Abstract
Transcription of the Salmonella enterica recA gene is negatively controlled by the LexA protein, the repressor of the SOS response. The introduction of a mutation (recAo6869) in the LexA binding site, in the promoter region of the S. enterica ATCC 14028 recA gene, allowed the analysis of the effect that RecA protein overproduction has on the fitness of this virulent strain. The fitness of orally but not intraperitoneally inoculated recAo6869 cells decreased dramatically. However, the SOS response of this mutant was induced normally, and there was no increase in the sensitivity of the strain toward DNA-damaging agents, bile salts, or alterations in pH. Nevertheless, S. enterica recAo6869 cells were unable to swarm and their capacity to cross the intestinal epithelium was significantly reduced. The swarming deficiency in recAo6869 cells is independent of the flagellar phase. Moreover, swimming activity of the recAo6869 strain was not diminished with respect to the wild type, indicating that the flagellar synthesis is not affected by RecA protein overproduction. In contrast, swarming was recovered in a recAo6869 derivative that overproduced CheW, a protein known to be essential for this function. These data demonstrate that an equilibrium between the intracellular concentrations of RecA and CheW is necessary for swarming in S. enterica. Our results are the first to point out that the SOS response plays a critical role in the prevention of DNA damage by abolishing bacterial swarming in the presence of a genotoxic compound.
In bacteria, RecA is a key protein in homologous recombination, enabling the alignment of DNA molecules prior to strand exchange (17). RecA is also the positive regulator of the SOS response, one of the most-well-studied DNA repair systems in bacteria (54). In Escherichia coli and other Enterobacteriaceae, the cellular SOS network consists of more than 40 genes whose products act together to ensure cell survival after DNA damage (25). The negative regulator of the SOS system, LexA, binds to a specific sequence, the SOS box, which is located in the promoter region of the controlled genes (54). In Gammaproteobacteria, the SOS box is an imperfect palindrome, comprising the sequence CTGTN8ACAG (54) and varying among the different bacterial phyla (22).
The induction pathway of the SOS response seems to be conserved in all bacteria in which this DNA repair system is found. The RecA protein is activated after binding to single-stranded DNA resulting from inhibition of chromosomal replication (48). In turn, the activated RecA triggers autocatalytic cleavage of the LexA repressor. In E. coli, this cleavage occurs in the Ala84-Gly85 bond of the regulator (36) in a process mediated by the residues Ser119 and Lys156. Autocleavage resembles that described for serine proteases (38), and it prevents the binding of LexA to its specific recognition motif in the promoter region of SOS genes.
In addition to DNA repair genes, the SOS response involves lytic-cycle repressors of temperate bacteriophages (54). In some cases, such as in λ or P22 phages, RecA promotes autocleavage of the repressor (45), whereas for another class of temperate bacteriophages, an anti-lytic-cycle repressor protein, encoded by the tum gene, is under the direct negative control of LexA (49). For instance, Salmonella enterica strains contain at least three prophages (Fels-2, Gifsy-1, and Gifsy-2) whose lytic cycle is regulated by a tum-encoded protein (7). In fact, constitutive expression of the LexA regulon is lethal in this bacterial species, as it results in spontaneous induction of the lytic cycle of these three resident phages (7). The loss of these prophages suppresses the lethality of S. enterica lexA(Def) (lexA defective) cells (7), but it is worth noting that some of these S. enterica resident phages harbor genes that are required for bacterial virulence (24), so they are useless for virulence studies.
It is known that high RecA protein levels increase the frequency of DNA recombination (16). Moreover, the virulence of recA-defective mutants is significantly decreased in S. enterica as well as in other bacterial pathogens (5, 6, 11, 26, 34). However, due to the dual role of RecA (i.e., in recombination and as a positive regulator of the SOS response), the decrease in virulence of recA-defective mutants may be attributed to a reduction of the recombination frequency, to the inhibition of the SOS system induction, or both. To further elucidate the relative importance of each of the roles of RecA in the virulence process, two recA mutants were constructed: an S. enterica ATCC 14028 recAo6869 mutant that constitutively expressed recA and a recAo6869 lexA3(Ind−) (lexA noninducible) strain derivative harboring a mutated LexA repressor that was unable to induce the SOS response. The fitness of these mutants in intraperitoneal (i.p.) and oral infection was analyzed compared with that of the wild-type strain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. S. enterica was grown at 37°C in Luria-Bertani (LB) broth or agar plates. When necessary, ampicillin (100 μg ml−1), kanamycin (150 μg ml−1), or chloramphenicol (34 μg ml−1) was added to the bacterial cultures. DNA techniques were performed as described elsewhere (46). For anoxic conditions, the cultures were grown in the appropriate atmosphere by using the GasPak EZ anaerobe container system from Becton Dickinson.
TABLE 1.
Bacterial strains and plasmids used in this work
| S. enterica strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| ATCC 14028 | Wild type | ATCC |
| UA1876 | As ATCC 14028, but recAo6869 Kmr | This study |
| UA1822 | As ATCC 14028, but lexA3(Ind−) Cmr | 9 |
| UA1877 | As UA1876, but lexA3(Ind−) Kmr Cmr | This study |
| UA1878 | As ATCC 14028, but ΔrecX::cat Cmr | This study |
| UA1879 | As UA1876, but ΔrecX::cat Kmr Cmr | This study |
| UA1885 | As ATCC 14028, but Kmr | This study |
| UA1886 | As ATCC 14028, but ΔruvC::cat Cmr | This study |
| UA1887 | As UA1876, but ΔruvC::cat Kmr Cmr | This study |
| Plasmids | ||
| pKOBEGA | Ampr, temperature sensitive | 14 |
| pKD3 | Ampr Cmr | 20 |
| pKD4 | Ampr Kmr | 20 |
| pGEM-T | Cloning vector; Ampr | Promega |
| pGEX 4T-1 | Glutathione S-transferase gene fusion vector carrying the Ptac promoter and the lacIq gene; Ampr | Amersham Biosciences |
| pUA1108 | pGEX 4T-1 derivative plasmid carrying the Ptac promoter and the lacIq gene; Ampr | This study |
| pUA1109 | pUA1108 derivative plasmid containing the recA gene under the control of the Ptac promoter | This study |
| pUA1110 | pUA1108 derivative plasmid containing the cheW gene under the control of the Ptac promoter | This study |
Construction of S. enterica ATCC 14028 mutant derivatives.
