Abstract
The RNA polymerase II core promoter is a diverse and complex regulatory element. To gain a better understanding of the core promoter, we examined the motif 10 element (MTE), which is located downstream of the transcription start site and acts in conjunction with the initiator (Inr). We found that the MTE promotes the binding of purified TFIID to the core promoter and that the TAF6 and TAF9 subunits of TFIID appear to be in close proximity to the MTE. To identify the specific nucleotides that contribute to MTE activity, we performed a detailed mutational analysis and determined a functional MTE consensus sequence. These studies identified favored as well as disfavored nucleotides and demonstrated the previously unrecognized importance of nucleotides in the subregion of nucleotides 27 to 29 (+27 to + 29 relative to A+1 in the Inr consensus) for MTE function. Further analysis led to the identification of three downstream subregions (nucleotides 18 to 22, 27 to 29, and 30 to 33) that contribute to core promoter activity. The three binary combinations of these subregions lead to the MTE (nucleotides 18 to 22 and 27 to 29), a downstream core promoter element (nucleotides 27 to 29 and 30 to 33), and a novel “bridge” core promoter motif (nucleotides 18 to 22 and 30 to 33). These studies have thus revealed a tripartite organization of key subregions in the downstream core promoter.
The expression of the tens of thousands of genes within a cell is regulated during growth, development, and response to environmental stimuli. In eukaryotes, transcription of protein-coding genes is mediated by RNA polymerase II. Transcription by RNA polymerase II is regulated by a wide variety of factors that include the basal transcription factors, sequence-specific enhancer- and promoter-binding proteins, coregulatory factors, and other chromatin remodeling and modifying factors. The signals from these factors ultimately converge at the core promoter during the process of transcription initiation (for reviews, see references 7, 12, 21, and 23).
The RNA polymerase II core promoter is the region of DNA that directs the initiation of transcription and generally spans from about nucleotide −40 to +40 relative to the transcription start site. The core promoter is diverse in terms of its structure and function as there are different mechanisms by which RNA polymerase II can be recruited to the promoter. For transcription that is directed by the TFIID transcription factor, there are several known sequences, termed core promoter elements, that mediate the recruitment of TFIID as well as other basal transcription factors to the DNA template. These core promoter elements include the TATA box, the initiator (Inr), the motif 10 element (MTE), the downstream core promoter element (DPE), the TFIIB recognition elements (BREu and BREd), the downstream core element (DCE), and the X core promoter element 1 (XCPE1) (for a recent review, see reference 12). In this work, we focus on the analysis of the MTE.
The study of the MTE began with the identification of motif 10 as an overrepresented sequence in a computational analysis of nearly 2,000 Drosophila promoters (19). The motif 10 consensus sequence (CSARCSSAACGS) was then found to be a functional core promoter element, termed the motif ten element (MTE) (18). The MTE functions cooperatively with the Inr, and there is a strict spacing requirement between the Inr and MTE motifs as the insertion or deletion of a single nucleotide between the Inr and MTE was observed to result in a 3- to 30-fold decrease in transcriptional activity. The addition of an MTE was found to compensate for the loss of basal transcription activity upon mutation of the TATA box or the DPE. Because MTE sequences from +18 to +27 were sufficient to confer MTE activity to heterologous promoters, the location of the MTE was designated to be from +18 to +27. In addition, there is synergism between the MTE and the TATA box as well as between the MTE and the DPE. This transcriptional synergism was exploited in the design of an unusually strong core promoter that contains TATA, Inr, MTE, and DPE motifs (10).
In the present study, we initially sought to gain a better understanding of the sequences that constitute the MTE as well as the role of the MTE in the binding of TFIID to the core promoter. In earlier work, the mutation of the MTE was found to result in an alteration of the weak binding of a partially purified preparation of Drosophila TFIID (18). Hence, it was necessary to examine the interaction of TFIID with the MTE. To this end, we developed a new method for the purification of TFIID and performed DNase I footprinting and photo-cross-linking experiments with the purified protein complex. It was also important to identify the specific sequences that are important for MTE function. We therefore carried out a systematic mutational analysis of the region encompassing the MTE. These studies unexpectedly led to a broader understanding of the downstream core promoter region. Specifically, we identified three key subregions that, in different binary combinations, yield the MTE, DPE, and novel “bridge” core promoter motifs.
MATERIALS AND METHODS
Purification of Drosophila TFIID.
The pCaSpeR-FLAG-TBP (where TBP is TATA-binding protein) plasmid used in stable transfection of Drosophila S2 cells was constructed by PCR of the TBP gene with Drosophila genomic DNA sequences, followed by subcloning into the pCaSpeR4 vector. The PCR was carried out in two steps and resulted in the introduction of sequences that add a double FLAG tag to the N terminus of TBP. Aside from the sequences encoding the N-terminal FLAG tag, pCaSpeR-FLAG contains wild-type TBP sequences from 650 bp upstream of the initiating Met codon to downstream of the polyadenylation site, which is 460 bp downstream of the translation termination codon. Stably transfected Drosophila S2 cell lines were established by cotransfecting 5 μg of pCaSpeR-Flag-TBP with 1 μg of the selection plasmid, pCoHygro (Invitrogen). The cells were cultured in Schneider's Drosophila medium (Gibco) supplemented with 300 μg/ml hygromycin B for the selection and maintenance of hygromycin resistance. For the purification of TFIID, the S2 cells expressing FLAG-TBP were cultured in suspension in 4 volumes of 400 ml of Schneider's Drosophila medium supplemented with 100 μg/ml hygromycin B and 0.05% Pluronic F68 (Invitrogen). The cells were harvested (at >6 × 106 cells/ml) by centrifugation in a Sorvall GSA rotor at 2,000 rpm for 5 min and yielded a cell pellet of approximately 20 g.
