals that changed 2-fold or greater in the respective knock down cells from control cell levels. The x-axis plots the log of the change in the array signal comparing control pLKO cells to shYY1-expressing cells; the y-axis plots the log of the change in the arrays from control cells to shYY2-expressing cells for each probe. (B–D) We then developed primer sets for qRT-PCR analyses for 57 mRNAs identified from our microarray data as showing significant responses to knock down of YY1, YY2 and/or their combined knock down. (B) We show a scatter plot comparing microarray (x-axis) and qRT-PCR measurements (y-axis) of the mean fold change in expression of each gene comparing LKO-transduced control cells and cells expressing the shYY1 construct. The array data shows the mean for array data from all probe sets for each gene. The qRT-PCR shows the mean for three determinations for each gene. A Pearson correlation analysis was performed comparing the mean qRT-PCR result and the mean for each individual probe set for each gene. Shown is the Pearson correlation coefficient and the corresponding P-value. (C) We show a similar scatter plot as in A but we compare the array and qRT-PCR results for changes between LKO-control transduced cells and shYY2-expressing cells. (D) We show a similar scatter plot comparing control LKO-transduced cells and cells expressing both shYY1 and shYY2.
Table 1.
Gene sets responding to both shYY1 and shYY2 using gene set analysis
| NAME | YY1 NES | YY1 NOM P-value | YY1 FDR Q-value | NAME | YY2 NES | YY2 NOM P-value | YY2 FDR Q-value | |
|---|---|---|---|---|---|---|---|---|
| Growth | ||||||||
| RIBOSOMAL PROTEINS | 3.508 | 0.000 | 0.000 | RIBOSOMAL PROTEINS | 2.685 | 0.000 | 0.000 | |
| MORF TPT1 | 3.432 | 0.000 | 0.000 | MORF TPT1 | 2.822 | 0.000 | 0.000 | |
| GCM TPT1 | 3.364 | 0.000 | 0.000 | GCM TPT1 | 2.783 | 0.000 | 0.000 | |
| GNF2 TPT1 | 3.267 | 0.000 | 0.000 | GNF2 TPT1 | 2.735 | 0.000 | 0.000 | |
| GNF2 DAP3 | 2.674 | 0.000 | 0.000 | GNF2 DAP3 | 2.327 | 0.000 | 0.000 | |
| GNF2 EIF3S6 | 3.307 | 0.000 | 0.000 | GNF2 EIF3S6 | 2.629 | 0.000 | 0.000 | |
| Myc | ||||||||
| SCHUMACHER MYC UP | −2.141 | 0.000 | 0.021 | IRITANI ADPROX DN | 2.352 | 0.000 | 0.000 | |
| LEE MYC UP | 1.800 | 0.000 | 0.060 | IRITANI ADPROX VASC | 2.286 | 0.000 | 0.000 | |
| Other transcription factors | ||||||||
| V$AR 02 | −1.583 | 0.026 | 0.326 | V$AR 02 | −1.422 | 0.075 | 0.596 | |
| V$HOX13 01 | −1.422 | 0.075 | 0.657 | V$HOX13 01 | −1.497 | 0.052 | 0.542 | |
| KTGGYRSGAA UNKNOWN | −1.797 | 0.004 | 0.169 | KTGGYRSGAA UNKNOWN | −1.543 | 0.013 | 1.000 | |
| MORF JUND | 2.984 | 0.000 | 0.000 | MORF JUND | 2.340 | 0.000 | 0.000 | |
| DNA damage | ||||||||
| ATRBRCA PATHWAY | −1.966 | 0.000 | 0.036 | ATRBRCAPATHWAY | −1.591 | 0.034 | 0.608 | |
| Growth factors | ||||||||
| IFN BETA GLIOMA DN | 2.182 | 0.000 | 0.001 | IFN BETA GLIOMA DN | 2.520 | 0.000 | 0.000 | |
| UV response | ||||||||
| UVB NHEK1 C1 | 2.309 | 0.000 | 0.001 | UVB NHEK1 C1 | 2.697 | 0.000 | 0.000 | |
| UVB NHEK2 UP | 1.854 | 0.000 | 0.056 | UVB NHEK2 UP | 2.488 | 0.000 | 0.000 | |
| Tissue type | ||||||||
| AGED MOUSE HYPOTH UP | 1.725 | 0.002 | 0.100 | AGED MOUSE HYPOTH UP | 2.109 | 0.000 | 0.001 | |
| Miscellaneous | ||||||||
| MORF ACTG1 | 3.455 | 0.000 | 0.000 | MORF ACTG1 | 2.857 | 0.000 | 0.000 | |
| MORF NPM1 | 3.320 | 0.000 | 0.000 | MORF NPM1 | 2.811 | 0.000 | 0.000 | |
| GNF2 ST13 | 3.147 | 0.000 | 0.000 | GNF2 ST13 | 2.415 | 0.000 | 0.000 | |
| MORF NME2 | 3.024 | 0.000 | 0.000 | MORF NME2 | 2.893 | 0.000 | 0.000 | |
| GCM NPM1 | 2.750 | 0.000 | 0.000 | GCM NPM1 | 2.502 | 0.000 | 0.000 | |
| GNF2 GLTSCR2 | 2.643 | 0.000 | 0.000 | GNF2 GLTSCR2 | 2.162 | 0.000 | 0.000 | |
| GCM ACTG1 | 2.353 | 0.000 | 0.000 | GCM ACTG1 | 2.294 | 0.000 | 0.000 |
NES are calculated by a running sum statistic, which increases for gene changes matching equivalent changes in compiled gene sets and decreases for gene changes absent from sets, normalized to the size of the set. The statistical nominal P-value (NOM P-value) compares the measured NES to a calculated null distribution. The false determination rate (FDR) is the estimated probability that a given ES represents a false positive given multiple hypotheses being tested. The molecular signature set annotations are found at: http://www.broad.mit.edu/gsea/msigdb/annotate.jsp.
Table 2.
