Abstract
Mammals compensate for unequal X-linked gene dosages between the sexes by inactivating one X chromosome in the female. In marsupials and in the early mouse embryo, X chromosome inactivation (XCI) is imprinted to occur selectively on the paternal X chromosome (XP). The mechanisms and events underlying XP imprinting remain unclear. Here, we find that the imprinted XP can be functionally divided into two domains, one comprising traditional coding genes (genic) and the other comprising intergenic repetitive elements. XP repetitive element silencing occurs by the two-cell stage, does not require Xist, and occurs several divisions prior to genic silencing. In contrast, genic silencing initiates at the morula-to-blastocyst stage and absolutely requires Xist. Genes translocate into the presilenced repeat region as they are inactivated, whereas active genes remain outside. Thus, during the gamete-embryo transition, imprinted XCI occurs in two steps, with repeat silencing preceding genic inactivation. Nucleolar association may underlie the epigenetic asymmetry of XP and XM. We hypothesize that transgenerational information (the imprint) is carried by repeats from the paternal germ line or that, alternatively, repetitive elements are silenced at the two-cell stage in a parent-of-origin-specific manner. Our model incorporates aspects of the so-called classical, de novo, and preinactivation hypotheses and suggests that Xist RNA functions relatively late during preimplantation mouse development.
Genomic imprinting refers to a parent-of-origin effect on gene expression in the developing embryo (3, 57). The existence of imprinting in the mammal means that male and female gametes contribute significantly different information to the zygote. One important difference is illustrated by X chromosome inactivation (XCI), the mechanism of dosage compensation in the mammal that results in the silencing of one X chromosome in the female embryo (2, 33, 34, 49, 64). While the eutherian form of XCI occurs randomly in the soma, the marsupial form is imprinted to occur exclusively on the paternal X (XP) (54). Imprinted XCI also occurs in some eutherians but is restricted to the preimplantation embryo and the extraembryonic tissues (25, 37, 47, 60). Imprinted XCI precedes random XCI in the early mouse embryo and continues through the placental lineages. In the epiblast (embryo proper), transient X reactivation is followed by random XCI, which accounts for the mosaic pattern of inactivation seen in all somatic tissues of the eutherian.
The mechanisms and developmental timing of imprinted XCI remain unclear and are much debated. In principle, the maternal or paternal germ line (or both) may differentially mark the X chromosomes, with the maternal imprint protecting the maternal X (XM) from inactivation and/or the paternal mark predestining XP for inactivation. The search for parent-specific regulators frequently has focused on the X inactivation center (Xic) (7), an X-linked region harboring several noncoding regulators for random XCI. Xist produces a 17-kb noncoding transcript whose accumulation on the X has been associated with the initiation of both random and imprinted XCI (6, 38, 50). In the preimplantation mouse embryo and in the extraembryonic lineages of the postimplantation embryo, Xist is imprinted to be paternally expressed in accordance with preferential XP inactivation (28). The randomization of Xist expression following X reactivation in the epiblast lineage results in mosaic XM and XP inactivation in the embryo proper.
The Xic also harbors Tsix, the 40-kb noncoding transcript that is complementary to and negatively regulates Xist (31). In contrast to Xist, Tsix is imprinted to be maternally expressed and therefore may be a maternal factor that protects XM from silencing in the early embryo and extraembryonic tissues (30, 53). Tsix expression also becomes randomized following X reactivation in the epiblast and is expressed exclusively from the future active X (Xa) in the developing embryo proper. In the eutherian embryo, the importance of Xist/Tsix in the imprinting of the X has been borne out by genetic analyses: deleting Xist from XP causes a loss of XP silencing in the placental lineages (38), whereas deleting Xist from XM (on which it is normally silent) has no consequence; conversely, deleting Tsix from XP (on which it is normally silent) has no consequence, whereas deleting Tsix from XM results in ectopic XCI on XM in the placental lineages (30, 53). Thus, for both imprinted and random XCI, Xist designates the future inactive X (Xi), while Tsix designates the future active X (Xa).
Although Xic clearly regulates imprinting in eutherians, XIC or an equivalent has yet to be identified in marsupials (11, 12, 22, 55). The absence of a marsupial Xist suggests that an alternative means of silencing the X must occur in mammals. Since the discovery of meiotic sex chromosome inactivation (MSCI) in the male germ line of both eutherian and marsupial mammals (14, 23, 32, 43, 58), several groups have hypothesized a link between MSCI and the imprinting of XP (10, 24, 26, 35, 39). Recent reports that XY silencing persists into the long postmeiotic period of spermatogenesis (16, 42, 62) support the idea that zygotic XP silencing is built in part on MSCI and its aftereffects in the paternal germ line. Because MSCI is Xist independent and Xist is not highly expressed during spermatogenesis (40, 61), germ line-driven silencing would provide an alternative imprinting mechanism that would not require an XIC in the marsupial and would dosage compensate the marsupial zygote from the time of conception.
The probability of an XIST-independent mechanism in the marsupial raises intriguing questions for imprinted XCI in eutherians. Did eutherian XCI evolve completely independently, or do vestiges of a marsupial mechanism still exist in the eutherians of today? Although the placental form of imprinted XCI in the mouse clearly depends on Xist (38), the role of Xist in the preimplantation embryo currently is unclear. Indeed, embryos deleted for Xist on XP are normal in the preimplantation stages and perish only after uterine implantation and the outgrowth of a placenta (38), suggesting that the early mouse embryo does not require Xist. There also is debate as to whether mouse XP is inherited from the male germ line in a partially inactive state, further raising the question of whether Xist is required to initiate imprinted XCI in the early mouse embryo (25, 45, 46). Here, we investigate the mechanism of XP silencing in the earliest stages following the gamete-to-embryo transition. We discover that imprinted XP silencing takes place in two sequential steps, one involving repetitive elements and the other involving coding genes, and implicate repeats in the transmission of parental information to the early embryo.
MATERIALS AND METHODS
Mice.
Mice carrying a deletion of Xist exons 1 to 6 (38) or an X-linked GFP transgene (D4/XEGFP) (17) have been described previously. To obtain XMXP;GFP,Xist− and XM;GFP,Xist−XP mice, we crossed Xist knockout mice to D4/XEGFP mice and obtained meiotic recombinants carrying the GFP transgene on the Xist-deficient X.
Embryo culture.
Embryonic day 3.5 (E3.5) blastocysts were flushed from uteri and cultured overnight to E4.5 in drops of potassium simplex optimized medium under mineral oil (Millipore). After that, they were transferred onto gelatin-coated Lab-Tek chamber slides (Nunc) and cultured in Dulbecco's modified essential medium (DMEM) with 10% fetal bovine serum (FBS) (Sigma) until E5.5 or E6.5 to obtain blastocyst outgrowths.
RNA and DNA FISH.
Cytologic analysis in the preimplantation embryo is exceedingly difficult, because the material is extremely limiting and the embryos have a lot of cytoplasm relative to nuclear volume, which makes probe penetration challenging. To improve the sensitivity of the fluorescent in situ hybridization (FISH) assay, we collected published protocols and systematically varied each parameter to identify conditions that would yield the highest signal-to-noise ratio. Our optimization process led us to conclude that the removal of the cytoplasmic background is the most critical determinant for the success of RNA/DNA FISH in early embryos. Specifically, the order of fixation versus permeabilization was crucial. When we fixed before permeabilizing (47), the cytoplasmic background always was high. In our optimal protocols, the permeabilization step had to either precede fixation or occur simultaneously. For all FISH experiments described in this study (except those in Fig. 1 A and B), permeabilization (using Tergitol detergent) and fixation (using paraformaldehyde) are performed simultaneously. Preimplantation embryos were recovered at appropriate stages in M2 medium and promptly treated with Tyrode solution to remove the zona pellucida. Embryos were incubated in 6 mg/ml bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for several minutes and briefly dried directly on glass slides. The slides then were fixed in ice-cold 1% paraformaldehyde in PBS with 0.05% NP-40 for 5 min and subsequently fixed again in ice-cold 1% paraformaldehyde in PBS for 5 min, and then they were stored in 70% ethanol. Prior to RNA FISH, the slides were dehydrated in a standard ethanol series (70, 80, and 100% ethanol, each for 2 min at room temperature).
FIG. 1.
Repeat silencing of XP in the early embryo. (A to C) Two-color RNA FISH reveals the relationship between Xist RNA and Cot-1 expression in three representative XX two-cell embryos, using published protocols (47) (A and B; both nuclei are shown in the right panels) or our newly developed protocols (C). All images are deconvolved single z sections. Blastomeres indicated by asterisks in leftmost panels are magnified. Results for the second blastomere are shown in Fig. S1 in the supplemental material. The intensity of Cot-1 expression is quantified by fluorimetry across the indicated path (′ to ″) and plotted in the relative intensity range of 0 to 1. Note that XP often resides next to the prenucleolus (N) and coincides with DAPI-intense regions. (D) X chromosome painting of a two-cell blastomere reveals that XP and XM occupy large nuclear territories. Note that XP is associated with the prenucleolus and is relatively lacking in Cot-1 compared to XM. The image represents merged z stacks (3D stacks projected onto a single plane) to capture X-chromosomal signals in multiple z sections. (E to H) Cot-1 RNA FISH of later stage XX embryos as indicated, presented as described for panel A to C. For panel G and H, serial z sections are merged (merged z's) to show the degree of Cot-1 expression throughout the nucleus. Following slide denaturation, X paint was performed to reveal the locations of XM and XP.
For Cot-1, Chic1, and G6pdx RNA FISH shown in Fig. 1A and B, we used wild-type two-cell embryos derived from B6CBAF1 crosses as described previously (47). All other RNA FISH experiments were performed using the embryo derived from crosses between B6D2F1 mice unless otherwise designated. For the RNA FISH analysis of the Xist mutant, embryos were derived from crosses between B6D2F1 females and Xist mutant males. Analysis of spermatogenesis was carried out as previously described (42).
