Abstract
Previous studies have identified novel modifications in the core fold domain of histone H2B, but relatively little is known about the function of these putative histone modification sites. We have mutated core modifiable residues that are conserved in Saccharomyces cerevisiae histone H2B and characterized the effects of the mutants on yeast silencing, gene expression, and the DNA damage response. We identified three histone H2B core modifiable residues as functionally important. We find that mutating H2B K49 in yeast confers a UV sensitivity phenotype, and we confirm that the homologous residue in human histone H2B is acetylated and methylated in human cells. Our results also indicate that mutating H2B K111 impairs the response to methyl methanesulfonate (MMS)-induced DNA lesions and disrupts telomeric silencing and Sir4 binding. In contrast, mutating H2B R102 enhances silencing at yeast telomeres and the HML silent mating loci and increases Sir4 binding to these regions. The H2B R102A mutant also represses the expression of endogenous genes adjacent to yeast telomeres, which is likely due to the ectopic spreading of the Sir complex in this mutant strain. We propose a structural model by which H2B R102 and K111 regulate the binding of the Sir complex to the nucleosome.
In eukaryotic cells, DNA is wound around a protein octamer consisting of two copies each of the histone proteins H2A, H2B, H3, and H4 to form a nucleosome core particle. Histone proteins are extensively posttranslationally modified in chromatin; common posttranslational modifications include lysine and arginine methylation, lysine acetylation, and lysine ubiquitylation (25). Accumulating evidence has highlighted the integral role of histone proteins and their corresponding posttranslational modifications in the regulation of many cellular processes. Until recently most studies had focused on modifications within the flexible N- and C-terminal histone tails; however, analysis of purified histone proteins by mass spectrometry has revealed a plethora of modifications existing within the core histone fold domains (reviewed in references 12 and 30). As detailed below, previous studies have shown that modifications in the core histone fold domains can regulate important cellular processes, including gene transcription, silencing, and DNA replication and repair.
To date, most work has focused on the histone H3 and H4 core residues. The first discovered modification in the core histone fold domain was methylation of histone H3 K79 (34, 49), a residue located on the solvent-accessible nucleosome face. Methylation of H3 K79 by Dot1 is implicated in the DNA damage response (18, 52) and regulates silencing in Saccharomyces cerevisiae (33, 34, 49). Other well-studied core histone modifications include histone H3 K56 acetylation and H4 K91 acetylation. Histone H3 K56 acetylation, which is catalyzed by the novel acetyltransferase Rtt109 (10, 17, 43), plays a critical role in genome stability, DNA replication and repair, and silencing in yeast (9, 10, 17, 19, 28, 38, 54). Altogether, at least 14 novel modifications have been discovered in the histone H3 and H4 core domains, many of which yield striking phenotypes when the homologous histone residue is mutated in yeast (12, 19, 30, 57).
Methylation of H3 K79 and acetylation of H3 K56 both regulate telomeric silencing in yeast, albeit through different mechanisms. Telomeric silencing is mediated by the Sir silencing complex (3), which consists of Sir2, Sir3, and Sir4 (42). Dot1-catalyzed methylation of H3 K79, which occurs predominately in euchromatin regions of the yeast genome, inhibits the binding of Sir3 and thus prevents the dispersal of the Sir complex from telomere regions (2, 33, 34, 49). In contrast, H3 K56 acetylation does not regulate Sir complex binding; instead, H3 K56 is deacetylated by the Sir2 histone deacetylase in telomeric regions (55). It has been suggested that H3 K56 deacetylation may directly regulate telomeric chromatin structure and accessibility (55). It is important to note that unmodified residues in the core fold domains of histone H3 and H4 also play important roles in gene silencing. For example, mutations in the LRS (loss of ribosomal DNA [rDNA] silencing) and SIN (switch independent) domains of the nucleosome have significant effects on silencing, although these mutations principally target unmodified histone H3 and H4 residues (8, 13). Intriguingly, recent studies suggest that the LRS domain directly regulates Sir3 binding to the nucleosome (35, 36).
The same mass spectrometry studies that discovered the core modifications in histone H3 and H4 also discovered novel posttranslational modifications in the core histone fold domains of histones H2A and H2B (5, 12, 24, 30, 51, 57). However, the role of these putative histone modification sites in gene expression, silencing, and DNA replication or repair has yet to be elucidated.
In this study, we have used a genetic approach to investigate the functional significance of modified residues in the core fold domain of histone H2B. We have systematically mutated homologous residues in yeast histone H2B and characterized the mutants for their effects on silencing, genome-wide expression, and DNA damage response. We show that mutating H2B K49 renders cells hypersensitive to UV irradiation and confirm that the homologous residue is acetylated and methylated in human cells. We find that a mutation in histone H2B K111, which is reported to be acetylated and methylated in mammalian histone H2B (5, 24, 51), disrupts telomeric silencing and renders yeast cells hypersensitive to the DNA-damaging agent methyl methanesulfonate (MMS). Chromatin immunoprecipitation (ChIP) experiments revealed that the level of Sir4p binding to telomeres and the HML locus was decreased in this mutant, although binding was not entirely abolished. In contrast, mutation of H2B R102, which is methylated in bovine histone H2B, enhanced silencing at yeast telomeres and the HML silent mating locus. Microarray analysis revealed that the H2B R102A mutant enhanced the repression of genes adjacent to yeast telomeres, and ChIP studies indicated an increase in Sir4 binding at silenced loci. Analysis of the H2B R102/K111 double mutant suggests that the H2B R102 and H2B K111 residues independently regulate Sir4 binding to yeast telomeres. We propose a structural model by which these H2B residues, which are located adjacent to one another on the solvent-accessible nucleosome face, directly regulate the binding of Sir3 to the nucleosome. In summary, these results highlight the important role that modifiable core residues within histone H2B play in gene expression, silencing, and the DNA damage response.
MATERIALS AND METHODS
Plasmid construction.
Histone H2B mutants were generated by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene). Plasmid pMP002 (39), which contains the wild-type histone H2A gene (HTA1) and histone H2B gene (HTB1), was used as a template for the mutagenesis reaction. Each mutant plasmid was verified by DNA sequencing. Mutagenic primer sequences are given at the website http://wyrick.sbs.wsu.edu/H2Bcore/.
The plasmids used in ChIP assays were all constructed as follows. To generate an ADE2 plasmid containing the wild-type HTA1-HTB1 genes, pMP002 was double digested with EcoRI and SpeI. The resulting 2.1-kb fragment was ligated into the pRS412 plasmid (6) to give plasmid pMK001. Plasmids pMK019 through pMK022, containing htb1 mutant alleles, were generated in the same manner by subcloning from pKV002, pJW625, pJW626, and pJW627.
Yeast strains.
Yeast strains are listed in Table 1. Yeast strains were derived from the PY013 (39), UCC6389 (14), or MKY059 (see below) parental strain. Briefly, plasmids containing the appropriate wild-type or mutant HTB1 alleles were transformed into the parent strains. The PY013-derived strains were constructed by plasmid shuffling on media containing 5-fluoroorotic acid (5-FOA; Zymo Research); the UCC6389-derived strains were cured of the wild-type pRG422 plasmid by growth on nonselective media and replica plating.
TABLE 1.
