Abstract
The genus Burkholderia includes over 60 species isolated from a wide range of environmental niches and can be tentatively divided into two major species clusters. The first cluster includes pathogens such as Burkholderia glumae, B. pseudomallei, and B. mallei and 17 well-studied species of the Burkholderia cepacia complex. The other recently established cluster comprises at least 29 nonpathogenic species, which in most cases have been found to be associated with plants. It was previously established that Burkholderia kururiensis, a member of the latter cluster, possesses an N-acyl homoserine lactone (AHL) quorum-sensing (QS) system designated “BraI/R,” which is found in all species of the plant-associated cluster. In the present study, two other BraI/R-like systems were characterized in B. xenovorans and B. unamae and were designated the BraI/RXEN and BraI/RUNA systems, respectively. Several phenotypes were analyzed, and it was determined that exopolysaccharide was positively regulated by the BraIR-like system in the species B. kururiensis, B. unamae, and B. xenovorans, highlighting commonality in targets. However, the three BraIR-like systems also revealed differences in targets since biofilm formation and plant colonization were differentially regulated. In addition, a second AHL QS system designated XenI2/R2 and an unpaired LuxR solo protein designated BxeR solo were also identified and characterized in B. xenovorans LB400T. The two AHL QS systems of B. xenovorans are not transcriptionally regulating each other, whereas BxeR solo negatively regulated xenI2. The XenI2/R2 and BxeR solo proteins are not widespread in the Burkholderia species cluster. In conclusion, the present study represents an extensive analysis of AHL QS in the Burkholderia plant-associated cluster demonstrating both commonalities and differences, probably reflecting environmental adaptations of the various species.
From its establishment in 1992, the genus Burkholderia has been extensively studied since its members are catabolically versatile and are found in many different environments and some are of medical importance (87). Validly described species have been isolated from a wide range of niches, including soil, water, wastes, plants, fungi, animals, and humans. Importantly, several species have been reported to have either a beneficial or a pathogenic interaction with plants, animals, or humans (62, 81). Currently available Burkholderia genome sequences suggest that this genus owes its niche versatility to its large genomes comprised of several large replicons, as well as to lateral gene transfer events and plasmid acquisition (13, 44).
Taxonomic analysis of more than 60 species described to date shows an internal division of the genus that can be viewed in two major clusters (11, 49). The first cluster includes pathogens such as Burkholderia glumae, B. pseudomallei, and B. mallei, as well as the 17 well-studied species of the Burkholderia cepacia complex (BCC) (83). The second and more recently established cluster comprises more than 25 related environmental nonpathogenic species, which in most cases have been found to be associated with plants. Several interesting properties found in members of the latter cluster include (i) their ability to colonize the rhizosphere or the internal intercellular spaces in several plants and promote plant growth, as is the case for B. kururiensis and B. phytofirmans (35, 50); (ii) the potential to increase plant nutrient availability via nitrogen fixation and/or phosphate solubilization, as demonstrated for B. unamae, B. tropica, and B. silvatlantica (10, 59, 62); (iii) the strong catabolic potential to degrade aromatic compounds, with B. xenovorans being the best known example (31); and (iv) their ability to form symbiotic interactions with plants as occurs, for example, with B. tuberum, B. phymatum, B. mimosarum, B. nodosa, and B. sabiae (15-17, 24, 80), and with mosses, as reported for B. megapolitana and B. bryophila (82). However, despite the increasing attention given to these species, the molecular mechanisms regulating most of their properties are still not addressed or poorly understood.
Bacteria communicate with neighbors and monitor their population density by producing and sensing signaling molecules in a process called quorum sensing (QS) (29). The concentration of the signaling molecule increases alongside the bacterial population density and, when it reaches a critical level, bacteria respond and modulate target gene expression. A typical QS system in Gram-negative bacteria involves the production and response to an acylated homoserine lactone (AHL) signal molecule produced by an AHL synthase which in most cases belongs to the LuxI protein family. A transcriptional regulator belonging to the LuxR family forms a complex with the cognate AHL at threshold (“quorum”) concentration and affects the transcriptional status of target genes. Traits under QS control in bacteria are most beneficial to a bacterial community and include biofilm formation, virulence, plant-growth promoting activity, and antibiotic production.
QS has been extensively studied in some Burkholderia species. For example, all species of the BCC complex share a conserved QS system designated CepI/R that produces and responds to N-octanoyl homoserine lactone (C8-AHL) regulating virulence and several important phenotypes such as biofilm formation and siderophore production (23, 36, 84). Several Burkholderia species have been shown to harbor more than one LuxI/R family pair and produce numerous AHL signal molecules, as reported for B. cenocepacia, B. vietnamiensis, B. mallei, B. pseudomallei, and B. thailandensis (21, 22, 39, 45, 47, 78). Furthermore, unpaired LuxR “solo” proteins that lack a cognate LuxI AHL synthase (74) have been characterized for B. cenocepacia (46). In contrast, little is known about the role of QS for members of the nonpathogenic plant-associated nitrogen-fixing Burkholderia cluster. In a previous study we reported the presence of an AHL-based QS system in the rice endophyte B. kururiensis designated BraI/RKUR, which produces and responds to 3-oxo-C12-HSL. It was determined that the BraI/RKUR system is stringently regulated by RsaL, and it is not involved in the regulation of nitrogen fixation or in other several important phenotypes (73). Importantly, BraI/R was found to be highly conserved in 20 species of the recently described cluster (73).
In the present study, the AHL QS systems of B. unamae and B. xenovorans, were identified and characterized in order to obtain more insight into AHL QS in the cluster of plant-associated Burkholderia species. We demonstrate that the genome of B. xenovorans LB400T possessed a BraI/R-like system (designated BraI/RXEN) but evidenced the presence of a second AHL QS system, which we designated XenI2/R2. This system was found to be present in several but not all of the species belonging to the cluster of plant-associated Burkholderia. We performed experiments aimed at elucidating whether the two systems in B. xenovorans are hierarchically organized or whether they act independently. In addition, we report the presence of a LuxR solo protein in several species of this cluster. Finally, we also identified gene targets for the conserved BraI/R-like system and studied other aspects of the AHL QS systems in three Burkholderia species.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The Burkholderia kururiensis, B. xenovorans, B. unamae, Pseudomonas spp., and E. coli strains and plasmids used in the present study are listed in Table 1. The list of primers used and the construction of most recombinant plasmids is given in Table S1 in the supplemental material. All other Burkholderia strains used in the present study are listed in Table S2 in the supplemental material. E. coli strains were grown in Luria-Bertani (LB) medium at 37°C. All Burkholderia strains were grown in King's medium (KB) (38) or LB at 30°C. Antibiotics were added when required at the following final concentrations: ampicillin, 100 μg ml−1; tetracycline, 15 μg ml−1 (E. coli) or 40 μg ml−1 (Burkholderia); gentamicin, 10 μg ml−1 (E. coli), 30 μg ml−1 (Agrobacterium), or 40 μg ml−1 (Burkholderia); kanamycin, 50 μg ml−1 (E. coli) or 100 μg ml−1 (Burkholderia); and nitrofurantoin, 50 μg ml−1. Conjugations in B. kururiensis, B. unamae, and B. xenovorans were performed by triparental mating using E. coli DH5α (pRK2013) as a helper (27) and incubated 22 h at 30°C. Transconjugants were counterselected in KB with the appropriate antibiotics.
TABLE 1.
B. kururiensis, B. xenovorans, B. unamae, P. putida, and E. coli strains and plasmids used in this study
| Strain or plasmida | Characteristicsb | Reference or source |
|---|---|---|
| Strains | ||
| B. kururiensis M130 | Ampr Rifr, isolated from surface-sterilized roots of rice | 5 |
| M130BRAI | braI::Km of B. kururiensis M130 | 73 |
| M130BRAR | braR::Km of B. kururiensis M130 | 73 |
| M130RSAL | rsaL::Km of B. kururiensis M130 | 73 |
| B. xenovorans LB400T | Type strain | 31 |
| LB400BRAI | braI::Km of B. xenovorans LB400T | This study |
| LB400BRAR | braI::Km of B. xenovorans LB400T | This study |
| LB400XENI2 | xenI2::Km of B. xenovorans LB400T | This study |
| LB400XENR2 | xenR2::Km of B. xenovorans LB400T | This study |
| LB400BXER | bxeR::Km of of B. xenovorans LB400T | This study |
| B. unamae MTl-641T | Maize rhizosphere isolate | 10 |
| UNABRAI | braI::Km of B. unamae MTl-641 | This study |
| UNABRAR | braR::Km of B unamae MTl-641 | This study |
| P. putida F117 | AHL-negative derivative of P. putida IsoF; PpuI− | 63 |
| E. coli M15 | NaI Str Rif Thi Lac− Ara+ Gal+ Mtl− F− RecA+ Uvr+ Lon+ | Qiagen |
| Plasmids | ||
| pRK2013 | Tra+ Mob+ ColE1 replicon; Kmr | 27 |
| pMOSBlue | Cloning vector; Ampr | Amersham-Pharmacia |
| pGEM-T Easy | Cloning vector; Ampr | Promega |
| pMP220 | Promoter probe vector, IncP, LacZ; Tetr | 70 |
| pLAFR3 | Broad-host-range cloning vector, IncP1; Tetr | 72 |
| pKNOCK-Km | Conjugative suicide vector; Kmr | 2 |
| pLZ1 | pLAFR3 containing B. unamae DNA | This study |
| pBBRMCS-5 | Broad-host-range vector Gmr | 42 |
| pKRC-12 | pBBR1MCS-5 carrying PlasB-gfp (ASV) Plac-lasR; Gmr | 63 |
| pMOS-LZ-1 | pMOSBlue carrying an 8-kb fragment containing partial B. kururiensis QS genes; Ampr | This study |
| pKNOCK-UNAI | Internal PCR braIUNA fragment of B. unamae cloned in pKNOCK-Km | This study |
| pKNOCK-UNAR | Internal PCR braRUNA fragment of B. unamae cloned in pKNOCK-Km | This study |
| pKNOCKXENR | Internal PCR braRXEN fragment of B. xenovorans cloned in pKNOCK-Km | This study |
| pKNOCKXENI | Internal PCR braIXEN fragment of B. xenovorans cloned in pKNOCK-Km | This study |
| pKNOCKXENI2 | Internal PCR xenI2 fragment of B. xenovorans cloned in pKNOCK-Km | This study |
| pKNOCKXENR2 | Internal PCR xenR2 fragment of B. xenovorans cloned in pKNOCK-Km | This study |
| pKNOCKBXE | Internal PCR bxeR fragment of B. xenovorans cloned in pKNOCK-Km | This study |
| PMPXENI1 | promoter of gene braIXEN cloned in pMP220 vector | This study |
| pMPX2I | Promoter of gene xenI2 cloned in pMP220 vector | This study |
| pMPBXE | Promoter of gene bxeR cloned in pMP220 vector | This study |
| pMP2786 | Promoter of gene Bxe_B2786 cloned in pMP220 vector | This study |
| pMP0016 | Promoter of gene Bxe_B0016 cloned in pMP220 vector | This study |
| pBBRXENI | braIXEN cloned into pBBR-MCS5 Gmr | This study |
| pMPUNAI | promoter of gene braIUNA cloned in pMP220 vector | This study |
| pGEMXENR1 | braRXEN cloned into pGEM; Ampr | This study |
| pGEMX2R | xenR2 cloned into pGEM; Ampr | This study |
| pQEXENR1 | braRXEN cloned into pQE30 expression vector; Ampr | This study |
| pQEXENR2 | xenR2 cloned into pQE30 expression vector; Ampr | This study |
| pQEBXER | bxeR cloned into pQE30 expression vector; Ampr | This study |
| pQEUNAR | braRUNA cloned into pQE30 expression vector; Ampr | This study |
| pQE30 | Expression vector, ColE1 replication origin, T5 promoter, His epitope; Ampr | Qiagen |
A superscript “T” indicates a type strain.
Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Tetr, tetracycline resistance; Ampr, ampicillin resistance; Rifr, rifampin resistance; Kmr, kanamycin resistance.
Recombinant DNA techniques.
Recombinant DNA techniques, including digestion with restriction enzymes, agarose gel electrophoresis, purification of DNA fragments, ligation with T4 ligase, end filling with the Klenow enzyme, hybridization, radioactive labeling by random priming, and transformation of Escherichia coli were performed as described previously (65). Southern hybridizations were performed using Amersham Hybond-XL membranes (Amersham, Biosciences); plasmids were purified by using EuroGold columns (EuroClone, Italy); total DNA from Burkholderia was isolated by Sarkosyl-pronase lysis as described previously (8). Generated plasmids were sequenced by Macrogen (South Korea). The fidelity of all marker exchange events was confirmed by Southern analysis.
β-Galactosidase activities were determined essentially as described by Miller (51) with the modifications of Stachel et al. (71); all experiments were performed in triplicate, and the mean value is given.
Identification, inactivation, and characterization of QS genes in B. unamae MTl-641T.
To identify the QS system of B. unamae MTl-641T, a cosmid library was constructed by using the cosmid pLAFR3 (72) as a vector. Insert DNA was prepared by partial EcoRI digestion of the genomic DNA and then ligated in the corresponding site in pLAFR3. The ligated DNA was then packaged into λ phage heads by using Gigapack III Gold packaging extract (Stratagene), and the phage particles were transduced to E. coli HB101 as recommended by the supplier. In order to identify the cosmid containing the AHL QS genes, the E. coli HB101 harboring the cosmid library was conjugated en masse into the AHL biosensor Pseudomonas putida F117(pKRC12) as acceptor (63). One transconjugant carrying the cosmid pLZ1 displayed green fluorescent protein (GFP) expression and was further studied. The braIUNA and braRUNA loci were located within a 8-kb EcoRV fragment and cloned in pMOSBlue to generate pMOS-pLZ-1, which was sequenced.
To generate genomic braRUNA- and braIUNA-null mutants, internal fragments from each gene were PCR amplified with primers described in Table S1 in the supplemental material and then cloned into pKNOCK-Km(2) to yield the suicide vectors pKNOCK-UNAI and pKNOCK-UNAR. Each plasmid was then mobilized into strain MTl-641T to generate B. unamae UNABRAI and B. unamae UNABRAR.
To identify the BraRUNA cognate AHL, braRUNA was PCR amplified from genomic DNA and cloned into pQE30 to generate pQEUNAR. The braIUNA promoter region was PCR amplified and cloned into pMP220 promoter probe vector to generate pMPUNAI (see the cloning details in Table S1 in the supplemental material). E. coli M15(pQEUNAR)(pMPUNAI) was then inoculated into 10 ml of LB-Amp-Km-Tet, grown overnight, and diluted to an optical density (OD) of 0.1 in 10 ml of prewarmed medium with 1 μM concentrations of each synthetic AHL. The β-galactosidase activity was determined after 4 h at 37°C.
Isolation of the braIXEN and braRXEN and the xenI2 and xenR2 AHL QS systems and of the bxeR solo gene of Burkholderia xenovorans LB400T and construction of derivative genomic mutants.
braIXEN, rsaLXEN, and braRXEN were identified by in silico comparison of the DNA and protein sequences with the genome of B. xenovorans LB400T(13) as the three loci Bxe_B0610, Bxe_B0609, and Bxe_B0608. Similarly, xenI2, xenR2, and bxeR solo were identified as the genomic loci Bxe_C0416, Bxe_C0415, and Bxe_B2275, respectively. To generate knockout mutants of braRXEN, braIXEN, xenI2, xenR2, and bxeR, internal fragments from each gene were PCR amplified (see Table S1 in the supplemental material), and cloned into pKNOCK-Km to yield the suicide plasmids pKNOCKXENR, pKNOCKXENI, pKNOCKXENI2, pKNOCKXENR2, and PKNOCKBXER. These plasmids were then conjugated into B. xenovorans LB400T to generate the genomic mutants LB400BRAR, LB400BRAI, LB400XENI2, LB400XENR2, and LB400BXER, respectively. Growth curves and CFU/ml for wild type and mutants were determined in KB media.
For complementation purposes, the braIXEN gene and its promoter region were PCR amplified and cloned into pMOS-Blue. A 623-bp fragment containing braRXEN was then excised using XbaI-KpnI and ligated into pBBR-MCS5 (42) to generate pBBRXENI. Similarly, the bxeR gene and its promoter region were PCR amplified and cloned into pGEM-T Easy. A 1,263-bp fragment was then excised using BamHI-SpeI and cloned into pBBRMCS-5 to generate pBBRBXER (see the primers in Table S1 in the supplemental material).
The presence of systems highly identical to that of xenI2 and xenR2 in four other B. xenovorans strains, as well as in 21 Burkholderia species, was determined by Southern analysis. This analysis was performed on EcoRI- or PstI-digested genomic DNAs and hybridized with a 794-bp probe comprising the xenR2 gene. This fragment was amplified by PCR from B. xenovorans genomic DNA and cloned into pGEM-T Easy to generate pGEMX2R (see Table S1 in the supplemental material). The plasmid was sequenced, and a probe was generated by random priming using the excised SpeI-NotI fragment as a template.
To generate pMP2786 and pMP0016, the promoter regions of Bxe_B2786 and Bxe_B0016 were PCR amplified and cloned as EcoRI-XbaI fragments in pMP220, respectively (primers are indicated in Table S1 in the supplemental material).
Determination of the cognate AHL for BraI/RXEN and XenI2/R2 AHL QS systems and cross-talk regulation between BraI/RXEN, XenI2/R2, and the unpaired BxeR solo.
The braRXEN and xenR2 genes were PCR amplified using genomic DNA as a template, and amplimers were cloned into pGEM-T Easy. Each gene was then excised with BamHI/HindIII and ligated into the corresponding sites of pQE30 to generate pQEXENR1 and pQEXENR2, respectively (for the primers, see Table S1 in the supplemental material). The gene promoters of braIXEN and xenI2 were amplified and cloned in the promoter probe vector pMP220 to generate pMPXENI1 and pMPX2I as described in Table S1 in the supplemental material.
E. coli M15(pQXENR1)(pMPXENI1) and E. coli M15(pQEXENR2)(pMPX2I) were inoculated into 10 ml of LB-Amp-Km-Tet, grown overnight, and then diluted to an OD of 0.1 in 10 ml of prewarmed medium with 1 μM concentrations of each synthetic AHL to be evaluated. The β-galactosidase activity was then determined after 4 h at 37°C. Possible regulation by BraRXEN of the xenI2 promoter and of the braIXEN promoter by XenR2 was evaluated by transforming the plasmid gene promoter fusion into E. coli M15(pQEXENR1) and E. coli M15(pQEXENR2), respectively. The β-galactosidase activity was measured in the presence of the cognate AHL after 4 h of growth.
BxeR and bxeR studies.
To overexpress BxeR, bxeR was PCR amplified and cloned into pGEM-T Easy (see the primers in Table S1 in the supplemental material). This fragment was then excised with BamHI-HindIII and cloned into pQE30 to generate pQEBXER. When required, the same fragment was used as a template to generate the radioactive DNA probe representing bxeR. Plasmid pMPBXE, containing the promoter of bxeR, was PCR amplified and then excised and cloned, using BamHI-XbaI, into pMP220 to generate pMPBXE (see the primers in Table S1 in the supplemental material).
Regulation by BxeR solo of the AHL QS systems of B. xenovorans was determined as follows. E. coli overexpressing BxeR via the pQEBXER plasmid was transformed with either pMPXENI1 or pMPX2I, as well as with the bxeR promoter transcriptional fusion pMPBXE. E. coli M15(pQEBXER)(pMPX2I), E. coli M15 (PQEBXER)(pMPXENI1), and E. coli M15 (PQEBXER)(pMPBXE) were then grown in the presence of three independent AHL mixtures. One contained 1 μM each of nine different unsubstituted AHLs at position 3, the second mixture contained 1 μM concentrations each of seven different 3-oxo-substituted AHLs, and a third cocktail contained 1 μM each of seven different 3-OH-substituted AHLs. In these three growth conditions, the β-galactosidase activities were determined.
AHL extraction, visualization, and quantification.
AHLs were purified from spent supernatant and separated by using a C18 reversed-phase thin-layer chromatography (TLC) plates as previously described (68). For visualization by TLC, the plate was overlaid with a thin layer of AB top agar seeded with Agrobacterium tumefaciens NTL4(pZLR4) in the presence of 100 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml, as described previously (68), or with LB top agar seeded with E. coli pSB1075 (86).
Motility assays, biofilm formation, EPS and siderophore production, and lipase and protease activities.
The lipolytic activity was determined in trybutyrin agar, as previously described (3). The protease activity was determined in the KB plates supplemented with 2% skim milk (36).
Swimming assays were performed on 0.25% KB agar plates and 0.25% nutrient agar. Swarming assays were performed using 0.5% nutrient agar plates and M8 medium plates (M9 salts without NH4Cl) (40) supplemented with 0.2% glucose, 0.05% glutamate, and 0.5% agar (52). The inoculation was performed by spotting 1 μl of a bacterial suspension having an OD at 600 nm (OD600) of 1. The swimming and swarming zones were measured after 48 h of incubation at 30°C for all wild-type species and their QS mutant derivatives.
