Abstract
The Msh2-Msh3 heterodimer recognizes various DNA mispairs, including loops of DNA ranging from 1 to 14 nucleotides and some base-base mispairs. Homology modeling of the mispair-binding domain (MBD) of Msh3 using the related Msh6 MBD revealed that mismatch recognition must be different, even though the MBD folds must be similar. Model-based point mutation alleles of Saccharomyces cerevisiae msh3 designed to disrupt mispair recognition fell into two classes. One class caused defects in repair of both small and large insertion/deletion mispairs, whereas the second class caused defects only in the repair of small insertion/deletion mispairs; mutations of the first class also caused defects in the removal of nonhomologous tails present at the ends of double-strand breaks (DSBs) during DSB repair, whereas mutations of the second class did not cause defects in the removal of nonhomologous tails during DSB repair. Thus, recognition of small insertion/deletion mispairs by Msh3 appears to require a greater degree of interactions with the DNA conformations induced by small insertion/deletion mispairs than with those induced by large insertion/deletions that are intrinsically bent and strand separated. Mapping of the two classes of mutations onto the Msh3 MBD model appears to distinguish mispair recognition regions from DNA stabilization regions.
The DNA mismatch repair (MMR) pathway recognizes and repairs mispaired and damaged bases in DNA, which primarily result from replication errors but which also result from recombination and chemical damage to DNA and DNA precursors (16, 22). Repairing mispairs improves the overall fidelity of DNA replication and is important for genome stability (24). Inherited defects in MMR are responsible for most cases of Lynch syndrome (hereditary nonpolyposis colorectal cancer [HNPCC]), and furthermore, the epigenetic silencing of one of the genes involved in MMR, MLH1, underlies most cases of sporadic MMR-defective cancer (19, 29).
MMR is initiated by the recognition of base-base mismatches or insertion/deletion mispairs. In bacteria, the homodimeric MutS complex directly binds mispairs, bending the mispair-containing DNA by almost 60 degrees and shifting one of the mispaired bases, such as the thymidine base from G-T or +T mispairs, out of the DNA base stack (17). The mispaired base is stabilized by π stacking with a conserved phenylalanine (17, 26, 26a). DNA binding induces a functional asymmetry to the MutS complex; one subunit directly recognizes the mispair via a mispair-binding domain (MBD), whereas the MBD of the second subunit is primarily involved in nonspecific backbone interactions (17, 26a).
In eukaryotes, mitotic MMR utilizes two heterodimeric complexes of MutS homologs: Msh2-Msh6 and Msh2-Msh3 (5, 16, 23, 41). In these asymmetric heterodimers, Msh6 and Msh3 directly recognize the mispair via their MBDs, whereas the Msh2 subunit appears to be functionally equivalent to the MutS subunit that nonspecifically binds the DNA backbone. In wild-type cells, the Msh2-Msh6 heterodimer is thought to primarily recognize and act in the repair of base-base mispairs and small 1- or 2-nucleotide insertion/deletions (12, 16, 20-24). The crystal structure of human Msh2-Msh6 revealed that mispair recognition by Msh6 shares many details with Escherichia coli MutS, including the π-stacking phenylalanine (17, 26a, 39). In contrast, in wild-type cells the Msh2-Msh3 heterodimer is thought to primarily recognize and act in the repair of insertions and deletions from 1 to 14 nucleotides in size (11, 20, 21, 27, 33, 37, 40), although we have previously shown that Msh2-Msh3 also recognizes some base-base mispairs with a preference for those that have weak hydrogen bonding (13). Msh2-Msh3 is also targeted to sites of DNA double-strand breaks (DSBs), potentially before a branched recombination intermediate is formed, where it acts in the processing of 3′ single-stranded tails (10, 28, 36).
While no structural information for any Msh3 homolog is available, several lines of evidence suggest that mispairs are recognized by Msh2-Msh3 in a substantially different way than mispairs are recognized by MutS and Msh2-Msh6. First, Msh3 lacks the conserved π-stacking phenylalanine present in both MutS and Msh6, which is required for MMR by these proteins in vivo (9, 18). In contrast, mutagenesis of the Saccharomyces cerevisiae Msh3 residue located at the position equivalent to that of the phenylalanine conserved in MutS and Msh6 (K158, called K187 prior to the identification of the correct start codon [13]) caused only a modest MMR defect (18). Second, when other conserved residues and predicted DNA-backbone-contacting residues in S. cerevisiae Msh3 were mutated to alanine, only msh3-R247A (previously called msh3-R276A) caused a significant defect in the repair of 1-, 2-, and 4-nucleotide-long insertion/deletion mispairs (18).
Despite these differences, the Msh3 MBD is likely related to the MBD of Msh6 and MutS. Replacement of the Msh6 MBD with the Msh3 MBD generated a functional chimera possessing Msh3 substrate specificity (32). Moreover, combining the msh3-K158A mutation with K160A gave rise to an msh3 mutant with an MMR defect greater than that for either single mutant alone (18). This double mutant caused a loss of specificity for mispaired DNA (18). Together these data indicate not only that mispair specificity is determined by the Msh3 MBD but also that the critical region of the Msh3 MBD mediating mispair recognition likely overlaps the same region as the MBDs of MutS and Msh6, even if the nature of the recognition is different. We have therefore used homology modeling and site-directed mutagenesis to gain insight into how Msh3 recognizes a diverse array of mispairs.