List of yeast strains and genotypes used this study
| Strain | Genotype | Reference or source |
|---|---|---|
| UCC6389 | MATalys2Δ0 trp1Δ63 his3Δ200 ade2Δ::hisG ura3Δ0 leu2Δ0 met15Δ0 ADE2-TEL-VR URA3-TEL-VIIL hta1-htb1::MET15 hta2-htb2::LEU2 plus pRG422 (CEN6 HIS3 HTA1-FLAG-HTB1) | 14 |
| WY186 | Isogenic to UCC6389, plus pKV002 (CEN6 TRP1 HTA1 htb1-K46A) | This study |
| WY188 | Isogenic to UCC6389, plus pMP002 (CEN6 TRP1 HTA1 HTB1) | This study |
| WY189 | Isogenic to UCC6389, plus pJW625 (CEN6 TRP1 HTA1 htb1-K88A) | This study |
| WY190 | Isogenic to UCC6389, plus pJW626 (CEN6 TRP1 HTA1 htb1-R102A) | This study |
| WY191 | Isogenic to UCC6389, plus pJW627 (CEN6 TRP1 HTA1 htb1-K111A) | This study |
| UCC6391 | MATalys2Δ0 trp1Δ63 his3Δ200 ade2Δ::hisG ura3Δ0 leu2Δ0 met15Δ0 ADE2-TEL-VR URA3-TEL-VIIL hta1-htb1::MET15 hta2-htb2::LEU2 SIR4::KanMX plus Prg422 (CEN6 HIS3 HTA1-FLAG-HTB1) | 14 |
| WY195 | Isogenic to UCC6391, plus pMP002 (CEN6 TRP1 HTA1 HTB1) | This study |
| WY196 | Isogenic to UCC6391, plus pJW626 (CEN6 TRP1 HTA1 htb1-R102A) | This study |
| WY204 | Isogenic to UCC6389, plus pJW632 (CEN6 TRP1 HTA1 htb1-K111R) | This study |
| WY205 | Isogenic to UCC6389, plus pJW633 (CEN6 TRP1 HTA1 htb1-R102K) | This study |
| WY210 | Isogenic to UCC6389, plus pJW635 (CEN6 TRP1 HTA1 htb1-K88R) | This study |
| WY211 | Isogenic to UCC6389, plus pJW636 (CEN6 TRP1 HTA1 htb1-K88Q) | This study |
| WY212 | Isogenic to UCC6389, plus pJW637 (CEN6 TRP1 HTA1 htb1-K111Q) | This study |
| WY213 | Isogenic to UCC6389, plus pJW638 (CEN6 TRP1 HTA1 htb1-R102A/K111A) | This study |
| WY214 | Isogenic to UCC6389, plus pJW640 (CEN6 TRP1 HTA1 htb1-K49A) | This study |
| WY215 | Isogenic to UCC6389, plus pJW641 (CEN6 TRP1 HTA1 htb1-K60A) | This study |
| WY216 | Isogenic to UCC6389, plus pJW642 (CEN6 TRP1 HTA1 htb1-K49R) | This study |
| WY217 | Isogenic to UCC6389, plus pJW643 (CEN6 TRP1 HTA1 htb1-K60R) | This study |
| PY014 | MATaade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 hta1-htb1::HIS3 hta2-htb2::LEU2 plus pMP002 (CEN6 TRP1 HTA1 HTB1) | 39 |
| WY313 | Isogenic to PY014, plus pJW622 (CEN6 TRP1 HTA1 htb1-K46Q) | This study |
| WY315 | Isogenic to PY014, plus pJW625 (CEN6 TRP1 HTA1 htb1-K88A) | This study |
| WY316 | Isogenic to PY014, plus pJW626 (CEN6 TRP1 HTA1 htb1-R102A) | This study |
| WY317 | Isogenic to PY014, plus pJW627 (CEN6 TRP1 HTA1 htb1-K111A) | This study |
| WY322 | Isogenic to PY014, plus pJW632 (CEN6 TRP1 HTA1 htb1-K111R) | This study |
| WY323 | Isogenic to PY014, plus pJW633 (CEN6 TRP1 HTA1 htb1-R102K) | This study |
| WY326 | Isogenic to PY014, plus pJW636 (CEN6 TRP1 HTA1 htb1-K88Q) | This study |
| WY328 | Isogenic to PY014, plus pJW638 (CEN6 TRP1 HTA1 htb1-R102A/K111A) | This study |
| WY329 | Isogenic to PY014, plus pJW640 (CEN6 TRP1 HTA1 htb1-K49A) | This study |
| WY330 | Isogenic to PY014, plus pJW641 (CEN6 TRP1 HTA1 htb1-K60A) | This study |
| WY331 | Isogenic to PY014, plus pJW642 (CEN6 TRP1 HTA1 htb1-K49R) | This study |
| WY332 | Isogenic to PY014, plus pJW643 (CEN6 TRP1 HTA1 htb1-K60R) | This study |
| WY340 | Isogenic to PY014, plus pKV002 (CEN6 TRP1 HTA1 htb1-K46A) | This study |
| WY341 | Isogenic to PY014, plus pJW635 (CEN6 TRP1 HTA1 htb1-K88R) | This study |
| WY342 | Isogenic to PY014, plus pJW637 (CEN6 TRP1 HTA1 htb1-K111Q) | This study |
| YJ122 | MATaade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 hta1-htb1::HIS3 hta2-htb2::LEU2 SIR4-9× Myc::TRP1 plus pMK001 (CEN6 ADE2 HTA1 HTB1) | This study |
| YJ123 | Isogenic to YJ122, plus pMK019 (CEN6 ADE2 HTA1 htb1-K88A) | This study |
| YJ124 | Isogenic to YJ122, plus pMK020 (CEN6 ADE2 HTA1 htb1-R102A) | This study |
| YJ125 | Isogenic to YJ122, plus pMK021 (CEN6 ADE2 HTA1 htb1-K111A) | This study |
| YJ126 | Isogenic to YJ122, plus pMK022 (CEN6 ADE2 HTA1 htb1-K46A) | This study |
| YJ127 | Isogenic to YJ122, plus pJW639 (CEN6 ADE2 HTA1 htb1-R102A/K111A) | This study |
| MKY006 | MATaade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 hta1-htb1::HIS3 hta2-htb2::LEU2 plus pMK001 (CEN6 ADE2 HTA1 HTB1) | This study |
| UCC3515 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 hml::URA3 | 44 |
| WY230 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 hml::URA3 hta1-htb1::HIS3 | This study |
| WY231 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 hml::URA3 hta1-htb1::HIS3 plus pMK001 (CEN6 ADE2 HTA1 HTB1) | This study |
| MKY059 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 hml::URA3 hta1-htb1::HIS3 hta2-htb2::KanMX plus pMK001 (CEN6 ADE2 HTA1 HTB1) | This study |
| MKY060 | Isogenic to MKY059, plus pMP002 (CEN6 TRP1 HTA1 HTB1) | This study |
| MKY061 | Isogenic to MKY059, plus pKV002 (CEN6 TRP1 HTA1 htb1-K46A) | This study |
| MKY062 | Isogenic to MKY059, plus pJW622 (CEN6 TRP1 HTA1 htb1-K46Q) | This study |
| MKY063 | Isogenic to MKY059, plus pJW625 (CEN6 TRP1 HTA1 htb1-K88A) | This study |
| MKY064 | Isogenic to MKY059, plus pJW626 (CEN6 TRP1 HTA1 htb1-R102A) | This study |
| MKY065 | Isogenic to MKY059, plus pJW627 (CEN6 TRP1 HTA1 htb1-K111A) | This study |
| MKY066 | Isogenic to MKY059, plus pJW632 (CEN6 TRP1 HTA1 htb1-K111R) | This study |
| MKY067 | Isogenic to MKY059, plus pJW633 (CEN6 TRP1 HTA1 htb1-R102K) | This study |
| MKY068 | Isogenic to MKY059, plus pJW635 (CEN6 TRP1 HTA1 htb1-K88R) | This study |
| MKY069 | Isogenic to MKY059, plus pJW636 (CEN6 TRP1 HTA1 htb1-K88Q) | This study |
| MKY070 | Isogenic to MKY059, plus pJW637 (CEN6 TRP1 HTA1 htb1-K111Q) | This study |
| MKY071 | Isogenic to MKY059, plus pJW638 (CEN6 TRP1 HTA1 htb1-R102A/K111A) | This study |
| MKY072 | Isogenic to MKY059, plus pJW640 (CEN6 TRP1 HTA1 htb1-K49A) | This study |
| MKY073 | Isogenic to MKY059, plus pJW641 (CEN6 TRP1 HTA1 htb1-K60A) | This study |
| MKY074 | Isogenic to MKY059, plus pJW642 (CEN6 TRP1 HTA1 htb1-K49R) | This study |
| MKY075 | Isogenic to MKY059, plus pJW643 (CEN6 TRP1 HTA1 htb1-K60R) | This study |
| JHY205 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hht1-hhf1::KAN hhf2-hht2::NAT hta1-htb1::HPH hta2-htb2::NAT pJH33 (CEN URA3 HTA1-HTB1 HHT2-HHF2) | 1 |
| WY504 | Isogenic to JHY205, plus pQQ18 (CEN LEU2 HTA1-HTB1 HHT2 HHF2) | 1 |
| WY505 | Isogenic to JHY205, plus pJW501 (CEN LEU2 HTA1-htb1-K111A HHT2 HHF2) | This study |
| WY506 | Isogenic to JHY205, plus pJW502 (CEN LEU2 HTA1-htb1-K111R HHT2 HHF2) | This study |
| WY507 | Isogenic to JHY205, plus pJW503 (CEN LEU2 HTA1-htb1-K111Q HHT2 HHF2) | This study |
All strains used to examine silencing within the HML locus were derived from the parental strain UCC3515 (44). Both copies of the chromosomal H2A and H2B genes were sequentially deleted from this strain to generate the HML silencing tester strains used in this study. Briefly, the HTA1-HTB1 gene pair was replaced with the HIS3 selectable marker via PCR-mediated gene disruption (4), and the resulting strain was transformed with plasmid pMK001. The HTA2-HTB2 gene pair was then replaced with the KanMX4 antibiotic resistance marker, generating strain MKY059. Plasmids containing wild-type or mutant HTB1 alleles were transformed into MKY059, and these strains were cured of the wild-type pMK001 plasmid by growth on nonselective media and replica plating.
Yeast phenotypic analysis.
All yeast spotting assays were performed essentially as described previously (20). Briefly, cells were grown overnight in yeast extract-peptone-dextrose (YPD) medium, and concentrations were normalized by optical density at 600 nm (OD600) to approximately 108 cells/ml. For initial phenotypic analysis, cells were 10-fold serially diluted and 5 μl of each dilution (approximately 5 × 105 cells were contained in the first spot) was spotted on synthetic complete (SC) medium or SC medium containing 0.015% or 0.025% methyl methanesulfonate (MMS; Sigma-Aldrich), 1.5 mg/ml caffeine (Sigma-Aldrich), 60 μg/ml or 100 μg/ml thiabendazole (Sigma-Aldrich), or 50 mg/ml or 100 mg/ml 6-azauracil (6AU; Sigma-Aldrich). Plates were incubated at 30°C for 2 to 5 days, depending on the growth rate on each test drug. Strains were also tested for sensitivity to heat shock by spotting on SC medium, followed by incubation at 37°C for 2 days. All spotting assays were performed a minimum of three times, and testing was performed in both the PY013 and UCC6389 parental strain backgrounds. Telomeric silencing spotting assays were performed in the same manner except cells were spotted on SC plates or SC plates containing 0.1% 5-FOA (Zymo Research) and incubated at 30°C. For all spotting assays, SC plates were incubated for 2 to 3 days and SC drug plates were incubated for 3 to 4 days prior to photographing.
UV sensitivity assay.
The UV sensitivity phenotype of the histone H2B alanine mutant strains (in the UCC6389 strain background) was examined using a quantitative phenotypic assay, as previously described (39, 40). The UV sensitivity data for histone H2B mutant strains were compared to data for the wild type using a one-tailed t test. Mutant strains were deemed sensitive to UV irradiation if the calculated P value was less than 0.05 for both UV doses (50 and 100 J/m2).
Telomere silencing assays.
Quantitative survival assays were used to determine the magnitude of the change in silencing of the URA3 marker gene caused by the H2B mutations. Cells were grown overnight in YPD medium, serially diluted, and plated on SC medium lacking only tryptophan (SC-Trp), lacking both tryptophan and uracil (SC-Trp-Ura), or lacking tryptophan and containing 0.1% 5-FOA (SC-Trp+FOA). All plates were incubated for 3 days at 30°C, and then colonies were counted to determine the survival rate on each type of medium. Three isolated colonies of each mutant were assayed in this manner. Survival rates of each mutant on SC-Trp medium were set to 100%, and survival rates on SC-Trp-Ura and SC-Trp+5-FOA were compared to the survival rate on SC-Trp. Statistically significant differences in survival rate between each mutant and the wild-type strain were determined using a two-tailed t test.