EPS production was tested by streaking single colonies in yeast extract mannitol medium, as described previously (88). EPS was extracted and quantified by the boiling phenol method, as described previously (20).
Biofilm formation was determined as previously described (36, 55). Briefly, single colonies were grown overnight in AB medium supplemented with 10 mM glucose, washed, and diluted to an OD600 of 0.01. Then, 100 μl of bacterial dilution was inoculated into the round-bottom wells of microtiter dishes, followed by incubation at 30°C. Biofilm formation was evaluated after 24, 48, 72, and 120 h. After the incubation time, the OD550 was determined prior to medium removal. Importantly, the mutants under these growth conditions did not display significant growth differences compared to the wild type (data not shown). Next, 100 μl of 1% (wt/vol) aqueous solution of crystal violet was added, followed by incubation at room temperature for 20 min, and the mixture was then washed thoroughly with water. For quantification of the attached cells, the crystal violet was solubilized in 120 μl of dimethyl sulfoxide, and the absorbance was determined at 570 nm.
The method used to detect siderophores was adapted from the universal chemical assay on chrome azurol S (CAS) agar plates (66, 73). The cultures were harvested, and the pellets washed and adjusted to an OD600 of 1. Then, 2-μl portions of culture were spotted on the surface of the plate, followed by incubation for 48 h at 30°C. Siderophore production was measured by the size of the orange halos formed around the colonies.
Analysis and identification of secreted proteins.
Overnight cultures were washed and diluted to an OD600 of 0.05 in 40 ml of prewarmed LB. After 12 h of incubation, the cultures were centrifuged for 15 min at 8,000 × g at 4°C. Culture supernatants were filtered through a 0.45-μm-pore-size filter (Millipore), and the proteins were precipitated overnight at 4°C with 10% (vol/vol) trichloroacetic acid (final concentration). The precipitates were separated by centrifugation at 15,000 × g for 20 min at 4°C, and the pellets were washed twice with ice-cold acetone. Another centrifugation was performed at 15,000 × g for 20 min, and protein pellets were air dried and resuspended in sample buffer. The suspension was then boiled for 10 min, and the proteins were separated by SDS-PAGE on gels containing 12% (wt/vol) polyacrylamide. The proteins were identified by mass spectroscopy as described by Tomaic et al. (76).
Growth of B. unamae MTl-641T and mutant derivatives using phenol as a carbon source.
B. unamae MTl-641T, UNABRAI, UNABRAR, and UNABRAR complemented with pLZ1 were grown in BSE liquid medium (25), and the cultures were incubated at 29°C with reciprocal shaking (200 rpm) for 18 h. The cultures were adjusted to ∼104 CFU/ml, and 1.0 ml was then inoculated into 99 ml of SAAC culture medium containing either phenol or mannitol 0.05% (11). Growth was tested at 24, 48, and 72 h.
Plant colonization assays.
Plant colonization was conducted as previously described (50). Briefly, rice seeds were dehusked, surface sterilized, and transferred to water-based 0.5% agar for seed pregermination, following incubation for 3 days at 28°C, in the absence of light. The pregerminated rice seeds were aseptically transferred to glass tubes (4 cm in diameter, 29 cm in height) containing 20 ml of a nitrogen-free Hoagland's nutrient solution (33). Plantlets infection assays were carried out by inoculation of 500 μl of bacterial culture (∼109 CFU) into each glass tube. After incubation for 12 days with a 12-h photoperiod at 28°C, plantlets were collected and cut (roots and aerial parts). The excised plant segments were subjected to surface sterilization with 1% sodium hypochlorite for 5 min, followed by several washes with sterile water. The plant segments were then weighed, transferred to microcentrifuge tubes containing 1 ml of sterile nutrient solution, and macerated with a pestle. From each of the obtained suspensions, a series of 10-fold dilutions were prepared using sterile saline, and aliquots of 100 μl were spread plated onto LB medium, followed by incubation for 4 days at 28°C. Bacterial quantification was expressed as CFU/g of fresh weight plant tissue.
B. unamae strains were grown in BSE liquid medium (25), and the cultures were incubated at 29°C with reciprocal shaking (200 rpm) for 18 h. Thereafter, the cultures were adjusted to an OD600 of 0.15 (∼1.5 × 107 CFU/ml). Three germinated seeds of maize (Zea mays) were sown per pot containing sterile river sand, and each seed was inoculated with 1.0 ml of bacterial culture. The plants were grown for 9 days (7 days after plant emergence). The three roots of each pot were cut into segments of ∼1 cm and mixed. Two grams of the roots mixture was macerated with 10 mM MgSO4·7H2O to get a dilution 10−1, and further dilutions were prepared and streaked onto BAc agar plates (25) containing the appropriate antibiotic; three replicates were made from each dilution.
Statistical analysis.
All experiments were performed at least three times, and mean values are given. Statistical analysis included unpaired t tests and analysis of variance (ANOVA) with Dunnett's post-test and were performed with Prism 4.0 software (GraphPad, San Diego, CA). A P value of <0.05 was considered significant.
DNA sequencing and nucleotide sequence accession numbers.
All DNA sequencing was performed either at the CRIBI center (University of Padua, Padua, Italy) or at Macrogen, and the nucleotide sequences were deposited in GenBank/EMBL/DDBJ. The BraI/RUNA QS loci of B. unamae MTl-641T is listed as a 1,977-bp fragment of pMOS-LZ1 under accession number FN640548.
RESULTS
AHL QS systems in B. xenovorans and B. unamae.
To identify the AHL QS system(s) of B. unamae, a cosmid gene library of strain MTl-641T was constructed and screened against the LasR-based P. putida F117(pRKC12) AHL biosensor (63). One cosmid (pLZ1) was identified and possessed an AHL QS system that consisted of three open reading frames (ORFs); two of them (braIUNA and braRUNA) displayed homology to luxI and luxR family genes, while a third ORF, located in between braIUNA and braRUNA, and divergently transcribed from braIUNA, displayed high similarity to the rsaL repressor. The BraRUNA protein consists of 235 amino acids 77% identical to BraRKUR, while the acyl-homoserine lactone synthase BraIUNA comprises 197 amino acids displaying 82% identity to BraIKUR. RsaLUNA from B. unamae is 103 amino acids long with 72% identity to RsaLKUR from B. kururiensis (Fig. 1 A). It was therefore concluded that BraI/RUNA was an ortholog to the B. kururiensis BraI/RKUR system.
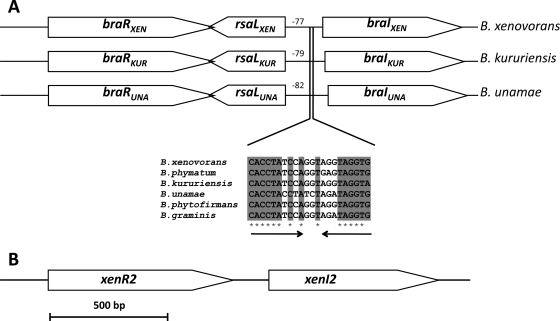
FIG. 1.
(A) Genetic maps of the B. xenovorans LB400T (braIXEN and braRXEN), B. kururiensis M130 (braIKUR and braRKUR), and B. unamae MTl-641T (braIUNA and braRUNA) QS systems. An alignment of putative lux boxes is shown in several members of the Burkholderia cluster; numbers indicate the positions upstream where the lux box is centered with the respect to the putative ATG start codon. (B) Map of the xenI2 and xenR2 system of B. xenovorans LB400T.
In silico analysis of the sequenced genome of B. xenovorans LB400T revealed that it possessed two complete luxI and luxR systems, referred to here as the braIXEN and braRXEN system (BraI/RXEN system) and the xenI2 and xenR2 system (13). The BraI/RXEN system was located in chromosome two, whereas the xenI2 and xenR2 system was located in the 1.47-Mb megaplasmid. The first system, the BraI/RXEN system, displays an organization similar to that of the BraI/RKUR and BraI/RUNA systems described above. In fact, BraRXEN, RsaLXEN, and BraIXEN (234, 105, and 197 amino acids, respectively) show high identity values (>75%) to BraI/RKUR and BraI/RUNA and, since they have very high identities and produce and respond to the same AHLs, these three systems may be considered orthologous (see below). Highly similar systems with the same gene organization were also found in the sequenced genomes of three other members of this species cluster, namely, B. phytofirmans (ORFs Bphyt_4277, Bphyt_4276 and Bphyt_4275), B. graminis (ORFs BgramDRAFT_3087, BgramDRAFT_3088, and BgramDRAFT3089), and B. phymatum (Bphy_4439, Bphy_4438, and Bphy_4437). When a nucleotide sequence analysis in the promoter regions of the five AHL synthase braI-like genes was performed, it was observed that a consensus putative lux-box was located between −77 and −82 with respect to the ATG translational start codon (Fig. 1A).
The XenI2/R2 system of B. xenovorans LB400T.
As mentioned above, in the genome of B. xenovorans LB400T a second putative AHL QS system was found and designated the xenI2 and xenR2 system. This system is located in the loci Bxe_C0415 and Bxe_C0416 of the 1.47-Mb megaplasmid (Fig. 1B). The AHL synthase XenI2 is composed of 249 amino acids, and the sensor/response regulator XenR2 is composed of 262 amino acids; both display ca. 70% identity to two uncharacterized putative LuxI/R-family members of the species cluster, namely, B. graminis (loci BgramDRAFT_4129 and BgramDRAFT 4128) and B. phytofirmans (loci Bphyt_0126 and Bphyt_0127). It was therefore concluded that, like B. xenovorans LB400T, the genomes of these two other species of the cluster also possess two AHL QS systems. Interestingly, XenI2 displays 49% identity to BtaI3 of Burkholderia thailandensis and BpsI3 of Burkholderia pseudomallei and BmaI3 of Burkholderia mallei; these three Burkholderia spp. do not belong to this newly described plant-associated species cluster and are more related to the Burkholderia cepacia complex (18). Similarly, XenR2 displays high identity (ca. 42%) to BtaR3 and BpsR3 of B. thailandensis and B pseudomallei, respectively. The two AHL QS systems of strain LB400 display a low level of relatedness since they have less than 25% similarity at protein level.
B. xenovorans LB400T possesses an unpaired LuxR solo protein.
Analysis of the genome sequence of B. xenovorans revealed also the presence of an unpaired LuxR solo protein having the typical modular structure of QS LuxR family proteins (58, 74). This ORF, which we designated BxeR (Bxe_B2275), contained the helix-turn-helix (HTH) DNA binding motif, as well as AHL-binding domain typical of LuxR QS family of proteins. BxeR encodes a 333-amino-acid protein with an autoinducer binding domain (Pfam03472) from positions 100 to 254 and an HTH domain (Pfam PF00196) from positions 271 to 328. BxeR contains the six well-conserved amino acids in the AHL-binding domain typically shared by the QS LuxR-family members (data not shown). BxeR is 95 and 90% similar to uncharacterized orthologs of B. phytofirmans (Bphyt_6042) and B. graminis (BgramDRAFT_2595), respectively.
