Abstract
The Mycobacterium tuberculosis cell envelope contains a wide variety of lipids and glycolipids, including mycolic acids, long-chain branched fatty acids that are decorated by cyclopropane rings. Genetic analysis of the mycolate methyltransferase family has been a powerful approach to assign functions to each of these enzymes but has failed to reveal the origin of cis cyclopropanation of the oxygenated mycolates. Here we examine potential redundancy between mycolic acid methyltransferases by generating and analyzing M. tuberculosis strains lacking mmaA2 and cmaA2, mmaA2 and cmaA1, or mmaA1 alone. M. tuberculosis lacking both cmaA2 and mmaA2 cannot cis cyclopropanate methoxymycolates or ketomycolates, phenotypes not shared by the mmaA2 and cmaA2 single mutants. In contrast, a combined loss of cmaA1 and mmaA2 had no effect on mycolic acid modification compared to results with a loss of mmaA2 alone. Deletion of mmaA1 from M. tuberculosis abolishes trans cyclopropanation without accumulation of trans-unsaturated oxygenated mycolates, placing MmaA1 in the biosynthetic pathway for trans-cyclopropanated oxygenated mycolates before CmaA2. These results define new functions for the mycolic acid methyltransferases of M. tuberculosis and indicate a substantial redundancy of function for MmaA2 and CmaA2, the latter of which can function as both a cis and trans cyclopropane synthase for the oxygenated mycolates.
Mycobacterium tuberculosis infection is an ongoing global health crisis. Alleviation of this crisis will require a multidisciplinary approach that must include new antibiotics active against M. tuberculosis. A growing body of literature implicates cell envelope lipids in the pathogenesis of M. tuberculosis infection (5-10, 14-15, 20). The enzymatic pathways that synthesize M. tuberculosis cell envelope lipids are the target of presently available antituberculosis antimicrobials and may be candidates for future antibiotic development.
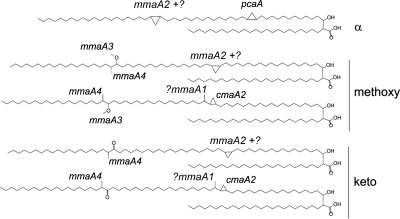
The mycolic acids of M. tuberculosis are alpha-alkyl, beta-hydroxy fatty acids which are 75 to 85 carbons in length (3). There are three classes of major mycolic acids: alpha-, methoxy-, and ketomycolates (Fig. 1). Whereas all mycobacteria synthesize mycolic acids, only pathogenic mycobacteria (for example, M. tuberculosis, M. leprae, M. avium, and M. bovis) produce significant quantities of mycolic acids with cyclopropane rings, three-member carbon rings which are added to the meromycolate chain (3). Alpha-mycolates have two cis cyclopropane rings, while methoxy- and ketomycolates have either a cis or trans cyclopropane ring at the proximal position, the latter with a distal methyl branch (Fig. 1). In contrast to the case with Escherichia coli, which encodes a single cyclopropane fatty acid synthase (CFAS) (16-17), the M. tuberculosis genome encodes a family of S-adenosyl methionine-dependent methyltransferases that modify cell envelope mycolic acids with methyl branches and cyclopropane rings. Despite substantial amino acid identity, systematic characterization of M. tuberculosis null mutants in each of these methyltransferases has revealed highly specific functions which were not revealed when the enzymes were overexpressed in M. smegmatis (12, 26, 28). Deletion of pcaA greatly reduces synthesis of the proximal cyclopropane ring of the alpha-mycolates (15), whereas deletion of mmaA2 greatly reduces the distal cyclopropane of the same lipid (13). Loss of mmaA2 also causes a mild impairment of methoxymycolate, but not ketomycolate, cis cyclopropanation (13). Similar genetic approaches established cmaA2 as the only trans cyclopropane synthase of oxygenated mycolates (14), while loss of mmaA3 abolishes methoxymycolates, a spontaneous mutation found in many M. bovis BCG strains (4, 11). Finally, deletion of mmaA4 abolishes synthesis of both methoxy- and ketomycolates (10). Recent chemical-genetic analysis of this enzyme family indicates that combined inhibition of their function is lethal to M. tuberculosis, strongly supporting an approach targeting this enzyme family for antimicrobial development (2, 8-19, 25).
FIG. 1.
Chemical structures of the major mycolic acids of M. tuberculosis. Cyclopropane rings and methyl branches are shown and annotated with the methyltransferase responsible for their synthesis.
In addition to this essential role in combination, recent evidence implicates individual cyclopropane modifications as important determinants of M. tuberculosis host-pathogen interactions. Inactivation of pcaA causes attenuation of M. tuberculosis in the mouse model of infection while stimulating less-severe granulomatous pathology (15, 23). In contrast, deletion of cmaA2 has no effect on bacterial loads during mouse infection but causes hypervirulence while inducing more-severe granulomatous pathology (24). Inactivation of mmaA4, which leads to an absence of methoxy- and ketomycolates, causes a severe growth defect during the first 3 weeks of infection (10). All of these studies implicate the fine structure of mycolic acids in the pathogenesis of M. tuberculosis infection. One mechanism by which cyclopropanation mediates pathogenesis is through altered inflammatory activity of trehalose dimycolate (TDM), an inflammatory glycolipid. The cyclopropane content of TDM is a major determinant of its inflammatory activity, and this altered TDM is responsible for the virulence phenotypes of cyclopropane-deficient M. tuberculosis strains (9, 23-24).