To examine changes in the silencing of the ADE2 marker gene, each telomeric silencing strain was grown overnight in YPD, serially diluted, and plated on SC medium lacking tryptophan. Plates were incubated at 30°C for 3 days, then transferred to 4°C for an additional 3 days. Each colony was examined and categorized by color as (i) completely white, (ii) completely dark pink, or (iii) light pink or sectored. Colony colors were calculated as a percentage of total colonies. Data shown are the averages of three independent samples, and error bars represent standard deviations. Each mutant strain was compared to the wild type using a two-tailed t test to determine statistically significant changes in the percentage of each colony color.
Semiquantitative analysis of HML silencing.
All strains were grown, diluted, and spotted onto SC medium or SC medium containing 0.1% 5-FOA in the same manner as for the telomere silencing assays. To determine the magnitude of the effect on silencing within this locus, results of the spotting assays were quantitated as previously described (22). Briefly, each mutant was examined to determine the last dilution at which any cells survived. To determine the relative silencing rate, the last surviving dilution on SC-Ura or SC+5-FOA was compared to the survival on SC. P values were determined using a one-tailed t test.
DNA microarray analysis.
Wild-type (PY014) and mutant (WY316 and WY317) yeast strains were grown in parallel in YPD medium to mid-log phase and harvested as previously described (39, 40). Total RNA was isolated by hot acid phenol extraction, as described previously (27). cDNA and biotin-cRNA were prepared from total RNA by following standard protocols (Affymetrix), and the biotin-cRNA was hybridized to the Yeast Genome 2.0 array (Affymetrix).
Microarray data were analyzed and normalized using GeneChip software (Affymetrix). A modified triplicate array error model was used to identify differentially expressed genes as previously described (39, 40), except that the variance of the X statistic was calculated directly from the microarray data sets instead of being estimated by permuting the data.
The chromosome plot of the microarray data for the histone H2B R102A mutant strain was performed as previously described (20). Genes were ordered based on their distance from a telomere end and grouped in consecutive 50-gene bins. The fractions of genes up- and downregulated in the H2B R102A mutant strain relative to the wild type were plotted for each bin. Statistical analysis of the distribution of downregulated genes adjacent to yeast telomeres was performed as previously described (20) using a hypergeometric probability distribution.
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation of myc-tagged Sir4 in the H2B wild-type and mutant strains (PY013 background) was performed as previously described (21) using an anti-myc antibody (catalog number AHO0052; Biosource) bound to magnetic beads (Dynal Biotech). Following cross-link reversal, the ChIP DNA was PCR amplified using primers to amplify a genomic region 372 bp distal from telomere V-L or 525 bp distal from telomere VI-R. Primer sequences are available at http://wyrick.sbs.wsu.edu/H2Bcore/. PCR products were quantified by analysis of ethidium bromide-stained agarose gels using the GelDoc EQ imager with Quantity One software (Bio-Rad). Sir4 binding ratios were normalized to ACT1 or SPS2 internal controls using the input DNA, as previously described (21). One-tailed t tests were used to test whether the observed changes in Sir4 binding ratios between the wild-type and H2B mutant yeast strains were statistically significant.
Sir4 binding within the HML-E silencer region was examined using the same method as that for the telomeric Sir4 chromatin immunoprecipitation assay. Due to the strong binding of the Sir complex to the HML locus, accurate quantification of the Sir4 binding ratio by comparison of the immunoprecipitated and whole-cell extract samples was not feasible. Hence, to determine Sir4 binding enrichment, only the amplification of the test and ACT1 control primers within the immunoprecipitated samples was analyzed. The ratio of product amplification with the HML-E experimental primers relative to that with the ACT1 control primers was calculated, giving a relative enrichment of Sir4 binding within each sample. The calculated enrichments are shown as percentages of the wild type. Error bars represent standard deviations calculated from the raw data, then normalized in the same manner as each average enrichment value.
Cell fractionation.
Cell fractionation and chromatin isolation were performed as previously described (23). The resulting total (data not shown) and chromatin fractions were analyzed by standard SDS-PAGE and immunoblotting techniques. Membranes were probed with an anti-H2B polyclonal antibody (Active Motif; 39237) or anti-H3 polyclonal antibody (Abcam; ab46765).
Yeast histone isolation.
Histones were isolated from yeast strain WY504 essentially as previously described (11). Briefly, 1 liter of yeast strain WY504 was grown in YPD to a density of approximately 108 cells per ml. Cells were collected by centrifugation, washed in double-distilled water, and resuspended in 90 ml room temperature buffer 1 (0.1 mM Tris [pH 9.4], 10 mM dithiothreitol). The cell suspension was incubated at 30°C with rotation for 15 min, and then cells were collected by centrifugation. The cell pellet was washed once with ice-cold buffer 2 (1.2 M sorbitol, 20 mM HEPES [pH 7.4]), collected by centrifugation, and resuspended in 90 ml ice-cold buffer 2. Zymolyase (MP Biomedicals; 320921) was added to a final concentration of 200 μg/ml, and cells were incubated at 30°C with rotation. To examine spheroplasting, 10-μl aliquots of the cell suspension were periodically added to 1 ml of 1% SDS and the absorbance at 600 nm was measured. Once the absorbance had dropped by more than 80% of the original value, the spheroplasts were collected by centrifugation, gently washed in 100 ml ice-cold buffer 3 [1.2 M sorbitol, 20 mM PIPES {piperazine-N,N′-bis(2-ethanesulfonic acid)}, 1 mM MgCl2 (pH 6.8)], and re-collected. The spheroplasts were resuspended in 50 ml ice-cold buffer 4 (nucleus isolation buffer [NIB]; 0.25 M sucrose, 60 mM KCl, 14 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 15 mM MES [morpholineethanesulfonic acid; pH 6.6], 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.8% Triton X-100) to isolate the nuclei and held in ice water for 20 min. The spheroplast suspension was collected by centrifugation, and the NIB wash and ice water incubation were repeated twice. The resulting pellet was washed three times in 50 ml ice-cold buffer 5 (10 mM Tris [pH 8.0], 0.5% NP-40, 75 mM NaCl, 30 mM sodium butyrate, 1 mM PMSF), holding the resuspended pellets in an ice water bath for 15 min following the first two washes and for 5 min following the third wash. The cell suspension was collected by centrifugation, resuspended in 50 ml ice-cold buffer 6 (10 mM Tris [pH 8.0], 0.4 M NaCl, 30 mM sodium butyrate, 1 mM PMSF), held in ice water for 10 min, collected by centrifugation, resuspended once more in buffer 6, and collected immediately. Histones were extracted by resuspension of the pellet in 10 ml cold 0.4 N H2SO4 and incubation at 4°C overnight with rotation. Debris was removed from the sample by centrifugation, and histones were precipitated from the supernatant (containing the extracted histones) by adding trichloroacetic acid to a final concentration of 20%. The sample was held in ice water for 30 min, and precipitated histones were collected by centrifugation. The pellet was washed once with acidified acetone (acetone with 1% HCl) and once with pure acetone, then air dried and resuspended in 10 mM Tris, pH 8.0. The histone concentration was determined by measuring the absorbance at 276 nm (assuming 1 OD unit = 0.45 mg/ml).
Western and dot blot analyses of novel histone H2B modifications.
MCF7 breast cancer cells were grown in Dulbecco's modified Eagle medium (DMEM), supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 10 mM HEPES, 100 units/ml penicillin G sodium, and 100 μg/ml streptomycin sulfate. Once cells had reached 70 to 80% confluence, the medium was replaced with fresh medium containing no drug or trichostatin A (TSA; Sigma; T8552) or valproic acid sodium salt (VPA; Sigma; P4543) at final concentrations of 300 nM and 1 mM, respectively. Cells were incubated for 12 h with no drug, VPA, or TSA, harvested in 100 μl of sample buffer (20 mM Tris [pH 8.0], 2 mM EDTA, 2% SDS, 1 mM dithiothreitol, and 20% glycerol), and briefly sonicated to reduce viscosity. Protein concentrations were determined by measuring the absorbance at 280 nm, and 40 μg of each sample or 750 ng of recombinant human histone H2B (New England BioLabs; M2505S) was examined using standard SDS-PAGE and Western blotting techniques. Membranes were probed with antitubulin monoclonal (Abcam; ab6160), anti-H2B polyclonal (Active Motif; 39125), anti-H2B acetyl K46 polyclonal (Active Motif; 39571), or anti-H2B dimethyl K46 (Active Motif; 39567) antibodies.
Custom antibodies against dimethylated H2B K111 peptide [PGELA-K(me2)-HAVSEGTC] or acetylated H2B K111 peptide [PGELA-K(Ac)-HAVSEGTC] were generated by YenZym Antibodies LLC. These antibodies were used for Western blotting of yeast whole-cell extracts, isolated using a standard glass bead lysis protocol, or purified yeast histones (see the yeast histone isolation protocol described above).
In order to determine the specificity of the anti-H2B K111me2 and anti-H2B K111ac antibodies, H2B K111 mutant strains were made in the BY4741 yeast strain background, in which H2B K111 dimethylation was previously reported (56). Histone H2B K111 mutations were generated on plasmid pQQ18 (1), which carries one wild-type copy of each histone gene, by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene). The resulting wild-type and H2B K111 mutant plasmids were transformed into parent strain JHY205, and the final test strains were constructed by plasmid shuffling on media containing 5-FOA. SDS-PAGE and Western blot analysis were conducted on whole-cell extracts isolated from wild-type or H2B K111 mutant strains using anti-H2B polyclonal (Active Motif; 39237), anti-H2B K111me2 (YenZym), or anti-H2B K111ac (YenZym) antibodies.
For the dot blot analyses of the anti-H2B K111me2 and anti-H2B K111ac antibodies, various amounts of unmodified H2B peptide (PGELAKHAVSEGTC), methylated or acetylated H2B peptide, and an H3 K4me2 peptide (Upstate; 12-460) were spotted directly onto a nitrocellulose membrane and probed with the appropriate antibody. Initial dot blot analysis of the anti-H2B K111ac antibody revealed a high degree of interaction with the unmodified H2B peptide, so this antibody was preincubated with the unmodified H2B peptide (3 μg/ml) to block nonspecific detection of the histone H2B peptide. Preincubation was also performed prior to using this antibody in Western blot analysis.
