Abstract
Denaturing gradient gel electrophoresis (DGGE) and quantitative real-time PCR (qPCR) were successfully developed to monitor functional aoxB genes as markers of aerobic arsenite oxidizers. DGGE profiles showed a shift in the structure of the aoxB-carrying bacterial population, composed of members of the Alpha-, Beta- and Gammaproteobacteria, depending on arsenic (As) and Eh levels in Upper Isle River Basin waters. The highest aoxB gene densities were found in the most As-polluted oxic surface waters but without any significant correlation with environmental factors. Arsenite oxidizers seem to play a key role in As mobility in As-impacted waters.
Arsenic (As) occurs naturally as a local geological constituent of the soils surrounding the Upper Isle River Basin (Massif Central, France) due to natural geochemical anomalies but is also released from Au/As deposits of disused gold mines (4, 11, 12, 33). Important variations in dissolved As concentrations are found in the Isle River and depend on the hydrogeological season, with maximum values in spring and summer generally detected during low-flow conditions (12, 22), and probably on temperature-controlled microbial As(V) reduction and/or microbial dissolution of solid As carrier phases (22). Two toxic inorganic forms of As are usually detected in aquatic system: arsenite, As(III), which is found mainly under anaerobic conditions and is more mobile than arsenate, As(V), which typically occurs under aerobic conditions and tends to associate with oxyhydroxides and clay minerals (11, 34). Although bacteria are known to play a key role in speciation, mobility, and bioavailability of As in the environment, they have never been considered in previous studies of As mobility in the Isle River system. Indeed, former investigations of As cycling were focused on geochemical studies (4, 11, 12, 22, 33).
As(III)-oxidizing bacteria can contribute to a natural attenuation of As pollution by decreasing its bioavailability and can help remove As from mine wastewaters through bioprocessing (1, 2). Many As(III) oxidizers have been isolated from various environments, especially mesophilic ecosystems (3, 5, 8, 16, 25, 27, 32, 38). They belong to more than 25 genera, mainly of the Proteobacteria phylum (3, 32, 38), and are related to organisms unable to oxidize As(III) based on 16S rRNA phylogeny. Diverse primer sets have been successfully developed to specifically target the functional aoxB gene (9, 14, 17, 25, 26), encoding the large molybdenum-bearing catalytic subunit of As(III)-oxidase (EC 1.20.98.1), an enzyme of the dimethyl sulfoxide (DMSO) reductase family. Using cloning-sequencing approaches, the aoxB gene has proven to be a reliable molecular marker for diversity studies of the polyphyletic aerobic As(III) oxidizers in As-impacted soil and water systems (17, 25). The genetic fingerprinting denaturing gradient gel electrophoresis (DGGE) technique is one useful tool for spatial, temporal, and geographical monitoring of complex bacterial population structure (23, 24). Quantitative real-time PCR (qPCR) provides reliable measurements of functional gene abundances and monitors their fluctuations in various ecosystems (6, 15, 21, 36). However, these tools need to be developed to study the aoxB-carrying As(III)-oxidizing community.
In this article, we demonstrate the feasibility of using qPCR and DGGE of aoxB genes for monitoring the structure, diversity, and abundance of As(III)-oxidizing bacterial populations in As-impacted waters and evaluate the impact of environmental factors on this community.
Surface water and groundwater were collected from eight locations upstream and downstream of disused mines of the Upper Isle River Basin. Sampling sites and physicochemical parameters of the waters are given in the supplemental material (see Fig. S1 and Table S1). DNA was extracted from filters (2 liters of water, 0.2 μm filtered) using the FastPrep spin kit for soil (Bio101).
