Abstract
Sporadic isolates of carbapenem-resistant KPC-2-producing Klebsiella pneumoniae were isolated in Tel Aviv Medical Center during 2005 and 2006, parallel to the emergence of the KPC-3-producing K. pneumoniae sequence type 258 (ST 258). We aimed to study the molecular epidemiology of these isolates and to characterize their blaKPC-carrying plasmids and their origin. Ten isolates (8 KPC-2 and 2 KPC-3 producing) were studied. All isolates were extremely drug resistant. They possessed the blaKPC gene and varied in their additional beta-lactamase contents. The KPC-2-producing strains belonged to three different sequence types: ST 340 (n = 2), ST 277 (n = 2), and a novel sequence type, ST 376 (n = 4). Among KPC-3-producing strains, a single isolate (ST 327) different from ST 258 was identified, but both strains carried the same plasmid (pKpQIL). The KPC-2-encoding plasmids varied in size (45 to 95 kb) and differed among each of the STs. Two of the Klebsiella blaKPC-2-carrying plasmids were identical to plasmids from Escherichia coli, suggesting a common origin of these plasmids. These data indicate that KPC evolution in K. pneumoniae is related to rare events of interspecies spread of blaKPC-2-carrying plasmids from E. coli followed by limited clonal spread, whereas KPC-3 carriage in this species is related almost strictly to clonal expansion of ST 258 carrying pKpQIL.
KPC-producing carbapenem-resistant Klebsiella pneumoniae strains have increasingly been reported worldwide. The first KPC-producing K. pneumoniae isolate identified in Tel Aviv Medical Center, Tel Aviv, Israel, isolated in October 2005, was a KPC-2-producing strain. During 2006 an extremely drug-resistant (XDR) KPC-3-producing carbapenem-resistant K. pneumoniae clone, sequence type (ST) 258 (pulsed-field gel electrophoresis [PFGE] type Q) emerged, causing a nationwide outbreak (9). Additional sporadic KPC-2-producing clinical isolates of XDR K. pneumoniae were identified during 2006 (9) and were recently reported from another hospital in northern Israel (7).
The epidemiology and clinical impact of Israeli KPC-producing K. pneumoniae were described previously in several studies (12, 16, 17). The major clone of K. pneumoniae (previously referred to as PFGE type Q) belongs to ST 258, which initially emerged in the United States (8). Since then, clinical isolates belonging to this clone have been detected in numerous geographic regions (13), carrying different blaKPC alleles. In the United States, for example, isolates belonging to ST 258 possess both blaKPC-2 and blaKPC-3 (8). Other places report the existence of a single allele carried by this clone, such as blaKPC-2 in Norway and Greece (15), Poland (1), and Finland (14) and blaKPC-3 in the United Kingdom (18), Sweden (15), and Italy (5).
Molecular studies of the blaKPC-carrying plasmid of K. pneumoniae ST 258 in Israel showed that it harbored a 105-kb blaKPC-3-carrying self-transmissible plasmid, pKpQIL (10), that differed from the plasmids carried by the genetically related K. pneumoniae ST 258 isolates in the United States (12).
The coexistence in our hospital of KPC-2-carrying K. pneumoniae isolates and KPC-3-carrying K. pneumoniae ST 258 isolates led to this study. We aimed to investigate the molecular epidemiology of these strains, examine their evolutionary relatedness, compare their blaKPC-carrying plasmids, and facilitate the understanding of the origin of these plasmids in K. pneumoniae by comparing them to blaKPC-2-carrying plasmids from other Enterobacteriaceae in our hospital, such as KPC-producing Escherichia coli.
(This work was performed by Azita Leavitt in partial fulfillment of the requirements for a Ph.D. from the Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel.)
(This study was presented in part at the 49th Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC], San Francisco, CA, 2009, abstr. C2-667.)
MATERIALS AND METHODS
Study isolates.