The S. enterica recAo6869, ruvC, and recX mutants were constructed using the one-step PCR-based gene replacement method (20). The kanamycin resistance (Kmr) and chloramphenicol resistance (Cmr) cassettes from plasmids pKD4 and pKD3, respectively, were amplified using suitable 100-nucleotide (nt)-long oligonucleotides containing 80-nt stretches homologous to the insertion sites and the point mutation in the SOS box when needed (S. enterica recAo6869 mutant). The PCR product was transformed into S. enterica ATCC 14028 (Table 1) containing plasmid pKOBEGA (14). After the transformant clones were selected, pKOBEGA was eliminated by taking advantage of its temperature sensitivity at 42°C. The presence of the deletions or the point mutation in the SOS box was confirmed by PCR and sequencing.
For the construction of the S. enterica recAo6869 recX double mutant, the same strategy was used, but the recipient strain of the Cmr cassette was the previously constructed recAo6869 strain harboring pKOBEGA (Table 1).
The S. enterica wild-type strain harboring the Kmr cassette insertion was obtained using the same method. In this case, the 100-nt-long oligonucleotides harboring the 80-nt stretches homologous to the insertion sites contained no point mutations in the SOS box.
In all cases, the construct resulting from this procedure was moved into a clean wild-type strain background by transduction using the P22int7(HT) bacteriophage (10). The absence of the prophage in the transductants was determined by streaking them onto green plates as previously described (21). The obtained strains were verified both by PCR using suitable oligonucleotides and by sequencing.
S. enterica recAo6869 lexA3(Ind−) and recAo6869 ruvC strains were obtained by transduction. The recAo6869 construct from S. enterica UA1876 strain was transduced to the previously described (9) lexA3(Ind−) strain UA1822, and the ruvC deletion from UA1886 was transduced to the S. enterica UA1876 strain. The presence of the mutations was confirmed by PCR and sequencing.
For the flagellar phase-reversal process, the Sven Gard method was used (15). Briefly, Salmonella enterica was inoculated into the center of swimming plates where sterile antiserum against the identified flagellar phase had been added. The antiserum immobilizes the bacterial cells with the homologous flagella, whereas the cells with the heterologous flagella swim from the center of the plate. After overnight (ON) incubation at 37°C, cells from the edge of the growing area were selected for further studies.
Real-time quantitative RT-PCR assays.
Reverse transcription (RT)-PCR analyses of gene expression were carried out for all bacterial strains as previously reported (8) and using the primer pairs suitable for each gene. The results were normalized with respect to those obtained for the hisG housekeeping gene, since the latter does not belong to the SOS response (23). The induction factor (IF) of each gene for a given strain was defined as the ratio between the studied gene expression level in the presence and in the absence of mitomycin C (80 ng ml−1) in the respective culture.
Quantification of RecA protein.
S. enterica wild-type or recAo6869, lexA3(Ind−), or recAo6869 lexA3(Ind−) mutant cells were grown in LB broth at 37°C to an optical density at 550 nm (OD550) of 0.4. Then, 4 ml of the culture was removed and treated as the preinduction control. An equal volume of cells was treated with mitomycin C (0.8 mg ml−1) for 2 h. Cells were lysed in buffer A (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 2 mM EDTA, 10% glycerol), and the samples were stored in buffer B (50 mM Tris-HCl, pH 6.8, 3% β-2-mercaptoethanol, 3% SDS, 30% glycerol, 0.1% bromophenol blue). Samples from each induction experiment were electrophoresed alongside purified RecA protein standard (10 to 500 ng).
Polyclonal rabbit antiserum was raised against purified RecA protein according to standard protocols. The IgG fraction of the sera was purified by affinity chromatography with protein A-Sepharose 4B (Pharmacia Biotech) as described by the suppliers. The protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Immunoblots were transferred and probed with anti-RecA antibodies as described previously (13). RecA protein bands on developed immunoblots were quantified with a scanning densitometer. Purified RecA protein standards yielded a linear relationship between antibody signal and the RecA protein concentration. The amount of RecA protein in each induced sample was interpolated from the purified RecA protein standard curve.
S. enterica in vivo and in vitro competition assays.
The competitive assays were performed as reported previously (4, 10). Briefly, wild-type and mutant strains were grown separately and then mixed together in a ratio of approximately 1:1. The initial concentration for each strain was checked by plating serial dilutions of the bacterial suspensions onto LB media. For in vitro competition assays, the bacterial mixture was inoculated onto fresh LB medium at a final concentration of 2 × 107 CFU ml−1 and the culture was grown overnight (ON) at 37°C under aerobic or anaerobic conditions. Afterwards, serial dilutions of the culture were plated onto LB broth. To determine the proportion of each strain, replica platings were performed on appropriate antibiotic-supplemented LB plates. All of the in vitro experiments were performed at least in triplicate. For in vivo studies, in each experiment 6 or 12 female (4 to 6 weeks old) BALB/c mice were inoculated either orally or intraperitoneally (i.p.) with 2 × 108 CFU or 2 × 103 CFU, respectively, of the bacterial mixture per animal. Blood samples taken from the heart immediately after the mice died due to septicemia (generally 4 to 5 days after inoculation) were cultured in LB medium. The proportion of each strain appearing in the cultures was determined by replica plating onto appropriate antibiotic-supplemented LB plates. In all cases, the competitive index (CI) was calculated for each mutant as the ratio between the mutant and the wild-type strain in the output (bacteria recovered from the ON cultures in in vitro assays or from the host after infection in in vivo studies) divided by their ratio in the input (initial inoculum) (4). Statistical analyses consisted of the two-tailed t test and the Wilcoxon signed-rank test, with a P of ≤0.05 considered significant.
pH, bile, H2O2, and UV survival assays.
S. enterica recAo6869, lexA3(Ind−), and recAo6869 lexA3(Ind−) survival assays were performed as previously described (50, 52, 53), but with some modifications. All bacterial strains were grown ON in LB medium, diluted to a final concentration of 3 × 107 CFU ml−1 in the same medium, and then incubated with the appropriate compound. For pH resistance assays, ON cultures were diluted and incubated in pH 3.3 LB broth for 300 min without previous adaptation (52). Survival was assessed in samples withdrawn at 0, 50, 100, 150, 200, 250, and 300 min. For bile salts survival assays, diluted ON cultures were incubated for 300 min in LB broth supplemented with sodium choleate (Sigma) at a final concentration of 30%; survival was assessed in samples withdrawn at 0, 200, 250, and 300 min (53). For H2O2 survival assays, diluted ON cultures were incubated for 120 min in LB medium containing 10 mM H2O2. Samples were removed at 0, 15, 30, 60, 90, and 120 min after H2O2 addition and diluted in LB broth containing catalase (10 μg ml−1) to neutralize the H2O2 (50). The UV resistance assays were performed as described previously (3). Briefly, bacterial cells in the exponential growth phase were washed by centrifugation, suspended in AB medium (3) to a concentration of about 2 × 108 cells ml−1, and then irradiated at several doses of UV (0, 6, 12, 18, 24, and 30 J m−2) in a glass petri dish in thin layers, with a General Electric GY1578 germicidal lamp, at a rate of 1 J m−2 s−1 (determined with a Latarjet dosimeter). Afterwards, for each UV dose, samples were taken, diluted and plated in LB media, and grown at 37°C. All experiments with UV-irradiated cells were performed under yellow light or in the darkness to prevent photoreactivation. For all treatments, cell survival was calculated as the survival ratio of the treated versus the untreated cells.