For qPCR of aoxB genes, a CODEHOP set (28) combined the aoxBM1-2F forward primer (5′-CCACTTCTGCATCGTGGGNTGYGGNTA-3′) (25) with a new reverse primer, aoxBM2-1R (5′-GGAGTTGTAGGCGGGCCKRTTRTGDAT-3′), targeting the conserved motif HNRPAYNE (25) of bacterial and archaeal AoxB protein reference sequences. Nondegenerated primers, aoxBM1-2F-ND (5′-CCACTTCTGCATCGTGGGCTGTGGCTA-3′) and aoxBM2-1R-ND (5′-GGAGTTGTAGGCGGGCCGGTTGTGGAT-3′), were designed for PCR-DGGE by replacing degeneracy of the primer pair aoxBM1-2F/aoxBM2-1R with the bases most frequently found in aligned aoxB nucleotide reference sequences in order to avoid the multiple band patterns often produced when degenerated primers are used for DGGE analysis. A GC clamp (5′-CCGCCGCGCGGCGGGCGGGGCGGGGGCACGGGC-3′) was added to the 5′ end of the reverse primer aoxBM2-1R-ND. Both primer sets gave a specific fragment of ca. 550 bp from the DNAs extracted from the eight waters and the As(III)-oxidizing Alpha-, Beta-, and Gammaproteobacteria and Chloroflexi strains, previously used as positive controls (25). No amplification was obtained from non-As(III)-oxidizing bacteria harboring a homologous enzyme of the DMSO reductase family.
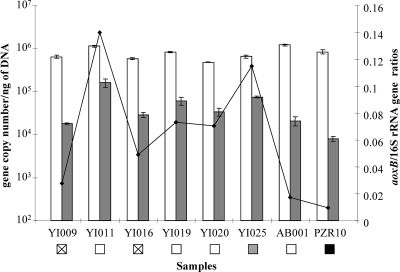
qPCR of aoxB and universal 16S rRNA genes and calculation of copy numbers were done as described in Cébron et al. (6). A linear calibration curve (r2 = 0.99) was obtained over 7 orders of magnitude, ranging from 102 to 108 aoxB gene copies of a linearized plasmid carrying a ca. 1,100-bp aoxB gene fragment (see Fig. S2 in the supplemental material). Similar data were reported for other functional genes, such as those involved in denitrification (15) and nitrate reduction (21). No PCR inhibitor was found in the water DNAs by the detection of 2.9 × 106 (±6.5 × 105) copies of aoxB genes, as against 1.1 × 106 (±1.1 × 105) expected copies, in a reaction mixture containing 106 copies of the plasmid added to 2 ng of water DNAs. Copy numbers of aoxB genes retrieved from the waters ranged from 8.1 × 103 (±1.1 × 103) to 1.6 × 105 (±3.6 × 104) per ng of DNA (Fig. 1). Their relative abundances, i.e., ratios of aoxB gene copies relative to universal bacterial 16S rRNA gene copies, ranged from 0.01 to 0.14 (Fig. 1). No significant relationship with environmental factors was observed, which may be due to the small number of studied samples. Nevertheless, as observed for the aoxB gene copy numbers, the lowest ratio (0.01) was measured in the anoxic PZR10 groundwater, which could be expected since these genes are carried by aerobic bacteria, while the highest ratios (0.14 and 0.11) were found in the most As-polluted YI011 and YI025 oxic surface waters (Fig. 1). These ratios are only estimations of the proportion of As(III) oxidizers in the overall bacterial community. Assuming that As(III) oxidizers harbored one to two aoxB gene copies, the copy number of aoxB genes obtained by qPCR is close to the number of cells. But copy numbers of 16S rRNA genes cannot be correlated to cell numbers due to their variation from 1 to 15 copies per genome (10, 20). Nevertheless, the proportions of bacteria harboring aoxB genes in waters of the Upper Isle River Basin are realistic compared to proportions stated by Salmassi et al. (31), where As(III)-oxidizing bacteria represented 6% to 56% of the cultivable members of a biofilm of a highly As-polluted tributary of the Owens River (Hot Creek, California).
FIG. 1.
Copy numbers of aoxB genes (gray bars) and 16S rRNA genes (white bars) in waters of the Upper Isle River Basin. Values are means (n = 3) ± standard errors (error bars) and are represented on a logarithmic scale. The aoxB/16S rRNA gene ratios (black curve) represent relative abundances of aoxB genes in the waters. White squares with a black cross, non-As-polluted (<10 μg·liter−1 As) or slightly As-polluted (15 μg·liter−1 As) waters; white squares, moderately (50 to 100 μg·liter−1 As) As(V)-polluted waters; gray squares, moderately (50 to 100 μg·liter−1 As) As(III)-polluted waters; black squares, highly (>1,000 μg·liter−1 As) As(III)-polluted water.