Eight carbapenem-resistant KPC-2-producing K. pneumoniae clinical isolates that comprise the entire collection of KPC-2 isolates isolated in the clinical laboratory of Tel Aviv Medical Center during 2005 and 2006 were studied; all of them showed non-Q pulsotypes (9). Two additional KPC-3-producing K. pneumoniae isolates were studied: a single non-Q strain and a representative ST 258 (PFGE type Q) KPC-3-producing isolate (Kpn557) (8). Seven KPC-2-producing E. coli isolates isolated during the same time period in our hospital (6) were used for plasmid comparison. E. coli GeneHogs (Invitrogene Corp., Dorset, United Kingdom) was used as a recipient strain in the transformation experiments.
Antibiotic susceptibility testing.
Antibiotic susceptibility testing was performed using a Vitek2 automated system (bioMerieux, Marcy l'Etoile, France). Resistance to imipenem, meropenem, and ertapenem was evaluated using agar dilution and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) (4). Susceptibility testing for colistin and tigecycline was performed by Etest according to the manufacturer's instructions (AB Biodisk, Solna, Sweden). The colistin breakpoint for susceptibility was ≤4 μg/ml according to the British Society of Antimicrobial and Chemotherapy (BSAC) criteria, and MICs for tigecycline were defined based on the U.S. Food and Drug Administration breakpoint criteria for Enterobacteriaceae (susceptible, ≤2 μg/ml; intermediate, 4 μg/ml; resistant, ≥8 μg/ml).
Determination of genetic relatedness.
The genetic relatedness of all carbapenem-resistant K. pneumoniae strains was determined by pulsed-field gel electrophoresis (PFGE) analysis and by multilocus sequence typing (MLST). PFGE was performed as previously described (9). Chromosomal restriction fragments were documented and compared using GelCompar II software (Applied Maths, Sint-Martens-Latem, Belgium). MLST was performed and analyzed using the K. pneumoniae MLST website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
PCR for determination of antibiotic resistance genes.
The identification of bla genes (including blaTEM, blaSHV, blaCTX-M, and blaOXA) and blaKPC genes in all isolates was determined by PCR as described previously (10).
Plasmid analysis and transformation.
Plasmid DNA was purified using a NucleoBond PC 100 plasmid midi-kit (Macherey-Nagel GmbH, Duren, Germany). Plasmids isolated from clinical isolates of K. pneumoniae and E. coli strains and their transformants were digested with different restriction endonucleases, such as BglII, SmaI, and EcoRV (New England Biolabs, Boston, MA), and their restriction patterns were compared. Plasmids were transformed by electroporation into the E. coli GeneHogs strain. Transformants possessing blaKPC were subjected to antibiotic susceptibility testing and further molecular characterization. Plasmid sizes were determined using S1 treatment following PFGE (10) and compared using GelCompar II software.
RESULTS AND DISCUSSION
Epidemiology and genetic relatedness of carbapenem-resistant K. pneumoniae isolates.
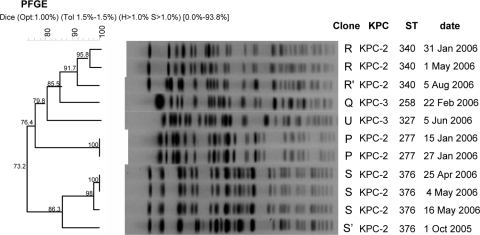
Genotyping of all the KPC-2-producing isolates revealed that they were genetically different (<85% identity) from KPC-3-producing ST 258 (PFGE type Q). These isolates belonged to three clusters: R (2 isolates), P (2 isolates), and S (4 isolates). Isolates 588 and 365 showed 91.7% and 86.3% identity with PFGE types R and S, respectively, and were designated R′ and S′, respectively (Fig. 1). Isolate 549, the KPC-3-producing isolate, belonged to PFGE type U, with 79.8% similarity with type Q. MLST data for these isolates were concordant with the PFGE data. Four STs were identified: ST 340 (PFGE type R), ST 277 (PFGE type P), a novel ST designated ST 376 (PFGE type S), and ST 327 (PFGE type U). The epidemic PFGE type Q belonged to ST 258 as reported earlier (8) (Fig. 1). ST 340 and ST 277 are genetically close variants of ST 258; ST 340 is a single locus variant (tonB allele) and ST 277 is a double locus variant (infB and tonB alleles) of ST 258. This close genetic relatedness may support the proposal of a common ancestor.