Presence of the xenI2 and xenR2 system and of the bxeR solo gene in the recently described Burkholderia cluster.
In order to determine the distribution of the xenI2 and xenR2 system and of bxeR solo in other B. xenovorans strains, as well as in the remaining members of the beneficial Burkholderia cluster, Southern hybridization under high-stringency conditions was performed with 26 strains representing 22 species using xenR2 and bxeR as probes (see Table S2 in the supplemental material). The xenR2 probe hybridized to six different species (B. graminis, B. phytofirmans, B. fungorum, B. terricola, B. ferrariae, and B. silvatlantica). Interestingly, however, no hybridization was observed in any B. xenovorans strain other than LB400T. In contrast, the bxeR solo gene was found to be present in all B. xenovorans strains, as well as in seven members of the cluster of plant-associated species, four of which also possessed the xenI2 and xenR2 system.
AHL production by B. unamae and B. xenovorans.
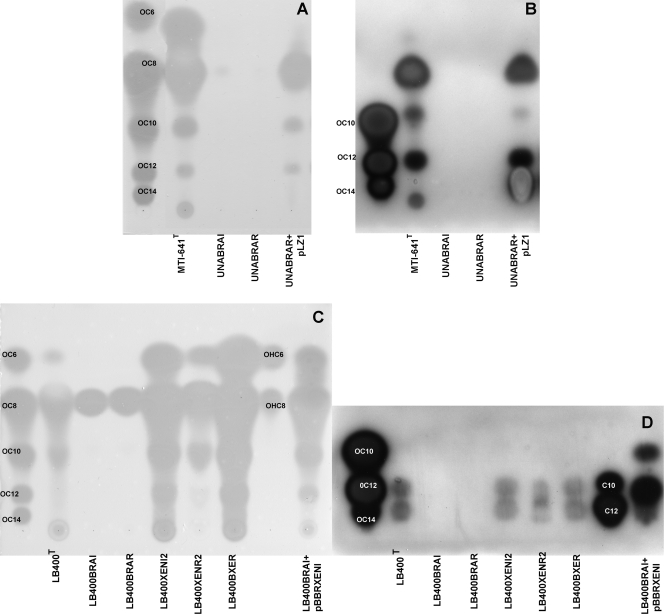
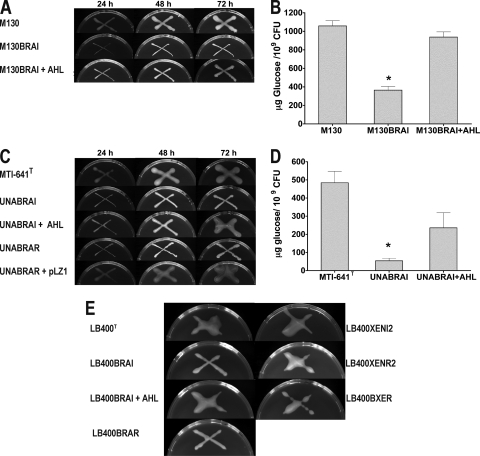
It was of interest to determine which AHLs were produced by B. unamae and by B. xenovorans. TLC analysis suggested that both strains most likely produced 3-oxo-C6-HSL, 3-oxo-C8-HSL, 3-oxo-C10-HSL, and 3-oxo-C12-HSL (Fig. 2 A and C). In B. xenovorans the most likely production of C14-3-oxo-HSL was also detected. In both species, the braI and braR orthologs were inactivated to generate knockout mutants B. unamae UNABRAI and UNABRAR and B. xenovorans LB400BRAI and LB400BRAR. It was established that both B. unamae UNABRAI and UNABRAR were unable to synthesize AHLs (Fig. 2A and B), indicating that the BraI/RUNA system is responsible for synthesizing all of the AHLs identified, and most probably no other AHL QS system was present in B. unamae. In addition, the lack of AHL production in UNABRAR suggests that BraRUNA positively regulates the braIUNA synthase through a positive autoinduction loop. The production of the putative AHLs synthesized by BraI/RUNA could be rescued by genetic complementation of the UNABRAR mutant by providing braRUNA in the cosmid pLZ1 (Fig. 2A and B).
FIG. 2.
TLC analysis of the AHLs produced by the Burkholderia and mutant derivatives. (A and B) B. unamae profiles. (C and D) B. xenovorans LB400 T profiles. AHL extraction was performed as described in Materials and Methods, and TLCs were performed in 70% methanol for 6 h; for each strain, the equivalent of 2.5 × 1010 cells was loaded. In panels A and C, A. tumefaciens (pNTL4) was used to detect the AHL signals, and in panels B and D, E. coli pSB1075 was used. Synthetic AHL compounds were used as a reference. OC6, 3-oxo-C6-HSL; OC8, 3-oxo-C8-HSL; OC10, 3-oxo-C10-HSL; OC12, 3-oxo-C12-HSL; OC14, 3-oxo-C14-HSL; OHC6, 3-OH-C6-HSL; OHC8, 3-OH-C8-HSL; C10, C10-HSL, C12; C12-HSL.
B. xenovorans mutants LB400BRAI and LB400BRAR were both unable to produce 3-oxo-C10-HSL, 3-oxo-C12-HSL, and 3-oxo-C14-HSL as shown by the AHL production profile observed using the LasR-based biosensor E. coli pSB1075 (Fig. 2D). AHL production profile of the two mutants using A. tumefaciens NTL4(pZLR4) evidenced the putative production of 3-OH-C8-HSL, indicating that the XenI2/R2 was most probably responsible for the synthesis of this AHL (Fig. 2C; see also below). As in B. unamae, the BraRXEN mutant of B. xenovorans did not produce 3-oxo-C10-HSL, 3-oxo-C12-HSL, or 3-oxo-C14-HSL, suggesting a positive-feedback loop (Fig. 2D). The synthesis of these AHLs could be complemented in the LB400BRAI mutant by providing braIXEN in trans via plasmid pBBRXENI1 (Fig. 2D). TLC analysis of the AHL molecules produced by the xenI2 and xenR2 mutants revealed the putative production of 3-oxo-C6-HSL, 3-oxo-C8-HSL, 3-oxo-C10-HSL, 3-oxo-C12-HSL, and 3-oxo-C14-HSL, confirming that BraIXEN is responsible for the production of this later group of AHLs. Finally, TLC analysis of the AHL molecules produced by the bxeR knockout solo mutant LB400BXER did not show significant differences between their AHL profile in comparison to the wild type. All of the generated knockout mutants did not exhibit any growth differences in the media that we have used.
BraIR-like systems respond to 3-oxo-C14-HSL.
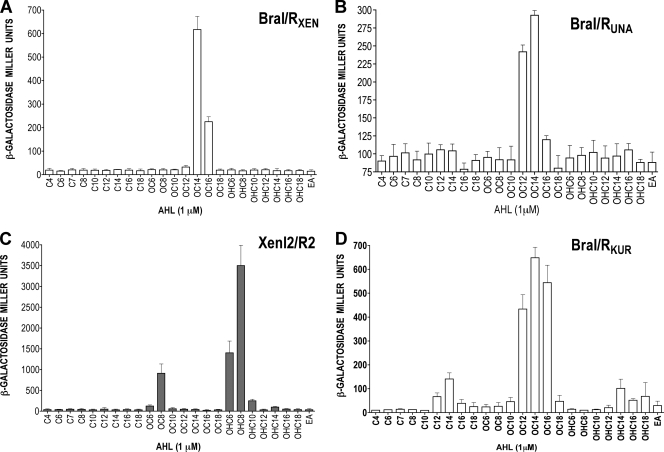
In order to determine to which AHL BraRXEN and BraRUNA best responded, the proteins BraRXEN and BraRUNA were overexpressed in E. coli M15 in the presence of different AHL molecules, and the cognate braIXEN and braIUNA promoter activities were determined. Testing the promoter activity in E. coli M15(pMPXENI1)(pQEXENR1) upon the presence of many different AHLs showed that the activity of the braIXEN promoter increased 30-fold in the presence of 3-oxo-C14-HSL, 10-fold in the presence of 3-oxo-C16-HSL, and only 2-fold in the presence of 3-oxo-C12-HSL, demonstrating a specific preference of BraRXEN for 3-oxo-C14-HSL (Fig. 3 A). An analog experiment performed in E. coli M15(pMPUNAI)(pQEUNAR1) suggested that braIUNA promoter activity increased 3.5-fold upon the presence of 3-oxo-C14-HSL and 2-fold when 3-oxo-C12 HSL was present in the media, indicating that 3-oxo-C14-HSL was most likely the cognate AHL for BraRUNA (Fig. 3B). We had previously concluded that BraRKUR responded to 3-oxo-C12-HSL by using a similar approach (73). In the present study, we tested whether AHLs with acyl chains longer than C12 could be recognized by BraRKUR and determined that it responded well to 3-oxo-C16-HSL and very well to 3-oxo-C14-HSL, (Fig. 3D). It was therefore concluded that 3-oxo-C14-HSL was the most likely cognate AHL for BraIR-like systems.
FIG. 3.
Determination of the biologically active AHL for BraRXEN, XenR2, BraRUNA, and BraRKUR AHL sensor/regulators. (A) Determination of the cognate AHL for the BraI/RXEN system of B. xenovorans. Bars correspond to β-galactosidase activities determined for E. coli harboring pQEXENR1 and pMPXENI1. (B) Determination of the cognate AHL for the BraI/RUNA system of B. unamae. Bars correspond to β-galactosidase activities determined for E. coli harboring pQEUNAR and pMPUNAI combination. (C) Determination of the cognate AHL for the XenI2/R2 system of B. xenovorans. Bars correspond to β-galactosidase activities determined for E. coli harboring pQEXENR2 and pMPX2I. (D) Determination of the cognate AHL for the BraI/RKUR system of B. kururiensis. Bars correspond to β-galactosidase activities determined for E. coli harboring pQEBRAR and PBRAI (73). Transcriptional fusions were harbored independently in E. coli expressing either BraRXEN or XenR2 proteins; various exogenous AHLs (1 μM) were provided as indicated, and the β-galactosidase activities were determined. The results are mean values ± the standard deviations of three independent biological replicates. EA, ethyl acetate.
XenR2 responds to 3-OH-C8-HSL and is not transcriptionally regulated by BraRXEN in B. xenovorans LB400T.