S. enterica invasion assays.
The protocol used to test the invasiveness of several S. enterica strains into Caco-2 cells was similar to that used by Kim and Wei (33), with some modifications. Caco-2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-GlutaMAX I medium (Difco) supplemented with 20% fetal bovine serum (Difco) at 37°C in 95% air and 5% CO2. Once the monolayer was formed, cells were trypsinized and the suspension was seeded at 106 cells ml−1 onto tissue culture-treated 6-well plates (Corning Incorporated, Corning, NY). The plates were incubated at 37°C in 95% air and 5% CO2 until confluence was reached in every well.
S. enterica wild-type and recAo6869 and recAo6869 lexA3(Ind−) mutant strains were inoculated into LB broth and incubated overnight at 37°C. The bacterial cultures were centrifuged at 5,000 × g for 10 min and washed twice with phosphate-buffered saline (PBS), and the OD600 value was adjusted to 0.4. Bacterial suspensions were inoculated to a final concentration of 106 CFU ml−1 into the confluent monolayers of Caco-2 cells in the 6-well plates. Plates were incubated for 1 h at 37°C in 5% CO2. The cells were then rinsed once with 1 ml of DMEM (Gibco) and incubated for 2 h at 37°C in 5% CO2 with 1 ml of cell culture medium containing 100 μg of gentamicin ml−1 to remove extracellular bacteria. The cells were finally rinsed 3 times with PBS and lysed at room temperature for 10 min with 0.1% Triton X-100 in PBS. Viable bacterial counts were determined in duplicate by plating serial dilutions on LB agar. The plates were incubated at 37°C for 24 h. The invasion assay was carried out in triplicate.
S. enterica coinfection analysis in murine ileal loops.
To examine the in vivo interaction of S. enterica wild-type and recAo6869 strains with murine intestinal epithelial cells, the ligated-ileal loop coinfection model was used as described previously (30), with some modifications. Six-week-old BALB/c female mice were starved for 24 h and then anesthetized by i.p. injection of ketamine (100 mg kg−1) and xylazine (10 mg kg−1) before surgery. A small incision was made to expose the small bowel, and an ileal loop was then created by ligating a section of intestine containing a grossly identifiable Peyer′s patch to a site proximal to the cecum. The blood supply to the loop was kept intact. Equal numbers of cells of the parental and recAo6869 mutant strains (∼107 CFU ml−1) in PBS (200 μl) were then injected through a 25-gauge needle, after which the bowel was returned to the abdomen and the incision stapled. The mice were kept alive for 60 min and then killed. The ileal loop was excised intact, opened longitudinally, and placed into a tube containing PBS. Extracellular bacteria were eliminated by three washes with PBS and a further incubation for 90 min in PBS containing gentamicin (100 μg ml−1). The samples were then rinsed with PBS to remove residual gentamicin, and the entire ileal segment was processed in a stomacher (Seward Medical) using 1 ml of PBS. To determine intracellular parental and mutant bacteria, appropriate dilutions of the homogenized samples were spread onto LB and LB-kanamycin plates.
Overexpression of S. enterica recA and cheW.
The overexpression plasmid was constructed by inverted PCR using the pGEX up (5′-CGGGATCCCGCATATGTACTGTTTCCTGTGTGAA-3′) and pGEX dw (5′-CGGGATCCCCGGAATTCCCG-3′) primers, containing the NdeI and BamHI restriction sites, together with the pGEX 4T-1 vector as a template. The constructed plasmid (pUA1108), containing the tac promoter and the lacI gene in pGEX 4T-1, was checked by sequencing. Both the recA and the cheW coding regions were amplified using suitable oligonucleotides containing the NdeI and BamHI restriction sites. The PCR products were cloned into the pGEM-T vector and confirmed by sequencing. Afterwards, the NdeI-BamHI recA and cheW fragments were obtained by digestion and cloned into pUA1108, yielding plasmids pUA1109 and pUA1110, respectively. In both cases, recA and cheW were under the control of the tac promoter. Plasmids pUA1109 and pUA1110 were transformed in the S. enterica wild-type strain and recAo6869 mutant derivative, respectively. IPTG (isopropyl-β-d-thiogalactopyranoside)-mediated overexpression of either RecA or CheW was confirmed in crude extracts by SDS-PAGE (data not shown). Briefly, the wild-type strain, harboring pUA1109, and the recAo6869 mutant, containing pUA1110, were grown on LB broth to an OD550 of 0.5, after which IPTG (1 mM) was added to the cultures followed by 3 h of incubation at 37°C. The crude protein extracts were loaded onto 15% acrylamide gels, and SDS-PAGE was conducted for 2 h. The protein profiles of the wild-type and mutant strains were visualized by Coomassie blue staining of the gels, which in each case confirmed the overexpression of either RecA or CheW.
Swimming and swarming assays.
Swimming and swarming assays were carried out as described previously (28). In short, the bacterial strains were grown in LB broth at 37°C. Then, 10 μl of the stationary cultures was inoculated in the middle of either swimming or swarming plates, supplemented when necessary with IPTG or mitomycin C. The plates were incubated ON at 37°C and then photographed.
RESULTS AND DISCUSSION
Construction of an S. enterica mutant constitutively expressing the recA gene.
The S. enterica recAo6869 mutant (UA1876), harboring a mutation in the LexA binding site of the recA gene promoter, was obtained using the one-step mutagenesis method (Fig. 1). As expected, this mutant presents a higher amount of RecA protein (∼25,000 protomers per cell) than that measured in the wild-type strain (∼5,000 protomers per cell) (Fig. 2 A). The basal level of the RecA protein was essentially the same in the wild-type and lexA3(Ind−) strains (Fig. 2A). Likewise, the basal level of RecA protein in the recAo6869 lexA3(Ind−) mutant was the same as that in the recAo6869 strain (Fig. 2A). It is worth noting that the basal number of molecules of S. enterica RecA protein per cell was similar to that obtained for E. coli (∼5,000) and Bacillus subtilis cells (∼4,500) (37).