Microarray data accession numbers.
The microarray data presented in this paper have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and can be accessed through GEO series accession numbers GSE15202 and GSE15203.
RESULTS
Phenotypic characterization of histone H2B proteins with mutations in core modifiable residues.
Mass spectrometry studies of human and bovine histones have identified multiple residues that are posttranslationally modified in the core fold domain of histone H2B (5, 12, 24, 30, 51, 57). Of these, six residues are conserved in yeast histone H2B (Fig. 1 A). These conserved, modifiable residues are located either on the DNA-associated lateral surface of the nucleosome (yeast H2B K46, K49, K60, and K88) or on the solvent-accessible nucleosome face (H2B R102 and K111) (Fig. 1B). In addition, there is evidence that bovine histone H2B K116 is acetylated (57), but this residue (equivalent to yeast H2B R119) is not conserved in S. cerevisiae and thus was not characterized in this study. As a first step toward understanding the function of these histone H2B residues, we systematically mutated each modifiable residue in yeast histone H2B (Fig. 1). Each residue was initially mutated to alanine. Lysine residues that were acetylated in mammalian cells (e.g., yeast H2B K88 and K111) were also mutated to arginine (to mimic the unmodified side chain) and glutamine (to mimic the acetylated side chain). Lysine and arginine residues that were methylated in mammalian cells were also mutated to arginine and lysine, respectively.
FIG. 1.
Identification of conserved residues in the core fold domain of histone H2B that are posttranslationally modified in mammalian cells. (A) Alignment of histone H2B sequences. Posttranslationally modified residues in the core fold domain are in color. Modifications that are not conserved in yeast histone H2B or that are located in the N- and C-terminal domains are not depicted. (B) Location of core modifiable histone H2B residues in the yeast nucleosome structure. Conserved sites of posttranslational modifications in the core fold domain of histone H2B are indicated. Histone H2B residues are depicted as aqua cartoons; other histone residues are shown as cartoons in white. Structural data (PDB code 1id3; www.rcsb.org) are from reference 50. The image was drawn using POLYVIEW (41).
We initially surveyed the phenotypes of the histone H2B point mutants in various growth conditions and in the presence of DNA-damaging agents. None of the H2B mutant strains exhibited detectable sensitivity to caffeine, thiabendazole, hydroxyurea, 6-azauracil, or incubation at 37°C (data not shown).
The histone H2B K111A mutant strain was sensitive to the DNA-damaging agent MMS (Fig. 2 A), suggesting that this histone H2B residue may play a role in the response to MMS-induced DNA lesions. We should note that the MMS sensitivity phenotype was considerably stronger in the S288C strain background (WY191) (Fig. 2A) than in the W303 strain background (WY317) (data not shown). The histone H2B K49A mutant strain showed significant sensitivity to UV irradiation compared to the wild type (Fig. 2B) (P < 0.01). In addition, the H2B R102A and H2B K111A mutant strains displayed a slight sensitivity to UV-induced irradiation (Fig. 2B). The UV and MMS sensitivity phenotypes were significantly enhanced in an H2B R102A/K111A double mutant strain (data not shown).
FIG. 2.
Mutations in histone H2B core residues render yeast cells sensitive to DNA-damaging agents. (A) Growth phenotypes of histone H2B alanine mutant strains in the presence of the DNA-damaging agent MMS. Tenfold serial dilutions of each strain (starting with approximately 5 × 105 cells) were spotted on SC-Trp medium or SC-Trp medium containing the indicated concentrations of MMS. (B) Histone H2B core mutant strains were examined for sensitivity to UV irradiation at doses of 0, 50, and 100 J/m2. The percentage of viable cells in each strain was plotted as a function of UV dose. Only the histone H2B alanine mutant strains with a significant UV sensitivity phenotype are depicted. WT, wild type.
Histone H2B core mutants affect silencing at yeast telomeres and the HML silent mating locus.
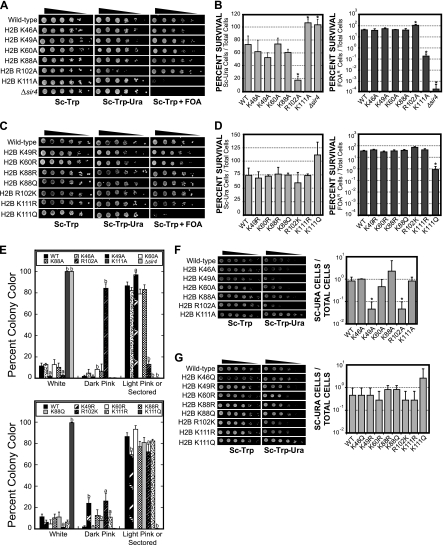
Previous studies have shown that core modifiable residues in histone H3 (e.g., H3 K56 and H3 K79) regulate the epigenetic silencing of yeast telomeres (19, 34, 49, 55). Hence, we tested whether mutations in histone H2B core modifiable residues affected telomeric silencing. To examine changes in telomeric gene silencing, we employed a reporter strain in which the URA3 and ADE2 genes were integrated adjacent to telomeres VII-L and V-R, respectively, and the chromosomal copies of the HTA1-HTB1 and HTA2-HTB2 genes had been deleted (14). Plasmids containing HTA1-htb1 alleles were transformed into the reporter strain, and the resulting strains were assayed for their effect on repression of the URA3 and ADE2 genes. Mutants that affected the level of repression of the ADE2 gene were identified by changes in colony color, and those that affected the URA3 gene were identified by their effect on the strain survival rate on medium either lacking uracil or containing 5-FOA. As the silencing status of heterochromatic regions is inherited in a semistable manner, a switch between transcriptionally active and silenced states may occur between generations (16). Therefore, a population of wild-type cells will contain a mixture of URA3+ and ura3 cells, and thus a fraction of the wild-type population will be able to grow in the presence of 5-FOA and a fraction will be able to grow in the absence of uracil. Thus, changes in the survival rate of a mutant strain on either type of medium, compared to that of the wild-type strain, indicate a change in the balance between the active and silenced states of the reporter genes.
We initially screened H2B proteins with alanine mutations in core modifiable residues. The data revealed that the H2B K111A mutant disrupted silencing of the telomere-located URA3 gene, rendering the mutant strain hypersensitive to 5-FOA (Fig. 3 A). Quantitative silencing assays indicated that the H2B K111A mutant strain displayed greater-than-200-fold-increased sensitivity to 5-FOA over the wild type (Fig. 3B), confirming that telomeric silencing was significantly disrupted. However, the H2B K111A mutant did not completely abolish silencing, as suggested by comparison with the Δsir4 negative control (Fig. 3B).
FIG. 3.
Effects of histone H2B core mutants on telomeric and HML silencing. (A) Tenfold serial dilutions (starting with approximately 5 × 105 cells) of each histone H2B alanine mutant strain were spotted on SC-Trp, Sc-Trp-Ura, or SC-Trp+5-FOA agar plates, as indicated. (B) Quantification of telomeric silencing phenotypes in the histone H2B mutant strains. Each data point represents the average of three replicate experiments. Error bars indicate the standard deviations, and strains that exhibit sensitivity significantly (P < 0.05) different from that of the wild type are indicated by asterisks. (C and D) Silencing phenotypes of lysine-to-arginine, lysine-to-glutamine, and arginine-to-lysine H2B mutants. Spotting and quantitative assays were performed as described for panels A and B. (E) Effects of histone H2B core mutants on silencing of the telomeric ADE2 reporter gene. Histone H2B mutants were serially diluted and grown for 3 days on SC-Trp plates, then placed at 4°C for an additional 3 days. The color of each colony was then categorized as white, dark pink, or light pink/sectored. Data shown are each colony color expressed as a percentage of total surviving colonies and represent the averages of three independent samples. Error bars represent standard deviations, and mutants that were significantly different from the wild type are indicated by “a” (P < 0.05) or “b” (P < 0.01) above the column. (F and G) Effects of histone H2B core mutants on silencing within the HML locus. Tenfold dilutions of the wild-type (WT) or histone H2B mutant HML silencing tester strains were spotted on SC-Trp or Sc-Trp-Ura agar plates. The silencing phenotypes of the histone H2B mutant strains were quantified by following a previously published method (22). Each data point represents the average of five replicate experiments, and error bars indicate the standard deviations. Strains that exhibited significantly (P < 0.05) different sensitivities than the wild type are indicated by asterisks.
In contrast, the H2B R102A mutant strain grew more robustly on 5-FOA but displayed a significant growth defect in media lacking uracil (Fig. 3A). These data suggest that the H2B R102A mutant enhanced silencing of the URA3 reporter gene. Quantitative analysis also demonstrated that the H2B R102A mutant strain was approximately 5-fold more sensitive to growth on media lacking uracil, indicating that this mutant enhanced telomeric silencing (Fig. 3B). Similar results were obtained for the H2B K111A and H2B R102A mutants using a colony color assay to measure expression of the ADE2 telomeric marker (Fig. 3E). The histone H2B K46A, H2B K49A, H2B K60A, and H2B K88A mutants did not cause a significant change in telomeric silencing of the URA3 locus (Fig. 3A and B), although a slight enhancement of silencing of the ADE2 telomere reporter gene was observed in the H2B K49A mutant (Fig. 3E).
Intriguingly, the silencing defect of the H2B K111A mutant strain was also observed in the H2B K111Q mutant strain but not in the H2B K111R mutant strain (Fig. 3C and D). The H2B K111Q mutant was approximately 40-fold more sensitive to 5-FOA than the wild type (Fig. 3C and D) and caused a similar defect in the silencing of the ADE2 telomeric reporter (Fig. 3E). In contrast, the H2B K111R mutant strain displayed wild-type levels of telomeric silencing (Fig. 3C to E). As the lysine-to-glutamine mutation is commonly used to mimic acetylation of a lysine residue, these results suggest that acetylation of histone H2B K111 could oppose the establishment or maintenance of telomeric silencing.