Gene sets responding to shYY1 using gene set analysis
| NAME | Type | SIZE | YY1 ES | YY1 NES | YY1 NOM P-value | YY1 FDR Q-value |
|---|---|---|---|---|---|---|
| E2F | ||||||
| REN E2F1 TARGETS | Gene set | 37 | −0.553 | −2.004 | 0.000 | 0.042 |
| CELL CYCLE CHECKPOINT | Gene set | 23 | −0.554 | −1.743 | 0.004 | 0.142 |
| Growth | ||||||
| MORF EIF4A2 | Computational | 136 | 0.491 | 2.205 | 0.000 | 0.000 |
| MORF EIF4E | Computational | 83 | 0.483 | 2.038 | 0.000 | 0.011 |
| Other transcription factors | ||||||
| SANSOM APC LOSS5 UP | Gene set | 63 | −0.498 | −2.010 | 0.000 | 0.055 |
| GAY YY1 DN | Gene set | 161 | −0.391 | −1.846 | 0.000 | 0.079 |
| Tissue type | ||||||
| HYPOPHYSECTOMY RAT UP | Gene set | 26 | 0.698 | 2.173 | 0.000 | 0.000 |
| STRIATED MUSCLE CONTRACTION | Gene set | 15 | 0.712 | 2.004 | 0.000 | 0.015 |
| MUNSHI MM UP | Gene set | 56 | 0.536 | 1.938 | 0.000 | 0.008 |
| Miscellaneous | ||||||
| CMV IE86 UP | Gene set | 49 | −0.522 | −1.990 | 0.000 | 0.038 |
| MOREAUX TACI HI IN PPC UP | Gene set | 70 | 0.479 | 1.967 | 0.000 | 0.043 |
| PARK MSCS DIFF | Gene set | 27 | 0.584 | 1.926 | 0.000 | 0.030 |
| GCM PSME1 | Computational | 83 | 0.492 | 2.063 | 0.000 | 0.001 |
| GNF2 BNIP3L | Computational | 44 | −0.489 | −1.819 | 0.000 | 0.042 |
| GNF2 CKS1B | Computational | 37 | −0.536 | −1.936 | 0.000 | 0.021 |
| GNF2 FBL | Computational | 145 | 0.593 | 2.710 | 0.000 | 0.000 |
| GNF2 FGR | Computational | 15 | 0.662 | 1.879 | 0.004 | 0.004 |
| GNF2 MBD4 | Computational | 24 | 0.598 | 1.932 | 0.002 | 0.003 |
| GNF2 NPM1 | Computational | 70 | 0.498 | 2.024 | 0.000 | 0.001 |
| GNF2 RFC3 | Computational | 41 | −0.518 | −1.911 | 0.000 | 0.016 |
| GNF2 RFC4 | Computational | 60 | −0.486 | −1.916 | 0.000 | 0.019 |
As in Table 1 except that type refers to the type of database used, size identifies the number of genes tested in the indicated set and the ES is the non-normalized enrichment score, without normalization to the size of the dataset.
Table 3.
Gene sets responding to shYY2 using gene set analysis
| Name | Type | Size | YY2 ES | YY2 NES | YY2 NOM P-value | YY2 FDR Q-value |
|---|---|---|---|---|---|---|
| UV damage | ||||||
| UVB NHEK1 UP | Gene set | 123 | 0.485 | 2.171 | 0.000 | 0.000 |
| UVB SCC UP | Gene set | 82 | 0.520 | 2.147 | 0.000 | 0.001 |
| UVC HIGH D2 DN | Gene set | 36 | −0.420 | −1.547 | 0.020 | 0.600 |
| Mitochondrial function | ||||||
| MOOTHA VOXPHOS | Gene set | 75 | 0.629 | 2.558 | 0.000 | 0.000 |
| ELECTRON TRANSPORT CHAIN | Gene set | 92 | 0.561 | 2.376 | 0.000 | 0.000 |
| Tissue type | ||||||
| PLATELET EXPRESSED | Gene set | 27 | 0.663 | 2.185 | 0.000 | 0.000 |
| GNATENKO PLATELET UP | Gene set | 36 | 0.654 | 2.282 | 0.000 | 0.000 |
| GNATENKO PLATELET | Gene set | 36 | 0.654 | 2.299 | 0.000 | 0.000 |
| Transcription factors | ||||||
| KRCTCNNNNMANAGC UNKNOWN | Cis | 20 | 0.645 | 1.943 | 0.000 | 0.032 |
| Growth factors | ||||||
| EGF HDMEC UP | Gene set | 38 | 0.598 | 2.131 | 0.000 | 0.001 |
| Miscellaneous | ||||||
| ET743 SARCOMA UP | Gene set | 57 | 0.540 | 2.105 | 0.000 | 0.001 |
| GUO HEX DN | Gene set | 40 | 0.602 | 2.193 | 0.000 | 0.000 |
| PROTEASOME PATHWAY | Gene set | 21 | 0.708 | 2.175 | 0.000 | 0.000 |
| MORF AP2M1 | Computational | 214 | 0.437 | 2.121 | 0.000 | 0.000 |
| MORF ATOX1 | Computational | 79 | 0.540 | 2.218 | 0.000 | 0.000 |
| MORF ERH | Computational | 113 | 0.482 | 2.132 | 0.000 | 0.000 |
| MORF PRDX3 | Computational | 84 | 0.519 | 2.160 | 0.000 | 0.000 |
| MORF RAD21 | Computational | 177 | 0.419 | 1.991 | 0.000 | 0.001 |
| MORF RAN | Computational | 263 | 0.484 | 2.404 | 0.000 | 0.000 |
| CTGAGCC,MIR-24 | cis | 140 | −0.417 | −1.976 | 0.000 | 0.027 |
As in Table 1 except that type refers to the type of database used, size identifies the number of genes tested in the indicated set and the ES is the non-normalized enrichment score, without normalization to the size of the dataset.
We validated the microarray data by comparing expression changes in microarrays to expression changes determined by qRT-PCR in the various knock down cell lines. We performed qRT-PCR to evaluate a total of 57 mRNAs, choosing 7–13 individual genes from each gene set that best represented changes occurring within those sets (Supplementary Table S2). Shown are scatter plots comparing the array fold change versus the qRT-PCR fold change for the shYY1, shYY2 and shYY1-shYY2 cells, comparing the mean array and qRT-PCR data for all 57 mRNAs (Figure 2B and C). Our array and qRT-PCR values were highly correlated using Pearson tests for significance.