DGGE, performed on 8% polyacrylamide gels in a 55% to 65% urea-formamide denaturant gradient, generally showed a single-band profile for each As(III)-oxidizing strain, indicating the presence of a single copy of the aoxB gene. Bands migrated at distinct positions in the gel (see Fig. S3 in the supplemental material), even for strains belonging to the same genus (i.e., strains affiliated with the Alcaligenes, Pseudomonas, or Thiomonas genus) or the same species (e.g., Alcaligenes sp. YI013 and T12RB [25]) based on 16S rRNA gene similarities. The DGGE profile of “Thiomonas arsenivorans” DSM 16361T (3) consisted of two bands corresponding to two distinct sequences, thus showing that “T. arsenivorans” carried two distinct copies of the aoxB gene. The deduced AoxB protein sequences were highly divergent (only 53.1% identity): one was properly positioned among the Thiomonas spp., while the other showed 91.7% identity with Pseudomonas sp. 72 (25). These data suggest the presence of multiple operons in “T. arsenivorans,” probably with different regulations and/or roles. Two almost identical copies of the aoxB gene have been found in Ancylobacter sp. OL1 (26).
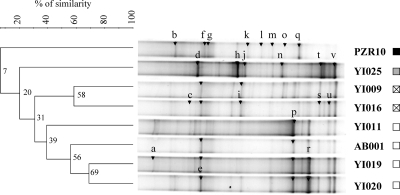
Different DGGE profiles of aoxB-carrying bacterial populations consisting of bands with various intensities and positions were obtained from the Upper Isle River Basin water DNAs (Fig. 2). Identical profiles were obtained from independent PCR runs on the same DNAs (data not shown), showing the reproducibility of this approach. Profile analyses were performed with the Quantity One software program (Bio-Rad) and the implemented unweighted pair group method using average linkages (UPGMA) clustering method (35). The most complex DGGE profile (22 visible bands) was obtained for the slightly As-polluted (15 μg·liter−1) YI009 surface water, whereas the highly As-polluted (>1,000 μg·liter−1) PZR10 groundwater exhibited the simplest DGGE profile (12 visible bands) and thus the lowest aoxB gene richness. The highest similarity (69%) was found between the moderately As(V)-polluted (50 to 100 μg·liter−1) YI019 and YI020 surface waters. A high similarity (58%) was also found between the slightly As-polluted and non-As-polluted (<10 μg·liter−1) YI009 and YI016 surface waters, despite the geographic distance that separated them. Here, the lowest As concentrations (15 μg·liter−1) may have contributed to the presence of similar As(III)-oxidizing populations. The highest divergence (only 7% similarity) occurred between the surface waters and the groundwater PZR10, which showed the highest As concentration (1,846 μg·liter−1). This level of As may not be the only explanation for the divergent structure, since all the As(III) oxidizers isolated up to now are capable of oxidizing the highest As concentrations (3, 5, 8, 25, 38). The low dissolved oxygen concentration (1.2 mg·liter−1) and a negative Eh (−131 mV) measured in the PZR10 groundwater could impact the structure of this aerobic aoxB-carrying bacterial group. Divergences (20% similarity) were also found between the DGGE profile of the YI025 surface water, corresponding to mine drainage water, and the other surface waters, and this despite similar dissolved oxygen and total As contents. Here, the low Eh (26 mV) measured in YI025 water may explain a divergent population structure. The remarkable molecular-fingerprint differences between (i) the YI025 and PZR10 aoxB-carrying bacteria and (ii) the others could also be due to the dominance of the As(III) form and/or their mine drainage origin. Both the YI025 and PZR10 waters have specific geochemical signatures, with the highest iron and manganese concentrations, nitrate values below the detection limit, and approximately 100 mg·liter−1 of HCO3− versus an average of 35 for the other waters. Changes in environmental conditions may impact the structure of bacterial populations in rivers, leading to a selection of distinct physiological and phylogenetic bacteria (19, 29, 39). Taken together, our results reveal DGGE profile similarities and differences associated with the Eh and the As pollution level, thus suggesting a link between the As concentration/speciation and the structure/diversity of the aoxB-carrying bacterial population.