FIG. 1.
The genetic relatedness of blaKPC-2- and blaKPC-3-producing carbapenem-resistant K. pneumoniae clones in Tel Aviv Medical Center during the years 2005 and 2006. blaKPC alleles, sequence types, and the date of isolation are presented on the right. Isolates were clustered into five different clusters based on GelCompar Dice algorithm coefficients, which range from 0% to 100%, and using a tolerance of 1.5%, as illustrated by the scale to the left of each dendrogram. Two of the three ST 340 isolates were further analyzed.
Except for the epidemic, worldwide-disseminated clone K. pneumoniae ST 258, the described KPC-producing clones differed from the previously reported KPC-producing isolates from the United States (8). Inspection of all the KPC-producing K. pneumoniae STs described in the literature, including the present study, indicates that the majority of the strains are scattered throughout the K. pneumoniae evolutionary tree rather than clustered into a specific genetic lineage (S. Brisse, personal communication), suggesting that dissemination of KPC resistance is due to horizontal gene transfer rather than clonal spread. The only strains that revealed a large degree of genetic relatedness were ST 277, ST 258, and ST 340, supporting the proposal of clonal relation between these strains.
Antibiotic susceptibilities and the presence of resistance genes.
All isolates studied were extremely drug resistant irrespective of ST type. Carbapenem MICs of the KPC-2-producing isolates were similar to the MICs of the KPC-3-producing ST 258 strain. ST 258 is typically resistant to amikacin and susceptible to gentamicin (9), whereas in the KPC-2-producing strains, resistances to amikacin and gentamicin vary (Table 1). Six of ten KPC-producing isolates were extended-spectrum beta-lactamase (ESBL) producers. In contrast to the Israeli ST 258 that was reported as a non-ESBL producer (10), ST 327, the other KPC-3-producing clone was an ESBL producer and so were five of the eight KPC-2-producing isolates. Multiple β-lactamases were identified and varied within the same ST type and between different ST types (Table 1). The plasmid-mediated quinolone resistance gene [aac(6′)-Ib-cr] was detected only in K. pneumoniae ST 376 and ST 277 (3).
TABLE 1.
Antibiotic susceptibilities, STs, blaKPC alleles, and bla genes of the studied K. pneumoniae isolatese
| K. pneumoniae isolate (ST type) or transformant | bla gene(s)b | MIC (μg/ml) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | GEN | CAZ | CRO | ATM | TZP | IPMc | MEMc | ERTc | CIP | LVX | CSTd | SXT | TGCd | ||
| KPC-2 producers | |||||||||||||||