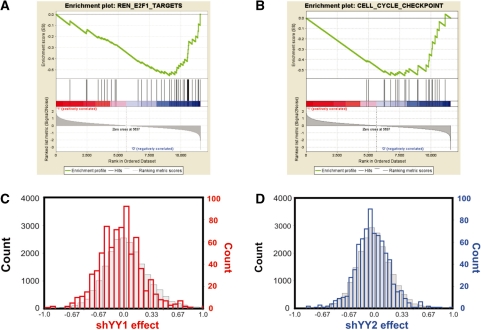
Many gene sets were altered in a similar manner by shYY1 and shYY2 (Table 1). The maximum ES for any individual gene set was found for ribosomal protein mRNAs, and enrichment plots are shown for shYY1 (Figure 3A) and shYY2 (Figure 3B). We also show histogram plots that compare expression changes for 198 ribosomal protein mRNAs to changes for all other mRNAs, which demonstrate that the YY1 knock down generally increased ribosomal protein mRNA levels (Figure 3C). Ribosomal protein mRNAs increased more in response to shYY1 than to shYY2 (Figure 3D), although this comparison also reflects the 90% decrease in YY1 expression versus the 50% decrease in YY2 in their respective knock down cells. A comparison of shRNA effects for ribosomal protein mRNAs generally shows that shYY1 and shYY2 both increased most ribosomal protein mRNAs (Figure 3A). However, unlike Pho and Phol whose combined mutations caused additive effects on homeotic gene expression, the combination of shYY1 and shYY2 was non-additive for ribosomal protein mRNAs (Figure 3E).
Figure 3.
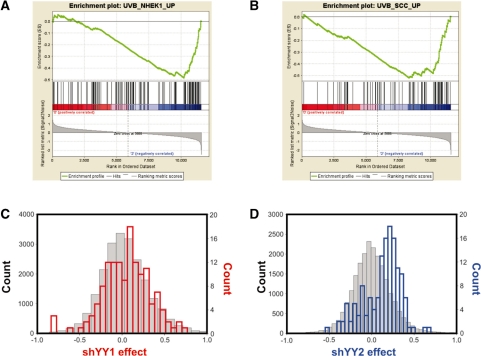
shYY1 and shYY2 effects on ribosomal protein mRNA expression. Gene Set Enrichment Analysis (GSEA) determines whether a defined set of genes shows concordant changes between two biological states. Enrichment scores (ES) are calculated by a running sum statistic, which increases for gene changes matching equivalent changes in compiled gene sets and decreases if changes were not seen for the given gene set. The enrichment plots shown here plot the concordance of ribosomal proteins mRNA changes with the ‘Ribosomal_Protein’ curated data set at: http://www.broad.mit.edu/gsea/msigdb/annotate.jsp. Shown are plots for shYY1 versus the LKO-vector-expressing cells (A). The enrichment score increments for every gene whose expression changes in response to shYY1 that is a ribosomal protein gene (hits). In this case, nearly all of the ribosomal protein mRNAs evaluated in these arrays increase in cells targeted by shYY1. In (B), cells expressing shYY2 show a similar response to YY2 knock down, although to a somewhat lesser extent. (C and D) Shown are histogram plots for the average change in the microarray signal for each of 198 ribosomal mRNAs compared with all other mRNA signals in the microarrays. The count (y-axis) shows the number of mRNAs deviating to the extent shown on the x-axis, which is the average deviation between the signal in the shYY arrays compared to the LKO vector control RNAs. [shYY1 and shYY2 effect shows fold change (ln) from control for the indicated bins.] The red histograms (C) show shYY1-induced changes and the blue histograms are for shYY2-induced changes (D). For the ribosomal protein mRNAs the y axes are on the right side of the plots. The grey histograms depict changes in all non-ribosomal protein mRNAs; their corresponding counts are in black on the left y axes. (E) qRT-PCR measurements of shYY1 and shYY2 effects on individual ribosomal protein mRNAs. We used qRT-PCR to evaluate the changes in 12 ribosomal mRNAS. Error bars demonstrate standard deviations, n = 3.
YY1 and YY2 differential functions
While YY1’s global effects on cell division, mitosis and DNA damage repair have been well-described, no YY2-specific functions or target genes have been identified. In order to determine whether YY1 and YY2 have any unique target genes, we examined genes most altered by shYY1 and shYY2 in microarray data and validated them using qRT-PCR (Figure 4). We identified genes most perturbed by shYY1 (Figure 4A and B), most perturbed by shYY2 (Figure 4C and D), most decreased by both shYY1 and shYY2 (Figure 4E and F), most increased by shYY2 and shYY1 (Figure 4G and H), most increased by shYY1 while decreased by shYY2 (Figure 4I and J) and vice versa (Figure 4K and L). Our array data were generally confirmed by the qRT-PCR results as shown in Figure 2B–D.
Figure 4.
YY1- and YY2-specific target genes identified by microarrays were validated by qRT-PCR. (A), (C), (E), (G), (I), (K) were derived from microarray; panels (B), (D), (F), (H), (J), (L) depict qRT-PCR results plotted as fold change in mRNA levels between shRNA-expressing and control vector-expressing cells. The mRNAs assayed are identified by their gene names along the x axes of each plot, including measurements of YY1 and YY2 mRNAs. Columns graph the fold-change for the identified genes in cells expressing shYY1 (light gray bars), cells expressing shYY2 (open bars) and cells expressing shYY1/2 (dark gray bars). Error bars = SD, n = 3. (A), (B) Genes most affected by shYY1. (C), (D) Genes most affected by shYY2. (E), (F) Genes most downregulated by shYY1 and shYY2. (G), (H) Genes most upregulated by shYY1 and shYY2. (I), (J) Genes increased by shYY1 but decreased by shYY2. (K), (L) Genes decreased by shYY1 but increased by shYY2.