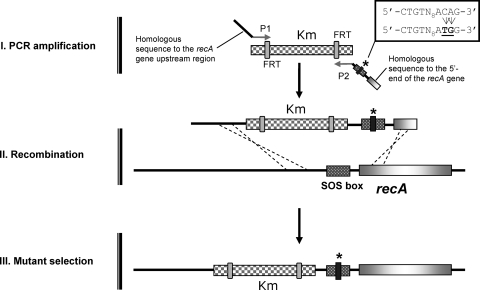
FIG. 1.
Construction of the S. enterica recAo6869 mutant using a one-step PCR-based gene replacement method. The kanamycin resistance gene (Km) was amplified from plasmid pKD4 using oligonucleotides containing sequences homologous to the chromosomal DNA of S. enterica (step I). This construct was used to transform the S. enterica ATCC 14028 wild-type strain (step II), and Kmr colonies were selected (step III). The presence of the mutation in the SOS box was verified by both PCR and sequencing. FRT, FLP recombination target. The asterisk indicates the point mutation in the LexA binding sequence that is boxed in the figure.
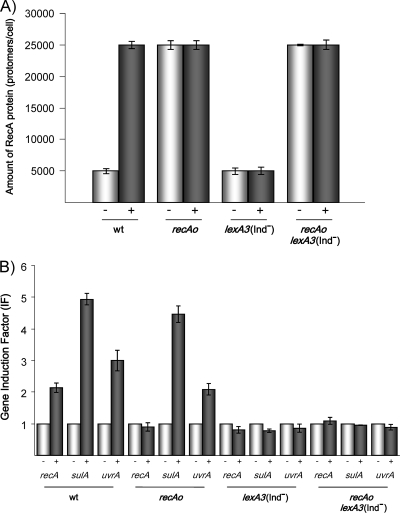
FIG. 2.
Mitomycin C-mediated induction of the RecA protein levels and of the expression of several SOS genes in S. enterica recAo6869 (recAo), lexA3(Ind−), and recAo6869 lexA3(Ind−) mutants. (A) Levels of RecA protein in the S. enterica ATCC 14028 wild type (wt) strain and in each mutant strain treated (+) or untreated (−) with mitomycin C. All determinations were performed at least three times. (B) Expression levels of the recA, uvrA, and sulA SOS genes in the S. enterica wild type or in each mutant derivative in the presence (+) or absence (−) of mitomycin C. In this case, the induction factor (IF) was defined as the ratio between the relative mRNA concentration of each gene in cells treated with mitomycin C and in untreated cells. The data are the means of two independent quantitative real-time RT-PCRs (each performed in triplicate), and the standard deviation of any value was never greater than 10%.
Furthermore, in mitomycin C-treated wild-type cells, the rate of RecA protein accumulation increased about 5-fold, whereas no induction was observed in the lexA3(Ind−) strain (Fig. 2A). Besides, the levels of RecA protein were essentially the same in recAo6869 and recAo6869 lexA3(Ind−) strains and both wild-type and mutant mitomycin C-treated cells (Fig. 2A). This rate probably represents maximum induction of the RecA protein by DNA damage because it was essentially the same as that from the recAo6869 mutant.
Nonetheless, the behavior of the S. enterica recAo6869 mutant was the same as that of the wild-type strain with respect to the DNA-damage-mediated induction of other SOS genes, such as sulA and uvrA (Fig. 2B). In contrast, in the S. enterica recAo6869 lexA3(Ind−) strain and the lexA3(Ind−) strain, these SOS genes were not induced after cellular DNA damage (Fig. 2B). In concordance with these results, and as previously reported in an E. coli recAo98 mutant (27), the survival of UV-irradiated S. enterica recAo6869 cells was the same as that of wild-type cells (data not shown). Altogether, these data indicated that constitutive expression of the recA gene in the recAo6869 strain affects neither the kinetics of the SOS response nor the resistance of S. enterica to DNA-damaging agents. Similar results were reported for E. coli recAo6869 cells (27).
To further characterize the S. enterica recAo98, lexA3(Ind−), and recAo6869 lexA3(Ind−) mutants and to determine whether their growth was affected, in vitro competitive assays were carried out and growth kinetics were measured in minimal medium and in rich medium. The results showed that neither the overproduction of RecA protein nor the inability to induce the SOS system seems to have an effect on S. enterica growth in vitro (data not shown). It must be noted that similar results have been reported for both E. coli recAo98 and lexA(Ind−) mutants (27, 42).
In vivo fitness of the S. enterica recAo6869 strain.
To elucidate whether RecA-mediated recombination or SOS activation is more important for the infection process, the S. enterica recAo6869, lexA3(Ind−), and recAo6869 lexA3(Ind−) mutants were examined in competition assays with the wild-type strain. Neither deregulation of the S. enterica recA gene nor the inability to induce the SOS response because of the presence of a lexA3(Ind−) mutation had any effect upon the virulence of intraperitoneally (i.p.) inoculated S. enterica (Table 2). Proteins encoded by ruvABC genes play an essential role in the late steps of homologous recombination (55), whereas RecA is involved in the early stages of this process (35). For this reason, an S. enterica recAo6869 ruvC mutant was constructed and its competitive index (CI) following i.p. inoculation, in comparison with that of the wild-type strain, was analyzed. Table 2 demonstrates that S. enterica recAo6869 ruvC cells present a 104-fold decrease in their CI. These results, together with those reported for recA mutants of S. enterica recA and other bacterial species (5, 6, 11, 26, 34), indicate that all stages of the DNA recombination process are important for a full bacterial infective capacity. Furthermore, and following oral inoculation, the lexA3(Ind−) strain exhibited the same behavior as the wild-type strain (Table 2), in concordance with results recently published (19) in which no defect in lexA3(Ind−) mutant is observed when it is i.p. inoculated. On the other hand, the fitness of the S. enterica recAo6869 and recAo6869 lexA3(Ind−) mutants was dramatically reduced when orally inoculated (Table 2). These results demonstrated that overexpression of recA has a negative effect on the oral fitness of S. enterica.
TABLE 2.