Despite major advances in our understanding of the biosynthesis and pathogenetic function of cyclopropanated mycolic acids through genetic approaches, the methyltransferase(s) that synthesizes the cis cyclopropane ring on the methoxy- and ketomycolates is unknown. In addition, the function of the MmaA1 methyltransferase has not been explored through construction of a null mutant. Prior experiments found that overexpression of mmaA1 in M. tuberculosis resulted in accumulation of trans-unsaturated and -cyclopropanated oxygenated mycolates (27). These data suggested that MmaA1 acts in the biosynthesis of trans-cyclopropanated oxygenated mycolates either by adding the methyl branch distal to the cyclopropane ring or as a cis-trans isomerase or both. In addition, although there was a defect in cis cyclopropanation of methoxymycolates in the ΔmmaA2 strain, this defect was mild, suggesting redundancy with another unidentified enzyme. In this article, we define novel functions for three cyclopropane synthases using a new selectable marker to construct M. tuberculosis strains deficient in multiple mycolic acid methyltransferases. Through this approach, we show that CmaA2 and MmaA2 are redundant for cis cyclopropanation of the proximal position of the methoxymycolates and ketomycolates and that MmaA1 is upstream of CmaA2 in trans cyclopropanation.
MATERIALS AND METHODS
Bacterial strains and media.
The Mycobacterium tuberculosis wild-type (WT) strain used in this study is Erdman EF2 and is an animal-passaged strain described previously (15). The mmaA2 (MGM104), cmaA1 (MGM47), and cmaA2 (MGM46) single mutants were described previously (13-14). M. tuberculosis strains were grown in 7H9 broth or 7H10 agar with 10% oleic acid-albumin-dextrose-catalase supplement (OADC), 0.5% glycerol, 0.05% Tween 80 (for 7H9), and where required hygromycin B (50 μg/ml), kanamycin (20 μg/ml), and zeocin (12.5 μg/ml).
Creation of ΔmmaA1, ΔcmaA1 ΔmmaA2, and ΔcmaA2 ΔmmaA2 by allelic exchange.
To create an mmaA1 null allele, the 5′ flanking region of mmaA1 (Rv0645c, MT0673) was amplified from M. tuberculosis genomic DNA using the primers OMSG1300-1 and OMSG1300-2. This 732-bp PCR product spanning a 600-bp portion of the upstream gene lipG and the first 53 bp of mmaA1 was purified, cloned, and sequenced. The 3′ flanking region containing 37 nucleotides (nt) of the 3′ end of mmaA1 and a portion of mmaA2 was excised as a BclI/StuI restriction fragment from the plasmid pMSG256 and cloned into BclI/EcoRV-digested pJSC407 to obtain the plasmid pMSG1300. The 5′ flanking PCR fragment was cloned into pMSG1300 as an Asp7181/XbaI fragment to get the final plasmid, pMSG1302. This plasmid contains both the 5′ and 3′ flanking regions of mmaA1 on either side of the hygromycin resistance gene and was used to construct a specialized transducing phage for allelic exchange of the native mmaA1 gene of M. tuberculosis as described previously (13-14). Hygromycin-resistant transductants were screened for allelic exchange by Southern blotting using the 5′ flanking PCR fragment as a probe. The mmaA1-disrupted strain was designated MGM1302.
To complement the ΔmmaA1 strain, the complete open reading frame (ORF) and upstream sequences of M. tuberculosis mmaA1 were excised from the plasmid pJSC252 as a 1,411-bp NotI/EcoRV fragment, which includes the entire mmaA1 open reading frame and 175 nt 5′ of the mmaA1 start codon, including 128 nt of the upstream gene lipG. This was inserted in pMV306kan (which integrates at the chromosomal attB site) and used to transform MGM1302 to assess for restoration of MmaA1 function.
To disrupt cmaA1 and cmaA2 in a ΔmmaA2 strain, we constructed a transducing phage with a zeocin resistance cassette in place of hygromycin. This was done by cloning the Zeor gene driven by the MOP promoter in place of the hygromycin resistance cassette in pMSG360, creating pMSG360Z. The flanking regions were taken from the previously described pMSG105 (for cmaA1 deletion) (13) and pMSG104 (for cmaA2 deletion) (14), creating pDB54 and pDB68, respectively. These two plasmids were used to construct the specialized transducing phages phDB4 and phDB10. Deletion of the target gene was confirmed by Southern blotting.
Preparation and analysis of mycolic acid methyl esters.
14C-labeled mycolic acid methyl esters (MAMEs) were prepared from logarithmic-phase cultures of M. tuberculosis with 50 μCi of [1-14C]acetic acid (58.9 mCi/mmol) (PerkinElmer Life sciences) for 24 h. MAMEs were prepared from whole bacilli and analyzed by two-dimensional argentation thin-layer chromatography (TLC) as described previously (13). Briefly, 50 ml of logarithmic-phase culture was pelleted, and bacteria were resuspended in water, diluted with 40% tetrabutylammonium hydroxide (TBAH) to a final concentration of 20% TBAH, and then heated overnight at 100°C. An equal volume of dichloromethane and 0.15 ml of methyl iodide were added, and the suspension was rotated at room temperature for 1 h. After phase separation, the aqueous phase was discarded and the organic fraction was evaporated under a nitrogen stream. The residue was extracted with ethyl ether and dried. Finally, MAMEs were precipitated from 2:1 toluene-acetonitrile by addition to two volumes of acetonitrile. The resulting MAMEs were resuspended in ethyl ether. For two-dimensional TLC separation, 90% of the TLC plate was immersed in 10% silver nitrate and then left to dry at 100°C for 15 min. The mycolic acid sample was first developed in the non-silver-immersed strip for 6 developments and then developed five times into the silver-immersed area. The solvent for both dimensions was hexane-ethyl acetate (95:5).