The E2F-related gene set contains YY1 targets
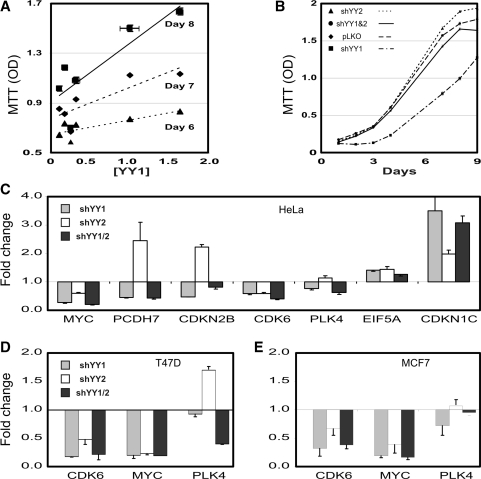
We identified gene sets responding more to shYY1 than to shYY2 using GSEA (Table 2). The most significant YY1 target sets were those involved in cell-cycle regulation, including an E2F target set (43) (Figure 5A and B). Previous reports of combinatorial interactions between YY1 and E2F (24,44) and its known effects on cell proliferation (20) validate our approach since YY1 has previously been shown to alter E2F targets. We could detect little effect of the shYY2 on these genes, as shown in histogram plots of mRNA changes (Figure 5C and D). To test the reproducibility of the shYY1 effects, we compared cell proliferation in cells transduced with all five shYY1 constructs available from the RNAi consortium. We plotted the cell density on Days 6, 7 and 8 after plating for each of the five knock down pools with respect to the levels of YY1 mRNA in each of the five pools. We found that cell proliferation decreased incrementally with decreasing YY1 levels (Figure 6A). We then tested the effects of shYY2 in proliferation assays using the most effective shYY2 knock down cells. Surprisingly, shYY2 increased proliferation. Moreover, when shYY2 was combined with shYY1, the shYY2 reversed shYY1 effects returning cells to a proliferation rate corresponding to the normal control cells (Figure 6B). Thus, we found that YY1 and YY2 actually serve antagonistic functions in cell proliferation. qRT-PCR assays generally confirmed the microarray results in HeLa cells (Figure 6C). We therefore tested the effects of shYY1, shYY2 and shYY1/2 on three candidate genes in T47D and MCF7 cells to evaluate the reproducibility of their effects (Figure 6D and E). CDK6 and c-myc were downregulated by both shYY1 and shYY2 in all cell lines, whereas polo-like kinase 4 (PLK4) was downregulated by shYY1, and upregulated by shYY2 in all three cell lines.
Figure 5.
shYY1 and shYY2 target E2F-regulated mRNAs. (A and B) Shown are enrichment plots for two E2F target data sets that were most significantly affected by shYY1. The datasets included E2F targets from Ren et al. (43) and a curated set of cell-cycle genes. No plots are shown for shYY2 since it did not alter either E2F or cell-cycle sets. (C and D) Histograms demonstrating shYY1 and shYY2 effects on E2F target gene mRNAs were plotted as described for the ribosomal protein mRNAs. C was plotted as described for 3C, and D was plotted as described for 3D.
Figure 6.
shYY1 and shYY2 altered cell proliferation rates. (A) Cell-proliferation rates correlated with YY1 expression level. Shown are cell densities (MTT OD) obtained on the indicated days versus the YY1 mRNA levels produced by each of five shYY1 vectors and the control vector. YY1 expression levels measured by qRT-PCR are plotted as fold change from the LKO vector control as [YY1]. Points for the various constructs were plotted using a linear regression plot to show the dose effect of YY1 loss versus the cell density achieved on each day. Error bars represent standard error of means (SEM), n = 16. All differences were significantly different by t-test. (B) Cell proliferation rates for cells expressing shRNA constructs. The cell densities achieved on the indicated days are plotted as the MTT OD value. Error bars = SEM, n = 24. Again differences were statistically significant on Days 7, 8 and 9 by t-test. (C) Fold change in mRNAs for selected E2F target genes affected by shYY1 and shYY2 were determined by qRT-PCR. Error bars represent SEM, n = 3. (D) T47D cells were transduced with the shYY1, shYY2 and combined shYY1 and shYY2 vectors. Gene-expression changes were determined using qRT-PCR by evaluating the fold-change in expression for three candidate genes comparing the knock down cells to control T47D cells transduced with the LKO vector alone. Shown are the mean and SEM, n = 3. (E) MCF7 cells were analyzed using qRT-PCR as described for the T47D cells and effects on the same three candidate target genes are graphed.
YY2 antagonizes YY1 in UV irradiation response
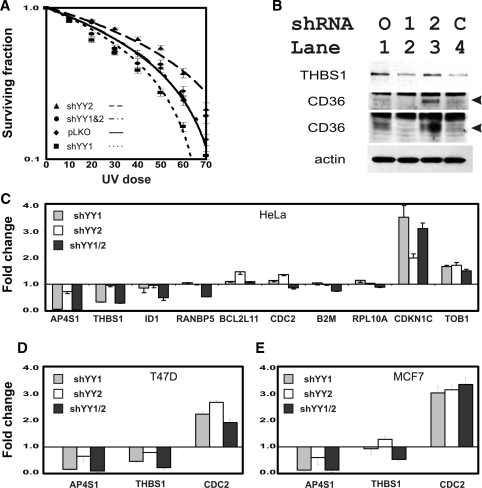
To identify potential YY2-specific functions, we examined gene sets especially affected by shYY2 in GSEA and found that ultraviolet irradiation (UV) damage is potentially targeted more by YY2 than YY1 (Table 3). YY1 knock down has previously been reported to sensitize UV response as part of its associations with the INO80 chromatin-remodeling complex (9). Thus, our finding of stronger effects for YY2 for some UV damage response expression sets was important. Two UV gene sets were particularly affected by shYY2 (45) (Figure 7A and B). The histogram comparison of shYY1 and shYY2 effects on these genes confirmed a more prominent shYY2 effect than shYY1 (Figure 7C and D). The differential effects of shYY1 and shYY2 were further demonstrated by analysis of cell survival after UV irradiation. Upon exposure to UV, shYY1 decreased the surviving fraction whereas shYY2 increased it. When the two shRNAs were co-expressed shYY2 reversed shYY1’s effects (Figure 8A) normalizing the surviving fraction at each dose.
Figure 7.
Gene sets that respond more to shYY2 than shYY1 include UV response genes. (A and B) Enrichment plots of two UV response gene sets targeted by shYY2 that were not affected by shYY1. These two data sets were derived from genome-wide comparisons of changes in gene expression in UV-irradiated keratinocytes (45). (C and D) Histogram counts of shYY1 (C) and shYY2 (D) effects on UV sensitive genes were plotted as in Figures 3 and 5.