| 475 (ST 277) | SHV-27, CTX-M-15, OXA-4 | <2 | >16 | 16 | 16 | >64 | >128 | 32 | 64 | 128 | 0.5 | 0.5 | 0.19 | >320 | 4 |
| 475-T | 8 | >16 | 16 | 32 | 32 | >128 | 8 | 4 | 8 | <0.25 | <0.12 | 0.032 | >320 | 0.125 | |
| 469 (ST 277) | SHV-27, CTX-M-15, OXA-4 | 4 | >16 | >64 | >64 | >64 | >128 | 32 | 32 | 64 | <0.25 | <0.25 | 0.125 | >320 | 1 |
| 469-T | 16 | >16 | >64 | >64 | 32 | >128 | 8 | 4 | 4 | <0.25 | <0.25 | 0.032 | >320 | 0.125 | |
| 523 (ST 340) | TEM-1, SHV-11,OXA-2 | 16 | >16 | 16 | 16 | >64 | 128 | 32 | 32 | 64 | >4 | >8 | 0.125 | 40 | 0.5 |
| 523-T | >64 | 2 | 16 | 32 | >64 | >128 | 8 | 4 | 8 | <0.25 | 0.25 | 0.047 | <20 | 0.125 | |
| 588 (ST 340) | TEM-1, SHV-11 | 32 | <1 | 16 | >64 | >64 | >128 | 32 | 64 | 128 | >4 | >8 | 0.19 | <20 | 2 |
| 588-T | 32 | 2 | 16 | 32 | >64 | >128 | 8 | 4 | 8 | <0.25 | <0.12 | 0.047 | <20 | 0.125 | |
| 365 (ST 376) | SHV-1, CTX-M-15, OXA-4 | 4 | >16 | >64 | >64 | >64 | >128 | 32 | 64 | 64 | >4 | >8 | 0.125 | >320 | 1 |
| 365-T | <2 | <1 | 16 | 16 | >64 | >128 | 8 | 4 | 8 | <0.25 | <0.12 | 0.032 | <20 | 0.125 | |
| 531 (ST 376) | SHV-1 | 8 | <1 | >64 | >64 | >64 | >128 | 32 | 64 | 128 | >4 | >8 | 0.125 | <20 | 1.5 |
| 531-T | 16 | <1 | 16 | 16 | 64 | 64 | 4 | 4 | 2 | <0.25 | <0.25 | 0.032 | <20 | 0.125 | |
| 525 (ST 376) | SHV-1 | 8 | <1 | >64 | >64 | >64 | >128 | 32 | 64 | 128 | >4 | >8 | 0.125 | <20 | 1 |
| 525-T | 16 | <1 | 16 | 8 | >64 | 64 | 4 | 4 | 4 | <0.25 | <0.25 | 0.032 | <20 | 0.125 | |
| 526 (ST 376) | SHV-1 | 4 | <1 | >64 | >64 | >64 | >128 | 32 | 64 | 64 | >4 | >8 | 0.125 | 40 | 1.5 |
| 526-T | 8 | <1 | 16 | 32 | >64 | >128 | 8 | 4 | 8 | <0.25 | <0.12 | 0.032 | <20 | 0.125 | |
| KPC-3 producers | |||||||||||||||
| 549 (ST 327) | TEM-1, CTX-M-2 | <2 | >16 | 16 | >64 | >64 | >320 | 0.75 | 8 | 32 | >4 | >8 | 0.125 | >320 | 0.75 |
| 549-T | <2 | <1 | >64 | 32 | >64 | 64 | 4 | 2 | 4 | <0.25 | <0.25 | 0.032 | <20 | 0.19 | |
| 557 (ST 258) | TEM-1 | 32 | <1 | >64 | >64 | >64 | >320 | 128 | 128 | 256 | >4 | >8 | 0.125 | >320 | 4 |
| 557-T | <2 | <1 | >64 | 32 | >64 | 64 | 4 | 4 | 4 | <0.25 | <0.25 | 0.032 | <20 | 0.125 | |
| E. coli Genehogsa | <2 | <1 | <1 | <1 | <1 | <4 | 0.094 | 0.012 | 0.012 | 0.002 | 0.006 | 0.125 | <20 | 0.094 | |
Transformants were in E. coli Genehogs.
Identified using ESBL confirmatory test (CLSI) and PCR.
Identified using agar dilution.
Identified using Etest.
T, transformants; AMK, amikacin; GEN, gentamicin; CAZ, ceftazidime; CRO, ceftriaxone; ATM, aztreonam; TZP, piperacillin-tazobactam; IPM, imipenem; MEM, meropenem; ERT, ertapenem; CIP, ciprofloxacin; LVX, levofloxacin; CST, colistin; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline.
KPC-encoding plasmid comparison and origin.