For preparation of mycolic acid classes, total MAMEs prepared from 2 liters of M. tuberculosis were applied to a 1-mm preparative silica gel TLC plate and developed 10 times with hexane-ethyl acetate (95:5). Lipids were scraped, and the silica was extracted three times with ethyl ether. Mycolates were then reprecipitated with toluene-acetonitrile before analysis. The purity of each isolated mycolic acid class was evaluated by TLC before structural characterization. For nuclear magnetic resonance (NMR) analysis, mycolates were dissolved in deuterochloroform (Cambridge Isotope Laboratories) and analyzed on a Bruker 500-MHz spectrophotometer.
Cording assays.
For cording assays, 10 μl of logarithmic cultures (A600 = 0.5 to 0.6) were spread on individual wells of a 4-well chamber slide. The cultures were then incubated at 37°C without shaking for 8 to 10 days in liquid medium containing either 0, 0.02, or 0.05% Tween 80. After removal of medium, the slides were fixed and stained with the TB auramine rhodamine staining kit (Difco) according to the manufacturer's recommendations and visualized under a fluorescence microscope.
RESULTS
Creation of ΔmmaA2 ΔcmaA2, ΔmmaA2 ΔcmaA1, and ΔmmaA1 M. tuberculosis.
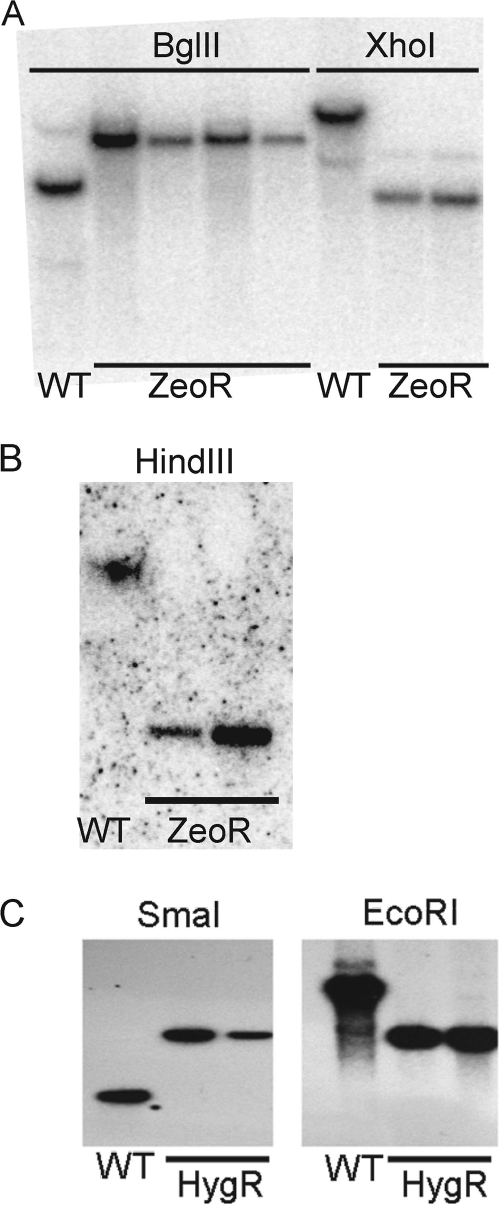
To examine if cmaA1 or cmaA2 may be redundant with mmaA2 in alpha- and methoxymycolate biosynthesis, we chose to delete each of them from our previously described mmaA2 deletion mutant. Since the ΔmmaA2 mutant carries a hygromycin resistance cassette, we constructed two specialized transducing mycobacteriophages carrying zeocin resistance cassettes, flanked by the 500-bp flanking regions of either cmaA1 or cmaA2. To our knowledge, this is the first time zeocin resistance has been used to construct deletion mutants in mycobacteria, although the marker has been used in M. smegmatis (22). The resulting colonies were screened by Southern blotting, identifying colonies in which the cmaA1 (Fig. 2A) or cmaA2 (Fig. 2B) coding sequence was replaced. The resulting double mutants were called MGM1967 (ΔmmaA2::hyg ΔcmaA1::zeo) and MGM1974 (ΔmmaA2::hyg ΔcmaA2::zeo).
FIG. 2.
Construction of M. tuberculosis ΔcmaA1 ΔmmaA2, ΔcmaA2 ΔmmaA2, and ΔmmaA1 strains. (A) Deletion of cmaA1 from MGM104 (ΔmmaA2). When probed with the 3′ flank of cmaA1, BglII-digested genomic DNA produces a 4.9-kb band in wild-type cells whereas the ΔcmaA1 strain has a 7.2-kb band. Two clones were also confirmed by XhoI digestion, where the WT strain is predicted to have an 11.7-kb band and the ΔcmaA1 strain is predicted to have a 4.4-kb band. (B) Deletion of cmaA2 from MGM104. When probed with the 3′ flank of cmaA2, HindIII-digested genomic DNA produces a 16.4-kb band in wild-type cells whereas the ΔcmaA2 strain is predicted to have a 3.5-kb band. (C) Deletion of mmaA1 from the WT background. When probed with the 5′ flank of mmaA1, SmaI-digested genomic DNA produces a 1.3-kb band in wild-type cells whereas the ΔmmaA1 strain is predicted to have a 3.1-kb band. EcoRI-digested genomic DNA produces 6.4- and 3.1-kb bands for the wild-type and ΔmmaA1 strains, respectively.