The histone H2B R102K mutant did not enhance or otherwise alter telomeric silencing of the URA3 reporter gene (Fig. 3C and D), unlike the H2B R102A mutant. However, this mutation did result in a slight increase in the silencing of the ADE2 telomeric reporter gene (Fig. 3E). Unfortunately, no mutation can adequately mimic methylation of an arginine residue. For this reason, we are unable to delineate the potential role of H2B R102 methylation in silencing. Finally, lysine-to-arginine or lysine-to-glutamine mutations in other histone H2B residues (e.g., H2B K88R and H2B K88Q) did not have a significant effect on telomeric silencing (Fig. 3C to E).
We also tested whether the histone H2B core mutants affected the silencing of a URA3 reporter gene integrated at the HML silent mating loci (44). We observed a significant enhancement in silencing at the HML locus by the H2B R102A and H2B K49A mutants (Fig. 3F). In contrast, the H2B K111A and H2B K111Q mutants did not cause a significant defect in HML silencing, as measured by growth on 5-FOA-containing media (data not shown). The URA3 gene at the HML locus is strongly silenced (45); hence, the H2B K111A or K111Q mutation may not be sufficient to disrupt the silencing at this locus. However, we should note that the H2B K111A and K111Q mutant strains grew qualitatively faster on SC-Ura media (Fig. 3F and G), suggesting that there may be a slight defect in HML silencing in these mutant strains.
Histone H2B core residues have opposing effects on Sir4 binding to telomeres and the HML locus.
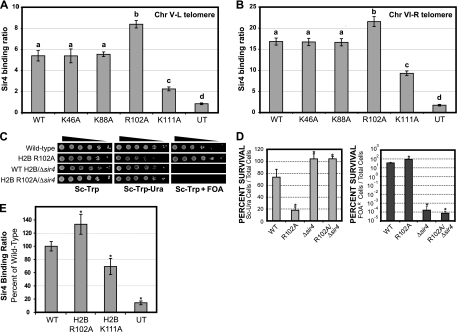
We hypothesized that the histone H2B core mutants might affect telomeric silencing by altering the binding of the Sir silencing complex to telomeric regions of yeast chromosomes. To test this hypothesis, we used ChIP assays to measure the binding of Sir4 in the histone H2B mutant and wild-type strains. Sir4 was used as a proxy for the binding of the entire Sir complex (i.e., Sir2, Sir3, and Sir4). Sir4 was tagged with a 9-myc epitope in the histone H2B mutant strain background, and anti-myc (9E10) antibodies were used to perform the ChIP experiments. Sir4 binding was measured at two distinct telomere regions: 372 bp distal from telomere V-L (Fig. 4 A) and 525 bp distal from telomere VI-R (Fig. 4B). These telomere regions were chosen as they have been previously used in Sir ChIP assays (20, 21, 26, 55).
FIG. 4.
Histone H2B R102 and K111 mutants affect Sir4 binding to yeast telomeres and HML loci. ChIP analysis was used to determine the level of Sir4 protein binding to telomere V-L (Chr V-L) (A) and telomere VI-R (Chr VI-R) (B). ChIP assays were performed for wild-type (WT) and histone H2B mutant strains. An untagged strain (UT) was used as a control. Sir4 binding ratios were normalized to ACT1 or SPS2 internal controls using the input DNA, as previously described (21). Error bars represent the standard deviations from three independent samples. Each group of statistically distinct samples (P < 0.05) is indicated by a separate letter above the data (a, b, c, or d). (C and D) The histone H2B R102A mutant enhances telomeric silencing in a SIR4-dependent manner. Telomeric silencing assays and quantitative analysis were performed on the H2B R102A Δsir4 double mutant, as described for Fig. 3A to D. The results were compared to those for the histone H2B R102A and Δsir4 individual mutants. Asterisks indicate samples that are statistically distinct from the wild type (P < 0.05). (E) Mutations in histone H2B R102 and K111 affect Sir4 binding to the yeast HML locus. ChIP assays were used to determine the level of Sir4 protein binding near the HML-E silencer region. ChIP analysis was performed for wild-type and histone H2B mutant strains, with an untagged strain as a control. The Sir4 binding ratios were normalized to ACT1 internal controls in the immunoprecipitated DNA samples. Error bars represent the standard deviations from three independent samples, and asterisks indicate samples that were significantly (P < 0.05) different from the wild type.
The results, shown in Fig. 4A and B, indicate that the histone H2B K111A mutant caused a significant decrease in Sir4 binding to both telomere V-L (2.4-fold decrease; P = 1.9 × 10−3) and telomere VI-R (1.8-fold decrease; P = 2.4 × 10−4). These results are in accordance with the decrease in telomeric silencing observed in this H2B mutant strain. However, comparison of these Sir4 binding data with those for the untagged control (Fig. 4A and B) suggests that the H2B K111A mutant weakened but did not abolish Sir4 binding to telomere regions. This was particularly apparent in the data for the telomere VI-R chromosomal region, in which the Sir4 binding ratio was ∼5-fold greater than that observed in the untagged control (Fig. 4B) (P = 3.0 × 10−4).
The histone H2B R102A mutant caused a significant increase in the binding of Sir4 to both telomere regions (Fig. 4A and B) (P < 0.005). These results suggest that the H2B R102A mutant enhances telomeric silencing by increasing the association of the Sir proteins with telomeric regions. This model is supported by our observation that deletion of the SIR4 gene completely abrogates telomeric silencing in the histone H2B R102A mutant background (Fig. 4C and D). Taken together, these results indicate that enhancement of telomeric silencing by the H2B R102A mutant is mediated by the Sir complex. Finally, the H2B K46A and H2B K88A mutants did not have a significant effect on Sir4 binding (Fig. 4A and B) or telomeric silencing (Fig. 3).
We also tested whether the H2B R102A and K111A mutants affected Sir complex binding to the HML silent mating locus. Sir4 ChIP experiments were performed as described above, except that Sir4 binding at a region adjacent to the HML-E silencer was measured (7, 31) and a different quantification method was used (see Materials and Methods). The results indicated that the H2B K111A mutant caused a significant decrease in Sir4 binding to the HML locus, while the H2B R102A mutant significantly enhanced Sir4 binding to the HML locus (Fig. 4E). These results parallel the effects observed at yeast telomeres, indicating that the H2B K111 and R102 residues have opposing effects on Sir4 binding at multiple silenced loci.
The histone H2B R102A mutant represses the transcription of genes adjacent to yeast telomeres.
We characterized the effects of the histone H2B R102A and K111A mutants on genome-wide expression in yeast using Affymetrix microarrays. Triplicate RNA samples from the histone H2B R102A mutant, H2B K111A mutant, and wild-type strains were profiled. The resulting microarray data were analyzed using a triple array error model (39, 40) in order to identify genes with a significant change in mRNA levels in the histone mutant strains. A significance (P value) threshold of 0.001 was used to identify differentially expressed genes.
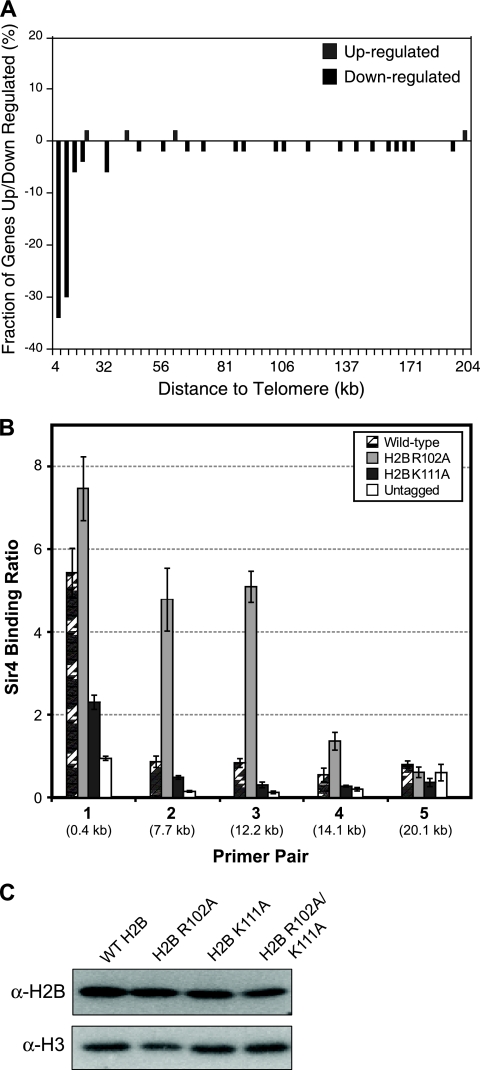
Analysis of the histone H2B R102A microarray data revealed that the mRNA levels of 9 genes were upregulated and the mRNA levels of 84 genes were downregulated in the histone mutant strain relative to the wild type. While the up- and downregulated genes had relatively little functional similarity (data not shown), we found that a large fraction of the downregulated genes were clustered adjacent to telomere regions of yeast chromosomes. A chromosome plot of the histone H2B R102A microarray data (Fig. 5A) indicated that the H2B R102A mutant repressed the expression of genes located within 10 kb of yeast telomeres. Indeed, 36% of the genes located in telomere-proximal regions (defined as within 10 kb of a telomere end) were downregulated in the H2B R102A mutant, compared to a genome-wide average of only ∼1.5%, a highly significant enrichment (P = 6.7 × 10−31). A similar result was obtained when we analyzed the data using the nonparametric Wilcoxon rank sum test. This test showed that there was a significant bias of downregulated genes adjacent to yeast telomeres in the H2B R102A mutant (P < 10−18). These data are in accordance with our observation that the H2B R102A mutant enhances the silencing of telomere-located reporter genes (Fig. 3) and suggest that the H2B R102A mutant may stimulate the spreading of the Sir complex into euchromatin regions adjacent to yeast telomeres.
FIG. 5.