Competitive indexes of several S. enterica mutants in murine model
| Strain comparison (mixed infection) | Challenge route | CIa |
|---|---|---|
| recAo6869 mutant vs wild type | i.p. | 0.42 |
| lexA3(Ind−) mutant vs wild type | i.p. | 0.42 |
| recAo6869 lexA3(Ind−) mutant vs wild type | i.p. | 0.48 |
| recAo6869 ruvC mutant vs wild type | i.p. | 0.00019* |
| recAo6869 mutant vs wild type | Oral | 0.00027* |
| lexA3(Ind−) mutant vs wild type | Oral | 2.73 |
| recAo6869 lexA3(Ind−) mutant vs wild type | Oral | 0.0054* |
CI is calculated as the ratio between the mutant and the wild-type strain in the output (bacteria recovered from the host after infection) divided by their ratio in the input. *, virulence of the mutants was significantly lower than that of the wild-type strain (in all cases with a P value of ≤0.05).
Overproduction of the RecA protein decreases invasiveness of S. enterica.
It should be noted that, following infection via the oral route, the bacterial cells cross the intestinal epithelium, and then the infection process proceeds as if they had been i.p. inoculated (40). Since no differences between the wild type and recAo6869 and recAo6869 lexA3(Ind−) mutants were observed in i.p. competition assays, our results suggested that the negative effect of RecA overexpression upon the oral fitness of the recAo6869 mutant derivatives must have occurred before S. enterica entered the bloodstream.
In this context, several factors that could explain the reduced fitness of the S. enterica recAo6869 mutant derivatives when orally inoculated were analyzed. First, in vitro competition assays carried out under anoxic conditions did not reveal any difference between the recAo6869 mutant and the wild-type strain (data not shown). Similar results were reported in E. coli recAo6869 cells (27). Second, the results of survival assays in which the bacteria were exposed to bile salts, acidic pH, and H2O2 likewise suggested that the sensitivity of the S. enterica recAo6869 cells to the above DNA-damaging agents was similar to that of the wild-type strain (data not shown). Third, as mentioned above, some S. enterica prophages have an important role in virulence (24). Accordingly, the maintenance of some resident prophages was examined in the S. enterica recAo6869 mutants. The results of PCRs showed that the reduced fitness of the recAo6869 cells could not be attributed to a loss of the prophages Gifsy-1 and Gifsy-2, since both were present in the recAo6869 mutant strains (data not shown).
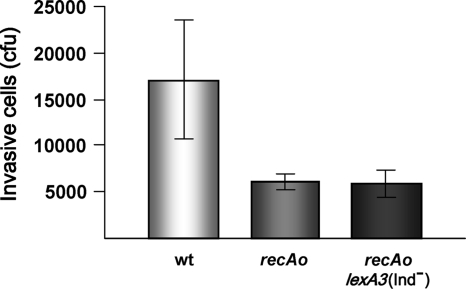
Another critical point during infection is the passing of the bacteria through the intestinal epithelium (40). Thus, invasiveness of S. enterica recAo6869 cells was tested using the human intestinal epithelial cell line Caco-2. Figure 3 indicates that both S. enterica recAo6869 and recAo6869 lexA3(Ind−) mutants have a decreased ability to invade Caco-2 cells (statistically significant at P = 0.0286). In concordance with this, coinfection studies in murine ileal loops using the S. enterica wild-type and recAo6869 and recAo6869 lexA3(Ind−) mutant strains show that the competitive indexes for both mutants in the ileal loops were 0.25 ± 0.04 and 0.376 ± 0.045, respectively. These data clearly indicate that the S. enterica recAo6869 and recAo6869 lexA3(Ind−) mutants were less able to reach the systemic compartment. It can therefore be concluded that the reduced fitness of S. enterica recAo6869 cells was associated with its deficiency in crossing the intestinal epithelium.
FIG. 3.
Invasiveness of S. enterica strains in Caco-2 cells. In all cases, a final concentration of 106 CFU ml−1 of either the wild type (wt) or the recAo6869 (recAo) mutant derivatives was inoculated into confluent monolayers of Caco-2 cells. After 1 h of incubation and a gentamicin treatment, Caco-2 cells were lysed; the number of invasive cells is shown. In all cases, the invasion assay was carried out in triplicate.
S. enterica recAo6869 mutants do not swarm.
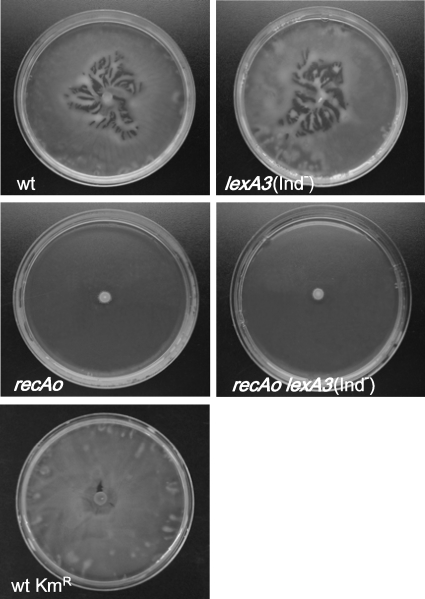
It has been demonstrated that S. enterica mutants defective in swarming as a consequence of mutations in either the cheW or the cheB gene show a lower invasiveness than the wild-type strain under anaerobic conditions (31). Furthermore, through comparative analyses among several S. enterica serovars, it has been suggested that a relationship between swarming and intestinal colonization must exist in this bacterial species (32). Likewise, swarming is known to be important for the virulence of other bacterial pathogens, such as Proteus mirabilis, Helicobacter pylori, Vibrio parahaemolyticus, and Campylobacter jejuni (43, 44, 51, 57). In addition, it has been reported that the RecA protein is necessary for the swarming motility of E. coli, since recA mutants of this species do not swarm (28). For all these reasons, the swarming behavior of the S. enterica recAo6869, lexA3(Ind−), and recAo6869 lexA3(Ind−) mutants was studied (Fig. 4). The data indicated that neither the S. enterica recAo6869 strain nor the recAo6869 lexA3(Ind−) strain swarmed, while the swarming capacity of lexA3(Ind−) cells was similar to that of the wild-type strain (Fig. 4). Thus, and in concordance with the results obtained in the competition assays, the inhibition of swarming was only associated with constitutive levels of recA and not with the presence of the lexA3(Ind−) mutation.
FIG. 4.
Swarming ability of the S. enterica ATCC 14028 wild-type strain (wt) and the wild-type Kmr (wt Kmr), recAo6869 (recAo), lexA3(Ind−), and recAo6869 lexA3(Ind−) mutant derivatives. Bacterial colony swarming patterns as they appeared on a semisolid (Difco) agar surface following incubation of the cultures for 24 h at 37°C are shown.