TFIID containing FLAG-TBP was purified from ∼20 g of cells as follows. The cells were washed twice with phosphate-buffered saline (PBS), pelleted by centrifugation in a Sorvall GSA rotor at 2,000 rpm for 5 min, resuspended in 5 cell pellet volumes (∼100 ml) of buffer H (10 mM Tris-HCl, pH 7.9, 10 mM KCl, 0.75 mM spermidine, 0.15 mM spermine, 0.1 mM EDTA, 0.1 mM EGTA, 2 mM dithiothreitol [DTT], 1 mM benzamidine-HCl, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], and 2 μg/ml each of aprotinin, leupeptin, and pepstatin), incubated on ice for 15 min, and lysed with 12 strokes of a 40-ml Dounce homogenizer with a loose (B) pestle. The resulting nuclei were pelleted by centrifugation in a GSA rotor at 7,000 rpm for 10 min, suspended in 15 ml of buffer H, repelleted by centrifugation in a Sorvall SS-34 rotor at 8,000 rpm for 10 min, and resuspended in 1 nucleus pellet volume (∼5 ml) of buffer E (50 mM Tris-HCl, pH 7.5, 0.6 M KCl, 20% [vol/vol] glycerol, 10% [wt/vol] sucrose, 5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 1 mM benzamidine-HCl, 0.2 mM PMSF). The mixture was stirred at 4°C for 30 min and then subjected to centrifugation in a Beckman SW28.1 rotor at 25,000 rpm for 1 h. The supernatant was dialyzed three times for 50 min each time against 2 liters (for each dialysis) of TM buffer (50 mM Tris-HCl, pH 7.9, 10% [vol/vol] glycerol, 12.5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 1 mM benzamidine-HCl, 0.1 mM PMSF, and 1 mM sodium metabisulfite) containing 0.1 M KCl. (When necessary, the extract was diluted with TM buffer without KCl until the mixture had the same conductivity as TM buffer with 0.1 M KCl.) The mixture was subjected to centrifugation in an SS-34 rotor at 10,000 rpm for 10 min. The supernatant was incubated with 0.01 volume of 1 mg/ml sonicated calf thymus DNA (to a final concentration of 10 μg/ml DNA) at 4°C for 30 min and then centrifuged in an SS-34 rotor at 10,000 rpm for 10 min to remove insoluble material. The supernatant, which was typically about 10 to 15 ml in volume, was applied to five parallel 1-ml sequence-specific DNA affinity columns containing the Drosophila G core promoter sequence (see, for example, reference 15) from −5 to +40 by using methods described previously (13). The 0.3 to 0.4 M KCl fractions were pooled and incubated with 150 μl (bed volume) of anti-FLAG M2 agarose resin (Sigma) for 8 to 10 h with rotation. The unbound proteins were removed by centrifugation at 2,000 rpm for 3 min in a clinical centrifuge. After five sequential washes in HEGN buffer (25 mM HEPES, K+ [pH 7.6], 0.1 mM EDTA, 10% [vol/vol] glycerol, 0.1% [vol/vol] NP-40) containing 0.1 M KCl and 5 mM β-mercaptoethanol, the agarose resin was transferred to an Amicon Ultrafree-MC spin column (0.45 μm), and residual buffer was removed by microcentrifugation at 3,000 rpm for 10 s. The bound TFIID complex was eluted from the anti-FLAG M2 resin by incubation with 100 μl of HEGN buffer containing 0.1 mg/ml of the peptide with three FLAG tags (3×FLAG) (Sigma), rotation of the sample at 4°C for 20 min, and collection by microcentrifugation at 3,000 rpm for 10 s. The elution procedure was repeated two more times. The concentration of purified TFIID was approximately 50 nM.
Core promoter constructs.
Plasmids containing minimal core promoters for the in vitro transcription and DNase I footprinting experiments were constructed by the insertion of double-stranded oligonucleotides into the XbaI and PstI sites of the multiple cloning region of pUC119. The specific nucleotide sequences are available upon request.
DNase I footprinting.
DNase I footprinting probes were prepared by PCR amplification of core promoter constructs with a 5′ 32P-labeled M13 reverse sequencing primer and unlabeled M13 forward primer. The PCR amplification products were purified by polyacrylamide gel electrophoresis. DNase I footprinting reactions were carried out as described previously (5).
In vitro transcription.
In vitro transcription reactions were carried out as described previously (24) by using 250 ng of supercoiled DNA templates with Drosophila high-salt nuclear extracts (22). The resulting transcripts were subjected to primer extension analysis with a 5′ 32P-labeled M13 reverse sequencing primer (AGCGGATAACAATTTCACACAGGA). Quantitation of reverse transcription products was carried out with a phosphorimager (GE Health Sciences). All experiments were carried out a minimum of three independent times to ensure reproducibility of the data.
Photo-cross-linking analysis.
The photoaffinity DNA probes were synthesized by minor modification of previously described methods (2, 3). Synthetic oligonucleotides with promoter sequences from −35 to +45 were used as templates instead of single-stranded M13 DNA. The 5-[N-(p-azidobenzoyl)-3-aminoallyl]-dUTP (AB-dUTP; also known as N3RdUTP) was generously provided by George Kassavetis and E. Peter Geiduschek (University of California, San Diego). The synthesized probes were subjected to 5% polyacrylamide gel electrophoresis and eluted passively (overnight at room temperature) into 0.5 ml of elution buffer (20 mM Tris-HCl [pH 8.0], 0.2 mM EDTA, 0.2% [wt/vol] SDS, 1 M LiCl). The DNA probes were precipitated with ethanol and resuspended in 30 μl of Tris-EDTA (TE) buffer. In a typical cross-linking experiment, photoaffinity DNA probe (50 fmol) was incubated with purified TFIID (approximately 80 ng) in a 10-μl reaction mixture containing 12.5 mM HEPES, K+ (pH 7.6), 50 mM KCl, 6.75 mM MgCl2, 0.05 mM EDTA, 2.5 mM 2-mercaptoethanol, 5% (vol/vol) glycerol, 0.05% (vol/vol) NP-40, and 2% (vol/vol) polyvinyl alcohol. The binding reactions were incubated on ice for 15 min and then at 25°C for 15 min. The samples were subjected to short-wavelength UV irradiation at 380 μW/cm2 for 5 min. Irradiated protein-DNA complexes were digested with DNase I at 37°C for 10 min. A solution of 10% (wt/vol) SDS was added to a final concentration of 0.5%, and the samples were heated at 90°C for 3 min. The DNA-protein complexes were additionally treated at 37°C for 10 min with S1 nuclease (Invitrogen). The cross-linked proteins were resolved by 10% polyacrylamide-SDS gel electrophoresis. The gel was dried and subjected to autoradiography. The identity of the TAF6 and TAF9 subunits was confirmed by Western blot analysis.