(A) The histone H2B R102A mutant represses the expression of telomere-proximal genes. Depicted is the chromosome plot of the genome-wide expression changes for the histone H2B R102A mutant relative to the wild type. A histogram of the fraction of genes whose mRNA levels are up- or downregulated is plotted as a function of their distance from a chromosome end. (B) The histone H2B R102A mutation causes spreading of Sir4 beyond the telomeres. ChIP analysis was used to examine the level of Sir4 protein binding to five regions on telomere V-L in wild-type and histone H2B R102A and K111A mutant strains. An untagged strain was used as a control, and Sir4 binding ratios were normalized to ACT1 using the input DNA, as previously described (21). Error bars represent the standard deviations from three independent samples. (C) Histone H2B core mutations do not alter the stability of the histone H2B protein or its incorporation into chromatin. Chromatin extracts from wild-type (WT) histone H2B and the R102A, K111A, and R102A/K111A mutants were prepared using a cell fractionation protocol previously described (23) and examined by Western blotting. The level of histone H2B protein was not significantly changed in the histone mutant strains compared to that in the wild type. Histone H3 was used as a loading control. α, anti.
Analysis of the histone H2B K111A microarray data indicated that the mRNA levels of 12 genes were upregulated and those of 38 genes downregulated in the histone H2B mutant strain compared to the wild type. Unlike those in the H2B R102A mutant strain, however, the differentially expressed genes in the H2B K111A mutant strain were not clustered adjacent to yeast telomeres (P = 0.47) (data not shown). It is possible that the H2B K111A silencing defect was not significant enough to be detected in the microarray data, at least among the set of genes categorized as differentially expressed. We should note that similar observations have been made for other histone mutants. For example, the histone H3 K9,14,18,23G mutant has a significant telomeric silencing phenotype (48), but this silencing defect was not observed in microarray profiling experiments (27).
The histone H2B R102A mutant elicits spreading of Sir4 into genomic regions adjacent to yeast telomeres.
The microarray results indicate that the H2B R102A mutant causes a decrease in the expression of many yeast genes located adjacent to yeast telomeres (Fig. 5A). We tested whether this was due to spreading of the Sir complex from telomere regions in the H2B R102A mutant strain. We tested Sir4 binding by ChIP assay using primer pairs located at increasing distances from the V-L telomere end (Fig. 5B), in accordance with our previously published protocol (20). In the wild-type strain, Sir4 binding was limited to the most telomere-proximal DNA region (primer set 1, located 0.37 kb from telomere V-L), as expected (20). In the H2B R102A mutant strain, increased Sir4 binding was observed up to 14 kb from the chromosome end (primer sets 2 to 4) (Fig. 5B). In contrast, the H2B K111A mutant caused a decrease in Sir4 binding relative to that in the wild-type strain (Fig. 5B). These results indicate that the H2B R102A mutant stimulates the spreading of Sir4 from telomere regions into the chromosome, which could explain the decreased levels of mRNA of telomere-proximal genes in the H2B R102A microarray data set.
It is important to note that that the expression levels of the SIR2, SIR3, and SIR4 genes were unchanged in the H2B R102A or K111A mutant strains (data not shown). In addition, we have found that the histone H2B R102A or K111A mutants do not affect histone H2B protein levels or its incorporation into chromatin (Fig. 5C). Taken together, these data suggest that the H2B R102 and K111 residues may play a direct role in regulating telomere silencing and Sir complex binding in wild-type chromatin.
Epistasis analysis of the histone H2B R102A and K111A mutants.
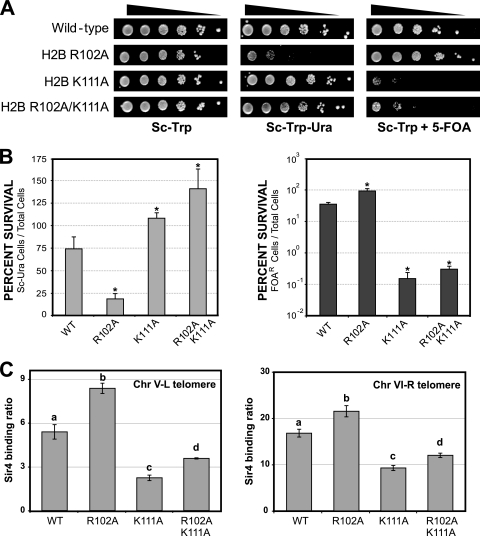
Our results indicate that the histone H2B R102 and K111 residues have opposing effects on telomeric silencing and Sir complex binding to telomeric regions. In order to test for epistasis between these two H2B residues, we constructed a histone H2B R102A/K111A double mutant and analyzed its effect on telomeric silencing. As shown in Fig. 6A, the H2B R102A/K111A double mutant caused a defect in telomeric silencing, as determined by measuring 5-FOA sensitivity. In addition, the H2B R102A/K111A double mutant did not show sensitivity to growth on media lacking uracil in the telomeric silencing tester strain (Fig. 6A).
FIG. 6.
Effects of the histone H2B R102A/K111A double mutant on telomeric silencing and Sir4 binding. (A and B) Telomeric silencing assays were performed as described for Fig. 3. Results for the H2B R102A/K111A double mutant were compared to the results for the H2B R102A and H2B K111A single mutants (Fig. 3). Asterisks indicate strains that are significantly (P < 0.05) different from the wild type (WT). (C) ChIP analysis of Sir4 binding to the telomere regions of telomeres V-L and VI-R. ChIP assays were performed as described for Fig. 4A and B. Data for the H2B R102A and H2B K111A single mutants are from Fig. 4A and B. Each group of statistically distinct samples (P < 0.05) is indicated by a separate letter above the data (a, b, c, or d).
We also examined the effects of the histone H2B R102A/K111A double mutant on the binding of Sir4 to telomere regions. Sir4 ChIP analysis of the H2B double mutant revealed that Sir4 binding to telomere regions was significantly decreased compared to that for the wild type (Fig. 6C). However, the Sir4 binding level was significantly greater in the H2B R102A/K111A double mutant strain than in the H2B K111A single mutant strain (Fig. 6C). Indeed, the relative increase in Sir4 binding between the H2B R102A/K111A double mutant and H2B K111A single mutant strains was roughly equivalent to the increase observed between the H2B R102A single mutant and the wild-type strains (data not shown). Taken together, these data indicate that the H2B R102A mutant enhances Sir4 binding to yeast telomeres in the silencing-defective H2B K111A mutant strain, suggesting that the H2B R102A and H2B K111A mutants may independently regulate silencing and Sir4 binding to yeast telomeres.
Screening for putative histone H2B core modifications.
Yeast histone H2B K111 is homologous to mammalian histone H2B K108 (Fig. 1), which has been reported to be acetylated in bovine histone H2B (57) and mono- and dimethylated and acetylated in human histone H2B (5, 24, 51). There is a recent report that yeast histone H2B K111 may be dimethylated in Saccharomyces cerevisiae, based on mass spectrometry studies of yeast histones (56).
To test whether histone H2B K111 is posttranslationally modified in yeast, we generated antibodies that recognized histone H2B peptides containing H2B K111-me2 or H2B K111Ac modifications (Fig. 7A). While we were able to detect a band of the appropriate size in yeast cell extracts using these antibodies, this band was not abolished in the histone H2B K111 mutants (Fig. 7B), suggesting that the antibodies were not specifically detecting the H2B K111 modifications. Indeed, we were unable to detect any signal in purified yeast histones using the anti-H2B K111me2 antibody (Fig. 7C). Hence, we were unable to validate the presence of these H2B K111 modifications in yeast.
FIG. 7.
Western blot analysis of potential histone H2B posttranslational modification sites. (A) Custom antibodies were designed to detect either dimethylation or acetylation of yeast histone H2B K111. Peptide dot blots confirm that each antibody has a higher specificity for the modified peptide than for the unmodified peptide and that the anti-H2B K111ac antibody does not nonspecifically bind to a histone H3 peptide dimethylated at lysine 4. (B) Custom anti-H2B K111me2 and anti-H2B K111Ac antibodies do not specifically detect modifications in yeast histone H2B. Wild-type and histone H2B K111 mutant yeast strain whole-cell extracts were examined by Western blotting using anti-H2B K111me2, anti-H2B K111Ac, or anti-H2B antibodies. (C) Histones were purified from wild-type yeast cells and analyzed by Western blotting using the H2B K111me2 antibody. (D) Human histone H2B K46 is acetylated and dimethylated. Anti-H2B K46Ac and anti-H2B K46me2 antibodies were used to confirm the presence of both modifications on human histone H2B (hH2B) K46 in MCF7 breast cancer cell extracts. Treatment with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) or valproic acid (VPA) resulted in an increase in histone H2B K46 acetylation. Neither acetylation nor dimethylation was detected on recombinant histone H2B (rH2B).
Mass spectrometry studies of human histone H2B (hH2B) have also identified a methylation site at hH2B K46 (5), corresponding to yeast histone H2B K49 (Fig. 1A). Using commercially available antibodies, we confirmed that hH2B K46 is dimethylated in the human MCF-7 breast cancer cell line (Fig. 7D). We have also found that hH2B K46 is acetylated in human cells and that this acetylation is stimulated by treatment of MCF-7 cells with the histone deacetylase inhibitors trichostatin A and valproic acid (Fig. 7D). Because the antibody recognition epitope is not conserved in yeast histone H2B (data not shown), we were unable to use these antibodies to test whether yeast histone H2B K49 is dimethylated or acetylated.
DISCUSSION
Previous studies have identified novel posttranslational modifications in the core fold domain of histone H2B (5, 12, 24, 30, 51, 57). However, the functional significance of these histone modification sites was unknown. In this study, we have mutated conserved modification sites in yeast histone H2B and characterized the effects of these mutants on yeast gene expression, silencing, and DNA damage response. We report that two conserved modifiable residues, histone H2B R102 and H2B K111, have important roles in these biological processes. In addition, we find that H2B K49 plays a potentially important role in the response to UV-induced DNA damage.
Role of histone H2B K49 in the DNA damage response and gene silencing.
Our data indicate that the H2B K49A mutant causes a significant growth defect following UV irradiation in yeast. This result suggests that H2B K49 may play a significant role in the cellular response to DNA damage. Our data also hint that this histone H2B residue regulates gene silencing. In the H2B K49A mutant strain, we observe a slight increase in the silencing of telomere-located reporter genes and a considerably greater increase in the silencing of a reporter gene located in the HML silent mating locus.