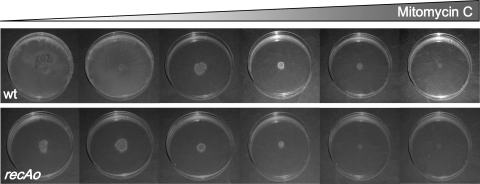
To ensure that the recAo6869 mutation was the only mutation responsible for this swarming-defective phenotype, a wild-type strain harboring the Kmr cassette in the same position as the recAo6869 mutants but without any mutation in the LexA binding site was constructed. This strain presents a swarming pattern identical to that of the S. enterica ATCC 14028 wild-type strain with no insertion (Fig. 4). Furthermore, it must be noted that in the wild-type strain, the increase in SOS-mediated recA expression following the addition of sublethal concentrations of mitomycin C abolished swarming (Fig. 5).
FIG. 5.
Swarming ability of the S. enterica wild-type (wt) and recAo6869 (recAo) mutant strains in the presence of an SOS inductor. Colony swarming patterns developed on a semisolid agar surface (Difco) in the presence of increasing concentrations of mitomycin C (0, 8, 80, 200, 800, and 1,600 ng ml−1) are shown.
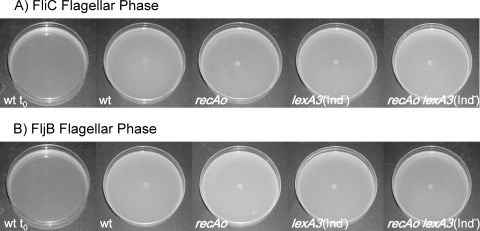
It is worth noting that the swarming deficiency shown by the S. enterica recAo6869 cells is not dependent upon the flagellar phase, since strains expressing either fliC or fljB genes do not swarm (data not shown). Moreover, swimming motility of S. enterica recAo6869 cells is not affected regardless of the flagellar phase which they present (Fig. 6), indicating that this mutant does not display any difficulties in synthesizing flagella.
FIG. 6.
Swimming ability of S. enterica wild-type strain (wt) and its recAo6869 (recAo), lexA3(Ind−), and recAo6869 lexA3(Ind−) mutant derivatives. Colony swimming patterns developed by strains presenting either the FliC (A) or the FljB (B) flagellar phase are shown. As a control, the wild-type plate immediately after inoculation (wt t0) is shown.
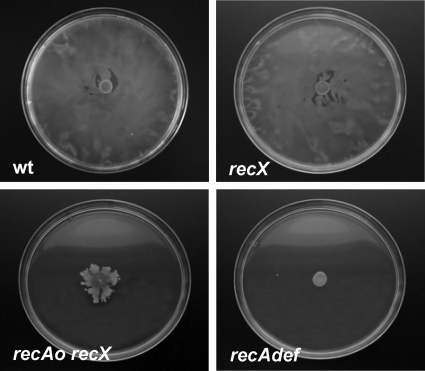
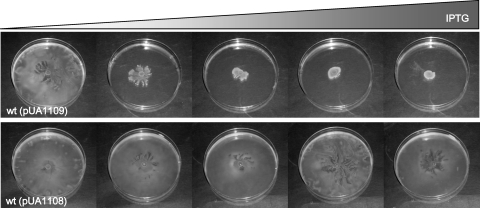
The S. enterica recA and recX genes form a single transcriptional unit (18, 41). To rule out the possibility that recX overexpression was responsible for the swarming defect, knockout mutant derivatives of this gene were constructed and the organism's ability to swarm was tested. The S. enterica recX mutant was able to swarm, while the recAo6869 recX double mutant was not (Fig. 7). These results again implicated recA constitutive expression in the swarming defect. To further confirm that the overexpression of RecA fully accounts for the inability of the mutant strain to swarm, the recA gene was cloned into plasmid pUA1108 under the control of an IPTG-inducible promoter (the tac promoter), giving rise to pUA1109. The pUA1109 plasmid was used to transform the S. enterica wild-type strain. The swarming phenotype of S. enterica carrying pUA1109 was then analyzed in the presence of increasing concentrations of IPTG. As expected, the swarming ability decreased in an IPTG-dependent manner (Fig. 8), unequivocally confirming that the overexpression of recA suppresses swarming.
FIG. 7.
Swarming ability of S. enterica recX and recAo6869 (recAo) recX mutant derivatives. Colony swarming patterns developed on a semisolid (Difco) agar surface following incubation of the cultures for 24 h at 37°C are shown. As a control, the negative swarming phenotype of the S. enterica recA-defective mutant (recAdef) is also presented.
FIG. 8.
Swarming ability of the S. enterica ATCC 14028 wild-type (wt) strain harboring the recA-overexpressing plasmid pUA1109. Colony swarming patterns developed on a semisolid (Difco) agar surface in the presence of increasing concentrations of IPTG (0, 5, 10, 20, and 30 μM) after incubation of the cultures for 24 h at 37°C. As a control, the swarming phenotype of the wild-type strain harboring the pUA1108 overexpression vector without the recA gene in the presence of IPTG is also presented.
CheW overexpression reestablishes swarming in the S. enterica recAo6869 mutant.
The swarming phenotype has been widely studied in several bacterial species (29, 44). In Salmonella species, swarming is under the control of the CheA-CheY two-component signal transduction system (39), and a critical role in this process has been also demonstrated for the CheW protein (2). Specifically, CheW is bound to CheA (a histidine protein kinase) and participates in its autophosphorylation, which leads to swarming activation (2). However, it is also known that the overexpression of CheW inhibits swarming (47) and that a precise quantitative association between the CheW and CheA proteins is required for this kind of cellular motility (12, 56). Furthermore, CheW and RecA have been shown to interact in vivo in E. coli (1), and, as mentioned above, E. coli mutants defective in recA do not swarm (28). Since both the absence of RecA and the overexpression of RecA result in an inability to swarm, we postulated that an equilibrium between the cellular amounts of RecA and CheW proteins must be critical for bacterial swarming.
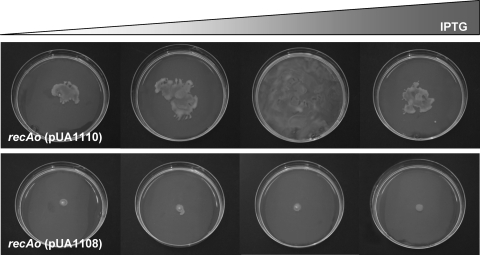
To test this hypothesis, the S. enterica cheW gene was cloned under the control of the tac promoter to obtain plasmid pUA1110, which was then transformed in the S. enterica recAo6869 mutant. The swarming phenotype of this strain was analyzed in the presence of increasing IPTG concentrations. As shown in Fig. 9, the S. enterica recAo6869 (pUA1110) strain recovered the swarming ability in an IPTG-dependent manner. However, when the amounts of CheW were large enough, and in accordance with previously published works (47), swarming was abolished (Fig. 9). These data confirmed that the in vivo equilibrium between CheW and RecA is important for bacterial swarming.