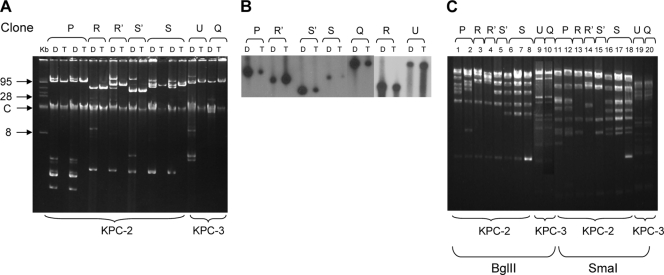
Carbapenem-resistant K. pneumoniae clones carried multiple plasmids. Plasmid analysis of transformants showed different-sized plasmids ranging from 45 to 95 kb for the blaKPC-2-carrying plasmids and 105 kb for the blaKPC-3-carrying plasmids (Fig. 2A).
FIG. 2.
(A) Plasmid analysis of 10 blaKPC-2- and blaKPC-3-carrying carbapenem-resistant K. pneumoniae clinical strains (lanes D) and their respective transformants (lanes T). C, chromosomal DNA. (B) Southern blot analysis of plasmid DNA of K. pneumoniae isolates, clinical strains (lanes D) and their respective transformants (lanes T). (C) The restriction pattern of plasmid DNA derived from K. pneumoniae transformants digested with BglII (lanes 1 to 10) and SmaI (lanes 11 to 20). PFGE types defined in Fig. 1 are presented in top parts of the panels.
Acquisition of blaKPC-carrying plasmids rendered resistance to piperacillin, cephalosporins, and aztreonam. MICs of carbapenems increased from 0.094, 0.012, and 0.012 μg/ml for imipenem, meropenem, and ertapenem to MIC50s of 8, 4, and 4 μg/ml, respectively. MICs of quinolones remained similar, and resistances to amikacin, gentamicin, and trimethoprim-sulfamethoxazole varied between transformants (Table 1).
Southern blot analysis using a labeled blaKPC probe proved the presence of a single KPC-encoding plasmid in each of the K. pneumoniae strains. The migration pattern of KPC-2-encoding plasmids varied, whereas for the two KPC-3-producing clones, the patterns were similar (Fig. 2B). Plasmid restriction analysis showed that blaKPC-2-carrying plasmids differed between clones but were similar or identical within the same clone and that they differed from the blaKPC-3-carrying plasmid. The two KPC-3-producing K. pneumoniae strains ST 258 and ST 327 carried the same plasmid, pKpQIL, reported previously (10). Interestingly, K. pneumoniae ST 327 was isolated from a patient that was coinfected with K. pneumoniae ST 258. This suggests the possibility of the horizontal transfer of pKpQIL from ST 258 to ST 327 within this patient.
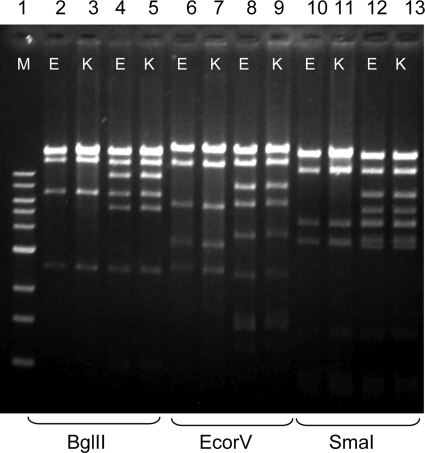
The origin of the KPC-encoding plasmids is unclear. pKpQIL, the Israeli blaKPC-3-carrying plasmid differs from blaKPC-2- and blaKPC-3-carrying plasmids of strains belonging to ST 258 from the United States (12). As for the blaKPC-2-carrying plasmids, they may have originated from other enteric pathogens. blaKPC-2 appeared initially among Enterobacter and E. coli isolates during 2004 and 2005 in our hospital (2, 6, 11). We may speculate that the gene originated from these organisms via horizontal transfer. blaKPC-2-carrying plasmids derived from Enterobacter strains (sized ∼200 kb) were significantly larger than the K. pneumoniae plasmids (2) and thus rule out the possibility that they had transferred to the latter organism. To verify the possibility of plasmid origin from E. coli, all Klebsiella blaKPC-2-carrying plasmids were compared to blaKPC-2-carrying plasmids from seven carbapenem-resistant E. coli strains isolated during the same period in which the K. pneumoniae isolates were identified (6, 11). Plasmid comparison indicated that two of the blaKPC-2-carrying plasmids of K. pneumoniae were identical to E. coli plasmids. The strains that shared common plasmids were K. pneumoniae ST 340 and E. coli strain 386 (plasmid size, 50 kb) (Fig. 3, lanes 2, 3, 6, 7, 10, and 11), K. pneumoniae ST 376, and E. coli 547 (75 kb) (Fig. 3, lanes 4, 5, 8, 9, 12, and 13). These data suggest the horizontal transfer of the intact blaKPC-2-carrying plasmid between these two species (Fig. 3). Based on restriction analysis, K. pneumoniae ST 277 did not show similarity with any of the E. coli plasmids, suggesting the possibility of acquisition of blaKPC-2 through Tn4401 transposition between plasmids followed by their independent horizontal transfer (13), or plasmid rearrangement that may have affected the restriction pattern obtained.