Figure 8.
shYY1 and shYY2 affect UV damage sensitivity. (A) Cell survival curves after UV exposure. Cells expressing vector control constructs, shYY1, shYY2 and combined shYY1/2 were exposed to UV radiation as described in methods. Irradiated cells were then plated in a standard survival curve analysis. Fraction survival at the indicated UV doses was calculated by dividing the cell density measured by MTT for the indicated dose by the cell density of untreated cells grown for the same time intervals. UV sensitivity is increased in shYY1 normalized in cells expressing both shYY1 and shYY2. Error bars = standard errors, n = 24 for each dose/construct combination. (B) Immunoblots for thrombospondin (THBS1) and CD36 confirmed that protein changes matched mRNA levels measured by qRT-PCR in Figure 4C and F. The CD36 immunoblot was performed twice using differing loading amounts and exposure times to highlight the differences between its levels in lanes 3 and 1 (top CD36) and lanes 4 and 2 (second CD36). Arrows indicate the CD36-specific band. An actin loading control is shown. (C) qRT-PCR confirms expression changes of UV-responsive genes affected by shYY1 and shYY2 in HeLa cells. Error bars = SEM, n = 3. (D and E) mRNA expression changes were confirmed using qRT-PCR for three candidate genes in the T47D and MCF7 cells transduced with the knock down constructs. Gene symbols are indicated along the x-axis and the fold-change for n = 3 are shown along the y-axis. We plot the means and SEM for each gene determination. Shown are T47D cells (D) and MCF7 cells (E).
Within our list of target genes, we were intrigued to note that thrombospondin (THBS1) was one member of the UV gene set, which was known to respond to UV exposure and enhance repair (46,47). We also noted that the thrombospondin receptor CD36 is a prominent YY2-responsive gene (Table 4, Figure 4K and L) (48). To confirm the functional relevance of our RNA analyses for these particular targets, we assessed protein levels of CD36 and thrombospondin (Figure 8B). As found for its mRNA, thrombospondin protein was decreased by shYY1 and shYY1/2, but not by shYY2 alone, which corresponds to the increased UV sensitivity of the shYY1-expressing cells. As found in our qRT-PCR results, CD36 protein was decreased by shYY1 and increased by shYY2. Importantly, combining shYY1 and shYY2 caused an increase in CD36 compared with shYY1 alone (Figure 8B, CD36 lanes 4 versus 2 and 3 versus 1). This change corresponds to the normalization of the UV response in cells expressing both shYY1 and shYY2. We validated several additional array results for genes in the UV-induced gene set using qRT-PCR measurements (Figure 8C). Finally, we tested the reproducibility of three of these candidate genes (APS4, thrombospondin 1 and CDC2) in additional cell lines. We found that their responses were similar in T47D, MCF7 and HeLa cells.
Table 4.
Genes that best discriminate between YY2 and YY1
| Gene name | Gene title | YY2 t-test | YY1 versus control | YY2 versus control | YY1 YY2 versus control | YY1–YY2 avg |
|---|---|---|---|---|---|---|
| CD36 | CD36 molecule (thrombospondin receptor) | 0.048 | −0.851 | 0.947 | −0.272 | −1.798 |
| ARRDC3 | Arrestin domain containing 3 | 0.018 | −0.320 | 0.908 | 0.214 | −1.228 |
| PCDH7 | Protocadherin 7 | 0.011 | −0.415 | 0.692 | −0.387 | −1.107 |
| AIM1 | Absent in melanoma 1 | 0.038 | −0.328 | 0.756 | −0.359 | −1.084 |
| PDZK1 | PDZ domain containing 1 | 0.013 | −0.241 | 0.830 | 0.570 | −1.072 |
| SSTR1 | Somatostatin receptor 1 | 0.006 | −0.067 | 1.000 | 0.557 | −1.067 |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 0.037 | −0.580 | 0.441 | −0.678 | −1.020 |
| ITGA5 | Integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | 0.000 | −0.207 | 0.811 | 0.453 | −1.018 |
| RERG | RAS-like, estrogen-regulated, growth inhibitor | 0.019 | −0.166 | 0.781 | 0.560 | −0.947 |
| PGM2L1 | Phosphoglucomutase 2-like 1 | 0.029 | −0.174 | 0.764 | 0.615 | −0.938 |
| Gene name | Gene title | YY2 t-test | YY1 versus control | YY2 versus control | YY1 YY2 versus control | YY1–YY2 avg |
|---|---|---|---|---|---|---|
| REXO2 | REX2, RNA exonuclease 2 homolog (S. cerevisiae) | 0.041 | 0.754 | −0.032 | −0.360 | 0.786 |
| PIAS1 | Protein inhibitor of activated STAT, 1 | 0.046 | 0.622 | −0.014 | 0.099 | 0.636 |
| ATP2C2 | ATPase, Ca++ transporting, type 2C, member 2 | 0.047 | 0.611 | −0.019 | 0.791 | 0.630 |
| TTMB | TTMB protein | 0.020 | 0.573 | −0.007 | 0.401 | 0.580 |
| ZFYVE9 | Zinc finger, FYVE domain containing 9 | 0.046 | 0.556 | −0.009 | 0.013 | 0.565 |
| PRCD | Progressive rod-cone degeneration | 0.011 | 0.550 | −0.002 | 0.317 | 0.553 |
| CNN3 | Calponin 3, acidic | 0.031 | 0.542 | −0.006 | −0.432 | 0.548 |
| ZBTB20 | Zinc finger and BTB domain containing 20 | 0.021 | 0.509 | −0.007 | −4.656 | 0.516 |
| PDCD6 | Programmed cell death 6 | 0.001 | 0.494 | −0.001 | 0.107 | 0.494 |
| SORBS2 | Sorbin and SH3 domain containing 2 | 0.036 | 0.472 | −0.013 | −0.344 | 0.485 |
DISCUSSION
Although gene duplication is thought to be the single most powerful factor in evolution (49), the retrotransposition of YY1-like/Pho-like genes presents an intriguing alternative case of convergent evolution (27,32). Processed copies of genes have emerged at the constant rate of 1 gene every 2 million years in Drosophila and primates (50). Compared with point mutations, retrotransposition is a rapid evolutionary process, which is a significant source of new coding sequences in gene evolution (51). The origin and significance of YY1 homologs may be an important example of this evolution (52). The parent genes YY1 and Pho consist of five exons encoding highly homologous domains. In contrast, Phol and YY2 are encoded by a single exon each. Notably, YY2 is located in the fifth intron of Membrane-Bound Transcription factor Peptidase, Site 2 (Mbtps2) of placental mammals but not other vertebrates, and its retrotransposition is a recent event in evolution (32). Phol and YY2’s sequences are also evolving more rapidly than YY1’s. Genetic analysis shows that Pho and Phol serve redundant roles in homeotic control in flies (10), whereas reporter constructs suggest that YY1 and YY2 can be antagonistic when over-expressed (33). Why convergent selection for an alternative YY1-like-coding sequence has occurred throughout evolution is intriguing and suggests that the alternative YY1-like proteins serve important regulatory functions that balance and modulate potent YY1 effects. Here we show that YY2 indeed contributes to overall gene-expression control.