Similarly for XenR2, we tested the response of many different AHLs using E. coli M15(pMPX2I)(pQEXENR2). The activity of xenI2 promoter increased 100-fold upon the presence of 3-OH-C8-HSL, 50-fold in the presence of 3OH-C6-HSL, and 25-fold in the presence of 3-oxo-C8-HSL (Fig. 3C). This result is in part in accordance with the AHL production profile detected for the xenI2 genomic mutant LB400XENI2, in which 3-OH-C8-HSL was detected as being produced by XenI2 (see above). The response to 3-oxo-C8-HSL suggested that XenR2, when exposed to high concentrations of AHLs, displayed a relaxed specificity, allowing it to respond to structurally related AHLs.
Experiments aimed at establishing whether there is any hierarchical transcriptional organization between the two AHL QS systems of B. xenovorans strongly suggested that the systems were not transcriptionally regulating each other (data not shown). The cognate AHL for XenI2/R2 system did not activate the braIXEN promoter, and the cognate AHL of the BraI/RXEN system did not activate the xenI2 promoter (data not shown). In addition, the AHL production profiles, as observed in TLC analysis, braIXEN- and braRXEN-deficient mutants continue to produce xenI2 and xenR2 AHLs, and vice versa, suggesting the systems do not regulate each other.
Regulation of the AHL systems in B. xenovorans by BxeR solo.
The role of BxeR solo in regulating the braIXEN and xenI2 promoters, when tested both in E. coli and in B. xenovorans, was determined as described in Materials and Methods. In E. coli, results indicated that BxeR solo did not drive the transcription of any of AHL synthase promoters of B. xenovorans in response to the AHLs tested (data not shown).
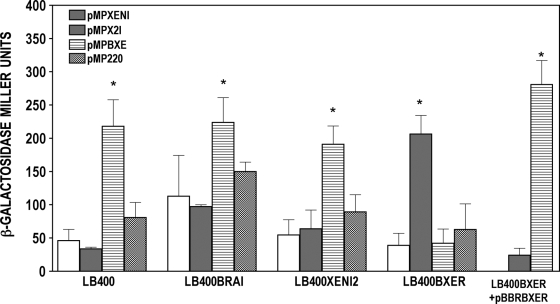
The activities of the braIXEN and xenI2 gene promoters were also determined in B. xenovorans wild-type and in the mutants LB400BRAI, LB400XENI2, and LB400BXER. The results demonstrated that the braIXEN promoter displayed low activity in the wild type, as well as in all mutants tested. The xenI2 promoter also displayed low activities in the braIXEN and xenI2 genomic knockout mutants but resulted in a 3-fold increase when harbored by the bxeR mutant, which suggested that the xenI2 synthase promoter was negatively regulated by the BxeR solo protein. Although no difference in the AHL production profile for the bxeR-deficient mutant was observed, xenI2 promoter activity was reduced when bxeR was provided in trans (Fig. 4). Finally, expression of bxeR was not regulated by the AHL systems but was found to be positively autoregulated, since the bxeR transcriptional promoter fusion decreased 3-fold in activity when harbored in the bxeR mutant LB400BXER, and was recovered when bxeR was provided in trans (Fig. 4). All measurements shown in Fig. 4 were also performed in four other different growth stages; the results have shown trends similar to the growth point depicted in Fig. 4 (data not shown).
FIG. 4.
braIXEN, xenI2, and bxeR promoter activities in wild-type and QS mutant strains of B. xenovorans LB400T. Bacterial cultures were started with an initial inoculum of 5 × 106 CFU in 20 ml of KB-Tc medium, and the β-galactosidase activities were measured over 12 h of growth. All experiments were performed in triplicate, and means values with standard deviations are indicated in the graph. ANOVA in combination with Dunnett's post-test and were performed with Prism 4.0 software (GraphPad). A P value of <0.05 was considered significant (*).
EPS production is regulated by QS in B. kururiensis, B. unamae, and B. xenovorans, whereas biofilm formation is QS regulated only in B. unamae.
Swimming, swarming, protease activity, lipase activity, siderophore production, exopolysaccharide (EPS) production, and biofilm formation were tested in the wild-type strains and compared to QS derivative mutants of B. xenovorans, B unamae, and B. kururiensis to determine whether these phenotypes were regulated by QS. Growth curves were determined for QS mutants, and no significant differences were observed with any of the mutants in their growth rates or in the CFU/ml compared to the wild type (data not shown).
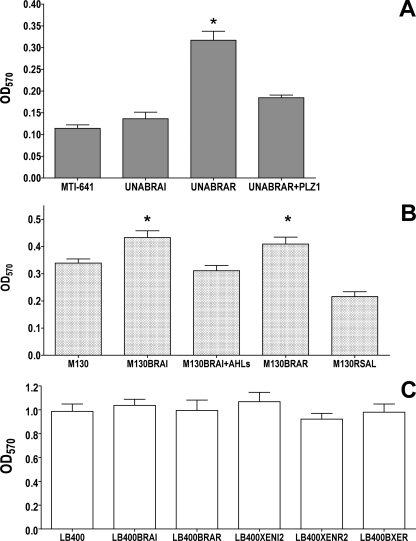
No differences between the Burkholderia wild-type strains and their QS mutants were observed with regard to motility, secreted proteolytic and lipolytic enzyme activities, or siderophore production, suggesting that these phenotypes were not QS regulated under the conditions tested (data not shown). However, in all three species, EPS production was found to be positively regulated by the BraI/R-like QS system, since the AHL-synthase mutants M130BRAI, LB400BRAI, and UNABRAI mutants were significantly less mucoid than the corresponding wild-type strains (Fig. 5). In all three cases, EPS production was restored by chemical complementation when a 1 μM concentration of the cognate AHL (3-oxo-C14-HSL) for each system was added to the medium. Quantification of the EPS produced for B. kururiensis showed that M130BRAI showed a 3-fold decrease in EPS produced compared to the wild type (Fig. 5B), whereas in B. unamae UNABRAI the reduction was 10-fold (Fig. 5D). In B. xenovorans, EPS production was reduced only in LB400BRAI and LB400BRAR mutants, while no difference was observed in LB400BXER and in the xenI2 and xenR2 mutants. These results suggested that EPS production was controlled only by braIXEN and braRXEN in B. xenovorans (Fig. 5E).
FIG. 5.
EPS Production of B. kururiensis M130 (A and B), B. unamae MTl-641T (C and D), and B. xenovorans LB400T wild-type and QS mutants (E). Single colonies were streaked in YEM agar plates. Chemical complementation was achieved by adding 1 μM AHL to the growth medium. Bar graphs show EPS quantification for B. kururiensis and B. unamae by using the boiling phenol method (described in Materials and Methods). Experiments were performed in triplicate, and means ± the standard deviations are plotted. ANOVA in combination with Dunnett's post-test was performed using Prism 4.0 software (GraphPad). A P value of <0.05 was considered significant.
Biofilm formation was found to be negatively regulated by QS in B. unamae, since UNABRAR mutants accumulated approximately three times more biofilm than the wild type. Biofilm accumulation was restored to wild-type levels in the UNABRAR mutant by providing in trans the braRUNA gene via cosmid pLZ1 (Fig. 6 A). Interestingly, the B. unamae UNABRAI mutant produced the same amount of biofilm as the wild type. In contrast, the B. kururiensis AHL-deficient mutants M130BRAI and M130BRAR displayed only slight increases in biofilm formation compared to the wild-type (P < 0.01); this increase was restored when 3-oxo-C14-HSL was provided to the medium containing M130BRAI (Fig. 6B). No major differences in biofilm formation were observed in any of the B. xenovorans QS mutants under the conditions tested (Fig. 6C). It was concluded that biofilm formation was regulated by QS in B. unamae and in B. kururiensis but was not QS dependent in B. xenovorans.
FIG. 6.
Biofilm production in B. unamae (A), B. kururiensis (B), and B. xenovorans (C) wild-type and QS mutants after 72 h of incubation. “+AHLs” refers to complementation by adding 1 μM 3-oxo-C14-HSL. Experiments were performed in triplicate, and means ± the standard error of the mean are plotted. ANOVA in combination with Dunnett's post-test were performed using Prism 4.0 software (GraphPad). A P value of <0.05 was considered significant compared to the wild type (*).
Phenol degradation is QS regulated in B. unamae.
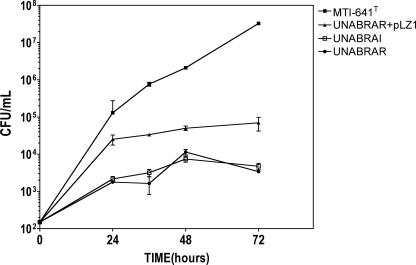
The ability of B. unamae to utilize phenol as carbon source was previously reported as a unique feature of this species among other N-fixing plant-associated Burkholderia spp. (11). AHL QS-deficient mutants (UNABRAR and UNABRAI) were found to have a decreased ability to utilize phenol compared to the wild type (Fig. 7). The ability to grow in the presence of phenol was partially restored in UNABRAR mutant when complemented in trans with the cosmid pLZ1. These results suggest that the phenol degradation is QS regulated in B. unamae. Importantly, using the same media and replacing phenol with mannitol as a carbon source showed that the mutants grew in a fashion similar to that of the wild type (data not shown).
FIG. 7.
Phenol degradation profile in B. unamae wild type and QS mutants. A growth profile was obtained with 103 CFU as starter the inoculum. As a control, phenol was replaced by mannitol as the carbon source, and all strains exhibited the same behavior.
BraI/RXEN negatively regulates production of a 40-kDa Porin-1 family protein in B. xenovorans LB400T.
It was of interest to find gene targets of the well-conserved BraI/R-like system among the recently described Burkholderia species cluster. It was decided to determine whether the levels of any of the secreted proteins were altered in the mutant compared to the wild type. For this experiment, B. xenovorans was used since, of the three species of the cluster studied here, it is the only one of which the genome has been sequenced and annotated. We analyzed the profile of secreted proteins of the wild-type strain LB400T versus the profile of the braIXEN mutant LB400BRAI. As depicted in Fig. S1 in the supplemental material, analysis revealed the presence of two proteins with apparent molecular masses of 40 and 21 kDa in the LB400BRAI mutant that were absent or present in very low amounts in the wild type. It was postulated that these proteins were negatively regulated by the BraI/RXEN system. Mass spectrometry analysis indicated that the 40-kDa protein corresponds to a putative 377-amino-acid outer membrane porin (OmpC family), encoded by the locus Bxe_B2786. In order to confirm whether this ORF was regulated by BraI/RXEN, the Bxe_B2786 gene promoter region was cloned into the promoterless probe vector pMP220 to generate pMP2786. This construct was then conjugated into the B. xenovorans LB400T wild-type and LB400BRAI mutant strains. Determination of the β-galactosidase activity showed a 50-fold increase in activity values obtained in the LB400BRAI mutant compared to the wild type, confirming that this ORF was negatively regulated by the BraI/RXEN system. Promoter activity was significantly reduced when braIXEN was provided in trans by conjugating the plasmid pBBRXENI1 into LB400BRAI (Fig. S1). The 21-kDa protein that was also abundantly present in the LB400BRAI mutant corresponded to a putative ABC-type transporter periplasmic ligand binding protein postulated to be involved in toluene resistance, encoded by the locus Bxe_B0016. By promoter activity analysis in the wild type versus the mutants, we were able to confirm the negative regulation by the BraI/RXEN system since the promoter activity increased 5-fold in the LB400BRAI mutants (see Fig. S1 in the supplemental material). In this case, however, the promoter activity was not restored to wild-type levels when we provided the braIXEN gene in trans via pBBRXENI1; the reason for this is currently not known.