FIG. 2.
UPGMA clustering analysis and DGGE profiles of aoxB gene fragments retrieved from waters of the Upper Isle River Basin. Arrows indicate excised and sequenced (primer aoxBM1-2F-ND) DGGE bands (for phylogenetic affiliation, refer to Fig. 3). As indicated in the legend to Fig. 1, the squares represent the levels of As concentration and speciation in water samples.
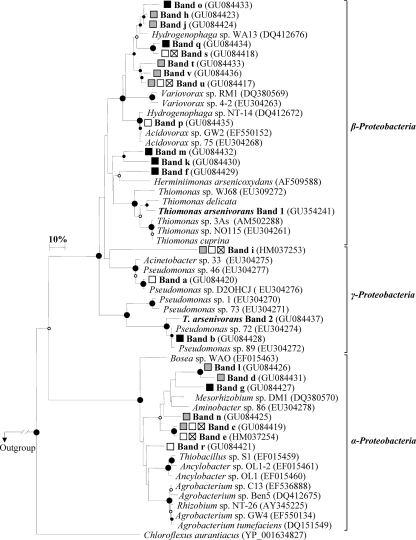
All nucleotide sequences obtained from dominant DGGE bands were affiliated with aoxB sequences, confirming the specificity of the developed DGGE approach. The neighbor-joining (30) tree based on Kimura two-parameter distances (18) of deduced AoxB sequences showed a large diversity of sequences related mainly to those from the Betaproteobacteria (55%) but also to those from the Alpha-, and Gammaproteobacteria (Fig. 3). Some were closely related to AoxB sequences retrieved from various geographically distant areas throughout the world, showing that the related aoxB-carrying bacteria are ubiquitous in As-polluted environments. The exclusive detection of proteobacterial AoxB sequences has been reported for mesophilic environments (9, 17, 25), while most of the nonproteobacterial AoxB sequences (Chloroflexi, Aquificae, Deinococcus-Thermus, and Crenarchaeota phyla) came from thermophilic strains or geothermal environment surveys (7, 14, 17).
FIG. 3.
Neighbor-joining phylogenetic tree of AoxB sequences (134 unambiguous amino acids) deduced from DGGE bands of the waters of the Upper Isle River Basin. The Thermus AoxB sequences (ABB17183 and BAD71923) were used as an outgroup. Circles at the branch nodes represent bootstrap confidence level percentages obtained from 1,000 replicates: large black circles, 95 to 100%; small black circles, 75 to 95%; small white circles, 50 to 75%. The scale bar corresponds to 10 mutations per 100 residues. As indicated in the legend to Fig. 1, the squares represent the levels of As concentration and speciation in water samples. Sequence manipulations and alignment were performed with the BioEdit software program (13). All phylogenetic programs are implemented in the Treecon software program (37).