FIG. 9.
Swarming ability of the S. enterica recAo6869 (recAo) mutant harboring the cheW-overexpressing plasmid pUA1110. Colony swarming patterns developed on a semisolid (Difco) agar surface in the presence of increasing concentrations of IPTG (0, 10, 20, and 30 μM) in cultures incubated for 24 h at 37°C. As a control, the swarming phenotype of the recAo6869 mutant harboring the pUA1108 overexpression vector without the cheW gene in the presence of IPTG is also presented.
In this context, it is tempting to speculate that bacterial colonies use the amplification of intracellular RecA as a tool to sense the presence of DNA-damaging compounds (antibiotics and bacteriocins, among other natural molecules) around them and, consequently, to cease swarming. In this scenario, the presence of a DNA-damaging agent would induce the SOS response, initiating an increase in the amount of RecA protein and thus an alteration of the equilibrium between RecA and CheW, which in turn would abolish the coordinated multicellular surface migration. Accordingly, exposure of the bacterial genetic material to even higher concentrations of the DNA-injuring agent is avoided. Thus, from a bacterial population point of view, the LexA regulon not only would play a fundamental role in DNA damage repair but also could serve to avoid DNA injuries by preventing bacterial colonies from being exposed to high concentrations of genotoxic agents.
Acknowledgments
This work was funded by grants BFU2008-01078 from the Ministerio de Ciencia e Innovación (MICINN) de España and 2009SGR1106 from the Generalitat de Catalunya to J.B. and BFU2009-07167 from MICINN to J.C.A. Laura Medina was the recipient of a predoctoral fellowship from the MICINN.
We are deeply indebted to G. M. Ghigo and to Francisco García del Portillo for the generous gift of plasmid pKOBEGA and the Caco-2 cell line, respectively. We are grateful to Montserrat Saco for helping us with the Salmonella flagellar phase reversal procedures and to Maria Pilar Cortés and Joan Ruiz for their excellent technical assistance.
Editor: A. Camilli
Footnotes
Published ahead of print on 10 May 2010.
REFERENCES
- 1.Arifuzzaman, M., M. Maeda, A. Itoh, K. Nishikata, C. Takita, R. Saito, T. Ara, K. Nakahigashi, H. C. Huang, A. Hirai, K. Tsuzuki, S. Nakamura, M. Altaf-Ul-Amin, T. Oshima, T. Baba, N. Yamamoto, T. Kawamura, T. Ioka-Nakamichi, M. Kitagawa, M. Tomita, S. Kanaya, C. Wada, and H. Mori. 2006. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 16:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, M. D., P. M. Wolanin, and J. B. Stock. 2006. Signal transduction in bacterial chemotaxis. Bioessays 28:9-22. [DOI] [PubMed] [Google Scholar]
- 3.Barbe, J., A. Villaverde, J. Cairo, and R. Guerrero. 1986. ATP hydrolysis during SOS induction in Escherichia coli. J. Bacteriol. 167:1055-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microb. Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 5.Buchmeier, N. A., S. J. Libby, Y. Xu, P. C. Loewen, J. Switala, D. G. Guiney, and F. C. Fang. 1995. DNA repair is more important than catalase for Salmonella virulence in mice. J. Clin. Invest. 95:1047-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchmeier, N. A., C. J. Lipps, M. Y. So, and F. Heffron. 1993. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol. Microbiol. 7:933-936. [DOI] [PubMed] [Google Scholar]
- 7.Bunny, K., J. Liu, and J. Roth. 2002. Phenotypes of lexA mutations in Salmonella enterica: evidence for a lethal lexA null phenotype due to the Fels-2 prophage. J. Bacteriol. 184:6235-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campoy, S., M. Fontes, S. Padmanabhan, P. Cortes, M. Llagostera, and J. Barbe. 2003. LexA-independent DNA damage-mediated induction of gene expression in Myxococcus xanthus. Mol. Microbiol. 49:769-781. [DOI] [PubMed] [Google Scholar]
- 9.Campoy, S., A. Hervas, N. Busquets, I. Erill, L. Teixido, and J. Barbe. 2006. Induction of the SOS response by bacteriophage lytic development in Salmonella enterica. Virology 351:360-367. [DOI] [PubMed] [Google Scholar]
- 10.Campoy, S., M. Jara, N. Busquets, A. M. Perez De Rozas, I. Badiola, and J. Barbe. 2002. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 70:4721-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardenas, M., A. R. Fernandez de Henestrosa, S. Campoy, A. M. Perez de Rozas, J. Barbe, I. Badiola, and M. Llagostera. 2001. Virulence of Pasteurella multocida recA mutants. Vet. Microbiol. 80:53-61. [DOI] [PubMed] [Google Scholar]
- 12.Cardozo, M. J., D. A. Massazza, J. S. Parkinson, and C. A. Studdert. 5 January 2010, posting date. Disruption of chemoreceptor signaling arrays by high level of CheW, the receptor-kinase coupling protein. Mol. Microbiol. [Epub ahead of print.] doi: 10.1111/j.1365-2958.2009.07032.x. [DOI] [PMC free article] [PubMed]
- 13.Carrasco, B., C. Cañas, G. J. Sharples, J. C. Alonso, and S. Ayora. 2009. The N-terminal region of the RecU Holliday junction resolvase is essential for homologous recombination. J. Mol. Biol. 390:1-9. [DOI] [PubMed] [Google Scholar]
- 14.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiou, C. S., J. F. Huang, L. H. Tsai, K. M. Hsu, C. S. Liao, and H. L. Chang. 2006. A simple and low-cost paper-bridged method for Salmonella phase reversal. Diagn. Microbiol. Infect. Dis. 54:315-317. [DOI] [PubMed] [Google Scholar]
- 16.Clark, A. J. 1982. recA operator mutations and their usefulness. Biochimie 64:669-675. [DOI] [PubMed] [Google Scholar]
- 17.Cox, M. M. 2007. Motoring along with the bacterial RecA protein. Nat. Rev. 8:127-138. [DOI] [PubMed] [Google Scholar]
- 18.Cox, M. M. 2007. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 42:41-63. [DOI] [PubMed] [Google Scholar]
- 19.Craig, M., and J. M. Slauch. 2009. Phagocytic superoxide specifically damages an extracytoplasmic target to inhibit or kill Salmonella. PLoS One 4:e4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Nat. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. A manual for genetic engineering. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 22.Erill, I., S. Campoy, and J. Barbe. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31:637-656. [DOI] [PubMed] [Google Scholar]
- 23.Erill, I., M. Escribano, S. Campoy, and J. Barbe. 2003. In silico analysis reveals substantial variability in the gene contents of the gamma proteobacteria LexA-regulon. Bioinformatics 19:2225-2236. [DOI] [PubMed] [Google Scholar]