MATERIALS AND METHODS
Molecular modeling.
An initial homology model for the S. cerevisiae Msh3 MBD (residues 133 to 255) was created using the human Msh6 MBD (Protein Data Bank accession number 2o8b [39]) and the SWISS-MODEL program (31). Two regions of the resulting model were treated as low-confidence regions. These regions were residues S230 to V244 (corresponding to a 3-fold crystal contact between the Msh6 MBDs in the Msh2-Msh6 crystal structure) and residues I175 to N193 (corresponding to a 14-amino-acid insertion not present in Msh6). Both low-confidence regions were outside the core recognition region of interest here and were rebuilt manually and refined with CNS software (4) to resolve steric problems in the original model built by SWISS-MODEL. The resulting model had reasonable stereochemical parameters (see Table S3 in the supplemental material), as revealed by the CNS and Procheck programs (25); however, our analysis of Msh3 MBD mutations using this model did not rely upon any detailed examination of particular side chain conformations or hydrogen-bonding interactions but, rather, relied on the position of interface residues on the hMsh6 MBD fold, which is less sensitive to errors in the homology-modeling process.
Plasmid construction.
Site-directed mutagenesis of a wild-type MSH3 low-copy-number LEU2 pRS315 plasmid (32, 35) was performed to generate mutations affecting the Msh3 MBD using the primers listed in Table S4 in the supplemental material. To measure Msh3 protein expression, the msh3 mutant alleles were C-terminally tagged with six copies of the HA epitope. The msh3 mutant plasmids were sequenced to confirm that only the desired mutation was present (see Table S5 in the supplemental material). All DNA sequencing was performed by using an Applied Biosystems 3730XL DNA sequencer and standard chemistry. Sequence analysis was performed using the Sequencher program (version 4.2.2; Gene Codes, Ann Arbor, MI).
General methods and strains.
All media have been described previously (2, 3, 30, 32). All strains used in the MMR studies were derivatives of S288c strain RDKY4234 MATα ura3-52 leu2Δ1 trp1Δ63 hom3-10 his3Δ200 lys2-10A msh3::hisG msh6::hisG (see Table S5 in the supplemental material) (3). All strains used in the DSB repair studies were derivatives of JKM146 Δho Δhml1::ADE1 Δhmr::ADE1 ade1 ade3::Gal::HO leu2-3 lys5 trp1::hisG ura3-52 (see Table S5 in the supplemental material) (28). Mutant derivatives were created using standard gene disruption and pop-in/pop-out gene replacement methods. The sequence of each mutant gene was verified by PCR amplification and sequencing.
Mutation assays.
Patches of cells from RDKY4234 containing various plasmid-borne msh3 alleles grown on plates lacking leucine were replica plated onto plates lacking leucine and threonine and grown at 30°C for 2 days to select for hom3-10 revertants. The microsatellite instability assay was performed by transforming a microsatellite-containing plasmid into the RDKY4234 strain containing a plasmid-borne msh3 allele. The microsatellite plasmid had a TRP1-selectable marker and contained the microsatellite repeat sequence (GT)16.5 or (CAGT)16 for 2 and 4 nucleotide repeats, respectively, in frame and prior to the URA3 gene (34). Strains containing both a plasmid with an msh3 allele and a plasmid required for a microsatellite stability assay were grown in patches on plates lacking leucine and tryptophan and then replica plated onto plates lacking leucine and tryptophan and containing uracil and 5-fluoroorotic acid and grown at 30°C for 2 to 3 days. Quantitative mutation rates were determined by fluctuation analysis using at least 14 independent colonies from each strain, as described previously (2, 3, 6, 30, 32).
Double-strand-break repair assays.
To analyze the DSB repair efficiency of the msh3 mutant alleles, several of the point mutations were made at the chromosomal locus of the MSH3 gene. These S. cerevisiae strains were then transformed with the DSB substrate plasmids pFP122 (mismatch), pFP140 (deletion of 30 nucleotides), and pFP120 (deletion of 300 nucleotides) (28). S. cerevisiae strains transformed with these plasmids were grown overnight in medium lacking uracil and were then serially diluted, plated onto yeast extract-peptone (YP)-glucose and YP-galactose plates at countable concentrations, and grown for 2 days. Colonies were then replica plated onto plates lacking uracil to check for plasmid retention, and after 2 days colonies from all plates were counted. For each DSB substrate, 7 to 10 independent experiments were performed using independent strain isolates. Galactose-induced expression of the HO endonuclease yielded a specific DSB in the substrate plasmids. The DSB repair efficiency is expressed as the percentage of plasmid retention under galactose conditions (DSB induction) relative to the level of plasmid retention under glucose conditions (no DSB induction).
Protein expression.
Cultures of RDKY4234 msh3Δ msh6Δ expressing the plasmid-borne tagged alleles were grown and harvested in log phase. Cell amounts were normalized by measuring the optical density and lysed in a standard buffer (50 mM Tris, pH 8.0, 5 mM dithiothreitol, 110 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail set IV [Calbiochem]) with glass beads (Sigma) by vortexing for 10 min. The supernatants from these strains were fractionated by SDS-PAGE and analyzed by Western blotting using an antibody to Cdc11 (Santa Cruz Biotechnology Inc., Santa Cruz, CA), a septin ring component to ensure that equivalent amounts of cellular protein were present in each extract. The supernatants were then used in an immunoprecipitation assay with anti-HA agarose resin (Sigma). The protein that eluted from the resin was fractionated by SDS-PAGE and analyzed by Western blotting using an anti-HA antibody (Roche) (see Fig. S5 in the supplemental material). The bands were quantitatively scanned with a Bio-Rad GS800 densitometer.