RESULTS
A new method for the purification of Drosophila TFIID.
To gain a better understanding of the role of the MTE in the binding of TFIID to the core promoter, we sought to develop a more rapid and reliable method for the generation and purification of TFIID. To this end, we stably transfected Drosophila S2 cells with a modified Drosophila TBP gene that encodes N-terminally FLAG-tagged TBP (Fig. 1 A). The TFIID complex containing FLAG-TBP was purified from the stably transfected cells as outlined in Fig. 1B. To enrich for TFIID relative to other TBP-containing complexes as well as free TBP, we partially purified TFIID by sequence-specific DNA affinity chromatography with the core promoter sequence of the TATA-less, DPE-containing Drosophila G promoter. By Western blot analysis with antibodies against Drosophila TBP and TAF1, we determined that most of the TFIID complex elutes from the DNA affinity column in the 0.3 to 0.4 M KCl fraction. The TFIID in the 0.3 to 0.4 M KCl fraction was then purified to near homogeneity by anti-FLAG immunoaffinity chromatography (Fig. 1C). The highly purified TFIID is similar to other preparations of TFIID obtained from Drosophila embryos (see references 4, 8, 9, and 14).
FIG. 1.
Purification of TFIID from Drosophila S2 cells containing FLAG-tagged TBP. (A) Synthesis of FLAG-tagged TBP in S2 cells. Whole-cell lysates derived from S2 cells and two different FLAG-TBP-containing S2 cell lines were subjected to Western blot analysis with antibodies against Drosophila TBP. (B) Scheme for the purification of TFIID from S2 cells containing FLAG-tagged TBP. (C) Purification of TFIID containing FLAG-tagged TBP. The polypeptides were resolved by 10% polyacrylamide-SDS gel electrophoresis and visualized by silver staining. In addition, by Western blotting and mass spectrometry, the purified TFIID was found to contain FLAG-tagged TBP as well as all TAFs from TAF1 through TAF14.
The MTE promotes transcription by increasing the affinity of TFIID for the core promoter.
The purification of TFIID enabled us to carry out DNase I footprinting experiments with the MTE-containing Drosophila Tollo and CG10479 core promoters (Fig. 2). The binding of TFIID to the Tollo promoter is much more distinct than that observed previously (18), possibly due to increased purity and activity of the TFIID. The wild-type versions of the Tollo and CG10479 promoters exhibit related patterns of alternating DNase I protection and hypersensitivity that extend from approximately −25 to +35 relative to the A+1 in the Inr consensus sequence. The most prominent DNase I protection and hypersensitivity span from the Inr to about the +15 position. These results indicate that TFIID binds throughout the core promoter region of MTE-containing promoters.
FIG. 2.
The MTE contributes to the binding of purified TFIID to the core promoter. The wild-type, m18-22 (MTE-inactivating), m30-33 (DPE-inactivating), and double mutant (m18-22 and m30-33) versions of the Drosophila Tollo (A) and CG10479 (B) core promoters were subjected to DNase I footprinting analysis with purified Drosophila TFIID. The positions of the Inr (−2 to + 4, relative to the position defined as A+1 in the Inr consensus), MTE (depicted as sequences from +18 to +27), and DPE (+28 to +33) motifs are indicated. In the wild-type promoters, regions of DNase I protection and hypersensitivity are indicated by brackets and filled dots, respectively.
To examine the role of the MTE in the binding of TFIID to the core promoter, we performed DNase I footprinting experiments with a series of core promoters that contain mutations in the MTE, the DPE, or both the MTE and DPE motifs. We employed mutations that had been previously demonstrated to inactivate the MTE and DPE motifs (18). Specifically, mutation m18-22 inactivates the MTE via mutation of the sequences from +18 to +22 (relative to the A+1 in the Inr) to ATCCA, whereas m30-33 inactivates the DPE by mutation of the sequences from +30 to +33 to CATA.
Mutation of the MTE or the DPE results in a reduction in the interaction of TFIID with the Tollo and CG10479 promoters, and mutation of both the MTE and the DPE further decreases the binding of TFIID (Fig. 2). The strength of TFIID binding to DNA observed in this work correlates with the efficiency of transcription that had been previously determined with the same promoter constructs (18). Thus, the MTE contributes to the affinity for TFIID and the transcriptional activity of the Tollo and CG10479 core promoters.
We additionally sought to determine whether the addition of MTE sequences to an MTE-deficient core promoter increases the binding of TFIID. To this end, we used the Drosophila E74B and Doc core promoters, both of which contain Inr and DPE motifs but lack an MTE (18). The addition of Tollo MTE sequences to the E74B and Doc promoters results in stronger binding of TFIID to the promoter (Fig. 3), which correlates with the increase in transcriptional activity that was observed with the same promoter constructs (18). Hence, studies involving either the loss or the gain of the MTE at the core promoter indicate that the MTE promotes transcription by increasing the affinity of the binding of TFIID to the core promoter.
FIG. 3.
The addition of an MTE increases the affinity of TFIID for the core promoter. The wild-type (WT) and WT+MTE (containing the Tollo MTE sequence from +18 to +27, relative to the A+1 position in the Inr) versions of the Drosophila E74B (A) and Doc (B) core promoters were subjected to DNase I footprinting analysis with purified Drosophila TFIID. The positions of the Inr (−2 to +4), MTE (Tollo core promoter sequence from +18 to +27), and DPE (+28 to +33) motifs are indicated. In the WT+MTE promoters, regions of DNase I protection and hypersensitivity are indicated by brackets and filled dots, respectively.