Cot-1 RNA FISH was performed as described previously (25). Xist RNA FISH was performed using a fluorescein isothiocyanate (FITC)-dUTP-labeled pSx9 probe generated by a nick translation kit (Roche). Cot-1 DNA (Invitrogen) was labeled with Cy3-dUTP (GE Healthcare) using the Prime-It kit (Stratagene). Cot-1 hybridization was performed at 42°C overnight with 100 ng of the Xist probe, 80 ng of Cot-1 probes, and 9 μg of herring sperm DNA (Invitrogen) in 20 μl of hybridization buffer (50% formamide, 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 2 mg/ml BSA, 10% dextran sulfate-500K) per slide. Slides were washed two times with 2× SSC, 50% formamide at 45°C for 5 min each and two times with 2× SSC at 45°C for 5 min each. Nascent RNA FISH was performed as described previously (5). The following bacterial artificial chromosome (BAC) clones were obtained from CHORI to generate probes labeled with FITC-dUTP (Stratagene), Cy3-dUTP (GE Healthcare), or Cy5-dUTP (GE Healthcare) via nick translation (Roche) to detect the following nascent transcripts: Usp9x, RP24-306P3; Utx, RP23-174N2; Lamp2, RP24-173A8; Hprt1, RP24-335G16; Chic1, CH29-617L21; Atrx, RP23-450B21; Atp7a, RP24-118E11; Jarid1c, RP24-148H21; and Pdha1, RP24-374N15. For the G6pdx probe, 5.6 kb of genomic DNA of g6pdx was cloned into pGEM-T-easy (Promega). The hybridization of the nascent transcript was performed at 37°C overnight with 100 ng of each gene-specific probe with 10 μg of herring sperm DNA (Invitrogen), 10 μg of yeast tRNA (Invitrogen), 10 μg of Cot-1 DNA (Invitrogen), and 20 mM ribonucleoside vanadyl complex (New England Biolabs) in 20 μl of hybridization buffer (50% formamide, 2× SSC, 2 mg/ml BSA, 10% dextran sulfate-500K) per slide. Slides were washed two times with 2× SSC, 50% formamide at 37°C for 5 min each and two times with 2× SSC at 37°C for 5 min each. Nascent RNA FISH signals were verified to be genuine by overlaying images from second-round DNA FISH performed using the same BAC probes. Coincident DNA FISH signals confirmed that nascent RNA signals were truly from the corresponding genes.
LINE and SINE RNA FISH probes were obtained by the PCR of conserved regions of each repeat class using consensus primers for LINEs and B1 and B2 SINEs (Repbase; http://www.girinst.org/repbase/index.html). B1 and B2 fragments were amplified using the following 5′-end-labeled TYE563 primers (Integrated DNA Technologies): SINE B1 (5′-GTGGCRCAYGCCTTTAAT-3′ and 5′-CGAGACAGGGTTTCTCTGTG-3′) and B2 (5′-GCTGGWGAGATGGCTCAGYG-3′ and 5′-AGCTGTCTTCAGACACACCAGA-3′). PCR products were <150 bp, purified following amplification, and used directly to perform RNA FISH in the presence of a 50-fold excess of herring sperm DNA (Invitrogen). For LINE RNA FISH, two overlapping fragments covering the consensus LINE region (4 kb) were amplified using the following primer pairs: 5′ primers, ATTACCATAGATGGAGAAACCAAA and TGACCATAGGTGTGTGGGTTC; 3′ primers, TTCTTTCCAGCTTCTGGCTATTA and GATTCAATGCAATCCCCATC. These fragments were combined in equal molar ratios, labeled by nick translation (Roche), and used in RNA FISH with a 50-fold excess of herring sperm DNA (Invitrogen).
For DNA FISH, Sx7 and πXE9 probes were labeled by nick translation (Roche) using FITC-dUTP (Stratagene), Cy3-dUTP (GE Healthcare), or Cy5-dUTP (GE Healthcare). Cy3-labeled X-painting probes were obtained from Cambrio (United Kingdom). For the analysis of specific X-linked repetitive domains, the following BAC clones were labeled by nick translation (Roche) using FITC-dUTP (Stratagene), Cy3-dUTP (GE Healthcare), or Cy5-dUTP (GE Healthcare): BAC79Mb, RP24-120A7; BAC90Mb, RP23-337O18; BAC116Mb, RP24-280J13; BAC124Mb, RP24-387A17; BAC21Mb, RP23-445I18; and BAC44Mb, RP23-311H6. DNA FISH was performed according to reference 66, with minor modifications. Slides were treated with 2 mg/ml RNase A at 37°C for 30 min. After being washed with PBS, slides were treated with 0.2 N HCl, 0.5% Triton X-100 on ice for 10 min and then were denatured in 70% formamide, 2× SSC at 80°C for 10 min. For the Xic probes, 100 ng of probes was suspended in hybridization buffer with 1.5 μg/μl mouse Cot-1 DNA (Invitrogen), denatured at 80°C for 10 min, and preannealed at 37°C for 10 to 20 min. For X paint analysis, 2 ml of 5× stock was used. Before hybridization, denatured slides were dehydrated in an ice-cold ethanol series (70, 80, and 100%), probe was applied overnight at 37°C, and the slides were washed two times with 2× SSC, 50% formamide at 37°C for 5 min each and two times with 2× SSC at 37°C for 5 min each.
Sequential RNA/DNA FISH.
RNA/DNA FISH was carried out serially. RNA FISH was carried out first, photographs were taken, and x-y coordinates were marked. Prior to DNA FISH, slides were fixed in 4% paraformaldehyde in PBS with 0.5% Tween 20 and 0.5% NP-40 for 10 min at room temperature to preserve the morphology of embryonic nuclei and to enhance the penetration of probes. Slides then were subjected to RNase treatment at 2 mg/ml. After being washed with PBS, slides were treated with 0.2 N HCl 0.5%, Triton X-100 on ice for 10 min and then denatured in 70% formamide, 2× SSC at 80°C for 10 min, and DNA FISH was performed as described above. Photographs then were taken, and RNA/DNA images of the same x-y coordinates were identified using Volocity software (Improvision) prior to being merged in Photoshop (Adobe). Note that the images were captured in each channel as gray-scale images, pseudocolored as red, green, and blue in RGB space, and then merged. To generate figure panels, each image was converted to CMYK mode.
Immunostaining.
Immunostaining was performed as described previously (47) using dilutions of the following antibodies: RNA polymerase II (Pol-II) (CTD8WG), 1:200; RNA Pol-II (H5), 1:50; and RNA Pol-III (RPC 53 subunit), 1:500.
Image acquisition and analysis.
For the Cot-1 RNA FISH analysis at the two-cell stage and in the male germ line (Fig. 1A and B; also see Fig. 9 and Fig. S1 in the supplemental material), images were acquired with the Axioplan2 microscope (Zeiss) and Openlab software (Improvision). All other images were acquired with the Eclipse 90i microscope (Nikon) and Volocity software (Improvision). Fifty z sections were taken at 0.2-μm intervals for each embryo and analyzed after deconvolution by Volocity software (Improvision). For the scoring of transcriptional activity and the nucleolar association of specific X-linked repeat elements, RNA FISH and DNA FISH were performed in series, and their corresponding images were serially captured using the XY stage function of the Nikon 90i microscope. Approximately 50 z sections were analyzed for each image. To find the corresponding z sections in the RNA and DNA FISH experiments, z sections of 4′,6′-diamidino-2-phenylindole (DAPI) images were converted to the black-white images, and the DAPI patterns were compared in all planes. Thereafter, using the DAPI images as guides, the RNA and DNA FISH signals were compared or merged in each z section.
FIG. 9.
Perinucleolar association present in two- and four-cell embryos is lost during the 8-cell stage. (A) Cot-1 and Xist RNA FISH with subsequent DNA FISH using the Xic probe, Sx7, in the wild-type four-cell embryo. (B) Cot-1 RNA FISH with subsequent DNA FISH in the Xist mutant four-cell embryo using a combination of Sx7 and πXE9 probes to distinguish XM (wild-type) from XP (Xist deficient). For panels A and B, two z sections (top and bottom) are shown for each blastomere to capture the XM and XP planes. (C) Xic nucleolar association of XP versus that of XM in 4- and 8-cell embryos. Cot-1 RNA FISH was performed on wild-type and Xist-deficient embryos as described for Fig. 7C with the following exception: for 4- and 8-cell embryos, nucleolar association correlated with a Cot-1− state of XP in 100% of blastomeres; however, for the wild-type embryos, a lack of nucleolar association was correlated with silencing in a fraction of blastomeres. P values were calculated using the unpaired student t test. WT, wild-type. Xist−, XMXP;Xist−. (D) X chromosome painting of a two-cell embryo of an Xist mutant reveals large X territories (circled) at this stage. (E) Cot-1 RNA FISH and subsequent X chromosome painting show no compaction of XP in the Xist mutant embryo at the blastocyst stage. (F) Pictorial representation of the deduced XP and XM structures in the early embryo. Repeat elements of XP lie in the silent perinucleolar compartment, while XM and active genic loci of XP reside in Cot-1+ regions.
For the line traces in Fig. 1, 2, 7, and 9, we captured the images in each channel as grayscale images in multiple z sections and then deconvolved the images using Volocity software. For quantitation, we chose the z section in which Xist RNA (green channel) is most intense (the center of the Xist focus) and extracted the corresponding z section in the Cot-1 (red) and DAPI (blue) channels. We exported these images from Volocity to Photoshop and adjusted the levels in each channel to increase our dynamic range. Because XM and XP lie in the same nucleus and their photographs were captured at the same time, level adjustments were performed on XM and XP together within the context of the same nucleus and quantitated across the same dynamic range (see the ImageJ plot in Fig. S6 in the supplemental material). To generate the line traces, we exported the adjusted images to the NIH's ImageJ software and performed the quantitative analysis along a single transect as shown.