To understand the biological function of the mmaA1 gene in M. tuberculosis mycolic acid modification, we deleted mmaA1 from the chromosome of M. tuberculosis using specialized transduction. We constructed a null allele of mmaA1 by replacing nucleotides 48 to 824 of the mmaA1 open reading frame with a hygromycin resistance gene. Our prior work indicated that the promoter for mmaA2 is located 3′ of the mmaA1 stop codon, making a polar effect on mmaA2 unlikely (13). A temperature-sensitive specialized transducing phage carrying this null allele transduced wild-type M. tuberculosis to hygromycin resistance. Hygromycin-resistant transductants were screened for allelic exchange at the mmaA1 locus by Southern blotting (Fig. 2C). To confirm that any phenotype observed in this mutant is due to loss of mmaA1 function, we constructed a genetically complemented strain. The intact mmaA1 gene with its native promoter was integrated into the chromosome at the attB site to create the complemented strain. This M. tuberculosis mmaA1 null mutant (MGM1302, or ΔmmaA1 mutant) and the corresponding complemented strain were characterized further.
CmaA2 but not CmaA1 is redundant with MmaA2 in cis cyclopropanation of methoxymycolates and ketomycolates.
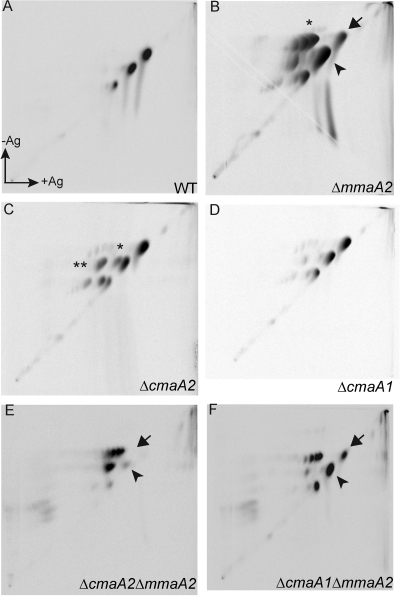
After obtaining the double mutants MGM1967 and MGM1974, we analyzed their mycolic acid profile by two-dimensional argentation thin-layer chromatography (TLC) of mycolic acid methyl esters. This system separates mycolates by polarity in the first dimension and then by degree of unsaturation in the second (silver-impregnated) dimension. In our prior studies of M. tuberculosis cyclopropane synthase-deficient strains, defective cyclopropane synthesis was visualized in the second dimension by the appearance of an unsaturated (retarded) derivative of a previously saturated lipid. The previously described mycolate profiles of the WT, ΔmmaA2, ΔcmaA1, and ΔcmaA2 strains are shown in Fig. 3 A to D. As previously reported, loss of mmaA2 causes accumulation of a distal monounsaturated derivative of the alpha-mycolate (Fig. 3B, asterisk), with persistence of a small fraction of mature alpha-mycolate (Fig. 3B, black arrow). Loss of cmaA2 causes accumulation of trans-unsaturated oxygenated mycolates (Fig. 3C, single asterisk) and a small amount of cis unsaturated mycolate (Fig. 3C, double asterisk), as previously reported (14). In contrast, loss of cmaA1 has no major effect (Fig. 3D). The strain carrying a double mutation in mmaA2 and cmaA1 has no additional phenotype beyond that with mmaA2 deletion alone (compare Fig. 3B and F). However, the cmaA2 mmaA2 double mutant had a mycolic acid pattern markedly distinct from that of either single mutant (Fig. 3E). First, the residual mature alpha-mycolates, visible in the ΔmmaA2 strain (Fig. 3B, arrow), are abolished by the cmaA2 mutation (Fig. 3E, arrow). This indicates that to a limited extent, CmaA2 can cyclopropanate the alpha-mycolate when mmaA2 is lost, but this function is not detectable in the ΔcmaA2 strain (which has mmaA2). Second, mature methoxymycolates are not synthesized in the ΔmmaA2 ΔcmaA2 strain (Fig. 3E), and all of the methoxymycolate produced is retarded by silver, indicating the presence of double bonds (Fig. 3E). This result indicates that cmaA2 and mmaA2 share a redundant function in cis cyclopropanation of the methoxymycolate. Clear conclusions about ketomycolate cyclopropanation in the ΔmmaA2 ΔcmaA2 strain could not be drawn. A small amount of ketomycolate is synthesized (Fig. 3E), but it was difficult to judge whether it was unsaturated or cyclopropanated due to its low abundance.
FIG. 3.
CmaA2 and MmaA2 are redundant for cis cyclopropanation of the oxygenated mycolates. Shown is two-dimensional argentation TLC of mycolic acid methyl esters from the M. tuberculosis wild type (A) or the ΔmmaA2 (B), ΔcmaA2 (C), or ΔcmaA1 (D) strain. Panel E shows mycolates from the ΔcmaA2 ΔmmaA2 strain (MGM1974), whereas panel F shows the ΔcmaA1 ΔmmaA2 strain (MGM1967). In panels B, E, and F, the arrow marks the position of the mature alpha-mycolates and the arrowhead marks mature methoxymycolate. In panel B, the asterisk indicates monounsaturated alpha-mycolate. In panel C, the double asterisk indicates cis-unsaturated methoxymycolates and the single asterisk indicates trans-unsaturated methoxymycolates.