FIG. 3.
A comparison of blaKPC-2-carrying plasmids originated from two K. pneumoniae clones and two E. coli clones isolated in the same time period. Plasmid restriction analysis of transformants carrying these plasmids showed identity between the K. pneumoniae plasmids (K) and the E. coli plasmids (E). Plasmids from both organisms were digested with BglII (lanes 2 to 5), EcoRV (lanes 6 to 9), and SmaI (lanes 10 to 13) prior to electrophoresis. GeneRuler 1-kb DNA ladder (Fermentas Life Sciences), lane 1 (M); E. coli 386, lanes 2, 6, and 10; K. pneumoniae 523 PFGE type R, lanes 3, 7, and 11; E. coli 547, lanes 4, 8, and 12); K. pneumoniae 531, PFGE type, lanes 5, 9, and 13.
Carbapenem-resistant KPC-producing K. pneumoniae is spreading worldwide, posing a real threat (13). Characterizing the strains and plasmids involved may aid in understanding the evolution and thereby control of the dissemination of this clinically important antibiotic-resistant phenotype. In the current study, we characterized all the KPC-2-harboring K. pneumoniae isolates from our hospital by MLST, plasmid mapping, and comparisons. We demonstrated a complex epidemiology that involves limited clonal and plasmid transmission via horizontal transfer either within the same species, like the blaKPC-3-carrying plasmid, or via rare events of intraspecies transmission, like the blaKPC-2-carrying plasmid from E. coli.
Acknowledgments
This work was supported in part by the European Commission Research grant FP7: SATURN—Impact of Specific Antibiotic Therapies on the Prevalence of Human Host Resistant Bacteria (grant no. 241796).
We would like to sincerely thank the Klebsiella pneumoniae MLST website researchers, particularly Sylvain Brisse, for their professional assistance and for providing the evolutionary tree of K. pneumoniae. We also thank Daphne Karfunkel for the critical reading of the manuscript.
Footnotes
Published ahead of print on 29 March 2010.