FIG. 2.
Characterization of the Cot-1− domain. A single representative blastomere is shown in all panels, and the intensity of immunostaining is quantified by densitometry across the indicated path (′ to ″) and plotted in the relative intensity range of 0 to 1. (A to C) Immunostaining using antibodies against RNA polymerases II and III as indicated. The frequencies of exclusion from XP were the following: Pol-II (CTD), 59% (n = 17); Pol-II (H5), 79% (n = 19); Pol-III, 58% (n = 19). (D to F) Expression analysis of the indicated repetitive elements by RNA FISH. Exclusion frequencies were the following: LINE, 62% (n = 13); B1, 67% (n = 12); and B2, 64% (n = 14).
To generate the interactive three-dimensional (3D) movies, we assembled ∼50 z sections taken at 0.2-μm intervals using the 3D opacity mode in Volocity. Volumes of RNA FISH images (Cot-1 and Xist) and DNA FISH images (BAC probes) were aligned using the DAPI stain as a guide, as the DAPI stain is common to both. Accurate DAPI alignments in 3D were made possible by the automated registration correction function in Volocity. The 3D volumes then were exported as QuickTime VR interactive movies and can be viewed using QuickTime player software.
GFP images of blastocysts and postimplantation embryos were acquired with the Eclipse TE2000-E microscope (Nikon) and Openlab software (Improvision). To compare GFP fluorescence intensities, images of equivalent stages were taken using identical microscope and software settings. Mean pixel intensity for each tissue and genotype was quantified using Volocity. Measurements of intranuclear distances and nuclear dimensions were taken using Volocity.
RT-PCR.
Allele-specific reverse transcription-PCR (RT-PCR) methods and primer pairs spanning single-nucleotide polymorphisms (SNPs) were described previously (25). Note that Xnp equals Atrx and Rlim equals Rnf12.
RESULTS
Repeat silencing in the preimplantation embryo.
To explore potential mechanisms of imprinted silencing, we first revisited the debate over how much of XP already is inactive at the two-cell stage when the major wave of zygotic gene activation (ZGA) takes place. According to the classical model, XCI first takes place during the implantation period (E3.5 to E5.5) (1, 13, 29). More recent models proposed that XCI takes place earlier, with the de novo hypothesis positing that XCI initiates at the four-cell stage (47) and the preinactivation hypothesis postulating that the embryo inherits a partially silent XP from the paternal germ line (25). The timing and extent of silencing have remained unclear, due in large part to the difficulty of carrying out cytological and expression analyses on preimplantation embryos because of their minute size. Here, we developed a more sensitive method of DNA and RNA fluorescence in situ hybridization (FISH) that reduces cytoplasmic background while improving signal detection using locus-specific, Cot-1, and chromosome-painting probes (see Materials and Methods). Thereafter, all images were captured, deconvolved using Volocity software (Improvision) to subtract out-of-focus light, and quantitated by fluorescence intensity scanning across the nucleus.
Critical to the analysis of nascent transcription on a global scale is the use of Cot-1 probes (9, 18, 25). The Cot-1 genomic fraction contains highly repetitive, non-coding-expressed elements such as retrotransposons, centromere-associated repeats, and other simple repeats that concentrate in intergenic and intronic regions. Therefore, Cot-1 probes identify domains of new transcription within both genic and intergenic regions. Using either previous methods (47) or our newly developed method of RNA FISH, we observed that 76% (n = 41) (Fig. 1A and B; also see Fig. S1 in the supplemental material) or 71% (n = 28) (Fig. 1C), respectively, of the Xist RNA domains excluded Cot-1 hybridization at the two-cell stage. At this stage, the size of Xist domains varied from a small RNA cluster (Fig. 1A and B; also see Fig. S1 in the supplemental material) to a large aggregate akin to what normally is seen in somatic cells when Xist starts to spread (Fig. 1C). Fluorimetric analysis revealed that Xist-coated XP domains not only resided in a neighborhood lacking Cot-1 transcription (the line plot shows that Xist peaks coincided with Cot-1 troughs) but they also are generally DAPI intense (the line plot shows that Xist peaks coincided with bright DAPI staining). X chromosome DNA painting showed that both XM and XP occupied large nuclear territories during the two-cell stage (Fig. 1D and data not shown). It is noteworthy that XP generally extended beyond the region of Xist accumulation. In eight out of eight blastomeres tested at the two-cell stage, XP lay within relatively Cot-1-poor regions and is closely associated with the nascent nucleolus (the nascent prenucleolus hereafter is referred to as the nucleolus for the sake of simplicity) (Fig. 1A to D; also see Fig. S1 in the supplemental material). Indeed, the core region of XP (as defined by X painting) frequently appeared to surround the nucleolus and lie within the domain of perinucleolar heterochromatin. The nucleolar association is consistent with recent work implicating physical association with the nucleolus as a defining feature of Xi (66). In contrast to XP, XM localized in relatively Cot-1-enriched regions and was not obviously associated with the nucleolus (see below for further analysis and quantitation).
During the next few cleavages, the Xist domain progressively enlarged and excluded Cot-1 hybridization in all blastomeres (Fig. 1E to H), indicating that the transcription of repeat elements on XP remained repressed through preimplantation development. From the 2- to 16-cell stages, the Xist+ domain coincided with a Cot-1− compartment, but neither the Xist+ domain nor the Cot-1− compartment covered all of XP. These data suggested that perhaps not all of XP is silent during the first four cleavage stages. Not until the blastocyst stage did XP, the Xist+ domain, and the Cot-1− compartment show substantial overlap at the cytological level (Fig. 1H). On the other hand, XM continued to reside in Cot-1+ regions throughout preimplantation development (Fig. 1D, G, and B; also see Fig. S2 in the supplemental material), indicating that the Cot-1− status is XP specific.
To obtain independent testing of XP's transcription state, we carried out combined immunofluorescence and FISH (immuno-FISH) using antibodies against RNA polymerase II and III (Pol-II and Pol-III, respectively), two polymerases involved in the transcription of repetitive elements. In the two-cell embryo, the patterns of RNA polymerase II staining, using antibodies directed against either the C terminus of Pol-II (CTD) or the active form of Pol-II (H5), were highly similar to that of Cot-1. Indeed, the Xist-coated portion of XP was not only Pol-II− but also was surrounded by Pol-II− chromatin (Fig. 2A and B), suggesting that silencing extends beyond the Xist+ domain. Immuno-FISH for Pol-III, which transcribes some repetitive elements such as B2 SINEs, likewise indicated that the Xist-coated XP was undertranscribed (Fig. 2C). Consistently with Cot-1 quantitation, the fluorimetric analysis of polymerase staining revealed an inverse correlation between the presence of Xist RNA and RNA polymerase (Fig. 2A to C, right).
To identify elements of the Cot-1 fraction that are undertranscribed from XP, we next generated consensus probes against LINEs, B1 SINEs, and B2 SINEs by using PCR primers that amplify the most highly conserved regions of each class of repeats (see Materials and Methods). Repetitive elements such as LINEs are highly enriched on the mouse X and consequently have been posited to play a role in XCI (36). In the two-cell embryo, distributions of each class of repeat RNA were similar to that of Cot-1 RNA (Fig. 2D to F). LINEs, B1s, and B2s generally were excluded from the nucleolus, the perinucleolar regions, and the Xist-coated XP. In addition to showing the inverse correlation between Xist RNA and repeat RNA staining, fluorimetric measurements showed that the Xist-coated XP resided in repeat RNA-poor areas (Fig. 2D to F, right). Taken together, these data support the idea that a portion of XP, at least the repeat-rich regions, is transcriptionally suppressed at the two-cell stage (25). They contrast with some earlier studies that found no evidence of Cot-1 or Pol-II exclusion from XP at the two-cell stage (45-48), most likely due to methodological differences (see Discussion).
Repeat silencing precedes coding gene repression in the early embryo.
To determine whether the transcription state of genic elements mirrored that of repeat elements, we next examined XP on a gene-by-gene basis using our optimized RNA/DNA FISH protocol. With BAC probes corresponding to nine X-linked genes, we queried nascent transcription from XM and XP during the two-cell to blastocyst stages. In each case, simultaneous hybridization to Xist probes enabled us to determine the embryo sex (Xist RNA+ equals female) and allelic origin of gene expression (Xist RNA indicates XP). Because genic loci could in principle be far from the Xic (and the Xist RNA signals), we confirmed that nascent RNA signals were specific by subsequently carrying out DNA FISH using a BAC probe labeled with a differently colored fluorophore. Such analysis demonstrated that nascent RNA signals indeed originated from the corresponding X-linked locus (a representative example is shown in Fig. 3 A).
FIG. 3.
Genic expression of XP and XM in the early embryo. (A) RNA/DNA FISH to confirm the specificity of RNA FISH signals. To ensure that nascent RNA signals originated from the corresponding genes, images were captured, cell coordinates marked, and slides denatured for subsequent DNA FISH using the same BAC probe (labeled with a different fluorophore). Note that RNA and DNA signals were perfectly coincident, confirming the specificity of RNA FISH. A representative experiment is shown. (B, D, G, and I) Nascent RNA FISH of indicated genes combined with Xist RNA FISH. Representative nuclei are shown. For all images, z sections were taken through the nucleus and merged into one plane to view all signals. (C, F, H, and J) Summary of all RNA FISH data from female (F) and male (M) embryos of the indicated stage. n, number of nuclei examined; the number of embryos analyzed is in parentheses. (E) Example of skewed expression in which the XP RNA shows fewer nascent transcripts at the four-cell stage.