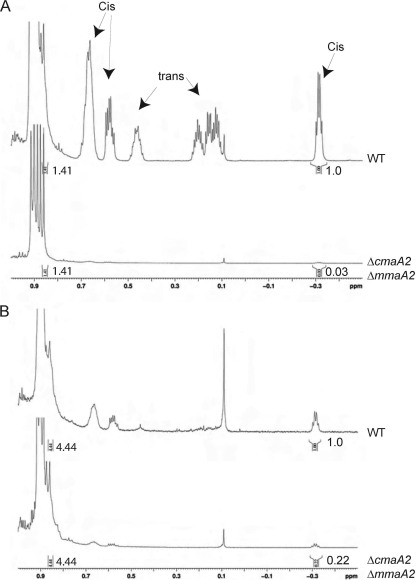
To clarify the structures of methoxy- and ketomycolates from the ΔmmaA2 ΔcmaA2 strain, we purified these lipids by preparative TLC and analyzed them by proton NMR. As expected from the TLC profile, ΔmmaA2 ΔcmaA2 methoxymycolates almost completely lacked proton resonances attributable to cis cyclopropane rings (peaks at −0.33, 0.54, and 0.62) (Fig. 4A). After normalization of the spectra to the terminal methyl groups (at 0.86 ppm), integration of cis cyclopropane hydrogen peak areas for each strain indicated that the residual cis-cyclopropanated methoxymycolate in the ΔmmaA2 ΔcmaA2 mutant was 3% compared to that in wild-type cells, consistent with the almost complete lack of mature methoxymycolate seen in TLC (Fig. 3E, arrowhead). Previous analysis of the ΔmmaA2 strain indicated that cis-cyclopropanated methoxymycolates were reduced by 50% compared to levels in wild-type cells (13), indicating that cmaA2 can substitute almost completely for mmaA2 in methoxymycolate modification. Analysis of purified ketomycolate from the ΔmmaA2 ΔcmaA2 strain indicated that cis cyclopropanation of the ketomycolate was 22% of that for wild-type cells (Fig. 4B). Both methoxy- and ketomycolates lacked trans cyclopropanes, as expected from the cmaA2 mutation, as previously reported (14). The residual cis-cyclopropanated ketomycolate indicates that this modification involves yet another enzyme in addition to MmaA2 and CmaA2.
FIG. 4.
Structural analysis of methoxy- and ketomycolates from the ΔcmaA2 ΔmmaA2 strain. NMR analysis of purified methoxymycolate (A) or ketomycolate (B) from WT M. tuberculosis or the ΔcmaA2 ΔmmaA2 double mutant. The relative peak area of the cis cyclopropane peak at −0.33 is indicated below each spectrum and is set to 1.0 for the wild type. Spectra are normalized to the resonance at 0.86 (equal for the WT and the mutant), representing the terminal methyl groups common to all mycolic acids. Note that the ΔcmaA2 ΔmmaA2 strain also lacks trans cyclopropyl protons due to the cmaA2 deletion, as previously reported (9).
These findings clearly identify CmaA2 as the second enzyme capable of cis cyclopropanation of methoxymycolates and as the enzyme capable of distal cyclopropanation of alpha-mycolates. Whereas the cis cyclopropanation of methoxymycolates is equally distributed between MmaA2 and CmaA2, the distal cyclopropanation of the alpha-mycolates seems to be preferentially done by MmaA2, with only some activity of CmaA2 when MmaA2 is absent, as evinced by the prominent spot of the monounsaturated alpha versus mature alpha in the mmaA2 single mutant (Fig. 3B).
MmaA1 is required for trans cyclopropane ring formation and trans double bond formation in M. tuberculosis.
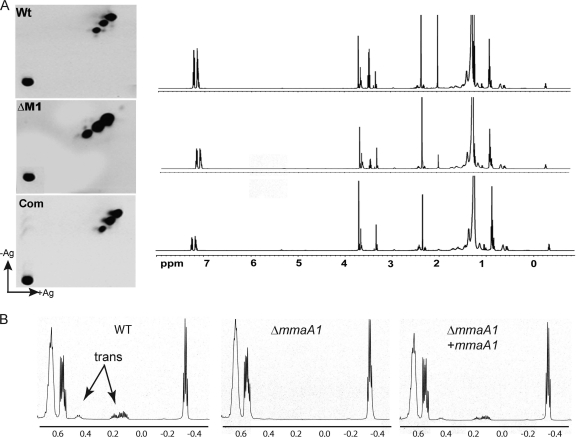
To investigate the function of mmaA1 in mycolic acid modification, we prepared total mycolic acids from the ΔmmaA1 and wild-type strains and examined these lipids by two-dimensional argentation TLC. When examined in this system, the mycolic acids of the wild type and the ΔmmaA1 strain were similar, without accumulation of silver-retarded lipids that would indicate the presence of double bonds (Fig. 5A).
FIG. 5.
mmaA1 is required for trans cyclopropane synthesis. (A) Two-dimensional TLC analysis of total mycolic acids from WT M. tuberculosis, the ΔmmaA1 mutant, and the complemented strain. The full-spectrum NMRs of total mycolic acids from each strain are shown immediately to the right of each corresponding TLC. (B) Magnification of the NMR spectra for the area between 0.7 and −0.5 ppm, showing the disappearance of the trans peaks at 0.15 and 0.45 in the mutant and their restoration in the complemented strain.