RESULTS
Homology model of the Msh3 MBD.
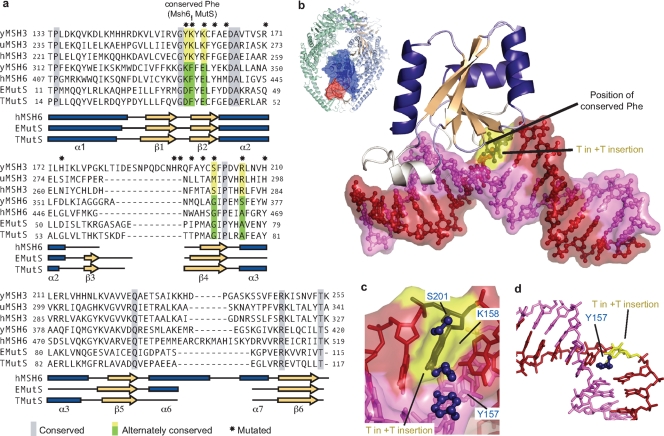
Several pieces of evidence argue that the overall fold of the Msh3 MBD is conserved with other MutS homologs: the extensive conservation between the MBD of MutS from bacteria, Msh6 from S. cerevisiae and humans, and Msh3 from S. cerevisiae and 28 other organisms (Fig. 1a; see Fig. S1 in the supplemental material); the similar patterns of predicted secondary structure (data not shown); and the ability to form a functional Msh6 chimera with an Msh3 MBD (32). We therefore generated a homology model of the S. cerevisiae Msh3 MBD (Fig. 1b) using the structure of the human Msh6 MBD (39). Superimposition of this model onto the structure of human Msh2-Msh6 complexed with a +T insertion revealed a number of clues to the differences between the DNA binding features of the Msh6 and Msh3 MBDs. Both K158, which is conserved in Msh3 and which aligns with the π-stacking phenylalanine in MutS and Msh6 (Fig. 1a), and S201, which is also conserved in Msh3 and which aligns with the conserved glycine in MutS and Msh6 that packs against the displaced nucleotide (Fig. 1a), sterically clash with the displaced thymidine in the Msh2-Msh6 complex (Fig. 1c). This model suggests that the displacement and stabilization of a single nucleotide from the base stack by MutS and Msh6 either do not occur or occur in a different fashion in Msh3.
FIG. 1.
Modeling of Msh3 MBD. (a) Alignment of the MutS homolog protein sequences: Msh3 from S. cerevisiae (y), Ustilago maydis (u), and Homo sapiens (h); Msh6 from S. cerevisiae (y) and H. sapiens (h); and MutS from Escherichia coli (E) and Thermus aquaticus (T). Gray boxes, conserved amino acid residues; green and yellow boxes, amino acid residues differentially conserved between Msh3, Msh6, and MutS; asterisks, residues that were mutated in this study. The secondary structures for E. coli MutS (Protein Data Bank accession number 1e3m [17]), T. aquaticus MutS (Protein Data Bank accession number 1fw6 [15]), and human Msh6 (Protein Data Bank accession number 2o8b [39]) are shown below the amino acid sequence. Blue bars, α helices; peach arrows, β sheets. (b) Model of Msh3 MBD on a +T insertion-containing DNA (red and pink) from the Msh2-Msh6 crystal structure (Protein Data Bank accession number 2o8f [39]). The +T insertion is shown in yellow and black. Regions of low confidence (see Materials and Methods) are shown in white. (Inset) Msh2-Msh6 heterodimer on +T insertion-containing DNA; the Msh6 MBD is in dark blue. (c) Model of Msh3 MBD residues on a +T insertion reveals a steric clash of K158, S201, and possibly, Y157 (blue) with the unpaired T (yellow and black). (d) Possible stacking of Y157 with the bases of the non-insertion-containing strand (pink). The molecular images were generated with the PyMOL program (7).
msh3 mutants differentially repair different DNA lesions.
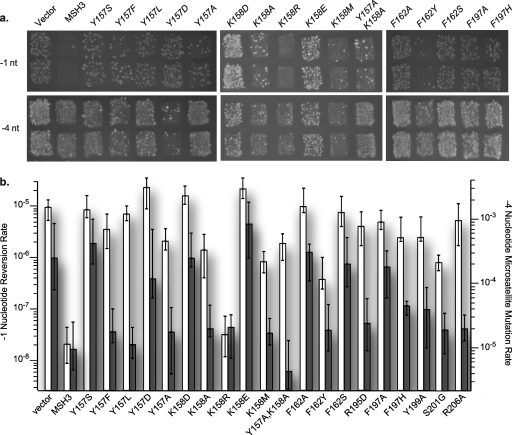
To experimentally probe the interactions between Msh3 and mispaired DNA, we designed a series of msh3 point mutation alleles in the MBD focusing on residues predicted by modeling to be at the MBD-DNA interface but also including residues from other regions of the MBD. These msh3 alleles were tested by expression from the native MSH3 promoter on a low-copy-number plasmid in an msh3Δ msh6Δ strain and evaluated for their effect on MMR proficiency using the −1 nucleotide hom3-10 frameshift reversion assay (Fig. 2; see Fig. S2 and Table S1 in the supplemental material) (20, 38). Four msh3 alleles had wild-type phenotypes, including E164A, R171A, H174A, and H194E (see Table S1 in the supplemental material). Of these mutations, only H174A and H194E affected residues with side chains predicted to be within 6 Å of the DNA. However, alleles predicted to affect amino acid residues at the MBD-DNA interface as well as some slightly removed from the interface had a defect in the repair of one nucleotide frameshift in the hom3-10 reversion assay, including Y157S, K158D, K160D, F162A, R195D, F197A, Y199A, S201G, R206A, and H210A.