At the two- and four-cell stages, biallelic expression was observed in almost all blastomeres of XX embryos, while monoallelic expression was detected in XY embryos (Fig. 3B to F; also see Fig. S3 in the supplemental material), which is consistent with previous observations made for some X-linked genes (45-48). In our hands, however, evidence of genic silencing on XP (which was distinctly different from that of XM in male embryos) did not appear until the 8- to 16-cell stage (morula) in a fraction of blastomeres (Fig. 3G and H), and not until the blastocyst stage did dosage compensation occur to any significant extent (Fig. 3I and J). Among the eight genes tested, only one (Utx) escaped silencing (Fig. 3I and J). Xic-proximal genes, such as Atrx and Atp7a, were silenced earlier than Xic-distal genes (Fig. 3H and J), which is consistent with a position-dependent order of silencing as shown by a previous study using allele-specific RT-PCR (25). This correlation suggests that Xist RNA is required in establishing genic silencing. In sum, these data argue that genic inactivation initiates in the morula stage and is not complete until the blastocyst stage or later (several divisions after the formation of an Xist+ Cot-1− silent compartment). This finding places the timing of genic silencing at a later stage than that proposed by contemporary models, which suggested either preinactivation (25) or inactivation initiating at the four-cell stage and being completed by the morula stage (46, 47). This finding, together with a more recent analysis of genic expression (48), is more consistent with the classical model, which proposed inactivation during the peri-implantation stage (E3.5 to E5.5) (1, 13, 29).
XP genes translocate into the repeat compartment during silencing.
We next performed two-color RNA FISH to examine the spatial and temporal aspects of genic silencing relative to the formation of the Xist+ Cot-1− compartment by measuring the distance between the Xist+ Cot-1− compartment and the nascent genic transcripts during genic silencing. We specifically examined the morula, where skewed allelic signals not only signified the onset of genic silencing but also enabled us to track both alleles simultaneously. Interestingly, while active XP genes tend to reside outside of the Xist+ Cot-1− compartment (Fig. 4 A), genes undergoing inactivation generally moved into or resided at the edge of the compartment (Fig. 4B). On the other hand, the Utx gene, the only one of the eight genes tested to escape imprinted XCI, continued to reside well outside of the Xist+ Cot-1− compartment (Fig. 4A and C), suggesting that translocation is specific to genes subject to inactivation. Thus, the process of silencing significantly correlated with the translocation of the corresponding genes into the Xist+ Cot-1− compartment (Fig. 4C), suggesting an overall contraction of XP during genic silencing. Indeed, our analysis showed that XM and XP were similar in size from the two-cell stage (Fig. 1D) to the 8-cell stage (Fig. 4D and data not shown). By the morula and blastocyst stages, however, XP occupied significantly less volume than XM (Fig. 4D).
FIG. 4.
Translocation of genic loci into the silent repeat compartment during silencing. (A) Active XP genes reside outside of the Xist+ Cot-1− repeat compartment before silencing. Three representative blastomeres from morulae are shown expressing three X-linked genes. Arrows indicate linear distances between the gene and the outer edge of the Xist+ Cot-1− compartment. (B) During the process of silencing (deduced by diminished XP expression), genes are translocated into the Xist+ Cot-1− compartment in the morula. The boxed region is magnified in the right panel. Arrows indicate linear distances between the gene and the outer edge of the Xist+ Cot-1− compartment. (C) Summary of linear distances between the gene and the silent compartment during silencing. The normalized distance is the linear distance from the center of the nascent RNA signal to the edge of the Xist+ Cot-1− compartment, each normalized to the nuclear diameter. Negative distances imply genic movement into the Xist+ Cot-1− compartment, whereas a zero distance implies localization at the edge of the Xist+ Cot-1− compartment. P values were calculated using an unpaired t test. (D) XP territory contracts over time. XP and XM territories were measured by Volocity software (Improvision) and normalized to total nuclear volume to yield the chromosome condensation index. P values were calculated using an unpaired t test. (E) Pictorial representation of genic localization into the preformed Xist+ Cot-1− compartment during silencing. The silent compartment is present by the two-cell stage, and it enlarges as genic loci are translocated into it as they are gradually inactivated, beginning at the morula stage. XP silencing is not complete until the blastocyst stage or later.
Taken together, these data argued that imprinted XCI is a multistep process that recruits repeat and genic elements at different developmental stages (Fig. 4E). Prior to coding gene silencing, repeat elements of XP are silenced in a process already evident by the two-cell stage. The silent region enlarges during preimplantation development as coding genes are recruited into this Cot-1− compartment during genic inactivation. Genic silencing does not reach its fullest extent until the blastocyst stage or later.
Xist is required for the initiation of genic silencing in the preimplantation embryo.
While Xist clearly is required for XP silencing in the placenta of the postimplantation embryo, the fact that its deletion has no consequence for preimplantation development has left open its involvement in the early mouse embryo (38). Using RNA FISH to examine nascent transcription from eight X-linked genes, we observed that genic silencing in the morula and blastocyst was abolished when Xist is deleted on XP (Fig. 5 A to H; also see Fig. S4 in the supplemental material). Nearly all blastomeres showed biallelic expression in XX embryos, whereas control male embryos showed monoallelic expression, as expected. In two- to four-cell embryos where genic silencing does not normally take place (Fig. 3; also see Fig. S3 in the supplemental material), the pattern of expression in XXXist− mutants was biallelic. Allele-specific RT-PCR confirmed these results using SNPs present in C57BL/6 and Mus castaneus mice for these X-linked genes (Fig. 5I). While wild-type embryos showed preferential XP silencing at the morula stage as previously reported (25), mutant embryos showed nearly equal biallelic expression. We conclude that Xist is required for the initiation of genic silencing in the preimplantation embryo.
FIG. 5.
Genic silencing depends on Xist. (A to D) Biallelic expression of indicated genes in Xist-deficient preimplantation embryos of different stages. Shown are merged z sections taken through the nucleus to capture signals in all focal planes. (E to H) Summary of genic expression in Xist mutant female (F) and male (M) embryos of the indicated stage. n, number of nuclei examined; the number of embryos analyzed is in parentheses. (I) Allele-specific RT-PCR of seven X-linked genes in wild-type (WT) and Xist mutant morulae produced by the indicated crosses. Mus, M. musculus; Cas, M. castaneus; M, maternal; P, paternal; PBS, negative control derived from the wash fluid after embryos are isolated to rule out contamination.
We further studied the role of Xist during genic silencing by the analysis of embryos carrying an X-linked GFP transgene (XMXP;GFP) (17). In E3.5 and E4.5 embryos, XMXP;GFP embryos showed indistinguishable green fluorescent protein (GFP) expression at the protein level compared to that of embryos for which Xist is deleted in cis (XMXP;GFP;Xist−) (Fig. 6 A to D and I), suggesting either the inefficient silencing of the GFP transgene in the blastocyst or the produrance of the GFP synthesized during earlier stages. However, in cultured E6.5 blastocyst outgrowths, we observed brighter GFP signals in the extraembryonic lineages of XMXP;GFP;Xist− embryos compared to that of XMXP;GFP embryos (Fig. 6G to I), confirming the requirement of the Xist gene at the onset of genic XP silencing. Furthermore, in cultured E5.5 to E6.5 blastocyst outgrowths and in E6.5 postimplantation embryos, mosaic GFP expression could be seen in the embryonic ectoderm regardless of whether the transgene is on XP or XM, which is consistent with random XCI in the embryo proper (Fig. 6E to K). When Xist is deleted on XP, higher-intensity GFP signals could be observed from the embryo proper, indicating the failure to inactivate XP as expected (38). Interestingly, the increase in GFP fluorescence in Xist mutants is substantially greater than the expected 2-fold increase (Fig. 6J and L). Although the reasons for this effect are not known, this result was quite reproducible. During the course of analysis, we also discovered that the GFP transgene was poorly expressed in the extraembryonic lineages on E6.5 (Fig. 6J to O), regardless of whether it is maternally or paternally inherited. Thus, the transgene is not a good marker to examine imprinted XCI in the Xist-deficient mutant during the early stages of postimplantation development.
FIG. 6.
Analysis of imprinted XCI using an X-linked GFP transgene. (A to D) Blastocysts isolated at E3.5 (A and B) or cultured for one additional day to E4.5. (C and D) The embryos carry a paternally transmitted X-linked GFP transgene on either a wild-type X (A and C) or Xist-deficient X (B and D). GFP expression is evident even when GFP is carried on a wild-type XP, consistently with a later onset of genic XP silencing than that expected and consistently with the produrance of GFP. Bright-field (left) and GFP fluorescence (right) images are shown for each embryo. (C) Male embryos without XP are nontrangenic for X-linked GFP and are therefore GFP−. ICM, inner cell mass; TE, trophectoderm. (E to H) Embryo outgrowths attached as blastocysts and cultured for 2 to 3 days until E5.5 (E and F) or E6.5 (G and H). Each embryo carries the paternally transmitted X-linked GFP transgene on a wild-type X (E and G) or Xist-deficient X (F and H). epi, epibast; ExE, extraembryonic ectoderm. (I) Quantitative assessment of X-linked GFP fluorescence for the stages shown in panels A to H. The mean fluorescence intensity was measured for each stage in the embryonic parts (ICM or epi) and in the extraembryonic parts (TE or ExE) as depicted in panels A and E. While no significant GFP signal differences can be quantified in E3.5 and E4.5 blastocysts, later on wild-type embryos show lower GFP signals than Xist-deficient embryos, which is consistent with the onset of genic XP silencing. Fluorescence differences between wild-type and Xist mutant embryos are statistically significant at E6.5 according to the nonpaired t test (epi, P = 0.0015; ExE, P = 0.0314). Error bars indicate standard errors of the means. (J to O) E6.5 embryos of the indicated genotypes as dissected from maternal deciduas (K to O) and the quantitation of the GFP signals depicted in panel L (J). GFP is almost silent in ExE regardless of whether it is carried on a wild-type or Xist mutant X chromosome and whether it is paternally or maternally transmitted, despite the fact that there is no imprinted inactivation of the maternal X in female (J, M, and N) or male (J and O) embryos. Therefore, the GFP transgene cannot be used as a reliable marker to analyze imprinted XCI. het, the mother is heterozygous for X-GFP; hom, the mother is homozygous for X-GFP.