A previous microarray study evaluated YY1 disruption in mouse embryonic fibroblasts, and identified YY1 targets (20). Importantly, our GSEA found that the gene set containing those targets in ES cells also responded to shYY1-knock downs in HeLa cells (Table 1), validating our approach. However, our YY1 and YY2 knock downs had their largest effects on ribosomal protein genes, which have not been noted in YY1 studies subsequent to its initial discovery. Transcriptional control elements are tightly conserved across all ribosomal protein genes (53). A highly conserved downstream-binding site (δ) was found in the earliest studies of ribosomal protein promoters, which was then shown to be the binding site for the factor that is now known as YY1 (12,54–58). Ribosomal protein genes have not emerged in subsequent analyses of YY1’s functions as critical targets, so our shRNA/GSEA strategy is the first to re-identify this previously described function for YY1. Both YY1 and YY2 work mainly as repressors of ribosomal gene transcription according to our results, but it is interesting to note that combined loss of both factors had no additive effects on their targets—a substantially different situation from Pho and Phol in flies.
Having established that YY1 and YY2 functions were non-additive for ribosomal gene expression, we then explored potentially different or antagonistic functions for other gene sets. Our shRNA/GSEA strategy agreed with other studies that E2F and cell proliferation are significant targets of YY1 (Table 2, Figure 5). We then analyzed the effects of shYY2 and combined shYY1/2 on cell proliferation and E2F target genes (Figure 6). Notably, YY2 downregulation actually enhanced proliferation in contrast to shYY1’s known effects on E2F functions and proliferation (24,25). In combination with YY1 knock down, decreased YY2 expression then normalized cell proliferation. This clear demonstration of antagonism on a significant cellular process shows that YY2 can serve important roles different from those of YY1. Moreover, YY1 and YY2 had clearly distinctive effects on PLK4 and on an important cell-cycle regulator, CDKN2B (p15INK4B).
We further explored YY2 responsive gene sets and discovered that UV responsive genes include YY2 targets that were not identified as YY1 targets. Importantly, YY1 participates in homologous recombination DNA repair (HRR) as a chromatin-remodeling complex INO80 component (9), and loss of YY1 sensitizes cells to UV damage. Importantly, we found that YY1 and YY2 had somewhat different effects on genes identified in two UV gene sets. Correspondingly, YY2 loss antagonized YY1 loss in survival after UV irradiation. This raises the possibility that YY2 may contribute to alternative DNA-repair complexes, similar to the case for Phol that nucleates distinctive polycomb complexes. Our data particularly confirm the importance of thrombospondin’s role in UV response regulation, and reveal a new mechanism for its regulation (46,59,60). Although CD36 is the thrombospondin receptor (48,61), to our knowledge its contribution to UV response has not been previously investigated. More importantly, CD36 is a particularly interesting YY1–YY2 target gene given its remarkable differential response to YY1 and YY2, and the importance of its roles in inflammation, malaria infection, tumor growth, uptake of dietary fat, atherogenesis and innate immune function (62–68). No doubt, all of these processes may be indirectly under control of YY1 and YY2, which greatly broadens the scope of physiological functions whose regulation by YY1 and YY2 deserve increased attention.
Taken together our data show that YY2 functions as a gene-expression regulator that may be as important as YY1 in target selection, which is involved in cellular activities ranging across proliferation, tissue type determination and UV response to oxidative phosphorylation. Unlike Phol, YY2 is not a fully redundant factor of YY1. It possesses several different regulatory target sets, and for some targets shared with YY1 it can exert opposite rather than complementary effects. Indeed, given that YY2 is less abundant than YY1 and shYY2 is less effective than shYY1in gene knock down, our data suggest that YY2 is a more potent regulator than might be initially expected. Moreover, our data re-assert a role for YY1 as a Myc regulator, which has received little attention since its initial description (40,69,70) and show that YY2 also potently regulates Myc. After the ribosomal protein set, the Myc-target gene set is the second most significantly altered gene set affected by both YY1 and YY2 (Table 1). Given that YY1 and YY2 decreased Myc mRNA by similar amounts (Figure 1B), this result should be expected. It further raises the question whether some YY1 and 2 target genes are actually indirect c-Myc targets. This question is especially important given that dMyc targets show remarkable overlap with those regulated by polycomb (71,72). To date this overlap has not been recapitulated in human cells, but our data show remarkable parallels to the fly result. In addition, this YY1–Myc interaction may provide the first explanation for Myc’s tendency to regulate ribosomal protein genes in the absence of genomic Myc-binding sites in those genes (73,74). Future interaction studies should begin to unravel this connection, and continue to identify new physiologic functions that may be differentially regulated by YY1 and its important homolog, YY2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
PHS RO1-CA63117 and UO1 AI070330; RO1-CA69069 to LC and EVS. The Molecular Profiling Laboratory of the Cancer Research Center at MGH provided service and technological support for microarray processing and data generation. Their support came from the Susan G. Komen for the Cure (FAS0703860) and the AstraZeneca-MGH Infrastructure Key Alliance Fund to TS. Funding for open access charge: UO1 AI070330.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Yang Shi of Harvard Medical School for providing the YY1 cDNA plasmid and Dr Ed Seto of University of South Florida for the YY2 cDNA plasmid. Primer sequences used in qRT-PCR were obtained from Massachusetts General Hospital Primer Bank. The Molecular Profiling Laboratory in Massachusetts General Hospital Cancer Research Center provided service and technical support for microarray analysis. The microarray data used for this work were deposited with the National Center for Biotechnology Information Gene Expression Omnibus database where they are listed under the accession number GSE14964.