FIG. 2.
Suppression of the msh3Δ phenotype by alternate amino acid substitutions in msh3 mutant alleles in MMR assays. (a) Patches of msh3Δ msh6Δ strains expressing msh3 alleles were replica plated onto plates lacking leucine and threonine for the −1-nucleotide (−1 nt) hom3-10 reversion assay. Patches of msh3Δ msh6Δ strains expressing msh3 alleles and containing a microsatellite plasmid with an in-frame 4-nucleotide repeat sequence upstream of the URA3 gene were replica plated onto plates lacking leucine and tryptophan and containing uracil and 5-fluoroorotic acid, as shown. (b) Mutation rates caused by msh3 mutant alleles in the frameshift repair assay (open bars) and the 4-nucleotide microsatellite assay (closed bars).
When alleles defective in the hom3-10 frameshift assay were tested for their effects in the repair of 2- and 4-nucleotide microsatellite stability assays (34), the alleles with MMR defects fell into two distinct classes (Fig. 2; see Fig. S2 and Table S1 in the supplemental material). One class also had defects in the repair of both 2-nucleotide and 4-nucleotide loops and included K158D, K160D, F162A, F197A, and H210A, in addition to defects in the repair of 1-nucleotide frameshifts. This class also included the ERN allele that replaced the S. cerevisiae-specific insertion between β3 and β4 (G180 to Q196; Fig. 1a) in the MBD, with the ERN sequence being found at the corresponding position in Msh3 from the fungus Ustilago maydis. The other class had no defect or nearly no defect in microsatellite stability (repair of 2-nucleotide and 4-nucleotide loops) and included Y157S, R195D, Y199A, S201G, and R206A. The four alleles that showed wild-type phenotypes in the hom3-10 frameshift assay also showed wild-type phenotypes in the 2- and 4-nucleotide microsatellite stability assays.
Two mutations that caused specific defects in frameshift repair when they were changed to the corresponding Msh6 or MutS residues, S201G and R206A, were used to design msh6 alleles encoding the corresponding Msh3 residues, G368S and S373R. These alleles were analyzed for their effects on Msh6-mediated 1-nucleotide frameshift repair. Neither msh6 allele enhanced the basal level of frameshift repair in the hom3-10 reversion assay; the msh6-G368S allele was completely defective, whereas the msh6-S373R allele did not cause any defect (see Fig. S3 in the supplemental material).
Additional mutations in the Msh3 MBD-DNA interface also fall into two classes.
To further investigate the msh3 Y157S, K158D, F162A, F197A, Y199A, and S201G alleles, we generated additional mutations that resulted in different amino acid substitutions at each position and tested them using the hom3-10 frameshift reversion assay and the 2-nucleotide and 4-nucleotide microsatellite stability assays.
In our Msh3 MBD model, Y157 is in the position to stack on bases in the strand opposite the +T insertion within the Msh6 structure (Fig. 1d). Consistent with this role, the Y157S, Y157F, and Y157A alleles were less defective for frameshift repair than Y157D and Y157L (Fig. 2a and b); however, all three alleles showed substantial defects relative to the wild-type sequence. In contrast to Y157D, alleles Y157S, Y157F, Y157A, Y157L, Y157A, and K158A were much more defective for frameshift repair than microsatellite stability (Fig. 2b; see Table S2 in the supplemental material).
Mutating Msh3 K158, which aligns with the π-stacking phenylalanine in MutS and Msh6, to aspartate or glutamate caused MMR defects in all three assays. In contrast, the K158R allele was indistinguishable from the wild type (95% confidence intervals) in the frameshift and microsatellite stability assays (Fig. 2a and b; see Table S2 in the supplemental material). Both the K158M and K158A alleles caused a slight defect, primarily in the frameshift repair assay.
The Msh3 F162Y allele caused an 18-fold defect in frameshift repair relative to that for the wild type, but the rate of microsatellite stability was indistinguishable from that for the wild type. In contrast, the F162S allele caused complete defects in both assays and the F162A allele caused partial defects in both assays (Fig. 2a and b; see Table S2 in the supplemental material). Importantly, the relative defect observed in the frameshift assay was similar to the relative defect observed in the microsatellite stability assay for each of the F162 alleles (Fig. 2b).
The Msh3 F197H allele caused a 114-fold defect in frameshift repair but a more modest defect in microsatellite stability. In contrast, the result for the F197A allele was indistinguishable from that for the empty-vector control for both the frameshift and microsatellite stability assays (Fig. 2a and b).