The DNase I footprinting patterns with the MTE-containing promoters are similar to those seen with DPE-containing promoters (see references 4, 5, and 15). There is alternating DNase I protection and hypersensitivity, which suggests a close interaction of TFIID with one face of the DNA helix. The upstream boundaries of the protected and/or hypersensitive sites are from about −15 to −24. When it is considered that each boundary of DNase I protection and/or hypersensitivity is several nucleotides larger than the actual site of protein binding, these data are consistent with the interaction of TFIID with the promoter from the Inr though the downstream promoter region and with cooperativity between the MTE and Inr for transcriptional activity (18).
Photo-cross-linking analysis of purified TFIID with MTE-containing promoters.
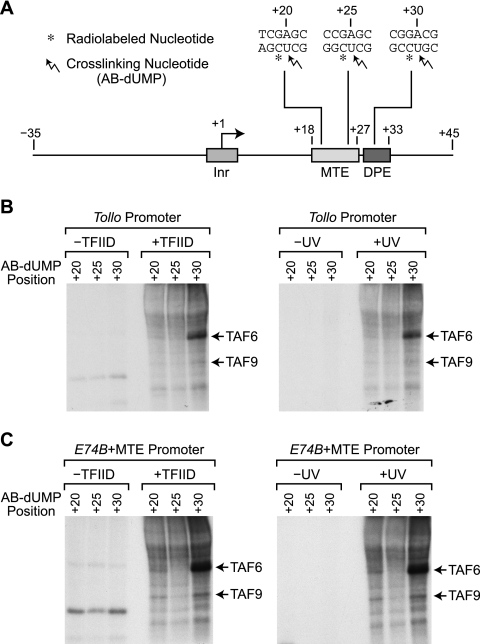
To ascertain which subunit or subunits of TFIID are in close proximity to the MTE, we performed photo-cross-linking studies of TFIID bound to MTE-containing promoters. In previous work on the DPE (5), we generated photoaffinity probes by the incorporation of 5-[N-(p-azidobenzoyl)-3-aminoallyl]-dUTP (AB-dUTP; also known as N3RdUTP) (2, 3), a photoreactive TTP analogue, into the DNA. The resulting photo-cross-linking experiments revealed that TAF6 and TAF9 (also known as TAFII60 and TAFII40, respectively) are in close proximity to the DPE. To enable the comparison of the new MTE data with the previous results on the DPE (5), we also used AB-dUTP to examine the interaction of TFIID with the MTE.
To investigate the interaction of TFIID subunits with the downstream core promoter region, we generated photoaffinity probes with the Tollo promoter and a hybrid promoter combining the MTE and E74B (E74B+MTE). The Tollo promoter is a natural core promoter that contains both MTE and DPE motifs (18). The E74B+MTE hybrid promoter also contains both MTE and DPE motifs and is identical to the construct used in the DNase I footprinting analysis shown in Fig. 3A. The MTE contributes to the binding of TFIID to each of these promoters (Fig. 2A and 3A). As depicted in Fig. 4 A, a photoreactive AB-dUMP nucleotide was introduced at the +20, +25, or +30 position (relative to the +1 start site) of each DNA probe alongside a radiolabeled nucleotide that enabled detection of the cross-linked polypeptides. Experiments were performed in the presence or absence of TFIID as well as with or without UV irradiation to allow the identification of polypeptides to which photo-cross-linking is dependent upon both TFIID and UV activation.
FIG. 4.
The TAF6 and TAF9 subunits of TFIID appear to be in close proximity to the MTE. (A) Diagram of photoaffinity probes containing AB-dUMP at the +20, +25, or +30 position relative to the +1 transcription start site. The Tollo core promoter sequences are shown. The diagram is roughly to scale. (B) Photo-cross-linking of purified TFIID with the Tollo core promoter. Reactions were performed in the presence or absence of TFIID as well as with or without UV irradiation, as indicated. (C) Photo-cross-linking of purified TFIID with the E74B core promoter containing the Tollo MTE. The E74B+MTE core promoter is identical to the construct that was used in the DNase I footprinting analysis shown in Fig. 3A. Reactions were performed as described for panel B.
With the Tollo promoter, we observed photo-cross-linking of a 60-kDa band at +30 (Fig. 4B). By Western blot analysis, this band was found to comigrate with TAF6, which is consistent with previous results on the DPE (5). We also observed a weak 60-kDa signal at +20 but not at +25. The 60-kDa signal at +20 is suggestive of an interaction of TAF6 with the MTE region. In addition, a weak band at 40 kDa, which corresponds to TAF9 by Western blot analysis, can be seen. The 40-kDa band is not sufficiently strong, however, for a conclusion to be drawn regarding the interaction of TAF9 with the Tollo promoter.
With the E74B+MTE promoter, we observed strong photo-cross-linking of the 60-kDa TAF6 band at +30 as well as a distinct TAF6 band at +20 but not at +25 (Fig. 4C). We also saw cross-linking of TAF9 at +20 and +30 but not at +25. The cross-linking of both TAF6 and TAF9 at +20 and +30 but not at +25 suggests that these proteins are in close proximity to one face of the DNA helix in this region. In this regard, it is relevant to note that TAF6 and TAF9 are related to histones H4 and H3 (6). It is therefore possible that a TAF6- and TAF9-containing subcomplex of TFIID interacts with the downstream core promoter region with key contacts at +20 and +30.
Single-nucleotide mutational analysis reveals sequences that are important for MTE activity.
To determine the key sequences that contribute to MTE function, we carried out a saturating single-nucleotide mutational analysis of the MTE region of the Tollo and CG10479 promoters. We performed these studies with two different core promoters because we sought to identify sequences that are important for MTE function in multiple sequence contexts. For each promoter, we generated a set of constructs containing A, C, G, or T at each position from +15 to +29 relative to the A+1 in the Inr. To study the function of the MTE in the absence of the DPE, all constructs contained the m30-33 mutation, which inactivates the DPE motif.
In vitro transcription analysis of the Tollo and CG10479 promoters revealed both similarities and differences in their responses to specific mutations (Fig. 5). For instance, at +17, both promoters exhibit a preference for C or T relative to A or G, whereas at +28, there is a preference for A or G relative to C or T. In contrast, at +22, +23, and +27, we observed different responses to mutations in the Tollo and CG10479 promoters, even though the wild-type nucleotides of the two promoters are the same at those positions. These findings indicate that the preferred nucleotides in the MTE are influenced by the surrounding sequence.