We next analyzed mycolic acids from wild-type, ΔmmaA1, and complemented strains by 500-MHz 1H NMR. Whereas both mutant and WT mycolates displayed the characteristic cis-cyclopropyl resonance at −0.33 ppm, mycolates from the ΔmmaA1 strain completely lacked the trans cyclopropyl resonances at 0.15 ppm and 0.45 ppm that were visible in the wild-type and complemented strains (Fig. 5B). Complementation with a single copy of mmaA1 restored these resonances, corresponding to formation of a trans ring (Fig. 5B). In addition, the mutant mycolates did not demonstrate trans double bond resonances at 5.3, indicating that a trans double bond does not accumulate in the ΔmmaA1 strain (Fig. 5A). These results demonstrate that mmaA1 is necessary for trans cyclopropane formation in M. tuberculosis and that a loss of MmaA1 function is not accompanied by an accumulation of trans double-bonded lipids, in contrast to our prior finding with the cmaA2 null mutant, in which a lack of trans cyclopropanation is accompanied by accumulation of a trans-unsaturated precursor lipid (14) (Fig. 3C).
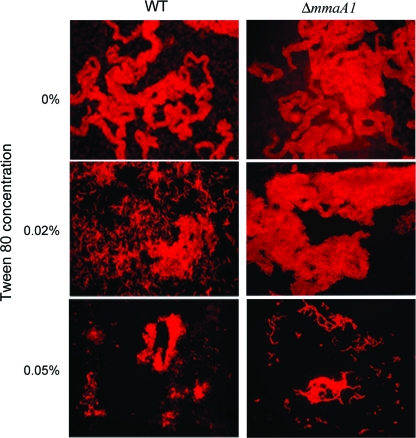
Loss of mmaA1 enhances cording in M. tuberculosis.
Previous observations with a pcaA mutant M. tuberculosis strain demonstrated that loss of cis cyclopropanation of the alpha-mycolate alters the cording morphology in slow-growing mycobacteria (15). To evaluate if mmaA1 played a role in cording of M. tuberculosis, we observed the cording morphology of ΔmmaA1 M. tuberculosis in the presence of escalating concentrations of the detergent Tween 80, a known inhibitor of the cording phenotype (21). In detergent-free medium, both WT and ΔmmaA1 strains formed long serpentine cords, characteristic of M. tuberculosis (Fig. 6). At a Tween concentration of 0.05%, although the wild-type strain lost the cording morphology (Fig. 6, lower left panel), the ΔmmaA1 strain retained cording (Fig. 6, lower right). These observations demonstrate that loss of mmaA1 increased the ability of M. tuberculosis to form cords, thus establishing mmaA1 as a negative regulator of cording in M. tuberculosis.
FIG. 6.
Loss of mmaA1 enhances cording in M. tuberculosis. Auramine-rhodamine-stained mycobacteria grown in the presence of the indicated concentrations of Tween 80 were observed by fluorescence microscopy. Representative areas of growth of WT and ΔmmaA1 M. tuberculosis are shown.
DISCUSSION
Mycolates are the major lipids of the M. tuberculosis cell envelope and are modified with a diversity of cyclopropane rings. The M. tuberculosis cell envelope, including mycolate modification, has emerged as an important determinant of pathogenesis. Cyclopropane modification is mediated by a family of S-adenosyl methionine-dependent methyltransferases expressed by M. tuberculosis. Many of these enzymes have a unique biosynthetic activity, which has been systematically elucidated by analysis of M. tuberculosis null mutants.
Our data demonstrate that although MmaA2 is the primary enzyme responsible for the distal cyclopropanation of the alpha-mycolates, CmaA2 has the ability to perform that function, and only deletion of these two genes together produces a complete abrogation of fully cyclopropanated alpha-mycolate. Similarly, we show that the cis cyclopropanation of the methoxymycolate is redundant between these two enzymes. Only a double deletion causes the almost total abrogation of methoxy-cis cyclopropanation. The ΔcmaA2 ΔmmaA2 strain also has a substantial reduction in keto cis cyclopropanation, but some low-level ketomycolate cis cyclopropanation remains in this strain, indicating yet a third enzyme that participates in this reaction. The identity of the third enzyme involved in cis cyclopropanation of the ketomycolates remains unknown. The two main candidates for this role are PcaA and CmaA1, both of which are capable of cis cyclopropanation. The minor role played by this third enzyme in ketomycolate modification makes it difficult to identify by a single deletion experiment, and its final identification will require creation of a ΔmmaA2 ΔcmaA2 ΔcmaA1 or ΔmmaA2 ΔcmaA2 ΔpcaA strain.
Our data provide some clarity about prior data indicating that CmaA2 is capable of cis cyclopropanation when expressed in M. smegmatis, a nonpathogenic mycobacterium that does not produce cyclopropane rings (12). Deletion of cmaA2 from M. tuberculosis had no effect on cis cyclopropanation evident on TLC but abolished trans cyclopropane formation (14), raising doubts about the in vivo relevance of the cis cyclopropanation activity of CmaA2. The data presented here uncover a major role for CmaA2 in cis cyclopropanation of the oxygenated mycolates, which is redundant with MmaA2 and therefore not evident in the ΔcmaA2 or ΔmmaA2 mutant. In contrast, our data do not reveal any role for CmaA1. The single deletion mutant previously described did not have any appreciable phenotype, and the ΔmmaA2 ΔcmaA1 double mutant did not have any phenotype compared to results for ΔmmaA2 alone.