We have used modification-specific antibodies to confirm that the homologous residue in human histone H2B (hH2B K46) is acetylated and methylated in human cells. The yeast histone H2B K49 residue is located on the nucleosome lateral surface, immediately adjacent to the nucleosomal DNA (Fig. 1). Hence, acetylation of this residue could potentially affect the stability or positioning of the DNA in the nucleosome. Intriguingly, our preliminary studies suggest that hH2B K46 acetylation levels may be altered in human cells in response to UV-induced DNA damage (data not shown). Future studies will examine the role of these novel modifications in regulating gene silencing and the DNA damage response.
Role of histone H2B R102 in gene silencing.
Our data indicate that the histone H2B R102A mutant enhances the silencing of reporter genes located in yeast telomeres or the HML locus and repressed the expression of endogenous genes located adjacent to yeast telomeres. The observed increase in telomeric silencing is likely mediated by the Sir silencing complex, as we find increased binding of Sir4 to yeast telomere regions in the H2B R102A mutant strain and deletion of the SIR4 gene abrogates telomeric silencing in the H2B R102A mutant strain. The simplest interpretation of these data is that the function of the histone H2B R102 residue in wild-type chromatin is to oppose telomeric silencing by inhibiting the binding of Sir4 to telomere regions. In accordance with this model, we observed that the H2B R102A mutant stimulates the spreading of Sir4 into regions adjacent to yeast telomeres.
The yeast histone H2B R102A mutation eliminates a conserved arginine residue that is methylated in bovine histone H2B (H2B R99 [57]), although the modification status of this residue in yeast histone H2B has yet to be reported. Intriguingly, the more conservative H2B R102K mutant does not significantly affect the silencing of the URA3 telomere reporter gene or the HML locus and causes only a slight increase in the telomeric silencing of the ADE2 reporter gene (Fig. 3E). These results suggest that the positively charged R102 side chain may be specifically required to inhibit gene silencing.
Previous studies have shown that mutations in conserved residues in the core fold domain of histone H4 (i.e., H4 R39K and H4 H75Y) can enhance telomeric silencing in a Sir-dependent manner (46, 53). However, these histone H4 residues are buried in the nucleosome core and likely regulate intranucleosomal histone-histone interactions. In contrast, the H2B R102 residue is located on the solvent-exposed nucleosome face and thus may directly regulate the recruitment of the Sir complex or other proteins to silenced regions in the yeast genome. Other studies have demonstrated that mutations that alter the charge in the LRS surface of the nucleosome (e.g., H3 Q76R, H3 D77N, and H3 D81G) can also enhance telomeric silencing and Sir complex binding by reducing electrostatic repulsion between Sir3 and the LRS domain (35, 36). We believe a similar mechanism may be responsible for the enhancement of silencing in the H2B R102A mutant strain (see below).
Functional analysis of the histone H2B K111 mutants.
The histone H2B K111A mutant disrupts the silencing of telomeric reporter genes and significantly diminishes the binding of Sir4 to yeast telomere regions. The K111 residue may play a direct role in telomeric silencing in wild-type chromatin. The H2B K111A mutant also causes an MMS sensitivity phenotype, suggesting that it may impair the response of these cells to DNA damage. We should note, however, that in a previous study (29) no MMS phenotype was observed for this histone mutant. Our microarray data indicate that the H2B K111A mutant affects the expression of 50 yeast genes, although the magnitude of the changes in mRNA levels was relatively modest. Additionally, a previous study has shown that the H2B K111A mutant has an Spt− phenotype (29), again suggesting that this histone H2B residue plays a role in transcriptional regulation.
Our results indicate that the H2B K111R mutant, which mimics the unmodified side chain, does not affect telomeric silencing. In contrast, the H2B K111Q mutant, which mimics an acetylated side chain, causes a disruption in telomeric silencing roughly comparable to the phenotype observed in the H2B K111A mutant strain. These results suggest that the unmodified side chain of H2B K111 is required for telomeric silencing and that acetylation of H2B K111 could potentially oppose silencing.
While previous studies have found that yeast H2B K111 is methylated in yeast (56) and that the homologous H2B residue (hH2B K108) is methylated and acetylated in mammalian cells (5, 24, 51), we have been unable to validate these findings in yeast using H2B K111 modification-specific antibodies. These experiments do not rule out the possibility that these posttranslational modifications are present in yeast histones, as the modifications may be present at too low abundance to be detected by Western blotting. It is also possible that the antibodies may not have sufficient specificity or affinity to detect these posttranslational modifications in yeast histones. Finally, it is possible that a different posttranslational modification may occur at H2B K111. For example, a recent study has reported that H2B K111 is ubiquitylated in yeast (15). It is possible that ubiquitylation may regulate the function of this histone H2B residue.
Regulation of gene silencing by histone H2B core residues.
Our data indicate that the histone H2B R102 and K111 residues have opposing roles in regulating silencing and Sir4 binding to telomere regions in yeast. A critical question that remains to be addressed is the mechanism by which these residues regulate silencing. While an intriguing model is that the posttranslational modification status of H2B R102 and K111 could directly regulate the binding of the Sir proteins, these modifications have not been confirmed in yeast (see above).
A second possible model is that H2B R102 and K111 could affect the levels of other histone modifications known to regulate telomeric silencing, such as histone H3 K4 methylation or H2B K123 monoubiquitylation. A previous study has demonstrated that the histone H2B R102A and H2B K111A mutants do not alter the levels of histone H3 K4 methylation in yeast (32). This result indicates that H2B K123 monoubiquitylation, which is required for H3 K4 methylation (47), is also unlikely to be altered in the mutant strains. Our data confirm that the H2B K111A mutant does not affect H2B K123 monoubiquitylation levels (data not shown).
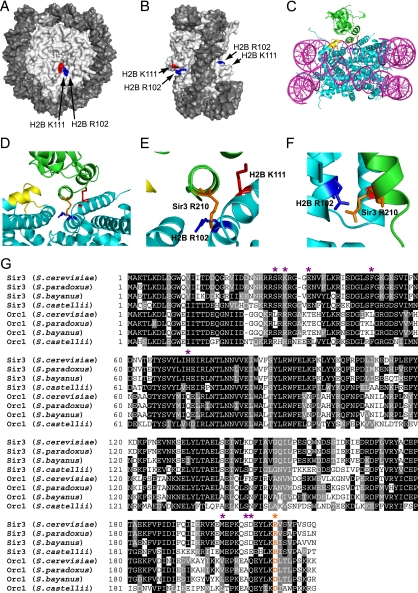
Although histone H2B R102 and H2B K111 are located in different H2B α-helices (α3 and αC, respectively), these residues are located in close juxtaposition to one another on the solvent-exposed face of the nucleosome (Fig. 8A and B). Their location in the nucleosome structure suggests that the H2B R102 and K111 residues could mediate internucleosome interactions or potentially regulate the binding of nucleosome-associated proteins. In the yeast nucleosome structure, the H2B αC helix was found to mediate internucleosome interactions that were critical for the crystallographic packing of yeast nucleosomes (50).
FIG. 8.
(A and B) Location of histone H2B R102 (blue) and H2B K111 (red) residues in the yeast nucleosome crystal structure (1id3 [50]). The image was drawn using POLYVIEW (41). (C to F) Model of histone H2B R102 and K111 interaction with nucleosome-bound Sir3. MacPyMOL (http://www.pymol.org) was used to generate images showing the interaction of histone H2B R102 and K111 with Sir3, based on a published model of Sir3 docking to the yeast nucleosome (35). DNA is in magenta, histone residues are in cyan, and Sir3 protein is in green. The LRS domain is highlighted in yellow, histone H2B R102 is depicted in dark blue, histone H2B K111 is shown in red, and Sir3 R210 is highlighted in orange. Panels D to F are magnified or rotated images of the Sir3-H2B interactions depicted in panel C, with the DNA hidden from view. (G) Sequence alignment of the BAH domain protein sequences of Sir3 and Orc1 from Saccharomyces cerevisiae, Saccharomyces paradoxus, Saccharomyces bayanus, and Saccharomyces castellii. Multiple sequence alignment was performed using CLUSTALW, with default parameters. Sites of orc-like and semi-orc-like suppressor mutants are highlighted with purple asterisks (data from reference 35). The Sir3 R210 residue is highlighted in orange and labeled with an orange asterisk.
A model of how the H2B R102 and K111 residues regulate Sir3 binding to the nucleosome.
We favor a model in which the H2B R102 and K111 residues directly regulate the binding of the Sir complex to the nucleosome. A previous study has developed a structural model of how the Sir3 bromoadjacent homology (BAH) domain binds to the yeast nucleosome (35). A key feature of this model is that electrostatic repulsion between similarly charged side chains in Sir3 and the LRS regions of histone H3 and H4 partially destabilizes the interaction of Sir3 with the nucleosome. Mutations in Sir3, histone H3, or H4 that eliminate this electrostatic repulsion stimulate Sir complex binding and increase telomeric silencing (35, 36).
Intriguingly, in this model the H2B R102 side chain is located less than 4 Å from the similarly charged Sir3 R210 side chain (Fig. 8C to F). If this model is correct, Sir3 binding to the nucleosome should be partially destabilized by the electrostatic repulsion between H2B R102 and Sir3 R210 residues. This model would explain why the H2B R102A mutant enhanced silencing and Sir complex binding, while the H2B R102K mutant, which maintains the positive charge of this side chain, had relatively little effect on silencing. In contrast, the H2B K111 residue contacts the Sir3 BAH domain but does not appear to be involved in electrostatic repulsion. Presumably, the interaction with H2B K111 facilitates Sir3 binding to the nucleosome, which would explain why H2B K111 mutants cause defects in silencing and Sir complex binding.