Identical AoxB sequences were retrieved at different locations of the Upper Isle River Basin. Bands e, u, and v were omnipresent in the surface water profiles. Bands u and v were affiliated to Hydrogenophaga WA13 AoxB sequences (81.9 to 89.4% identity), and band e was phylogenetically located in a distinct branch of an Alphaproteobacteria-like AoxB sequence group formed only by sequences retrieved in this study (Fig. 3). Band i, a divergent AoxB sequence, was present in all but one (YI019) of the surface waters. Specific bands were common to the profiles of moderately As(V)-polluted AB001, YI020, YI019, and YI011 surface waters. Bands p and r were always present in these waters but were not detected in As(III)-polluted waters (Y1025 and PZR10). Band p was related to AoxB sequences of Acidovorax sp. GW2 and sp. 75 (97.1% identity), respectively, found in As-impacted mining sediments (China) (9) and soil (France) (25), and to operational taxonomic unit (OTU) LYB3 (99% identity), retrieved from Yangebup lake sediments impacted by As-containing industrial waste (Australia) (17). Band r clustered with sequences of Ancylobacter sp. OL1 (82.2 to 82.7% identity), a chemoautotrophic bacterium growing even under aerobic and denitrifying conditions. Band a appeared only in the profiles of the YI019 and YI011 tributary surface waters but was not detected in AB001 and YI020 surface waters collected in the river itself, suggesting that bacteria harboring this gene cannot survive or are found only at low density under the river conditions. It was closely related to AoxB sequences of Gammaproteobacteria-related OTU T12RSOL13 (97.3% identity) and Pseudomonas sp. D2OHCJ (97.7% identity), originating from two geographically distant As-impacted industrial soils of the Meuse Basin (Belgium) (25), and to AM clones (>90% identity) retrieved from an agricultural soil treated with As(III) in a column experiment (17), which suggests that bacteria harboring this gene could have originated from soils with a high natural As background level surrounding the river tributaries. The two most intense bands of the YI025 profile were specific to that water and could represent indigenous aoxB-carrying organisms: band d was affiliated with AoxB sequences of Aminobacter sp. 86 (81% identity), isolated from a disused mine soil (France) (25), and band h was affiliated with Hydrogenophaga spp. (82.7 to 92.4% identity).
The developed aoxB gene-targeting qPCR and DGGE tools proved to be efficient for evaluating diversity pattern and abundance of aerobic As(III)-oxidizing bacteria. Considering their abundance, broad distribution, and large-scale diversity, these bacteria are likely to play key roles in As transformation and mobility in the Upper Isle River Basin. Diversity and structure changes were highlighted, with dissolved As, Eh, and oxygen levels perhaps being important variables among several possible factors influencing As(III) oxidizer structure. A large-scale study integrating contrasted situations is now needed to demonstrate the impact of various environmental factors (organic carbon, dissolved oxygen levels, metal, and nutrient concentrations) on the structure and abundance of this functional group. If relationships can be established, aoxB gene-targeting tools may help to predict biological As(III) oxidation potential during bioremediation processes and serve as bioindicators of As bioavailability.
Nucleotide sequence accession numbers.
aoxB gene sequences have been deposited under GenBank accession numbers HM037253, HM037254, GU354241, and GU084417 to GU084437 (DGGE experiments), GU133618 (Thiomonas cuprina DSM 5495T), and GU133619 (Thiomonas delicata DSM 17897T).
Supplementary Material
Acknowledgments
We thank the Agence de l'Environnement et de la Maîtrise de l'Energie (ADEME) for financial support. The work was done partly within the European Union FP6 Integrated Project AquaTerra (project no. GOCE 505428).
We are grateful to Michel Jauzein for helpful discussions. We acknowledge Kevin B. Hallberg for providing Thiomonas sp. WJ68 and NO115 and Marie-Claire Lett, Daniel Muller, Didier Lièvremont, and Philippe Bertin for providing Herminiimonas arsenicoxydans, Acidovorax sp. 75, Acinetobacter sp. 33, Aminobacter sp. 86, Limnobacter sp. 83, Pseudomonas sp. 1, 46, 72, 73, and 89, and Ralstonia sp. 22. We thank Catherine Crouzet from BRGM for the physicochemical water analysis.
This is BRGM contribution no. 6257.