REFERENCES
- 1.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 2.Kohler C, Villar CB. Programming of gene expression by Polycomb group proteins. Trends Cell. Biol. 2008;18:236–243. doi: 10.1016/j.tcb.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Denell RE. Homoeosis in Drosophila. I. Complementation studies with revertants of Nasobemia. Genetics. 1973;75:279–297. doi: 10.1093/genetics/75.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hormaeche I, Licht JD. Chromatin modulation by oncogenic transcription factors: new complexity, new therapeutic targets. Cancer Cell. 2007;11:475–478. doi: 10.1016/j.ccr.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Alkema MJ, Bronk M, Verhoeven E, Otte A, van 't Veer LJ, Berns A, van Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- 7.Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 8.Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H, Lu J, Qi HH, Wang W, Nickoloff JA, et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct. Mol. Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JL, Fritsch C, Mueller J, Kassis JA. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen N, Zhang X, Olashaw N, Seto E. Molecular cloning and functional characterization of the transcription factor YY2. J. Biol. Chem. 2004;279:25927–25934. doi: 10.1074/jbc.M402525200. [DOI] [PubMed] [Google Scholar]
- 12.Hariharan N, Kelley DE, Perry RP. Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc. Natl Acad. Sci. USA. 1991;88:9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 14.Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, Gyory I, Wright K, Seto E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003;17:1019–1029. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, et al. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- 16.Austen M, Cerni C, Luscher-Firzlaff JM, Luscher B. YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene. 1998;17:511–520. doi: 10.1038/sj.onc.1201968. [DOI] [PubMed] [Google Scholar]
- 17.Bain M, Sinclair J. Targeted inhibition of the transcription factor YY1 in an embryonal carcinoma cell line results in retarded cell growth, elevated levels of p53 but no increase in apoptotic cell death. Eur. J. Cell. Biol. 2005;84:543–553. doi: 10.1016/j.ejcb.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 18.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorente M, Perez C, Sanchez C, Donohoe M, Shi Y, Vidal M. Homeotic transformations of the axial skeleton of YY1 mutant mice and genetic interaction with the Polycomb group gene Ring1/Ring1A. Mech. Dev. 2006;123:312–320. doi: 10.1016/j.mod.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giangrande PH, Zhu W, Rempel RE, Laakso N, Nevins JR. Combinatorial gene control involving E2F and E Box family members. EMBO J. 2004;23:1336–1347. doi: 10.1038/sj.emboj.7600134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JD, Faulk C, Kim J. Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1. Nucleic Acids Res. 2007;35:3442–3452. doi: 10.1093/nar/gkm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mongan NP, Martin KM, Gudas LJ. The putative human stem cell marker, Rex-1 (Zfp42): Structural classification and expression in normal human epithelial and carcinoma cell cultures. Mol. Carcinog. 2006;45:887–900. doi: 10.1002/mc.20186. [DOI] [PubMed] [Google Scholar]
- 29.Hosler BA, LaRosa GJ, Grippo JF, Gudas LJ. Expression of REX-1, a gene containing zinc finger motifs, is rapidly reduced by retinoic acid in F9 teratocarcinoma cells. Mol. Cell. Biol. 1989;9:5623–5629. doi: 10.1128/mcb.9.12.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JZ, Gao W, Yang HB, Zhang B, Zhu ZY, Xue YF. Screening for genes essential for mouse embryonic stem cell self-renewal using a subtractive RNA interference library. Stem cells (Dayton, Ohio) 2006:24, 2661–2668. doi: 10.1634/stemcells.2006-0017. [DOI] [PubMed] [Google Scholar]
- 31.Shi W, Wang H, Pan G, Geng Y, Guo Y, Pei D. Regulation of the pluripotency marker Rex-1 by Nanog and Sox2. J. Biol. Chem. 2006;281:23319–23325. doi: 10.1074/jbc.M601811200. [DOI] [PubMed] [Google Scholar]
- 32.Luo C, Lu X, Stubbs L, Kim J. Rapid evolution of a recently retroposed transcription factor YY2 in mammalian genomes. Genomics. 2006;87:348–355. doi: 10.1016/j.ygeno.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Klar M, Bode J. Enhanceosome Formation over the beta interferon promoter underlies a remote-control mechanism mediated by YY1 and YY2. Mol. Cell. Biol. 2005;25:10159–10170. doi: 10.1128/MCB.25.22.10159-10170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knowles LM, Smith JW. Genome-wide changes accompanying knockdown of fatty acid synthase in breast cancer. BMC Genomics. 2007;8:168. doi: 10.1186/1471-2164-8-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 38.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Meth. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt EV, Christoph G, Zeller R, Leder P. The cytomegalovirus enhancer: a pan-active control element in transgenic mice. Mol. Cell. Biol. 1990;10:4406–4411. doi: 10.1128/mcb.10.8.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riggs KJ, Saleque S, Wong KK, Merrell KT, Lee JS, Shi Y, Calame K. Yin-yang 1 activates the c-myc promoter. Mol. Cell. Biol. 1993;13:7487–7495. doi: 10.1128/mcb.13.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoshida Y, Brunet JP, Tamayo P, Golub TR, Mesirov JP. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS ONE. 2007;2:e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Ginkel PR, Hsiao KM, Schjerven H, Farnham PJ. E2F-mediated growth regulation requires transcription factor cooperation. J. Biol. Chem. 1997;272:18367–18374. doi: 10.1074/jbc.272.29.18367. [DOI] [PubMed] [Google Scholar]
- 45.Dazard JE, Gal H, Amariglio N, Rechavi G, Domany E, Givol D. Genome-wide comparison of human keratinocyte and squamous cell carcinoma responses to UVB irradiation: implications for skin and epithelial cancer. Oncogene. 2003;22:2993–3006. doi: 10.1038/sj.onc.1206537. [DOI] [PubMed] [Google Scholar]