The H2B/Sir3 electrostatic repulsion model is supported by sequence analysis of the Sir3 BAH domain. Previous studies have shown that LRS suppressor mutations in the Sir3 BAH domain, which presumably strengthen Sir3 binding to the nucleosome, are often similar to those of corresponding residues in the Orc1 BAH domain (i.e., the orc-like and semi-orc-like suppressors [see reference 35 for more details]). The Orc1 BAH domain, whose sequence is very similar to the Sir3 BAH domain protein sequence, appears to bind histones more tightly than the Sir3 BAH domain, based on pulldown experiments in yeast (37). Intriguingly, sequence alignment of these BAH domains (Fig. 8G) indicates that the Sir3 R210 residue, which we propose has a repulsive electrostatic interaction with H2B R102, is altered to a more favorable aspartic acid residue in the Orc1 BAH domain (Orc1 D210). This would correlate with the stronger nucleosome binding observed in the Orc1 BAH domain and the previously identified orc-like Sir3 suppressor mutants. Future studies will be required to rigorously test whether the postulated electrostatic repulsion between the H2B R102 and Sir3 R210 residues regulates the stability and extent of Sir3 binding to yeast nucleosomes.
Acknowledgments
We thank Robert Morris and Brandon Kyriss for helpful comments on the manuscript. We are grateful to Kasey Vargo for assistance in constructing a few of the histone H2B mutants, and we thank Stacy Hathcox and Katie Bergstrom for their assistance with the deletion of the endogenous H2A and H2B genes in the HML silencing tester strain. We also thank Stacy Hathcox for her help in testing the anti-H2B K111me2 antibodies, and we thank Erna Agmata for her assistance with the peptide dot blots. We are grateful to Derek Pouchnik for technical assistance with DNA microarray experiments. We thank Ray Reeves for the MCF-7 cells and Julie Stanton and Li Mao for preparing the MCF-7 cell protein extracts. We thank Daniel Gottschling for the gift of the yeast silencing strains and David Allis for the gift of yeast strain JHY205 and plasmid pQQ18.
We gratefully acknowledge support from the Washington State University College of Sciences Undergraduate Minigrant program (Kasey Vargo and J.A.S.), from a NIH Biotechnology Graduate Training grant (M.N.M.K and I.J.G.), and from the Washington State Fraternal Order of Eagles.
Footnotes
Published ahead of print on 17 May 2010.
REFERENCES
- 1.Ahn, S. H., W. L. Cheung, J. Y. Hsu, R. L. Diaz, M. M. Smith, and C. D. Allis. 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120:25-36. [DOI] [PubMed] [Google Scholar]
- 2.Altaf, M., R. T. Utley, N. Lacoste, S. Tan, S. D. Briggs, and J. Cote. 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28:1002-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio, O. M., B. L. Billington, and D. E. Gottschling. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66:1279-1287. [DOI] [PubMed] [Google Scholar]
- 4.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck, H. C., E. C. Nielsen, R. Matthiesen, L. H. Jensen, M. Sehested, P. Finn, M. Grauslund, A. M. Hansen, and O. N. Jensen. 2006. Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteomics 5:1314-1325. [DOI] [PubMed] [Google Scholar]
- 6.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri, S., J. J. Wyrick, and M. J. Smerdon. 2009. Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae. Nucleic Acids Res. 37:1690-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, J., E. M. Hyland, D. S. Yuan, H. Huang, J. S. Bader, and J. D. Boeke. 2008. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134:1066-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Downs, J. A. 2008. Histone H3 K56 acetylation, chromatin assembly, and the DNA damage checkpoint. DNA Repair (Amst.). 7:2020-2024. [DOI] [PubMed] [Google Scholar]
- 10.Driscoll, R., A. Hudson, and S. P. Jackson. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315:649-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 12.Freitas, M. A., A. R. Sklenar, and M. R. Parthun. 2004. Application of mass spectrometry to the identification and quantification of histone post-translational modifications. J. Cell. Biochem. 92:691-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fry, C. J., A. Norris, M. Cosgrove, J. D. Boeke, and C. L. Peterson. 2006. The LRS and SIN domains: two structurally equivalent but functionally distinct nucleosomal surfaces required for transcriptional silencing. Mol. Cell. Biol. 26:9045-9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner, R. G., Z. W. Nelson, and D. E. Gottschling. 2005. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell. Biol. 25:6123-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng, F., and W. P. Tansey. 2008. Polyubiquitylation of histone H2B. Mol. Biol. Cell 19:3616-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751-762. [DOI] [PubMed] [Google Scholar]
- 17.Han, J., H. Zhou, B. Horazdovsky, K. Zhang, R. M. Xu, and Z. Zhang. 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315:653-655. [DOI] [PubMed] [Google Scholar]
- 18.Huyen, Y., O. Zgheib, R. A. Ditullio, Jr., V. G. Gorgoulis, P. Zacharatos, T. J. Petty, E. A. Sheston, H. S. Mellert, E. S. Stavridi, and T. D. Halazonetis. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406-411. [DOI] [PubMed] [Google Scholar]
- 19.Hyland, E. M., M. S. Cosgrove, H. Molina, D. Wang, A. Pandey, R. J. Cottee, and J. D. Boeke. 2005. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:10060-10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin, Y., A. M. Rodriguez, J. D. Stanton, A. A. Kitazono, and J. J. Wyrick. 2007. Simultaneous mutation of methylated lysine residues in histone H3 causes enhanced gene silencing, cell cycle defects, and cell lethality in Saccharomyces cerevisiae. Mol. Cell. Biol. 27:6832-6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, Y., A. M. Rodriguez, and J. J. Wyrick. 2009. Genetic and genomewide analysis of simultaneous mutations in acetylated and methylated lysine residues in histone H3 in Saccharomyces cerevisiae. Genetics 181:461-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, T. J., S. Qin, D. E. Gottschling, and M. R. Parthun. 2000. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol. Cell. Biol. 20:7051-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keogh, M. C., T. A. Mennella, C. Sawa, S. Berthelet, N. J. Krogan, A. Wolek, V. Podolny, L. R. Carpenter, J. F. Greenblatt, K. Baetz, and S. Buratowski. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20:660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, S. C., R. Sprung, Y. Chen, Y. Xu, H. Ball, J. Pei, T. Cheng, Y. Kho, H. Xiao, L. Xiao, N. V. Grishin, M. White, X. J. Yang, and Y. Zhao. 2006. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23:607-618. [DOI] [PubMed] [Google Scholar]
- 25.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128:693-705. [DOI] [PubMed] [Google Scholar]
- 26.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 27.Martin, A. M., D. J. Pouchnik, J. L. Walker, and J. J. Wyrick. 2004. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics 167:1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masumoto, H., D. Hawke, R. Kobayashi, and A. Verreault. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436:294-298. [DOI] [PubMed] [Google Scholar]
- 29.Matsubara, K., N. Sano, T. Umehara, and M. Horikoshi. 2007. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells 12:13-33. [DOI] [PubMed] [Google Scholar]
- 30.Mersfelder, E. L., and M. R. Parthun. 2006. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 34:2653-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nag, R., M. Kyriss, J. W. Smerdon, J. J. Wyrick, and M. J. Smerdon. 2010. A cassette of N-terminal amino acids of histone H2B are required for efficient cell survival, DNA repair and Swi/Snf binding in UV irradiated yeast. Nucleic Acids Res. 38:1450-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakanishi, S., B. W. Sanderson, K. M. Delventhal, W. D. Bradford, K. Staehling-Hampton, and A. Shilatifard. 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15:881-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. U. S. A. 100:1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst, Y. Zhang, and K. Struhl. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norris, A., M. A. Bianchet, and J. D. Boeke. 2008. Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet. 4:e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norris, A., and J. D. Boeke. 2010. Silent information regulator 3: the Goldilocks of the silencing complex. Genes Dev. 24:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onishi, M., G. G. Liou, J. R. Buchberger, T. Walz, and D. Moazed. 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 28:1015-1028. [DOI] [PubMed] [Google Scholar]
- 38.Ozdemir, A., S. Spicuglia, E. Lasonder, M. Vermeulen, C. Campsteijn, H. G. Stunnenberg, and C. Logie. 2005. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J. Biol. Chem. 280:25949-25952. [DOI] [PubMed] [Google Scholar]
- 39.Parra, M. A., D. Kerr, D. Fahy, D. J. Pouchnik, and J. J. Wyrick. 2006. Deciphering the roles of the histone H2B N-terminal domain in genome-wide transcription. Mol. Cell. Biol. 26:3842-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parra, M. A., and J. J. Wyrick. 2007. Regulation of gene transcription by the histone H2A N-terminal domain. Mol. Cell. Biol. 27:7641-7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porollo, A. A., R. Adamczak, and J. Meller. 2004. POLYVIEW: a flexible visualization tool for structural and functional annotations of proteins. Bioinformatics 20:2460-2462. [DOI] [PubMed] [Google Scholar]
- 42.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 43.Schneider, J., P. Bajwa, F. C. Johnson, S. R. Bhaumik, and A. Shilatifard. 2006. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 281:37270-37274. [DOI] [PubMed] [Google Scholar]
- 44.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 45.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Mahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, C. M., Z. W. Haimberger, C. O. Johnson, A. J. Wolf, P. R. Gafken, Z. Zhang, M. R. Parthun, and D. E. Gottschling. 2002. Heritable chromatin structure: mapping “memory” in histones H3 and H4. Proc. Natl. Acad. Sci. U. S. A. 99(Suppl. 4):16454-16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. S., X. Ling, and M. Grunstein. 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369:245-247. [DOI] [PubMed] [Google Scholar]
- 49.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745-756. [DOI] [PubMed] [Google Scholar]
- 50.White, C. L., R. K. Suto, and K. Luger. 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20:5207-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wisniewski, J. R., A. Zougman, and M. Mann. 2008. Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 36:570-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wysocki, R., A. Javaheri, S. Allard, F. Sha, J. Cote, and S. J. Kron. 2005. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 25:8430-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, E. Y., X. Bi, M. J. Holland, D. E. Gottschling, and J. R. Broach. 2005. Mutations in the nucleosome core enhance transcriptional silencing. Mol. Cell. Biol. 25:1846-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, F., K. Zhang, and M. Grunstein. 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121:375-385. [DOI] [PubMed] [Google Scholar]
- 55.Xu, F., Q. Zhang, K. Zhang, W. Xie, and M. Grunstein. 2007. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell 27:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, K., Y. Chen, Z. Zhang, and Y. Zhao. 2009. Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. J. Proteome Res. 8:900-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, L., E. E. Eugeni, M. R. Parthun, and M. A. Freitas. 2003. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 112:77-86. [DOI] [PubMed] [Google Scholar]