Footnotes
Published ahead of print on 7 May 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Battaglia-Brunet, F., M. C. Dictor, F. Garrido, C. Crouzet, D. Morin, K. Dekeyser, M. Clarens, and P. Baranger. 2002. An arsenic(III)-oxidizing bacterial population: selection, characterization, and performance in reactors. J. Appl. Microbiol. 93:656-667. [DOI] [PubMed] [Google Scholar]
- 2.Battaglia-Brunet, F., Y. Itard, F. Garrido, F. Delorme, C. Crouzet, C. Greffie, and C. Joulian. 2006. A simple biogeochemical process removing arsenic from a mine drainage water. Geomicrobiol. J. 23:201-211. [Google Scholar]
- 3.Battaglia-Brunet, F., C. Joulian, F. Garrido, M. C. Dictor, D. Morin, K. Coupland, D. B. Johnson, K. B. Hallberg, and P. Baranger. 2006. Oxidation of arsenite by Thiomonas strains and characterization of Thiomonas arsenivorans sp. nov. Antonie Van Leeuwenhoek 89:99-108. [DOI] [PubMed] [Google Scholar]
- 4.Bodenan, F., P. Baranger, P. Piantone, A. Lassin, M. Azaroual, E. Gaucher, and G. Braibant. 2004. Arsenic behaviour in gold-ore mill tailings, Massif Central, France: hydrogeochemical study and investigation of in situ redox signatures. Appl. Geochem. 19:1785-1800. [Google Scholar]
- 5.Bruneel, O., J. C. Personne, C. Casiot, M. Leblanc, F. Elbaz-Poulichet, B. J. Mahler, A. Le Fleche, and P. A. D. Grimont. 2003. Mediation of arsenic oxidation by Thiomonas sp. in acid-mine drainage (Carnoulès, France). J. Appl. Microbiol. 95:492-499. [DOI] [PubMed] [Google Scholar]
- 6.Cébron, A., M. P. Norini, T. Beguiristain, and C. Leyval. 2008. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHD alpha) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 73:148-159. [DOI] [PubMed] [Google Scholar]
- 7.Clingenpeel, S. R., S. D'Imperio, H. Oduro, G. K. Druschel, and T. R. McDermott. 2009. Cloning and in situ expression studies of the Hydrogenobaculum arsenite oxidase genes. Appl. Environ. Microbiol. 75:3362-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duquesne, K., A. Lieutaud, J. Ratouchniak, A. Yarzabal, and V. Bonnefoy. 2007. Mechanisms of arsenite elimination by Thiomonas sp. isolated from Carnoulès acid mine drainage. Eur. J. Soil Biol. 43:351-355. [Google Scholar]
- 9.Fan, H., C. Su, Y. Wang, J. Yao, K. Zhao, and G. Wang. 2008. Sedimentary arsenite-oxidizing and arsenate-reducing bacteria associated with high arsenic groundwater from Shanyin, northwestern China. J. Appl. Microbiol. 105:529-539. [DOI] [PubMed] [Google Scholar]
- 10.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 11.Grosbois, C., A. Courtin-Nomade, F. Martin, and H. Bril. 2007. Transportation and evolution of trace element bearing phases in stream sediments in a mining-influenced basin (Upper Isle River, France). Appl. Geochem. 22:2362-2374. [Google Scholar]
- 12.Grosbois, C., J. Schafer, H. Bril, G. Blanc, and A. Bossy. 2009. Deconvolution of trace element (As, Cr, Mo, Th, U) sources and pathways to surface waters of a gold mining-influenced watershed. Sci. Total Environ. 407:2063-2076. [DOI] [PubMed] [Google Scholar]
- 13.Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 14.Hamamura, N., R. E. Macur, S. Korf, G. Ackerman, W. P. Taylor, M. Kozubal, A. L. Reysenbach, and W. P. Inskeep. 2009. Linking microbial oxidation of arsenic with detection and phylogenetic analysis of arsenite oxidase genes in diverse geothermal environments. Environ. Microbiol. 11:421-431. [DOI] [PubMed] [Google Scholar]
- 15.Henry, S., E. Baudoin, J. C. Lopez-Gutierrez, F. Martin-Laurent, A. Baumann, and L. Philippot. 2004. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59:327-335. [DOI] [PubMed] [Google Scholar]
- 16.Hoeft, S. E., J. S. Blum, J. F. Stolz, F. R. Tabita, B. Witte, G. M. King, J. M. Santini, and R. S. Oremland. 2007. Alkalilimnicola ehrlichii sp. nov., a novel, arsenite-oxidizing haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int. J. Syst. Evol. Microbiol. 57:504-512. [DOI] [PubMed] [Google Scholar]
- 17.Inskeep, W. P., R. E. Macur, N. Hamamura, T. P. Warelow, S. A. Ward, and J. M. Santini. 2007. Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ. Microbiol. 9:934-943. [DOI] [PubMed] [Google Scholar]