FIG. 5.
Single-nucleotide substitution analysis reveals sequences that are important for MTE activity. (A and C) Single-nucleotide substitution analysis of the MTE in the Drosophila Tollo (A) and CG10479 (C) core promoters. Mutant promoters containing every possible single-nucleotide substitution from +15 to +29 (relative to the A+1 in the Inr) were generated. To eliminate the contribution of the DPE in these experiments, all of the constructs contain the m30-33 mutation (CATA from +30 to +33), which inactivates DPE function. The promoters were subjected to in vitro transcription analysis with a Drosophila embryo nuclear extract, and the resulting transcripts were detected by primer extension-reverse transcription analysis. The primer extension data that correspond to the wild-type (WT) promoter are boxed. (B and D) The relative transcriptional activities of wild-type and mutant MTE sequences in the Tollo (B) and CG10479 (D) core promoters. Quantitation of the data from three independent experiments is shown. The data are normalized to promoters containing the wild-type MTE sequence. The error bars represent the standard deviations.
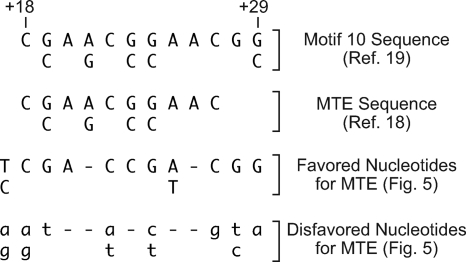
As a guide for the analysis of the MTE, we categorized the single-nucleotide substitution data in Fig. 5 as follows. For each position, nucleotides that result in >90% of the activity of the wild-type nucleotide (which is defined to be 100%) in both the Tollo and CG10479 promoters are designated “favored nucleotides,” whereas nucleotides that result in <60% of the wild-type nucleotide in both promoters are designated “disfavored nucleotides” (Fig. 6). The favored nucleotide sequence is a generally more restrictive subset of the computationally derived motif 10 consensus (19). It is also useful to note that the m18-22 sequence, which is ATCCA from +18 to +22, correlates well with the disfavored nucleotides at those positions.
FIG. 6.
Identification of sequences that are important for MTE activity. The data shown in Fig. 5 were analyzed as follows. For each position, nucleotides that resulted in >90% of the transcriptional activity of the wild-type nucleotide (defined to be 100%) for both the Tollo and CG10479 promoters were designated favored nucleotides, whereas nucleotides that resulted in <60% of the transcriptional activity of the wild-type nucleotide for both promoters were designated disfavored nucleotides. The computationally derived motif 10 sequence (19) and the MTE sequence based on the initial characterization of the element (18) are also shown.
Three downstream subregions are important for MTE and DPE activity.
Inspection of the single-nucleotide substitution data (Fig. 5) reveals that there is a strong nucleotide preference at positions +27 to +29 of both the Tollo and CG10479 promoters containing the m30-33 mutation. The +28 and +29 positions overlap with the DPE consensus (12). Because the single-nucleotide mutational analysis was performed with constructs containing the m30-33 mutation, which inactivates the DPE, it appeared that the +28 and +29 positions are important for both the MTE and DPE motifs.
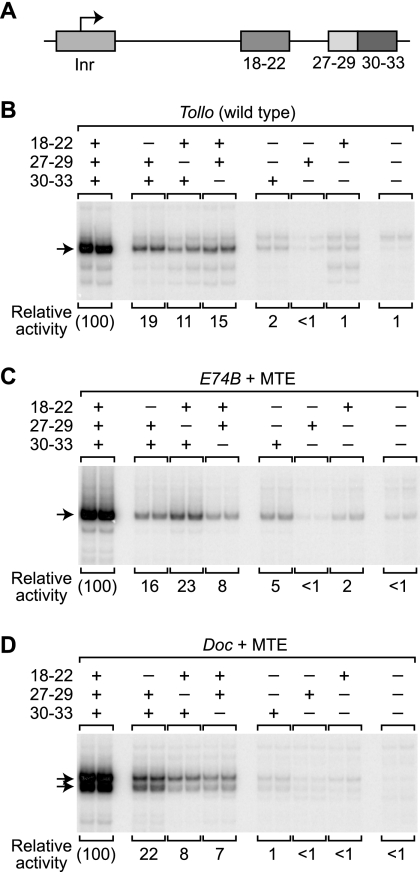
We therefore examined the role of the subregion from nucleotides 27 to 29 (27-29 subregion) in the function of the MTE and DPE motifs. To this end, we constructed and analyzed three series of core promoter constructs that contain all possible combinations of mutations in the 18-22, 27-29, and 30-33 subregions. The promoters used in this analysis each contain Inr, MTE, and DPE motifs and are as follows: (i) the wild-type Tollo promoter, which contains Inr, MTE, and DPE motifs; (ii) an E74B+MTE hybrid promoter that contains the Inr and DPE motifs from E74B and the Tollo MTE (from +18 to +27); and (iii) a Doc+MTE hybrid promoter that contains the Inr and DPE motifs from Doc and the Tollo MTE (from +18 to +27). The E74B+MTE and Doc+MTE hybrid promoters are identical to those used in the experiment shown in Fig. 3. We employed the m18-22 and m30-33 mutations to inactivate the MTE and DPE, as shown in Fig. 2 and Lim et al. (18), and used the disfavored sequence of GTA from +27 to +29 (Fig. 6) to inactivate the 27-29 subregion.
In vitro transcription analysis of the three promoter series revealed that mutation of any one of the three downstream subregions (subregions 18-22, 27-29, and 30-33) results in a substantial reduction in transcription relative to the reference promoters containing optimal sequences in the three subregions (Fig. 7). Mutation of any two of the three subregions leads to a further decrease in transcriptional activity. Finally, constructs lacking all three subregions are essentially inactive.
FIG. 7.