- 24.Figueroa-Bossi, N., and L. Bossi. 1999. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33:167-176. [DOI] [PubMed] [Google Scholar]
- 25.Fry, R. C., T. J. Begley, and L. D. Samson. 2005. Genome-wide responses to DNA-damaging agents. Annu. Rev. Microbiol. 59:357-377. [DOI] [PubMed] [Google Scholar]
- 26.Fuchs, S., I. Muhldorfer, A. Donohue-Rolfe, M. Kerenyi, L. Emody, R. Alexiev, P. Nenkov, and J. Hacker. 1999. Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb. Pathog. 27:13-23. [DOI] [PubMed] [Google Scholar]
- 27.Ginsburg, H., S. H. Edmiston, J. Harper, and D. W. Mount. 1982. Isolation and characterization of an operator-constitutive mutation in the recA gene of E. coli K-12. Mol. Gen. Genet. 187:4-11. [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Gomez, J. M., C. Manfredi, J. C. Alonso, and J. Blazquez. 2007. A novel role for RecA under non-stress: promotion of swarming motility in Escherichia coli K-12. BMC Biol. 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harshey, R. M., and T. Matsuyama. 1994. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Nat. Acad. Sci. U. S. A. 91:8631-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, B. D., C. A. Lee, and S. Falkow. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katribe, E., L. M. Bogomolnaya, H. Wingert, and H. Andrews-Polymenis. 2009. Subspecies IIIa and IIIb salmonellae are defective for colonization of murine models of salmonellosis compared to Salmonella enterica subsp. I serovar Typhimurium. J. Bacteriol. 191:2843-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, S. H., and C. I. Wei. 2007. Invasiveness and intracellular growth of multidrug-resistant Salmonella and other pathogens in Caco-2 cells. J. Food Sci. 72:M72-M78. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, K. K., R. Srivastava, V. B. Sinha, J. Michalski, J. B. Kaper, and B. S. Srivastava. 1994. recA mutations reduce adherence and colonization by classical and El Tor strains of Vibrio cholerae. Microbiology 140:1217-1222. [DOI] [PubMed] [Google Scholar]
- 35.Kuzminov, A. 1996. Unraveling the late stages of recombinational repair: metabolism of DNA junctions in Escherichia coli. Bioessays 18:757-765. [DOI] [PubMed] [Google Scholar]
- 36.Little, J. W. 1991. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie 73:411-421. [DOI] [PubMed] [Google Scholar]
- 37.Lovett, C. M., Jr., P. E. Love, R. E. Yasbin, and J. W. Roberts. 1988. SOS-like induction in Bacillus subtilis: induction of the RecA protein analog and a damage-inducible operon by DNA damage in Rec+ and DNA repair-deficient strains. J. Bacteriol. 170:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo, Y., R. A. Pfuetzner, S. Mosimann, M. Paetzel, E. A. Frey, M. Cherney, B. Kim, J. W. Little, and N. C. Strynadka. 2001. Crystal structure of LexA: a conformational switch for regulation of self-cleavage. Cell 106:585-594. [DOI] [PubMed] [Google Scholar]
- 39.Mariconda, S., Q. Wang, and R. M. Harshey. 2006. A mechanical role for the chemotaxis system in swarming motility. Mol. Microbiol. 60:1590-1602. [DOI] [PubMed] [Google Scholar]
- 40.Mastroeni, P., A. Grant, O. Restif, and D. Maskell. 2009. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat. Rev. Microbiol. 7:73-80. [DOI] [PubMed] [Google Scholar]
- 41.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 42.Mount, D. W., K. B. Low, and S. J. Edmiston. 1972. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet light-induced mutations. J. Bacteriol. 112:886-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakajima, K., S. Inatsu, T. Mizote, Y. Nagata, K. Aoyama, Y. Fukuda, and K. Nagata. 2008. Possible involvement of putA gene in Helicobacter pylori colonization in the stomach and motility. Biomed. Res. 29:9-18. [DOI] [PubMed] [Google Scholar]
- 44.Rather, P. N. 2005. Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7:1065-1073. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, J. W., and R. Devoret. 1983. Lysogenic induction, p. 123-144. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 46.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Sanders, D. A., B. Mendez, and D. E. Koshland, Jr. 1989. Role of the CheW protein in bacterial chemotaxis: overexpression is equivalent to absence. J. Bacteriol. 171:6271-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sassanfar, M., and J. W. Roberts. 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 212:79-96. [DOI] [PubMed] [Google Scholar]
- 49.Shearwin, K. E., A. M. Brumby, and J. B. Egan. 1998. The Tum protein of coliphage 186 is an antirepressor. J. Biol. Chem. 273:5708-5715. [DOI] [PubMed] [Google Scholar]
- 50.Stohl, E. A., A. K. Criss, and H. S. Seifert. 2005. The transcriptome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection. Mol. Microbiol. 58:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takata, T., S. Fujimoto, and K. Amako. 1992. Isolation of nonchemotactic mutants of Campylobacter jejuni and their colonization of the mouse intestinal tract. Infect. Immun. 60:3596-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiganitas, A., N. Zeaki, A. S. Gounadaki, E. H. Drosinos, and P. N. Skandamis. 2009. Study of the effect of lethal and sublethal pH and a(w) stresses on the inactivation or growth of Listeria monocytogenes and Salmonella Typhimurium. Int. J. Food Microbiol. 134:104-112. [DOI] [PubMed] [Google Scholar]
- 53.van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker, G. C. 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev. 48:60-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West, S. C. 1997. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31:213-244. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, P., C. M. Khursigara, L. M. Hartnell, and S. Subramaniam. 2007. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Nat. Acad. Sci. U. S. A. 104:3777-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zorrilla, I., S. Arijo, M. Chabrillon, P. Diaz, E. Martinez-Manzanares, M. C. Balebona, and M. A. Morinigo. 2003. Vibrio species isolated from diseased farmed sole, Solea senegalensis (Kaup), and evaluation of the potential virulence role of their extracellular products. J. Fish Dis. 26:103-108. [DOI] [PubMed] [Google Scholar]