- 18.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J. Mol. Biol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 19.Kirchman, D. L., A. I. Dittel, S. E. G. Findlay, and D. Fischer. 2004. Changes in bacterial activity and community structure in response to dissolved organic matter in the Hudson River, New York. Aquat. Microb. Ecol. 35:243-257. [Google Scholar]
- 20.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Gutierrez, J. C., S. Henry, S. Hallet, F. Martin-Laurent, G. Catroux, and L. Philippot. 2004. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 57:399-407. [DOI] [PubMed] [Google Scholar]
- 22.Masson, M., J. Schafer, G. Blanc, and A. Pierre. 2007. Seasonal variations and annual fluxes of arsenic in the Garonne, Dordogne and Isle Rivers, France. Sci. Total Environ. 373:196-207. [DOI] [PubMed] [Google Scholar]
- 23.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 24.Nakatsu, C. H. 2007. Soil microbial community analysis using denaturing gradient gel electrophoresis. Soil Sci. Soc. Am. J. 71:562-571. [Google Scholar]
- 25.Quéméneur, M., A. Heinrich-Salmeron, D. Muller, D. Lièvremont, M. Jauzein, P. N. Bertin, F. Garrido, and C. Joulian. 2008. Diversity surveys and evolutionary relationships of aoxB genes in aerobic arsenite-oxidizing bacteria. Appl. Environ. Microbiol. 74:4567-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhine, E. D., S. M. Ni Chadhain, G. J. Zylstra, and L. Y. Young. 2007. The arsenite oxidase genes (aroAB) in novel chemoautotrophic arsenite oxidizers. Biochem. Biophys. Res. Commun. 354:662-667. [DOI] [PubMed] [Google Scholar]
- 27.Rhine, E. D., C. D. Phelps, and L. Y. Young. 2006. Anaerobic arsenite oxidation by novel denitrifying isolates. Environ. Microbiol. 8:899-908. [DOI] [PubMed] [Google Scholar]
- 28.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubin, M. A., and L. G. Leff. 2007. Nutrients and other abiotic factors affecting bacterial communities in an Ohio river (USA). Microb. Ecol. 54:374-383. [DOI] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Salmassi, T. M., J. J. Walker, D. K. Newman, J. R. Leadbetter, N. R. Pace, and J. G. Hering. 2006. Community and cultivation analysis of arsenite oxidizing biofilms at Hot Creek. Environ. Microbiol. 8:50-59. [DOI] [PubMed] [Google Scholar]
- 32.Santini, J. M., L. I. Sly, R. D. Schnagl, and J. M. Macy. 2000. A new chemolithoautotrophic arsenite-oxidizing bacterium isolated from a gold mine: phylogenetic, physiological, and preliminary biochemical studies. Appl. Environ. Microbiol. 66:92-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schafer, J., and G. Blanc. 2002. Relationship between ore deposits in river catchments and geochemistry of suspended particulate matter from six rivers in southwest France. Sci. Total Environ. 298:103-118. [DOI] [PubMed] [Google Scholar]
- 34.Smedley, P. L., and D. G. Kinniburgh. 2002. A review of the source, behaviour and distribution of arsenic in natural waters. Appl. Geochem. 17:517-568. [Google Scholar]
- 35.Sokal, R. R., and C. D. Michener. 1958. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 38:1409-1438. [Google Scholar]
- 36.Sun, Y. M., E. A. Polishchuk, U. Radoja, and W. R. Cullen. 2004. Identification and quantification of arsC genes in environmental samples by using real-time PCR. J. Microbiol. Methods 58:335-349. [DOI] [PubMed] [Google Scholar]
- 37.Van de Peer, Y., and R. Dewachter. 1994. Treecon for Windows—a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 38.Weeger, W., D. Lievremont, M. Perret, F. Lagarde, J. C. Hubert, M. Leroy, and M. C. Lett. 1999. Oxidation of arsenite to arsenate by a bacterium isolated from an aquatic environment. Biometals 12:141-149. [DOI] [PubMed] [Google Scholar]
- 39.Winter, C., T. Hein, G. Kavka, R. L. Mach, and A. H. Farnleitner. 2007. Longitudinal changes in the bacterial community composition of the Danube River: a whole-river approach. Appl. Environ. Microbiol. 73:421-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.