QS regulates in planta growth in B. kururiensis M130 but does not affect rhizosphere colonization in B. unamae MTl-641T.
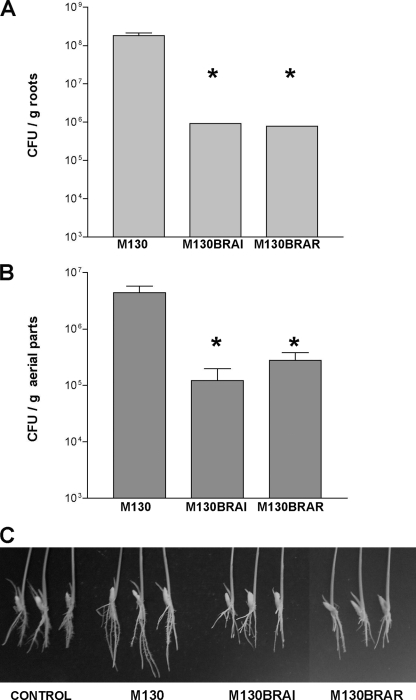
Since members of this recently described Burkholderia cluster are very often associated with plants, it was of interest to study the role of QS in planta. Since B. kururiensis M130 was isolated as a rice endophyte, we tested the role of QS in rice colonization and growth. A significant decrease in colonization of the QS mutants was observed in comparison to that obtained for the wild type, both in the roots and in the aerial parts of the plant (Fig. 8 A and B). In addition, the roots of plantlets colonized with the wild type exhibited an increase in length and branching compared to roots colonized with the QS mutants (Fig. 8C). These data suggested that, under the conditions tested, QS positively regulates endophytic rice colonization by B. kururiensis M130.
FIG. 8.
Rice colonization assays performed with B. kururiensis M130 wild type and impaired QS mutants M130BRAI and M130BRAR. Plantlets were surface sterilized and inoculated as described in Materials and Methods. Bacterial colonization was measured by grinding and plating surface-sterilized plants after 12 days, and CFU/g levels are plotted. A P value of <0.05 was considered significant compared to the wild type (*). (A) Endophytic root colonization levels. (B) Endophytic aerial colonization. (C) Enhanced root development promoted by B. kururiensis M130 wild-type inoculation compared to QS mutants and noninoculated plants.
Since B. unamae MTI-641T was isolated from the maize rhizosphere (12), we determined the role of QS in maize rhizosphere colonization and growth. Experiments revealed that the wild-type strain and the UNABRAI and UNABRAR genomic mutants colonize the maize roots at similar levels, indicating that in this strain AHL QS does not play a major role in root colonization (data not shown).
DISCUSSION
A new cluster composed of beneficial Burkholderia species has been consolidated in the last years following the rapid increase in the number of described species. Despite the metabolic versatility and agrobiotechnological potential of this new group, the regulatory mechanisms underlying their interaction with the environment are largely unknown. In the present study, we investigated the AHL QS systems of two species of the cluster: the maize rhizosphere isolate B. unamae MTI-641T and the B. xenovorans LB400 T, which has become a model system for the bacterial breakdown of highly persistent contaminants (31, 67, 69). In addition, we extended our studies on the role of BraI/RKUR of rice endophyte B. kururiensis M130 (73).
It was established that the BraI/RXEN, BraI/RUNA, and BraI/RKUR systems respond to C14-3-oxo-HSL. This is in accordance with previous studies which demonstrated that other members of the cluster, namely, B. phytofirmans (strains PsJNT and RG6-12) and B. graminis (strains M1 and M14), produce C14-3-oxo-HSL (7, 77). In summary, these results suggest that 3-oxo-C14-HSL is the most probable cognate AHL for the BraI/R-like systems in this Burkholderia cluster. As expected, BraI/R ortholog systems are also present in the sequenced genomes of cluster mates B. phytofirmans, B. graminis, and B. phymatum. The existence of a conserved QS system could facilitate interspecies communication within this Burkholderia cluster, possibly providing advantages in multispecies niche adaptation. The relatedness between the BraI/R-like systems to the LasI/R and PpuI/R from Pseudomonas spp. could suggest that their coding genes might have been involved in lateral gene transfer (12, 45).
The BraI/RUNA and BraI/RXEN systems were not involved in the regulation of motility, siderophore production, or lipolytic or proteolytic activity under the conditions tested. EPS production, however, is positively regulated by the BraI/R-like systems in B. kururiensis, B xenovorans, and B. unamae. It has been reported that EPS production is also subject to QS regulation in other plant-associated bacteria such as Pantoea stewartii (85), Ralstonia solanacearum (28), Pseudomonas syringae (61) and Sinorhizobium meliloti (48). Significantly, in these species EPS production affected host invasion and the pathogenic or symbiotic interaction with the plant was diminished via alteration in surface attachment (biofilm formation) as described for P. stewartii (41) and S. meliloti (34, 64). Importantly, in B. kururiensis M130 both rice colonization and EPS production were reduced in the QS mutants, which could indicate that EPS production is involved in endophytic colonization. It has been reported that one of the EPS polymers produced by B. kururiensis (EPS B) has structural similarity to the EPS produced by B. cepacia (cepacian) (32), which is considered a virulence factor required to the formation of thick and mature biofilms (19). A similar polymer has been recently detected in members of the plant-associated Burkholderia cluster, such as B. graminis, B. phytofirmans, B. phymatum, and B. xenovorans, and is believed to be involved in the plant-bacterium interaction (26). Future understanding of the genes involved in the EPS synthesis in these species may help in identifying the gene targets of QS in this pathway.
Although EPS production was a common QS regulated trait in the three species studied, biofilm formation was found to be negatively regulated by BraI/RUNA in B. unamae, whereas it was only slightly modulated in B. kururiensis, and no QS regulation of biofilm formation was observed for B. xenovorans. The reasons for this differential regulation among the three species are currently unknown, but it is worth noting that the maize rhizospheric colonization of B. unamae was not affected by the reduced EPS production in QS mutants, which suggests that EPS does not affect plant colonization in this strain, whereas it does in B. kururiensis M130.
Several species from the new plant-associated Burkholderia cluster have shown extraordinary ability to degrade phenolic compounds, and this potential has been extensively reviewed (37, 54, 56). The presence of a phenol monooxygenase and the ability to degrade phenol have been previously demonstrated in B. unamae (11). Here, it is demonstrated that this trait is regulated by the BraI/RUNA QS system in B. unamae. The ability to degrade phenol was reported to be associated with AHL production by members of the microbial community in activated sludges, and supplementation with AHL was able to sustain phenol-degrading activity beyond the point of starvation. It was hypothesized that this event was associated to surfactant production and composition rearrangement within the community (79). Interestingly, B. kururiensis was recently reported to be able to grow in consortia that degrade 2,4,6-trichlorophenol, using phenol as a primary intermediate (30). From these findings, it cannot be excluded that QS might have a role in the consortial behavior of Burkholderia, leading to increased degradation of aromatic compounds.
Maize colonization experiments performed with B. unamae indicated that the BraI/RUNA QS system does not regulate ability to colonize this host under the conditions used, whereas rice colonization by B. kururiensis was positively regulated by QS. These results suggest that plant-bacterium interaction may be subject to regulation in a cell density manner in plant-associated Burkholderia. However, more experiments need to be performed in order to determine whether plant colonization (at both the endophytic and the rhizospheric levels) may be dependent on the plant genotype. In fact, B. phytofirmans PsJN has been shown to have variable colonization levels in different cultivars in several plant species (60, 77).
Secreted proteins profiling for B. xenovorans evidenced several bands present in the AHL synthase mutant LB400BRAI profile and absent in wild-type profile. However, transcriptional analysis of their promoters indicated that only two of them were transcriptionally regulated by BraI/RXEN, and only one of them could be fully complemented by supplying the braIXEN gene in trans. This latter gene encoded for a porin belonging to the OmpC family (pFAM00267), and a putative lux box centered at −96 was identified in its promoter region. A similar protein was identified in B. cenocepacia, and it was suggested that it may function as a pore for small molecules involved in osmoregulatory control (6). Several porins have been reported to be regulated by QS in several species, including B. cepacia (1), Azospirillum lipoferum (9) and P. aeruginosa (4), but the implications of such regulation remains to be explored.
We report that B. xenovorans xenI2 and xenR2 are found in only one-third of the members of the Burkholderia plant-associated cluster. Surprisingly, xenI2 and xenR2 are not present in four other B. xenovorans strains, which implies that the occurrence of XenI2/R2 is strain dependent rather than species dependent. In fact, in strain LB400T xenI2 and xenR2 are located in a megaplasmid, indicating that it is not part of the core chromosome of B. xenovorans. Interestingly, B. phytofirmans strain PsJN was also reported to produce C8-3OH-HSL, but these AHLs are not produced by strain RG6-12, indicating that xenI2 and xenR2 may not be present in all B. phytofirmans strains (77). A similar scenario occurs for the CciIR system, which is found only in some BCC B. cenocepacia strains and has been shown to be associated with a pathogenicity island (45). Homology studies demonstrated that XenI2/R2 is closely related to the CciIR system and to the BtaIR3, BmaIR3, and BpsIR3 systems of B. mallei, B. pseudomallei, and B. thailandensis, respectively (12, 45), possibly indicating a probable common ancestor for these systems (14, 22). The cciI and cciR system has recently been characterized as a global regulator exerting an important regulatory control of the cepI and cepR, the other AHL QS present in B. cenocepacia K-52 (53).
Our studies showed that under the conditions we tested, no transcriptional regulatory hierarchies are present between BraI/RXEN and XenI2/R2. In addition, transcriptional studies of braIXEN and xenI2 gene promoters in B. xenovorans showed that both genes are expressed at very low levels, indicating that they are most likely under transcriptional regulation. The BraI/RXEN system is most probably regulated by the intergenically located rsaL repressor, as previously described for the BraI/RKUR system (73).