Msh3 Y199 was changed to leucine, aspartate, and lysine. When they were qualitatively tested for MMR proficiency using patch tests, the Y199D allele was completely defective in both assays and the Y199K allele was partially defective in both assays, similar to the original Y199A allele. The Msh3 Y199L allele caused a greater defect in the frameshift repair assay than the 4-nucleotide microsatellite stability assay (see Fig. S4 in the supplemental material).
Msh3 S201 was changed to leucine, aspartate, and arginine residues. The S201L allele was partially defective in both the frameshift and microsatellite assays. The S201D and S201R alleles caused null phenotypes in both the frameshift repair and microsatellite stability assays (see Fig. S4 in the supplemental material).
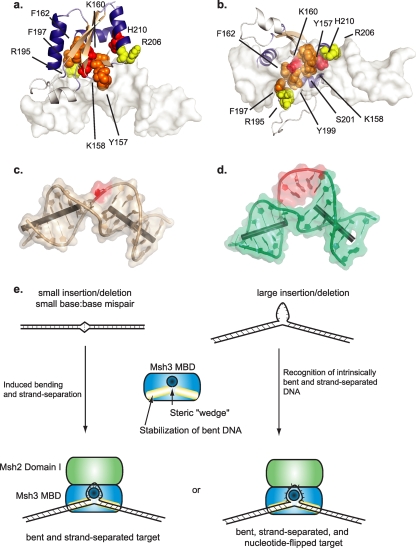
Mapping alleles causing MMR defects onto the Msh3 MBD model (see Fig. 4a and b) revealed that a central region, likely directly involved in mispair recognition, contains positions that, when they were mutagenized, caused equivalent defects in all of the MMR assays; other positions that, when they were mutagenized, caused greater defects in frameshift repair than in the microsatellite stability assays; and yet other positions that, when they were mutated, caused one or the other class of defect depending on the specific amino acid substitution tested. Remarkably, most of the central positions can be mutated to alleles that either equally affect frameshift repair and microsatellite stability or primarily affect frameshift repair. The amino acid positions associated with frameshift-specific defects tend to be on the periphery of the core recognition region.
FIG. 4.
Differential effects of msh3 mutant alleles in frameshift repair versus microsatellite stability assays. (a and b) Mutations mapped onto the model of the Msh3 MBD placed on a +T-containing DNA from the Msh2-Msh6 crystal structure (white; Protein Data Bank accession number 2o8f [39]). Red residues, positions that, when they are mutated, cause relative defects that are similar in all MMR assays; yellow residues, positions that cause more severe defects in the frameshift reversion assay than the microsatellite stability assay; orange residues, positions that, depending on the specific amino acid substitution, can cause equivalent defects in all MMR assays or greater defects in the frameshift assay than the microsatellite stability assay. (c) Structure of a DNA containing a +T insertion whose bend is induced by Msh2-Msh6 binding (Protein Data Bank accession number 2o8f [39]). Red, T mispair. (d) Structure of an intrinsically bent DNA containing a +5 A insertion (red; Protein Data Bank accession number 1qsk [8]). The molecular images were generated with the PyMOL program (7). (e) Model of the Msh3 MBD binding to intrinsically bent DNA containing large insertions or inducing and stabilizing nonbent DNA containing small DNA insertions. Recognition likely involves a steric wedge inserting between the DNA strands and stabilization of the DNA bend.
Double-strand-break repair defects of msh3 mutants mirror MMR defects.
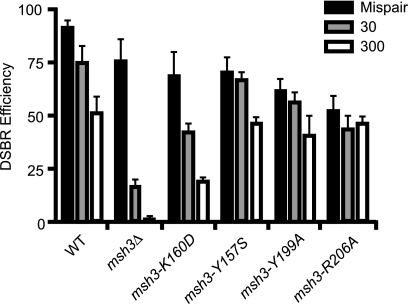
To investigate whether the msh3 mutants can perform other known functions of MSH3, we examined the repair of DNA DSBs. We used a previously characterized assay (28, 36) in which galactose induction of HO endonuclease leads to a single-site-specific DSB in the lacZ gene on the substrate plasmid that is subsequently a substrate for DSB repair. Retention of the substrate plasmid requires DSB repair by gene conversion using as a template the second copy of the lacZ gene on the plasmid that was not cut by HO due to a single nucleotide change in the HO recognition sequence. Consistent with previous results (28, 36), the msh3Δ null strain and also the msh3-K160D strain were able to repair a DSB nearly as well as the wild-type strain when the homologous target sequence was available (Fig. 3). However, substrate plasmids in which the DSB contained either 30- or 300-nucleotide regions of flanking nonhomology showed a significant reduction in plasmid retention in the msh3Δ and msh3-K160D mutants relative to that in the wild-type strains. In contrast, the msh3-Y157S, msh3-Y199A, and msh3-R206A mutants exhibited 55% to 70% retention of the plasmid with homology flanking the DSB site and exhibited no relative reduction in plasmid retention for the plasmids with either 30- or 300-nucleotide regions of nonhomology flanking the DSB compared to that for the wild type. These results parallel the results of MMR assays, where the mutant that was defective for the repair of all classes of substrates was fully defective in the repair of DSBs containing nonhomologies flanking the DSB, whereas the mutants that were defective for the repair of small mispairs (+1 frameshift) and proficient for the repair of larger loop mispairs (2- and 4-nucleotide insertion/deletions) had no defect in the repair of DSBs containing nonhomologies flanking the DSB. It is not clear why the msh3-Y157S, msh3-Y199A, and msh3-R206A mutants exhibited a modest decrease in retention of the plasmid substrate with homology at the DSB; however, this could be related to the fact that the Msh2-Msh3 complex is targeted to DSBs in vivo independently of the formation of recombination intermediates, combined with the fact that these mutants express separation-of-function mutant Msh2-Msh3 complexes that would interact with the induced DSB prior to the initiation of recombination (10).