In this study, we have extended our analysis to the MmaA1 cyclopropane synthase to show that mmaA1 is required for trans cyclopropane formation. A prior study demonstrated that the overexpression of mmaA1 led to an overabundance of trans-unsaturated and trans-cyclopropanated methoxy- and ketomycolates (27). These data suggested that the methyltransferase activity of MmaA1 adds the methyl branch adjacent to the trans cyclopropane ring, followed by CmaA2-mediated trans cyclopropanation (27). The data presented here provide genetic evidence that supports this model. The lack of unsaturated oxygenated lipids in the mmaA1 mutant is consistent with this reported activity, since MmaA1 is required for formation of the double-bonded precursor, which is subsequently cyclopropanated by CmaA2. Thus, although both mmaA1 and cmaA2 are required for trans cyclopropane formation, unsaturated oxygenated mycolates accumulate only in the cmaA2 mutant because the action of MmaA1 precedes the formation of the double bond. A summary of our present understanding of the mycolate modification pathway is presented in Table 1, which synthesizes data from this study and prior work.
TABLE 1.
Biosynthetic origin of each cyclopropane ring or methyl branch found in the M. tuberculosis mycolates pictured in Fig. 1a
| Lipid structure | Methyltransferase(s) | Reference(s) |
|---|---|---|
| Cyclopropane ring | ||
| α proximal (cis) | PcaA | 15 |
| α distal (cis) | MmaA2 (CmaA2) | 13; this study |
| Methoxy cis | MmaA2, CmaA2 | This study |
| Methoxy trans | CmaA2 | 14 |
| Keto cis | MmaA2, CmaA2 (?b) | This study |
| Keto trans | CmaA2 | 14 |
| Methyl groups | ||
| Methoxy group | MmaA3 | 4, 11 |
| Oxygenated mycolates; distal methyl branch | MmaA4 | 9-10, 26 |
| trans-cyclopropanated oxygenated mycolates, proximal methyl branch | MmaA1 | 27; this study |
| Unknown function | ||
| None | CmaA1 | 13; this study |
The first column lists each cyclopropane ring or methyl branch (see Fig. 1 for structures). The second column lists the enzymes that synthesize each modification. Two enzymes are listed without parentheses when the modification is lost only in a double mutant of the genes encoding the listed enzymes and not in either of the single mutants. When two enzymes are listed with one in parentheses, the parenthetical enzyme plays a secondary role that is evident only when the gene encoding the primary enzyme is deleted.
A third, as-yet-unidentified enzyme is also capable of this modification.
In the last 10 years, convincing data have emerged implicating the different M. tuberculosis cyclopropane synthases and methyltransferases in the pathogenicity of M. tuberculosis. More recently, data suggesting these enzymes play an important role in mycobacterial viability were published, and chemical inhibitors of the whole class, which kill mycobacteria, were described (2, 25). In addition, the activity of thiacetazone, a second-line antitubercular drug, depends on an intact mmaA4 gene (1). Exact knowledge of the biosynthetic role of each enzyme and the specific origin of each modification/cyclopropanation is therefore of high importance. This study defines the biosynthetic origin of the cis cyclopropanation of the oxygenated mycolates, the last remaining major mycolic acid modification of unknown origin. The data gathered in this study will also inform construction of an M. tuberculosis strain lacking all cyclopropanation (ΔcmaA2 ΔmmaA2 ΔpcaA) but with intact major mycolate classes. Characterization of this strain in mouse infection models will further elucidate the pathogenic role of cyclopropanation in M. tuberculosis infection.
Acknowledgments
This work was supported by NIH grant AI53417 to M.S.G. D.B. was partially supported by the Michael and Ethel L. Cohen Foundation.
We thank Zully Feliciano for assistance with preparation of the manuscript.
Footnotes
Published ahead of print on 14 May 2010.