Analysis of three downstream subregions that are important for MTE and DPE activity. (A) Diagram of the Inr and downstream core promoter subregions. The relative locations of the elements are drawn approximately to scale. (B) Systematic analysis of three downstream subregions of the wild-type Tollo core promoter. A set of promoters with all possible combinations of the m18-22, m27-29, and m30-33 mutations in the Tollo core promoter was constructed. For each promoter, the wild-type (+) and mutant (−) versions of each sequence are indicated. The numbers below each promoter construct are the mean of three or four independent experiments relative to the wild-type Tollo promoter. (C and D) Analysis of three downstream subregions in E74B- and Doc-based core promoters. The relative contributions of the 18-22, 27-29, and 30-33 subregion sequences were tested by using the hybrid E74B+MTE and Doc+MTE core promoters, which consist of the natural E74B and Doc promoters containing the Tollo MTE sequences from +18 to +27 relative to the A+1 in the Inr. Thus, the hybrid promoters contain Inr, MTE, and DPE motifs. Beginning with each hybrid promoter, a set of promoters that comprises all possible combinations of the m18-22, m27-29, and m30-33 mutations was constructed. For each promoter, the wild-type (+) and mutant (−) versions of each sequence are indicated. The numbers below each promoter construct indicate the mean of three or four independent experiments relative to the hybrid core promoter containing the optimal sequences in the Inr, MTE, and DPE motifs.
Even though the 27-29 subregion is shared by the MTE and DPE motifs, the loss of the 27-29 subregion is not more deleterious to promoter activity than the loss of the 18-22 or the 30-33 subregion. Instead, it appears that the 18-22, 27-29, and 30-33 subregions each make roughly equivalent contributions to core promoter activity. These findings, in combination with the single-nucleotide mutational data (Fig. 5) and our current understanding of the MTE and DPE, suggest that the MTE comprises both the 18-22 and 27-29 subregions, whereas the DPE contains the 27-29 and 30-33 subregions. It is also interesting that core promoter constructs containing a novel configuration of the 18-22 and 30-33 subregions (with the m27-29 mutation) have comparable activity to those containing MTE (subregions 18-22 and 27-29) or DPE (subregions 27-29 and 30-33) motifs (Fig. 7).
Use of the downstream subregions in natural core promoters.
The results of the mutational analysis of the three downstream subregions led us to consider whether all possible binary combinations of these subregions occur in natural core promoters. In previous studies, we found that the Drosophila E74B and Doc core promoters have a DPE (subregions 27-29 and 30-33) and lack substantial MTE (18-22 subregion) activity (18). On the other hand, we have not yet identified MTE-containing promoters that lack substantial DPE (30-33 subregion) activity. In addition, core promoters that are predominantly driven by the novel combination of the 18-22 and 30-33 subregions, which we term the bridge configuration, have not yet been characterized.
We therefore sought to identify naturally occurring core promoters that are predominantly driven by MTE (18-22 and 27-29 subregions) or bridge (18-22 and 30-33 subregions) sequences in conjunction with an Inr (Inr+MTE or Inr+bridge, respectively). To this end, we searched a database of predicted Drosophila melanogaster transcription start sites (kindly provided by C. Benner and C. K. Glass, University of California, San Diego, La Jolla, CA). We found two Inr+MTE core promoters that are more sensitive to mutation of the 18-22 subregion than to mutation of the 30-33 subregion (Fig. 8 A and B) as well as an Inr+bridge core promoter that is more sensitive to mutation of the 18-22 or 30-33 subregion than to mutation of the 27-29 subregion (Fig. 8C). For the Inr+MTE promoters (CG5397 and CG6980), we further confirmed the absence of a strong 30-33 subregion by converting it to a consensus sequence at nucleotides 30 to 33 and found that the addition of the consensus sequences yielded a 2.5- to 3.7-fold increase in transcriptional activity (Fig. 8A and B). Analogously, we constructed a version of the CG15253 promoter that contains a favorable sequence at nucleotides 27 to 29 and observed a 2-fold increase in transcription. It thus appears that the Inr+MTE and Inr+bridge structures are used in natural core promoters.
FIG. 8.
Natural core promoters that are driven predominantly by the MTE or bridge core promoter motifs. (A and B) Natural TATA-less, MTE-containing core promoters that lack a strong DPE motif. Wild-type and mutant versions of the CG5397 (A) and CG6980 (B) core promoters were subjected to in vitro transcription and primer extension analyses. In addition to the standard m18-22 and m30-33 mutations, we analyzed promoters in which a consensus DPE sequence was introduced from +30 to +33 (Consensus 30-33). The relative activity values are the means of three or four independent experiments normalized to the cognate wild-type promoter. (C) Natural TATA-less, bridge (subregion 18-22 plus 30-33)-containing core promoter that lacks a strong sequence at nucleotides +27 to +29. Wild-type and mutant versions of the CG15253 core promoter were subjected to in vitro transcription and primer extension analyses. In addition to the m18-22, m27-29, and m30-33 mutations, we tested a mutant promoter containing an optimal sequence at positions 27 to 29 (Optimal 27-29) (Fig. 6). The relative activity values are the means of three or four independent experiments normalized to the value of the cognate wild-type promoter.
These findings led us to consider the general cooccurrence of the DPE (subregion 27-29 plus 30-33), MTE (subregion 18-22 plus 27-29), and bridge (subregion 18-22 plus 30-33) sequences with the Inr in natural core promoters. To this end, we analyzed core promoter sequences derived from the transcription start site data in the Drosophila MachiBase database (1). As shown in Table 1, the Inr is present in about 27% of all core promoters but is seen in only about 21% of core promoters that lack all three downstream subregions. On the other hand, the Inr is present in about two-thirds of core promoters that contain MTE (67%), DPE (65%), or bridge (63%) sequence. In this analysis, we used somewhat stringent criteria for the definition of the Inr, and the 18-22, 27-28, and 30-33 subregion sequences (Table 1), but similar relative values are observed if the criteria are relaxed. Hence, the presence versus the absence of any of the three binary combinations of the three downstream subregions results in about a 3-fold (∼65% versus ∼21%) increase in the occurrence of the Inr. These observations support the notion that the MTE, DPE, and bridge sequences are functionally important core promoter motifs.