FIG. 3.
Differential effect of msh3 mutations on the removal of nonhomologous tails during double-strand-break repair (DSBR). A wild-type strain and derivatives containing the indicated chromosomal msh3 mutations were analyzed for their ability to repair linear plasmids containing a single-base mismatch (black bars), a 30-base nonhomology tail (gray bars), or a 300-base nonhomology tail (white bars) produced by cleavage of plasmid DNAs by HO endonuclease in vivo. Repair is indicated by retention of the plasmids, and defects in repair are indicated by reduced retention of the plasmids. Each experiment was performed 7 to 10 times, and the error bars indicate the standard deviations of the measurements. The msh3-K160D mutation caused defects in the repair of 1-, 2-, and 4-base insertion/deletion mispairs; and the msh3-Y157S, msh3-Y199A, and msh3-R206A mutations caused defects only in the repair of 1-base insertion/deletion mispairs.
msh3 mutant proteins are equally expressed.
To eliminate the possibility that the differential MMR phenotype exhibited by some msh3 mutants could be due to the differential expression of the mutant proteins, we measured the Msh3 protein levels in selected mutants, including those containing alleles that cause differential MMR defects. Plasmid-borne msh3 alleles were C-terminally tagged with 6 copies of HA, immunoprecipitated from S. cerevisiae lysates, and detected by Western blotting. Comparable levels of expression were seen for all proteins analyzed, including Msh3, Msh3-Y157S, Msh3-K158D, Msh3-K160D, Msh3-Y199A, Msh3-S201G, and Msh3-R206A (see Fig. S5 in the supplemental material). Controls demonstrated that the supernatants had equivalent overall protein levels, as judged by analyzing the level of a septin ring component, Cdc11, by Western blot assay using a specific antibody. These results indicate that the phenotypes observed in this study are unlikely to be the result of the reduced expression of the mutant Msh3 proteins.
DISCUSSION
Here we have demonstrated by theoretical modeling and analysis of point mutations that mismatch recognition by Msh3 differs from that by MutS and Msh6. Unlike MutS and Msh6, in Msh3 there is no clear equivalent to the π-stacking phenylalanine residue involved in stabilizing the bases in the mismatch, as at least some alternative amino acids could be tolerated at each of the positions tested. Additionally, swapping individual amino acid residues or short stretches of residues between the Msh3 and Msh6 MBDs has not successfully altered mispair specificity, as demonstrated here and previously (18, 32). We have also shown that mutations affecting the Msh3 MBD fall into two classes. One class, including the Y157D, K158D, K158R, K158E, F162A, F162S, F197A, Y199D, Y199K, S201D, S201L, S201R, and H210A alleles, caused similar defects for all Msh3-based repairs. The second class, including the Y157S, Y157F, Y157A, Y157L, Y157A, K158A, K158A, K158M, F162Y, R195D, F197H, Y199A, Y199L, S201G, and R206A alleles, selectively disrupted 1-nucleotide frameshift repair but not 2- and 4-base loop repair; we would also anticipate that these mutations would prevent repair of the A-A, A-C, C-C, and C-T base-base mismatches that are recognized and repaired by Msh3; but currently, a simple, quantitative assay is not available to test the repair of specific single base-base mispairs (13). At present, analysis of the repair of Msh3-dependent base-base mispairs is performed by mutation spectrum analysis, which is tedious and not quantitative, and genetic assays to measure the in vivo repair of specific base-base mismatches by msh3-induced reversion have not yet been developed. While we did not test all of the msh3 alleles in the DSB repair assays, our results also indicate that the first class of alleles but not the second class of alleles causes defects in the removal of nonhomologous tails during DSB repair. This suggests that recognition of large loops during MMR and nonhomologous tails during DSB repair share common recognition properties. Importantly, we have not identified any mutations that specifically cause defects in 2- and 4-base loop repair but that are still proficient for 1-base frameshift repair, suggesting that loop repair may not specifically require any structural features of Msh3 that are not required for frameshift repair.
Why should repair of the DNA loops present in large insertion/deletion mispairs and DSB repair intermediates be less sensitive to mutation of the Msh3 MBD than repair of small frameshift mispairs? The structures of DNAs containing insertions of several nucleotides (+5 A insertions) demonstrate that these insertions form loops that cause the DNA helix to bend and force the inserted nucleotides to separate from the opposite strand (Fig. 4d) (8). The overall orientation and bend of the DNA strands in a +5 A insertion are highly reminiscent of those of the bend of the G-T mispair and the +T insertion containing DNAs bound by Msh2-Msh6 (Fig. 4c) (39). On the other hand, structures of small mispairs, such as 1-base insertion/deletions, are substantially less bent and the loop-containing strand is not as separated as DNAs containing large loops (Fig. 4e) (26, 39). Thus, we propose that 1-nucleotide frameshift mispairs require additional stabilization relative to the amount needed by large loops in order for the DNA substrates to be bent and recognized by the Msh3 MBD. This hypothesis would explain why we observe a class of mutations that is specifically defective in the repair of 1-base frameshift insertions and why we do not observe mutations that are specifically defective in the repair of larger loops. This hypothesis is also consistent with the fact that positions that affect frameshift repair only when they are mutated are outside the central mispair recognition region (Fig. 4a and b). The fact that the central region typically contains positions that, when they are mutated, affect both 1-nucleotide frameshift and 2- and 4-nucleotide loop repairs or primarily 1-nucleotide frameshift repair suggests that the loop recognition features of Msh2-Msh3 can also be the same features that stabilize induced conformations in small insertion/deletion mispairs.