Repeat silencing initiates without Xist and is associated with the nucleolus.
We next asked if Xist is required for the formation of the silent Cot-1− compartment at the two-cell stage. We carried out Cot-1 hybridization on undenatured slides and then followed with slide denaturation and DNA FISH to examine the expression state of X-linked repeat elements when Xist is deleted on XP. Because Xist RNA no longer could be used to distinguish XM and XP, we used a combination of two DNA probes, πXE9 (which is deleted on the mutant chromosome) and Sx7 (which is present on both X chromosomes), to identify XP. Intriguingly, deleting Xist on XP did not appreciably change its Cot-1− status in two-cell embryos, at least in the region around the Xic (Fig. 7 A and B). As is the case in wild-type embryos, fluorimetric analysis showed that the Xic and surrounding regions of XM in mutant preimplantation embryos tend to reside in Cot-1+ regions, while those of XP resided in a Cot-1− neighborhood in the nucleolar region or surrounding perinucleolar heterochromation (Fig. 7A to D). RNA FISH using LINE and SINE probes demonstrated that perinucleolar localization is perfectly correlated with repeat element silencing on XP (Fig. 7D). Thus, Xist is not required for the repeat silencing of XP that already is present at the two-cell stage.
FIG. 7.
Repeat silencing in the early preimplantation embryo does not require Xist. Cot-1 and Xist RNA FISH with subsequent DNA FISH using the Xic probe, Sx7, in the wild-type two-cell embryo (A), and Cot-1 RNA FISH with subsequent DNA FISH in the Xist mutant two-cell embryo using a combination of Sx7 and πXE9 probes to distinguish XM (wild-type) from XP (Xist deficient) (B). (C) Pictorial representation of the DAPI staining pattern and corresponding Cot-1 RNA FISH pattern at the two-cell stage. Because the nucleolus and perinucleolar heterochromatin are devoid of Cot-1 signal, the Cot-1 hole is always larger than the DAPI hole left by the nucleolus itself. When the DNA FISH signal in question localizes to the perinucleolar Cot-1 hole, the signal is scored as nucleolar association inactive. On the other hand, when the signal localizes in Cot-1+ regions, it is scored as no association active. (D) Xic nucleolar association of XP versus XM. Cot-1, LINE, B1, and B2 RNA FISH were performed on wild-type and Xist-deficient two-cell embryos in combination with an Xic probe (Sx7). DNA FISH was conducted subsequently to compare the frequency with which XP and XM come in direct contact with the nucleolus. The combination of Sx7 and πXE9 probes was used to distinguish XM (wild-type) from XP (Xist deficient). When the Sx7 signal was directly adjacent to the nucleolus or located in the perinucleolar heterochromatic ring, the chromosome was judged to be nucleolus associated and Cot-1−. For the two-cell embryo, there was 100% correlation between nucleolus-associated and Cot-1−, LINE−, and SINE B2− states of XP; there was a 95% correlation between the nucleolus-associated and the SINE B1− state. In contrast, chromosomes that were not nucleolus associated were Cot-1+. P values were calculated using the student t test. WT, wild-type. Xist−, XMXP;Xist−.
We observed that deleting is Xist did not change the predisposition of XP toward the heterochromatic perinucleolar region (Fig. 1 and 7D). Cot-1 RNA FISH together with DAPI staining showed that the region around the nucleolus was almost always Cot-1− and DAPI intense (Fig. 7C). The XP in both wild-type and XMXP;Xist− embryos showed enrichment in this perinucleolar region, with the Xic of XP making direct contact with the nucleolar edge (Fig. 1). In the wild-type embryo, 71.4% of two-cell blastomeres showed XP-nucleolus contact (n = 28). Likewise in the Xist mutant, 70% of two-cell blastomeres showed XP-nucleolus contact (n = 20). Analysis using LINE and SINE probes yielded similar results (Fig. 7D). In contrast, the Xic of XM showed a relatively low frequency of association in both wild-type and mutant embryos. We conclude that the perinucleolar localization of XP in the early preimplantation embryo also does not depend on Xist. In light of Xi's association with the perinucleolar compartment during random XCI (66), these results raised the possibility that repeat silencing during imprinted XCI depends less on Xist and more on other epigenetic mechanisms, such as nuclear compartmentalization.
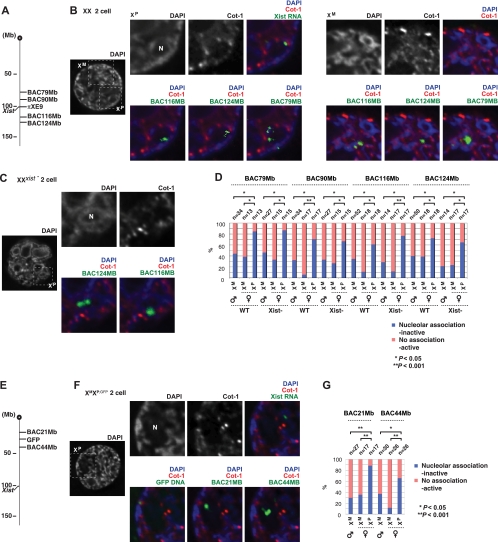
To investigate further, we asked to what extent repeat silencing along XP was unaffected by the Xist deletion. Heretofore, our analyses examined only the portion of XP coated by Xist RNA, which remains small at the two-cell stage. To examine the behavior of X-linked regions outside of the Xist domain, we carried out serial RNA-DNA FISH. Because both XP and XM occupy large nuclear territories at the two-cell stage (Fig. 1D), X-painting probes could not provide the desired spatial resolution. Therefore, we carried out Cot-1/Xist RNA FISH in combination with DNA FISH using BAC probes against specific repeat regions along XP (Fig. 8). When examining regions close to the Xic in wild-type embryos, XM versus XP alleles could easily be distinguished by proximity to Xist RNA. We first tested four Xic-proximal BACs that are highly enriched for repetitive elements and do not contain any known coding genes (Fig. 8A). Consistently with repeat silencing, all four X-linked regions of XP localized to the heterochromatic ring around the nucleolus within a Cot-1− hole, whereas the corresponding alleles of XM were relatively Cot-1 enriched (Fig. 8B and D).
FIG. 8.
Repetitive elements outside of the Xist RNA domain on XP also are silenced in an Xist-independent manner (A) Locations of repeat-rich X-linked BAC sequences examined in panels B and C. Each region is located outside of the Xist RNA-coated domain of XP in the two-cell embryo. (B and C) Cot-1 and Xist RNA FISH with subsequent DNA FISH of repetitive elements outside of the Xist RNA-coated domain in the wild-type two-cell embryo (B) or the XMXP;Xist− two-cell embryo (C). One z section is shown. Note that some BAC probes yielded multiple signals, possibly reflecting sister chromatids of a cell in the G2 stage of the cell cycle and/or the longer probe lengths of some BACs (200 to 300 kb), which would yield a linear track of signals. (D) Quantitation of nucleolar association and repeat silencing for the experiments shown in panel B and C. Cot-1 RNA FISH was performed on wild-type and Xist-deficient embryos as described for Fig. 7D. P values were calculated using the χ2 test. (E) Locations of repeat-rich X-linked BAC probes for the analysis shown in panels F and G. (F) Cot-1 and Xist RNA FISH with subsequent DNA FISH in the wild-type two-cell embryo with a paternally transmitted GFP transgene. One z section is shown. (G) Quantitation of nucleolar association and repeat silencing for the experiments shown in panel F. P values were calculated using the unpaired student t test.
To visualize relative spatial relationships between the alleles of XM and XP, we compiled an interactive 3D movie from ∼50 z sections taken at 0.2-μm intervals for the nucleus shown in Fig. 8B (see Fig. S5A, B, and C in the supplemental material). From this depiction, it is clear that (i) Xist RNA does not merely form a small pinpoint but partially spreads around the presumptive prenucleolus, (ii) XP signals from the nongenic BACs closely follow the contours of the presumptive prenucleolus, and (iii) the XP signals are unmistakably in Cot-1− space (but are in DAPI+ regions), whereas XM signals are in a relatively Cot-1-enriched domain. During the quantitative analysis of signal intensities, all XM and XP signals were evaluated across the same dynamic range, as indicated by ImageJ analysis (see Fig. S6 in the supplemental material).
When Xist was deleted from XP, all four regions remained associated with the perinucleolar heterochromatin in the Cot-1− compartment, and the homologous regions on XM continued to associate with Cot-1+ regions (Fig. 8C and D). We next examined two X-linked regions farther from the Xic, using the X-linked GFP transgene as a marker for XP (Fig. 8E). These distal regions also showed a similar tendency toward nucleolar association and silencing (Fig. 8F and G). Interestingly, the GFP transgene, which is presumptively expressed on XP at the two-cell stage, localized to a Cot-1+ (active) region, further supporting the dichotomy between genic and nongenic compartments on XP. It also is noteworthy that nongenic regions occasionally could be observed within the nucleolus rather than around it (Fig. 8B and F), which is consistent with X-painting results that which showed that the XP territory often partially overlaps the nucleolus in the two-cell embryo (Fig. 1D) (note that the DAPI staining of regions that protrude into the nucleolus would not be easily detectable due to the low concentration of DNA in the nucleolus relative to that of the extranucleolar regions). Taken together, these data show that repeat elements on XP within domains not coated by Xist RNA also are silenced at the two-cell stage, and this form of silencing occurs independently of Xist RNA.