TABLE 1.
Cooccurrence of downstream promoter elements with the initiator (Inr) in D. melanogaster
| Core promoter group or motif (subregions)a | Cooccurrence of Inr (no. of positive sequences/no. of sequences tested [%])b |
|---|---|
| All core promoters | 3,122/11,638 (27) |
| Core promoters lacking | |
| downstream subregions | 1,880/8,930 (21) |
| MTE (18-22, 27-29) | 49/73 (67) |
| DPE (27-29, 30-33) | 141/217 (65) |
| Bridge (18-22, 30-33) | 99/157 (63) |
Core promoter sequences were based on the transcription start site data from the Drosophila MachiBase database (1).
The presence of an Inr motif was based on at least a 5/6 match with the Inr consensus sequence of TCA+1KTY while not allowing for deviation from CA+1. The presence of the 18-22 subregion was based on at least a 3/4 match to the favored nucleotides shown in Fig. 6 while not allowing for any of the disfavored nucleotides in this region. The presence of the 27-29 subregion was based on a 3/3 match to the favored nucleotides shown in Fig. 6. The presence of the 30-33 subregion was based on at least a 3/4 match to the favored nucleotides WYGT while not allowing for any disfavored nucleotides (SRW from +30 to +32).
DISCUSSION
In this work, we have analyzed the interaction of TFIID with the MTE and investigated the sequences that are important for MTE function. These studies led to the identification of three downstream subregions (subregions 18-22, 27-29, and 30-33) that contribute to core promoter activity. Notably, the three different binary combinations of these subregions create the MTE (subregions 18-22 and 27-29), DPE (subregions 27-29 and 30-33), and novel bridge (subregions 18-22 and 30-33) core promoter motifs. Thus, these findings have resulted in a broader understanding of the downstream core promoter region (Fig. 9).
FIG. 9.
Model of a tripartite organization of key interaction points of TFIID with downstream core promoter sequences. In this model, the 18-22, 27-29, and 30-33 subregion sequences are three key downstream points of interaction of TFIID with the core promoter. Based on the photo-cross-linking studies (Fig. 4), the TAF6 and TAF9 subunits of TFIID are shown in close proximity to the downstream core promoter region. The Inr is included because it has been previously shown that the MTE as well as the DPE acts in a cooperative manner with the Inr (4, 18). The MTE comprises the 18-22 and 27-29 subregion sequences, whereas the DPE contains the 27-29 and 30-33 subregion sequences. In addition, the 18-22 and 30-33 subregion sequences can function synergistically in the absence of an optimal 27-29 sequence and form the bridge element.
Interaction of TFIID with the downstream core promoter.
To study the binding of TFIID to the MTE, we developed a new and reliable method for the purification of Drosophila TFIID (Fig. 1). One key feature of this method is the use of sequence-specific DNA affinity chromatography with the TATA-less, DPE-containing core promoter sequence from the G long interspersed nuclear element (LINE). We had previously found that the G core promoter has an unusually high affinity for TFIID (15). Therefore, the use of this strong TATA-less core promoter for affinity chromatography enabled the purification of TFIID relative to free TBP and other TBP-containing species.
The DNase I footprinting (Fig. 2 and 3) and photo-cross-linking (Fig. 4) data of this study, combined with the previous analysis of the binding of TFIID to the DPE (5, 20), suggest that the TAF6 and TAF9 subunits of TFIID are in close proximity to the downstream core promoter region. As noted above, TAF6 and TAF9 are related to histones H4 and H3, respectively; thus, TAF6 and TAF9 may form a subcomplex that interacts with downstream core promoter sequences. In addition, examination of the DNase I footprinting data reveals an extended alternating pattern of DNase I protection and hypersensitivity that suggests a close interaction between promoter DNA and TFIID. A schematic diagram of this model is depicted in Fig. 9.
Tripartite organization of the downstream core promoter.
In the single-nucleotide mutational analysis of the MTE, we found that the region of nucleotides +27 to +29 was particularly important for transcriptional activity (Fig. 5). These experiments were performed in constructs that contained the m30−33 (DPE-inactivating) mutation; thus, the 27-29 subregion is important for MTE-driven transcription in the absence of a DPE. We then further examined the relation between the 18-22, 27-29, and 30-33 subregions and the MTE and DPE motifs by analyzing all possible combinations of mutations of the subregions in three different promoters (Fig. 7). These experiments led to the model that the MTE comprises the 18-22 and 27-29 subregions, whereas the DPE contains the 27-29 and 30-33 subregions (Fig. 9). The Drosophila E74B and Doc core promoters are examples of natural downstream core promoters that predominantly use the DPE motif (18). The Drosophila CG5397 and CG6980 promoters are predominantly MTE-driven downstream core promoters (Fig. 8A and B). We also found that the bridge combination of the 18-22 and 30-33 subregions can yield core promoters with comparable activity to MTE-driven (18-22 and 27-29 subregions) or DPE-driven (27-29 and 30-33 subregions) core promoters (Fig. 7). The Drosophila CG15253 core promoter is a natural core promoter that is driven predominantly by a bridge (18-22 and 30-33 subregions) motif (Fig. 8C). These observations suggest that an Inr along with any binary combination of the 18-22, 27-29, and 30-33 subregions can yield a strong core promoter. We additionally determined the general cooccurrence of the Inr with core promoters containing MTE, DPE, or bridge motifs (Table 1). This analysis revealed that the presence of an MTE, DPE, or bridge motif results in a substantial increase in the frequency of occurrence of an Inr in the core promoter. Moreover, it is interesting that the novel bridge motif appears to occur more frequently than the MTE.
We had previously found that the addition of Tollo MTE sequences from +18 to +27 can compensate for the loss of DPE activity (18) and had therefore designated the MTE to be sequences from +18 to +27. In those experiments, however, the constructs contained the favored nucleotide G at position +29 downstream of the Tollo MTE sequences. The G at +29 probably contributed to the activity of the Tollo MTE sequences from +18 to +27. Based on our more extensive analysis of this motif and the importance of the region of +27 to +29 (Fig. 7), it is probably more accurate to describe the MTE as encompassing the sequence from +18 (or +17) to +29 (Fig. 9).