Analysis of individual mutations in the context of the homology model also suggests that strand separation is important for mispair recognition by Msh2-Msh3, which is distinct from how Msh2-Msh6 and MutS recognize mispairs. Msh3 Y157 is well positioned to stack with bases of the non-loop-containing strand (Fig. 1d), whereas Msh3 K158, K160, and S201 could be part of either a steric wedge separating the two strands and hydrogen bonding to bases at the insertion/deletion site or a specific surface that interacts with and stabilizes the phosphates of a displaced and nucleotide-flipped loop-containing strand (Fig. 1c and 4e). Charge and size seem to be critical for the role of K158: K158R was mostly functional; K158A and K158M had increased defects, primarily in frameshift repair; and the negatively charged K158D or K158E alleles caused a substantial MMR defect, as did the negatively charged K160D allele. If Msh3 binds to and stabilizes a strand-separated substrate, then residues like F197 might π stack with bases in the loop. We note that the more conservative F197H allele that could retain some π-stacking ability was less defective for Msh3 repair than F197A.
While the E. coli E38 residue of the F-X-E motif is absolutely required for interaction with mispaired bases or 1-nucleotide insertions, or the corresponding residue E339 in S. cerevisiae Msh6 is dispensable for the repair of insertion/deletions and most base-base mispairs (14). While a similar F-X-E pattern is present in the Msh3 homologs, S. cerevisiae Msh3 E164 is not predicted to interact with the mispaired base or +1 frameshift mispair in the homology model and the E164A mutant appears to be wild type in the frameshift and loop repair assays.
Recognition of a bent and strand-separated substrate could easily allow recognition of a range of different loop sizes, consistent with the wide range of sizes recognized by Msh2-Msh3 (from 1 to 14 nucleotides) and the fact that Msh2-Msh3 binds to 1-base and larger insertion/deletion mispairs with similar affinities (1, 11, 13, 18, 27, 37, 40). This model is also consistent with the fact that Msh2-Msh3 has been observed to bind and distort some DNA substrates containing secondary structures, including substrates with 3′ single-stranded DNA overhangs and a splayed Y structure (37). The large loop-containing strand would also be positioned close to Msh2 domain I (S. cerevisiae Msh2 residues 2 to 133), which is equivalent to the Msh3 and Msh6 MBDs. Intriguingly, Msh3, but not Msh6, requires Msh2 domain I for repair (18), although this is not a fundamental requirement of the Msh3 MBD, as an Msh6 chimera containing the Msh3 MBD was independent of Msh2 domain I (32). The model presented here explains the flexibility exhibited by Msh3 during recognition of such varied substrates as weakly hydrogen bonded base-base mispairs and large insertion/deletion loops; however, analysis of the precise details of the interface await structural determination of Msh2-Msh3 complexed with various substrates at atomic resolution.
Supplementary Material
Acknowledgments
We thank Tom Petes (Duke Medical Center) for microsatellite instability plasmids. We also thank James Haber (Brandeis University) for the DSB repair plasmids.
This work was funded by NIH grant GM50006.
We declare that we have no competing financial interests.
Footnotes
Published ahead of print on 26 April 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Acharya, S., T. Wilson, S. Gradia, M. F. Kane, S. Guerrette, G. T. Marsischky, R. Kolodner, and R. Fishel. 1996. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. U. S. A. 93:13629-13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani, E., R. A. Reenan, and R. D. Kolodner. 1994. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics 137:19-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amin, N. S., M. N. Nguyen, S. Oh, and R. D. Kolodner. 2001. exo1-dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 21:5142-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 5.Constantin, N., L. Dzantiev, F. A. Kadyrov, and P. Modrich. 2005. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 280:39752-39761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das Gupta, R., and R. D. Kolodner. 2000. Novel dominant mutations in Saccharomyces cerevisiae MSH6. Nat. Genet. 24:53-56. [DOI] [PubMed] [Google Scholar]
- 7.DeLano, W. L. 2002. The PyMol molecular graphics system. DeLano Scientific, South San Francisco, CA.