REFERENCES
- 1.Alahari, A., L. Alibaud, X. Trivelli, R. Gupta, G. Lamichhane, R. C. Reynolds, W. R. Bishai, Y. Guerardel, and L. Kremer. 2009. Mycolic acid methyltransferase, MmaA4, is necessary for thiacetazone susceptibility in Mycobacterium tuberculosis. Mol. Microbiol. 71:1263-1277. [DOI] [PubMed] [Google Scholar]
- 2.Barkan, D., Z. Liu, J. C. Sacchettini, and M. S. Glickman. 2009. Mycolic acid cyclopropanation is essential for viability, drug resistance, and cell wall integrity of Mycobacterium tuberculosis. Chem. Biol. 16:499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry, C. E., III, R. E. Lee, K. Mdluli, A. E. Sampson, B. G. Schroeder, R. A. Slayden, and Y. Yuan. 1998. Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37:143-179. [DOI] [PubMed] [Google Scholar]
- 4.Behr, M. A., B. G. Schroeder, J. N. Brinkman, R. A. Slayden, and C. E. Barry III. 2000. A point mutation in the mma3 gene is responsible for impaired methoxymycolic acid production in Mycobacterium bovis BCG strains obtained after 1927. J. Bacteriol. 182:3394-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatt, A., N. Fujiwara, K. Bhatt, S. S. Gurcha, L. Kremer, B. Chen, J. Chan, S. A. Porcelli, K. Kobayashi, G. S. Besra, and W. R. Jacobs, Jr. 2007. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. U. S. A. 104:5157-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 7.Converse, S. E., J. D. Mougous, M. D. Leavell, J. A. Leary, C. R. Bertozzi, and J. S. Cox. 2003. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. U. S. A. 100:6121-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs, Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 9.Dao, D. N., K. Sweeney, T. Hsu, S. S. Gurcha, I. P. Nascimento, D. Roshevsky, G. S. Besra, J. Chan, S. A. Porcelli, and W. R. Jacobs. 2008. Mycolic acid modification by the mmaA4 gene of M. tuberculosis modulates IL-12 production. PLoS Pathog. 4:e1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubnau, E., J. Chan, C. Raynaud, V. P. Mohan, M. A. Laneelle, K. Yu, A. Quemard, I. Smith, and M. Daffe. 2000. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 36:630-637. [DOI] [PubMed] [Google Scholar]
- 11.Dubnau, E., H. Marrakchi, I. Smith, M. Daffe, and A. Quemard. 1998. Mutations in the cmaB gene are responsible for the absence of methoxymycolic acid in Mycobacterium bovis BCG Pasteur. Mol. Microbiol. 29:1526-1528. [PubMed] [Google Scholar]
- 12.George, K. M., Y. Yuan, D. R. Sherman, and C. E. Barry III. 1995. The biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Identification and functional analysis of CMAS-2. J. Biol. Chem. 270:27292-27298. [DOI] [PubMed] [Google Scholar]
- 13.Glickman, M. S. 2003. The mmaA2 gene of Mycobacterium tuberculosis encodes the distal cyclopropane synthase of the alpha-mycolic acid. J. Biol. Chem. 278:7844-7849. [DOI] [PubMed] [Google Scholar]
- 14.Glickman, M. S., S. M. Cahill, and W. R. Jacobs, Jr. 2001. The Mycobacterium tuberculosis cmaA2 gene encodes a mycolic acid trans-cyclopropane synthetase. J. Biol. Chem. 276:2228-2233. [DOI] [PubMed] [Google Scholar]
- 15.Glickman, M. S., J. S. Cox, and W. R. Jacobs, Jr. 2000. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5:717-727. [DOI] [PubMed] [Google Scholar]
- 16.Grogan, D. W., and J. E. Cronan, Jr. 1984. Cloning and manipulation of the Escherichia coli cyclopropane fatty acid synthase gene: physiological aspects of enzyme overproduction. J. Bacteriol. 158:286-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grogan, D. W., and J. E. Cronan, Jr. 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 61:429-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guianvarc'h, D., T. Drujon, T. E. Leang, F. Courtois, and O. Ploux. 2006. Identification of new inhibitors of E. coli cyclopropane fatty acid synthase using a colorimetric assay. Biochim. Biophys. Acta 1764:1381-1388. [DOI] [PubMed] [Google Scholar]
- 19.Guianvarc'h, D., E. Guangqi, T. Drujon, C. Rey, Q. Wang, and O. Ploux. 2008. Identification of inhibitors of the E. coli cyclopropane fatty acid synthase from the screening of a chemical library: in vitro and in vivo studies. Biochim. Biophys. Acta 1784:1652-1658. [DOI] [PubMed] [Google Scholar]
- 20.MacGurn, J. A., and J. S. Cox. 2007. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect. Immun. 75:2668-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Middlebrook, G., R. J. Dobos, and C. Pierce. 1947. Virulence and morphological characteristics of mammalian tubercle bacilli. J. Exp. Med. 86:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raghunand, T. R., W. R. Bishai, and P. Chen. 2006. Towards establishing a method to screen for inhibitors of essential genes in mycobacteria: evaluation of the acetamidase promoter. Int. J. Antimicrob. Agents 28:36-41. [DOI] [PubMed] [Google Scholar]
- 23.Rao, V., N. Fujiwara, S. A. Porcelli, and M. S. Glickman. 2005. Mycobacterium tuberculosis controls host innate immune activation through cyclopropane modification of a glycolipid effector molecule. J. Exp. Med. 201:535-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao, V., F. Gao, B. Chen, W. R. Jacobs, Jr., and M. S. Glickman. 2006. Trans-cyclopropanation of mycolic acids on trehalose dimycolate suppresses Mycobacterium tuberculosis-induced inflammation and virulence. J. Clin. Invest. 116:1660-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaubourgeix, J., F. Bardou, F. Boissier, S. Julien, P. Constant, O. Ploux, M. Daffe, A. Quemard, and L. Mourey. 2009. S-adenosyl-N-decyl-aminoethyl, a potent bisubstrate inhibitor of Mycobacterium tuberculosis mycolic acid methyltransferases. J. Biol. Chem. 284:19321-19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan, Y., and C. E. Barry III. 1996. A common mechanism for the biosynthesis of methoxy and cyclopropyl mycolic acids in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 93:12828-12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan, Y., D. C. Crane, J. M. Musser, S. Sreevatsan, and C. E. Barry III. 1997. MMAS-1, the branch point between cis- and trans-cyclopropane-containing oxygenated mycolates in Mycobacterium tuberculosis. J. Biol. Chem. 272:10041-10049. [DOI] [PubMed] [Google Scholar]
- 28.Yuan, Y., R. E. Lee, G. S. Besra, J. T. Belisle, and C. E. Barry III. 1995. Identification of a gene involved in the biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 92:6630-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]