- 46.Yano K, Oura H, Detmar M. Targeted overexpression of the angiogenesis inhibitor thrombospondin-1 in the epidermis of transgenic mice prevents ultraviolet-B-induced angiogenesis and cutaneous photo-damage. J. Investigative Dermatol. 2002;118:800–805. doi: 10.1046/j.1523-1747.2002.01752.x. [DOI] [PubMed] [Google Scholar]
- 47.Yano K, Kajiya K, Ishiwata M, Hong YK, Miyakawa T, Detmar M. Ultraviolet B-induced skin angiogenesis is associated with a switch in the balance of vascular endothelial growth factor and thrombospondin-1 expression. J. Investigative Dermatol. 2004;122:201–208. doi: 10.1046/j.0022-202X.2003.22101.x. [DOI] [PubMed] [Google Scholar]
- 48.Asch AS, Barnwell J, Silverstein RL, Nachman RL. Isolation of the thrombospondin membrane receptor. J. Clin. Invest. 1987;79:1054–1061. doi: 10.1172/JCI112918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 50.Bai Y, Casola C, Feschotte C, Betran E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 2007;8:R11. doi: 10.1186/gb-2007-8-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Z, Morais D, Ivanga M, Harrison PM. Analysis of the role of retrotransposition in gene evolution in vertebrates. BMC Bioinformatics. 2007;8:308. doi: 10.1186/1471-2105-8-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strano S, Rossi M, Fontemaggi G, Munarriz E, Soddu S, Sacchi A, Blandino G. From p63 to p53 across p73. FEBS Lett. 2001;490:163–170. doi: 10.1016/s0014-5793(01)02119-6. [DOI] [PubMed] [Google Scholar]
- 53.Perry RP. The architecture of mammalian ribosomal protein promoters. BMC Evol. Biol. 2005;5:15. doi: 10.1186/1471-2148-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung S, Perry RP. The importance of downstream delta-factor binding elements for the activity of the rpL32 promoter. Nucleic Acids Res. 1993;21:3301–3308. doi: 10.1093/nar/21.14.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moura-Neto R, Dudov KP, Perry RP. An element downstream of the cap site is required for transcription of the gene encoding mouse ribosomal protein L32. Proc. Natl Acad. Sci. USA. 1989;86:3997–4001. doi: 10.1073/pnas.86.11.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hariharan N, Kelley DE, Perry RP. Equipotent mouse ribosomal protein promoters have a similar architecture that includes internal sequence elements. Genes Dev. 1989;3:1789–1800. doi: 10.1101/gad.3.11.1789. [DOI] [PubMed] [Google Scholar]
- 57.Chung S, Perry RP. Importance of introns for expression of mouse ribosomal protein gene rpL32. Mol. Cell. Biol. 1989;9:2075–2082. doi: 10.1128/mcb.9.5.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dudov KP, Perry RP. Properties of a mouse ribosomal protein promoter. Proc. Natl Acad. Sci. USA. 1986;83:8545–8549. doi: 10.1073/pnas.83.22.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howell BG, Wang B, Freed I, Mamelak AJ, Watanabe H, Sauder DN. Microarray analysis of UVB-regulated genes in keratinocytes: downregulation of angiogenesis inhibitor thrombospondin-1. J. Dermatol. Sci. 2004;34:185–194. doi: 10.1016/j.jdermsci.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 60.Kim MS, Oh YJ, Lee S, Kim JE, Kim KH, Chung JH. Ultraviolet radiation attenuates thrombospondin 1 expression via PI3K-Akt activation in human keratinocytes. Photochem. Photobiol. 2006;82:645–650. doi: 10.1562/2005-09-29-RA-702. [DOI] [PubMed] [Google Scholar]
- 61.Armesilla AL, Vega MA. Structural organization of the gene for human CD36 glycoprotein. J. Biol. Chem. 1994;269:18985–18991. [PubMed] [Google Scholar]
- 62.Hajjar DP, Gotto AM. Targeting CD36: modulating inflammation and atherogenesis. Curr. Atherosclerosis Rep. 2003;5:155–156. doi: 10.1007/s11883-003-0016-z. [DOI] [PubMed] [Google Scholar]
- 63.Simantov R, Silverstein RL. CD36: a critical anti-angiogenic receptor. Front Biosci. 2003;8:s874–s882. doi: 10.2741/1168. [DOI] [PubMed] [Google Scholar]
- 64.Sid B, Sartelet H, Bellon G, El Btaouri H, Rath G, Delorme N, Haye B, Martiny L. Thrombospondin 1: a multifunctional protein implicated in the regulation of tumor growth. Crit. Rev. Oncol. Hematol. 2004;49:245–258. doi: 10.1016/j.critrevonc.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Abumrad NA. CD36 may determine our desire for dietary fats. J. Clin. Invest. 2005;115:2965–2967. doi: 10.1172/JCI26955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howlett GJ, Moore KJ. Untangling the role of amyloid in atherosclerosis. Curr. Opin. Lipidology. 2006;17:541–547. doi: 10.1097/01.mol.0000245260.63505.4f. [DOI] [PubMed] [Google Scholar]
- 67.Hazen SL. Oxidized phospholipids as endogenous pattern recognition ligands in innate immunity. J. Biol. Chem. 2008;283:15527–15531. doi: 10.1074/jbc.R700054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parsons MS, Barrett L, Little C, Grant MD. Harnessing CD36 to rein in inflammation. Endocr. Metab. Immune Disorders Drug Targets. 2008;8:184–191. doi: 10.2174/187153008785700073. [DOI] [PubMed] [Google Scholar]
- 69.Shrivastava A, Saleque S, Kalpana GV, Artandi S, Goff SP, Calame K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science. 1993;262:1889–1892. doi: 10.1126/science.8266081. [DOI] [PubMed] [Google Scholar]
- 70.Shrivastava A, Yu J, Artandi S, Calame K. YY1 and c-Myc associate in vivo in a manner that depends on c-Myc levels. Proc. Natl Acad. Sci. USA. 1996;93:10638–10641. doi: 10.1073/pnas.93.20.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goodliffe JM, Wieschaus E, Cole MD. Polycomb mediates Myc autorepression and its transcriptional control of many loci in Drosophila. Genes Dev. 2005;19:2941–2946. doi: 10.1101/gad.1352305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goodliffe JM, Cole MD, Wieschaus E. Coordinated regulation of Myc trans-activation targets by Polycomb and the Trithorax group protein Ash1. BMC Mol. Biol. 2007;8:40. doi: 10.1186/1471-2199-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–3221. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 74.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat. Rev. Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.