The genome of B. xenovorans contains an unpaired LuxR family solo, designated BxeR, with the typical N-terminal AHL-binding domain and the C-terminus HTH motif. Protein alignment showed that BxeR possesses the six conserved amino acids of the AHL-binding domain, while one of the three conserved residues in the HTH motif presents an E178Q substitution, which in fact was also substituted in the DNA-binding domain of XenR2 protein. BxeR positively autoregulated its own transcription and negatively regulated the expression of xenI2. BxeR was present in all B. xenovorans strains, as well as in most of the type strains of the B. graminis clade, and in the species B. tuberum. It is not known whether BxeR binds to AHLs, but the conservation of the residues in the AHL binding domain suggests that BxeR might bind AHL molecules. The role of LuxR-type solos in AHL producers has been reviewed recently (58, 74); several examples demonstrate that LuxR-type solos present in AHL producing bacteria are most often integrated with the resident AHL QS regulatory networks, as are the cases of ExpR and NesR from S. meliloti (34, 57), PpoR from P. putida (75), and QscR from P. aeruginosa (43). Our results suggest that such integration might exist for BxeR, since the negative regulation of xenI2 promoter was observed in the bxeR-deficient background.
In the present study we describe the characterization of the QS systems in species from the new plant-associated Burkholderia cluster isolated from three different environmental niches. It was confirmed that the cluster shares a highly conserved BraI/R-like QS system, and the existence of a common core of targets is proposed, although species-specific targets may have been acquired as a response to niche adaptation. A second QS system and a LuxR solo protein were found to be present in a few members of the cluster that are independent of the BraI/R-like system. Future work will focus on determining the complete regulons of the two systems and of the LuxR type solo BxeR in this important recently described cluster of Burkholderia species.
Supplementary Material
Acknowledgments
Z.R.S.-M. is supported by an ICGEB fellowship.
We thank coresearcher L. Gigli for support in this work. We thank I. Bertani, S. Subramoni, L. Steindler, J. Gonzalez, and G. Degrassi for encouragement and critical reading of the manuscript. We are grateful to L. Martínez-Aguilar (CCG-UNAM) for technical assistance in the colonization of maize assays.
Footnotes
Published ahead of print on 30 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aguilar, C., A. Friscina, G. Devescovi, M. Kojic, and V. Venturi. 2003. Identification of quorum-sensing-regulated genes of Burkholderia cepacia. J. Bacteriol. 185:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26:824-828. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. A. 1939. The use of tributyrin agar in dairy bacteriology. Berlin Int. Mikrobiol. Kongress 3:726-728. [Google Scholar]
- 4.Arevalo-Ferro, C., M. Hentzer, G. Reil, A. Gorg, S. Kjelleberg, M. Givskov, K. Riedel, and L. Eberl. 2003. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5:1350-1369. [DOI] [PubMed] [Google Scholar]
- 5.Baldani, V. L., E. Oliveira, E. Balota, J. I. Baldani, G. Kirchhof, and J. Döbereiner. 1997. Burkholderia brasilensis sp. nov. uma nova espécie de bactéria diazotrófica endofítica. An. Acad. Bras. Cienc. 69:1. [Google Scholar]
- 6.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barriuso, J., B. Ramos Solano, R. G. Fray, M. Camara, A. Hartmann, and F. J. Gutierrez Manero. 2008. Transgenic tomato plants alter quorum sensing in plant growth-promoting rhizobacteria. Plant Biotechnol. J. 6:442-452. [DOI] [PubMed] [Google Scholar]
- 8.Better, M., B. Lewis, D. Corbin, G. Ditta, and D. Helinski. 1983. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell 35:479-485. [DOI] [PubMed] [Google Scholar]
- 9.Boyer, M., R. Bally, S. Perrotto, C. Chaintreuil, and F. Wisniewski-Dye. 2008. A quorum-quenching approach to identify quorum-sensing-regulated functions in Azospirillum lipoferum. Res. Microbiol. 159:699-708. [DOI] [PubMed] [Google Scholar]
- 10.Caballero-Mellado, J., L. Martinez-Aguilar, G. Paredes-Valdez, and P. E. Santos. 2004. Burkholderia unamae sp. nov., an N2-fixing rhizospheric and endophytic species. Int. J. Syst. Evol. Microbiol. 54:1165-1172. [DOI] [PubMed] [Google Scholar]
- 11.Caballero-Mellado, J., J. Onofre-Lemus, P. Estrada-de Los Santos, and L. Martinez-Aguilar. 2007. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 73:5308-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Case, R. J., M. Labbate, and S. Kjelleberg. 2008. AHL-driven quorum-sensing circuits: their frequency and function among the proteobacteria. ISME J. 2:345-349. [DOI] [PubMed] [Google Scholar]
- 13.Chain, P. S., V. J. Denef, K. T. Konstantinidis, L. M. Vergez, L. Agullo, V. L. Reyes, L. Hauser, M. Cordova, L. Gomez, M. Gonzalez, M. Land, V. Lao, F. Larimer, J. J. LiPuma, E. Mahenthiralingam, S. A. Malfatti, C. J. Marx, J. J. Parnell, A. Ramette, P. Richardson, M. Seeger, D. Smith, T. Spilker, W. J. Sul, T. V. Tsoi, L. E. Ulrich, I. B. Zhulin, and J. M. Tiedje. 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. U. S. A. 103:15280-15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler, J. R., B. A. Duerkop, A. Hinz, T. E. West, J. P. Herman, M. E. Churchill, S. J. Skerrett, and E. P. Greenberg. 2009. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J. Bacteriol. 191:5901-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, W.-M., S. M. de Faria, J.-H. Chou, E. K. James, G. N. Elliott, J. I. Sprent, C. Bontemps, J. P. W. Young, and P. Vandamme. 2008. Burkholderia sabiae sp. nov., isolated from root nodules of Mimosa caesalpiniifolia. Int. J. Syst. Evol. Microbiol. 58:2174-2179. [DOI] [PubMed] [Google Scholar]
- 16.Chen, W.-M., S. M. de Faria, E. K. James, G. N. Elliott, K.-Y. Lin, J.-H. Chou, S.-Y. Sheu, M. Cnockaert, J. I. Sprent, and P. Vandamme. 2007. Burkholderia nodosa sp. nov., isolated from root nodules of the woody Brazilian legumes Mimosa bimucronata and Mimosa scabrella. Int. J. Syst. Evol. Microbiol. 57:1055-1059. [DOI] [PubMed] [Google Scholar]
- 17.Chen, W.-M., E. K. James, T. Coenye, J.-H. Chou, E. Barrios, S. M. de Faria, G. N. Elliott, S.-Y. Sheu, J. I. Sprent, and P. Vandamme. 2006. Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int. J. Syst. Evol. Microbiol. 56:1847-1851. [DOI] [PubMed] [Google Scholar]
- 18.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 19.Cunha, M. V., S. A. Sousa, J. H. Leitao, L. M. Moreira, P. A. Videira, and I. Sa-Correia. 2004. Studies on the involvement of the exopolysaccharide produced by cystic fibrosis-associated isolates of the Burkholderia cepacia complex in biofilm formation and in persistence of respiratory infections. J. Clin. Microbiol. 42:3052-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dharmapuri, S., and R. V. Sonti. 1999. A transposon insertion in the gumG homologue of Xanthomonas oryzae pv. oryzae causes loss of extracellular polysaccharide production and virulence. FEMS Microbiol. Lett. 179:53-59. [DOI] [PubMed] [Google Scholar]
- 21.Duerkop, B. A., R. L. Ulrich, and E. P. Greenberg. 2007. Octanoyl-homoserine lactone is the cognate signal for Burkholderia mallei BmaR1-BmaI1 quorum sensing. J. Bacteriol. 189:5034-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duerkop, B. A., J. Varga, J. R. Chandler, S. B. Peterson, J. P. Herman, M. E. Churchill, M. R. Parsek, W. C. Nierman, and E. P. Greenberg. 2009. Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis. J. Bacteriol. 191:3909-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberl, L. 2006. Quorum sensing in the genus Burkholderia. Int. J. Med. Microbiol. 296:103-110. [DOI] [PubMed] [Google Scholar]
- 24.Elliott, G. N., W. M. Chen, C. Bontemps, J. H. Chou, J. P. Young, J. I. Sprent, and E. K. James. 2007. Nodulation of Cyclopia spp. (Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann. Bot. 100:1403-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Estrada-de Los Santos, P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira, A. S., J. H. Leitão, I. N. Silva, P. F. Pinheiro, S. A. Sousa, C. G. Ramos, and L. M. Moreira. 2010. Distribution of cepacian biosynthesis genes among environmental and clinical Burkholderia strains and role of cepacian exopolysaccharide in resistance to stress conditions. Appl. Environ. Microbiol. 76:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flavier, A. B., S. J. Clough, M. A. Schell, and T. P. Denny. 1997. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26:251-259. [DOI] [PubMed] [Google Scholar]
- 29.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-De Jesus, A., F. Romano-Baez, L. Leyva-Amezcua, C. Juarez-Ramirez, N. Ruiz-Ordaz, and N. Galindez-Mayer. 2009. Biodegradation of 2,4,6-trichlorophenol in a packed-bed biofilm reactor equipped with an internal net draft tube riser for aeration and liquid circulation. J. Hazard. Mater. 161:1140-1149. [DOI] [PubMed] [Google Scholar]
- 31.Goris, J., P. De Vos, J. Caballero-Mellado, J. Park, E. Falsen, J. F. Quensen III, J. M. Tiedje, and P. Vandamme. 2004. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400 and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 32.Hallack, L. F., D. S. Passos, K. A. Mattos, O. A. Agrellos, C. Jones, L. Mendonca-Previato, J. O. Previato, and A. R. Todeschini. 2010. Structural elucidation of the repeat unit in highly branched acidic exopolysaccharides produced by nitrogen fixing Burkholderia. Glycobiology 20:348-447. [DOI] [PubMed] [Google Scholar]
- 33.Hoagland, D. 1975. Mineral nutrition, p. 129-134. In Laboratory experiments in plant physiology, vol. 1. Macmillan Publishing Co., Inc., New York, NY. [Google Scholar]
- 34.Hoang, H. H., N. Gurich, and J. E. Gonzalez. 2008. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J. Bacteriol. 190:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hontzeas, N., S. S. Saleh, and B. R. Glick. 2004. Changes in gene expression in canola roots induced by ACC-deaminase-containing plant-growth-promoting bacteria. Mol. Plant-Microbe Interact. 17:865-871. [DOI] [PubMed] [Google Scholar]
- 36.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 37.Iwaki, H., A. Kazuya, and H. Yoshie. 2007. Isolation and characterization of a new 2,4-dinitrophenol-degrading bacterium Burkholderia sp. strain KU-46 and its degradation pathway. FEMS Microbiol. Lett. 274:112-117. [DOI] [PubMed] [Google Scholar]
- 38.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 39.Kiratisin, P., and S. Sanmee. 2008. Roles and interactions of Burkholderia pseudomallei BpsIR quorum-sensing system determinants. J. Bacteriol. 190:7291-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohler, T., L. K. Curty, F. Barja, C. van Delden, and J.-C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koutsoudis, M. D., D. Tsaltas, T. D. Minogue, and S. B. von Bodman. 2006. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. U. S. A. 103:5983-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovach, M., P. Elzer, D. Hill, G. Robertson, M. Farris, R. Roop, and K. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 43.Lequette, Y., J. H. Lee, F. Ledgham, A. Lazdunski, and E. P. Greenberg. 2006. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J. Bacteriol. 188:3365-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, C. H., G. Bourque, and P. Tan. 2008. A comparative synteny map of Burkholderia species links large-scale genome rearrangements to fine-scale nucleotide variation in prokaryotes. Mol. Biol. Evol. 25:549-558. [DOI] [PubMed] [Google Scholar]
- 45.Malott, R. J., A. Baldwin, E. Mahenthiralingam, and P. A. Sokol. 2005. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect. Immun. 73:4982-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malott, R. J., E. P. O'Grady, J. Toller, S. Inhulsen, L. Eberl, and P. A. Sokol. 2009. A Burkholderia cenocepacia orphan LuxR homolog is involved in quorum-sensing regulation. J. Bacteriol. 191:2447-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malott, R. J., and P. A. Sokol. 2007. Expression of the bviIR and cepIR quorum-sensing systems of Burkholderia vietnamiensis. J. Bacteriol. 189:3006-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marketon, M. M., S. A. Glenn, A. Eberhard, and J. E. Gonzalez. 2003. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 185:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez-Aguilar, L., R. Diaz, J. J. Pena-Cabriales, P. Estrada-de Los Santos, M. F. Dunn, and J. Caballero-Mellado. 2008. Multichromosomal genome structure and confirmation of diazotrophy in novel plant-associated Burkholderia species. Appl. Environ. Microbiol. 74:4574-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattos, K. A., V. L. Padua, A. Romeiro, L. F. Hallack, B. C. Neves, T. M. Ulisses, C. F. Barros, A. R. Todeschini, J. O. Previato, and L. Mendonca-Previato. 2008. Endophytic colonization of rice (Oryza sativa L.) by the diazotrophic bacterium Burkholderia kururiensis and its ability to enhance plant growth. An. Acad. Bras. Cienc. 80:477-493. [DOI] [PubMed] [Google Scholar]
- 51.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Laboratory Press, Cold Spring Harbor, NY.