To determine whether the Xist-independent repeat-silencing mechanism persists through preimplantation development, we examined 4- and 8-cell mutant embryos. The quantitative analysis of RNA/DNA FISH experiments showed that an Xist deficiency on XP also had no effect on its localization in a Cot-1− perinucleolar region relative to that of XM (Fig. 9 A to C). Interestingly, the preferential perinucleolar localization of XP was lost at the 8-cell stage for both wild-type and mutant embryos. However, whereas the wild-type XP maintained the Cot-1− status, the XP lacking Xist became Cot-1+. This outcome indicated that repeat silencing in later preimplantation embryos could not be maintained without Xist. Furthermore, X chromosome painting revealed that XP in XMXP;Xist− embryos could not adopt the compact configuration that was observed in wild-type embryos (compare Fig. 9E to Fig. 1H and 4D). Thus, the silencing of XP initially is repeat based and Xist independent, but it becomes Xist dependent at the 8-cell stage. These data further support the idea that imprinted XCI in the early mouse embryo is a two-step process and raise the hypothesis that imprinting information is carried by repeats from the paternal germ line, with the perinucleolar region playing a crucial role in maintaining the imprint specifically on XP (Fig. 9F).
Xist is not required for postmeiotic silencing in the male germ line.
Given that MSCI (40, 61) and repeat XP silencing in the early embryo are Xist independent, we were especially curious to learn if sex chromosome silencing in the transitional period (spermiogenesis) also is Xist independent. In comparing the postmeiotic sex chromatin (PMSC) of wild-type and Xist-deficient spermatids, we observed no obvious differences between them when analyzing Cot-1 expression and epigenetic markers associated with PMSC (Fig. 10 A to H). In the mutant germ line, the postmeiotic X (and Y) strongly stained with DAPI, localized next to the chromocenter, and excluded Cot-1 hybridization, as previously described for the wild-type germ line (16, 42, 62). Thus, repeat silencing in the postmeiotic period also does not depend on Xist. Furthermore, heterochromatic modifications, such as HP1β and methylation at lysine 9 of histone H3 (H3-2meK9), on the PMSC were intact in Xist mutant spermatids. We conclude that sex chromosome silencing throughout spermatogenesis is Xist independent. Therefore, during the gamete-to-embryo transition, Xist is not required for X chromosome repression until late in preimplantation development.
FIG. 10.
Postmeiotic silencing also does not require Xist. (A and B) Cot-1 RNA FISH and the chromosome-specific painting of X and Y in the round spermatids of wild-type mice. Arrow, PMSC. Single z sections are shown for all panels. (C and D) Immunofluorescence for HP1β (C) and H3-2meK9 (D) in wild-type round spermatids. (E and F) Cot-1 RNA FISH and chromosome-specific painting of X and Y in the round spermatids of Xist-deficient mice. (G and H) Immunofluorescence for HP1β (G) and H3-2meK9 (H) in mutant round spermatids. (I) A working hypothesis for the developmental history of the X chromosome from gamete to embryo.
DISCUSSION
Could transgenerational information be carried by repeat elements of XP?
Our study shows that, during the transition from gamete to embryo in the mouse species, imprinted XCI occurs in two genetically separable steps, with repeat silencing preceding genic inactivation (Fig. 10I). The repeat elements of XP form a Cot-1−/Pol-II−/Pol-III− silent compartment next to the nucleolus by the two-cell stage. Although Xist RNA localizes within it, the initial formation of the silent compartment does not actually require Xist. Genic silencing does not follow until several divisions later, occurring predominantly in the morula-blastocyst stages. In contrast to the initiation of repeat silencing, the initiation of genic repression strictly depends on Xist. Thus, imprinted XCI in the early embryo is biphasic, divisible not only on the basis of genic content but also by the relationship to Xist. In the mouse, maternal and paternal pronuclei do not fuse in the 1-cell embryo and come together only during the first mitotic division to form the two-cell embryo. At this stage, XP already can be distinguished from XM, not only by XP's association with Cot-1−/Pol-II−/III− regions but also by its preferential localization to the perinucleolar compartment. Interestingly, a class of repeat elements (LINEs) has been suggested to play a crucial role in another context: the spreading of silencing during random XCI (36). Therefore, one possible contribution of repeat silencing during imprinted XCI is to facilitate the spreading of Xist RNA and genic silencing. Consistently with this idea, we observed that Xist RNA spreading takes place around the nucleolus (see Fig. S1 in the supplemental material and data not shown). The nucleolus (more accurately, the nascent prenucleolus) might help scaffold repeat- and Xist RNA-mediated silencing.
These observations suggest one of two scenarios. First, XP repetitive elements may become silenced at the two-cell stage. In this scenario, XP silencing occurs de novo in the early embryo and takes place in two consecutive waves, involving repetitive elements before affecting coding genes. An alternative scenario is that the repeats arrive in the zygote in a preinactivated condition. In this model, transgenerational instruction (the imprint) may be carried by repeat elements of XP from the paternal germ line (Fig. 10I). Several observations lead us to favor the latter scenario. For example, the exclusion of repeat RNA hybridization is evident from the pachytene stage of meiosis I (MSCI) through the postmeiotic period and into mature sperm (16, 42, 62), and Xist is dispensable for both postmeiotic germ line silencing and the zygotic silencing of repeat sequences (Fig. 10). Thus, the heterochromatic state acquired during MSCI might more than predispose XP for XCI in the embryo, possibly indicating a mechanistic continuity of repeat-based XCI from gamete to embryo. In this context, it is interesting that recent studies link MSCI with the meiotic silencing of unpaired chromatin/DNA (MSUC/MSUD), a mechanism originating in lower eukaryotes to protect the genome from the proliferation of foreign DNA, such as transposons and retroviruses (4, 56, 63). It is known that repeat elements (such as LINEs) are especially enriched on the X compared to that on autosomes (36). We therefore speculate that chromatin marks placed specifically onto such repeats in the male germ line, perhaps during MSCI, could comprise the imprint and be responsible for the first of two steps in the inactivation of XP in the early embryo. As DNA methylation has been shown to play a lesser role in imprinted XCI (52), we suggest a chromatin-based mechanism dependent on inherited nucleosomes and their associated factors. Interestingly, a recent study indicates that sperm chromatin profiles correlate with embryonic developmental programs (19). A second imprint, placed independently on Xist, would be responsible for the second phase of imprinted XCI.
Relevant to this hypothesis, another study has proposed that autosomal Xist transgenes can recapitulate imprinted XCI when inherited through the paternal germ line, apparently without going through MSCI (45). This outcome has been interpreted as evidence that XP silencing in the mouse is strictly Xist dependent and unrelated to meiotic silencing. However, the data have not addressed the efficiency and stability of autosomal silencing in the transgenic mice. Indeed, the transgenic animals are viable and have no overt phenotype (20). Furthermore, a lacZ reporter gene could not be silenced in cis in transgenic pre- or postimplantation embryos (20). These data therefore exclude the possibility that the transgene-bearing autosome is stably silenced. We suggest that the published data and our current analysis together support the idea of two synergistic mechanisms for imprinted XCI, a repeat-based silencing mechanism (potentially originating in the paternal germ line via MSCI) and a gene-based mechanism dependent on zygotic Xist expression (established without MSCI) later during preimplantation development.
Our data imply that imprinted XCI is biparentally controlled. The existence of a strong maternally expressed factor in the oocyte is well accepted (15, 28, 59). So robust is this maternal factor that embryos cannot inactivate XM even in the presence of multiple XM chromosomes. One study has proposed the maternally expressed Tsix gene as a candidate for the maternal factor (30). Under debate is whether there also exists a paternal regulator or if XP inactivation is a default consequence of XM's resistance to inactivation. In light of our present findings, we argue that the male germ line (through MSCI/PMSC) actively participates in imprinted XCI by contributing a preformed inactive XP compartment (repeat based in nature) to the zygote.
Our data show that a repeat-rich silent compartment recruits specific genes into it during the process of genic silencing. The notion of a preinactivated repeat region argues that imprinted XCI bears some mechanistic resemblance to random XCI (8, 9). The Xi nuclear territory has been shown to be organized into a gene-rich outer rim and a gene-poor inner core enriched for repetitive elements of the Cot-1 fraction (8, 9). One study suggests that the silencing of the Cot-1 fraction does not require Xist's repeat A motif in mouse embryonic stem (ES) cells (8), an element crucial for random XCI in mice (21, 65, 67). Taken together, these findings indicate that genic elements on both the XP of the early embryo and the future Xi of embryonic stem cells are translocated into the Cot-1− silent compartment upon repression. For both imprinted XP and somatic Xi silencing, Xist is absolutely required for the translocation and silencing of genic loci. Therefore, although imprinted and random XCI differ significantly in the control of their initiation, the former being parentally controlled and the latter being zygotically controlled by a counting mechanism, some aspects of the inactivation processes themselves apparently are conserved.
In the zygote, the nucleolus may play a key role in establishing and maintaining the epigenetic asymmetry of XP and XM. Initially, only the repeat elements of XP associate with the Cot-1−/Pol-II−/Pol-III− ring around the nucleolus. Recent studies showed that nucleoli may play a general role in epigenetic regulation. Intriguingly, although the nucleolus itself is inherited from the mother (44), pericentromic heterochromatin in the sperm is continuously decorated with PRC1 after fertilization in the perinucleolar region during preimplantation development (51), suggesting that the nucleolus continuously retains heterochromatic memory from the male germ line to the zygote. In random XCI, the perinucleolar compartment is thought to play a key role in maintaining the heterochromatic state of Xi (66). Taken together, these observations raise the possibility that the perinucleolar region is critical not only for somatic silencing but also for parent-of-origin effects in the early embryo. One crucial difference that distinguishes somatic Xi from imprinted XP is the role of Xist in perinucleolar targeting. The fact that XP targeting occurs independently of Xist further suggests that although imprinted and random XCI share some aspects of the inactivation processes, they significantly diverge in how silencing is initiated.
A requirement for Xist in the initiation of coding gene silencing.