REFERENCES
- 1.Baraniak, A., R. Izdebski, M. Herda, J. Fiett, W. Hryniewicz, M. Gniadkowski, I. Kern-Zdanowicz, K. Filczak, and U. Lopaciuk. 2009. Emergence of Klebsiella pneumoniae ST258 with KPC-2 in Poland. Antimicrob. Agents Chemother. 53:4565-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chmelnitsky, I., S. Navon-Venezia, J. Strahilevitz, and Y. Carmeli. 2008. Plasmid-mediated qnrB2 and carbapenemase gene blaKPC-2 carried on the same plasmid in carbapenem-resistant ciprofloxacin-susceptible Enterobacter cloacae isolates. Antimicrob. Agents Chemother. 52:2962-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chmelnitsky, I., O. Hermesh, S. Navon-Venezia, J. Strahilevitz, and Y. Carmeli. 2009. First detection of aac (6′)-Ib-cr in KPC-producing Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 64:718-722. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Giani, T., M. M. D'Andrea, P. Pecile, L. Borgianni, P. Nicoletti, F. Tonelli, A. Bartoloni, and G. M. Rossolini. 2009. Emergence in Italy of Klebsiella pneumoniae sequence type 258 producing KPC-3 carbapenemase. J. Clin. Microbiol. 47:3793-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goren, M. G., S. Navon-Venezia, I. Chmelnitsky, and Y. Carmeli. 2010. Carbapenem-resistant KPC-2-producing Escherichia coli in a Tel Aviv Medical Center, 2005 to 2008. Antimicrob. Agents Chemother. 54:2687-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussein, K., H. Sprecher, T. Mashiach, I. Oren, I. Kassis, and R. Finkelstein. 2009. Carbapenem resistance among Klebsiella pneumoniae isolates: risk factors, molecular characteristics, and susceptibility patterns. Infect. Control Hosp. Epidemiol. 30:666-671. [DOI] [PubMed] [Google Scholar]
- 8.Kitchel, B., J. K. Rasheed, J. B. Patel, A. Srinivasan, S. Navon-Venezia, Y. Carmeli, A. Brolund, and C. G. Giske. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leavitt, A., S. Navon-Venezia, I. Chmelnitsky, M. J. Schwaber, and Y. Carmeli. 2007. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob. Agents Chemother. 51:3026-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leavitt, A., I. Chmelnitsky, I. Ofek, Y. Carmeli, and S. Navon-Venezia. 24 November 2009. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug resistant epidemic Klebsiella pneumoniae strain. J. Antimicrob. Chemother. 65:243-248. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 11.Navon-Venezia, S., I. Chmelnitsky, A. Leavitt, M. J. Schwaber, D. Schwartz, and Y. Carmeli. 2006. Plasmid-mediated imipenem-hydrolyzing enzyme KPC-2 among multiple carbapenem-resistant Escherichia coli clones in Israel. Antimicrob. Agents Chemother. 50:3098-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navon-Venezia, S., A. Leavitt, M. J. Schwaber, J. K. Rasheed, A. Srinivasan, J. B. Patel, Y. Carmeli, and Israeli KPC Kpn Study Group. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob. Agents Chemother. 53:818-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nordmann, P., G. Cuzon, and T. Naas. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228-236. [DOI] [PubMed] [Google Scholar]
- 14.Osterblad, M., J. Kirveskari, S. Koskela, P. Tissari, K. Vuorenoja1, A. J. Hakanen, M. Vaara, and J. Jalava1. 2009. First isolations of KPC-2-carrying ST258 Klebsiella pneumoniae strains in Finland, June and August 2009. Euro Surveill. 14(40):pii=19349. [PubMed] [Google Scholar]
- 15.Samuelsen, Ø., U. Naseer, S. Tofteland, D. H. Skutlaberg, A. Onken, R. Hjetland, A. Sundsfjord, and G. G. Giske. 2009. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J. Antimicrob. Chemother. 63:654-658. [DOI] [PubMed] [Google Scholar]
- 16.Schwaber, M. J., and Y. Carmeli. 2008. Carbapenem-resistant Enterobacteriaceae: a potential threat. JAMA 300:2911-2913. [DOI] [PubMed] [Google Scholar]
- 17.Schwaber, M. J., S. Klarfeld-Lidji, S. Navon-Venezia, D. Schwartz, A. Leavitt, and Y. Carmeli. 2008. Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob. Agents Chemother. 52:1028-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodford, N., J. Zhang, M. Warner, M. E. Kaufmann, J. Matos, A. MacDonald, D. Brudney, D. Sompolinsky, S. Navon-Venezia, and D. M. Livermore. 2008. Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J. Antimicrob. Chemother. 62:1261-1264. [DOI] [PubMed] [Google Scholar]