- 52.Murray, T. S., and B. I. Kazmierczak. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 188:6995-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Grady, E. P., D. F. Viteri, R. J. Malott, and P. A. Sokol. 2009. Reciprocal regulation by the CepIR and CciIR quorum sensing systems in Burkholderia cenocepacia. BMC Genomics 10:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Sullivan, L. A., and E. Mahenthiralingam. 2005. Biotechnological potential within the genus Burkholderia. Lett. Appl. Microbiol. 41:8-11. [DOI] [PubMed] [Google Scholar]
- 55.O'Toole, G., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 56.Parnell, J. J., J. Park, V. Denef, T. Tsoi, S. Hashsham, J. Quensen III, and J. M. Tiedje. 2006. Coping with polychlorinated biphenyl (PCB) toxicity: physiological and genome-wide responses of Burkholderia xenovorans LB400 to PCB-mediated stress. Appl. Environ. Microbiol. 72:6607-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patankar, A. V., and J. E. Gonzalez. 2009. An orphan LuxR homolog of Sinorhizobium meliloti affects stress adaptation and competition for nodulation. Appl. Environ. Microbiol. 75:946-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patankar, A. V., and J. E. Gonzalez. 2009. Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 33:739-756. [DOI] [PubMed] [Google Scholar]
- 59.Perin, L., L. Martinez-Aguilar, G. Paredes-Valdez, J. I. Baldani, P. Estrada-de Los Santos, V. M. Reis, and J. Caballero-Mellado. 2006. Burkholderia silvatlantica sp. nov., a diazotrophic bacterium associated with sugar cane and maize. Int. J. Syst. Evol. Microbiol. 56:1931-1937. [DOI] [PubMed] [Google Scholar]
- 60.Pillay, V. K., and J. Nowak. 1997. Inoculum density, temperature, and genotype effects on in vitro growth promotion and epiphytic and endophytic colonization of tomato (Lycopersicon esculentum L.) seedlings inoculated with a pseudomonad bacterium. Can. J. Microbiol. 43:354-361. [Google Scholar]
- 61.Quiñones, B., G. Dulla, and S. E. Lindow. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:682-693. [DOI] [PubMed] [Google Scholar]
- 62.Reis, V. M., P. Estrada-de los Santos, S. Tenorio-Salgado, J. Vogel, M. Stoffels, S. Guyon, P. Mavingui, V. L. Baldani, M. Schmid, J. I. Baldani, J. Balandreau, A. Hartmann, and J. Caballero-Mellado. 2004. Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int. J. Syst. Evol. Microbiol. 54:2155-2162. [DOI] [PubMed] [Google Scholar]
- 63.Riedel, K., M. Hentzer, O. Geisenberger, B. Huber, A. Steidle, H. Wu, N. Hoiby, M. Givskov, S. Molin, and L. Eberl. 2001. N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249-3262. [DOI] [PubMed] [Google Scholar]
- 64.Rinaudi, L. V., and J. E. Gonzalez. 2009. The low-molecular-weight fraction of exopolysaccharide II from Sinorhizobium meliloti is a crucial determinant of biofilm formation. J. Bacteriol. 191:7216-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 66.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 67.Seeger, M., M. Gonzalez, B. Camara, L. Munoz, E. Ponce, L. Mejias, C. Mascayano, Y. Vasquez, and S. Sepulveda-Boza. 2003. Biotransformation of natural and synthetic isoflavonoids by two recombinant microbial enzymes. Appl. Environ. Microbiol. 69:5045-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shaw, P., G. Ping, S. Daly, C. Cha, J. Cronan, K. Rinehart, and S. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. U. S. A. 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith, D. J., J. Park, J. M. Tiedje, and W. W. Mohn. 2007. A large gene cluster in Burkholderia xenovorans encoding abietane diterpenoid catabolism. J. Bacteriol. 189:6195-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spaink, H., R. Okker, C. Wijffelmann, E. Pees, and B. Lugtemberg. 1987. Promoter in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 71.Stachel, S., G. An, C. Flores, and E. Nester. 1985. A Tn3 lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 4:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suarez-Moreno, Z. R., J. Caballero-Mellado, and V. Venturi. 2008. The new group of non-pathogenic plant-associated nitrogen-fixing Burkholderia spp. shares a conserved quorum-sensing system, which is tightly regulated by the RsaL repressor. Microbiology 154:2048-2059. [DOI] [PubMed] [Google Scholar]
- 74.Subramoni, S., and V. Venturi. 2009. LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology 155:1377-1385. [DOI] [PubMed] [Google Scholar]
- 75.Subramoni, S., and V. Venturi. 2009. PpoR is a conserved unpaired LuxR solo of Pseudomonas putida which binds N-acyl homoserine lactones. BMC Microbiol. 9:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomaic, V., D. Gardiol, P. Massimi, M. Ozbun, M. Myers, and L. Banks. 2009. Human and primate tumour viruses use PDZ binding as an evolutionarily conserved mechanism of targeting cell polarity regulators. Oncogene 28:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trognitz, F., K. Scherwinski, A. Fekete, S. Schmidt, L. Eberl, J. Rodewald, M. Schmid, S. Compant, A. Hartmann, P. Schmitt-Kopplin, B. Trognitz, and A. Sessitsch. 2009. Interaction between potato and the endophyte Burkholderia phytofirmans. Tagung Vereinigung Pflanzenzüchter Saatgutkaufl Os̈terreichs 59:63-66. [Google Scholar]
- 78.Ulrich, R. L., H. B. Hines, N. Parthasarathy, and J. A. Jeddeloh. 2004. Mutational analysis and biochemical characterization of the Burkholderia thailandensis DW503 quorum-sensing network. J. Bacteriol. 186:4350-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Valle, A., M. Bailey, A. Whiteley, and M. Manefield. 2004. N-Acyl-l-homoserine lactones (AHLs) affect microbial community composition and function in activated sludge. Environ. Microbiol. 6:424-433. [DOI] [PubMed] [Google Scholar]
- 80.Vandamme, P., J. Goris, W. M. Chen, P. de Vos, and A. Willems. 2002. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov. nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25:507-512. [DOI] [PubMed] [Google Scholar]
- 81.Vandamme, P., and E. Mahenthiralingam. 2003. Strains from the Burkholderia cepacia complex: relationship to opportunistic pathogens. J. Nematol. 35:208-211. [PMC free article] [PubMed] [Google Scholar]
- 82.Vandamme, P., K. Opelt, N. Knochel, C. Berg, S. Schonmann, E. De Brandt, L. Eberl, E. Falsen, and G. Berg. 2007. Burkholderia bryophila sp. nov. and Burkholderia megapolitana sp. nov., moss-associated species with antifungal and plant-growth-promoting properties. Int. J. Syst. Evol. Microbiol. 57:2228-2235. [DOI] [PubMed] [Google Scholar]
- 83.Vanlaere, E., A. Baldwin, D. Gevers, D. Henry, E. De Brandt, J. J. LiPuma, E. Mahenthiralingam, D. P. Speert, C. Dowson, and P. Vandamme. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59:102-111. [DOI] [PubMed] [Google Scholar]
- 84.Venturi, V., A. Friscina, I. Bertani, G. Devescovi, and C. Aguilar. 2004. Quorum sensing in the Burkholderia cepacia complex. Res. Microbiol. 155:238-244. [DOI] [PubMed] [Google Scholar]
- 85.Von Bodman, S. B., D. R. Majerczak, and D. L. Coplin. 1998. A negative regulator mediates quorum-sensing control of exopolysaccharide production in Pantoea stewartii subsp. stewartii. Proc. Natl. Acad. Sci. U. S. A. 95:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. A. B. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
- 87.Yabuuchi, E., Y. Kosako, H. Oyaizu, I. Yano, H. Hotta, Y. Hashimoto, T. Ezaki, and M. Arakawa. 1992. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36:1251-1275. [DOI] [PubMed] [Google Scholar]
- 88.Zlosnik, J. E., T. J. Hird, M. C. Fraenkel, L. M. Moreira, D. A. Henry, and D. P. Speert. 2008. Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J. Clin. Microbiol. 46:1470-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.