It is also relevant to compare the MTE and DPE subregions with those of the downstream core element (DCE), which is another downstream core promoter element (16, 17). The DCE was found to consist of three subelements: subelement I is CTTC in the region from +6 to +11, subelement II is CTGT located from +16 to +21, and subelement III is AGC in the region from +30 to +34 (16). Comparison of the DCE sequences with the motif 10 consensus (19), the MTE favored nucleotides (Fig. 6), and the DPE consensus (12) reveals no relation between the DCE and the downstream sequences associated with the MTE and DPE.
The results presented here suggest a tripartite organization of the downstream sequences that contributes to the binding of TFIID to the core promoter (Fig. 9). These studies clarify the downstream interactions of TFIID with the core promoter. These interactions may be an important feature of the mechanisms by which sequence-specific activators, such as Caudal protein, activate transcription in a core promoter-specific manner (11). In the future, it is likely that many additional dimensions of complexity will be uncovered in the core promoter. For instance, recent data have revealed transcription systems that do not involve TFIID (for a review, see reference 7). These other transcription systems may have their own variations in core promoter sequences and functions and therefore suggest the existence of a vast range of regulatory phenomena that are mediated via the core promoter.
Acknowledgments
We are very grateful to Uwe Ohler, Tammy Juven-Gershon, Timur Yusufzai, and Sharon Torigoe for critical reading of the manuscript. We thank Chris Benner and Chris Glass (University of California, San Diego, La Jolla, CA) for sharing the Drosophila transcription start site data. We also thank George Kassavetis and E. Peter Geiduschek (University of California, San Diego, La Jolla, CA) for the gift of the AB-dUTP photo-cross-linking reagent.
This work was supported by a grant from the National Institutes of Health (GM041249) to J.T.K.
Footnotes
Published ahead of print on 10 May 2010.
REFERENCES
- 1.Ahsan, B., T. L. Saito, S. Hashimoto, K. Muramatsu, M. Tsuda, A. Sasaki, K. Matsushima, T. Aigaki, and S. Morishita. 2009. Machibase: a Drosophila melanogaster 5′-end mRNA transcription database. Nucleic Acids Res. 37:D49-D53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartholomew, B., G. A. Kassavetis, B. R. Braun, and E. P. Geiduschek. 1990. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 9:2197-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartholomew, B., G. A. Kassavetis, and E. P. Geiduschek. 1991. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol. Cell. Biol. 11:5181-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke, T. W., and J. T. Kadonaga. 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10:711-724. [DOI] [PubMed] [Google Scholar]
- 5.Burke, T. W., and J. T. Kadonaga. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burley, S. K., and R. G. Roeder. 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65:769-799. [DOI] [PubMed] [Google Scholar]
- 7.D'Alessio, J. A., K. J. Wright, and R. Tjian. 2009. Shifting players and paradigms in cell-specific transcription. Mol. Cell 36:924-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dynlacht, B. D., T. Hoey, and R. Tjian. 1991. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell 66:563-576. [DOI] [PubMed] [Google Scholar]
- 9.Hoey, T., R. O. Weinzierl, G. Gill, J. L. Chen, B. D. Dynlacht, and R. Tjian. 1993. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell 72:247-260. [DOI] [PubMed] [Google Scholar]
- 10.Juven-Gershon, T., S. Cheng, and J. T. Kadonaga. 2006. Rational design of a super core promoter that enhances gene expression. Nat. Methods. 3:917-922. [DOI] [PubMed] [Google Scholar]
- 11.Juven-Gershon, T., J.-Y. Hsu, and J. T. Kadonaga. 2008. Caudal, a key developmental regulator, is a DPE-specific transcription factor. Genes Dev. 22:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juven-Gershon, T., and J. T. Kadonaga. 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadonaga, J. T. 1991. Purification of sequence-specific DNA binding proteins by DNA affinity chromatography. Methods Enzymol. 208:10-23. [DOI] [PubMed] [Google Scholar]
- 14.Kokubo, T., D.-W. Gong, J. C. Wootton, M. Horikoshi, R. G. Roeder, and Y. Nakatani. 1994. Molecular cloning of Drosophila TFIID subunits. Nature 367:484-487. [DOI] [PubMed] [Google Scholar]
- 15.Kutach, A. K., and J. T. Kadonaga. 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20:4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, D. H., N. Gershenzon, M. Gupta, I. P. Ioshikhes, D. Reinberg, and B. A. Lewis. 2005. Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol. Cell. Biol. 25:9674-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis, B. A., T. K. Kim, and S. H. Orkin. 2000. A downstream element in the human beta-globin promoter: evidence of extended sequence-specific transcription factor IID contacts. Proc. Natl. Acad. Sci. U. S. A. 97:7172-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim, C. Y., B. Santoso, T. Boulay, E. Dong, U. Ohler, and J. T. Kadonaga. 2004. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 18:1606-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohler, U., G. C. Liao, H. Niemann, and G. M. Rubin. 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3:RESEARCH0087. http://genomebiology.com/2002/3/12/RESEARCH/0087. [DOI] [PMC free article] [PubMed]
- 20.Shao, H., M. Revach, S. Moshonov, Y. Tzuman, K. Gazit, S. Albeck, T. Unger, and R. Dikstein. 2005. Core promoter binding by histone-like TAF complexes. Mol. Cell Biol. 25:206-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 22.Soeller, W. C., S. J. Poole, and T. Kornberg. 1988. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 2:68-81. [DOI] [PubMed] [Google Scholar]
- 23.Thomas, M. C., and C. M. Chiang. 2006. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41:105-178. [DOI] [PubMed] [Google Scholar]
- 24.Wampler, S. L., C. M. Tyree, and J. T. Kadonaga. 1990. Fractionation of the general RNA polymerase II transcription factors from Drosophila embryos. J. Biol. Chem. 265:21223-21231. [PubMed] [Google Scholar]