The two-step process indicated by our study invokes Xist RNA only in the second step, when coding genes become inactivated on XP. We therefore have reached a different conclusion from that of a recent study, which proposed that the initiation of genic silencing does not require Xist (27). While our studies agree that coding gene silencing occurs in the morula-blastocyst stages, that of Kalantry et al. detects no loss of gene silencing in the morula when Xist is deleted from XP. These contrasting results could arise from methodological differences in the RNA FISH and allele-specific RT-PCR protocols. Our RNA FISH protocol has a sensitivity of >90% in morulae and blastocysts, and our allele-specific RT-PCR was quantitated by Southern hybridization to end-labeled oligonucleotide probes (Fig. 5). Because Kalantry et al. did not perform DNA FISH on the Xist mutants, the origins of XP and XM could not be concluded with certainty. Kalantry et al. also drew conclusions about Xist-independent gene silencing from the X-linked GFP assay, which does not appear to be a reliable X-linked reporter during the peri-implantation period (Fig. 6). Although we believe that these technical differences explain our contrasting findings, it is possible that some genes analyzed by Kalantry et al. behaved like repeats and were inactivated in an Xist-independent manner. Significantly, our studies do agree on the point that at least some elements on XP can undergo imprinted XCI in an Xist-independent manner.
Implications for developmental and evolutionary models.
The findings in this study shed new light on the ongoing debate regarding the timing and mechanism of imprinted XCI in the mouse embryo. On the basis of isoenzyme analyses, the classical model proposed that XP initially is active and is silenced only during the implantation period (1, 13, 29; also reviewed in reference 24). Based mostly on the appearance of heterochromatic signatures on XP at the four-cell stage and the presence of Pol-II and Cot-1 staining on XP at the two-cell stage, the more recent de novo model proposed that XP silencing initiates at the four-cell stage (45, 47, 48). Finally, based on Cot-1 hybridization patterns in the early embryo, the preinactivation model proposed that XP is partially preinactivated by the paternal germ line and arrives in the zygote in a semisilent state (25). Our present work queried chromosomal expression states using a combination of approaches and found that genic and repeat sequences of XP undergo silencing at different times. Interpretive differences of the three models therefore may be partially attributable to the use of different assays, with the preinactivation model based on repeat expression profiles and the de novo and classical models based primarily on genic profiles. Methodological differences also may contribute. For example, we have found that fixing cells prior to permeabilization yields higher background, which may prevent the visualization of Cot-1/Pol-II holes (45, 47, 48). Our optimized protocols either permeabilize before fixation (25) or permeabilize and fix simultaneously (this study). In sum, the present analysis indicates that the three models each are partially correct and represent different aspects of the dynamic process of imprinted silencing.
We also might speculate on the extent to which the two-step process carries over into random XCI in eutherian mammals. The work of Chaumeil et al. showed that the X excludes transcription machinery and forms a Pol-II hole prior to the inactivation of specific genic elements in mouse ES cells and that the genes lie outside of the Pol-II hole during this time (8). It is thought that the exclusion of the transcriptional machinery depends on Xist, but the observation indicating a dispensability of RepA, the repeat A RNA motif required to initiate silencing (65, 67), raises questions as to whether random XCI also is separable into repeat versus genic phases.
For imprinted XCI, the idea of an Xist-independent mechanism based on repeat silencing raises the intriguing possibility that dosage compensation in marsupials, which seem to lack XIST, operates on a similar basis. Indeed, the Cot-1 analysis of the opossum male germ line has shown that repeat elements of the X are transcriptionally suppressed in the postmeiotic period (43). With the complete sequencing of the opossum genome (41) and the development of the laboratory opossum model (23), the ancestral mechanism of imprinted XCI will not be long in coming.
Supplementary Material
Acknowledgments
We thank all members of the laboratory for valuable discussion throughout this work, R. G. Roeder for the kind gift of RNA polymerase III antibody (RPC 53), and J. Dennis and M. Borowsky for helpful suggestions during probe design for LINE and SINE RNA FISH. S.H.N. and J.T.L. designed the study and wrote the paper. For independent confirmation, multiple authors performed experiments in parallel. S.H.N. optimized immuno-FISH protocols and performed all RNA/DNA FISH and immunofluorescence experiments. B.P. and S.H.N. carried out X-GFP analyses, and K.D.H. carried out allele-specific RT-PCR. B.P. generated the interactive movies. R.J. contributed the Xist mutant mice. The authors thoroughly discussed the data and contributed to data interpretation.
S.H.N. is supported by research fellowships of the Japan Society for the Promotion of Science (JSPS) and the Charles King Trust; B.P. by a fellowship from the Human Frontier Science Program (HFSP); K.D.H. by an NIH KO1 award; R.J. by NIH grants RO1-HDO45022 and R37-CA084198; and J.T.L. by NIH RO1-GM58839. J.T.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 19 April 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adler, D., J. West, and V. Chapman. 1977. Expression of alpha-galactosidase in preimplantation mouse embryos. Nature 267:838-839. [DOI] [PubMed] [Google Scholar]
- 2.Avner, P., and E. Heard. 2001. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2:59-67. [DOI] [PubMed] [Google Scholar]
- 3.Bartolomei, M. S., and S. M. Tilghman. 1997. Genomic imprinting in mammals. Annu. Rev. Genet. 81:493-525. [DOI] [PubMed] [Google Scholar]
- 4.Bean, C. J., C. E. Schaner, and W. G. Kelly. 2004. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 36:100-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beatty, B., S. Mai, and J. Squire. 2002. FISH. A practical approach. Oxford University Press, Oxford, United Kingdom.
- 6.Brown, C. J., B. D. Hendrich, J. L. Rupert, R. G. Lafreniere, Y. Xing, J. Lawrence, and H. F. Willard. 1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71:527-542. [DOI] [PubMed] [Google Scholar]
- 7.Brown, C. J., R. G. Lafreniere, V. E. Powers, G. Sebastio, A. Ballabio, A. L. Pettigrew, D. H. Ledbetter, E. Levy, I. W. Craig, and H. F. Willard. 1991. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 349:82-84. [DOI] [PubMed]
- 8.Chaumeil, J., P. Le Baccon, A. Wutz, and E. Heard. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20:2223-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemson, C. M., L. L. Hall, M. Byron, J. McNeil, and J. B. Lawrence. 2006. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl. Acad. Sci. U. S. A. 103:7688-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, D. W. 1971. Directed genetic change model for X chromosome inactivation in eutherian mammals. Nature 230:292-294. [DOI] [PubMed] [Google Scholar]
- 11.Davidow, L. S., M. Breen, S. E. Duke, P. B. Samollow, J. R. McCarrey, and J. T. Lee. 2007. The search for a marsupial XIC reveals a break with vertebrate synteny. Chromosome Res. 15:137-146. [DOI] [PubMed] [Google Scholar]
- 12.Duret, L., C. Chureau, S. Samain, J. Weissenbach, and P. Avner. 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312:1653-1655. [DOI] [PubMed] [Google Scholar]
- 13.Epstein, C. J., S. Smith, B. Travis, and G. Tucker. 1978. Both X chromosomes function before visible X-chromosome inactivation in female mouse embryos. Nature 274:500-503. [DOI] [PubMed] [Google Scholar]
- 14.Franco, M. J., R. B. Sciurano, and A. J. Solari. 2007. Protein immunolocalization supports the presence of identical mechanisms of XY body formation in eutherians and marsupials. Chromosome Res. 15:815-824. [DOI] [PubMed] [Google Scholar]
- 15.Goto, Y., and N. Takagi. 2000. Maternally inherited X chromosome is not inactivated in mouse blastocysts due to parental imprinting. Chromosome Res. 8:101-109. [DOI] [PubMed] [Google Scholar]
- 16.Greaves, I. K., D. Rangasamy, M. Devoy, J. A. Marshall Graves, and D. J. Tremethick. 2006. The X and Y chromosomes assemble into H2A.Z, containing facultative heterochromatin, following meiosis. Mol. Cell. Biol. 26:5394-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadjantonakis, A. K., M. Gertsenstein, M. Ikawa, M. Okabe, and A. Nagy. 1998. Non-invasive sexing of preimplantation stage mammalian embryos. Nat. Genet. 19:220-222. [DOI] [PubMed] [Google Scholar]
- 18.Hall, L. L., M. Byron, K. Sakai, L. Carrel, H. F. Willard, and J. B. Lawrence. 2002. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc. Natl. Acad. Sci. U. S. A. 99:8677-8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammoud, S. S., D. A. Nix, H. Zhang, J. Purwar, D. T. Carrell, and B. R. Cairns. 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heard, E., C. Kress, F. Mongelard, B. Courtier, C. Rougeulle, A. Ashworth, C. Vourc'h, C. Babinet, and P. Avner. 1996. Transgenic mice carrying an Xist-containing YAC. Hum. Mol. Genet. 5:441-450. [DOI] [PubMed] [Google Scholar]
- 21.Hoki, Y., N. Kimura, M. Kanbayashi, Y. Amakawa, T. Ohhata, H. Sasaki, and T. Sado. 2009. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 136:139-146. [DOI] [PubMed] [Google Scholar]
- 22.Hore, T. A., E. Koina, M. J. Wakefield, and J. A. Marshall Graves. 2007. The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Res. 15:147-161. [DOI] [PubMed] [Google Scholar]
- 23.Hornecker, J. L., P. B. Samollow, E. S. Robinson, J. L. Vandeberg, and J. R. McCarrey. 2007. Meiotic sex chromosome inactivation in the marsupial Monodelphis domestica. Genesis 45:696-708. [DOI] [PubMed] [Google Scholar]
- 24.Huynh, K. D., and J. T. Lee. 2001. Imprinted X inactivation in eutherians: a model of gametic execution and zygotic relaxation. Curr. Opin. Cell. Biol. 13:690-697. [DOI] [PubMed] [Google Scholar]
- 25.Huynh, K. D., and J. T. Lee. 2003. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426:857-862. [DOI] [PubMed] [Google Scholar]
- 26.Huynh, K. D., and J. T. Lee. 2005. X-chromosome inactivation: a hypothesis linking ontogeny and phylogeny. Nat. Rev. Genet. 6:410-418. [DOI] [PubMed] [Google Scholar]
- 27.Kalantry, S., S. Purushothaman, R. B. Bowen, J. Starmer, and T. Magnuson. 2009. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature 460:647-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kay, G. F., S. C. Barton, M. A. Surani, and S. Rastan. 1994. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell 77:639-650. [DOI] [PubMed] [Google Scholar]
- 29.Kratzer, P. G., and S. M. Gartler. 1978. HGPRT activity changes in preimplantation mouse embryos. Nature 274:503-504. [DOI] [PubMed] [Google Scholar]
- 30.Lee, J. T. 2000. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103:17-27. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. T., and N. Lu. 1999. Targeted mutagenesis of Tsix leads to nonrandom X-inactivation. Cell 99:47-57. [DOI] [PubMed] [Google Scholar]
- 32.Lifschytz, E., and D. L. Lindsley. 1972. The role of X-chromosome inactivation during spermatogenesis (Drosophila-allocycly-chromosome evolution-male sterility-dosage compensation). Proc. Natl. Acad. Sci. U. S. A. 69:182-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucchesi, J. C., W. G. Kelly, and B. Panning. 2005. Chromatin remodeling in dosage compensation. Annu. Rev. Genet. 39:615-651. [DOI] [PubMed] [Google Scholar]
- 34.Lyon, M. F. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1 90:372-373. 13764598 First author name does not match (LYON). [DOI] [PubMed] [Google Scholar]
- 35.Lyon, M. F. 1999. Imprinting and X chromosome inactivation, p. 73-90. In R. Ohlsson (ed.), Results and problems in cell differentiation. Springer-Verlag, Heidelberg, Germany. [DOI] [PubMed]
- 36.Lyon, M. F. 2003. The Lyon and the LINE hypothesis. Semin. Cell Dev. Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 37.Mak, W., T. B. Nesterova, M. de Napoles, R. Appanah, S. Yamanaka, A. P. Otte, and N. Brockdorff. 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303:666-669. [DOI] [PubMed] [Google Scholar]
- 38.Marahrens, Y., B. Panning, J. Dausman, W. Strauss, and R. Jaenisch. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11:156-166. [DOI] [PubMed] [Google Scholar]
- 39.McCarrey, J. R. 2001. X-chromosome inactivation during spermatogenesis: the original dosage compensation mechanism in mammals?, p. 59-72. In G. Xue, Z. Xue, R. Xu, R. Holmes, G. L. Hammond, and H. A. Lim (ed.), Gene families: studies of DNA, RNA, enzymes, and proteins. World Scientific Publishing Co., Hackensack, NJ.