- 8.Dornberger, U., A. Hillisch, F. A. Gollmick, H. Fritzsche, and S. Diekmann. 1999. Solution structure of a five-adenine bulge loop within a DNA duplex. Biochemistry 38:12860-12868. [DOI] [PubMed] [Google Scholar]
- 9.Drotschmann, K., W. Yang, F. E. Brownewell, E. T. Kool, and T. A. Kunkel. 2001. Asymmetric recognition of DNA local distortion. Structure-based functional studies of eukaryotic Msh2-Msh6. J. Biol. Chem. 276:46225-46229. [DOI] [PubMed] [Google Scholar]
- 10.Evans, E., N. Sugawara, J. E. Haber, and E. Alani. 2000. The Saccharomyces cerevisiae Msh2 mismatch repair protein localizes to recombination intermediates in vivo. Mol. Cell 5:789-799. [DOI] [PubMed] [Google Scholar]
- 11.Habraken, Y., P. Sung, L. Prakash, and S. Prakash. 1996. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr. Biol. 6:1185-1187. [DOI] [PubMed] [Google Scholar]
- 12.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34:359-399. [DOI] [PubMed] [Google Scholar]
- 13.Harrington, J. M., and R. D. Kolodner. 2007. Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base-base mispairs. Mol. Cell. Biol. 27:6546-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes, S. F., K. D. Scarpinato, S. D. McCulloch, R. M. Schaaper, and T. A. Kunkel. 2007. Specialized mismatch repair function of Glu339 in the Phe-X-Glu motif of yeast Msh6. DNA Repair (Amst) 6:293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Junop, M. S., G. Obmolova, K. Rausch, P. Hsieh, and W. Yang. 2001. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell 7:1-12. [DOI] [PubMed] [Google Scholar]
- 16.Kolodner, R. D., and G. T. Marsischky. 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9:89-96. [DOI] [PubMed] [Google Scholar]
- 17.Lamers, M. H., A. Perrakis, J. H. Enzlin, H. H. Winterwerp, N. de Wind, and T. K. Sixma. 2000. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature 407:711-717. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. D., J. A. Surtees, and E. Alani. 2007. Saccharomyces cerevisiae MSH2-MSH3 and MSH2-MSH6 complexes display distinct requirements for DNA binding domain I in mismatch recognition. J. Mol. Biol. 366:53-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch, H. T., and A. de la Chapelle. 2003. Hereditary colorectal cancer. N. Engl. J. Med. 348:919-932. [DOI] [PubMed] [Google Scholar]
- 20.Marsischky, G. T., N. Filosi, M. F. Kane, and R. Kolodner. 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10:407-420. [DOI] [PubMed] [Google Scholar]
- 21.Marsischky, G. T., and R. D. Kolodner. 1999. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J. Biol. Chem. 274:26668-26682. [DOI] [PubMed] [Google Scholar]
- 22.Modrich, P. 1991. Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 25:229-253. [DOI] [PubMed] [Google Scholar]
- 23.Modrich, P. 2006. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 281:30305-30309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Modrich, P., and R. Lahue. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65:101-133. [DOI] [PubMed] [Google Scholar]
- 25.Morris, A. L., M. W. MacArthur, E. G. Hutchinson, and J. M. Thornton. 1992. Stereochemical quality of protein structure coordinates. Proteins 12:345-364. [DOI] [PubMed] [Google Scholar]
- 26.Natrajan, G., M. H. Lamers, J. H. Enzlin, H. H. Winterwerp, A. Perrakis, and T. K. Sixma. 2003. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucleic Acids Res. 31:4814-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Obmolova, G., C. Ban, P. Hsieh, and W. Yang. 2000. Crystal structure of the mismatch repair protein MutS and its complex with a substrate DNA. Nature 407:703-710. [DOI] [PubMed] [Google Scholar]
- 27.Palombo, F., I. Iaccarino, E. Nakajima, M. Ikejima, T. Shimada, and J. Jiricny. 1996. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 6:1181-1184. [DOI] [PubMed] [Google Scholar]
- 28.Paques, F., and J. E. Haber. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peltomaki, P. 2003. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 21:1174-1179. [DOI] [PubMed] [Google Scholar]
- 30.Reenan, R. A., and R. D. Kolodner. 1992. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics 132:975-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shell, S. S., C. D. Putnam, and R. D. Kolodner. 2007. Chimeric Saccharomyces cerevisiae Msh6 protein with an Msh3 mispair-binding domain combines properties of both proteins. Proc. Natl. Acad. Sci. U. S. A. 104:10956-10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sia, E. A., M. Dominska, L. Stefanovic, and T. D. Petes. 2001. Isolation and characterization of point mutations in mismatch repair genes that destabilize microsatellites in yeast. Mol. Cell. Biol. 21:8157-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sia, E. A., R. J. Kokoska, M. Dominska, P. Greenwell, and T. D. Petes. 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17:2851-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugawara, N., F. Paques, M. Colaiacovo, and J. E. Haber. 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. U. S. A. 94:9214-9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Surtees, J. A., and E. Alani. 2006. Mismatch repair factor MSH2-MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J. Mol. Biol. 360:523-536. [DOI] [PubMed] [Google Scholar]
- 38.Wang, Q., U. G. G. Hennig, R. G. Ritzel, E. A. Savage, and R. C. von Borstel. 1990. Double-stranded base sequencing confirms the genetic evidence that the hom3-10 allele of Saccharomyces cerevisiae is a frameshift mutation. Yeast 6:S76. (Abstract.) [Google Scholar]
- 39.Warren, J. J., T. J. Pohlhaus, A. Changela, R. R. Iyer, P. L. Modrich, and L. S. Beese. 2007. Structure of the human MutSalpha DNA lesion recognition complex. Mol. Cell 26:579-592. [DOI] [PubMed] [Google Scholar]
- 40.Wilson, T., S. Guerrette, and R. Fishel. 1999. Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J. Biol. Chem. 274:21659-21664. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, Y., F. Yuan, S. R. Presnell, K. Tian, Y. Gao, A. E. Tomkinson, L. Gu, and G. M. Li. 2005. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122:693-705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.