- 40.McCarrey, J. R., C. Watson, J. Atencio, G. C. Ostermeier, Y. Marahrens, R. Jaenisch, and S. A. Krawetz. 2002. X-chromosome inactivation during spermatogenesis is regulated by an Xist/Tsix-independent mechanism in the mouse. Genesis 34:257-266. [DOI] [PubMed] [Google Scholar]
- 41.Mikkelsen, T. S., M. J. Wakefield, B. Aken, C. T. Amemiya, J. L. Chang, S. Duke, M. Garber, A. J. Gentles, L. Goodstadt, A. Heger, J. Jurka, M. Kamal, E. Mauceli, S. M. Searle, T. Sharpe, M. L. Baker, M. A. Batzer, P. V. Benos, K. Belov, M. Clamp, A. Cook, J. Cuff, R. Das, L. Davidow, J. E. Deakin, M. J. Fazzari, J. L. Glass, M. Grabherr, J. M. Greally, W. Gu, T. A. Hore, G. A. Huttley, M. Kleber, R. L. Jirtle, E. Koina, J. T. Lee, S. Mahony, M. A. Marra, R. D. Miller, R. D. Nicholls, M. Oda, A. T. Papenfuss, Z. E. Parra, D. D. Pollock, D. A. Ray, J. E. Schein, T. P. Speed, K. Thompson, J. L. VandeBerg, C. M. Wade, J. A. Walker, P. D. Waters, C. Webber, J. R. Weidman, X. Xie, M. C. Zody, J. A. Graves, C. P. Ponting, M. Breen, P. B. Samollow, E. S. Lander, and K. Lindblad-Toh. 2007. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447:167-177. [DOI] [PubMed] [Google Scholar]
- 42.Namekawa, S. H., P. J. Park, L. F. Zhang, J. E. Shima, J. R. McCarrey, M. D. Griswold, and J. T. Lee. 2006. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16:660-667. [DOI] [PubMed] [Google Scholar]
- 43.Namekawa, S. H., J. L. VandeBerg, J. R. McCarrey, and J. T. Lee. 2007. Sex chromosome silencing in the marsupial male germ line. Proc. Natl. Acad. Sci. U. S. A. 104:9730-9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogushi, S., C. Palmieri, H. Fulka, M. Saitou, T. Miyano, and J. Fulka, Jr. 2008. The maternal nucleolus is essential for early embryonic development in mammals. Science 319:613-616. [DOI] [PubMed] [Google Scholar]
- 45.Okamoto, I., D. Arnaud, P. Le Baccon, A. P. Otte, C. M. Disteche, P. Avner, and E. Heard. 2005. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature 438:369-373. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto, I., and E. Heard. 2006. The dynamics of imprinted X inactivation during preimplantation development in mice. Cytogenet. Genome Res. 113:318-324. [DOI] [PubMed] [Google Scholar]
- 47.Okamoto, I., A. P. Otte, C. D. Allis, D. Reinberg, and E. Heard. 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303:644-649. [DOI] [PubMed] [Google Scholar]
- 48.Patrat, C., I. Okamoto, P. Diabangouaya, V. Vialon, P. Le Baccon, J. Chow, and E. Heard. 2009. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc. Natl. Acad. Sci. U. S. A. 106:5198-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Payer, B., and J. T. Lee. 2008. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 42:733-772. [DOI] [PubMed] [Google Scholar]
- 50.Penny, G. D., G. F. Kay, S. A. Sheardown, S. Rastan, and N. Brockdorff. 1996. Requirement for Xist in X chromosome inactivation. Nature 379:131-137. [DOI] [PubMed] [Google Scholar]
- 51.Puschendorf, M., R. Terranova, E. Boutsma, X. Mao, K. Isono, U. Brykczynska, C. Kolb, A. P. Otte, H. Koseki, S. H. Orkin, M. van Lohuizen, and A. H. Peters. 2008. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 40:411-420. [DOI] [PubMed] [Google Scholar]
- 52.Sado, T., M. Okano, E. Li, and H. Sasaki. 2004. De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development 131:975-982. [DOI] [PubMed] [Google Scholar]
- 53.Sado, T., Z. Wang, H. Sasaki, and E. Li. 2001. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128:1275-1286. [DOI] [PubMed] [Google Scholar]
- 54.Sharman, G. B. 1971. Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature 230:231-232. [DOI] [PubMed] [Google Scholar]
- 55.Shevchenko, A. I., I. S. Zakharova, E. A. Elisaphenko, N. N. Kolesnikov, S. Whitehead, C. Bird, M. Ross, J. R. Weidman, R. L. Jirtle, T. V. Karamysheva, N. B. Rubtsov, J. L. Vandeberg, N. A. Mazurok, T. B. Nesterova, N. Brockdorff, and S. M. Zakian. 2007. Genes flanking Xist in mouse and human are separated on the X chromosome in American marsupials. Chromosome Res. 15:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shiu, P. K., N. B. Raju, D. Zickler, and R. L. Metzenberg. 2001. Meiotic silencing by unpaired DNA. Cell 107:905-916. [DOI] [PubMed] [Google Scholar]
- 57.Sleutels, F., and D. P. Barlow. 2002. The origins of genomic imprinting in mammals, p. 119-154. In J. C. Dunlap and C.-T. Wu (ed.), Homology effects. Academic Press, San Diego, CA. [DOI] [PubMed]
- 58.Solari, A. J., and N. O. Bianchi. 1975. The synaptic behaviour of the X and Y chromosomes in the marsupial Monodelphis dimidiata. Chromosoma 52:11-25. [DOI] [PubMed]
- 59.Tada, T., Y. Obata, M. Tada, Y. Goto, N. Nakatsuji, S. S. Tan, T. Kono, and N. Takagi. 2000. Imprint switching for non-random X-chromosome inactivation during mouse oocyte growth. Development 127:3101-3105. [DOI] [PubMed] [Google Scholar]
- 60.Takagi, N., and M. Sasaki. 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256:640-642. [DOI] [PubMed] [Google Scholar]
- 61.Turner, J. M., S. K. Mahadevaiah, D. J. Elliott, H. J. Garchon, J. R. Pehrson, R. Jaenisch, and P. S. Burgoyne. 2002. Meiotic sex chromosome inactivation in male mice with targeted disruptions of Xist. J. Cell Sci. 115:4097-4105. [DOI] [PubMed] [Google Scholar]
- 62.Turner, J. M., S. K. Mahadevaiah, P. J. Ellis, M. J. Mitchell, and P. S. Burgoyne. 2006. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev. Cell 10:521-529. [DOI] [PubMed] [Google Scholar]
- 63.Turner, J. M., S. K. Mahadevaiah, O. Fernandez-Capetillo, A. Nussenzweig, X. Xu, C. X. Deng, and P. S. Burgoyne. 2005. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 37:41-47. [DOI] [PubMed] [Google Scholar]
- 64.Wutz, A., and J. Gribnau. 2007. X inactivation xplained. Curr. Opin. Genet. Dev. 17:387-393. [DOI] [PubMed] [Google Scholar]
- 65.Wutz, A., T. P. Rasmussen, and R. Jaenisch. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30:167-174. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, L. F., K. D. Huynh, and J. T. Lee. 2007. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129:693-706. [DOI] [PubMed] [Google Scholar]
- 67.Zhao, J., B. K. Sun, J. A. Erwin, J. J. Song, and J. T. Lee. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322:750-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.